ABSTRACT
Although the canonical Wnt pathway and β-catenin have been extensively studied, less is known about the role of p120-catenin (also known as δ1-catenin) in the nuclear compartment. Here, we report that p120-catenin binds and negatively regulates REST and CoREST (also known as Rcor1), a repressive transcriptional complex that has diverse developmental and pathological roles. Using mouse embryonic stem cells (mESCs), mammalian cell lines, Xenopus embryos and in vitro systems, we find that p120-catenin directly binds the REST–CoREST complex, displacing it from established gene targets to permit their transcriptional activation. Importantly, p120-catenin levels further modulate the mRNA and protein levels of Oct4 (also known as POU5F1), Nanog and Sox2, and have an impact upon the differentiation of mESCs towards neural fates. In assessing potential upstream inputs to this new p120-catenin–REST–CoREST pathway, REST gene targets were found to respond to the level of E-cadherin, with evidence suggesting that p120-catenin transduces signals between E-cadherin and the nucleus. In summary, we provide the first evidence for a direct upstream modulator and/or pathway regulating REST–CoREST, and reveal a substantial role for p120-catenin in the modulation of stem cell differentiation.
KEY WORDS: p120-catenin, REST, CoREST, E-cadherin, Mouse embryonic stem cell
INTRODUCTION
Stem cell maintenance and lineage-specific differentiation are modulated by intrinsic and extrinsic signals acting upon mediators including transcription factors. Core transcriptional regulators required for mouse and human embryonic stem cell (ESC) maintenance include Oct4 (also known as POU5F1), Sox2 and Nanog (Young, 2011). Repressor complexes, such as that organized about REST, likewise regulate embryonic and neural stem cells and their derivatives (Aoki et al., 2012; Gao et al., 2011; Sun et al., 2008; Sun et al., 2005). Whereas debate surrounds the role of REST in maintaining ESC self-renewal (Buckley et al., 2009; Jørgensen et al., 2009a; Jørgensen and Fisher, 2010; Singh et al., 2008), it is agreed that it participates in the self-renewal of neural stem cells (NSCs) (Gao et al., 2011), as well as differentiation from ESCs and NSCs, such as that towards neural lineages (Aoki et al., 2012; Chen et al., 1998; Yamada et al., 2010).
REST recognizes and binds to specific genomic regions containing RE1 consensus sites within target gene promoters. There, it recruits a broad array of epigenetic and transcriptional modulators, often with the assistance of scaffold proteins, such as CoREST (also known as Rcor1) (Gopalakrishnan, 2009; Qureshi et al., 2010). REST is itself transcriptionally and post-transcriptionally modulated (Ballas et al., 2005; Westbrook et al., 2008). Despite this knowledge, there remains a large gap in our understanding of the roles of the micro-environment and upstream signals that regulate REST–CoREST (Gopalakrishnan, 2009).
Cell–cell adhesion is crucial for early vertebrate development and stemness (Boggetti and Niessen, 2012; Pieters and van Roy, 2014; Stepniak et al., 2009). E-cadherin, for example, enhances the self-renewal and survival of human ESCs (hESCs) (Li et al., 2010b), and its expression improves the derivation of induced pluripotent stem cells from mouse embryonic fibroblasts (Chen et al., 2010). Indeed, the heightened expression of E-cadherin can compensate for reductions in Oct4 during reprogramming, whereas in the absence of E-cadherin, the reprogramming of mouse embryonic fibroblasts is prevented (Redmer et al., 2011). At adherence junctions, the cytoplasmic tail of E-cadherin (membrane-distal region) associates not only with β-catenin but also with catenin proteins of the p120-catenin subfamily (Daniel and Reynolds, 1995; Reynolds et al., 1994). This subfamily includes p120-catenin itself (also known as δ1-catenin), and its members have roles in cadherin stabilization, small GTPase and cytoskeletal modulation and gene regulation (Ireton et al., 2002; McCrea and Park, 2007; Pieters et al., 2012a; Reynolds, 2007).
p120-catenin can translocate into the nucleus to modulate genes (Daniel et al., 2002; Hosking et al., 2007; Kim et al., 2004). We have previously reported that shared mechanisms (e.g. components of the destruction complex) regulate the stability of p120-catenin isoform 1 and β-catenin (Hong et al., 2010; Miller et al., 2013). In addition to some shared regulation at the protein level, p120-catenin and β-catenin act upon a number of the same gene targets in the nucleus (Crawford et al., 1999; Kim et al., 2004; Park et al., 2005; Spring et al., 2005). Distinct from β-catenin, which contains a transactivation domain (Gordon and Nusse, 2006; Gottardi and Peifer, 2008; MacDonald et al., 2009), p120-catenin appears to activate gene expression by competitively displacing repressors from gene promoter sites.
Here, we extend our knowledge of the nuclear role of p120-catenin and of stem cell biology by revealing that p120-catenin binds and derepresses the REST–CoREST complex to activate gene targets. Our results suggest that the p120-catenin–REST–CoREST pathway is coupled to the cadherin–catenin complex at cell–cell junctions, and, taken together, point to a new role for p120-catenin in the decision process of stem cells advancing to a differentiated state.
RESULTS
p120-catenin associates with REST and CoREST
In an earlier work, we determined that the p120-catenin family member ARVCF binds to kazrin (Cho et al., 2010), a protein with both cytoskeletal and nuclear roles (Groot et al., 2004; Sevilla et al., 2008). To identify nuclear binding partners of kazrin, we used yeast two-hybrid screening of a mouse adult brain library and found that kazrin associates with CoREST (data not shown). Central to this report, we went on to test whether ARVCF and the related p120-catenin bind to CoREST and its partner, the DNA-binding transcriptional repressor REST.
In this study, we focused on p120-catenin and its association with the REST–CoREST complex. We first confirmed, however, that ARVCF associated with CoREST and REST (supplementary material Fig. S1A), suggesting that it might share nuclear functions of p120-catenin as outlined below.
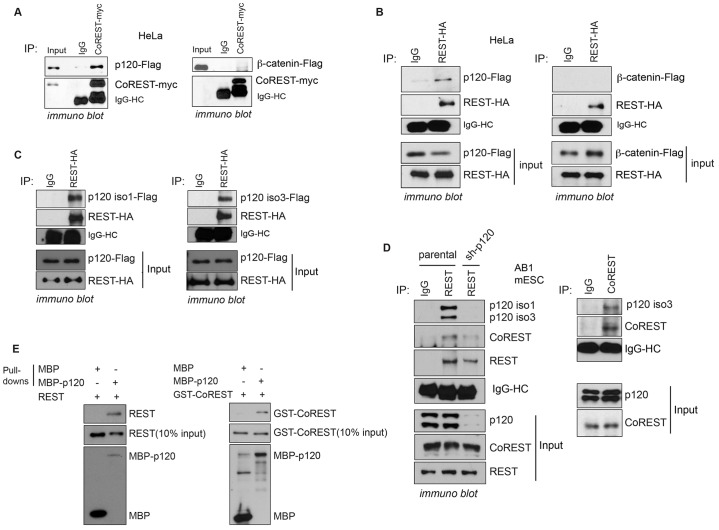
Often associating with the larger REST complex, CoREST contributes to gene target repression (Andrés et al., 1999). To test for an association between p120-catenin and CoREST, we initially used co-expression in HeLa cells followed by co-immunoprecipitation, which supported their interaction (Fig. 1A). β-Catenin, which is structurally similar to p120-catenin and ARVCF but falls within the distinct β-catenin subfamily displayed little if any interaction with CoREST, even when expressed at higher levels than p120-catenin, suggesting specificity in the p120-catenin–CoREST interaction.
Fig. 1.
p120-catenin associates with REST and CoREST. (A) p120-catenin, but not β-catenin, associates with CoREST in HeLa cells. CoREST–Myc was co-expressed with p120-catenin or β-catenin in HeLa cells, and CoREST–Myc was immunoprecipitated (IP). Upon longer film exposures (data not shown), CoREST is detected as expected in the input control within the right-hand panel, as appears in the left panel. (B) p120-catenin associates with REST in HeLa cells. REST was co-expressed with p120 or β-catenin in HeLa cells, and REST–HA was precipitated. (C) Both p120-catenin isoform 1 (iso1) and isoform 3 (iso3) associate with REST using an in vitro transcription and translation system. p120-catenin isoforms 1 and 3 were incubated with REST–HA, followed by REST–HA precipitation. (D) Endogenous p120-catenin associates with endogenous REST and CoREST in mESCs. Note that both p120-catenin isoform 1 and 3 associate with CoREST (see supplementary material Fig. S1C). The nuclear fraction was isolated and used for co-immunoprecipitation. The sh-p120 lane (cells expressing shRNA against p120-catenin) shows that we obtained specific immuno-blot detection of p120. (E) p120-catenin directly associates with REST and CoREST. Bacterially purified proteins were used for direct binding assays. MBP–p120-catenin was pulled down using amylose magnetic beads, followed by blotting with anti-GST or anti-REST antibodies, respectively, to detect GST–CoREST and REST.
We next determined whether exogenous p120-catenin associated with REST in HeLa cells. As with CoREST, we observed a reproducible p120-catenin–REST association, relative to IgG precipitations or REST precipitations with co-expressed β-catenin (Fig. 1B). This association was further observed in other cell lines, including HEK293 (supplementary material Fig. S1B,D) and mESCs (Fig. 1D; supplementary material Fig. S1C). To further assess the p120-catenin–REST association, we conducted binding assays using standard in vitro transcription-translation. Depending on context, p120-catenin exists in a number of isoforms generated from distinct translational start sites (in the same transcript) (Pieters et al., 2012b). Isoform 3 begins at a more downstream translational start site than isoform 1, and we found that REST associates with both isoforms (Fig. 1C).
We next tested whether endogenous p120-catenin associated with REST and CoREST in mESCs, where the REST–CoREST complex has been well studied, and found that both isoform 1 and isoform 3 associate with REST (Fig. 1D). When p120-catenin was depleted from mESCs, the p120-catenin–REST interaction was not observed (Fig. 1D). Endogenous CoREST also associated with endogenous p120-catenin in mESCs (Fig. 1D; supplementary material Fig. S1C). Although isoforms 1 and 3 of p120-catenin associate with CoREST in mESCs (supplementary material Fig. S1C), the p120-catenin isoform 3 interaction was more consistently evident (Fig. 1D). As in mESCs, we observed that endogenous p120-catenin associates with endogenous REST and CoREST in HEK293 cells (supplementary material Fig. S1D). We also evaluated the localization of p120-catenin relative to CoREST and REST using immunofluorescence. REST and CoREST, if expressed alone, were largely nuclear in the absence of p120-catenin (supplementary material Fig. S1F), whereas exogenous p120-catenin was localized in both the cytoplasm and nucleus (data not shown). Although less evident for REST (conceivably owing to its more rapid cytoplasmic destruction), p120-catenin expression caused the re-localization of a proportion of CoREST to the cytoplasmic compartment following their co-expression, consistent with a functional interaction. Given that p120-catenin associates with the transcriptional repressor kaiso (Kim et al., 2004), which binds N-CoR (Yoon et al., 2003), we tested whether kaiso associates with REST. In AB1 mESCs expressing the indicated exogenous proteins, CoREST (positive control) but not kaiso co-precipitated with REST in mESCs (supplementary material Fig. S1E), suggesting that kaiso is not part of the larger REST complex.
To assess whether p120-catenin directly binds REST and CoREST, we mixed purified maltose-binding protein (MBP)-tagged p120-catenin with purified GST–CoREST or GST–REST, followed by MBP pulldown. Both purified REST and CoREST directly associated with MBP–p120-catenin, whereas MBP alone interacted to an almost undetectable extent (Fig. 1E). Taken together, our findings suggest that p120-catenin binds both REST and CoREST in various cell types.
The zinc finger DNA-binding domain of REST and the central region of CoREST associate with the armadillo domain of p120
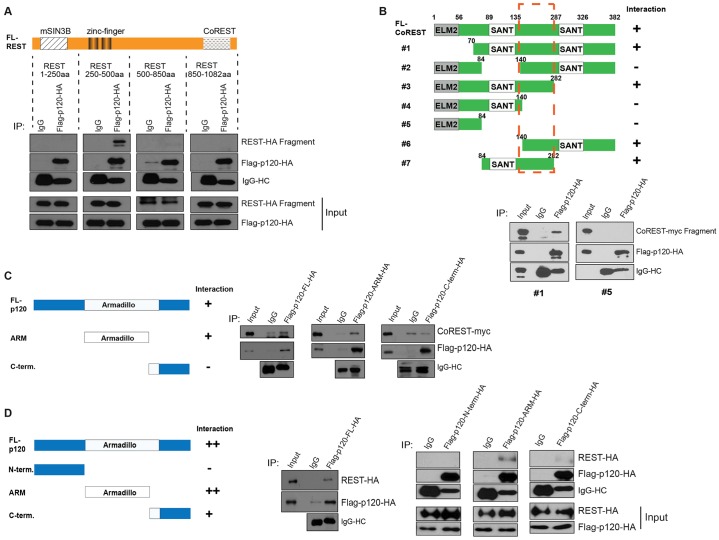
To help model the interactions of p120-catenin with REST and CoREST, we mapped the p120-catenin–REST and p120-catenin–CoREST associations using an in vitro transcription-translation system. REST contains regions required for partner binding [e.g. N-terminal versus C-terminal regions for mouse SIN3B (mSIN3B) and CoREST, respectively], whereas its zinc finger domain is used for sequence-specific DNA interactions (e.g. RE1 binding sites) (Ballas et al., 2001). Intriguingly, relative to IgG precipitations and other REST fragments, p120-catenin associated selectively with the zinc finger DNA-binding region of REST (Fig. 2A). This is an indication that p120-catenin might affect REST function.
Fig. 2.
Mapping REST and CoREST domains that bind p120-catenin and vice versa. (A) The zinc finger region of REST is necessary for p120-catenin association. REST fragments were incubated with full-length Flag–p120-catenin–HA, followed by p120-catenin immunoprecipitation (IP). (B) The region between the two SANT domains of CoREST associates with p120-catenin. Two example blots are provided; see also supplementary material Fig. S2A. CoREST fragments were incubated with p120-catenin and precipitated using the indicated antibodies. A dashed rectangle indicates that the CoREST region needed to associate with p120. (C,D) The armadillo repeat (ARM) domain of p120-catenin is needed for CoREST association (C), and for REST association (D). p120-catenin fragments (FL-p120, full-length p120-catenin; N-terminal, N-terminal region; C-term; C-terminal region) were incubated with CoREST (C) or REST (D), followed by precipitation of p120-catenin. +, some interaction, ++, strong interaction; –, no interaction. All proteins used in Fig. 2 were generated using an in vitro transcription and translation system. Note that the C-terminal fragment of p120-catenin includes a small portion of the armadillo repeat domain (repeat region 8.5–9).
By conducting p120-catenin–CoREST interaction mapping, we found that the region between the two SANT domains of CoREST is required for p120-catenin association (Fig. 2B; supplementary material Fig. S2A). The first SANT domain of CoREST binds partners including REST and HDAC1 (You et al., 2001), whereas the second SANT domain binds Brg1-associated factor 57 (BAF57) (Battaglioli et al., 2002). Finally, we mapped the p120-catenin regions needed for REST and CoREST association. We found that the armadillo domain of p120-catenin associates with both CoREST (Fig. 2C) and REST (Fig. 2D). Relative to the armadillo domain of p120-catenin, its N- and C-terminal domains did not associate with REST or only did so weakly (Fig. 2D). For example, the C-terminal domain of p120-catenin displayed a minor association with REST, perhaps owing to the fact that this construct had retained 1.5 armadillo repeats. Our mapping suggested that p120-catenin might modulate the REST–CoREST interaction or the REST–DNA interaction, affecting the gene-regulatory functions of the complex.
To assess whether p120-catenin forms distinct complexes with REST or CoREST or whether p120-catenin simultaneously binds to REST and CoREST, we generated a REST C-terminus deletion mutant incapable of associating with CoREST (Andrés et al., 1999) and conducted binding assays in vitro (supplementary material Fig. S2B). When CoREST was precipitated in the presence of p120-catenin, C-terminal-deleted REST could be co-precipitated. Full-length REST could be co-precipitated with CoREST in the absence or presence of p120-catenin, as expected. These findings suggest that p120-catenin not only binds to REST and CoREST individually as previously noted (Fig. 1E), but that p120-catenin can simultaneously associate with REST and CoREST.
p120-catenin modulates REST–CoREST gene targets in mESCs, in mammalian cell lines and in Xenopus laevis embryos
Mapping indicated that p120-catenin associates with the zinc finger DNA-binding region of REST. By manipulating p120-catenin levels in mESCs, we tested the hypothesis that p120-catenin modulates the function of REST–CoREST protein to de-repress gene targets.
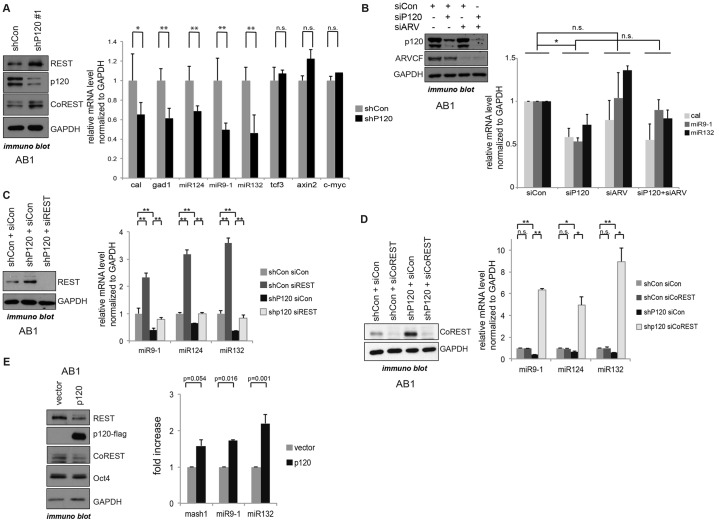
Indeed, we found that the short hairpin RNA (shRNA)-mediated depletion of p120-catenin in mESCs reproducibly led to increased REST and CoREST protein levels (REST, Fig. 3A,C and supplementary material Fig. S3A; CoREST, Fig. 3A). (This effect was not seen in Fig. 1D given the shorter duration of p120-catenin depletion prior to cell harvest). Conversely, the exogenous expression of p120-catenin led to decreased REST protein levels (Fig. 3E). Given that the p120-catenin expression effect was rescued upon the addition of a proteasome inhibitor (MG-132; supplementary material Fig. S2C), we expect that p120-catenin might promote REST protein destabilization through proteasome-dependent mechanisms.
Fig. 3.
p120-catenin activates REST–CoREST gene targets in mESCs. (A) REST–CoREST gene targets are repressed upon p120-catenin depletion (mediated by an shRNA against p120-catenin, shP120#1; shCon, control shRNA). See also supplementary material Fig. S3A, for the modulation of REST–CoREST gene targets by two independent shRNA constructs against p120-catenin. p120-catenin-depleted mESCs were generated using shRNAs directed against distinct regions of the p120-catenin transcript. At 72 h after puromycin selection, p120-catenin and REST protein levels were tested by immunoblotting, and the indicated REST–CoREST gene target transcripts were tested by real-time PCR. GAPDH was used as an internal control. Data are presented as mean±s.d. *P<0.05; **P<0.01; n.s., not statistically significant. (B) ARVCF co-knockdown does not increase p120-catenin depletion effects on REST–CoREST gene targets. The indicated siRNAs (siCon, control siRNA; siP120, siRNA against p120-catenin; siARV, siRNA against ARVCF) were transfected into AB1 cells, followed by incubation for 72 h and harvest for immunoblotting and real-time PCR. Data are presented as mean±s.d. *P<0.05; n.s., not statistically significant. (C,D) REST (C) and CoREST (D) knockdown rescues p120-depletion effects. Using siRNA, REST (siREST) and CoREST (siCoREST) were respectively knocked down in control-depleted and p120-catenin-depleted mESCs, which was confirmed by immunoblotting. REST–CoREST target gene expression was tested by real-time PCR. All transcripts were normalized to expression of GAPDH. In all graphs, data are presented as mean±s.e.m. (E) REST–CoREST gene target transcripts are activated upon p120-catenin expression. p120-catenin was transiently expressed in mESCs using liposome-mediated transfection. At 48 h after transfection, cells were lysed and used for immunoblotting with the indicated antibodies. The REST–CoREST gene transcripts were tested by real-time PCR and normalized to GAPDH and data are presented as mean±s.e.m.
Given that REST protein levels are increased in p120-depleted cells, we predicted that there would be decreased transcription from the REST gene targets calbindin, Gad1, miR-124, miR-9-1, and miR-132. Using two independent shRNA constructs directed against p120-catenin (supplementary material Fig. S3A), decreased expression was found in most but not all cases (Fig. 3A). For example, p120-catenin depletion did not lead to increased repression of the gene encoding TCF3 (Fig. 3A). In addition, non-REST targets, such as Axin2 and c-Myc, were not responsive to p120-catenin depletion, suggesting that p120-catenin depletion has clear but selective effects on REST gene target transcription.
Given that REST and CoREST further associate directly (in vitro) with ARVCF (supplementary material Fig. S1A), we tested whether the co-depletion of ARVCF and p120-catenin exhibited an additive effect on REST targets. Their co-depletion, however, did not enhance the previously observed partial p120-depletion effects upon REST targets (Fig. 3B). The partial effect of p120-catenin depletion on REST targets is presumably due to the involvement of other cis-acting regulators, or to the involvement of yet other catenins, such as δ-catenin.
To determine whether the observed p120-depletion effect occurs as predicted through REST or CoREST, we conducted REST and CoREST knockdowns with short interfering RNA (siRNA). Prior to such REST- or CoREST-knockdown tests, we confirmed that the expression of a non-targetable p120-catenin construct could rescue p120-catenin-depletion effects (supplementary material Fig. S3B). As expected, REST depletion led to a partial, but statistically significant, rescue of the p120-catenin knockdown effect on REST–CoREST target genes (Fig. 3C). Interestingly, CoREST depletion in mESCs increased REST–CoREST target gene expression beyond that seen in the control conditions (4–10-fold ‘over-rescue’; Fig. 3D). This result appears to be in keeping with findings that CoREST has substantial roles in REST-independent gene repression, including at genes modulated by REST–CoREST (Ballas et al., 2005).
We next determined whether the expression of p120-catenin had a positive effect on REST–CoREST target gene transcription in mESCs. Indeed, as a function of the identity of the transcript, we observed 1.5–2-fold increases in gene target levels (Fig. 3E). This effect might be due to p120-catenin-promoted reductions in REST–CoREST protein levels and/or p120-catenin-mediated competitive displacement of REST–CoREST from DNA (see below).
To determine whether p120-catenin-mediated modulation of REST–CoREST target genes extends beyond mESCs, we used NIH3T3 and HEK293 (differentiated) cell lines. We found that p120-catenin depletion in these cells likewise reduced the expression of REST–CoREST gene targets (supplementary material Fig. S3C), with effects similarly rescued upon CoREST depletion (supplementary material Fig. S3D). p120-catenin expression, conversely, increased the transcript levels of REST–CoREST gene targets (Mash1, also known as Ascl1, and synaptotagmin 4) (supplementary material Fig. S3E), as was observed in HEK293 cells (data not shown). These increased transcript levels were rescued by the expression of CoREST, or to a partial extent REST (supplementary material Fig. S3E), suggesting that p120-catenin-mediated modulation of REST–CoREST gene targets occurs through the REST–CoREST complex, consistent with the results obtained in mESCs.
Finally, we tested for functional affects in vivo using Xenopus laevis. We observed that knockdown of p120-catenin using an established morpholino antisense oligonucleotide (Fang et al., 2004) decreased transcripts that are regulated by REST–CoREST, such as Xenopus calbindin and Mash1 (supplementary material Fig. S3G). These results suggest that, with respect to REST–CoREST gene target modulation, p120-catenin has a shared role across differing types of mammalian cells and in amphibians.
p120-catenin modulates REST binding to RE1 sites at cis-regulatory regions of target genes
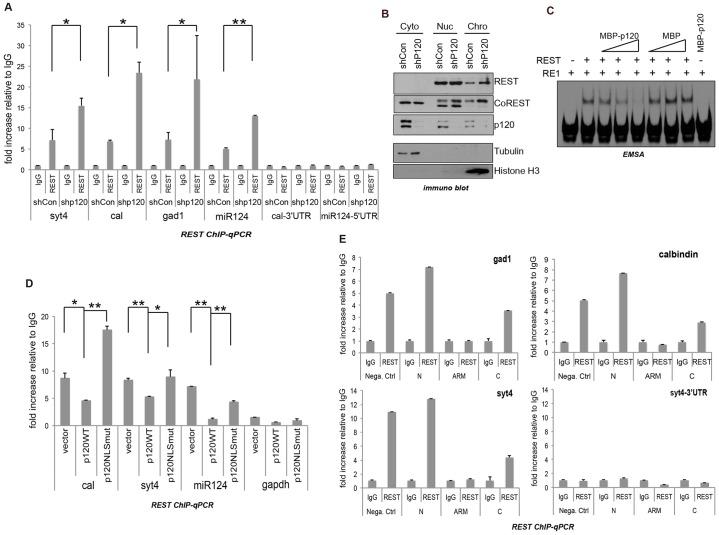
To determine whether p120-catenin has an effect on REST occupancy at RE1 sites in known gene targets (e.g. the type II voltage-dependent Na+ channel), we conducted chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) using normal and p120-depleted mESCs. We kept in mind our above evidence (Fig. 2A) that suggested that p120-catenin might compete with DNA (RE1) for association with the zinc-finger domain of REST.
Using ChIP-qPCR in mESCs, we tested whether p120-catenin depletion led to enhanced REST association with DNA, and indeed observed increased REST–DNA ChIP (2–4-fold) across four established REST–CoREST gene targets (synaptotagmin 4, calbindin, gad1 and miR-124) (Fig. 4A). Negative controls included IgG precipitations, and ChIP of 3′ or 5′ untranslated regions (UTRs) lacking RE1 consensus binding sites. We next examined REST–CoREST protein localization. In p120-catenin-depleted mESCs, the levels of REST and CoREST increased in the chromatin-bound and nucleoplasmic fractions (Fig. 4B). Although we do not know the basis, the cytoplasmic, nucleoplasmic and chromatin fractions of CoREST exhibited slight differences in their gel migration patterns. In all cases, our ChIP and other findings continued to suggest that p120-catenin and REST are functionally linked in mESCs.
Fig. 4.
p120-catenin modulates REST association with RE1 regions. (A) p120-catenin knockdown increases REST binding to RE1 regions. ChIP-qPCR was conducted using p120-depleted (shp120) and control-depleted (shCon) mESCs (see Materials and Methods). Cross-linked DNA-protein complexes were precipitated using anti-REST or control (IgG) antibody. The Cal-3′UTR and miR-124-5′UTR regions were used as internal-transcript negative controls. Data are normalized to the IgG input value and presented as mean±s.d. *P<0.05; **P<0.01. (B) p120-catenin knockdown increases the REST and CoREST association with chromatin. Control and p120-depleted mESCs were fractionated into cytosolic (Cyto), nuclear (Nuc), and chromatin (Chro) fractions. The indicated proteins were tested using immunoblotting. Tubulin was used as a cytoplasmic fraction marker, and histone H3 was used as a chromatin fraction marker. (C) p120-catenin decreases REST binding to RE1 region in vitro. REST (thrombin-cleaved from GST–REST), MBP–p120-catenin and MBP were purified from bacteria. In two different pair-wise manners, protein–protein or protein–DNA combinations were made, generating similar experimental results: either REST and MBP–p120-catenin (or MBP alone) were incubated overnight at 4°C followed by addition of the oligonucleotide; or REST was incubated with the oligonucleotide for 10 min, followed by addition of increasing amounts of MBP–p120-catenin (or MBP alone) for 10 min at room temperature (as shown). See Materials and Methods for more details. (D) p120-catenin expression partially decreases the binding of REST to RE1 regions, whereas expression of the p120-catenin NLS mutant (NLSmut) had little effect. At 48 h after transfection of wild-type (WT) or NLS-mutant p120, mESCs were fixed and harvested for ChIP-qPCR. The bars indicate the fold increase relative to the IgG value. GAPDH was used as a negative control. Data are normalized to the IgG input value and presented as mean±s.d. *P<0.05; **P<0.01. (E) Expression of the armadillo or C-terminal tail domains of p120-catenin resulted in reduced REST–DNA association. The p120-catenin fragments of the N-terminal (N), armadillo repeat (ARM), or C-terminal tail (C) were expressed in mESCs; these were then processed for ChIP-qPCR. The Syt4 3′UTR was used as an internal-transcript negative control. Data are normalized to the IgG input value and presented as mean ±s.d.
This was further supported by complementary experiments in which p120-catenin was overexpressed, where the expectation would be decreased REST occupancy at RE1 regions. Consistent with electrophoretic mobility shift assay (EMSA) results indicating that p120-catenin prevents REST from binding to RE1 consensus sites (Fig. 4C), our REST ChIP-qPCR findings indicated that wild-type p120-catenin expression significantly reduced binding of REST to the RE1 gene-control region of miR-124, and partially decreased REST binding to the RE1 regions of calbindin and synaptotagmin 4 (Fig. 4D). In contrast, the expression of a previously characterized nuclear localization sequence (NLS) mutant of p120-catenin, which is known to mostly localize in the cytoplasm (Kelly et al., 2004), reduced REST–DNA binding to a lesser extent, suggesting that p120-catenin nuclear translocation might precede the detachment of REST from DNA (RE1 binding sites) (Fig. 4D). In the case of calbindin, the p120-catenin NLS-mutant unexpectedly increased the endogenous REST–DNA association, conceivably by sequestering a negative modulator of REST in the context of the calbindin promoter.
Given that the central armadillo domain of p120-catenin associates with REST and CoREST (Fig. 2), we tested whether the expression of the isolated p120-catenin armadillo domain affected REST–DNA occupancy. Indeed, relative to the more N-terminal domain of p120-catenin, the p120-catenin armadillo domain decreased REST–DNA binding at known REST gene-control (RE1) regions. This was observed for Gad1, calbindin, and synaptotagmin 4, but not at the 3′ UTR of synaptotagmin 4, which does not contain an RE1 site (Fig. 4E). In addition to the armadillo domain of p120-catenin, expression of its C-terminal region partially decreased REST–DNA binding, consistent with its weak REST interaction (Fig. 2D). Controls demonstrated that each Flag–HA epitope construct was expressed at similar levels (data not shown). These results suggest that p120-catenin de-represses REST–CoREST gene targets through inhibitory interactions with REST and/or CoREST.
Expression of mESC stemness markers is increased upon p120-catenin depletion
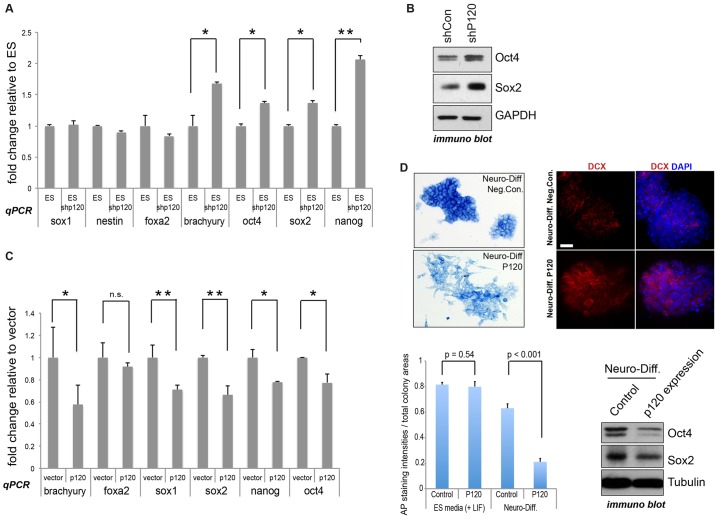
Among other functions, the REST–CoREST complex has a role in preventing the precocious differentiation of neural stem cells and, more contentiously, perhaps also of ESCs (see Introduction). We examined the expression of stemness and differentiation markers in mESCs in which p120-catenin was depleted. Up to 2-fold increases were found in the transcript level of the core pluripotency gene Nanog, with more modest increases occurring for Oct4 and Sox2 (Fig. 5A). Importantly, we saw substantial increases in the level of Oct4 and Sox2 at the protein level on p120-catenin depletion (Fig. 5B). Given that Oct4 and Sox2 contribute to Nanog expression in mESCs (Kuroda et al., 2005; Rodda et al., 2005), the increased level of Oct4 and Sox2 expression following p120-catenin depletion might contribute to the observed increased levels of Nanog transcript. Anti-Nanog antibodies performed poorly even in control mESCs, so we omitted the Nanog protein data. Consistent with our molecular findings, the depletion of p120-catenin did not lead to increased mESC differentiation on the basis of alkaline phosphatase readouts (data not shown). Likewise, the transcript levels of differentiation markers such as Sox1 and nestin (neuro-ectodermal markers), Foxa2 (endodermal marker) and brachyury (mesodermal marker) did not show notable responses to p120-catenin depletion (Fig. 5A). Complementing p120-depletion effects on pluripotency markers (Oct4, Sox2 and Nanog), p120-catenin expression instead led to their reduction by ∼20–30% (Fig. 5C). Although we do not know its basis, in this overexpression setup, modest decreases were seen in the transcript levels of two differentiation markers (brachyury and Sox1) (Fig. 5C). p120-catenin expression had little effect upon alkaline phosphatase readouts of mESC stemness under self-renewal conditions (data not shown); however, under neuronal differentiation conditions, we found that p120-catenin expression noticeably accelerated differentiation, as evident from alkaline phosphatase staining, increased doublecortin (DCX) expression (as assessed by immunostaining), and a reduction in Oct4 and Sox2 protein levels (visualized following immunoblotting) (Fig. 5D). In summary, our findings suggest that p120-catenin modulates established pluripotency markers at both the mRNA and protein levels. Given that we reveal that p120-catenin binds REST and CoREST and that a functional relationship is observed at REST–CoREST gene targets, p120-catenin-mediated effects upon pluripotency genes might arise in part as a consequence of the relationship between p120-catenin and REST–CoREST.
Fig. 5.
mESC stemness markers are increased upon p120-catenin depletion, and decreased upon p120-catenin expression. (A) p120-catenin knockdown (ES shp120) increases mRNA levels of pluripotency markers. Pluripotency and differentiation marker transcripts of p120-depleted and control mESCs were tested by real-time PCR. Oct4, Sox2 and Nanog serve as pluripotency markers; Sox1 and nestin as neuro-ectodermal differentiation markers; Foxa2 as an endoderm marker; and brachyury as a mesoderm marker. Results are mean±s.d. *P<0.05; **P<0.01. (B) p120-catenin knockdown results in increased protein expression of the pluripotency markers Oct4 and Sox2. The p120-depleted (shP120) and control (shCon) mESC lysates were used for immunoblotting. GAPDH was used as an internal control. (C) mESC stemness markers are decreased upon p120-catenin expression. Pluripotency and differentiation marker transcripts were tested by real-time PCR. All transcripts were normalized to GAPDH. Results are mean±s.d. *P<0.05; **P<0.01; n.s., not significant. (D) p120-catenin expression enhances pluripotency loss and neuronal differentiation of mESCs under differentiation conditions. p120-catenin-expressing and control mESCs were maintained in regular stem cell medium or with N2B27 neuronal differentiation medium (direct differentiation method) for 48 h. Pluripotency was tested by alkaline phosphatase (AP) staining and by immunoblotting for Oct4 and Sox2. Results are mean±s.d. P-values were obtained by Student's t-test. Prior to immunostaining with anti-DCX antibody, cells were maintained with N2B27 for 72 h. DAPI was used for counter-staining. Scale bar: 30 µm.
p120-catenin modulates the neuronal differentiation of mESCs
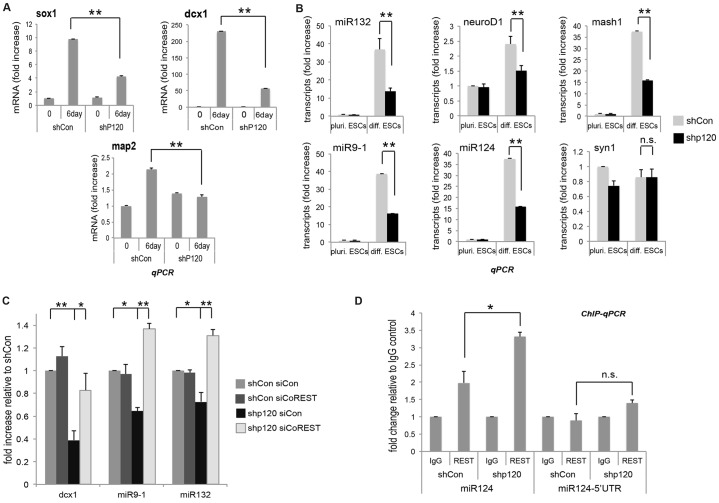
Although members of the p120-catenin subfamily contribute to neuronal development (Abu-Elneel et al., 2008; Arikkath et al., 2009; Elia et al., 2006; Ho et al., 2000; Matter et al., 2009), the nuclear roles of p120-catenin in this context are largely unknown. Given that REST represses multiple neuronal differentiation genes (Abrajano et al., 2009b; Chen et al., 1998), we determined whether p120-catenin has an effect on the neural differentiation of mESCs. Under differentiation conditions (using the direct or monolayer method), neural-differentiation marker transcripts (Sox1, Dcx1 and Map2) were increased in control cells. However, in response to p120-catenin depletion (and thereby maintained or enhanced gene target repression by REST–CoREST), the expression of these same neuronal transcripts was reduced (Fig. 6A). Dcx1 expression was reduced more than 70% in p120-depleted cells compared to control cells, and the reductions in Sox1 and Map2 were likewise significant. Consistent with these findings, p120-catenin expression instead increased Dcx1 expression as compared to controls under neuro-differentiation conditions (Fig. 5D). In parallel, we performed immunostaining with anti-nestin antibody. However, in this case, we did not observe a significant difference between control and p120-catenin expression, perhaps because we only tested at one extended time point (3-day differentiating AB1 cells; data not shown). In all cases these findings suggest that p120-catenin plays a role in the neurogenesis of mESCs.
Fig. 6.
p120-catenin modulates neuronal differentiation of mESCs. (A) p120-catenin knockdown reduces the neural differentiation of mESCs (direct differentiation method). p120-catenin-depleted (shP120) and control (shCon) mESCs were differentiated to neuronal cells by incubation over 6 days with N2B27 neuronal differentiation medium. Neuronal differentiation markers were tested by real-time PCR. Results are mean±s.d. *P<0.05; **P<0.01. Note that Sox1 serves as a neural stem and progenitor marker; Dcx1 as an early neuronal differentiation marker; and Map2 as a late neuronal differentiation marker. (B) p120-catenin knockdown decreases the mRNA levels of known REST–CoREST targets in mESCs undergoing neural differentiation. At 4 days after differentiation, p120-depleted and control mESCs were harvested and the indicated REST–CoREST gene targets were tested by real-time PCR. All transcripts were normalized to GAPDH. Results are mean±s.d. *P<0.05; **P<0.01; n.s., not statistically significant. ‘pluri. ESC’, growth in stem cell medium containing LIF and a high concentration of FBS to maintain stemness; ‘diff. ESC’, growth under differentiation conditions. (C) CoREST knockdown rescues p120-catenin knockdown effects. CoREST was knocked down using siRNA (siCoREST) in p120-depleted mESCs. The indicated REST–CoREST target transcripts were tested by real-time PCR and normalized to GAPDH. Results are mean±s.d. *P<0.05; **P<0.01. (D) p120-catenin knockdown increases REST binding to the RE1 region of miR-124 in neuronal differentiating mESCs. ChIP-qPCR was conducted using p120-depleted versus control mESCs, differentiated over 4 days in N2B27 neuronal differentiation medium. Results are mean±s.d. *P<0.05; n.s., not statistically significant.
To further test the effects of p120-catenin on neurogenesis and whether they are mediated through REST–CoREST, we measured the mRNA levels of REST–CoREST gene targets known to have a role in early neural development. In keeping with our findings above involving general neural markers, the mESC transcript levels of miR-132, Mash1, miR-9-1, miR-124, NeuroD1 (Fig. 6B) and Dcx1 (Fig. 6A) were decreased upon p120-catenin depletion. This decrease was effectively rescued upon CoREST depletion (Fig. 6C; ‘over-rescue’ effects were addressed earlier). Furthermore, ChIP-qPCR findings showed enhanced REST occupancy at the miR-124 RE1 region in p120-depleted cells, whereas the negative control 5′UTR region of miR-124 showed little, if any, REST binding (Fig. 6D). To support the results described above, in which we used the direct monolayer neural differentiation method, we used another established neural differentiation method that involves retinoic acid (Fraichard et al., 1995), and obtained similar results (supplementary material Fig. S4). These findings are consistent with the view that reduced neural differentiation in p120-depleted mESCs arises through functional effects on the REST–CoREST complex.
Upstream pathway regulation: E-cadherin appears to modulate REST–CoREST gene targets through p120-catenin in mESCs
In this report, we have concentrated on the relationship of p120-catenin with REST–CoREST, and thereby the stemness and differentiation of mESCs (downstream effects). However, a remaining question concerns the upstream modulator(s) of p120-catenin in this pathway. Given that canonical Wnt signaling contributes to both stemness and differentiation in mESCs, and we have previously shown that p120-catenin isoform 1 positively responds to Wnt signals through mechanisms shared with β-catenin (Hong et al., 2010), we considered that Wnt signals might be upstream participants. We instead found, however, that mESC exposure to Wnt1 and Wnt3a (Wnt ligands) lowered the expression of neural differentiation markers such as Sox1 and nestin (data not shown), consistent with prior reports that canonical Wnt signals enhance mESC pluripotency (Nusse, 2008; Sokol, 2011).
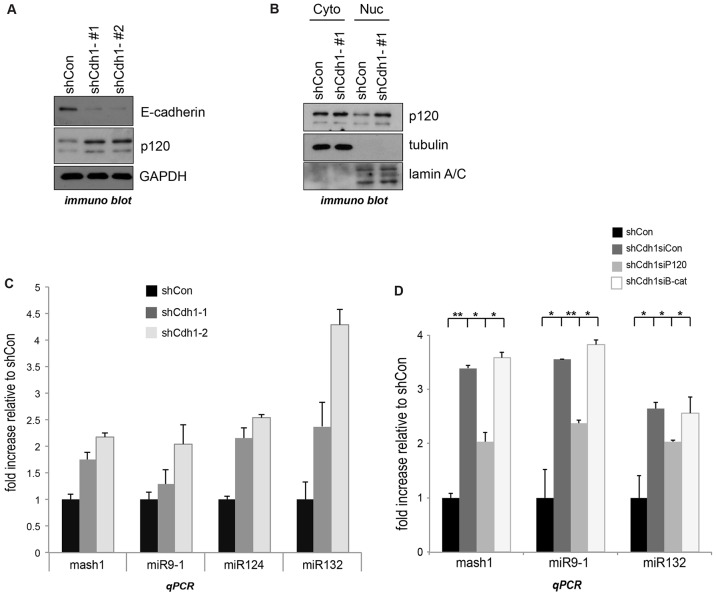
We next turned our attention to a second important player in both stem cell and catenin biology, namely cadherins. The relationship between p120-catenin and E-cadherin is characterized in contexts such as cadherin stability (see Introduction) (Hatzfeld, 2005; Humphries and Reynolds, 2009; McCrea and Park, 2007; McEwen et al., 2012; Reynolds, 2007; Boggetti and Niessen, 2012). Intriguingly, E-cadherin modulates hESC differentiation in vitro, with reductions in E-cadherin levels associated with a departure from stemness (Li et al., 2010a). We thus tested whether low E-cadherin levels are functionally associated with p120-catenin-mediated REST–CoREST modulation of target genes. We generated mESCs with E-cadherin depletion through RNAi. For reasons we do not understand, total p120-catenin protein was moderately increased in E-cadherin-knockdown mESCs (Fig. 7A), and this effect was also evident in the nuclear fraction of p120-catenin (Fig. 7B). Of interest, E-cadherin depletion was correlated with the elevation of REST–CoREST gene target transcripts, such as Mash1, miR-9-1, miR-124 and miR-132 (Fig. 7C). Furthermore, p120-catenin knockdown, but not β-catenin knockdown (specificity control), partially counteracted these effects in E-cadherin-depleted ESCs (Fig. 7D). Given that the physical interaction p120-catenin with E-cadherin is relevant to cadherin–catenin functions and to associated downstream signaling (Ireton et al., 2002; Ratheesh et al., 2013; Schackmann et al., 2011; Van den Bossche et al., 2012), we tested whether a p120-catenin point-mutant unable to bind E-cadherin retained rescuing activity, in line with our focus upon the nuclear effects of p120-catenin. Indeed, to a similar extent as p120-catenin wild-type, the established p120-catenin N478A mutant (Ishiyama et al., 2010) retained partial rescuing effects upon REST gene targets (supplementary material Fig. S3F). These findings together suggest that E-cadherin participates, in an upstream capacity, in regulating p120-transduced effects upon the nuclear REST–CoREST complex, and thereby differentiation processes in mESCs.
Fig. 7.
E-cadherin regulates REST–CoREST gene targets through p120-catenin. (A) E-cadherin-depleted mESCs were generated using an shRNA approach (two shRNAs, shCdh1-#1 and shCdh1-#2), with knockdown efficacy confirmed by immunoblotting. (B) E-cadherin-depleted versus negative control-depleted (shCon) mESCs were separated into cytoplasmic (Cyto) and nucleoplasmic (Nuc) fractions that were assayed for p120-catenin using immunoblotting. Note that tubulin serves as a cytoplasmic marker and nuclear lamin A/C as a nucleoplasmic marker. (C) The indicated REST–CoREST target transcripts from control and E-cadherin-depleted mESCs were tested by real-time PCR. Results are mean±s.d. (D) p120- and β-catenins were knocked down using a siRNA approach (siP120 and siB-cat, respectively) in E-cadherin-depleted mESCs. The REST–CoREST gene target transcripts were tested by real-time PCR. All transcripts were normalized to GAPDH. Results are mean±s.d. *P<0.05; **P<0.01.
DISCUSSION
Numerous regulatory mechanisms contribute to stemness or lineage-specific differentiation. In the nucleus of embryonic and neural stem cells, the REST–CoREST complex is thought to restrict the precocious expression of genes that promote differentiation as well as assist in lineage differentiation. In particular, in embryonic and neural stem cells, as well as in non-neural cells that have already differentiated, REST–CoREST appears to be crucial to suppressing the neural gene program (Abrajano et al., 2009a; Aoki et al., 2012; Chen et al., 1998; Gao et al., 2011; Jørgensen et al., 2009b; Sun et al., 2005). Less is known about the upstream mechanisms regulating REST–CoREST itself. One of the best characterized mechanisms is that REST is post-translationally downregulated in NSCs through ubiquitylation (β-TRCP E3 ligase) and proteasomal destruction (Westbrook et al., 2008), as occurs in ESCs differentiating towards neuronal lineages (Ballas et al., 2005; Su et al., 2004). Here, we find that p120-catenin binds REST and CoREST, to regulate the REST–CoREST complex in pluripotent mESCs, as well as in mESCs undergoing neural differentiation. We also provide suggestive evidence, requiring a separate study to address fully, that E-cadherin plays an upstream role in this catenin pathway.
p120-catenin modulates REST–CoREST association with DNA
We find that for p120-catenin to modulate gene targets of REST–CoREST, it must be capable of associating with REST or CoREST. Expression of the central armadillo p120-catenin domain, but not its N-terminal domain, resulted in reduced REST occupancy at gene targets (RE1 binding sites). Expression of the armadillo domain also reduced endogenous REST protein levels (data not shown). We found that the armadillo domain of p120-catenin associates with REST and CoREST. On the basis of REST ChIP experiments, p120-catenin lowers the ability of REST to bind DNA.
p120-catenin might lower REST binding to DNA by competition for REST (a direct effect), or alternatively, this might occur through lowering REST binding to CoREST (an indirect effect). Indeed, we found that expression of the armadillo domain of p120-catenin diminished the REST–CoREST association at the protein level in vitro (data not shown). Although future studies will be required, we surmise that p120-catenin directly (through REST) or indirectly (through CoREST) reduces REST–CoREST associations with DNA and/or with chromatin-regulatory factors. This correlates with increased protein turnover of REST–CoREST. Such a mechanism would be analogous to p120-catenin-mediated displacement of kaiso, another zinc finger transcriptional repressor (Daniel and Reynolds, 1999; Kelly et al., 2004; Kim et al., 2004).
p120-catenin modulation of gene targets
We observed that p120-catenin modulates several, but not all, REST target genes (Fig. 3). This result is consistent with those of a previous report using mESCs, in which neuronal genes that contained canonical RE1 regions responded preferentially to REST depletion (Jørgensen et al., 2009b). Proteasome inhibitors helped to maintain REST and CoREST levels in the face of p120-catenin expression (supplementary material Fig. S2C), suggesting that p120-catenin modulates REST–CoREST destruction, which fits with published studies of REST ubiquitylation (Westbrook et al., 2008). Given that p120-catenin isoform 1 is likewise negatively regulated by ubiquitylation (Hong et al., 2010), and both it and REST share the same E3 ligase (β-TrCP) (Westbrook et al., 2008), REST destruction might be enhanced when associated with p120-catenin.
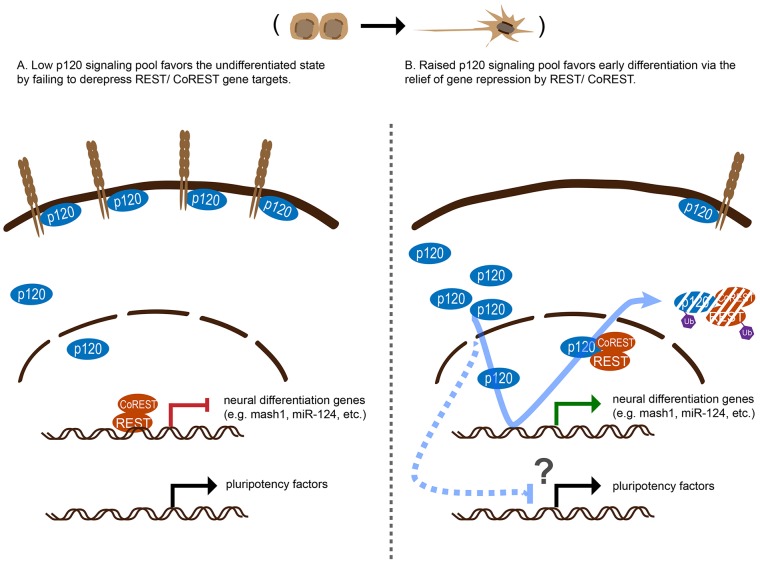
Regardless of the mechanism by which p120-catenin influences REST–CoREST protein levels in mESCs, our p120-catenin–REST and p120-catenin–CoREST interaction results, EMSA and REST ChIP findings support p120-catenin directly or indirectly displacing REST, resulting in the relief of repression (activation) of REST–CoREST gene targets (a model is shown in Fig. 8).
Fig. 8.
Model of the roles of p120-catenin in early neural differentiation of mESCs. (A) In the pluripotent state, ESCs express E-cadherin at higher levels which sequesters p120-catenin to junctional regions. Although a p120-catenin ‘signaling pool’ exists in the cytoplasm and nucleus, it is small. REST–CoREST represses neural differentiation genes, preventing precocious expression. (B) During early differentiation, E-cadherin levels are decreased, such that the p120-catenin signaling pool is increased. The enlarged p120-catenin signaling pool contributes to ESC differentiation through the de-repression (activation) of REST–CoREST gene targets (e.g. Mash1, miR-124 and miR-132). Furthermore, increased p120-catenin signaling pools reduce the levels some pluripotency factors (Nanog, Oct4 and Sox2) by an unknown mechanism, to favor differentiation. This conceivably occurs in part through p120-catenin-mediated de-repression of REST–CoREST gene targets (see text, given there is debate on the role of REST role maintaining ESC stemness) or through other mechanisms.
p120-catenin in modulating stemness and differentiation
Recognizing controversies in the literature, principally with respect to the role of REST in maintaining ESC pluripotency (Buckley et al., 2009; Jørgensen et al., 2009a; Jørgensen and Fisher, 2010; Singh et al., 2008; Yamada et al., 2010), REST has been proposed to modulate the balance of stemness versus differentiation and lineage specification of both embryonic and neural stem cells (Aoki et al., 2012; Gao et al., 2011; Su et al., 2004; Sun et al., 2005). These activities involve the direct repression (or in some cases activation) of differentiation-promoting genes, especially of the neural lineage. Elucidating the means by which REST is itself regulated is thus important to understanding both normal and pathological biological processes. In disease, the overly repressive activity of the REST–CoREST complex promotes excessive gene silencing due to effects upon DNA methylation or histone modifications. Among other things, pathological expression of REST alters the level and activity of non-coding RNAs (Rossbach, 2011), some of which are associated with glioblastoma multiforme (Conti et al., 2012) and poor prognosis (Kamal et al., 2012; Wagoner and Roopra, 2012).
In homeostasis, ESCs maintain a ‘poised’ state to enable genes that are crucial to differentiation to respond quickly and precisely to differentiation signals (Young, 2011). Although speculative, our data leave open the possibility that p120-catenin helps poise the REST complex for the balanced modulation of its many gene targets in response to upstream signals. Alternatively, p120-catenin might modulate REST–CoREST in an acute manner that is responsive to upstream signals that act on p120-catenin itself. We discuss one possibility below, which involves the E-cadherin–catenin complex.
Although p120-catenin knockdown lowered the expression of REST–CoREST direct gene targets (e.g. pro-neural genes and neuronal miRNAs) and indirect neural markers (e.g. Map2), mESCs still underwent partial neural differentiation when subjected to neural differentiation conditions. Because knockdowns are often limited in mammalian cells, gene expression from REST–CoREST targets might simply have remained adequate to permit differentiation. In addition, it is possible that a more impressive block in differentiation would occur upon the co-depletion of p120-catenin and other catenins, such as δ2-catenin, which is enriched in neural tissues (Kosik et al., 2005; Paffenholz and Franke, 1997; Zhou et al., 1997).
Our mESC findings appear at first to contradict those in an earlier report using hESCs, in which p120-catenin knockdown led to stemness loss under hESC maintenance conditions (Li et al., 2010a). Although mESCs share similarities with hESCs, they differ in several major aspects (Hirai et al., 2011). This includes that stemness in mESCs is insensitive to cell–cell dissociation (Mohamet et al., 2010). p120-catenin depletion destabilizes E-cadherin (Davis et al., 2003), often resulting in altered cell–cell interactions. In contrast to hESCs, p120-catenin depletion did not induce differentiation of the mESCs used in our study (Fig. 5). Furthermore, consistent with a prior mESC report (Soncin et al., 2009), when we directly targeted E-cadherin for depletion in mESCs (Fig. 7), we observed smaller colony sizes but not enhanced differentiation. Our findings, therefore, appear to be compatible given some known differences between mouse and human ESCs.
E-cadherin, a potential upstream modulator of p120-catenin nuclear activity
We turned our attention to the cadherin–catenin complex, as it offered a potential means to modulate p120-catenin and thereby REST–CoREST. Although cell–cell adhesive interactions affect stemness and differentiation in hESCs, but less so mESCs (Li et al., 2010a), we remained interested in testing whether the E-cadherin present in mESCs contributes to p120-catenin-mediated modulation of REST–CoREST. Given that E-cadherin levels become lessened in the neuroectoderm of mouse embryos (Stemmler, 2008), p120-catenin should be less sequestered (an enlarged ‘signaling pool’) (Bellovin et al., 2005; Shibata et al., 2004), to potentially act on REST–CoREST to de-repress (activate) neuronal gene targets. Indeed, E-cadherin depletion in mESCs increased REST–CoREST gene target expression. Importantly, as assessed by p120-catenin co-depletion, this effect was at least in part dependent on p120-catenin. Thus, our results are consistent with the possibility that the E-cadherin–catenin complex has a role in gene regulation, where cadherin loss results in enhanced p120-mediated relief of REST–CoREST-mediated repression.
Although E-cadherin levels steadily decrease upon the initiation of neuronal differentiation, we observed N-cadherin protein appearing at 4 days after initiating differentiation and then increasing during differentiation (data not shown). N-cadherin expression is important for neuronal differentiation (Brusés, 2006; Takeichi, 2007), and p120-catenin knockdown might impair neuronal differentiation, because N-cadherin is lowered at the protein level (our unpublished data). However, we find that a p120-catenin mutant incapable of binding cadherin (N478A) is still able to partially rescue REST gene targets in mESCs depleted for p120-catenin (supplementary material Fig. S3F). Thus, it appears that it is the signaling function of p120-catenin, and not so much functions related to cell adhesion (e.g. E- or N-cadherin) that is largely responsible for the observed effects. Thus, neuronal differentiation in response to p120-catenin might be modulated in both REST-dependent and REST-independent manners.
Given that the REST–CoREST complex acts in stemness, differentiation and disease, our finding that it both associates with and is modulated by p120-catenin provides some insight. For example, we previously showed that the kinase Dyrk1a stabilizes p120-catenin (Hong et al., 2012). Dyrk1a is implicated in reducing REST expression by an unknown mechanism in a Down syndrome mouse model, where ESC pluripotency and differentiation are dysregulated (Canzonetta et al., 2008). It is thus conceivable that Dyrk1a modulates REST through p120-catenin, contributing to aberrant pluripotency and differentiation. Our discovery of a relationship between p120-catenin and REST–CoREST in mESCs suggests that this pathway should also be examined in vivo. For example, REST is central to neurogenic processes such as eye formation (Kuwabara et al., 2004; Mao et al., 2011; Olguin et al., 2006). It should be informative to probe for p120-catenin–REST or p120-catenin–CoREST developmental contributions in these contexts.
Given the association of REST–CoREST with chromatin regulatory factors such as lysine-specific histone demethylase 1A and histone deacetylases (e.g. HDAC1 and HDAC2), later studies will likewise be needed to flush out molecular details that account for observed outcomes. Furthermore, although not examined here, CoREST modulates many genes in which REST is not involved. Thus, the effects p120-catenin on gene regulation might be wider still and might be coordinated with its previously reported regulation of other transcription factors, such as kaiso and Glis2 (Hosking et al., 2007; Kelly et al., 2004; Kim et al., 2004).
MATERIALS AND METHODS
Cell culture and transfection
HEK293 and NIH3T3 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) following standard protocols. AB1 mESCs (McMahon and Bradley, 1990) are a kind gift from Michelle Barton (MD Anderson Cancer Center, Houston, TX) and were grown on feeder-cell-free and gelatinized dishes in DMEM containing 1000 U/ml leukemia inhibitory factor (LIF), 20% FBS (ES-qualified, Invitrogen) and β-mercaptoethanol. Cells were transfected using Lipofectamine 2000 reagent (Invitrogen) or electroporated for cDNA-construct transfection. Lipofectamine RNAiMAX reagent (Invitrogen) was used for siRNA oligonucleotide transfections. SMARTpool-siGENOME siRNAs against mouse p120, ARVCF, CoREST and REST were purchased from Dharmacon.
cDNA cloning and plasmids
Mouse p120-catenin (isoform p120-1B), mouse CoREST and mouse REST cDNA constructs were purchased from Openbiosystems, and sub-cloned into the pCS2 vector. To generate NLS-mutant p120, KK residues (K622 and K623) of mouse p120-catenin were mutated to AA using site-directed mutagenesis (Invitrogen). For interaction mapping studies, the N-terminal fragment of p120-catenin corresponds to all amino acids prior to armadillo repeat 1; the armadillo repeat fragment contains repeats 1 through 8.5; and the C-terminal tail fragment was generated from armadillo repeat 8.5 through to the C-terminal amino acid. All fragments of p120-catenin, REST and CoREST for interaction mapping were generated by PCR-based cloning into pCS2.
Viral transduction and selection
For viral packaging, 293T cells were transfected with psPAX2, pMD2.G, and selected lentiviral shRNA constructs. During infection, polybrene was added to the plated cells, and 48 h after infection, cells were subjected to puromycin selection for 48–72 h. The indicated shRNAs were purchased from the shRNA and ORFeome Core (MD Anderson Cancer Center, Houston, TX).
In vitro neuronal differentiation
To induce mESC neuronal differentiation in vitro, we followed published protocols (Ying et al., 2003). AB1 cells were plated on gelatinized dishes in N2B27 neuronal differentiation medium (1∶1, DMEM/F12 with N2 supplement and L-glutamine neurobasal medium with B27 supplement; Invitrogen). Differentiating cells were harvested at the indicated times for further experiments.
In vitro transcription and translation and in vitro binding assay
In vitro binding assays were performed as described previously (Gu et al., 2011). In brief, proteins were synthesized using the TNT SP6 high-yield wheat germ protein expression system (Promega). To conduct in vitro binding assays, we incubated synthesized proteins with the indicated antibodies in PBS containing protease inhibitors and 0.5% NP-40, and precipitated them using Protein A/G plus agarose (Santa Cruz Biotechnology). Immuno-complexes were washed three times with PBS containing 0.5% NP-40.
GST pulldown assays
GST pulldown assays were conducted using a described protocol (Cho et al., 2010). Bacterially purified GST–CoREST or REST proteins (GST was cleaved from REST using thrombin protease) were incubated with bacterially purified MBP–p120-catenin fusion proteins and precipitated using glutathione–Sepharose-4B resin (GE Healthcare), for immunoblot analysis.
Nuclear fractionation, endogenous co-immunoprecipitation and immunoblotting
Conventional protocols were used for nuclear fractionation (Schreiber et al., 1989). We followed standard immunoprecipitation protocols, and diluted nuclear fractions or whole-cell lysates were used for co-immunoprecipitation (2 µg of anti-CoREST or anti-REST antibodies, Millipore). Antibodies for immunoblotting were against: p120-catenin and Cdh1 (BD Transduction Laboratory), ARVCF (Abnova), GAPDH (Santa Cruz Biotechnology), Myc (9E10) and the HA epitope (Developmental Studies Hybridoma Bank), Flag and actin (Sigma), and lamin A/C, Oct4 and Sox2 (Thermo Scientific).
Chromatin immunoprecipitation
Cells were plated on 100-mm tissue culture dishes and fixed with 1% formaldehyde for 10–15 min. Isolated nuclei from fixed cells were sonicated using a Diagenode Bioruptor to obtain mean DNA fragmentation sizes of less than 500 base pairs. After centrifugation, lysates were pre-cleared and then incubated overnight at 4°C with 2 µg of antibody directed against REST (Millipore) or rabbit IgG, followed by protein-A/G–agarose incubation for 2 h. After precipitation and washing, the immune complex was incubated with RNase and proteinase K at 37°C and de-crosslinked overnight at 65°C. DNA regions of interest were tested using qPCR (Open Biosystems SyBr green Mastermix).
Quantitative real-time PCR and RT-PCR
Total RNA was prepared as previously described (Hong et al., 2010). Primer sequences for the detection of calbindin, synaptotagmin 4 and Mash1 transcripts were obtained from a previous study (Ballas et al., 2005). At least three independent experiments were performed. See supplementary material Table S1 for primer sequence information.
Alkaline phosphatase staining
mESC colonies were stained with Vector Blue alkaline phosphatase substrate kit I (Vector Labs), following the manufacturer's instructions. To quantify alkaline phosphatase staining readouts, colonies were picked from three random areas and images taken using a 4× microscope objective. Alkaline phosphatase readouts were analyzed using Image J software, and the experiment repeated three times.
Electrophoretic mobility shift assay
Using a Lightshift Chemiluminescent EMSA Kit (Thermo Scientific), EMSA was performed according to the indicated instructions. The indicated recombinant proteins of REST (thrombin-cleaved from GST–REST), MBP–p120-catenin and MBP were bacterially produced and then purified. In two different pair-wise manners, protein–protein or protein–DNA combinations were made, generating similar experimental results: either REST and MBP–p120-catenin (or MBP alone) were incubated overnight at 4°C followed by addition of the oligonucleotide; or REST was incubated with the oligonucleotide for 10 min, followed by addition of MBP–p120-catenin (or MBP alone) for 10 min at room temperature. The complementary 5′ biotin-labeled oligonucleotides were hybridized and encoded a single RE1 consensus sequence (Otto et al., 2007), and were used as probe (5′-biotin-CTCTATCGATAGTTC AGCACCAAAGGACAGCGCCGGTACCGAGCTCTTA-3′ and 5′-biotin-TAAGAGCTCGGTACCGGCGCTGTCCTTTGGTGCTGAACTATCGATAGAG-3′).
Immuno-staining
Differentiating mESC colonies were fixed with 4% formaldehyde and permeabilized using 0.3% Triton X-100. Anti-doublecortin (DCX) antibody (Abcam) was used to detect endogenous DCX. For immunostaining of the indicated tagged proteins (supplementary material Fig. S1F) expressed in pluripotent mESC colonies, anti-HA (Y-11, Santa Cruz Biotechnology) and anti-Myc (9E10, Developmental Studies Hybridoma Bank) antibodies were used. DAPI was used for counter-staining. Images were acquired using 3i Confocal microscopy (Zeiss).
Statistical analysis
To analyze the significance of data and obtain P-values, we used the Student's t-test within the Microsoft Excel software program, and one-way ANOVA within GraphPad Prism Verson 6.0a.
Supplementary Material
Acknowledgments
We thank Amy Sater (U. Houston), Michelle Barton, Zhimin Lu and Xiaobing Shi (all UT MDACC), for their advice during the graduate training of M.L. For critical reading of the manuscript we thank Chai-An Mao, William Klein and Sadhan Majumder (all UT MDACC). For helpful discussions, we thank laboratory members William Munoz and Rachel Miller, our departmental colleagues Ahbinov Jain, Ngoc Bui, Sabrina Stratton, Kendra Alton, Kadir Akedemir and Zeynep Coban Akedemir (all UT MDACC), and Noboru Ishiyama (U Toronto). For sharing findings prior to publication, we thank Frans van Roy, Jolanda van Hengel and Tim Pieters (all U. Ghent). Appreciated constructs were provided by Xiaomin Chen (Pfizer, Inc.), and Gail Mandel (HHMI, Oregon Health and Science University).
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
M.L., Y.F., J.-I.P. and P.D.M. participated in designing or advising upon experiments; M.L. and H.J. conducted experiments; M.L and P.D.M. interpreted the results and wrote the paper.
Funding
We acknowledge support for this work from the National Institutes of Health [grant number RO1 GM52112 to P.D.M.]; University of Texas MDACC Bridge Funding to P.D.M.; a Cancer Prevention and Research Institute of Texas Research Training Award [grant number RP101502 to M.L.]; University of Texas MDACC Center for Stem Cell and Developmental Biology Pilot Grant to Y.F. and P.D.M.; and assistance from a University of Texas M.D. Anderson Cancer Center National Cancer Institute Core Grant [grant number CA-16672]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.151944/-/DC1
References
- Abrajano J. J., Qureshi I. A., Gokhan S., Zheng D., Bergman A., Mehler M. F. (2009a). Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS ONE 4, e7665 10.1371/journal.pone.0007665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrajano J. J., Qureshi I. A., Gokhan S., Zheng D., Bergman A., Mehler M. F. (2009b). REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS ONE 4, e7936 10.1371/journal.pone.0007936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Elneel K., Ochiishi T., Medina M., Remedi M., Gastaldi L., Caceres A., Kosik K. S. (2008). A delta-catenin signaling pathway leading to dendritic protrusions. J. Biol. Chem. 283, 32781–32791 10.1074/jbc.M804688200 [DOI] [PubMed] [Google Scholar]
- Andrés M. E., Burger C., Peral-Rubio M. J., Battaglioli E., Anderson M. E., Grimes J., Dallman J., Ballas N., Mandel G. (1999). CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96, 9873–9878 10.1073/pnas.96.17.9873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H., Hara A., Era T., Kunisada T., Yamada Y. (2012). Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development 139, 667–677 10.1242/dev.072272 [DOI] [PubMed] [Google Scholar]
- Arikkath J., Peng I. F., Ng Y. G., Israely I., Liu X., Ullian E. M., Reichardt L. F. (2009). Delta-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J. Neurosci. 29, 5435–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Battaglioli E., Atouf F., Andres M. E., Chenoweth J., Anderson M. E., Burger C., Moniwa M., Davie J. R., Bowers W. J. et al. (2001). Regulation of neuronal traits by a novel transcriptional complex. Neuron 31, 353–365 10.1016/S0896-6273(01)00371-3 [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D. D., Speh J. C., Mandel G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 10.1016/j.cell.2005.03.013 [DOI] [PubMed] [Google Scholar]
- Battaglioli E., Andrés M. E., Rose D. W., Chenoweth J. G., Rosenfeld M. G., Anderson M. E., Mandel G. (2002). REST repression of neuronal genes requires components of the hSWI.SNF complex. J. Biol. Chem. 277, 41038–41045 10.1074/jbc.M205691200 [DOI] [PubMed] [Google Scholar]
- Bellovin D. I., Bates R. C., Muzikansky A., Rimm D. L., Mercurio A. M. (2005). Altered localization of p120 catenin during epithelial to mesenchymal transition of colon carcinoma is prognostic for aggressive disease. Cancer Res. 65, 10938–10945 10.1158/0008-5472.CAN-05-1947 [DOI] [PubMed] [Google Scholar]
- Boggetti B., Niessen C. M. (2012). Adherens junctions in mammalian development, homeostasis and disease: lessons from mice. Subcell. Biochem. 60, 321–355 10.1007/978-94-007-4186-7_14 [DOI] [PubMed] [Google Scholar]
- Brusés J. L. (2006). N-cadherin signaling in synapse formation and neuronal physiology. Mol. Neurobiol. 33, 237–252 10.1385/MN:33:3:237 [DOI] [PubMed] [Google Scholar]
- Buckley N. J., Johnson R., Sun Y. M., Stanton L. W. (2009). Is REST a regulator of pluripotency? Nature 457, E5–E6discussion E7 [DOI] [PubMed] [Google Scholar]
- Canzonetta C., Mulligan C., Deutsch S., Ruf S., O'Doherty A., Lyle R., Borel C., Lin-Marq N., Delom F., Groet J. et al. (2008). DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am. J. Hum. Genet. 83, 388–400 10.1016/j.ajhg.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. F., Paquette A. J., Anderson D. J. (1998). NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136–142 10.1038/2431 [DOI] [PubMed] [Google Scholar]
- Chen T., Yuan D., Wei B., Jiang J., Kang J., Ling K., Gu Y., Li J., Xiao L., Pei G. (2010). E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells 28, 1315–1325 10.1002/stem.456 [DOI] [PubMed] [Google Scholar]
- Cho K., Vaught T. G., Ji H., Gu D., Papasakelariou-Yared C., Horstmann N., Jennings J. M., Lee M., Sevilla L. M., Kloc M. et al. (2010). Xenopus Kazrin interacts with ARVCF-catenin, spectrin and p190B RhoGAP, and modulates RhoA activity and epithelial integrity. J. Cell Sci. 123, 4128–4144 10.1242/jcs.072041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Crisafulli L., Caldera V., Tortoreto M., Brilli E., Conforti P., Zunino F., Magrassi L., Schiffer D., Cattaneo E. (2012). REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS ONE 7, e38486 10.1371/journal.pone.0038486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H. C., Fingleton B. M., Rudolph-Owen L. A., Goss K. J., Rubinfeld B., Polakis P., Matrisian L. M. (1999). The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene 18, 2883–2891 10.1038/sj.onc.1202627 [DOI] [PubMed] [Google Scholar]
- Daniel J. M., Reynolds A. B. (1995). The tyrosine kinase substrate p120cas binds directly to E-cadherin but not to the adenomatous polyposis coli protein or alpha-catenin. Mol. Cell. Biol. 15, 4819–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. M., Reynolds A. B. (1999). The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 19, 3614–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. M., Spring C. M., Crawford H. C., Reynolds A. B., Baig A. (2002). The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 30, 2911–2919 10.1093/nar/gkf398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Ireton R. C., Reynolds A. B. (2003). A core function for p120-catenin in cadherin turnover. J. Cell Biol. 163, 525–534 10.1083/jcb.200307111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L. P., Yamamoto M., Zang K., Reichardt L. F. (2006). p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron 51, 43–56 10.1016/j.neuron.2006.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Ji H., Kim S. W., Park J. I., Vaught T. G., Anastasiadis P. Z., Ciesiolka M., McCrea P. D. (2004). Vertebrate development requires ARVCF and p120 catenins and their interplay with RhoA and Rac. J. Cell Biol. 165, 87–98 10.1083/jcb.200307109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraichard A., Chassande O., Bilbaut G., Dehay C., Savatier P., Samarut J. (1995). In vitro differentiation of embryonic stem cells into glial cells and functional neurons. J. Cell Sci. 108, 3181–3188 [DOI] [PubMed] [Google Scholar]
- Gao Z., Ure K., Ding P., Nashaat M., Yuan L., Ma J., Hammer R. E., Hsieh J. (2011). The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772–9786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V. (2009). REST and the RESTless: in stem cells and beyond. Future Neurology 4, 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. D., Nusse R. (2006). Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 281, 22429–22433 10.1074/jbc.R600015200 [DOI] [PubMed] [Google Scholar]
- Gottardi C. J., Peifer M. (2008). Terminal regions of beta-catenin come into view. Structure 16, 336–338 10.1016/j.str.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot K. R., Sevilla L. M., Nishi K., DiColandrea T., Watt F. M. (2004). Kazrin, a novel periplakin-interacting protein associated with desmosomes and the keratinocyte plasma membrane. J. Cell Biol. 166, 653–659 10.1083/jcb.200312123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D., Tonthat N. K., Lee M., Ji H., Bhat K. P., Hollingsworth F., Aldape K. D., Schumacher M. A., Zwaka T. P., McCrea P. D. (2011). Caspase-3 cleavage links delta-catenin to the novel nuclear protein ZIFCAT. J. Biol. Chem. 286, 23178–23188 10.1074/jbc.M110.167544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M. (2005). The p120 family of cell adhesion molecules. Eur. J. Cell Biol. 84, 205–214 10.1016/j.ejcb.2004.12.016 [DOI] [PubMed] [Google Scholar]
- Hirai H., Karian P., Kikyo N. (2011). Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem. J. 438, 11–23 10.1042/BJ20102152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C., Zhou J., Medina M., Goto T., Jacobson M., Bhide P. G., Kosik K. S. (2000). delta-catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J. Comp. Neurol. 420, 261–276 [DOI] [PubMed] [Google Scholar]
- Hong J. Y., Park J. I., Cho K., Gu D., Ji H., Artandi S. E., McCrea P. D. (2010). Shared molecular mechanisms regulate multiple catenin proteins: canonical Wnt signals and components modulate p120-catenin isoform-1 and additional p120 subfamily members. J. Cell Sci. 123, 4351–4365 10.1242/jcs.067199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. Y., Park J. I., Lee M., Muñoz W. A., Miller R. K., Ji H., Gu D., Ezan J., Sokol S. Y., McCrea P. D. (2012). Down's-syndrome-related kinase Dyrk1A modulates the p120-catenin-Kaiso trajectory of the Wnt signaling pathway. J. Cell Sci. 125, 561–569 10.1242/jcs.086173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosking C. R., Ulloa F., Hogan C., Ferber E. C., Figueroa A., Gevaert K., Birchmeier W., Briscoe J., Fujita Y. (2007). The transcriptional repressor Glis2 is a novel binding partner for p120 catenin. Mol. Biol. Cell 18, 1918–1927 10.1091/mbc.E06-10-0941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries M. J., Reynolds A. (2009). Cell-to-cell contact and extracellular matrix. Curr. Opin. Cell Biol. 21, 613–615 10.1016/j.ceb.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Ireton R. C., Davis M. A., van Hengel J., Mariner D. J., Barnes K., Thoreson M. A., Anastasiadis P. Z., Matrisian L., Bundy L. M., Sealy L. et al. (2002). A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 159, 465–476 10.1083/jcb.200205115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N., Lee S. H., Liu S., Li G. Y., Smith M. J., Reichardt L. F., Ikura M. (2010). Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 141, 117–128 10.1016/j.cell.2010.01.017 [DOI] [PubMed] [Google Scholar]
- Jørgensen H. F., Fisher A. G. (2010). Can controversies be put to REST? Nature 467, E3–E4discussion E5 [DOI] [PubMed] [Google Scholar]
- Jorgensen H. F., Chen Z. F., Merkenschlager M., Fisher A. G. (2009a). Is REST required for ESC pluripotency? Nature 457, E4–E5discussion E7 [DOI] [PubMed] [Google Scholar]
- Jørgensen H. F., Terry A., Beretta C., Pereira C. F., Leleu M., Chen Z. F., Kelly C., Merkenschlager M., Fisher A. G. (2009b). REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development 136, 715–721 10.1242/dev.028548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal M. M., Sathyan P., Singh S. K., Zinn P. O., Marisetty A. L., Liang S., Gumin J., El-Mesallamy H. O., Suki D., Colman H. et al. (2012). REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells 30, 405–414 10.1002/stem.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. F., Spring C. M., Otchere A. A., Daniel J. M. (2004). NLS-dependent nuclear localization of p120ctn is necessary to relieve Kaiso-mediated transcriptional repression. J. Cell Sci. 117, 2675–2686 10.1242/jcs.01101 [DOI] [PubMed] [Google Scholar]
- Kim S. W., Park J. I., Spring C. M., Sater A. K., Ji H., Otchere A. A., Daniel J. M., McCrea P. D. (2004). Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat. Cell Biol. 6, 1212–1220 10.1038/ncb1191 [DOI] [PubMed] [Google Scholar]
- Kosik K. S., Donahue C. P., Israely I., Liu X., Ochiishi T. (2005). Delta-catenin at the synaptic-adherens junction. Trends Cell Biol. 15, 172–178 10.1016/j.tcb.2005.01.004 [DOI] [PubMed] [Google Scholar]
- Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005). Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25, 2475–2485 10.1128/MCB.25.6.2475-2485.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T., Hsieh J., Nakashima K., Taira K., Gage F. H. (2004). A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793 10.1016/S0092-8674(04)00248-X [DOI] [PubMed] [Google Scholar]
- Li D., Zhou J., Wang L., Shin M. E., Su P., Lei X., Kuang H., Guo W., Yang H., Cheng L. et al. (2010a). Integrated biochemical and mechanical signals regulate multifaceted human embryonic stem cell functions. J. Cell Biol. 191, 631–644 10.1083/jcb.201006094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang B. H., Wang S., Moalim-Nour L., Mohib K., Lohnes D., Wang L. (2010b). Individual cell movement, asymmetric colony expansion, rho-associated kinase, and E-cadherin impact the clonogenicity of human embryonic stem cells. Biophys. J. 98, 2442–2451 10.1016/j.bpj.2010.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B. T., Tamai K., He X. (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C. A., Tsai W. W., Cho J. H., Pan P., Barton M. C., Klein W. H. (2011). Neuronal transcriptional repressor REST suppresses an Atoh7-independent program for initiating retinal ganglion cell development. Dev. Biol. 349, 90–99 10.1016/j.ydbio.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter C., Pribadi M., Liu X., Trachtenberg J. T. (2009). Delta-catenin is required for the maintenance of neural structure and function in mature cortex in vivo. Neuron 64, 320–327 10.1016/j.neuron.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea P. D., Park J. I. (2007). Developmental functions of the P120-catenin sub-family. Biochim. Biophys. Acta 1773, 17–33 10.1016/j.bbamcr.2006.06.009 [DOI] [PubMed] [Google Scholar]
- McEwen A. E., Escobar D. E., Gottardi C. J. (2012). Signaling from the adherens junction. Subcell. Biochem. 60, 171–196 10.1007/978-94-007-4186-7_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A. P., Bradley A. (1990). The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62, 1073–1085 10.1016/0092-8674(90)90385-R [DOI] [PubMed] [Google Scholar]
- Miller R. K., Hong J. Y., Muñoz W. A., McCrea P. D. (2013). Beta-catenin versus the other armadillo catenins: assessing our current view of canonical Wnt signaling. Prog. Mol. Biol. Transl. Sci. 116, 387–407 10.1016/B978-0-12-394311-8.00017-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamet L., Lea M. L., Ward C. M. (2010). Abrogation of E-cadherin-mediated cellular aggregation allows proliferation of pluripotent mouse embryonic stem cells in shake flask bioreactors. PLoS ONE 5, e12921 10.1371/journal.pone.0012921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R. (2008). Wnt signaling and stem cell control. Cell Res. 18, 523–527 10.1038/cr.2008.47 [DOI] [PubMed] [Google Scholar]
- Olguin P., Oteiza P., Gamboa E., Gomez-Skarmeta J. L., Kukuljan M. (2006). RE-1 silencer of transcription/neural restrictive silencer factor modulates ectodermal patterning during Xenopus development. J. Neurosci. 26, 2820–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. J., McCorkle S. R., Hover J., Conaco C., Han J. J., Impey S., Yochum G. S., Dunn J. J., Goodman R. H., Mandel G. (2007). A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27, 6729–6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffenholz R., Franke W. W. (1997). Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation 61, 293–304 [DOI] [PubMed] [Google Scholar]
- Park J. I., Kim S. W., Lyons J. P., Ji H., Nguyen T. T., Cho K., Barton M. C., Deroo T., Vleminckx K., Moon R. T. et al. (2005). Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell 8, 843–854 10.1016/j.devcel.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Pieters T., van Roy F. (2014). Role of cell-cell adhesion complexes in embryonic stem cell biology. J. Cell Sci. 127, 2603–2613 10.1242/jcs.146720 [DOI] [PubMed] [Google Scholar]
- Pieters T., van Hengel J., van Roy F. (2012a). Functions of p120ctn in development and disease. Front. Biosci. 17, 760–783 10.2741/3956 [DOI] [PubMed] [Google Scholar]
- Pieters T., van Roy F., van Hengel J. (2012b). Functions of p120ctn isoforms in cell-cell adhesion and intracellular signaling. Front. Biosci. 17, 1669–1694 10.2741/4012 [DOI] [PubMed] [Google Scholar]
- Qureshi I. A., Gokhan S., Mehler M. F. (2010). REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle 9, 4477–4486 10.4161/cc.9.22.13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A., Priya R., Yap A. S. (2013). Coordinating Rho and Rac: the regulation of Rho GTPase signaling and cadherin junctions. Prog. Mol. Biol. Transl. Sci. 116, 49–68 10.1016/B978-0-12-394311-8.00003-0 [DOI] [PubMed] [Google Scholar]
- Redmer T., Diecke S., Grigoryan T., Quiroga-Negreira A., Birchmeier W., Besser D. (2011). E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 12, 720–726 10.1038/embor.2011.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. B. (2007). p120-catenin: Past and present. Biochim. Biophys. Acta 1773, 2–7 10.1016/j.bbamcr.2006.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. B., Daniel J., McCrea P. D., Wheelock M. J., Wu J., Zhang Z. (1994). Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol. Cell. Biol. 14, 8333–8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005). Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 10.1074/jbc.M502573200 [DOI] [PubMed] [Google Scholar]
- Rossbach M. (2011). Non-Coding RNAs in Neural Networks, REST-Assured. Front. Genet. 2, 8 10.3389/fgene.2011.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackmann R. C., van Amersfoort M., Haarhuis J. H., Vlug E. J., Halim V. A., Roodhart J. M., Vermaat J. S., Voest E. E., van der Groep P., van Diest P. J. et al. (2011). Cytosolic p120-catenin regulates growth of metastatic lobular carcinoma through Rock1-mediated anoikis resistance. J. Clin. Invest. 121, 3176–3188 10.1172/JCI41695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989). Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17, 6419 10.1093/nar/17.15.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla L. M., Nachat R., Groot K. R., Watt F. M. (2008). Kazrin regulates keratinocyte cytoskeletal networks, intercellular junctions and differentiation. J. Cell Sci. 121, 3561–3569 10.1242/jcs.029538 [DOI] [PubMed] [Google Scholar]
- Shibata T., Kokubu A., Sekine S., Kanai Y., Hirohashi S. (2004). Cytoplasmic p120ctn regulates the invasive phenotypes of E-cadherin-deficient breast cancer. Am. J. Pathol. 164, 2269–2278 10.1016/S0002-9440(10)63783-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Kagalwala M. N., Parker-Thornburg J., Adams H., Majumder S. (2008). REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453, 223–227 10.1038/nature06863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S. Y. (2011). Maintaining embryonic stem cell pluripotency with Wnt signaling. Development 138, 4341–4350 10.1242/dev.066209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncin F., Mohamet L., Eckardt D., Ritson S., Eastham A. M., Bobola N., Russell A., Davies S., Kemler R., Merry C. L. et al. (2009). Abrogation of E-cadherin-mediated cell-cell contact in mouse embryonic stem cells results in reversible LIF-independent self-renewal. Stem Cells 27, 2069–2080 10.1002/stem.134 [DOI] [PubMed] [Google Scholar]
- Spring C. M., Kelly K. F., O'Kelly I., Graham M., Crawford H. C., Daniel J. M. (2005). The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp. Cell Res. 305, 253–265 10.1016/j.yexcr.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Stemmler M. P. (2008). Cadherins in development and cancer. Mol. Biosyst. 4, 835–850 10.1039/b719215k [DOI] [PubMed] [Google Scholar]
- Stepniak E., Radice G. L., Vasioukhin V. (2009). Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb. Perspect. Biol. 1, a002949 10.1101/cshperspect.a002949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Kameoka S., Lentz S., Majumder S. (2004). Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol. Cell. Biol. 24, 8018–8025 10.1128/MCB.24.18.8018-8025.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. M., Greenway D. J., Johnson R., Street M., Belyaev N. D., Deuchars J., Bee T., Wilde S., Buckley N. J. (2005). Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol. Biol. Cell 16, 5630–5638 10.1091/mbc.E05-07-0687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. M., Cooper M., Finch S., Lin H. H., Chen Z. F., Williams B. P., Buckley N. J. (2008). Rest-mediated regulation of extracellular matrix is crucial for neural development. PLoS ONE 3, e3656 10.1371/journal.pone.0003656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. (2007). The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20 10.1038/nrn2043 [DOI] [PubMed] [Google Scholar]
- Van den Bossche J., Malissen B., Mantovani A., De Baetselier P., Van Ginderachter J. A. (2012). Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood 119, 1623–1633 10.1182/blood-2011-10-384289 [DOI] [PubMed] [Google Scholar]
- Wagoner M. P., Roopra A. (2012). A REST derived gene signature stratifies glioblastomas into chemotherapy resistant and responsive disease. BMC Genomics 13, 686 10.1186/1471-2164-13-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook T. F., Hu G., Ang X. L., Mulligan P., Pavlova N. N., Liang A., Leng Y., Maehr R., Shi Y., Harper J. W. et al. (2008). SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 10.1038/nature06780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Aoki H., Kunisada T., Hara A. (2010). Rest promotes the early differentiation of mouse ESCs but is not required for their maintenance. Cell Stem Cell 6, 10–15 10.1016/j.stem.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. (2003). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 10.1038/nbt780 [DOI] [PubMed] [Google Scholar]
- Yoon H. G., Chan D. W., Reynolds A. B., Qin J., Wong J. (2003). N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12, 723–734 10.1016/j.molcel.2003.08.008 [DOI] [PubMed] [Google Scholar]
- You A., Tong J. K., Grozinger C. M., Schreiber S. L. (2001). CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98, 1454–1458 10.1073/pnas.98.4.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A. (2011). Control of the embryonic stem cell state. Cell 144, 940–954 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liyanage U., Medina M., Ho C., Simmons A. D., Lovett M., Kosik K. S. (1997). Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport 8, 2085–2090 10.1097/00001756-199705260-00054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.