Abstract
Peptidoglycan (PG) is a polysaccharide matrix that protects bacteria from osmotic lysis. Inhibition of its biogenesis is a proven strategy for killing bacteria with antibiotics. The assembly of PG requires disaccharide-pentapeptide building blocks attached to a polyisoprene lipid carrier called lipid II. Although the stages of lipid II synthesis are known, the identity of the essential flippase that translocates it across the cytoplasmic membrane for PG polymerization is unclear. We developed an assay for lipid II flippase activity and used a chemical genetic strategy to rapidly and specifically block flippase function. We combined these approaches to demonstrate that MurJ is the lipid II flippase in Escherichia coli.
Bacteria use polyisoprenoid-linked oligosaccharides to assemble the essential peptidoglycan (PG) matrix that surrounds their cytoplasmic membrane and fortifies their cell envelope against high internal osmotic pressure (1). The building block of PG (cell wall) is a disaccharide-pentapeptide that is synthesized at the cytoplasmic leaflet of the inner membrane (IM) as a precursor known as lipid II (Fig. 1A) (1, 2). This precursor must be flipped across the membrane for cell wall synthesis.
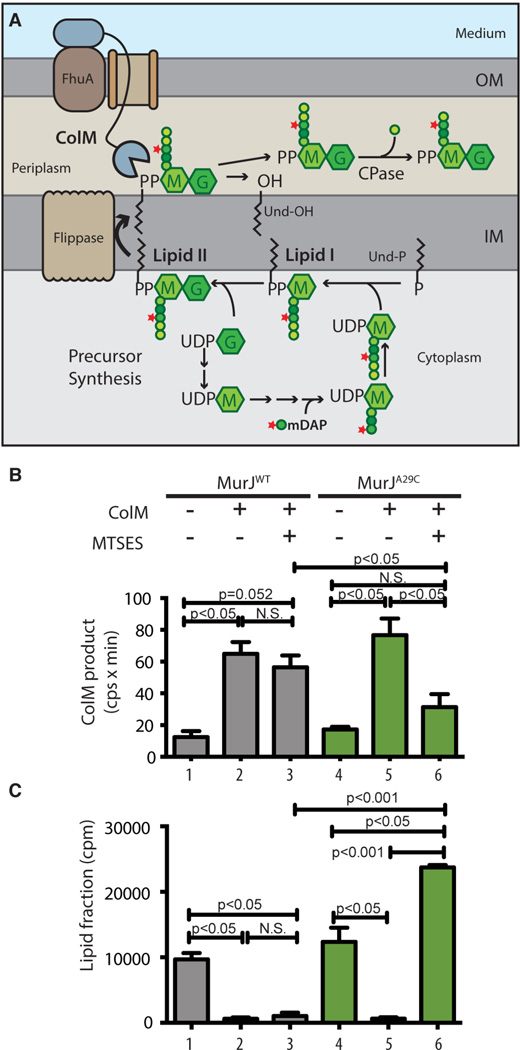
Fig. 1. In vivo assay for lipid II flippase activity.
(A) PG precursor synthesis starts with the conversion of UDP-N-acetylglucosamine (UDP-G) to UDP-N-acetylmuramic acid (UDP-M), followed by the addition of amino acids (represented by colored spheres) to UDP-M to form the pentapeptide (pep5) stem (L-Ala-γ-D-Glu-m-DAP-D-Ala-D-Ala). [3H]-mDAP label is indicated by the star. The UDP-sugars are transferred to undecaprenol-P (Und-P) in the IM to form lipid II, which is flipped across the IM to expose the disaccharide-pep5 (Mpep5-G) for polymerization and crosslinking into PG (not illustrated). Exogenous ColM binds to FhuA and is translocated across the OM presumably through a porin. In the periplasm, ColM cleaves lipid II into undecaprenol (Und-OH) and soluble PP-Mpep5-G, which is further processed by carboxypeptidases (CPase) to produce PP-Mpep4-G. (B-C) Cells of ΔmurJ ΔlysA strains producing FLAGMurJ lacking endogenous Cys residues referred to as MurJWT (NR2592) or its derivative MurJA29C (NR2593) were labeled with [3H]-mDAP. After 15 min, ColM and/or MTSES were added as indicated and growth was continued for 10 min. Samples were then withdrawn and either extracted with hot water alone or sequentially with water then butanol. Hot water extracts were subjected to high-performance liquid chromatography (HPLC) and radiodetection to quantify the labeled ColM product (B); scintillation counting was used to quantify label in the lipid (butanol) fraction (C). See figs. S1–S4 for experimental details and peak identification. Shown are the mean ± SD; N = 3. P value determined with Student’s t test. N.S., not significant.
The identity of the lipid II flippase has been controversial with the debate centered on two candidates: MurJ-like and FtsW/RodA-like proteins (3–6). MurJ is a polytopic IM protein and member of the MOP (multidrug/oligo-saccharidyl-lipid/polysaccharide) exporter superfamily (7). It is essential in Escherichia coli. Cells depleted of MurJ fail to complete PG biogenesis, accumulate PG precursors, and lyse (4, 6). A 3-D structural model and corresponding transmembrane topology of MurJ is similar to that of MOP exporters of amphipathic drugs and undecaprenyl-PP-linked oligosaccharides (8). Furthermore, a hydrophilic central cavity in MurJ is essential for function. FtsW and its paralog RodA are polytopic IM proteins that belong to the SEDS (shape, elongation, division, and sporulation) superfamily and are required for PG synthesis during division or elongation, respectively (3, 5, 9). Support for SEDS proteins functioning as flippases is based on in vitro studies where lipid II flippase activity was detected for purified FtsW incorporated into liposomes (10). The identity of the lipid II flippase has been sought after for decades (2). Determining which proteins flip lipid II in vivo requires a sensitive method to detect lipid II flippase activity and a method to connect this activity to a specific protein within the cell. When added to E. coli, the protein toxin colicin M (ColM) is translocated into the periplasm where it cleaves lipid II (Fig. 1A) (11, 12). We therefore reasoned that ColM could be used in an assay to detect freshly flipped lipid II. To evaluate this possibility, cells were metabolically labeled with [3H]-mDAP, an amino acid unique to the PG peptide, and either left untreated or incubated with purified ColM. Cells were then extracted with hot water followed by butanol to separate soluble PG intermediates and ColM-derived products from lipid-linked PG precursors, respectively. HPLC analysis of the water-soluble extract revealed a new peak in the ColM-treated samples (Figs. 1B and S1) and its appearance correlated with the loss of radiolabel in the butanol extract (Fig. 1C). Moreover, increasing the cellular lipid II concentration by overproducing the lipid-carrier synthase UppS (2) enhanced the production of the ColM-specific peak (Fig. S2). The ColM-specific product was identified as PP-Mpep4-G (Figs. S3–S4), which presumably results from the processing of the ColM product, PP-Mpep5-G, by a carboxypeptidase (Fig. 1A). Because carboxypeptidases function only in the periplasm (1), this result confirms that ColM acts on flipped lipid II.
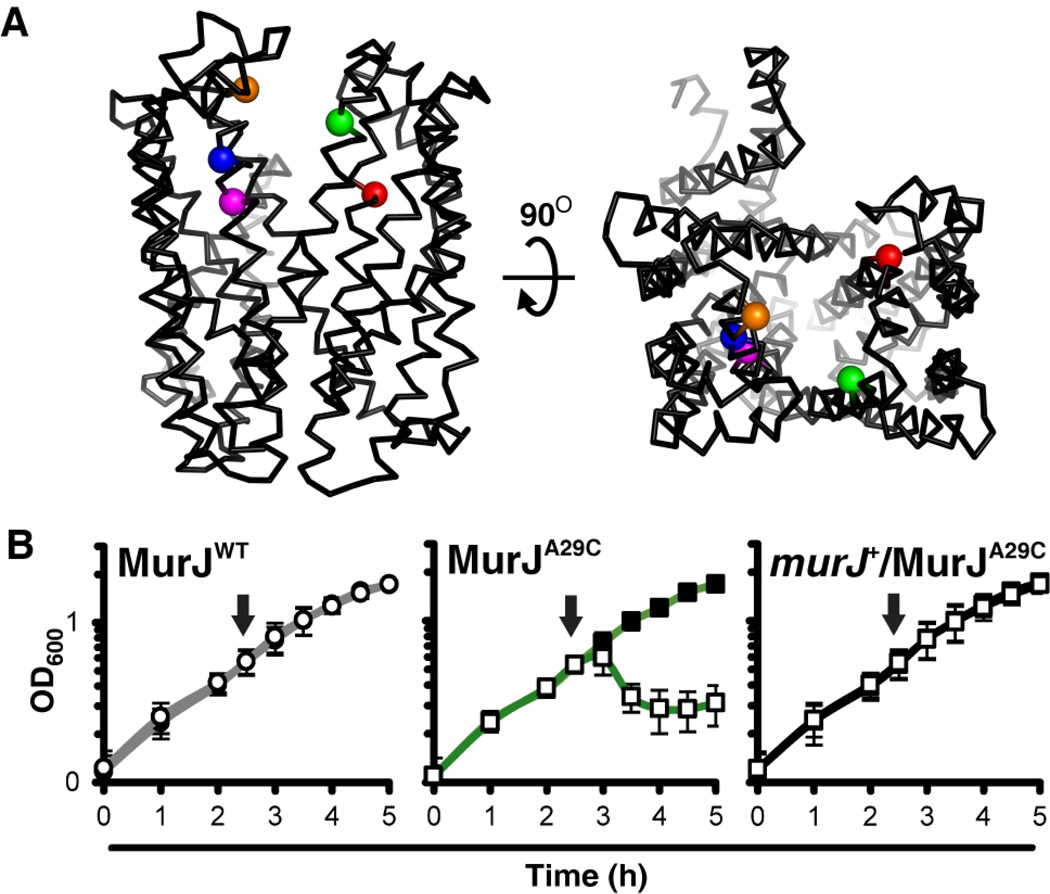
To test whether MurJ flips lipid II, a method to rapidly and specifically inactivate it was needed. A collection of 39 functional single-Cys MurJ variants modifiable by the Cys-reacting molecule MTSES were previously used to determine the membrane topology of MurJ (8). We asked whether any of these mutant proteins were rendered non-functional by derivitization with MTSES. Treatment of Cys-free MurJ (MurJWT) cells with MTSES had no effect on growth, but addition of MTSES to cells producing derivatives with Cys substitutions at positions 29, 49, 263, and 269 rapidly induced lysis, suggesting that MurJ function, and thus PG synthesis, was inhibited (Fig. 2 and S5). In contrast, treatment of MurJE273C cells with MTSES caused cell shape defects and limited lysis indicative of an incomplete PG synthesis block due to partial MurJ inhibition. The toxicity of MTSES labeling was suppressed in all five strains by the presence of the wild-type murJ allele (Fig. 2 and S5). Thus, MTSES specifically and rapidly inhibits these single-Cys MurJ variants. We chose MurJA29C (Figs. 2, S6 and S7) to assess the effect of MurJ inactivation on lipid II flipping.
Fig. 2. MTSES specifically inhibits the function of MurJA29C.
(A) Structural model of MurJ (8). Sensitivity to MTSES is limited to specific residues within the MurJ cavity: residues 29 (green) in transmembrane domain (TMD) 1, 49 (red) in TMD 2 , and 263 (orange), 269 (blue) and 273 (magenta) in TMD 8. (B) Effect of MTSES on the growth of haploid cells producing MurJWT (left) or MurJA29C in glucose M63 medium. Lysis of MurJA29C cells is suppressed by the presence of a wild-type murJ allele (right). Arrows indicate time of MTSES addition; filled marker, no MTSES; empty marker, MTSES treated. Data represent mean ± SD; N = 3. See fig. S5 for MTSES sensitivity of other variants.
This chemical genetic method for MurJ inactivation was compatible with the in vivo flippase assay. MTSES treatment of MurJWT cells did not affect lipid II processing by ColM (Figs. 1B–C and S1). Additionally, in the absence of MTSES, MurJA29C cells behaved like MurJWT cells (Figs. 1B–C and S1). However, simultaneous addition of MTSES and ColM to MurJA29C cells failed to produce significant quantities of the ColM-dependent product PP-Mpep4-G. In fact, radiolabel in the lipid fraction increased in these samples (Figs. 1B–C and S1). Thus, when MurJA29C was inactivated with MTSES, lipid II was protected from ColM cleavage and label accumulated in the lipid fraction as observed previously for MurJ-depletion strains (4, 6).
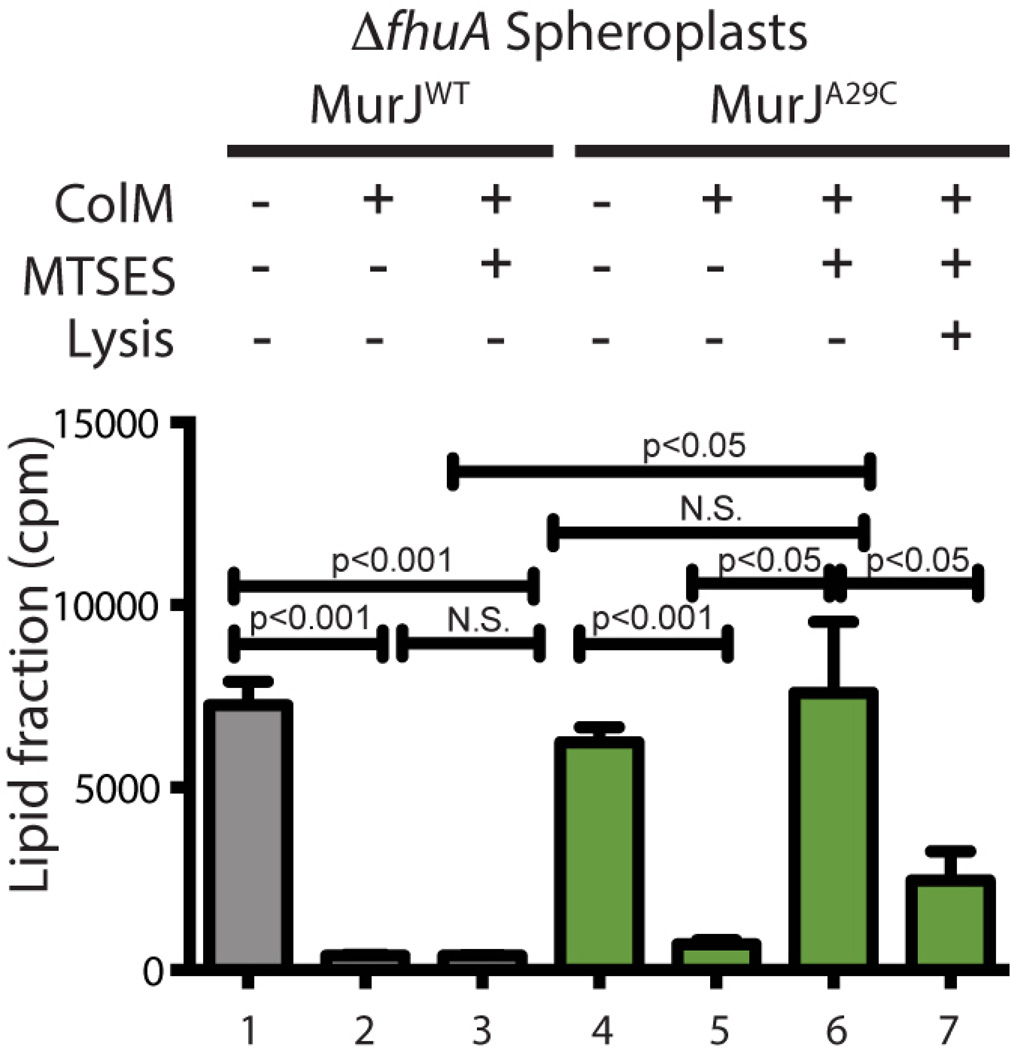
The protection of lipid II from ColM cleavage upon MurJA29C inactivation suggests that either lipid II is not flipped or that inhibiting MurJA29C somehow interferes with ColM import or activity. To investigate this, we performed our assay using spheroplasting to remove the OM barrier (13) and provide ColM with direct access to flipped lipid II. In the absence of MTSES, ColM treatment of MurJWT or MurJA29C spheroplasts reduced the amount of label in the lipid fraction (Fig. 3), indicating that lipid II was actively flipped and thus cleaved by ColM. Although MTSES did not affect ColM activity on MurJWT spheroplasts, it completely abolished lipid II processing by ColM in MurJA29C spheroplasts (Fig. 3). Moreover, lysis of MTSES-treated MurJA29C spheroplasts restored lipid II processing, indicating that the intact IM impeded access of ColM to lipid II. Thus, MurJ appears to act as a lipid II flippase.
Fig. 3. MurJ activity is required for ColM-dependent cleavage of lipid II in spheroplasts.
Cells lacking the ColM receptor FhuA and producing the indicated MurJ variants were grown, labeled, and treated with MTSES as for figure 1. Spheroplasts were then prepared. In all but one case, spheroplasts were pelleted and resuspended in ColM reaction buffer with sucrose and added MTSES (0.8 mM) as indicated. The Lysis + sample was resuspended in buffer lacking sucrose to lyse the spheroplasts. ColM (100 µg) was added to the prepared spheroplasts as indicated and they were incubated for 15 min at 37°C. Lipid intermediates were detected after butanol extraction by scintillation counting. Statistics are as for figure 1.
When MurJA29C was inactivated with MTSES, flippase activity was reduced to a level that was barely detectable and incompatible with life. This observation indicates that the essential function of MurJ is to translocate lipid II and that other factors catalyzing lipid II flipping are unlikely to exist in E. coli. Nevertheless, we investigated the requirement of SEDS proteins for flippase activity by depleting FtsW in a ΔrodA strain. We found that lipid II flipping remained robust in this background (Figs. S8–S9). Although it is possible that residual FtsW in these cells was sufficient for the observed activity, this result suggests that SEDS proteins are not responsible for lipid II flippase activity in vivo. Alternatively, the decrease in levels of PG lipid intermediates upon FtsW depletion (Fig. S9) suggests that either synthesis of PG precursors or recycling of undecaprenyl-P might be affected by the loss of SEDS activity. From these data and the fact that MurJ contains a central, solvent-exposed cavity that is essential for function (8), we conclude that MurJ is the lipid II flippase in E. coli.
Supplementary Material
Acknowledgments
We thank D. Mengin-Lecreulx for the generous gift of plasmids for ColM production, and H. Joseph and R. M. Davis for their technical assistance. Research was supported by funds from the American Heart Association (L.S.) and the National Institutes of Health (NIH) under award numbers F32GM103056 (M.D.L.), R01GM100951 (N.R.), R01AI099144 (T.G.B.), and R01GM76710 (D.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
For additional data, see Supporting Online Material.
Supplementary Materials:
Materials and Methods
Figs. S1-S9
Tables S1-S2
References (14–29)
References and Notes
- 1.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012 Feb;10:123. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008 Mar;32:208. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 3.de Pedro MA, Donachie WD, Holtje JV, Schwarz H. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J Bacteriol. 2001 Jul;183:4115. doi: 10.1128/JB.183.14.4115-4126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue A, et al. Involvement of an essential gene mviN, in murein synthesis in Escherichia coli. J Bacteriol. 2008 Nov;190:7298. doi: 10.1128/JB.00551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khattar MM, Begg KJ, Donachie WD. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol. 1994 Dec;176:7140. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A. 2008 Oct 7;105:15553. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hvorup RN, et al. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem / FEBS. 2003 Mar;270:799. doi: 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- 8.Butler EK, Davis RM, Bari V, Nicholson PA, Ruiz N. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J Bacteriol. 2013 Oct;195:4639. doi: 10.1128/JB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henriques AO, Glaser P, Piggot PJ, Moran CP., Jr Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol. 1998 Apr;28:235. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- 10.Mohammadi T, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011 Mar 8;30:1425. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Ghachi M, et al. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem. 2006 Aug 11;281:22761. doi: 10.1074/jbc.M602834200. [DOI] [PubMed] [Google Scholar]
- 12.Touze T, et al. Colicin M, a peptidoglycan lipid-II-degrading enzyme: potential use for antibacterial means? Biochem Soc Trans. 2012 Dec 1;40:1522. doi: 10.1042/BST20120189. [DOI] [PubMed] [Google Scholar]
- 13.Heppel LA. Selective release of enzymes from bacteria. Science. 1967 Jun 16;156:1451. doi: 10.1126/science.156.3781.1451. [DOI] [PubMed] [Google Scholar]
- 14.Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 15.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 16.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 18.Mercer KL, Weiss DS. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J Bacteriol. 2002 Feb;184:904. doi: 10.1128/jb.184.4.904-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004 Jun;52:1255. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Begg KJ, et al. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol. 1990 Dec;172:6697. doi: 10.1128/jb.172.12.6697-6703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendezu FO, de Boer PA. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol. 2008 Mar;190:1792. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barreteau H, et al. Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme. J Biol Chem. 2010 Apr 16;285:12378. doi: 10.1074/jbc.M109.093583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Heijenoort Y, Gomez M, Derrien M, Ayala J, van Heijenoort J. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J Bacteriol. 1992 Jun;174:3549. doi: 10.1128/jb.174.11.3549-3557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernhardt TG, de Boer PA. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol. 2003 Jun;48:1171. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebar MD, et al. Forming cross-linked peptidoglycan from synthetic gram-negative Lipid II. J Am Chem Soc. 2013 Mar 27;135:4632. doi: 10.1021/ja312510m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MS, Demsey AF, Qi J, Linsdell P. Cysteine-independent inhibition of the CFTR chloride channel by the cysteine-reactive reagent sodium (2-sulphonatoethyl) methanethiosulphonate. Br J Pharmacol. 2009 Jul;157:1065. doi: 10.1111/j.1476-5381.2009.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson JE, Lackner LL, Hale CA, de Boer PA. ZipA is required for targeting of DMinC/DicB, but not DMinC/MinD, complexes to septal ring assemblies in Escherichia coli. J Bacteriol. 2004 Apr;186:2418. doi: 10.1128/JB.186.8.2418-2429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2008 Apr 8;105:5537. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.