Abstract
Exposure to ionizing radiation can result in delayed effects that can be detected in the progeny of an irradiated cell multiple generations after the initial exposure. These effects are described under the rubric of radiation-induced genomic instability and encompass multiple genotoxic endpoints. We have developed a green fluorescence protein (GFP)-based assay and demonstrated that ionizing radiation induces genomic instability in human RKO-derived cells and in human hamster hybrid GM10115 cells, manifested as increased homologous recombination (HR). Up to 10% of cells cultured after irradiation produce mixed GFP+/− colonies indicative of delayed HR or, in the case of RKO-derived cells, mutation and deletion. Consistent with prior studies, delayed chromosomal instability correlated with delayed reproductive cell death. In contrast, cells displaying delayed HR showed no evidence of delayed reproductive cell death, and there was no correlation between delayed chromosomal instability and delayed HR, indicating that these forms of genome instability arise by distinct mechanisms. Because delayed hyperrecombination can be induced at doses of ionizing radiation that are not associated with significantly reduced cell viability, these data may have important implications for assessment of radiation risk and understanding the mechanisms of radiation carcinogenesis.
Ionizing radiation induces many forms of DNA damage directly by energy absorption and indirectly by producing highly reactive free radicals, including base damage, DNA-protein cross-links, single-strand breaks, and double-strand breaks (40). Ionizing radiation can directly induce mutations (18), chromosome aberrations (10), and homologous recombination (HR) (2, 6). In recent years, it has become evident that radiation also induces delayed genomic instability, defined as an increased rate of genetic alterations in the genome of progeny of irradiated cells multiple generations after the initial insult. Delayed effects include chromosomal rearrangements and aberrations (chromosomal instability), micronuclei, gene mutations, microsatellite instability, changes in ploidy, and decreased plating efficiency (PE) (32, 33). Prior studies have also established an association between delayed chromosomal instability and delayed reproductive cell death (23, 28).
Loss of genomic integrity has been proposed as a common feature of cancer (9, 17). Understanding the mechanism of radiation-induced instability is critical to radiation risk assessment and for determining its potential role in carcinogenesis. Of the many delayed effects of radiation, chromosomal instability is perhaps the best described. However, the cytogenetic assays employed to measure chromosomal instability are time and labor intensive, and they are not practical for investigating potential detrimental effects over a range of doses of ionizing radiation. Furthermore, a rapid and sensitive assay to measure radiation-induced genomic instability would greatly facilitate mechanistic studies.
Chromosomal aberrations induced in first-division metaphase cells after irradiation include classical breakage and rejoining events that produce dicentric chromosomes and their associated fragments (10). In contrast, radiation-induced delayed chromosomal instability frequently involves duplication and insertion of genomic regions suggestive of recombination (11, 14, 28). Little et al. (25) reported that most delayed mutations after exposure to radiation were point mutations, but a significant fraction (∼20%) were associated with partial or total gene deletions suggestive of delayed recombination. However, cytogenetic analysis cannot distinguish between homologous and nonhomologous recombination, so it has been unclear whether HR contributes to delayed genome instability. Although HR is often characterized as a mechanism for accurate repair of DNA damage, certain HR events can result in deleterious chromosomal rearrangements. For example, gene conversion produces local loss of heterozygosity (LOH), and gene conversion is sometimes associated with crossovers that can lead to large-scale LOH in the next cell division, as well as deletions and inversions at linked repeats and translocations (35).
To determine whether ionizing radiation induces delayed genomic instability manifested as enhanced HR, we have developed two cell lines carrying a green fluorescence protein (GFP)-based HR substrate. We show that radiation-induced delayed HR is a sensitive measure of delayed radiation effects. Because HR can be measured rapidly and unambiguously, the system is well suited for studies of delayed instability where biological effects might be rare. We further show that delayed HR is not associated with delayed chromosomal instability or with delayed reproductive cell death, indicating that these delayed effects of radiation arise by at least two distinct mechanisms. Because the majority of cells survive radiation exposure at lower doses, delayed effects occurring in the progeny of irradiated cells may have significant implications for evaluating risks associated with radiation exposure and provide mechanistic insights into radiation carcinogenesis.
MATERIALS AND METHODS
Plasmid DNAs.
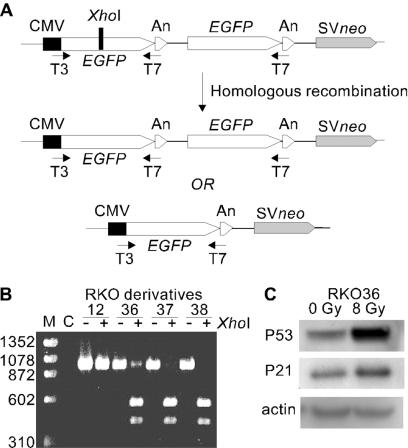
The XhoI and HindIII sites in pCMV-Script (Stratagene, La Jolla, Calif.) were destroyed sequentially by cleavage, fill-in with T4 DNA polymerase, and ligation. A 700-bp BamHI/ApaI fragment carrying the EGFP gene (hereafter referred to as GFP) was excised from pEGFP (Clonetech, Palo Alto, Calif.) and inserted into the modified pCMV-Script vector, creating pCMV-EGFP. A custom linker with BglII and HindIII sites was inserted into the DraIII site, and a +5-bp XhoI frameshift insertion was introduced 310 bp downstream of the GFP start codon by site-directed mutagenesis (12) using a 39-base primer (5′-GCGGGTCTTGTAGTTGCCTCGAGGTCGTCCTTGAAGAAG-3′; inserted bases are underlined, XhoI site is boldface), creating pCMV-EGFPXhoBglH3. GFP plus the SV40 poly(A) signal sequence (An) was excised from pCMV-EGFP as a 1.2-kbp MluI/BamHI fragment (with the MluI site filled in) and inserted into the HincII/BamHI sites of pUC19, creating pUC-EGFPAn. GFPAn was then transferred from this plasmid as a BamHI/HindIII fragment into the BglII/HindIII sites in pCMV-EGFPXhoBglH3, creating pCMV-EGFP2Xho (Fig. 1A).
FIG. 1.
Experimental system. (A) The recombination substrate consists of 1,137-bp direct repeats [700-bp GFP and 437-bp poly(A) signal sequences] separated by 225 bp and SV40 promoter-driven neo. The XhoI frameshift mutation (black bar) inactivates the upstream GFP. HR includes gene conversion of the XhoI site without an associated crossover or deletion of one GFP copy by crossover or single-strand annealing. PCR primers targeted to T3 and T7 promoter sequences amplify GFP in both types of recombination products. (B) RT-PCR analysis of recombination substrate in RKO derivatives. RT-PCR products from RKO derivatives (with or without XhoI) are indicated; control (nontransfected) RKO cells are shown in lane C, and size markers (M) are ΦX-174/HaeIII. (C) Western analysis of p53 and p21 in RKO36 cells with or without 8-Gy X-ray exposure, with β-actin loading controls below.
Cell culture and transfection.
Two mammalian cell lines were used in this study: RKO cells, because they have a wild-type functional p53 gene and a near diploid karyotype, and GM10115 cells, which have a mutant p53 gene (21) and have been used extensively for studies of radiation-induced genomic instability (20-23, 28). The human colorectal carcinoma cell line, RKO, and its derivatives were grown in monolayer cultures in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (HyClone, Logan, Utah) at 37°C in 5% CO2 in air. The hamster human hybrid GM10115 cells (Human Genetic Mutant Cell Repository, Camden, N.J.) contain one copy of human chromosome 4 in a background of 20 to 24 Chinese hamster ovary chromosomes. GM10115 cells and the derivatives were grown as monolayers in Dulbecco's minimal essential medium containing 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 0.2 mM l-proline and cultured at 34°C in humidified incubators containing 5% CO2. Both cell lines were routinely monitored for mycoplasma (MycoFluor mycoplasma detection kit; Molecular Probes, Eugene, Oreg.) and showed no evidence of infection.
Cells were grown to 30 to ∼50% confluence in 100-mm-diameter dishes and were transfected with 4 μg of pCMV-EGFP2Xho linearized with ApaLI (between neo and the cytomegalovirus [CMV] promoter) by using Lipofectamine reagent (Life Technologies) according to the manufacturer's protocol. G418 (1 mg/ml) was added 2 days after transfection, and 2 to 3 weeks later colonies were isolated and expanded, and GFP expression was monitored by fluorescence microscopy or reverse transcription (RT)-PCR as described below.
One clone from the RKO cell line (RKO36) and three clones from the GM10115 cell line (TG9, GH1, and GH24) were selected for detailed analysis. The RKO36 clone contained a mixture of GFP+ and GFP− cells, whereas all three of the GM10115 clones contained the pCMV-EGFP2Xho HR substrate but were almost exclusively GFP− cells. These three cell lines were selected because they showed a measurable spontaneous HR frequency, and it is possible that exposure to a DNA-damaging agent might reduce this background level of HR (30, 37).
Characterization of GFP gene structure and expression by PCR and RT-PCR.
Genomic DNA was extracted from clonally expanded cells using a GenElute mammalian genomic DNA kit (Sigma, St. Louis, Mo.). The GFP gene was amplified by PCR using primers targeted to the T3 sequence upstream of the CMV promoter (5′-CTCGAAATTAACCCTCACTAAAGG-3′) and to the T7 sequence between the GFP coding sequence and the An signal sequence (5′-GAATTGTAATACGACTCACTATAG-3′). The PCR conditions consisted of an initial 2-min incubation at 95°C, followed by 40 cycles at 94°C for 30 s, 50°C for 30 s, 73°C for 1 min, and a final extension at 73°C for 2 min. The resulting 880-bp PCR products were visualized on a 1% agarose gel with or without prior digestion with XhoI to identify the frameshift mutation in the upstream copy of GFP. Total RNA was isolated using an RNeasy mini kit (QIAGEN, Valencia, Calif.) following the manufacturer's protocol. Total RNA (2 μg) was reverse transcribed into cDNA using the T7 primer and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, Wis.). The resultant GFP cDNA was amplified by PCR using the T3 and T7 primers and characterized on agarose gel as described above.
Measurement of p53 and p21 protein levels in RKO cells.
RKO and its derived RKO36 cells treated with X rays (0 or 8 Gy) were harvested 6 h after exposure. Cells were washed once with cold phosphate-buffered saline, scraped, and collected by centrifugation. Cell lysates were prepared in 250 μl of lysis buffer [50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1% (octylphenoxy)polyethoxyethanol octylphenyl-polyethylene glycol, 10% glycerol, 20 mM β-mercaptoethanol, and protease inhibitors]. After 15 min on ice, lysates were centrifuged and supernatants were transferred to new tubes. Protein concentrations were determined by using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.). Proteins (50 μg) from each lysate were mixed with NuPAGE LDS sample buffer (4×; Invitrogen, Carlsbad, Calif.) and dithiothreitol to a final concentration of 50 mM and denatured at 70°C for 10 min. Samples were electrophoresed on a NuPAGE Novex bis-Tris gel (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Invitrogen). The membrane was incubated in 5% nonfat dry milk in TBST (60 mM Tris-base, 120 mM NaCl, 0.2% Tween 20) for 1 h and probed with mouse anti-p53 (Upstate Biotechnologies Inc., Lake Placid, N.Y.), rabbit polyclonal anti-p21 (Santa Cruz Biotechnology, Santa Cruz, Calif.), or mouse anti-β-actin (Sigma) for 1 h at dilutions of 1:1,000, 1:250, and 1:1,000, respectively. Blots were washed three times in TBST and incubated for 30 min with a 1:5,000 dilution of horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) and then washed three times in TBST and developed using a LumiGlo chemiluminescent detection system (KPL Inc., Gaithersburg, Md.) and visualized by autoradiography.
It has previously been established that the GM10115 cell line has a mutant p53 gene that is not inducible by X-irradiation (21), and measurement of p53 was not pursued further in these cells.
X-ray-induced delayed instability at the GFP locus.
Cells carrying the pCMV-EGFP2Xho HR substrate were irradiated at a dose rate of 2.4 Gy/min at ambient temperature using a Pantak HF320 X-ray machine (250-kV peak, 13 mA; half-value layer, 1.65-mm copper). Appropriate numbers of cells were seeded in 100-mm-diameter dishes to yield ∼50 surviving colonies per dish at each dose. Unirradiated cells served as control. Ten to fourteen days after radiation, colonies (>50 cells) were examined for GFP expression using an inverted Nikon Eclipse TE200 fluorescence microscope. In the RKO36 clone that comprised a mixture of GFP+ and GFP− cells, preexisting GFP+ or GFP− cells, if unchanged, will produce uniform GFP+ or GFP− colonies, respectively. Preexisting GFP+ cells can be converted to GFP− cells directly by a radiation-induced point mutation and/or small or large deletions, and preexisting GFP− cells can be converted to GFP+ cells directly by radiation-induced HR. Both these events are expected to be rare because of the small target of the GFP cassette and would result in uniform GFP+ or GFP− colonies, respectively. These uniform GFP+ or GFP− colonies reflect a stable GFP substrate. However, if radiation induces delayed instability at the GFP substrate, this will be reflected as mixed GFP+/− colonies. Such colonies can arise by delayed HR (GFP− → GFP+/−) or delayed mutation and deletion (GFP+ → GFP+/−). On the other hand, the three GM10115 clones were almost exclusively made up of GFP− cells, and only a delayed HR event would produce a mixture of GFP+/− cells in a colony surviving irradiation. Our criterion for radiation-induced genomic instability in surviving colonies is defined as the presence of both GFP+ cells and GFP− cells in a colony, with >4 cells of each type in each mixed colony.
X-ray-induced delayed chromosomal instability.
Genomic instability was also assessed cytogenetically by fluorescence in situ hybridization (FISH). One hundred RKO36 colonies and at least 60 independent colonies from each of the three GM10115 clones were selected 10 to 14 days after irradiation; 30 colonies from nonirradiated transfected cells served as controls. Isolated colonies were picked at random by using sterile Dacron swabs dipped in trypsin, transferred to individual 25-cm2 tissue culture flasks, and grown to confluence. The total growth time postirradiation was 5 to 6 weeks for each of these clonal isolates. Metaphase cells were collected by mitotic shake-off after a 3-h incubation in the presence of Colcemid (0.2 μM final concentration). Cells were treated with prewarmed hypotonic solution (0.2% KCl, 0.2% sodium citrate, 1% fetal bovine serum) for 45 min at 37°C, dehydrated in 100% cold methanol, and then fixed in methanol-acetic acid (5:1, vol/vol). The cell suspension was dropped onto precleaned glass microscope slides and incubated at 68°C for 1 day before processing for FISH or storage at −20°C.
To investigate chromosomal instability in RKO36 clones, human chromosome-specific probes for chromosomes 4, 13, and 16 were prepared from plasmids pBS4, pBS13, and pBS16, respectively (kindly provided by J. Gray and D. Pinkel, University of California, San Francisco, San Francisco). pBS4 and pBS16 were labeled with biotin by using a BioNick labeling kit (Life Technologies). pBS4 and pBS13 were labeled with digoxigenin-11-dUTP (digoxigenin) by using a Nick translation kit (Life Technologies). Prior to FISH, slides were treated with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min at 37°C and then dehydrated in a cold ethanol series (70, 80, and 100%) for 2 min each. After drying, slides were denatured for 4 min at 72°C in 70% formamide-2× SSC and dehydrated in a cold ethanol series (70, 80, and 100% ethanol) for 2 min each. Denatured slides were kept in a 50°C slide warmer after air drying. Each slide was treated with 35 μl of hybridization mix containing 70% formamide-15% dextran sulfate-2× SSC and 40 ng of each labeled probe (biotinylated pBS4 and pBS16, digoxigenin-labeled pBS4 and pBS13). The slides were then covered with glass coverslips, sealed, and incubated for 2 days in a humidified chamber at 37°C. For simultaneous detection of biotinylated and digoxigenin-labeled probes, slides were incubated sequentially for 20-min periods with fluorescein isothiocyanate-avidin plus mouse-antidigoxigenin antibody, anti-avidin plus digoxigenin anti-mouse antibody, fluorescein isothiocyanate-avidin plus mouse antidigoxigenin antibody, and cyc3 anti-mouse antibody, interspersed with 2-min washes in phosphate-buffered detergent; during these treatments, each slide was covered with a plastic coverslip. Chromosomes were counterstained with 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI) in Antifade (Oncor Inc., Gaithersburg, Md.). Slides were covered with glass coverslips and stored at 4°C. Slides were coded, and two hundred metaphase spreads per clonal isolate were scored. A clone was defined as unstable when ≥3 unique subpopulations of metaphase cells were found showing rearrangements involving chromosome 4, 13, or 16, which made up ≥9% of the total metaphases examined.
Chromosomal instability in surviving colonies from the three GM10115 clones was examined by two-color FISH by using fluorescence-labeled probes targeted to the single copy of human chromosome 4 and the telomeric T2AG3(n) sequence as previously described (20-22, 28). In these clones, our criterion for instability was that a clone demonstrate ≥3 unique subpopulations of metaphase cells showing rearrangements involving chromosome 4, which made up ≥5% of the total metaphases examined (20).
Delayed reproductive cell death.
Delayed reproductive cell death (7, 8) was determined by measuring PE of clonal isolates of cells displaying delayed instability at the GFP substrate (GFP+/− colony), or delayed chromosomal instability following irradiation, and unirradiated controls. For each determination, 200 RKO cells or 100 GM10115 cells per dish were seeded in five 60-mm-diameter dishes, the dishes were incubated for 10 to 14 days, and the cells were stained with a 2% crystal violet in 40% methanol. PE was calculated as the percentage of surviving colonies containing more than 50 cells, and the values reported were normalized to controls.
RESULTS
Experimental design.
Radiation directly induces gene mutation, chromosome aberrations, HR, and cell death. Radiation also induces delayed effects, including gene mutation, chromosomal instability, and delayed reproductive cell death (reviewed in references 32 and 33). Given that HR can result in deleterious genetic rearrangements (35), and given the possible involvement of HR in delayed chromosomal instability and mutagenesis (11, 14, 25, 28), we asked whether radiation induces delayed HR in mammalian cells. To address this question, we constructed a plasmid carrying GFP direct repeats linked to an SV40 promoter-driven neo gene. One copy of GFP, driven by a CMV promoter, was inactivated by a +5 frameshift mutation that created an XhoI site; the second copy had a wild-type coding sequence but lacked a promoter (Fig. 1A). This plasmid was transfected into two mammalian cell lines, firstly, the human colon carcinoma RKO cell line, and forty G418r transfectants were isolated. Of these, 37 were GFP−, as they showed no evidence of green under fluorescence microscopy, and RT-PCR products were fully cleaved by XhoI (e.g., RKO37 and RKO38; Fig. 1B). Two colonies were uniformly green, and RT-PCR products were completely resistant to XhoI digestion (e.g., RKO12; Fig. 1B), indicating spontaneous gain of GFP function at the time of transfection. One colony, RKO36, had both GFP− and GFP+ cells, indicating spontaneous gain of functional GFP during clonal expansion. This mixed phenotype was confirmed by RT-PCR analysis, as only a fraction of the RT-PCR product was cleaved by XhoI (Fig. 1B). However, when RKO36 cells were diluted and plated, the colony from a single cell was usually either homogeneously green or colorless; only 1% of RKO36 cells yielded GFP+/− colonies, indicating a low but measurable background of spontaneous HR in this cell line. We focused our attention on RKO36 cells, as the appearance of both GFP− cells and GFP+ cells provided us with the opportunity to assay radiation-induced delayed effects manifested as either gain or loss of GFP function. In addition, the measurable spontaneous frequency of induced HR provided a unique opportunity to evaluate exposures to DNA-damaging agents that might result in a reduction in the spontaneously occurring HR frequency (30, 37). Like the parent RKO cell line, RKO36 cells express p53 and display normal p53 stabilization and p21 induction after irradiation (Fig. 1C), indicating that the DNA damage response network in RKO36 cells is intact and functional.
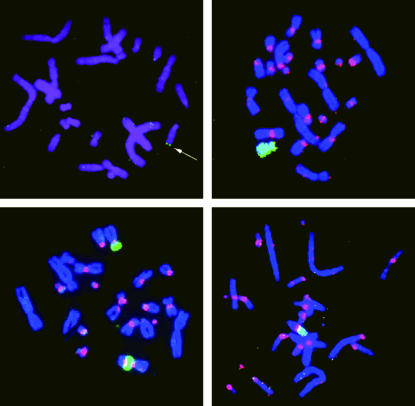
Fifty G418r GM10115 transfectants were isolated and screened for the pCMV-EGFP2Xho HR substrate as described above. None of those isolated showed evidence of a high frequency of both GFP+ and GFP− cells, and the majority (48/50) were GFP− isolates. Three of these, TG9, GH1, and GH24, were selected, as they showed evidence of a single GFP integration site as determined by FISH with the fluorescence-labeled pCMV-EGFP2Xho vector as a probe (Fig. 2, top left). These clones grow well in culture and maintained the human marker chromosome in a significant number of cells.
FIG. 2.
Metaphase chromosomes from the GM10115-derived clone GH24. (Top left) FISH with a biotinylated probe to the pCMV-EGFP2Xho HR substrate. A single hybridization signal is arrowed. (Top right, bottom left, and bottom right) FISH with fluorescence-labeled probes to the T2AG3(n) interstitial telomeric-like repeat (digoxigenin labeled, red) and human chromosome 4 (biotinylated labeled, green). (Top right) Control unirradiated metaphase cell. (Bottom left and right). Two metaphase cells from a clonally expanded cell surviving 1 Gy of X-irradiation, illustrating instability of the human chromosome and how it can recombine with the hamster chromosomes at the interstitial telomeric sites.
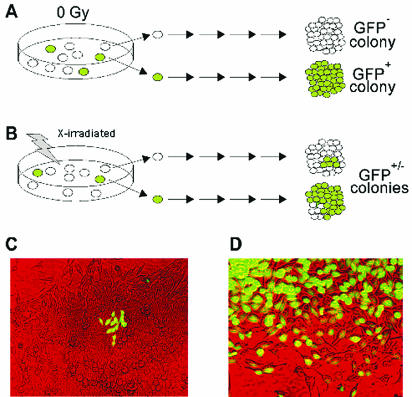
Our general strategy for detecting delayed instability at the GFP locus is outlined in Fig. 3. In unirradiated cultures, the majority of RKO36 parental cells were GFP− and will remain GFP−. Similarly, preexisting GFP+ cells will produce GFP+ colonies (Fig. 3A). For irradiated cultures, however, cells may be directly converted from GFP− to GFP+ by radiation-induced HR, in which case all cells within a colony would be GFP+, or GFP+ cells may be directly converted by radiation to GFP− by a variety of processes, including point mutagenesis and induction of small- or large-scale deletions. Thus, all uniform GFP+ or GFP− colonies reflect either a prior stable phenotype or a new stable phenotype that was directly induced by radiation. However, if radiation induces delayed instability at the GFP locus, this will give rise to mixed GFP+/− colonies (Fig. 3B). Such colonies can arise by delayed HR (GFP− → GFP+/−; Fig. 3C) or delayed mutation and deletion (GFP+ → GFP+/−; Fig. 3D). These GFP+/−colonies are scored as unstable based on our criterion. Note that for GM10115 clones, only GFP− to mixed GFP+/− colonies were considered unstable.
FIG. 3.
Strategy for detecting delayed instability at the GFP locus. (A) Unirradiated cells are stable and give uniform GFP+ or GFP− colonies. (B) Irradiated cells showing delayed instability involving HR (top) or delayed mutation (bottom). (C and D) Photomicrographs of GFP+/− colonies presumably displaying delayed HR and delayed mutation, respectively.
Radiation-induced delayed effects manifested as enhanced HR.
Approximately 1% of RKO36 and 2 to 5% of the three GM10115 derivative cells produce GFP+/− colonies when subcloned. However, after exposure to ionizing radiation, there was an increase in the numbers of surviving colonies that were GFP+/− (Tables 1 and 2). Among these GFP+/− colonies, the majority had >4 GFP+ cells in a background of GFP− cells (Fig. 3C). These results indicate that radiation induces delayed HR events. Some RKO36 GFP+/− colonies had >4 GFP− cells in colonies that were predominantly GFP+ (Fig. 3D), suggesting that the GFP gene was lost through delayed mutation in cells arising from a single GFP+ cell. Alternatively, induced instability was an early event occurring soon after radiation exposure. Thus, RKO36 cells allow the simultaneous analysis of radiation-induced delayed instability reflected as increased HR and increased mutation and deletion. It is likely that most or all GFP+/− colonies that are largely GFP− result from delayed HR rather than mutation and deletion (and vice versa for GFP+/− colonies that are largely GFP+). However, because of uncertainties regarding the timing of these phenotypic conversions, and potential differences in growth rates owing to radiation exposure, all subsequent analyses were performed by combining these GFP+/− classes.
TABLE 1.
Radiation-induced genomic instability as measured by GFP-based assay in RKO36 cellsa
| X-ray dose (Gy) | No. of expts | Total no. of colonies scored | GFP+/− colonies (%) | SE (%) | Surviving fraction |
|---|---|---|---|---|---|
| 0 | 4 | 1,286 | 0.97 | 0.11 | 1 |
| 1.0 | 2 | 951 | 3.68 | 0.22 | 0.68 |
| 2.5 | 3 | 1,427 | 4.59* | 0.58 | 0.334 |
| 5.0 | 4 | 2,256 | 9.76* | 1.43 | 0.028 |
| 10.0 | 3 | 2,458 | 7.69* | 0.73 | 0.0009 |
Surviving fraction is the percentage of RKO36 cells surviving the given dose of radiation as determined by clonogenic survival. *, P < 0.05.
TABLE 2.
Radiation-induced genomic instability as measured by the GFP-based assay in three transfected clones of GM10115 cellsa
| Clone and X-ray dose (Gy) | Total no. of colonies scored | GFP+/− colonies (%) | SE (%) |
|---|---|---|---|
| TG9 | |||
| 0 | 449 | 2.07 | 0.92 |
| 1.0 | 587 | 2.18 | 0.64 |
| 2.5 | 559 | 5.46* | 0.88 |
| GH1 | |||
| 0 | 485 | 5.32 | 0.81 |
| 1.0 | 432 | 9.53* | 0.71 |
| 2.5 | 395 | 14.67* | 1.86 |
| GH24 | |||
| 0 | 646 | 2.45 | 1.10 |
| 1.0 | 727 | 6.30 | 1.10 |
| 2.5 | 690 | 9.09* | 1.40 |
Surviving fraction is the percentage of GM10115 cells surviving the given dose of radiation as determined by clonogenic survival. *, P < 0.05. Ten plates were used at each dose.
The frequency of GFP+/− colonies in both RKO36 cells (Table 1) and GM10115 cells (Table 2) increased after increasing doses of X-irradiation. In RKO36 cells exposed to high doses of radiation, 5 or 10 Gy, GFP+/− colonies were 4- to 10-fold more frequent than in unirradiated controls. There was a significantly greater frequency of GFP+/− colonies with 5 and 10 Gy than with 1 Gy, suggesting that delayed instability increases with dose. However, this was not a typical dose-response profile consistent with other reports of delayed effects in clonally expanded cells surviving irradiation (39). GFP+/− colonies were not more frequent at 10 Gy than at 5 Gy, perhaps reflecting saturation at higher doses or the relative lack of survivors at the highest dose. At the radiation doses used, there was no reduction in the spontaneous frequency of HR, but there was a dose-dependent increase in GFP+/− colonies, with the exception of clone TG9 exposed to 1 Gy of X rays, where no change over background was observed.
Linear mixed-effects models were used to assess the impact of X-irradiation on the percentage of unstable colonies. Logit transformation was used to achieve approximate normality of the response (percentage of GFP+/− colonies). The number of cells seeded for each experiment had no statistically significant effect on the percentage of GFP+/− colonies (P = 0.56) and was excluded from the subsequent analyses. Dose of X-irradiation was used as a continuous variable to estimate the rate of change in the percentage of GFP+/− colonies per unit increment in the dose of irradiation. As the dose increased, so did the percentage of GFP+/− colonies (P = 0.0008). We also compared the percentage of unstable colonies in each dose group versus the control group. The P values were adjusted for multiple comparisons using Dunnett's procedure (Tables 1 and 2).
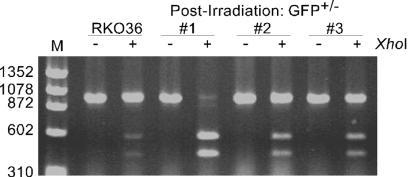
We analyzed genomic DNA isolated from radiation-induced GFP+/− colonies by using PCR with or without XhoI digestion and found no size differences between the parental RKO36 and GFP+/− colonies or between parental GM10115 cells and GFP+/− colonies (representative data are shown in Fig. 4). These observations indicate that the delayed effect(s) of radiation does not typically lead to large-scale deletions at the GFP locus, a result consistent with data from Little and colleagues (25). Thus, these results indicate that radiation can induce delayed HR, and importantly, that delayed HR is a convenient and reliable endpoint for studying the delayed effects of ionizing radiation. In both cell lines, an increase in hyperrecombination could be observed at the lowest radiation dose used, 1 Gy. At this dose, 68 and 78% of the irradiated RKO36 and GM10115 cells, respectively, survive relative to the unirradiated controls. Furthermore, analysis of HR only requires cells be cultured for 10 to 14 days for colony formation rather than the 6 to 8 weeks required for cytogenetic analysis.
FIG. 4.
Analysis of genomic DNA isolated from GFP+/− colonies. The GFP locus was amplified from genomic DNA from parental RKO36 cells and three radiation-induced GFP+/− colonies. PCR products were separated on an agarose gel directly or following XhoI digestion as indicated.
Radiation-induced delayed chromosomal instability.
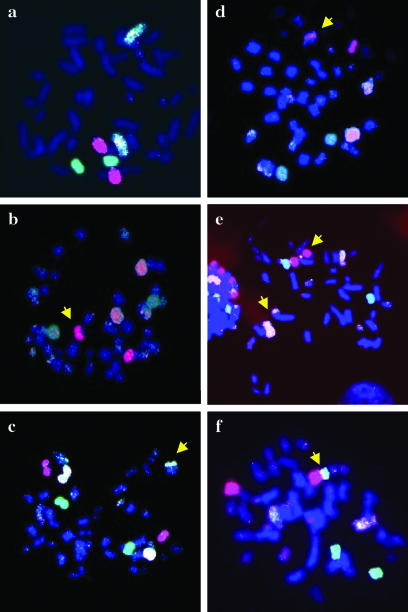
RKO36 cells have a near-diploid chromosome complement. FISH with probes for chromosomes 4, 13, and 16 in metaphase spreads from 30 unirradiated colonies of RKO36 showed normal patterns in all cases. Of 100 colonies surviving a 10-Gy X-ray dose, 8% showed ≥3 metaphase populations with a rearranged chromosome 4, 13, or 16. The stringent ≥3 limit eliminates clones with rearrangements formed by direct effects of radiation, as well as possible accidental isolation of two nearby colonies each having a single population of rearranged chromosomes. These eight clones displaying delayed chromosomal instability had a wide range of cytogenetic phenotypes (Table 3). For example, in clones B-4 and C-6, 91% of metaphase spreads were normal, with the remaining 9% distributed among five unique populations with rearrangements in chromosomes 4, 13, or 16. In clone C-32, only 60.5% of metaphase spreads were normal (shown in Fig. 5a), and in the remainder there were seven unique populations with different rearrangements such as insertion and reciprocal and nonreciprocal translocations; representative examples are shown in Fig. 5b to f. The chromosome aberrations observed in these chromosomally unstable clones are similar to those seen previously in GM10115 cells (Fig. 2, top right and bottom panels, and references 11 and 28).
TABLE 3.
Metaphase populations with rearranged chromosome 4, 13, or 16 in 10-Gy-irradiated unstable colony isolates
| Clone | % of normal metaphasesa | No. of populations with rearrangementsb |
|---|---|---|
| B-2 | 64.0 | 8 |
| B-4 | 91.0 | 5 |
| C-6 | 91.0 | 5 |
| C-17 | 14.0 | 4 |
| C-18 | 57.0 | 4 |
| C-32 | 60.5 | 7 |
| C-41 | 33.0 | 3 |
| C-89 | 87.0 | 6 |
Percentage of metaphase cells showing no rearrangements of chromosome 4, 13, and 16. Two hundred metaphase cells from each clone were analyzed.
Number of metaphase spreads showing unique rearrangements of chromosome 4, 13, or 16.
FIG. 5.
Representative examples of chromosomal instability on metaphases from an unstable clone, C-32, surviving 10 Gy of X-radiation. Metaphases were hybridized with labeled whole-chromosome probes specific for chromosome 4 (biotinylated and digoxigenin labeled, yellow), chromosome 13 (digoxigenin labeled, red), or chromosome 16 (biotinylated labeled, green). (a) Subpopulation with normal karyotype, containing two copies of chromosome 4, 13, and 16. (b to f) Different subpopulations of metaphase chromosomes showing different kinds of rearrangements (arrowhead).
As delayed HR and delayed chromosomal instability are both induced in ∼8% of colonies that survive 10-Gy doses, we examined six RKO36 subclones that survived 10-Gy radiation and displayed chromosomal instability to determine the relationship between these two endpoints. We found that GFP+/− colonies were no more frequent than in unirradiated RKO36 cells. Similarly, 12 radiation-induced GFP+/− colonies were expanded and assayed by FISH and none displayed chromosomal instability (data not shown).
The frequency of delayed hyperrecombination was very similar to the frequency of radiation-induced chromosomal instability that has traditionally been observed in GM10115 cells, i.e., ∼3% of clones surviving per gray of radiation were chromosomally unstable (20). Analysis of a random sample of GFP+/− colonies from each of the three independent GM10115 transfectants clearly demonstrated no relationship between genomic instability, as measured by our HR reporter construct, and chromosomal instability (Table 4).
TABLE 4.
Chromosomal instability in transfected GM10115 cells showing GFP mosaicisma
| Transfectant clone | X-ray dose (Gy) | GFP expression | No. of subclones analyzed | No. of stable subclones | No. of unstable subclonesb |
|---|---|---|---|---|---|
| TG9 | 0 | +/− | 7 | 7 | 0 |
| 1.0 | +/− | 12 | 12 | 0 | |
| 2.5 | +/− | 29 | 29 | 0 | |
| GH1 | 0 | +/− | 27 | 27 | 0 |
| 1.0 | +/− | 36 | 36 | 0 | |
| 2.5 | +/− | 47 | 45 | 2 | |
| GH24 | 0 | +/− | 12 | 12 | 0 |
| 1.0 | +/− | 43 | 43 | 0 | |
| 2.5 | +/− | 55 | 55 | 0 |
Two hundred metaphase cells from each clone were analyzed.
As defined by at least three unstable arrangements per 200 counted in each subclone.
Delayed HR is not associated with delayed reproductive death or delayed chromosomal instability.
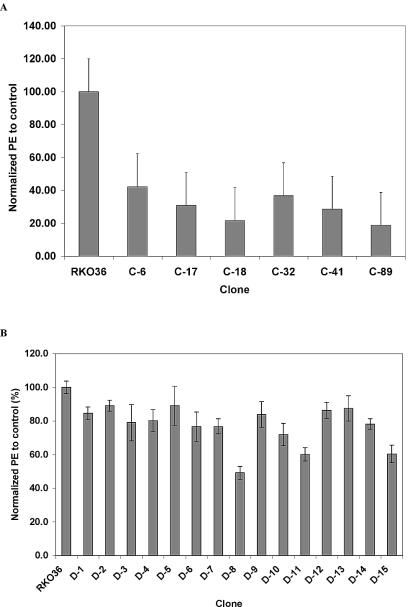
Radiation induces delayed reproductive cell death, reflected as reduced PE (7, 8), and this phenotype has been associated with delayed chromosomal instability (23, 28). We measured PEs of six subclones of RKO36 that survived a 10-Gy radiation dose and exhibited chromosomal instability. Each of these six subclones had markedly reduced PE compared to that of parental RKO36 cells (Fig. 6A). In contrast, among 12 radiation-induced GFP+/− colonies of RKO36, only three clones, D-8, D-11, and D-15, showed a reduced PE, while the others were indistinguishable from parental cells (Fig. 6B).
FIG. 6.
(A) PE of parental RKO36 cells and chromosomally unstable clones. (B) PE of parental RKO36 cells and clones expanded from nonirradiated (D-1, D-2, and D-3) and radiation-induced GFP+/− (D-4 to ∼D-15) colonies. These data have been normalized to 100% for control. Error bars, standard deviations based on five determinations.
A similar result was observed with the GM10115 clones. Analysis of 50 radiation-induced GFP+/− colonies chosen at random showed a PE that was not significantly different from that of the nonirradiated control clones (data not shown). In contrast, 14 of 20 clones (70%) showing chromosomal instability exhibited a PE that was significantly lower than that observed in the nonirradiated controls. This observation is consistent with but slightly lower than (81 versus 70%) that which has been historically observed in GM10115 cells (22, 28).
Thus, while delayed chromosomal instability and delayed reproductive cell death are strongly linked, delayed HR does not correlate with either of these phenotypes. This observation is consistent with our previous studies in GM10115 human-hamster hybrid cells indicating differential manifestation of radiation-induced genomic instability in clones surviving irradiation (23).
DISCUSSION
Radiation directly induces mutations, chromosome aberrations, and recombination (2, 6, 10, 18), reflecting repair or misrepair of DNA damage within or near the locus under study. The delayed effects of radiation, on the other hand, probably reflect dysregulation of genome-stabilizing factors and would therefore be expected to have more global effects. Prior studies of radiation-induced delayed genome instability in mammalian cells have focused on genotoxic endpoints such as delayed reproductive cell death, mutation, and chromosomal instability. Fabre and Roman (13) showed that irradiated haploid yeast cells could induce HR competence when mated with an unirradiated diploid, providing the first evidence for radiation-induced delayed HR, and a recent study demonstrated HR induction in the progeny of irradiated yeast (3). Our study provides the first direct evidence of delayed HR in irradiated human cells. Genomic instability, in any form, is likely to be important in tumor initiation and/or progression (4, 9, 17), thus it is important to understand the various mechanisms by which instability arises and is perpetuated.
A key finding in the present study is that, with the exception of GM10115 clone TG9, delayed HR is induced at doses of radiation (1 Gy) that produce relatively little cytotoxicity (Tables 1 and 2). This was the first hint that radiation-induced delayed HR and chromosomal instability arise by distinct mechanisms. Additional support for this idea comes from the observation that cells displaying delayed HR are not chromosomally unstable, and vice versa. Moreover, chromosomal instability is typically associated with delayed reproductive cell death (this study and references 23 and 28), but almost all of the clones displaying delayed HR show normal PE (Fig. 6). This result also differentiates delayed HR in human cells from that seen in yeast, as delayed HR in irradiated yeast is associated with reduced PE (3). There is evidence that DNA is the target for at least a fraction of delayed chromosomal instability (16, 20). However, because delayed chromosomal instability can be induced at high frequencies (up to 8% of cells surviving 10-Gy X-rays in RKO cells [Table 3] and as high as 33 to 50% in GM10115 cells [20, 24, 28]), it has been argued that the target is quite large, perhaps the entire cell, and that for this reason, it may reflect an epigenetic or nontargeted effect (38, 42). We note that delayed HR is induced at high frequencies in both cell lines. However, in general, chromosomal instability typically does not show classical dose dependence (39), similar to that observed for delayed HR (Tables 1 and 2). Hence, the target for delayed HR is also likely to be large and may reflect epigenetic and/or nontargeted changes.
Prior studies have shown that most cell types are susceptible to radiation-induced chromosomal instability, including normal human lymphocytes and human bone marrow cells, plus a host of human and rodent cell lines in vitro (reviewed in reference 32). Radiation-induced genomic instability has also been demonstrated in vivo, but because the genetic background can modulate induced instability (36, 41), the in vivo literature is more confusing (reviewed in reference 33). Our prior studies of radiation-induced chromosomal instability utilized the GM10115 cell line with a single copy of human chromosome 4 and mutant p53. As an in vitro model, cell lines derived from human RKO cells are superior for mechanistic studies. In particular, the GFP-based assay in RKO36 provides a rapid and sensitive assay for genome instability, with both HR and mutation and deletion events scored by direct visual inspection of colonies within 2 weeks of irradiation. This contrasts with 5 to 6 weeks of postirradiation expansion of individual colonies (to obtain sufficient numbers of cells) and the far more labor-intensive cytogenetic analysis required to analyze chromosomal instability. Unfortunately, we were unable to identify a comparable cell line in GM10115 cells. Nevertheless, it is noteworthy that in both RKO and GM10115 cells, the frequency of induced instability as measured by either mixed GFP+/− colonies or cytogenetic analysis was very similar, even though they measure different endpoints.
RKO has wild-type p53 and displays normal radiation responses, including p53 stabilization and p21 induction. The radiation-induced chromosomal instability and delayed HR observed in irradiated p53+ RKO36 cells are consistent with prior studies showing that delayed instability is not restricted to p53-defective cells (15, 29).
Our data indicate that ionizing radiation can disrupt one or more systems that regulate HR, yet this effect is not necessarily associated with significantly reduced cell viability. These results are important for several reasons. First, genome instability associated with hyperrecombination is, in effect, a mutator phenotype, as it increases the probability of uncovering recessive mutations via LOH. This is analogous to mutator phenotypes associated with defects in mismatch repair. As such, hyperrecombination may facilitate the accumulation of mutations important for tumor initiation and/or progression. Second, hyperrecombination can be induced by doses of radiation that cause little cytotoxicity, and this increases the fraction of surviving cells at risk. Third, because delayed hyperrecombination is not associated with reduced cell viability, as with delayed chromosomal instability, the former might represent a less severe instability phenotype, and such cells are hence more likely to survive and accumulate mutations necessary for immortalization and cellular transformation. In this regard, hyperrecombination may be an important determinant of secondary tumors in patients treated with radiation. Finally, because HR is important for the repair of DNA double-strand breaks, cells with a hyperrecombination phenotype may display enhanced resistance to radiation or other agents that induce strand breakage (19, 34). Thus, tumor cells with a hyperrecombination phenotype may have enhanced resistance to killing by ionizing radiation or chemotherapeutic agents, and such tumors may thus be more difficult to treat.
There has been much speculation regarding the mechanism(s) of radiation-induced genomic instability. A number of recent reports suggest a role for nontargeted “bystander”-like responses both in vitro (1, 26, 27, 31) and in vivo (5, 43). Whatever the mechanism, our results showing that radiation can elicit delayed responses in the progeny of irradiated cells have important implications for radiation risk assessment and support current perceptions of detrimental health risks associated with exposure to low-dose ionizing radiation.
Acknowledgments
We thank all the members of the Radiation Oncology Research Laboratory for their critical reading of the manuscript.
This work was supported by the Biological and Environmental Research Program (BER), U.S. Department of Energy, grant no. DE-FG02-01-ER63230, and National Institutes of Health Awards CA73924 and CA83872 to W.F.M and CA77693 to J.A.N.
REFERENCES
- 1.Barcellos-Hoff, M. H., and A. L. Brooks. 2001. Extracellular signaling through the microenvironment: a hypothesis relating carcinogenesis, bystander effects, and genomic instability. Radiat. Res. 156:618-627. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin, M. B., and J. B. Little. 1992. X rays induce interallelic homologous recombination at the human thymidine kinase gene. Mol. Cell. Biol. 12:2730-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, R. J., and R. H. Schiestl. 2001. Persistent genomic instability in the yeast Saccharomyces cerevisiae induced by ionizing radiation and DNA-damaging agents. Radiat. Res. 155:768-777. [DOI] [PubMed] [Google Scholar]
- 4.Cahill, D. P., K. W. Kinzler, B. Vogelstein, and C. Lengauer. 1999. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 9:M57-M60. [PubMed] [Google Scholar]
- 5.Camphausen, K., M. A. Moses, C. Menard, M. Sproull, W. D. Beecken, J. Folkman, and M. S. O'Reilly. 2003. Radiation abscopal antitumor effect is mediated through p53. Cancer Res. 63:1990-1993. [PubMed] [Google Scholar]
- 6.Cao, J., S. E. DePrimo, and J. R. Stringer. 1997. Cell cycle dependence of radiation-induced homologous recombination in cultured monkey cells. Mutat. Res. 374:233-243. [DOI] [PubMed] [Google Scholar]
- 7.Chang, W. P., and J. B. Little. 1991. Delayed reproductive cell death in X-irradiated Chinese hamster ovary cells. Int. J. Radiat. Biol. 60:483-496. [DOI] [PubMed] [Google Scholar]
- 8.Chang, W. P., and J. B. Little. 1992. Persistently elevated frequency of spontaneous mutations in progeny of CHO clones surviving X-irradiation: association with delayed reproductive death phenotype. Mutat. Res. 270:191-199. [DOI] [PubMed] [Google Scholar]
- 9.Coleman, W. B., and G. J. Tsongalis. 1999. The role of genomic instability in human carcinogenesis. Anticancer Res. 19:4645-4664. [PubMed] [Google Scholar]
- 10.Cornforth, M. N. 1998. Radiation-induced damage and the formation of chromosomal aberrations, p. 559-585. In J. A. Nickoloff and M. F. Hoekstra (ed.), DNA damage and repair, vol. II. DNA repair in higher eukaryotes. Humana Press, Totowa, N.J. [Google Scholar]
- 11.Day, J. P., C. L. Limoli, and W. F. Morgan. 1998. Recombination involving interstitial telomere repeat-like sequences promotes chromosomal instability in Chinese hamster cells. Carcinogenesis 19:259-265. [DOI] [PubMed] [Google Scholar]
- 12.Deng, W. P., and J. A. Nickoloff. 1992. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200:81-88. [DOI] [PubMed] [Google Scholar]
- 13.Fabre, F., and H. Roman. 1977. Genetic evidence for inducibility of recombination competence in yeast. Proc. Natl. Acad. Sci. USA 74:1667-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosovsky, A. J., K. K. Parks, C. R. Giver, and S. L. Nelson. 1996. Clonal analysis of delayed karyotypic abnormalities and gene mutations in radiation-induced genetic instability. Mol. Cell. Biol. 16:6252-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadhim, M. A., C. A. Walker, M. A. Plumb, and E. G. Wright. 1996. No association between p53 status and alpha-particle-induced chromosomal instability in human lymphoblastoid cells. Int. J. Radiat. Biol. 69:167-174. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan, M. I., and W. F. Morgan. 1998. The nucleus is the target for radiation-induced chromosomal instability. Radiat. Res. 150:382-390. [PubMed] [Google Scholar]
- 17.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 18.Liber, H. L., and E. N. Phillips. 1998. Interrelationships between radiation-induced mutations and modifications in gene expression linked to cancer. Crit. Rev. Eukaryot. Gene Expr. 8:257-276. [DOI] [PubMed] [Google Scholar]
- 19.Limoli, C. L., J. J. Corcoran, R. Jordan, W. F. Morgan, and J. L. Schwartz. 2001. A role for chromosomal instability in the development of and selection for radioresistant cell variants. Br. J. Cancer 84:489-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limoli, C. L., J. J. Corcoran, J. R. Milligan, J. F. Ward, and W. F. Morgan. 1999. Critical target and dose and dose-rate responses for the induction of chromosomal instability by ionizing radiation. Radiat. Res. 151:677-685. [PubMed] [Google Scholar]
- 21.Limoli, C. L., A. Hartmann, L. Shephard, C. R. Yang, D. A. Boothman, J. Bartholomew, and W. F. Morgan. 1998. Apoptosis, reproductive failure, and oxidative stress in Chinese hamster ovary cells with compromised genomic integrity. Cancer Res. 58:3712-3718. [PubMed] [Google Scholar]
- 22.Limoli, C. L., M. I. Kaplan, J. Corcoran, M. Meyers, D. A. Boothman, and W. F. Morgan. 1997. Chromosomal instability and its relationship to other end points of genomic instability. Cancer Res. 57:5557-5563. [PubMed] [Google Scholar]
- 23.Limoli, C. L., M. I. Kaplan, J. W. Phillips, G. M. Adair, and W. F. Morgan. 1997. Differential induction of chromosomal instability by DNA strand-breaking agents. Cancer Res. 57:4048-4056. [PubMed] [Google Scholar]
- 24.Limoli, C. L., B. Ponnaiya, J. J. Corcoran, E. Giedzinski, M. I. Kaplan, A. Hartmann, and W. F. Morgan. 2000. Genomic instability induced by high and low LET ionizing radiation. Adv. Space Res. 25:2107-2117. [DOI] [PubMed] [Google Scholar]
- 25.Little, J. B., H. Nagasawa, T. Pfenning, and H. Vetrovs. 1997. Radiation-induced genomic instability: delayed mutagenic and cytogenetic effects of X rays and alpha particles. Radiat. Res. 148:299-307. [PubMed] [Google Scholar]
- 26.Lorimore, S. A., M. A. Kadhim, D. A. Pocock, D. Papworth, D. L. Stevens, D. T. Goodhead, and E. G. Wright. 1998. Chromosomal instability in the descendants of unirradiated surviving cells after alpha-particle irradiation. Proc. Natl. Acad. Sci. USA 95:5730-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorimore, S. A., and E. G. Wright. 2003. Radiation-induced genomic instability and bystander effects: related inflammatory-type responses to radiation-induced stress and injury? A review. Int. J. Radiat. Biol. 79:15-25. [PubMed] [Google Scholar]
- 28.Marder, B. A., and W. F. Morgan. 1993. Delayed chromosomal instability induced by DNA damage. Mol. Cell. Biol. 13:6667-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIlrath, J., S. A. Lorimore, P. J. Coates, and E. G. Wright. 2003. Radiation-induced genomic instability in immortalized haemopoietic stem cells. Int. J. Radiat. Biol. 79:27-34. [PubMed] [Google Scholar]
- 30.Mitchel, R. E., J. S. Jackson, D. P. Morrison, and S. M. Carlisle. 2003. Low doses of radiation increase the latency of spontaneous lymphomas and spinal osteosarcomas in cancer-prone, radiation-sensitive Trp53 heterozygous mice. Radiat. Res. 159:320-327. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, W. F. 2003. Is there a common mechanism underlying genomic instability, bystander effects and other nontargeted effects of exposure to ionizing radiation? Oncogene 22:7094-7099. [DOI] [PubMed] [Google Scholar]
- 32.Morgan, W. F. 2003. Non-targeted and delayed effects of exposure to ionizing radiation. I. Radiation-induced genomic instability and bystander effects in vitro. Radiat. Res. 159:567-580. [DOI] [PubMed] [Google Scholar]
- 33.Morgan, W. F. 2003. Non-targeted and delayed effects of exposure to ionizing radiation. II. Radiation-induced genomic instability and bystander effects in vivo, clastogenic factors and transgenerational effects. Radiat. Res. 159:581-596. [DOI] [PubMed] [Google Scholar]
- 34.Morgan, W. F., and J. P. Murnane. 1995. A role for genomic instability in cellular radioresistance? Cancer Metastasis Rev. 14:49-58. [DOI] [PubMed] [Google Scholar]
- 35.Nickoloff, J. A. 2002. Recombination: mechanisms and roles in tumorigenesis, p. 49-59. In J. R. Bertino (ed.), Encyclopedia of cancer, 2nd ed., vol. 4. Elsevier Science, San Diego, Calif. [Google Scholar]
- 36.Ponnaiya, B., M. N. Cornforth, and R. L. Ullrich. 1997. Radiation-induced chromosomal instability in BALB/c and C57BL/6 mice: the difference is as clear as black and white. Radiat. Res. 147:121-125. [PubMed] [Google Scholar]
- 37.Redpath, J. L., S. C. Short, M. Woodcock, and P. J. Johnston. 2003. Low-dose reduction in transformation frequency compared to unirradiated controls: the role of hyper-radiosensitivity to cell death. Radiat. Res. 159:433-436. [DOI] [PubMed] [Google Scholar]
- 38.Seymour, C. B., and C. Mothersill. 1997. Delayed expression of lethal mutations and genomic instability in the progeny of human epithelial cells that survived in a bystander-killing environment. Radiat. Oncol. Investig. 5:106-110. [DOI] [PubMed] [Google Scholar]
- 39.Smith, L. E., S. Nagar, G. J. Kim, and W. F. Morgan. 2003. Radiation-induced genomic instability: radiation quality and dose response. Health Phys. 85:23-29. [DOI] [PubMed] [Google Scholar]
- 40.Ward, J. F. 1998. Nature of lesions formed by ionizing radiation, p. 65-84. In J. A. Nickoloff and M. F. Hoekstra (ed.), DNA damage and repair, vol. II. DNA repair in higher eukaryotes. Humana Press, Totowa, N.J. [Google Scholar]
- 41.Watson, G. E., S. A. Lorimore, S. M. Clutton, M. A. Kadhim, and E. G. Wright. 1997. Genetic factors influencing alpha-particle-induced chromosomal instability. Int. J. Radiat. Biol. 71:497-503. [DOI] [PubMed] [Google Scholar]
- 42.Watson, G. E., S. A. Lorimore, D. A. Macdonald, and E. G. Wright. 2000. Chromosomal instability in unirradiated cells induced in vivo by a bystander effect of ionizing radiation. Cancer Res. 60:5608-5611. [PubMed] [Google Scholar]
- 43.Watson, G. E., D. A. Pocock, D. Papworth, S. A. Lorimore, and E. G. Wright. 2001. In vivo chromosomal instability and transmissible aberrations in the progeny of haemopoietic stem cells induced by high- and low-LET radiations. Int. J. Radiat. Biol. 77:409-417. [DOI] [PubMed] [Google Scholar]