Abstract
Background and objective
Small cell lung cancer (SCLC) is one of the most challenging tumors to treat due to high proliferation rate, early metastatic dissemination and rapid development of chemotherapy resistance. The current treatment protocols involve the use of topoisomerase 1 (TOP1) poisons such as irinotecan and topotecan in combination with platinum-based compounds. TOP1 poisons kill cancer cells by trapping TOP1 on DNA, generating lethal DNA double-strand breaks. A potential mechanism employed by cancer cells to resist killing by TOP1 poisons is to overexpress enzymes involved in the repair of TOP1-DNA breaks. Tyrosyl DNA phosphodiesterase 1 (TDP1) is a key player in this process and despite its importance, no data is currently available to correlate TDP1 protein and mRNA levels with catalytic activity in SCLC. In addition, it is not known if TDP1 and TOP1 protein levels correlate with the cellular response of SCLC to TOP1 based therapies.
Methods and results
We report a remarkable variation in TDP1 and TOP1 protein levels in a panel of SCLC cell lines. TDP1 protein level correlates well with TDP1 mRNA and TDP1 catalytic activity, as measured by two newly developed independent activity assays, suggesting the potential utility of immunohistochemistry in assessing TDP1 levels in SCLC tissues. We further demonstrate that whilst TDP1 protein level alone does not correlate with topotecan sensitivity, TDP1/TOP1 ratio correlates well with sensitivity in 8 out of 10 cell lines examined.
Conclusion
This study provides the first cellular analyses of TDP1 and TOP1 in SCLC and suggests the potential utility of TDP1/TOP1 ratio to assess the response of SCLC to topotecan. The establishment and validation of an easy-to-use TDP1 enzymatic assay in cell extracts could be exploited as a diagnostic tool in the clinic. These findings may help in stratifying patients that are likely to benefit from TOP1 poisons and TDP1 inhibitors currently under development.
Keywords: Topotecan, Small cell lung cancer, TOP1-DNA, TDP1 protein level, Enzymatic assay
Introduction
Small cell lung cancer (SCLC) accounts for approximately 10-20% of all lung cancers and is frequently associated with rapid tumor proliferation and a median survival of less than 3 months in untreated patients [1,2]. At diagnosis, the majority of patients present with extensive-stage disease (ED). SCLC is highly sensitive to chemotherapy, and a combination therapy with cisplatin/carboplatin and etoposide or the camptothecin drug, irinotecan is currently considered as the standard first-line treatment [3,4]. In patients with ED SCLC, chemotherapy produces response rates of 50% to 70% and median survival times of 7 to 9 months. However, most tumors will relapse after an initial response and nearly all patients will rapidly die from chemotherapy-resistant disease. The second-line treatment for relapsed SCLC further involves the use of another camptothecin-based drug, topotecan, which was shown to improve survival from 14 weeks to 26 weeks [1,5-7].
Both irinotecan and topotecan are camptothecin-based drugs. Camptothecins (CPT) promote cancer cell death by locking topoisomerase I (TOP1) on a DNA single-strand break intermediate [8,9]. An unrepaired TOP1-mediated DNA single-strand break is converted into the potentially toxic DNA double-strand break upon collision with the replication machinery [10]. The repair of these hampered TOP1 complexes involve proteasomal degradation of TOP1 to leave a 3′-phosphotyrosyl peptide that is further processed by the 3′-phosphodiesterase activity of TDP1 during chromosomal single-strand break repair [11,12].
The ability of camptothecins (CPT) to kill cancer cells is dependent on both TOP1 activity on DNA and the rate of repair of TOP1-DNA breaks. Inhibition of TDP1 repair activity is therefore regarded as a promising mean for improving the efficacy of camptothecin-based drugs in the clinic [12-15]. Indeed, multiple studies have shown that inactivation or loss of TDP1 sensitises cells to CPT whereas overexpression of TDP1 promotes CPT tolerance [16-20]. Such findings additionally imply that high cellular levels of TDP1 may contribute to CPT drug resistance in cancer. Multiple drug discovery projects attempting to develop TDP1 inhibitors are currently underway and though some inhibitor structures have been identified, there are no validated cellular or in vivo models for confirming inhibitor hits or clinical trials in progress [21-24].
TOP1 levels are widely accepted to predict sensitivity to camptothecin in a large number of cancer types [25,26]. In contrast, little is known about the correlation between TDP1 or TDP1/TOP1 ratio and sensitivity to camptothecin-based therapies. Such an understanding would be particularly informative in selecting cancer types that could benefit from TDP1 inhibition in future clinical trials. Suitable candidates include cancers such as SCLC that are currently treated with camptothecin derivatives. Therefore, in this study we aimed to characterise both TDP1 and TOP1 status in a panel of SCLC cell lines and to examine the relationship between TDP1 transcript level, protein level, activity, and sensitivity to the camptothecin derivative, topotecan.
We report a remarkable variation in both TDP1 and TOP1 levels in SCLC lines. TDP1 protein levels correlate well with both TDP1 mRNA and TDP1 catalytic activity. We further show that whilst TDP1 protein level alone does not significantly correlate with topotecan sensitivity, TDP1/TOP1 ratio correlates well in 8 out of 10 SCLC cell lines examined, suggesting the potential utility of this ratio to assess topotecan sensitivity in SCLC.
Materials and Methods
Cells
The small cell lung cancer lines HCC33, H69, N417, H2171, H209 and H510 and the non-small cell lung cancer lines Hop62, Hop92, HCC78, H322M and H23 were obtained from Professor Peter Schmid. The remaining SCLC lines, H82, H187, H146 and H345 were bought from ATCC (ATCC, Virginia, USA). Mammalian cells were grown in a 37°C, 5% CO2 incubator. The small cell lung cancer lines HCC33, H69, N417, H2171, H209, H510, H82 and H187 were grown in Gibco RPMI media (Life Technologies, Paisley, UK) supplemented with 10% FCS and 2 mM L-glutamine. The H146 cell line was grown in Gibco RPMI supplemented with 10% FCS, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 4.5 g/L glucose and 1.5 g/L sodium bicarbonate. The H345 cell line was grown in Gibco RPMI supplemented with 20% FCS, 4 mM L-glutamine, 10 mM HEPES and 1.5 g/L sodium bicarbonate. The non-small cell lung cancer lines Hop62, Hop92, HCC78, H322M and H23 were maintained in Gibco RPMI (Life Technologies, Paisley, UK) supplemented with 10% FCS and 2 mM L-glutamine and the HCC193 cell line was maintained in Gibco DMEM (Life Technologies, Paisley, UK) containing 10% FCS and 2 mM L-glutamine. The generation and characterisation of the DT40 Tdp1−/− chicken cells and the DT40 Tdp1−/− chicken cells stably expressing human TDP1 have been described recently [12,16,17,20,22-31]. The DT40 chicken cells were maintained at 39°C, 5% CO2, in Gibco RPMI (Life Technologies, Paisley, UK) supplemented with 10% FCS, 1% chicken serum, 100 U Penicillin, 100 μg Streptomycin, 2 mM L-glutamine and 50 μM β-mercaptoethanol.
Whole cell extract preparation
Cells were collected by centrifugation (DT40 and small cell lung cancer suspension lines) or by scraping (non-small cell lung cancer adherent lines) and washed twice in ice cold phosphate buffered saline (PBS). Cell pellets were resuspended in ice-cold lysis buffer (20 mM Tris-HCl pH 7.5, 10 mM EDTA pH 8.0, 100 mM NaCl, 1% Triton X-100) containing the recommended final concentration of Complete Mini EDTA-free Protease Inhibitor Cocktail (Roche Applied Science, Burgess Hill, UK). Samples were briefly vortexed and extraction carried out for 30 min on ice. The lysate was cleared by centrifugation at 13,000 rpm for 10 min and the supernatant collected as whole cell extract (WCE). Protein concentration was calculated using Bradford assay and samples stored at −80°C.
Western blotting
Protein separation of WCE (40 μg) was carried out by 10% SDS-polyacrylamide gel electrophoresis (PAGE) at 125 V for 2 hr. The separated proteins were subsequently transferred onto a Hybond-C Extra Nitrocellulose membrane (Fisher Scientific UK, Loughborough, UK) at 25 V for 90 min. Membranes were blocked in 5% PBS-milk for 1 hr prior to immunoblotting overnight with antibodies against TDP1 (ab4166; Abcam, Cambridge, UK), TOP1 (SC-32736; Santa Cruz Biotechnologies, California, USA) and actin (A4700; Sigma-Aldrich, Gillingham, UK) diluted in 5% PBST-milk to 1:2000, 1:1000 and 1:3000, respectively. HRP-labelled polyclonal rabbit anti-mouse and polyclonal goat anti-rabbit secondary antibodies obtained from Dako (Ely, UK) were used at a 1:3000 dilution in 5% PBST-milk. Blots were developed using the chemiluminescent detection reagent, SuperSignal West Pico Chemiluminescent Substrate (Fisher Scientific UK, Loughborough, UK).
TDP1 activity assay: Gel-based assay
The in vitro 3′-tyrosyl-DNA phosphodiesterase activity of SCLC whole cell extracts (WCE) was determined using a gel-based 3′-tyrosyl-DNA phosphodiesterase activity assay. Reactions were performed in 10 μL reaction volumes containing assay buffer (25 mM HEPES, pH 8.0, 130 mM KCl, 1 mM DTT), WCE (20 ng and 25 ng) and 50 nM Cy5.5 labelled substrate oligomer containing a 3′-phosphotyrosyl group, 5′-(Cy5.5)GATCTAAAAGACT(pY)-3′ (Midland Certified Reagent Company, Texas, USA). Reactions were carried out at 37°C for 1 hr and stopped by addition of 10 μL loading buffer (44% deionized formamide, 2.25 mM Tris-borate, 0.05 mM EDTA, 0.01% xylene cyanol, 1% bromophenol blue). Samples were then heated at 90°C for 10 min prior to separation on a 20% Urea SequaGel (Fisher Scientific, Loughborough, UK) by gel electrophoresis at 190 V for 2 hr. Reaction products were visualised by gel imaging using the Fuji Film Fluor Imager FLA-5100 at 635 nm and bands quantified using Image J.
TDP1 activity assay: Fluorescence assay
In vitro 3′-tyrosyl-DNA phosphodiesterase activity of whole cell extracts (WCE) was determined using our recently developed fluorescence assay [21]. Briefly, assays were performed in a 15 μl reaction volume containing assay buffer (50 mM Tris pH 8.0, 5 mM MgCl2, 80 mM KCl, 0.05% Tween 20, 1 mM DTT), 10 nM 13-mer substrate with a 3′-phosphotyrosyl oligonucleotide conjugated to a FITC molecule (Midland Certified Reagent Company, Texas, USA) and 2 μl of the indicated amounts of WCE. Reactions were carried out at room temperature for 10 min prior to quenching with a mix of 30 μl enhancer and 2 μl sensor (Gyrasol Technologies, Kansas, USA). Fluorescence intensity was measured using a BMG Labtech Pherastar plate reader at excitation and emission wavelengths of λex 490 nm and λem 520 nm. Recombinant human TDP1 (6.5 pM) [21] diluted in assay buffer was used as a 3′-tyrosyl-DNA phosphodiesterase activity control for the reactions between independent experiments.
Quantitative PCR
Total mRNA was purified from cells using the RNeasy kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions. The mRNA concentration was determined using the NanoDrop 1000 spectrophotometer (Nanodrop, Willmington, USA) at OD 230 nm. cDNA was generated from 1 μg mRNA using the Superscript First Strand RT-PCR system (Invitrogen, Paisley, UK) and real time PCR analysis was carried out on the MX3005p qPCR system (Agilent Technologies, Santa Clara, USA) using Blue qPCR Sybr low ROX mix (Fisher Scientific, Loughborough, UK) for cDNA amplification. The average cycle threshold for three readings per each of three repeat experiments was used in further calculations to determine the relative quantities of mRNA (RQ) according to the equation RQ=2ΔΔCT.
CellTiter-Blue cell viability assay
The CellTiter-Blue cell viability assay (Promega, Southampton, UK) was used to determine SCLC cell sensitivity to topotecan hydrochloride hydrate (Sigma-Aldrich, Gillingham, UK). Briefly, cells were seeded in triplicate at 10,000 cells per well of a 96-well plate in media containing topotecan (0 nM to 500 nM). The contents were mixed thoroughly and the plates incubated at 37°C for 4 days. At day 4, CellTiter-Blue reagent (20 μl) was added to each well, the plates incubated at 37°C for 2 hr and fluorescence intensity determined using the GloMax Multi Detection System (Promega, Southamption, UK) at excitation and emission wavelengths of λex 560 nm and λem 590 nm. Percentage viability (%) was calculated using background-subtracted readings as:
Results
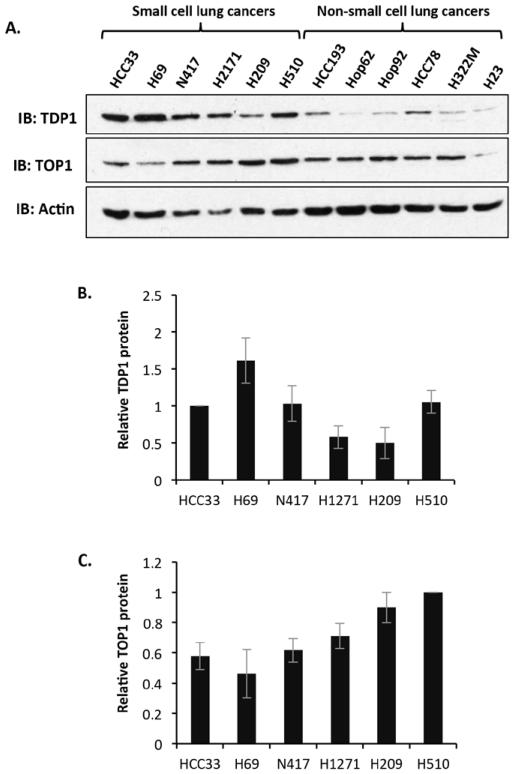
TDP1 is viewed as a promising target for inhibition in cancers such as SCLC that are currently treated with camptothecin-based therapies. Since topoisomerase I (TOP1) is generally thought to dictate the sensitivity of cancers to camptothecin, we sought to determine the cellular level of both TDP1 and TOP1 in a panel of SCLC cell lines (HCC33, H69, N417, H2171, H209 and H510). Separation of 40 μg of whole cell extracts (WCE) for each of the SCLC cell lines by 10% SDS-PAGE and analysis by Western blotting using TDP1 and TOP1 specific antibodies revealed a significant variation in both TDP1 and TOP1 protein level (Figure 1A-C). Interestingly, we noticed that SCLC lines possess higher levels of TDP1 compared to non-small cell lung cancer lines (HCC193, Hop62, Hop92, HCC78, H322M and H23; Figure 1A), suggesting that TDP1 may play a role in the biology, progression and, in particular, the therapeutic response of SCLC to camptothecin-based treatments.
Figure 1. Small cell lung cancer cell lines display remarkable variation in both TDP1 and TOP1 protein level.
(A) Whole cell extracts (40 μg) from the indicated small cell lung cancer cell lines and other non-small cell lung cancer cells were separated by 10 % SDS-PAGE and immunoblotted using antibodies against TDP1, TOP1 and actin. A quantitation of relative TDP1 (B) and TOP1 (C) protein level is depicted. Data are the average of 3 independent experiments ± STD normalised to HCC33 for TDP1 and H510 for TOP1.
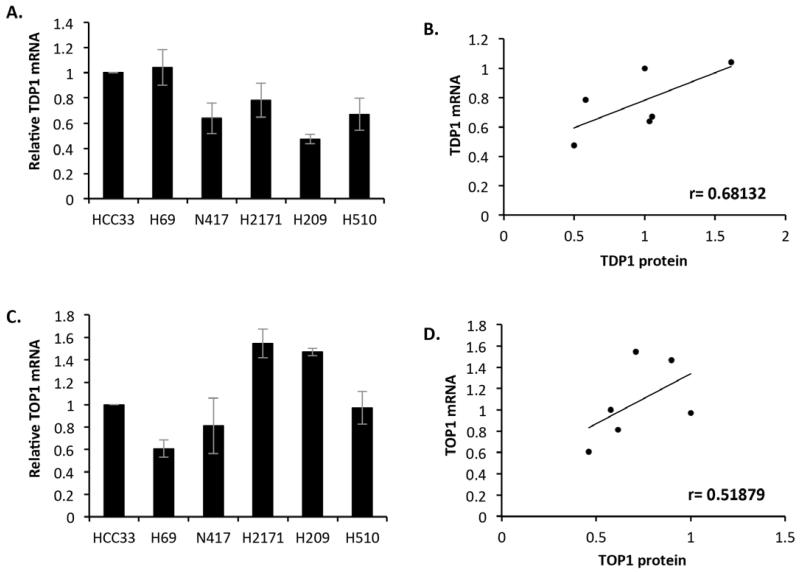
Cellular protein levels are maintained by both transcriptional and post-translational regulation. To determine the nature of the variation in cellular TDP1 and TOP1 protein levels in SCLC, cellular mRNA levels for both TDP1 and TOP1 were determined by quantitative real-time PCR using primers designed against exon-crossing regions for TDP1 and TOP1 cDNA (Figure 2A and 2C). Since there is no real isogenic control gene between the cell lines, the relative quantifications were obtained by normalising mRNA levels to that obtained for HCC33 for TDP1 mRNA measurements, or H510 for TOP1 mRNA. We further determined the Pearson’s correlation coefficient between mRNA and protein levels for both TDP1 and TOP1 to examine the relationship between mRNA and protein levels (Figure 2B and 2D). We observed a positive correlation between TDP1 mRNA and TDP1 protein (r=0.681, p=0.136) and a slightly weaker correlation between TOP1 mRNA and TOP1 protein (r=0.519, p=0.292).
Figure 2. Small cell lung cancer cell lines display good correlation between TDP1 or TOP1 protein level and mRNA.
cDNA from each of the SCLC lines was generated from 1 μg purified mRNA using the Superscript First Strand RT-PCR and real time PCR analysis was conducted using TDP1 primers (A) or TOP1 primers (C) and an MX3005p qPCR system. The relative quantity of mRNA (RQ value) for each repeat experiment was calculated according to the equation RQ = 2ΔΔCT and the RQ values normalised to that obtained for the HCC33 cell line. The average of normalised RQ values from 3 independent experiments ± STD is depicted (A&C). A graphical representation of the Pearson’s correlation coefficient (r) between SCLC TDP1 protein levels and TDP1 mRNA is shown in (B) and that for SCLC TOP1 protein level and TOP1 mRNA is shown in (D).
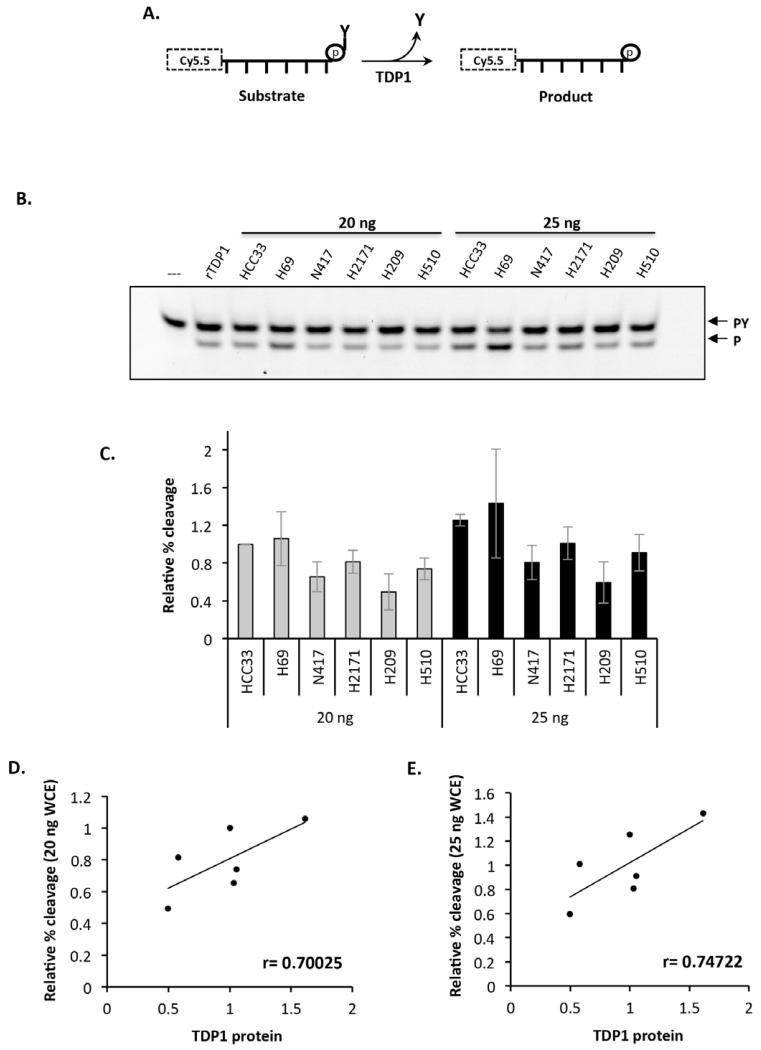
Post-translational modifications may further dictate the cellular activity of TDP1 [11,16,18,19,32]. It is therefore important to examine whether cellular TDP1 protein levels correlate with its activity, which is critical for designing a suitable diagnostic tool for selecting tumors that could benefit from TDP1 inhibitor-based therapies. To test this, we employed two orthogonal enzymatic assays; a gel-based assay and a fluorescence-based assay that was recently developed in our labs [21]. For the gel-based assay we engineered a 5′-Cy5.5 labelled substrate containing a 3′-phosphotyrosine modification (Figure 3A), thus introducing a unique imaging opportunity that does not involve the bothersome radioactive based methods that we used previously [11,17,18]. Whole cell extracts prepared from the SCLC cell lines were used as a source for TDP1 activity and reaction products were subsequently separated on a 20% denaturing PAGE by gel electrophoresis, and then visualised using a FujiFilm Fluor Imager FLA-5100 at 635 nm (Figure 3B). The relative activity was calculated by normalising the percentage cleavage to that obtained for 20 ng HCC33 whole cell extract and results obtained from three biological replicates are shown in Figure 3C. The results demonstrate a positive correlation between TDP1 protein level and TDP1 activity in SCLC cell lines (r=0.700, p=0.121 for 20 ng WCE; and r=0.747, p=0.088 for 25 ng WCE) (Figure 3D and 3E). These observations are in agreement with a recent study by the Pommier group on NSCLC reporting a remarkably similar positive correlation (r=0.75) between TDP1 protein level and catalytic activity [33].
Figure 3. TDP1 catalytic activity (Gel-based assay) correlates with the level of TDP1 protein in SCLC cells.
(A) Diagrammatic representation of TDP1 processing of a Cy5.5 labelled oligonucleotide harbouring a 3′-phosphotyrosine modification. The product of a TDP1 reaction is a Cy5.5 labelled oligonucleotide with 3′-phosphate. (B) The indicated amounts of WCE (20 ng and 25 ng) were incubated with 50 nM substrate in assay buffer at 37°C for 1 hr. Reaction products were separated on a 20% Urea SequaGel by gel electrophoresis at 190 V for 2 hr and visualised using a FujiFilm Fluor Imager FLA-5100 at 635 nm. Arrows denote the position of the substrate (PY) and cleaved product (P). (C) Quantification of the average relative TDP1 activity normalised to that obtained from HCC33 cell line from 3 independent experiments ± STD. Recombinant TDP1 (8 pM) was employed as a positive control. (D) A graphical representation of the Pearson’s correlation coefficient (r) between TDP1 protein level and activity.
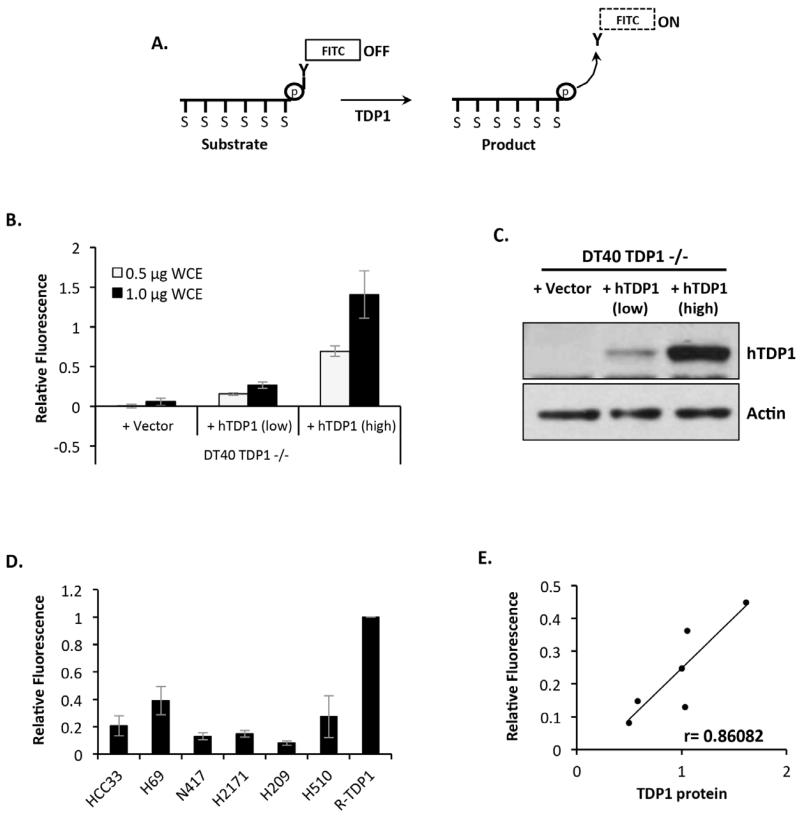
To better understand the relationship between TDP1 protein level and cellular activity, we additionally employed our recently developed fluorescence assay [21]. This 384-well plate format assay uses a FITC-conjugated 13-mer 3′-phosphotyrosyl substrate that releases a fluorescence signal upon processing (Figure 4A). We first examined whether this new assay would be reliable in determining TDP1 enzymatic activity in whole cell extracts. To test this, we generated chicken DT40 Tdp1−/− cells stably expressing low and high levels of human TDP1 and examined whether TDP1 level correlates with enzymatic activity. Whilst extracts from DT40 Tdp1−/− cells had no detectable TDP1 activity, incubation of the TDP1 substrate with increasing concentrations of whole cell extract prepared from DT40 Tdp1−/− cells expressing human TDP1 led to a proportional increase in TDP1 activity as measured by relative fluorescence (Figure 4B and 4C). These observations validate the assay and suggest its robustness in determining TDP1 activity in cellular extracts. We next examined whole cell extracts (0.25 ng) generated from the SCLC cell lines using a recombinant TDP1 protein (6.5 pM) as a control. The relative activity obtained for each cell line (Figure 4D) was calculated by normalising the fluorescence intensity against that obtained for recombinant TDP1 for three independently generated extracts. These results illustrate a clear variation in TDP1 activity in SCLC and, importantly, the Pearson’s correlation coefficient (r=0.86, p=0.028) suggests a significant positive correlation between cellular TDP1 activity and protein level (Figure 4E).
Figure 4. TDP1 catalytic activity (Fluorescence assay) correlates with the level of TDP1 protein in SCLC cells.
(A) A Metal ion sensor (indicated S) along the phosphate backbone of the substrate oligomer keeps the bound FITC-conjugated tyrosine in a non-fluorescent state (OFF). During a TDP1 driven reaction, the FITC conjugated tyrosine is released and distance generated between the released FITC-tyrosine and the oligonucleotide increases, allowing for FITC fluorescence (ON) that can be detected and quantified using a fluorescence imager. (B) A 15 μl final volume reaction containing 0.5 μg or 1.0 μg WCE from the indicated DT40 cell lines and 10 nM substrate was incubated for 10 min at room temperature. Reactions were stopped by addition of a reaction quencher and fluorescence intensity was measured using a BMG Labtech Pherastar plate reader at excitation and emission wavelengths of λex 490 nm and λem 520 nm. Fluorescence was normalised to that obtained for recombinant human TDP1 (6.5 pM) and data represent the average of 3 independent experiments ± STD. (C) Whole cell extracts (40 μg) from the indicated DT40 cell lines were separated by 10% SDS-PAGE and immunblotted using antibodies against TDP1 and actin. (D) A 15 μl final volume reaction containing 0.5 μg SCLC WCE and 10 nM substrate was incubated for 10 min at room temperature. Reactions were stopped by addition of a reaction quencher and fluorescence intensity was measured using a BMG Labtech Pherastar plate reader at excitation and emission wavelengths of λex 490 nm and λem 520 nm. Average fluorescence was normalised to that obtained for recombinant human TDP1 (6.5 pM), and data represent the average of 3 independent experiments ± STD. (E) A graphical representation of the Pearson’s correlation coefficient (r) between SCLC TDP1 protein level and TDP1 activity from 0.5 μg WCE.
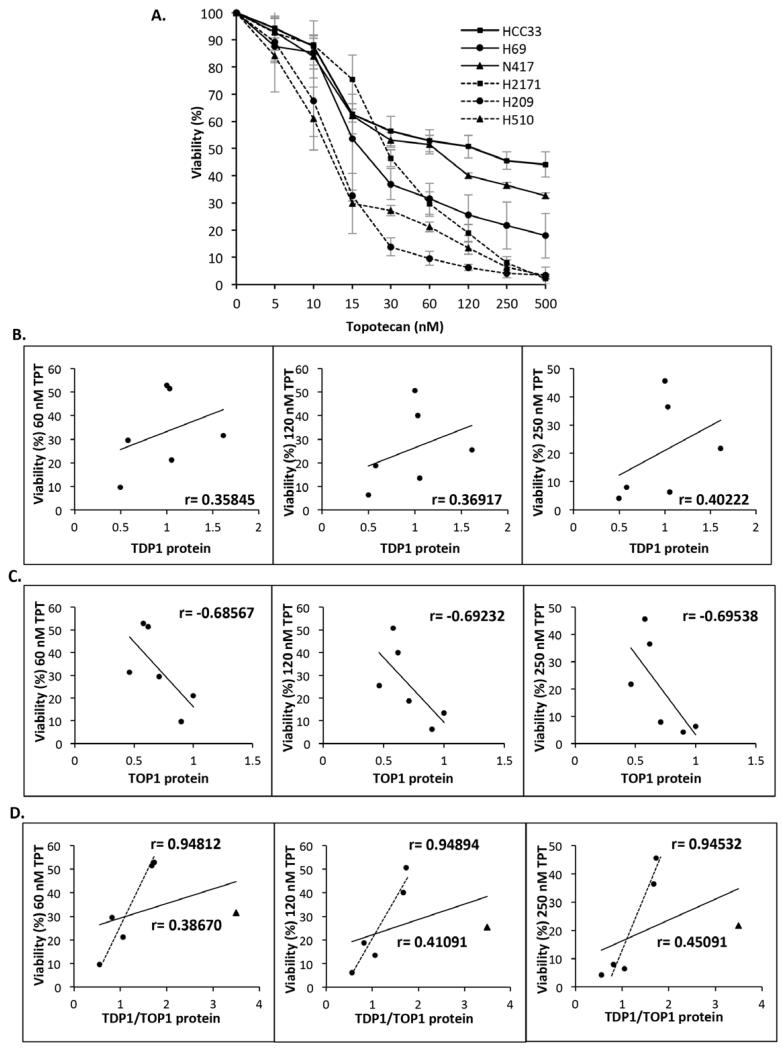
TDP1 is a key repair factor responsible for resolving TOP1-linked DNA breaks generated by camptothecin and its derivatives [17,20]. It was therefore of interest to understand the relationship between TDP1 levels and sensitivity of SCLC to the camptothecin drug, topotecan that is currently used in the clinic to treat relapsed SCLC. Cell viability in the presence or absence of topotecan (TPT) was determined using the CellTiter-Blue viability reagent. Cells were treated with a concentration range of topotecan (0 nM to 500 nM) and grown for four days prior to analysis from which percentage viability (Figure 5A) and the Pearson’s correlation coefficient between TDP1 protein level and topotecan sensitivity (Figure 5B) were calculated. We observed a weak positive correlation between TDP1 level and viability at all topotecan concentrations analysed (Figure 5B and Table 1). Following topotecan treatment, TDP1 exclusively repairs TOP1 mediated DNA breaks, thus we reasoned that the weak correlation might be due to variations in TOP1 levels. Indeed, subsequent analyses revealed a stronger negative correlation between TOP1 levels and topotecan sensitivity (Figure 5C), demonstrating that TOP1 is an important player in determining the response of SCLC to topotecan. To account for variations in TOP1 levels that may explain the weak correlation between TDP1 and topotecan sensitivity, we normalised TDP1 to TOP1 protein levels (TDP1/TOP1) for further analysis. This normalisation revealed a modest increase in the positive correlation between TDP1 and topotecan sensitivity (Figure 5D, solid line). Interestingly, the cell line H69 that expresses the highest level of TDP1 and the lowest level of TOP1 unexpectedly exhibited relatively high topotecan sensitivity. Repeating the analyses without this outlier cell line revealed a strong positive correlation between TDP1/TOP1 and topotecan sensitivity in the five remaining cell lines (Figure 5D, dotted line and Table 1).
Figure 5. TDP1 and TOP1 levels modulate the cellular response of SCLC to topotecan.
(A) The indicated SCLC cell lines were seeded in triplicate at 10,000 cells per well in a 96-well plate in 100 μl media containing the indicated concentration of topotecan (TPT) and incubated at 37°C for 4 days. Viability was assessed by adding 20 μL of the CellTiter-Blue viability assay reagent to wells on day 4, followed by 2 hour incubation at 37°C. Fluorescence was determined using a GloMax Multi Detection System (Promega, Southamption, UK) at excitation and emission wavelengths of λex 560 nm and λem 590 nm, respectively. For each cell line, percentage viability (%) was calculated as [(Fluorescence readtreatment/Fluorescence readuntreated)*100%] and the graphical representation shows the results obtained from three independent experiments ± STD. Graphical representations depicting Pearson’s correlation coefficient (r) between viability at the indicated TPT concentrations and TDP1 (B), TOP1 (C), or TDP1/TOP1 levels (D). The solid line represents the correlation coefficient for all six SCLC cell lines whereas the dotted line represents the correlation coefficient for the five SCLC cell lines HCC33, N417, H2171, H209 and H510, excluding H69 (triangle), which exhibits high levels of TDP1 and an unexpected hypersensitivity to topotecan.
Table 1.
Summary of the Pearson’s correlation coefficient (r) and respective probability for all correlation analyses performed in the study.
| Pearson’s Correlation Coefficient (r) | Probability (p) (2-tailed) | |
|---|---|---|
| TDP1/TOP1 and Viability (%) 120 nM TPT, n=8 (No H69 or H187) | 0.941 | 0.000491 |
| TDP1/TOP1 and Viability (%) 120 nM TPT, n= 5 (No H69) | 0.949 | 0.014 |
| TDP1/TOP1 and Viability (%) 60 nM TPT, n=5 (No H69) | 0.948 | 0.014 |
| TDP1/TOP1 and Viability (%) 250 nM TPT, n=5 (No H69) | 0.945 | 0.154 |
| TDP1 protein and TDP1 ac,vity-Gyrasol, n=6 | 0.861 | 0.028 |
| TDP1 protein and TDP1 ac,vity-GB assay 25 ng, n=6 | 0.747 | 0.088 |
| TDP1 protein and TDP1 ac,vity-GB assay 20 ng, n=6 | 0.703 | 0.121 |
| TOP1 protein and Viability (%) 250 nM TPT, n=6 | −0.695 | 0.125 |
| TOP1 protein and Viability (%) 120 nM TPT, n=6 | −0.692 | 0.127 |
| TOP1 protein and Viability (%) 60 nM TPT, n=6 | −0.686 | 0.133 |
| TDP1 protein and TDP1 mRNA, n=6 | 0.681 | 0.136 |
| TOP1 protein and Top1 mRNA, n=6 | 0.519 | 0.292 |
| TDP1/TOP1 and Viability (%) 250 nM TPT, n=6 | 0.451 | 0.369 |
| TDP1/TOP1 and Viability (%) 120 nM TPT, n=6 | 0.411 | 0.418 |
| TDP1/TOP1 and Viability (%) 60 nM TPT, n=6 | 0.387 | 0.448 |
| TDP1 protein and Viability (%) 250 nM TPT, n=6 | 0.402 | 0.429 |
| TDP1 protein and Viability (%) 120 nM TPT, n=6 | 0.369 | 0.471 |
| TDP1 protein and Viability (%) 60 nM TPT, n=6 | 0.358 | 0.485 |
| TDP1/TOP1 and Viability (%) 120 nM TPT, n=10 | 0.199 | 0.582 |
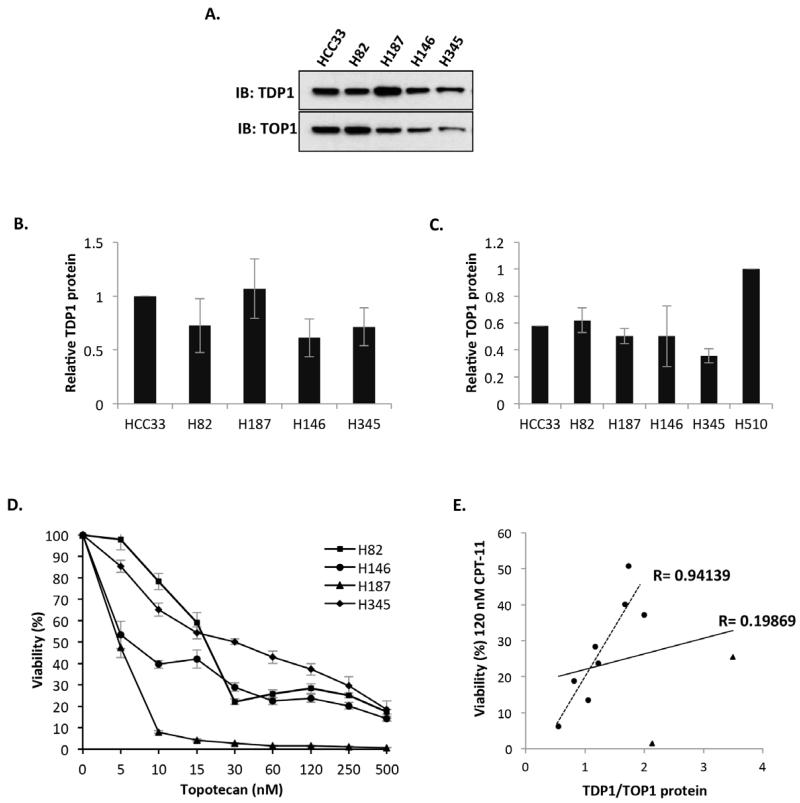
To examine if these findings are restricted to the 5 SCLC lines employed above, we obtained and analysed four additional SCLC lines. TDP1 and TOP1 protein level for each cell line was determined by Western blotting (Figure 6A-C) and the corresponding topotecan sensitivity was measured as percentage viability using the CellTiter-Blue cell viability assay (Figure 6D). These analyses revealed that the majority of SCLC cell lines (8 out of 10) showed a strong and significant positive correlation (r=0.94139, p=0.000491) between TDP1/TOP1 ratio and topotecan sensitivity (Figure 6E, dotted line and Table 1) with 2 hypersensitive outliers, H69 and H187 that intriguingly possess the highest level of TDP1 (Figure 6E, solid line). These data suggest TDP1/TOP1 ratio as a useful indicator for the response of SCLC to topotecan and identify H69 and H187 cell lines as potential resources to further study the molecular basis of the repair of TOP1 breaks in SCLC.
Figure 6. TDP1/TOP1 ratio predicts the cellular response of SCLC to topotecan.
(A) Whole cell extracts (40 μg) from the indicated SCLC cell lines were separated by 10 % SDS-PAGE and immunblotted using antibodies against TDP1, TOP1 and actin. A quantitation of relative TDP1 (B) and TOP1 (C) protein level is shown. Data are the average of three independent experiments ± STD normalised to HCC33 for TDP1 and H510 for TOP1. (D) The indicated SCLC cell lines were seeded in triplicate at 10,000 cells per well in a 96-well plate in 100 μl media containing the indicated concentration of topotecan (TPT) and incubated at 37 °C for 4 days. Viability was assessed by the CellTiter-Blue viability assay. Fluorescence was determined using a GloMax Multi Detection System (Promega, Southamption, UK) at excitation and emission wavelengths of λex 560 nm and λem 590 nm, respectively. For each cell line, percentage viability (%) was calculated as [(Fluorescence readtreatment/Fluorescence readuntreated)*100%] and the graphical representation shows the results obtained from three independent experiments ± STD. (E) Graphical representations depicting Pearson’s correlation coefficient (r) between SCLC viability at 120 nM TPT treatment and TDP1/TOP1 levels. The solid line represents the correlation coefficient for all 10 SCLC cell lines whereas the dotted line represents the correlation coefficient for the eight SCLC cell lines HCC33, N417, H2171, H209, H510, H82, H146 and H345, excluding H69 and H187 (triangles), which exhibit high levels of TDP1 and an unexpected hypersensitivity to topotecan.
Discussion
The highly proliferative and metastatic small cell lung cancers are routinely treated in the clinic with the camptothecin derivatives irinotecan and topotecan [7]. Though most patients initially respond to chemotherapy, virtually all patients experience rapid relapse and bad prognosis. Camptothecins induce damage to cancer cells by locking topoisomerase I (TOP1) on to DNA at single-strand breaks (SSBs) [9,12,17,34-36]. These SSBs can be converted to toxic DNA double-strand breaks by collision with the DNA replication machinery. Tyrosyl DNA phosphodiesterase 1 (TDP1) is a key player in the repair of TOP1 associated DNA breaks [16,18,19,37]. Loss of TDP1 has been shown to prolong the persistence of TOP1-SSBs and increases the likelihood of DNA double-strand break formation, thereby potentiating camptothecin-induced cytotoxicity. Many research groups are therefore keen to uncover small molecule TDP1 inhibitors to improve the clinical response to camptothecin-based therapies. Despite the common use of camptothecin derivatives to treat SCLC in the clinic, the impact of TOP1 and TDP1 on the response of SCLC to camptothecins remains largely unknown. Furthermore, an understanding of the relationship between cellular TDP1 protein level, transcript level and catalytic activity in SCLC is also lacking. This knowledge is important for the design of practical diagnostic tools for patient tumor characterisation.
In this study, we report a higher level of TDP1 expression in SCLC compared to other lung cancer cell lines and a remarkable variation in both TDP1 and TOP1 protein levels in SCLC. These data highlight SCLC as a key cancer type that may benefit from TDP1 inhibition. TDP1 protein level correlates well with both TDP1 mRNA and cellular TDP1 activity, as determined by two independent TDP1 activity assay methods. Although we found a good negative correlation between TOP1 levels and SCLC sensitivity to the camptothecin drug topotecan, we noted that TDP1 protein level alone did not correlate well with the sensitivity of SCLC cell lines to topotecan. Normalisation of TDP1 levels to that of TOP1 revealed a strong positive correlation to topotecan in 80 % of the SCLC cell lines employed in the study. Interestingly, the two outlier cell lines (H69 and H187) that display relatively high sensitivity to topotecan possess remarkably higher levels of TDP1 than the rest. It is worth noting that TDP1 is not the only factor involved in the cellular response to topotecan and that redundant and partially overlapping pathways driven by TDP2, MUS81, XPF-ERCC1, and MRE11 are also implicated in this process [27,38,39]. It is possible that the H69 and H187 cell lines lack one or more TDP1 alternative pathway for dealing with TOP1-breaks, which might account for their unexpected high sensitivity to topotecan. TDP1 posttranslational modification by phosphorylation, SUMOylation, and PARylation is important for recruitment and enrichment of TDP1 at sites of DNA damage [11,18,19,32], and it is thus possible that a lack of one or more of these modifications renders TDP1 less active in vivo in H69 and H187 cells, despite its higher expression level compared to the rest of the panel. A complete understanding of why these two cell lines are hypersensitive to CPT may furthermore reveal novel synthetic lethal targets for drug design and combination therapy with camptothecin treatments and will be the focus of further studies.
The data presented here suggest the potential for utilising TDP1/TOP1 ratio as an indicator for the response of SCLC to topotecan, however it will be important to additionally determine if this will hold true in patient SCLC tissues in future studies. Another important outcome of this study is the development and validation of an easy-to-use TDP1 enzymatic assay in cell extracts, which could be exploited as a diagnostic tool in the clinic to stratify patients undergoing TOP1 based therapies. Furthermore, the strong correlation between TDP1 protein level, enzymatic activity, and mRNA level, suggest the potential utility of immunohistochemistry and microarray analysis in assessing TDP1 levels in SCLC tissues.
In summary, we report a remarkable variation in TDP1 and TOP1 levels in a panel of SCLC cell lines and suggest that SCLC could be an ideal model for confirming TDP1 inhibitor hits emerging from existing drug discovery projects and in future clinical trials. We additionally highlight the potential for TDP1/TOP1 ratio as a promising indicator for the response of SCLC to topotecan, though further work is required to confirm these findings in primary SCLC tissues.
Acknowledgement
This work is funded by a Cancer Research UK grant to Sherif El-Khamisy and Simon Ward. El-Khamisy lab is additionally funded by a Wellcome Trust grant number WT091043 and a Fellowship from the Lister Institute of Preventative Medicine. The authors would like to thank members of the Sussex Translational Drug Discovery Group for helpful discussions. We thank Owen Wells for help with the DT40 cells and other members of the El-Khamisy lab for useful discussions.
References
- 1.Rosti G, Bevilacqua G, Bidoli P, Portalone L, Santo A, et al. Small cell lung cancer. Ann Oncol. 2006;17(Suppl 2):ii5–10. doi: 10.1093/annonc/mdj910. [DOI] [PubMed] [Google Scholar]
- 2.van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet. 2011;378:1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 3.Jiang J, Liang X, Zhou X, Huang L, Huang R, et al. A meta-analysis of randomized controlled trials comparing irinotecan/platinum with etoposide/platinum in patients with previously untreated extensive-stage small cell lung cancer. J Thorac Oncol. 2010;5:867–873. doi: 10.1097/jto.0b013e3181d95c87. [DOI] [PubMed] [Google Scholar]
- 4.Lima JP, dos Santos LV, Sasse EC, Lima CS, Sasse AD. Camptothecins compared with etoposide in combination with platinum analog in extensive stage small cell lung cancer: systematic review with meta-analysis. J Thorac Oncol. 2010;5:1986–1993. doi: 10.1097/JTO.0b013e3181f2451c. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien ME, Ciuleanu TE, Tsekov H, Shparyk Y, Cuceviá B, et al. Phase III trial comparing supportive care alone with supportive care with oral topotecan in patients with relapsed small-cell lung cancer. J Clin Oncol. 2006;24:5441–5447. doi: 10.1200/JCO.2006.06.5821. [DOI] [PubMed] [Google Scholar]
- 6.Eckardt JR, von Pawel J, Pujol JL, Papai Z, Quoix E, et al. Phase III study of oral compared with intravenous topotecan as second-line therapy in small-cell lung cancer. J Clin Oncol. 2007;25:2086–2092. doi: 10.1200/JCO.2006.08.3998. [DOI] [PubMed] [Google Scholar]
- 7.Kalemkerian GP, Akerley W, Bogner P, Borghaei H, Chow LQ, et al. Small cell lung cancer. J Natl Compr Canc Netw. 2013;11:78–98. doi: 10.6004/jnccn.2013.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Wang LK, Kingsbury WD, Johnson RK, Hecht SM. Differential effects of camptothecin derivatives on topoisomerase I-mediated DNA structure modification. Biochemistry. 1998;37:9399–9408. doi: 10.1021/bi980451k. [DOI] [PubMed] [Google Scholar]
- 10.Sordet O, Nakamura AJ, Redon CE, Pommier Y. DNA double-strand breaks and ATM activation by transcription-blocking DNA lesions. Cell Cycle. 2010;9:274–278. doi: 10.4161/cc.9.2.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudson JJR, Chiang S-C, Wells OS, Rookyard C, El-Khamisy SF. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nature Communications. 2012;3:733–813. doi: 10.1038/ncomms1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alagoz M, Chiang SC, Sharma A, El-Khamisy SF. ATM deficiency results in accumulation of DNA-topoisomerase I covalent intermediates in neural cells. PLoS One. 2013;8:e58239. doi: 10.1371/journal.pone.0058239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert DC, Chalmers AJ, El-Khamisy SF. Topoisomerase I inhibition in colorectal cancer: biomarkers and therapeutic targets. Br J Cancer. 2012;106:18–24. doi: 10.1038/bjc.2011.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S-YN, Murai J, Dalla Rosa I, Dexheimer TS, Naumova A, et al. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Res. 2013;41:7793–7803. doi: 10.1093/nar/gkt483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pommier Y, Huang SY, Gao R, Das BB, Murai J, et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA Repair (Amst) 2014;19:114–129. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alagoz M, Wells OS, El-Khamisy SF. TDP1 deficiency sensitizes human cells to base damage via distinct topoisomerase I and PARP mechanisms with potential applications for cancer therapy. Nucleic Acids Res. 2014;42:3089–3103. doi: 10.1093/nar/gkt1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 18.Chiang SC, Carroll J, El-Khamisy SF. TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage. Cell Cycle. 2010;9:588–595. doi: 10.4161/cc.9.3.10598. [DOI] [PubMed] [Google Scholar]
- 19.Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, et al. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009;28:3667–3680. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J Biol Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker S, Meisenberg C, Bibby RA, Askwith T, Williams G, et al. Development of an oligonucleotide-based fluorescence assay for the identification of tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors. Anal Biochem. 2014;454:17–22. doi: 10.1016/j.ab.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang SN, Pommier Y, Marchand C. Tyrosyl-DNA Phosphodiesterase 1 (Tdp1) inhibitors. Expert Opin Ther Pat. 2011;21:1285–1292. doi: 10.1517/13543776.2011.604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidlich IE, Dexheimer T, Marchand C, Antony S, Pommier Y, et al. Inhibitors of human tyrosyl-DNA phospodiesterase (hTdp1) developed by virtual screening using ligand-based pharmacophores. Bioorg Med Chem. 2010;18:182–189. doi: 10.1016/j.bmc.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchand C, Lea WA, Jadhav A, Dexheimer TS, Austin CP, et al. Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay. Mol Cancer Ther. 2009;8:240–248. doi: 10.1158/1535-7163.MCT-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomicic MT, Kaina B. Topoisomerase degradation, DSB repair, p53 and IAPs in cancer cell resistance to camptothecin-like topoisomerase I inhibitors. Biochim Biophys Acta. 2013;1835:11–27. doi: 10.1016/j.bbcan.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Nitiss JL, Wang JC. Mechanisms of cell killing by drugs that trap covalent complexes between DNA topoisomerases and DNA. Mol Pharmacol. 1996;50:1095–1102. [PubMed] [Google Scholar]
- 27.Zeng Z, Sharma A, Ju L, Murai J, Umans L, et al. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 2012;40:8371–8380. doi: 10.1093/nar/gks622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Khamisy SF. To live or to die: a matter of processing damaged DNA termini in neurons. EMBO Mol Med. 2011;3:78–88. doi: 10.1002/emmm.201000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barthelmes HU, Habermeyer M, Christensen MO, Mielke C, Interthal H, et al. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J Biol Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 30.El-Khamisy SF, Katyal S, Patel P, Ju L, McKinnon PJ, et al. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair (Amst) 2009;8:760–806. doi: 10.1016/j.dnarep.2009.02.002. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das BB, Huang SY, Murai J, Rehman I, Amé JC, et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014;42:4435–4449. doi: 10.1093/nar/gku088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao R, Das BB, Chatterjee R, Abaan OD, Agama K, et al. Epigenetic and genetic inactivation of tyrosyl-DNA-phosphodiesterase 1 (TDP1) in human lung cancer cells from the NCI-60 panel. DNA Repair (Amst) 2014;13:1–9. doi: 10.1016/j.dnarep.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, et al. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, et al. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst) 2006;5:1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alagoz M, Gilbert DC, El-Khamisy S, Chalmers AJ. DNA repair and resistance to topoisomerase I inhibitors: mechanisms, biomarkers and therapeutic targets. Curr Med Chem. 2012;19:3874–3885. doi: 10.2174/092986712802002590. [DOI] [PubMed] [Google Scholar]
- 37.Hamilton NK, Maizels N. MRE11 function in response to topoisomerase poisons is independent of its function in double-strand break repair in Saccharomyces cerevisiae. PLoS One. 2010;5:e15387. doi: 10.1371/journal.pone.0015387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng C, Brown JA, You D, Brown JM. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci U S A. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]