Summary
Antibiotic resistance is a key medical concern, with antibiotic use likely being an important cause. However, here we describe an alternative route to clinically-relevant antibiotic resistance that occurs solely due to competitive interactions between bacterial cells. We consistently observe that isolates of Methicillin-resistant Staphylococcus aureus diversify spontaneously into two distinct, sequentially arising strains. The first evolved strain outgrows the parent strain via secretion of surfactants and a toxic bacteriocin. The second is resistant to the bacteriocin. Importantly, this second strain is also resistant to intermediate levels of vancomycin. This so-called VISA (vancomycin-intermediate S. aureus) phenotype is seen in many hard-to-treat clinical isolates. This strain diversification also occurs during in vivo infection in a mouse model, consistent with the fact that both coevolved phenotypes resemble strains commonly found in clinic. Our study shows how competition between coevolving bacterial strains can generate antibiotic resistance and recapitulate key clinical phenotypes.
Introduction
Antibiotics are the primary treatment for bacterial infections. However, the number of effective antibiotics is decreasing due to the rising numbers of multi-drug resistant pathogens. The path to resistance generally involves the acquisition of specific mutations that enable bacteria to grow in the presence of the antibiotics recommended for their treatment (Canton and Morosini, 2011). However, the development of antibiotic resistance is also a naturally occurring process in bacteria, which is not exclusively restricted to clinically relevant species (Benveniste and Davies, 1973). For example, bacteria in the environment may secrete antibiotics to remove niche competitors. These bacteria themselves harbor genes enabling them to resist their own antimicrobials (Allen et al., 2010) and in turn they exert a selective pressure on neighboring bacteria to acquire mutations or transferred genes that are protective (Hibbing et al., 2010). This raises the possibility that the rising levels of antibiotic resistance in pathogens may also be influenced by competitive microbial interactions, similar to what occurs in natural environments.
Multi-drug resistant strains are becoming more and more prevalent in the hospital environment and as a consequence they are increasingly responsible for nosocomial infections. One of the most threatening hospital-associated pathogens is the bacterium Staphylococcus aureus, which currently represents a major problem in both the clinical and community settings. However, S. aureus is not exclusively a pathogen and commonly colonizes the nasopharynx and skin (Kluytmans et al., 1997). Nevertheless, most of these strains have the capacity to cause severe infections when associated with bones and soft tissues, which occasionally progress to life-threatening diseases such as necrotizing fasciitis or pneumonia (Otto, 2012). Moreover, due to the widespread use of the antibiotic methicillin in the 1960s, several strains exist that are resistant to a wide range of β-lactam antibiotics, known as methicillin-resistant S. aureus or MRSA (Kreiswirth et al., 1993). 6More recently, a subset of community-associated MRSA (CA-MRSA) strains have emerged that, in addition to being resistant to multiple antibiotics, are no longer restricted to clinical settings and have the ability to cause severe and pandemic 888infections in healthy individuals (Diep et al., 2008). The traditional approach to treat MRSA and CA-MRSA infections has been the glycopeptide vancomycin and therefore it is a common treatment for staphylococcal infections. However, the general use of the antibiotic has led to the emergence of several strains that display reduced susceptibility to vancomycin (Hiramatsu et al., 1997). These strains are commonly referred to as vancomycin-intermediate S. aureus (VISA) and, although their level of resistance is relatively moderate (MIC = 4-8 μg/ml) in comparison to strains that are fully resistant to vancomycin (VRSA) (MIC ≥ 16 μg/ml), due to their prevalence, VISA strains represent a major threat to overcoming staphylococcal infections (Howden et al., 2010).
The antibacterial activity of vancomycin relies on its capacity to bind to and inactivate certain precursors of cell wall synthesis, which exclusively localize at the division septum in S. aureus (Pereira et al., 2007). S. aureus strains acquire intermediate resistance to vancomycin treatment via the acquisition of point mutations in regulatory genes that lead to the thickening of the cell wall. This provides a protective barrier against the diffusion of vancomycin to its target at the division septum (Howden et al., 2010; Pereira et al., 2007). VISA strains emerge from vancomycin susceptible strains during infections treated with the antibiotic (Hiramatsu et al., 1997) and, in contrast to the rare cases of VRSA infections, VISA strains cause numerous hard-to-treat, hospital-associated infections. However, it is also puzzling that VISA strains have been detected in patients with renal failure or a prosthetic joint that have not been treated with vancomycin (Charles et al., 2004; Howden et al., 2010). These infections are usually associated with high cell density bacterial colonization, which is currently considered one of the risk factors that promote the occurrence of VISA (Charles et al., 2004; Howden et al., 2010).
Bacterial colonization at high cell density involves prolonged periods of infection and the formation of surface-associated aggregates or biofilms (Lopez et al., 2010). Biofilm formation is a key feature of staphylococcal infections, in addition to the secretion of virulence factors (Herbert et al., 2010). The agr quorum-sensing (QS) system of S. aureus controls the infection process by upregulating virulence factors, such as membrane-disrupting toxins (Hla, Hlb and Hlg), proteases, lipases, enterotoxins, superantigens and urease while at the same time indirectly downregulating the extracellular matrix and adhesin proteins responsible for cell aggregation and biofilm formation (Boles and Horswill, 2008; Peng et al., 1988; Recsei et al., 1986). Importantly, agr is repressed by the σB stress-induced sigma factor. σB is activated during biofilm formation in response to cellular stress and triggers the expression of a general-stress response regulon that includes genes associated with pigmentation and biofilm formation (Bischoff et al., 2001; Kullik et al., 1998),
A growing body of empirical and theoretical work suggests that biofilm-encased cells are subjected to strong natural selection, as they compete for space and nutrients, which can shape microbial phenotypes and diversity (Nadell et al., 2009). These conditions give rise to a heterogeneous population of genetically different bacteria that display characteristics that are relevant to understanding the progression of an infection. We hypothesized that a biofilm environment of a single staphylococcal clone could be a factor shaping the diversity of MRSA strains, in which the pathogen could evolve new phenotypes that resist antibiotic treatment. This report presents evidence for an unexpected route to the evolution of antibiotic resistance in staphylococcal communities that occurs without the selective pressure of antibiotic treatment but solely via intra-clonal competitive interactions between bacterial cells. Microbial aggregates of a single staphylococcal isolate spontaneously evolved new competitive phenotypes in a coevolutionary arms-race that ultimately led to the emergence of an intermediate resistant phenotype similar to VISA clinical isolates. Our study provides evidence for how bacterial interactions play an important role in microbial evolution and may serve to explain the diversification of key clinical phenotypes.
Results
Strain diversification in staphylococcal biofilms
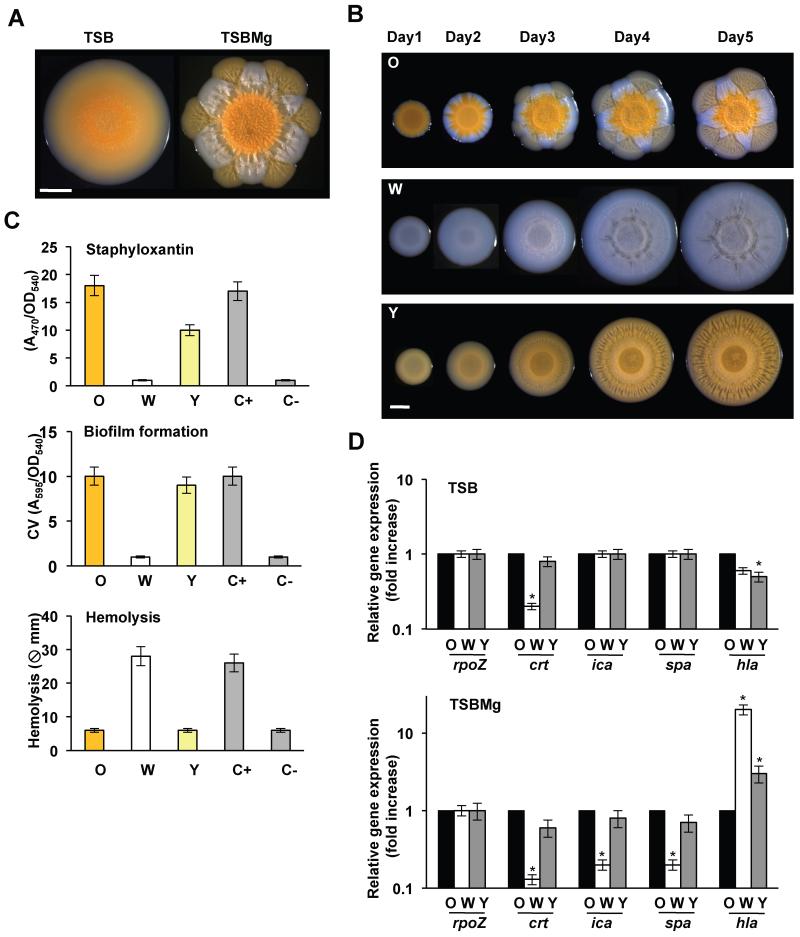
The genetic basis of biofilm formation in S. aureus has been routinely explored using submerged surfaces in TSB medium + NaCl 500 mM (Beenken et al., 2003; Gotz, 2002). However, to study the diversification and evolution of new strains, we established a biofilm formation assay in S. aureus using solid agar (Branda et al., 2001). The assay utilized Mg2+-enriched conditions, since previous studies have shown that chronic staphylococcal infections due to biofilm formation occurs in niches contain a high concentration of Mg2+ (e.g. joints or bones) (Gunther, 2011; Jahnen-Dechent, 2012) while tissues unintentionally depleted of Mg2+ are prone to acute staphylococcal infections (Schlievert, 1985). S. aureus strains were grown on TSB + 100mM MgCl2 solid agar (TSBMg) for 5 days. After incubation, staphylococcal communities developed into robust aggregates with a complex architecture in which immobilized cells became encased within an extracellular matrix, as reported for other species (Branda et al., 2001; Serra et al., 2013) (Figs. 1A and S1A, B). When extending this assay to clinical isolates, we observed morphological diversity (Figs. S1C) and a number of strains (SC01, RN1, N315, LAC and HG003 strains) reproducibly developed distinct sectors that were phenotypically different to the parental strain as the colonies grew across the plate (Figs. 1A and S1C) (Servin-Massieu, 1961). The development of the distinct sectors is consistent with the appearance of S. aureus variants that display a competitive advantage during growth.
Figure 1. Strain diversification in communities of S. aureus.
(A) S. aureus SC01 derivative strain grown in TSB and TSBMg for 5 days at 37°C. (B) Progression of O, W and Y strains when growing in TSBMg for 5 days at 37°C. Scale bars 1 mm. (C) Measurements of Staphyloxanthin production, biofilm formation and secretion of hemolytic toxins in O, W and Y strains. C+ is LAC strain and C− is LAC ΔσB mutant. Hemolysis efficiency used LAC ΔσB mutant as a C+ and C− is LAC strain. (D) qRT-PCR of the expression staphyloxanthin production (crt), biofilm formation (ica and spa) and hemolytic toxins (hla) in O, W and Y strains in TSB and TSBMg growing media (Student’s t-test p ≤ 0.05). See figs. S1, S2.
To gain insight into the evolution of these new variants and their potential roles in biofilm formation during clinically-relevant infections, we focused on a community-associated MRSA (CA-MRSA) clinical isolate (SC01 derivative) that displayed the most evident sectoring phenotype among all strains tested. The wildtype strain initially formed an orange-colored center region or “origin” (O). The orange color comes from the carotenoid pigment staphyloxanthin, which is responsible for the golden coloration of S. aureus (Marshall and Wilmoth, 1981). Over time, a second unpigmented strain emerged that is white (W) and rapidly surrounded the origin. Subsequently, a third strain emerged, forming yellow flares (Y) that radiated from the origin and through the white sector to generate a distinctive flower-like distribution pattern of cells within the bacterial community (Figs. 1B and S1D). Diversification of W and Y phenotypes strongly depended on the presence of Mg2+ in the medium and occurred in both liquid culture and solid agar biofilm assays (Figs. S1E and S2A).
When isolated and re-inoculated, O cells developed a similar distribution of alternating sectors of non-pigmented and pale-yellow pigmented cells. However, isolation and re-inoculation of W and Y cells resulted in a homogeneous community (Fig. 1B). In addition to the differences in pigmentation, O cells displayed an increased ability to form biofilms in the conventional biofilm formation assay and a low ability to secrete hemolytic toxins (Fig. 1C). W cells showed the opposite biological activities and Y cells displayed an intermediate response in the collection of assays that we performed. Furthermore, qRT-PCR analyses verified that these differences were associated with changes in expression of key genes involved in these processes (Fig. 1D and S2B). Pigment production, biofilm formation and hemolysis directly correlated with the expression of staphyloxanthin (crt) (Pelz et al., 2005), biofilm-related genes (ica and spa genes) (O’Gara, 2007; Otto, 2008) and hemolytic toxin genes (hla) (Brown and Pattee, 1980), respectively, suggesting that O, W and Y strains are physiologically distinct.
W strain displays a hyperactive QS phenotype
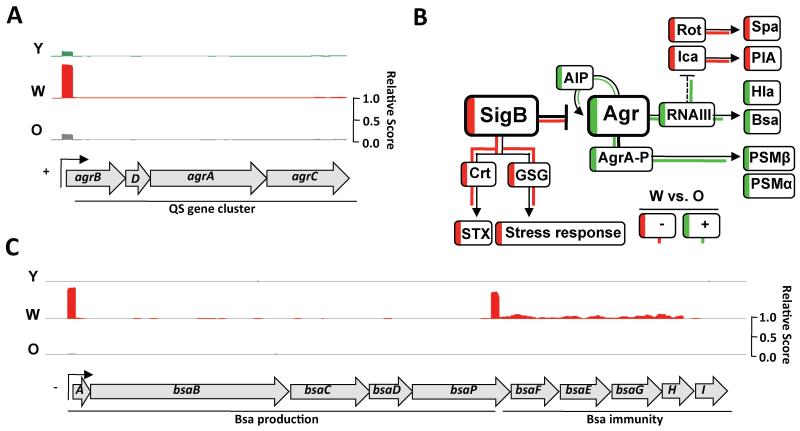
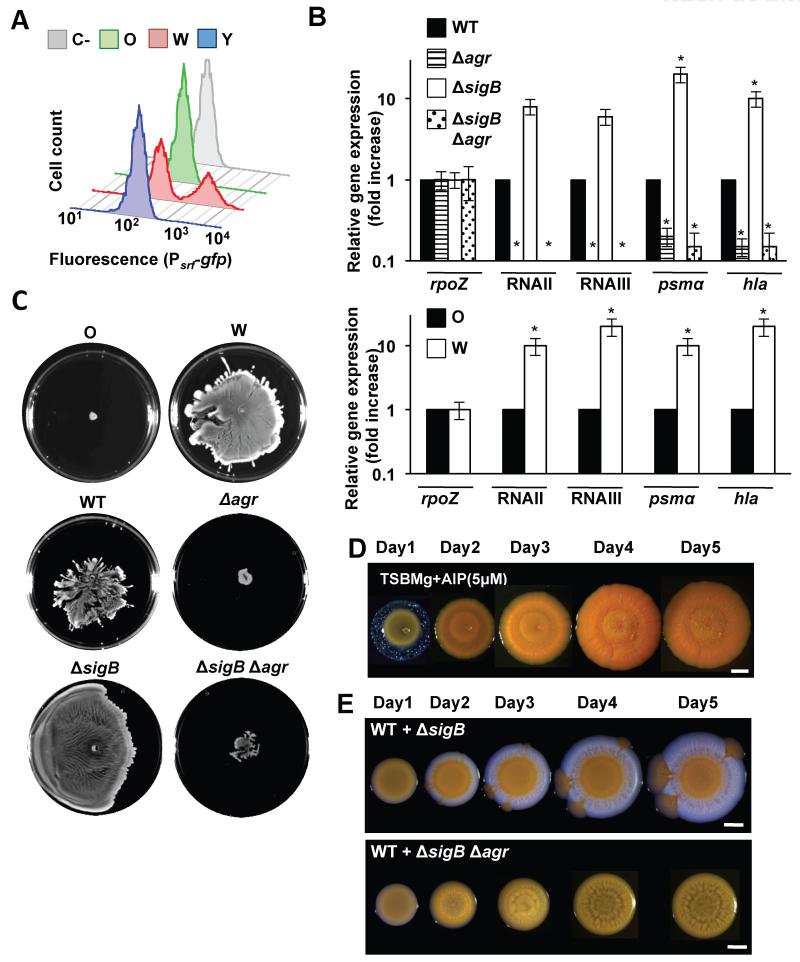
The phenotypic characterization of the initial O strain displaying increased biofilm production and decreased toxin secretion, suggested that the QS system was repressed, since agr quorum-sensing (QS) system activates the expression of secreted toxins and inhibits genes involved in biofilm formation. Yet, the decreased biofilm formation and increased toxin secretion in W suggested that the agr pathway had become active (See fig. 2B). The differential activation of the QS system was explored by the global gene expression profiling using RNA-sequencing (RNA-seq) for the O, W and Y strains (Fig. S3A and Table S3). Comparison of the O and W gene expression signatures revealed hyperactivated expression of the agr QS system in W compared to O (Fig. 2A and S3B), which occurred concomitantly with enhanced expression of a number of agr-regulated genes. Activation of QS in W is associated with hyperproduction of the autoinducing peptidic pheromone (AIP). At the threshold concentration, AIP binds to its membrane-bound receptor AgrC and triggers a QS response in S. aureus (Novick and Geisinger, 2008; P et al., 2001; Thoendel et al., 2011). We genetically engineered a synthetic Bacillus subtilis strain to express a fluorescent AIP-responsive reporter (Fig. S3C-G). Diluted supernatants of O, W and Y strains were added to cultures of the B. subtilis reporter strain. Supernatants from W contained more AIP than those from O or Y (Fig. 3A).
Figure 2. Correlation of genome-wide gene expression levels quantified by RNA-seq.
(A) Read coverage of the agr gene cluster in O, W and Y. The expression of agr in W is increased approx. 20-fold in relation to O. (B) Schematic overview of the genetic circuitry that antagonistically regulates toxin secretion and biofilm formation in S. aureus. Arrows are positive regulation and T-bars are repression. Dashed line is indirect regulation. Staphyloxanthin production is STX and GSG are general stress genes. Transcriptional activation in W is represented in green and downregulation is represented in red. (D) Read coverage of the bsa gene cluster in O, W and Y cells. The expression of the bsa in W strain is increased approx. 300-fold in relation to O strain. See fig. S3 and tables S3-5.
Figure 3. Adaptive abilities of the W strain.
(A) Quantification of AIP in O, W and Y supernatants using a B. subtilis transducer strain and monitored by flow cytometry (n = 50,000 events). Grey profile is control with no supernatant added. (B) qRT-PCR analysis of RNAII, RNAIII and regulated genes in different genetic backgrounds (Student’s t-test p ≤ 0.05). (C) Spreading assay to monitor the ability of distinct strains to expand on solid surfaces. Samples were spotted and incubated for 24h at 37°C. (C) Muticellular communities of distinct strains growing in TSBMg for 5 days at 37°C. (D) Complementation of TSBMg with 5μM AIP. Plates were incubated at 37°C during 5 days. (E) Synthetic communities of different strain mixtures (ratio 5:1) were spotted in TSBMg and incubated at 37°C for 5 days YA strain differentiates. Scale bars are 1 mm. See figs. S3, S4, S6.
The phenotypic and transcriptional changes found in the W strain were consistent with a σB loss-of-function mutation. The stress-induced sigma factor σB strongly represses agr and induces staphyloxanthin production (Bischoff et al., 2001; Kullik et al., 1998), which would explain the high expression agr and the lack of pigmentation in W strain (Fig. 2B). To test this, we used qRT-PCR analysis to monitor two agr-dependent RNAII and RNAIII transcripts (Novick and Geisinger, 2008; Thoendel et al., 2011). RNAII and RNAIII were highly expressed in the W strain compared with the O strain, consistent with increased agr expression in W (Fig. 3B). RNAII activates the agrBDCA operon that encodes the agr regulatory cascade (Bischoff et al., 2004; Bischoff et al., 2001), which regulates the expression of the surfactant Phenol-Soluble Modulins (PSMs) (Queck et al., 2008). RNAIII activates agr-dependent genes such as hla (Morfeldt et al., 1995; Morfeldt et al., 1996) (Fig. 2B). Consistent with a σB-deficient phenotype, both psm’s and hla were highly expressed in the W strain (Fig. 3B). We therefore, searched for adaptive mutations that would produce a σB-deficient phenotype by sequencing the sigB operon (rsbU-rsbV-rsbW-sigB) (Fig. S4A) in nine different W strains derived from three independent evolution experiments. In each case, we found key point mutations in an essential residue for the kinase activity of the RsbW anti-sigma factor (Fig. S4Aii, B). RsbW binds to and prevents σB binding to the RNA polymerase. RsbW is also sequestered by RsbV. Phosphorylation of RsbV by RsbW releases RsbW to bind to and inhibit σB (Marles-Wright and Lewis, 2010) (Fig. S4A). The mutation in RsbW inhibits its binding to RsbV generating a non-functional σB complex with RsbW (Dufour and Haldenwang, 1994), possibly targeting it for degradation (Graham et al., 2013) (Fig. S4C, D).
QS upregulates the production of secondary metabolites, including antibiotics and PSMs surfactants (Table S4). This included increased production of antibiotics that inhibited the growth of O strain (see below) and PSMs surfactants that enabled W to efficiently expand and rapidly colonize the plates in a spreading assay (Fig. 3C) (Tsompanidou et al., 2013) and also in the biofilm formation assay (Fig. 3E). The PSMs were responsible for the increased spreading (Fig. S4E-G). Complementation of a σB-deficient laboratory strain with the σB operon from W strains did not restore the WT phenotype, which was rescued only by the σB operon from the O strain (Fig. S4H). To demonstrate the evolutionary advantage of QS hyperactivation in W, we purified the AIP signal that triggers QS in S. aureus from W cultures and added to the growth medium of the O strain (P et al., 2001). Activation of QS in W is associated with hyperproduction of the autoinducing peptidic pheromone (AIP). Under these conditions, the W or Y phenotypes were not observed (Fig. 3D). To summarize, W arises through a mutation that inhibits σB and causes hyperactivation of QS enabling the strain to both spread from and inhibit the growth of the parental strain.
W strain produces Bsa bacteriocin that selects for the evolution of the Y strain
To provide evidence for a growth advantage of W over O, a growth assay was performed using a tractable CA-MRSA strain (LAC) as a proxy O strain and a ΔsigB mutant as W strain. The WT and ΔsigB strain were combined in a 5:1 ratio and grown in TSBMg (5:1 is the empirically determined O:W ratio observed upon initiation of diversification. Fig. S2A). After incubation, the synthetic communities recreated the phenotype previously observed in the CA-MRSA colonies, with the WT strain restricted to the origin and the QS-high strain (ΔsigB strain) rapidly expanding and forming a white ring surrounding the colony (Fig. 3E). Thus, hyperactivation of the agr QS in the σB-deficient strain provided a growth advantage over the WT strain enabling it to rapidly spread. Importantly, the third yellow phenotype strain that was not present in the original mixture also evolved during the time course of the experiment. This third yellow strain radiated from the WT sector in the center and grew through the σB-deficient strain sector, similar to behavior of the Y phenotype of the original CA-MRSA colonies.
The emergence of the Y strain after the W strain suggests that the presence of W may be important for the evolution of Y strain. As described above, phenotypic and transcriptomic data revealed that the W strain produced several extracellular active compounds. We detected high expression of a CA-MRSA associated gene cluster, responsible for the production of the lantibiotic Bsa (bacteriocin of S. aureus) and its resistance machinery (Fig. 2C and S3B). Antibiotic-producing bacteria use antibiotics to remove niche competitors and Bsa enables CA-MRSA to compete with the natural flora of healthy individuals and facilitates the growth of CA-MRSA when coexisting with other bacteria (Daly et al., 2010) Thus, indicating the potential of Bsa to act as a selective pressure in the evolution of microbes.
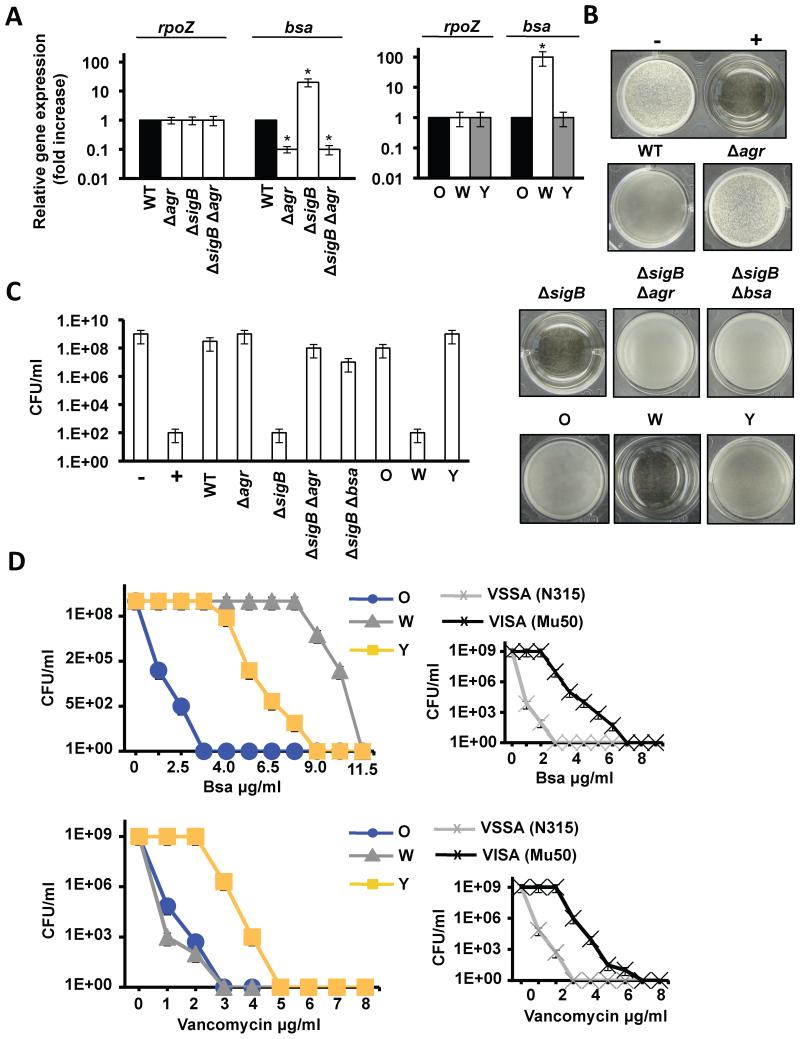
Expression analysis of σB-deficient strain revealed an increase in bsa expression, which was also dependent on agr (Kies et al., 2003). As expected, W also showed higher expression of bsa-related genes compared to O and Y (Fig. 4A). Next, supernatants of O, W and Y cultures were tested for antimicrobial activity against B. subtilis (Fig. 4B, C). Supernatants from W inhibited B. subtilis growth. Likewise, supernatants from σB-deficient strains were toxic and this toxicity was attenuated in the absence of the agr or bsa gene clusters (Fig. 4B, C).
Figure 4. Adaptive abilities of the Y strain.
(A) qRT-PCR analysis of the expression of bsa in distinct genetic backgrounds (Student’s t-test p ≤ 0.05). (B) Antimicrobial activity of filter-sterilized supernatants. 50 μl was used to supplement 1 ml cultures of B. subtilis. Growth inhibition prevented biofilm growth. Negative and positive controls (− and +) represent B. subtilis cultures without and with kanamycin 5 μM. (C) Quantification microbial survival (CFU/ml) in B. subtilis cultures conditioned with Bsa. (D) Growth curves of O, W and Y in TSB medium with different concentrations of Bsa or vancomycin (CFU/ml). Right panels show control growth curves of isogenic VSSA/VISA strains (N315/Mu50) in the presence of different concentrations of Bsa or vancomycin. See fig. S5.
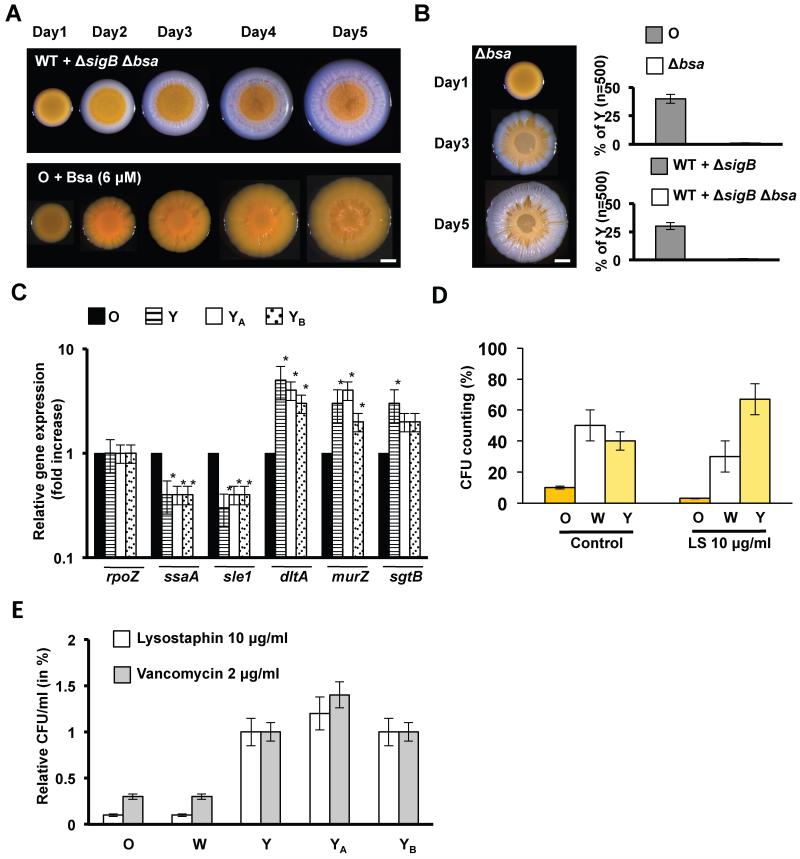
The above experiments confirmed that the W strain produced more Bsa antibiotic than O and Y strains (Table S4 and S5), but it was unclear if Bsa was responsible for the selective pressure to generate the Y genotype. Addition of exogenous Bsa to O, W and Y cultures in TSB liquid medium strongly inhibited the growth of O. Importantly, the addition of Bsa did not affect the growth of W and minimally affected the growth of Y (Fig. 4D upper row). Furthermore, in the synthetic mixed populations (described above) deletion of the bsa cassette in a σB-deficient strain did not prevent the rapid expansion of the σB-deficient strain but prevented the emergence of the Y strain (Fig. 5A upper row). Likewise, the Y strain did not evolve from an O strain lacking the bsa cluster (Fig. 5B). Importantly, the addition of exogenous Bsa to the O strain (grown on TSBMg agar) bypassed the requirement for the W phenotype and Y flares directly evolved from a growth-restricted O community in the absence of W sectors (Fig. 5A bottom row). These results suggest an important role for Bsa in the evolution of the Y phenotype.
Figure 5. Y strain is a VISA-like strain.
(A) Progression of mixed LAC wild type strain + LAC ΔsigB Δbsa mutant in relation 5:1 (upper row). The communities grew in TSBMg at 37°C during 5 days. After incubation, the community failed to develop Y flares through the W sector. Progression of the O strain in TSBMg at 37°C during 5 days when supplemented with previously purified Bsa (~ 6 μM) (lower row). YB strain differentiates. Scale bar is 1mm. (B) Progression of the O strain Δbsa mutant in TSBMg at 37°C during 5 days. After incubation, the community failed to develop Y flares through the W sector. (C) qRT-PCR analysis of several VISA-related genes (ssaA, sle1, dltA murZ and sgtB) in O, Y strains and YA and YB strains derived from the artificial mixture WT+ΔsigB (fig. 3G) and Bsa supplementation (fig. 5A), respectively (Student’s t-test p ≤ 0.05). (D) Quantification of bacterial survival in response to lysostaphin treatment (LS 10 μg/ml during 15 min at 37°C). (E) Quantification of bacterial survival in response to lysostaphin and vancomycin treatment of different strains in relation to Y strain. See figs. S5, S7 and table S6.
Y strain shows a VISA-like phenotype that tolerates the presence of Bsa
Similar to many other antibiotics, Bsa is an epidermin-like antibiotic that targets lipid II during bacterial cell wall synthesis. Vancomycin also inhibits cell wall synthesis by binding to the D-Ala-D-Ala residues in the Lipid II-linked pentapeptide precursor of peptidoglycan (Lipid-II-AA) (See fig. S7A) (Howden et al., 2010). Vancomycin has been consistently used to treat MRSA infections. However, recently the emergence of strains that are intermediate-resistant to vancomycin (VISA) (Hiramatsu et al., 1997) as well as other cell wall antibiotics has been observed (Camargo et al., 2008; Peleg et al., 2012). VISA strains display increased cell wall thickening, which reduces the access of vancomycin to its lethal target (Lipid-II-AA) at the division septum (Fig. S7A, B). Interestingly, we find that most sequenced VISA strains derived from susceptible parental strains that do not contain the bsa gene cluster (Fig. S5A, B), similar to what is seen in VRSA (Kos et al., 2012). Thus, the acquisition of the VISA phenotype potentially enables Bsa-defective strains to tolerate the presence of Bsa in mixed communities (Fig. S5C), since it has been recently reported that approximately 30% of staphylococcal infections involves colonization by multiple-strains (Votintseva et al., 2014).
We explored the possibility that Y cells have acquired a VISA like phenotype to tolerate the presence of Bsa. Addition of vancomycin to Y cultures revealed intermediate resistance (IR) to vancomycin similar to their IR to Bsa (Fig. 4D bottom row). In contrast, IR to vancomycin was not detected in W cultures, which indicated that the VISA-phenotype was specific to Y strains. There are multiple pathways by which cells can acquire a VISA phenotype. Nevertheless, mutations in graRS, vraRS and walKR two-component systems have been consistently found in VISA isolates (Hafer et al., 2012; Howden et al., 2010). To detect the existence of mutations that may confer a VISA-like phenotype to Y, we performed a whole-genome sequence comparison between Y and O. This revealed mutations in the GraRS and WalKR two-component systems (Fig. S6B), which were further confirmed by Sanger sequencing of nine independent evolutionary events in different colonies. Importantly, amino acid substitutions detected in the these two-component systems have been previously described in VISA isolates (Matsuo et al., 2013). The agr operon was sequenced as a control to confirm that no mutations occurred in this locus. In addition, the Y strains that evolved from the experiments of mixed communities (YA strain shown in fig. 3E) or after exposure to Bsa (YB strain shown in fig. 5A) were also sequenced. VISA-like mutations were also found in the graRS, vraRS and walKR operons in both strains (Fig. S6B).
GraRS, VraRS and WalKR synergistically regulate a number of genes involved in cell wall synthesis. This stimulon is permanently activated in VISA strains, which results in cell wall thickening that reduces the access of vancomycin to its lethal target Lipid-II-AA at the division septum (Howden et al., 2010) (Fig. S7A, B). Gene expression analyses of VISA-related genes confirmed the upregulation of cell-wall genes and downregulation of autolysins in the different Y strains (Fig. 5C and S5D). Reduced diffusion of vancomycin to the septum was confirmed in Y cells coupled to a thickening of the cell wall using electron microscopy (Fig. S7C-E) and reduced membrane susceptibility to the endopeptidase lysostaphin (Sieradzki and Tomasz, 2003) (Fig. 5D). In addition to this, the YA and YB strains were more tolerance to the action of lysostaphin than W cells and comparable to the natural Y strain (Fig. 5E). As expected, YA and YB strains showed IR to vancomycin similar to the Y strain suggesting a VISA-like phenotype (Fig. 5E). Complementation of a VSSA laboratory strain with the graRS, vraRS and walKR operons from different Y strains also resulted in IR to vancomycin (Fig. S5D). In sum, the production of Bsa by W drives the evolution of the Y strain, which bears a striking similarity to VISA isolates that have been described in the clinics.
O, W and Y strains also evolve in in vivo infections
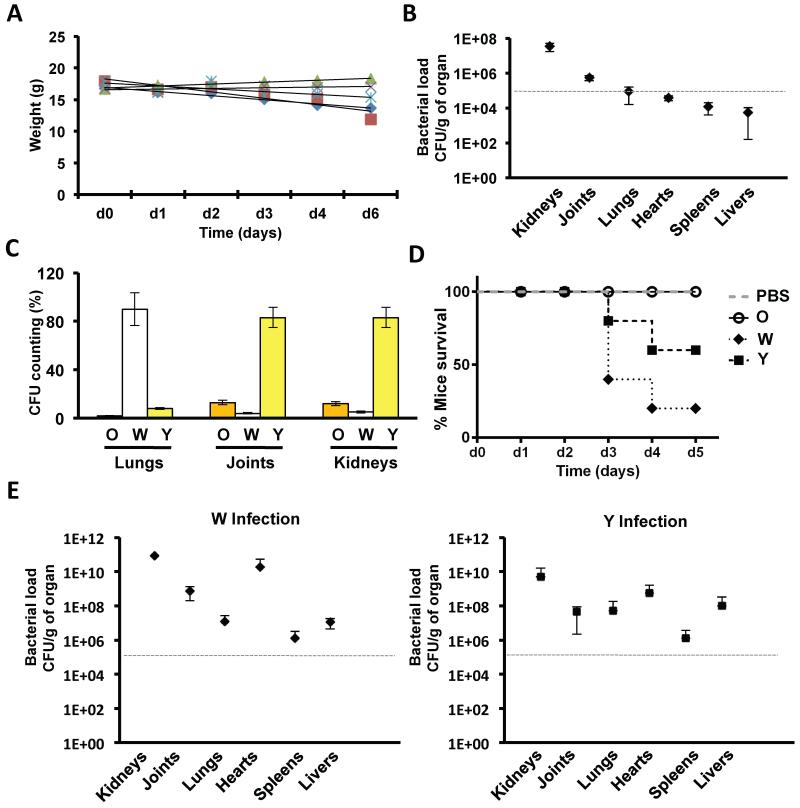
The emergence of two phenotypes from a MRSA isolate in vitro demonstrates that clinically relevant properties can evolve solely via competitive interactions within an MRSA colony biofilm. However, it was unclear if such biofilm-associated evolutionary events could drive similar diversification in an in vivo infection situation. Based on our in vitro data, we predicted that regions of the body that are efficiently colonized by S. aureus (Cut-off ≥ 105 CFU/g of organ) (Table S7) would coincide with the reservoirs of magnesium of the body (i.e. bones and kidneys) (Gunther, 2011; Jahnen-Dechent, 2012), which would favor biofilm-like growth and the emergence of the W and Y strains. Hence, a cohort of five mice was infected with 107 CA-MRSA cells that were previously grown in the absence of Mg2+ to prevent strain diversification (Fig. S2A). After 5 days of infection, mice were sacrificed (Fig. 6A), their organs collected and the bacterial load counted and examined for strain diversification (Fig. 6B). We detected low bacterial load (≤ 105 CFU/g) in livers, spleens and hearts. Importantly, in these organs, the microbial community was a homogenous population of the parental O phenotype (data not shown). By contrast, the bacterial load in samples from the bones, lungs and kidneys were higher (≥ 105 CFU/g) and contained a heterogeneous mixture of O, W and Y cells (Fig. 6C). Y was the most abundant strain collected from kidneys and bones, which are the two most important organs involved in magnesium homeostasis (Jahnen-Dechent, 2012). The lungs contained a higher proportion of W cells, which is consistent with the high number of σB-deficient strains generally isolated from lung infections (Karlsson-Kanth et al., 2006).
Figure 6. Diversification of S. aureus communities in vivo.
(A) Weight loss of infected BALB/c mice (n = 5) during the course of the infection. Mice were infected with 107 SC01 derivative previously grown in TSB medium (B) Bacterial loads in the different organs counted as CFU/g of organ. (C) Quantification of the colony forming units (CFU) of the distinct strains in the organs that showed higher bacterial load (≥ 105 CFU/g). (D) Survival curve of mice infected with O, W or Y strains. Cohorts of 5 mice were infected with 107 cells (in the case of O, W and Y infections) or PBS buffer as control. Mice were sacrificed when showing severe symptoms of infections (description of symptoms is in SI). (E) Quantification of CFU/g of W and Y strains in the organs of infected mice. A grey, dashed line defines a cut-off number of 105 CFU/g to define high and low bacterial load. See table S7.
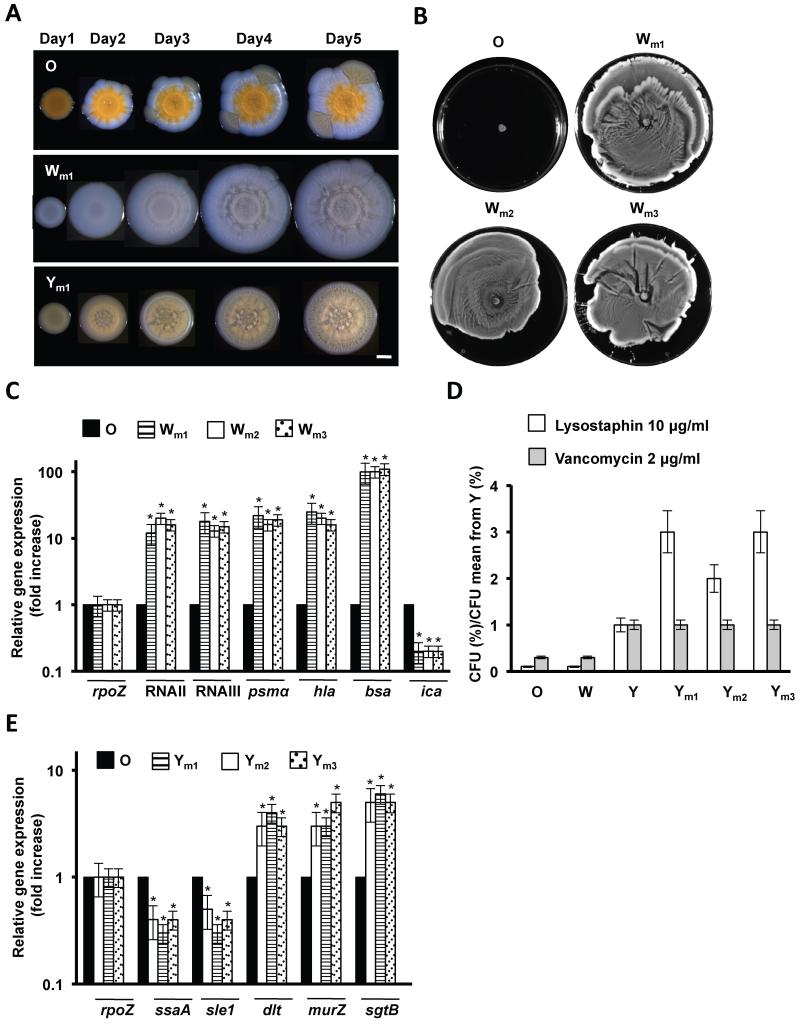
The O, W and Y strains isolated from mice developed similar biofilm morphologies to the strains from the diversification experiments (Fig. 7A). We tested three different W isolates from lungs (Wm1), bones (Wm2) and kidneys (Wm3) for their ability to spread, which occurred as previously described for the W strain (Fig. 7B). Further qRT-PCR analyses verified the upregulation of RNAII and RNAIII and related genes (Fig. 7C), which is consistent with a σB-deficient phenotype in the Wm strains. We sequenced the sigB operon in three different Wm isolates from each organ (lungs, bones and kidneys). One bone isolate and one kidney isolate contained the same mutation in rsbW that was identified in our diversification experiment (D105N) (Fig. S6A). A third bone isolate contained I11L mutation that generates an unstable SigB protein (Karlsson-Kanth et al., 2006) (Fig. S6A). The other Wm strains contained a 11-bp deletion of the 5′ end of the rsbU gene, a frequently observed mutation known to confer a σB-deficient phenotype to diverse staphylococci (Herbert et al., 2010) (Fig. S6A). Similarly, three different Y strains isolated from lungs (Ym1), bones (Ym2) and kidneys (Ym3) displayed IR to vancomycin and lysostaphin treatment (Fig. 7D). Further qRT-PCR analyses provided additional evidence for a gene expression profile consistent with a VISA-like phenotype (Fig. 7E) and sequencing of graRS, vraRS and walKR operons from three different Ym isolates from lungs, bones and kidneys detected several mutations in graRS and vraRS (Fig. S6B). Altogether, these data provide multiple lines of evidence that strain diversification also occurs in vivo in a reproducible manner and generated similar phenotypes to those observed in the laboratory.
Figure 7. Adaptive abilities of Wm and Ym strains diversified in vivo.
(A) Progression of the O, Wm and Ym strains when growing in TSBMg for 5 days at 37°C. Scale bar is 1 mm. (B) Spreading assay of distinct Wm strains. (C) qRT-PCR of RNAII and RNAIII, RNAII- and RNAIII-regulated genes psmα and hla, bsa and repressed ica gene. (D) Quantification of bacterial survival in response to lysostaphin and vancomycin treatment of different Ym strains. Values are presented in relation to Y strain. (E) qRT-PCR analysis to monitor the differential expression of the VISA-related genes ssaA and sle1 autolysins, dltA and cell wall turnover murZ and sgtB genes (Student’s t-test p ≤ 0.05). See figs. S6, S7.
Finally, we compared the infective potential of the O, W and Y strains that evolved in the laboratory experiment. To do this, two cohorts of five mice were infected with 107 W or Y cells previously grown in TSB medium and the progression of the infection was monitored. Remarkably, we observed higher virulence in W and Y strains, manifested by the lower survival rate of infected mice in comparison to infections with the parental O strain (Fig. 6D). The organs were collected and their bacterial load was counted. Accordingly, high bacterial load (≥ 105 CFU/g) was detected in almost all organs, suggesting that, in contrast to the origin O strain, W or Y strains are more prone to give rise to severe bacteremia in our infection model (Fig. 6E).
Discussion
There is a growing recognition of the importance of social interactions for microbial communities (Nadell et al., 2009; West et al., 2006). Growth under biofilm-like conditions places cells in close proximity, which increases the potential for the phenotype of one cell to influence the evolution of other cells. In particular, there is often strong competition for resources and space that will favor strategies that enable one strain to suppress or outgrow another. Here we have shown that these processes can have a major impact on one of the world’s major pathogens. Growing CA-MRSA cells under biofilm-promoting conditions leads to an evolutionary arms race that begins when the W strain evolves and utilizes the increased secretion of PSMs and antibiotics to outcompete the wildtype O strain. However, the wildtype counter-adapts to this challenge and generates the Y strain that can resist the secreted products of W. Our assay utilizes a similar Mg2+ concentration to that found in tissues where staphylococcal infections become chronic due to biofilm formation, such as bones (50 mM) or kidneys (30 mM) and is equivalent to other conventional assays (Beenken et al., 2003; Beenken et al., 2004; Jahnen-Dechent, 2012). In addition to this, it is known that the sequestration of Mg2+ from tissues due to the use of tampons led to an outbreak of S. aureus induced toxic shock syndrome in women in the USA (Schlievert, 1985). Given these correlations between Mg2+ and S. aureus infections, it is possible that Mg2+ acts upon the agr QS system that reciprocally regulates biofilm formation and the secretion of hemolytic toxins by a yet unknown mechanism.
This assay revealed genetic diversification of a number of isolates, being especially evident in a CA-MRSA strain isolated from a hip wound (Tenover et al., 1994). The bone-associated origin of this strain is possibly relevant in its predisposition to diversify in our assay, which reflects the overwhelming inter-strain variation that exists within staphylococcal isolates (Herbert et al., 2010).
The evolved W and Y phenotypes resemble those seen in the clinics. σB-deficient strains such as the W strain are detected in 10% of staphylococcal infections (Lennette, 1985) and the Y phenotype closely resembles VISA strains, which are currently a major health problem and suggests that VISA phenotypes can emerge in a manner that is unrelated to vancomycin treatment. Consistent with this, VISA strains have been observed in patients with renal failure or infections of a prosthetic joint that have not received vancomycin (Charles et al., 2004; Howden et al., 2010; Jahnen-Dechent, 2012). Our work suggests competition within biofilms can be central to understanding pathogenic phenotypes and emphasizes the need to consider not only the impact of treatment of bacterial infections but also the interactions between the bacteria themselves.
Experimental procedures
Strains, Media and Culture Conditions
The strain used for diversification was S. aureus derivative of Sc01 (Beenken et al., 2003). Complete strain and plasmid lists are shown in Tables S1 and S2. For biofilm formation in agar, an inoculum TSB plate was grown for 12h at 37°C. 2 μl of a cell suspension were spotted on the surface of TSBMg and incubated at 37°C for 5 days. When specified, 5μM AIP was added. Bsa was added to different concentrations that are specified in the paper. Specific growth conditions are presented in figure legends.
Conventional liquid-surface biofilm formation assays was performed in TSB + Glucose 0.5% + NaCl 3% (Beenken et al., 2003; Gotz, 2002). Secretion of hemolytic toxins was monitored by spotting 3 μl of cultures in TSB 5% blood agar plates and measuring the diameter size after incubating at 37°C for 24 h. A protocol for staphyloxanthin purification was obtained from (Pelz et al., 2005). To assay spreading on solid surfaces, 2 μl of an overnight culture was spotted on TSB 0.24% agar plates and incubated at 37°C for 24 h (Tsompanidou et al., 2011). To assess resistance of cells to lysostaphin, cells were resuspended in 1 ml PBS and incubated for 15 minutes at 37°C in the presence of lysostaphin (10 μg/ml) (Cui et al., 2006) prior dilution plating on TSB agar to determine CFU/ml.
RNA extraction and analysis
O, W and Y cultures were resuspended in RNA Protect (Qiagen®, USA) and were mechanically lysed using glass beads in a Fast Prep Shaker (2 times, 45 s, speed: 6.5). The supernatants were used for RNA isolation using the RNeasy mini kit (Qiagen®, USA). The isolated RNA was treated with RNase-free DNase I to remove DNA traces. The cDNA libraries were generated as described previously but omitting the RNA size-fractionation step prior to cDNA synthesis (Dugar et al., 2013). The libraries were sequenced with an Illumina HiSeq machine with 100 cycles in single end mode. The demultiplexed files and coverages files have been deposited in NCBI’s GEO database (GSE49636). For qRT-PCR analysis, total RNA was reverse transcribed using hexameric random primers, followed by qRT-PCR using SsoAdvanced SYBR Green Supermix (BioRad, USA) (Primers listed in table S2).
Mouse infection studies
All animal studies were approved by the local government of Franconia, Germany (license number 55.2-2531.01-06/12) and performed in strict accordance with the guidelines for animal care and experimentation. We used female BALB/c mice (16–18 g) (Charles River®, Germany). S. aureus was cultured for 18 h at 37°C on a TSB plate. Cells were diluted to the desired concentration and validated by dilution plating in TSB agar. 100 μl S. aureus culture was injected into the tail vein. 6 days after bacterial challenge, organs were aseptically harvested and the CFU determined. Organs were homogenized in 2 ml of sterile PBS using Dispomix (Bio-Budget Technologies GmbH, Germany). Joints were ground in a mortar prior homogenization. Serial dilutions of each organ were plated on mannitol salt-phenol red agar plates and TSB plates and incubated at 37°C for at least 48 h. CFUs were counted and the bacterial burden calculated as CFU/g of organ. To compare the infective potential of O, W and Y strains, 3 cohorts of 5 mice were infected 100 μl S. aureus culture containing 107 cells injected into the tail vein. The infections were allowed to progress until severe infections symptoms occurred or to an endpoint of 5 days. Organs were aseptically harvested and the CFU determined.
Supplementary Material
Highlights.
Biofilm competition leads MRSA to evolve new strains.
New strains resemble clinically relevant multi-drug resistant isolates.
In vivo experiments show similar strain diversification pattern.
Biofilm evolution generates antibiotic resistance in the absence of antibiotic.
Acknowledgments
We thank IMIB, especially Stan Gorski for his help in editing the manuscript. Dr. S. Engelmann (Univ. Greifswald, Germany) for providing antibodies and AG Ohlsen for guidance with animal experiments. This work was funded by ZINF (DL) and the DFG grants LO 1804/2-1 (DL), SFB-TR34 Z3 (KO) and ERC grants 242670 (KF) and 335568 (DL). GK is recipient of FWF fellowship.
References
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nature reviews Microbiology. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste R, Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci U S A. 1973;70:2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bachi B, Projan S. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol. 2004;186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M, Entenza JM, Giachino P. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol. 2001;183:5171–5179. doi: 10.1128/JB.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Pattee PA. Identification of a chromosomal determinant of alpha-toxin production in Staphylococcus aureus. Infection and immunity. 1980;30:36–42. doi: 10.1128/iai.30.1.36-42.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo IL, Neoh HM, Cui L, Hiramatsu K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother. 2008;52:4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton R, Morosini M-I. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS microbiology reviews. 2011;35:977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- Charles PGP, Ward PB, Johnson PDR, Howden BP, Grayson ML. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clinical infectious diseases. 2004;38:448–451. doi: 10.1086/381093. [DOI] [PubMed] [Google Scholar]
- Cui L, Iwamoto A, Lian JQ, Neoh HM, Maruyama T, Horikawa Y, Hiramatsu K. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:428–438. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly KM, Upton M, Sandiford SK, Draper LA, Wescombe PA, Jack RW, O’Connor PM, Rossney A, Gotz F, Hill C, et al. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J Bacteriol. 2010;192:1131–1142. doi: 10.1128/JB.01375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, Chambers HF, Graber CJ, Szumowski JD, Miller LG, Han LL, Chen JH, Lin F, Lin J, Phan TH, et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med. 2008;148:249–257. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]
- Dufour A, Haldenwang WG. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugar G, Herbig A, Forstner KU, Heidrich N, Reinhardt R, Nieselt K, Sharma CM. High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Campylobacter jejuni Isolates. PLoS Genet. 2013;9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz F. Staphylococcus and biofilms. Molecular microbiology. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Graham JW, Lei MG, Lee CY. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J Bacteriol. 2013;195:4506–4516. doi: 10.1128/JB.00758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T. Magnesium in bone and the magnesium load test. Magnes Res. 2011;24:223–224. doi: 10.1684/mrh.2011.0297. [DOI] [PubMed] [Google Scholar]
- Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:5845–5851. doi: 10.1128/AAC.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clinical microbiology reviews. 2010;23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnen-Dechent WKM. Magnesium basics. Clinical Kidney Journal. 2012;5:2. doi: 10.1093/ndtplus/sfr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson-Kanth A, Tegmark-Wisell K, Arvidson S, Oscarsson J. Natural human isolates of Staphylococcus aureus selected for high production of proteases and alpha-hemolysin are sigmaB deficient. Int J Med Microbiol. 2006;296:229–236. doi: 10.1016/j.ijmm.2006.01.067. [DOI] [PubMed] [Google Scholar]
- Kies S, Vuong C, Hille M, Peschel A, Meyer C, Gotz F, Otto M. Control of antimicrobial peptide synthesis by the agr quorum sensing system in Staphylococcus epidermidis: activity of the lantibiotic epidermin is regulated at the level of precursor peptide processing. Peptides. 2003;24:329–338. doi: 10.1016/s0196-9781(03)00046-9. [DOI] [PubMed] [Google Scholar]
- Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, Godfrey P, Palmer KL, Bodi K, Mongodin EF, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;3 doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B, Kornblum J, Arbeit RD, Eisner W, Maslow JN, McGeer A, Low DE, Novick RP. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennette EH, Balows A, Hausler WJ, Jr., Shadomy HJ. Manual of clinical microbiology. 1985 [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marles-Wright J, Lewis RJ. The stressosome: molecular architecture of a signalling hub. Biochem Soc Trans. 2010;38:928–933. doi: 10.1042/BST0380928. [DOI] [PubMed] [Google Scholar]
- Marshall JH, Wilmoth GJ. Proposed pathway of triterpenoid carotenoid biosynthesis in Staphylococcus aureus: evidence from a study of mutants. J Bacteriol. 1981;147:914–919. doi: 10.1128/jb.147.3.914-919.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Cui L, Kim J, Hiramatsu K. Comprehensive Identification of Mutations Responsible for Heterogeneous Vancomycin-Intermediate Staphylococcus aureus (hVISA)-to-VISA Conversion in Laboratory-Generated VISA Strains Derived from hVISA Clinical Strain Mu3. Antimicrob Agents Chemother. 2013;57:5843–5853. doi: 10.1128/AAC.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E, Taylor D, Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- O’Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS microbiology letters. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. MRSA virulence and spread. Cell Microbiol. 2012;14:1513–1521. doi: 10.1111/j.1462-5822.2012.01832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MDowell P, Affas Z, Reynolds C, Holden MT, Wood SJ, Saint S, Cockayne A, Hill PJ, Dodd CE, Bycroft BW, et al. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC, Jr., Eliopoulos GM. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One. 2012;7:e28316. doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz A, Wieland KP, Putzbach K, Hentschel P, Albert K, Gotz F. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem. 2005;280:32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PM, Filipe SR, Tomasz A, Pinho MG. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:3627–3633. doi: 10.1128/AAC.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck SY, Jameson-Lee M, Villaruz AE, Bach T-HL, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Molecular cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- Schlievert PM. Effect of magnesium on production of toxic-shock-syndrome toxin-1 by Staphylococcus aureus. J Infect Dis. 1985;152:618–620. doi: 10.1093/infdis/152.3.618. [DOI] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Klauck G, Mika F, Hengge R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio. 2013;4:e00103–00113. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin-Massieu M. Spontaneous appearance of sectored colonies in Staphylococcus aureus cultures. J Bacteriol. 1961;82:316–317. doi: 10.1128/jb.82.2.316-317.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieradzki K, Tomasz A. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J Bacteriol. 2003;185:7103–7110. doi: 10.1128/JB.185.24.7103-7110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert GA, Hill B, Hollis R. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. Journal of clinical microbiology. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompanidou E, Denham EL, Becher D, de Jong A, Buist G, van Oosten M, Manson WL, Back JW, van Dijl JM, Dreisbach A. Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl Environ Microbiol. 2013;79:886–895. doi: 10.1128/AEM.03157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompanidou E, Sibbald MJ, Chlebowicz MA, Dreisbach A, Back JW, van Dijl JM, Buist G, Denham EL. Requirement of the agr locus for colony spreading of Staphylococcus aureus. J Bacteriol. 2011;193:1267–1272. doi: 10.1128/JB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votintseva AA, Miller RR, Fung R, Knox K, Godwin H, Peto TE, Crook DW, Bowden R, Walker AS. Multiple-Strain Colonization in Nasal Carriers of Staphylococcus aureus. J Clin Microbiol. 2014;52:1192–1200. doi: 10.1128/JCM.03254-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.