Abstract
Objective
Preeclampsia (PE) has been sub-divided into early- and late-onset phenotypes. The pathogenesis of these two phenotypes has not been elucidated. To gain insight into the mechanisms of disease, the transcriptional profiles of whole blood from women with early- and late-onset PE were examined.
Methods
A cross-sectional study was conducted to include women with: 1) early-onset PE (diagnosed prior to 34 weeks, n=25); 2) late-onset PE (after 34 weeks, n=47); and 3) uncomplicated pregnancy (n=61). Microarray analysis of mRNA expression in peripheral whole blood was undertaken using Affymetrix microarrays. Differential gene expression was evaluated using a moderated t-test (false discovery rate <0.1 and fold change >1.5), adjusting for maternal WBC count and gestational age. Validation by real-time qRT-PCR was performed in a larger sample size [early PE (n=31), late PE (n=72) and controls (n=99)] in all differentially expressed genes. Gene Ontology analysis and pathway analysis were performed.
Results
1) 43 and 28 genes were differentially expressed in early- and late-onset PE compared to the control group respectively; 2) qRT-PCR confirmed the microarray results for early and late-onset PE in 77% (33/43) and 71% (20/28) of genes, respectively; 3) 20 genes that are involved in coagulation (SERPINI2), immune regulation (VSIG4, CD24), developmental process (H19) and inflammation (S100A10) were differentially expressed in early-onset PE alone. In contrast, only seven genes that encoded proteins involved in innate immunity (LTF, ELANE) and cell-to-cell recognition in the nervous system (CNTNAP3) were differentially expressed in late-onset PE alone. Thirteen genes that encode proteins involved in host defense (DEFA4, BPI, CTSG, LCN2), tight junctions in blood-brain barrier (EMP1) and liver regeneration (ECT2) were differentially expressed in both early- and late-onset PE.
Conclusion
Early- and late-onset PE are characterized by a common signature in the transcriptional profile of whole blood. A small set of genes were differentially regulated in early- and late-onset PE. Future studies of the biological function, expression timetable and protein expression of these genes may provide insight into the pathophysiology of PE.
Keywords: Gene expression, transcriptomic, pregnancy, microarray, PAX gene, H19, white blood cell count, Affymetrix, PCR
Introduction
Preeclampsia (PE), one of the great obstetrical syndromes [1-53], is a leading cause of maternal death and perinatal morbidity and mortality [54-60]. Women with a previous history of PE are at increased risk of death as a result of cardiovascular disease [61-65]
The mechanisms of disease responsible for PE remain elusive. High dimensional biology techniques including genomics [66-69], transcriptomics [70-73], proteomics [74-85], metabolomics [86-90] and others -omic sciences are being utilized to gain insight into the pathophysiology of disease state and the identification of biomarkers in many disciplines including obstetrics [91-98].
Transcriptome analysis has been used to classify lymphomas [99,100] and cancer [101-113]. Several investigators have used this approach to examine the placental transcriptome in PE [114-119]. The gene expression profiles of peripheral blood have shown promising results in elucidating the mechanisms of other disorders [120-127]. This approach may be helpful in gaining understanding of the mechanism of disease in early- and late-onset PE.
The objective of this study was to characterize the transcriptional profiles of whole blood from women with early- and late-onset PE.
Patients and Methods
Study Design
A prospective cross-sectional study was conducted and included women in the following groups; 1) early-onset PE (diagnosed prior to 34 weeks, n=25); 2) late-onset PE (after 34 weeks, n=47); and 3) uncomplicated pregnancy (n=61) (for microarray experiment). Exclusion criteria were the following: 1) chronic hypertension; 2) known major fetal or chromosomal anomaly; 3) multiple gestations; and 4) received medications other than iron, stool softener, or vitamins prior to venipuncture. All women were enrolled at Hutzel Women's Hospital, Detroit, MI and followed until delivery.
Clinical definition
PE was defined as hypertension (systolic blood pressure ≥ 140 mmHg or diastolic blood pressure ≥ 90 mmHg on at least two occasions, 4 hours to 1 week apart) and proteinuria [61,128]. Proteinuria was defined as protein of greater than 300 mg in a 24-hour urine specimen. If a 24-hour urine collection was not available, the diagnosis of PE required ≥1+ proteinuria on two separate occasions 4 hours to 1 week apart or ≥2+ proteinuria on a urine dipstick. Early- and late-onset PE was defined as a diagnosis of PE at ≤ 34 and >34 weeks of gestation respectively [129,130]. Pregnancy was considered uncomplicated if women had no major medical, obstetrical or surgical complications, and delivered a normal term (≥ 37 weeks) infant whose birthweight was appropriate for gestational age (10th-90th percentile) [131].
All patients provided written informed consent for the collection and use of samples for research purposes under the protocols approved by the Institutional Review Boards of Wayne State University and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services (NICHD/NIH/DHHS).
Sample Collection and Preparation
Venipunctures were performed and 2.5 milliliters (ml) of whole blood was collected into PAXgene Blood RNA tubes (PreAnalytiX GmbH, distributed by Becton, Dickinson and Company, New Jersey, NY), which contain a proprietary cell lysis and RNA stabilizing solution. PAXgene Blood RNA tubes were kept at room temperature for 24 hours to ensure complete cellular lysis, then frozen at -70 degrees C until further processing. Blood samples were also collected to determine WBC count (WBC). For the patients with PE, maternal whole blood was collected at the time of diagnosis. Samples for the control group were collected at the prenatal clinic where patients had regular prenatal care, at the labor reception center where patients visited for minor complaints (eg. headache, asymptomatic short cervix, itching, pelvic pressure, minor accident, etc.) or at the labor-delivery unit for scheduled cesarean section at term. All patients were followed until delivery.
RNA Isolation
Intracellular total RNA was isolated from whole blood using the PAXgene Blood RNA Kit (Qiagen, Valencia, CA). Blood lysates were reduced to pellets by centrifugation, washed, and re-suspended in buffer. Proteins were removed by proteinase K digestion and cellular debris removed by centrifugation through a PAXgene Shredder spin column. RNA was semi-precipitated with ethanol and selectively bound to the silica membrane of a PAXgene spin column. The membrane was treated with DNase I to remove any residual DNA, washed, and the purified total RNA was eluted in nuclease-free water. Puified total RNA was quantified by UV spectrophotometry using the DropSense96 Microplate Spectrophotometer (Trinean, Micronic North America LLC, McMurray PA) and the purity assessed based on the A260/A280 and A260/A230 ratios. An aliquot of the RNA was assessed using the RNA 6000 Nano Assay for the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). The electrophoretogram, RNA Integrity Number (RIN), and the ratio of the 28S:18S RNA bands were examined to determine overall quality of the RNA.
Whole blood transcriptome
The transcriptome of peripheral blood samples was profiled using Affymetrix GeneChip HG-U133 PLUS 2.0 arrays (Affymetrix Inc., Santa Clara, CA). Briefly, isolated RNA was amplified using the Ovation RNA Amplication System V2 (NuGEN Technologies, Inc., San Carlos, CA). Complementary DNA (cDNA) was synthesized using the Ovation buffer mix, first strand enzyme mix, and first strand primer mix with 5μL (∼20 ng) of total RNA in specified thermal cycler protocols according to the manufacturer's instructions. Amplification and purification of the generated cDNA were performed by combining SPIA Buffer Mix, Enzyme Mix, and water with the products of the second strand cDNA synthesis reactions in pre-specified thermal cycler programs. Optical density of the amplified cDNA product was obtained to demonstrate product yield and verify purity. Fragmentation and labeling was done using the FL-Ovation cDNA Biotin Module V2 (NuGEN Technologies, Inc. San Carlos, CA). In the primary step, a combined chemical and enzymatic fragmentation process was used to produce cDNA products in the 50 to 100 bases range. Fragmented cDNA products were then biotin-labeled using the Encore Biotin Module (NuGEN Technologies, Inc). All reactions were carried out according to the manufacturer's protocols. Amplified, fragmented and biotin-labeled cDNAs were used for hybridization cocktail assembly, then hybridized to the Affymetrix GeneChip HG-U133 PLUS 2.0 arrays according to the Affymetrix standard protocol.
Real-time quantitative polymerase chain reaction (qRT-PCR)
Validation of mRNA levels of selected genes was performed on a larger group of cases and controls using real-time qRT-PCR. The catalog number for each RNA primer is presented in supplementary Table I. Total RNA was isolated as described above. Biomark™ System (Fluidigm, San Francisco, CA) was used for high-throughput PCR. Briefly, Invitrogen Superscript III 1st Strand Kit was used to generate cDNA. Pre-amplification procedures included combining 1.25uL cDNA with 2.5uL TaqMan PreAMP Mastermix and 1.25 uL pooled assay mix. The reaction was performed with a thermal cycler for one cycle at 95 °C for 10 minutes and 14 cycles at 95°C for 15 seconds and 60°C for 4 minutes. After cycling, the reaction was diluted 1:5 by double-distilled water to a final volume of 25μl. Fluidigm 96.96 Dynamic Array chip was used to perform the next step qRT-PCR assays. The 96.96 array chip was primed in an Integrated Fluidic Circuit (IFC) controller with control fluid. After priming, 2.5μl 20× TaqMan gene expression assays (Applied Biosystems) were mixed with 2.5 μl 2× assay loading reagent (Fluidigm) and loaded into the assay inlet on the 96.96 array chip. 2.25 μl preamplified cDNA was mixed with 2.5μl TaqMan Universal PCR master mix (Applied Biosystems) and 0.25 μl 20× sample loading reagent (Fluidigm) and loaded into the sample inlet on the chip. The chip was returned to the IFC controller for loading. After loading, the chip was placed in the Biomark System to run the reactions. The cycle threshold (Ct) value of each reaction was obtained with the Fluidigm RT-PCR analysis software.
Analysis for microarray and qRT-PCR data
For microarray analysis, 25 patients with early-onset PE and 47 patients with late-onset PE were compared with 61 controls. To confirm the results from microarray, all differentially expressed genes which met the criteria for significances in microarray experiment were tested with qRT-PCR assays in a subset of the original sample set (early-onset PE n =25; late-onset PE n=44 and controls n=61) and in a new set of samples (early-onset PE n = 6; late-onset PE n=28 and controls n= 38). The selection criteria for the new specimens were the same as those used to select patients for the microarray analysis. Four patients in the late-onset PE group did not have enough RNA available after the microarray experiment and were not included in the qRT-PCR experiments.
Statistical Analysis
Gene expression data pre-processing, that included background correction, normalization and summarization, was performed with the Robust Multi-array Average algorithm (RMA) [61] implemented in the affy package of Bioconductor [128,132]. Further analysis was performed only on the probe sets that were called “present” (expressed) in at least half of the number of samples in the smaller groups that were compared. The Affymetrix MAS 5.0 detection call method implemented in the affy package was used for determining the “present” calls of a given probe set in a given sample.
Comparisons between early-onset PE and control samples as well as between late-onset PE and controls were performed by fitting a linear model on the gene expression levels using disease status, gestational age and WBC count of patients as variables in the model. The significance of the disease status coefficient was tested via a moderated t-statistic. The resulting p-values were adjusted to account for multiple testing, using the False Discovery Rate (FDR) algorithm. Genes with adjusted p-values (q-values) <0.1 and fold change >1.5 were considered significant.
Gene Ontology analysis was conducted to determine the “biological processes”, which are enriched in the list of differentially expressed genes as described elsewhere [120] and implemented in the GO stats package [133,134]. Pathway analysis was performed on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database using an overrepresentation analysis as well as the Signaling Pathway Impact Analysis [99].
The differential mRNA expression between early-onset PE and control samples and between late-onset PE and controls were compared using qRT-PCR data. The -ΔCt values, surrogates for log gene expression, were fitted using the same linear model described for the microarray analysis. A nominal p-value <0.05 for the disease status coefficient in the linear model was considered as significant (validated).
The Mann-Whitney U and Chi-square tests were used to compare the differences in demographics and clinical characteristics between PE and controls. SPSS (version 12.0; SPSS Inc, Chicago, IL) was used for the analysis of demographic and clinical characteristic data. A probability value of <0.05 was considered significant.
Results
Table 1 displays the demographic and clinical characteristics of the study population for both microarray and qRT-PCR procedures. Patients with early-onset PE had a higher median WBC count than those in the control groups (p=0.002). Among patients with uncomplicated pregnancy, there were 56 genes (70 probe sets) whose expression levels were correlated with WBC count after adjustment for gestational age (supplementary Table II) and only 2 genes [FPR (formyl peptide receptor)-3 and AGPAT (1-acylglycerol-3-phosphate O-acyltransferase)-3] whose expression levels were correlated with gestational age after adjustment for WBC count.
Table 1. Demographics and clinical characteristics of the study populations.
| Microarray | qRT-PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control Patients (n=61) |
Early-onset Preeclampsia (n=25) |
p-value | Late-onset Preeclampsia (n=47) |
p-value | Control Patients (n=99) |
Early-onset Preeclampsia (n=31) |
p-value | Late-onset Preeclampsia (n=72) |
p-value | |
| Maternal Age (years) | 24.1 (16-39) | 24.0 (16-37) | 0.38 | 21.7 (16-36) | 0.07 | 24.3 (16-39) | 22.7 (16-37) | 0.21 | 21.8 (16-36) | 0.01 |
| Nulliparity | 19 (31.1) | 14 (56.0) | 0.03 | 27 (57.4) | 0.006 | 29 (29.3) | 19 (61.3) | 0.001 | 41 (56.9) | <0.001 |
| African American | 56 (91.8) | 21 (84.0) | 0.28 | 41 (87.2) | 0.44 | 85 (85.9) | 25 (80.6) | 0.48 | 66 (91.7) | 0.24 |
| Tobacco Use | 4 (6.6) | 4 (16.0) | 0.17 | 9 (19.1) | 0.046 | 17 (17.2) | 5 (16.1) | 0.89 | 10 (13.9) | 0.56 |
| BMI (kg/m2) | 27.1 (16.6-47.7) | 24.6 (13.8-35.9) | 0.04 | 26.4 (17.7-48.3) | 0.69 | 26.2 (16.4-54.9) | 24.7 (13.8-36.0) | 0.22 | 27.9 (16.5-52.4) | 0.37 |
| Gestational Age at blood Draw (weeks) | 33.7 (20.0-40.1) | 31.1 (25.0-33.9) | 0.004 | 37.9 (34.3-41.4) | <0.001 | 34.7 (20.0-40.1) | 31.1 (24.7-34.0) | <0.001 | 38.1 (34.3-42.0) | <0.001 |
| WBC Cells/mm3 | 8.0 (4.2-13.8) | 9.7 (6.7-18.7) | 0.002 | 8.3 (4.8-15.1) | 0.19 | 8.8 (2.7-18.7) | 9.7 (3.8-18.7) | 0.04 | 8.5 (4.8-23.6) | 0.63 |
| Severe Preeclampsia | ---- | 22 (88.0) | ---- | 28 (59.6) | ---- | ---- | 26 (83.9) | ---- | 39 (54.2) | ---- |
| Gestational Age at Delivery (weeks) | 39.1 (37.0-41.7) | 31.7 (26.1-34.0) | <0.001 | 38.0 (35.0-41.4) | <0.001 | 39.1 (37.0-42.1) | 31.7 (24.9-35.4) | <0.001 | 38.3 (35.0-42.1) | <0.001 |
| Birthweight (grams) | 3390 (2575-4010) | 1235 (550-1910) | <0.001 | 2880 (1670-4180) | <0.001 | 3340 (2575-4010) | 1330 (474-2730) | <0.001 | 2968 (1670-4180) | <0.001 |
| Birthweight <10%ile | 0 | 11 (44.0) | <0.001 | 15 (31.9) | <0.001 | 0 | 14 (45.2) | <0.001 | 18 (25.0) | <0.001 |
Data are expressed as median (range).
Data are expressed as count (% of group)
BMI: Body mass index
WBC: White blood cell counts
Microarray Analysis
Early-onset PE vs. controls
Microarray analysis demonstrated that 49 probe sets (corresponding with 43 unique genes) were differentially expressed in maternal whole blood between early-onset PE and uncomplicated pregnancy after adjustment for WBC count and gestational age (Table 2; without adjustment, there were 31 genes differentially expressed). A total of 18 genes had higher expression and 25 genes had lower expression in the early-onset disease group than in the control group. A “volcano plot” shows the differential expression of all the annotated probe sets on the Affymetrix GeneChip HG-U133 PLUS 2.0 array with the log (base10) of the FDR-adjusted probability values (y-axis) plotted against the log (base 2) fold changes (x-axis) between the early-onset PE and control groups (Figure 1).
Table 2.
Differentially expressed genes between early-onset preeclampsia and controls based on microarray data.
| Entrez Gene | Symbol | Gene Name | Fold Change | Adjusted p-value |
|---|---|---|---|---|
| Higher Expression | ||||
| 2012 | EMP1 | epithelial membrane protein 1 | 2.33 | 0.0001 |
| 2354 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 2.06 | 0.0000 |
| 23243 | ANKRD28 | ankyrin repeat domain 28 | 1.72 | 0.0233 |
| 1894 | ECT2 | epithelial cell transforming sequence 2 oncogene | 1.72 | 0.0077 |
| 6281 | S100A10 | S100 calcium binding protein A10 | 1.69 | 0.0000 |
| 7252 | TSHB | thyroid stimulating hormone, beta | 1.68 | 0.0002 |
| 283120 | H19 | H19, imprinted maternally expressed transcript (non-protein coding) | 1.66 | 0.0011 |
| 283120 | H19 | H19, imprinted maternally expressed transcript (non-protein coding) | 1.66 | 0.0011 |
| 11326 | VSIG4 | V-set and immunoglobulin domain containing 4 | 1.63 | 0.0026 |
| 2359 | FPR3 | formyl peptide receptor 3 | 1.63 | 0.0139 |
| 84617 | TUBB6 | tubulin, beta 6 | 1.62 | 0.0106 |
| 2335 | FN1 | fibronectin 1 | 1.6 | 0.0074 |
| 1396 | CRIP1 | cysteine-rich protein 1 (intestinal) | 1.59 | 0.0000 |
| 2335 | FN1 | fibronectin 1 | 1.55 | 0.0204 |
| 388610 | TRNP1 | TMF1-regulated nuclear protein 1 | 1.55 | 0.0508 |
| 257177 | C1orf192 | chromosome 1 open reading frame 192 | 1.54 | 0.0008 |
| 79992 | C6orf59 | chromosome 6 open reading frame 59 | 1.53 | 0.0008 |
| 5276 | SERPINI2 | serpin peptidase inhibitor, clade I (pancpin), member 2 | 1.53 | 0.0077 |
| 4000 | LMNA | lamin A/C | 1.52 | 0.0011 |
| 114327 | EFHC1 | EF-hand domain (C-terminal) containing 1 | 1.52 | 0.0047 |
| 4000 | LMNA | lamin A/C | 1.51 | 0.0012 |
| Lower Expression | ||||
| 1669 | DEFA4 | defensin, alpha 4, corticostatin | 2.36 | 0.0110 |
| 4680 | CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | 2.1 | 0.0409 |
| 4680 | CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | 2.06 | 0.0334 |
| 671 | BPI | bactericidal/permeability-increasing protein | 2 | 0.0214 |
| 23254 | RP1-21O18.1 | kazrin | 1.97 | 0.0166 |
| 1511 | CTSG | cathepsin G | 1.93 | 0.0993 |
| 820 | CAMP | cathelicidin antimicrobial peptide | 1.93 | 0.0002 |
| 1088 | CEACAM8 | carcinoembryonic antigen-related cell adhesion molecule 8 | 1.9 | 0.0508 |
| 1232 | CCR3 | chemokine (C-C motif) receptor 3 | 1.84 | 0.0002 |
| 932 | MS4A3 | membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | 1.83 | 0.0962 |
| 23254 | RP1-21O18.1 | kazrin | 1.82 | 0.0051 |
| 6947 | TCN1 | transcobalamin I (vitamin B12 binding protein, R binder family) | 1.75 | 0.0120 |
| 7053 | TGM3 | transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase) | 1.7 | 0.0539 |
| 23462 | HEY1 | hairy/enhancer-of-split related with YRPW motif 1 | 1.66 | 0.0543 |
| 100133941 | CD24 | CD24 molecule | 1.63 | 0.0387 |
| 6518 | SLC2A5 | solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | 1.63 | 0.0614 |
| 4318 | MMP9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 1.62 | 0.0390 |
| 8993 | PGLYRP1 | peptidoglycan recognition protein 1 | 1.59 | 0.0065 |
| 2280 | FKBP1A | FK506 binding protein 1A, 12kDa | 1.59 | 0.0104 |
| 1053 | CEBPE | CCAAT/enhancer binding protein (C/EBP), epsilon | 1.59 | 0.0036 |
| 100133941 | CD24 | CD24 molecule | 1.59 | 0.0508 |
| 3934 | LCN2 | lipocalin 2 | 1.58 | 0.0984 |
| 5806 | PTX3 | pentraxin-related gene, rapidly induced by IL-1 beta | 1.57 | 0.0448 |
| 154664 | ABCA13 | ATP-binding cassette, sub-family A (ABC1), member 13 | 1.56 | 0.0855 |
| 116448 | OLIG1 | oligodendrocyte transcription factor 1 | 1.54 | 0.0166 |
| 762 | CA4 | carbonic anhydrase IV | 1.52 | 0.0398 |
| 8851 | CDK5R1 | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 1.51 | 0.0315 |
| 660 | BMX | BMX non-receptor tyrosine kinase | 1.5 | 0.0332 |
Figure 1.
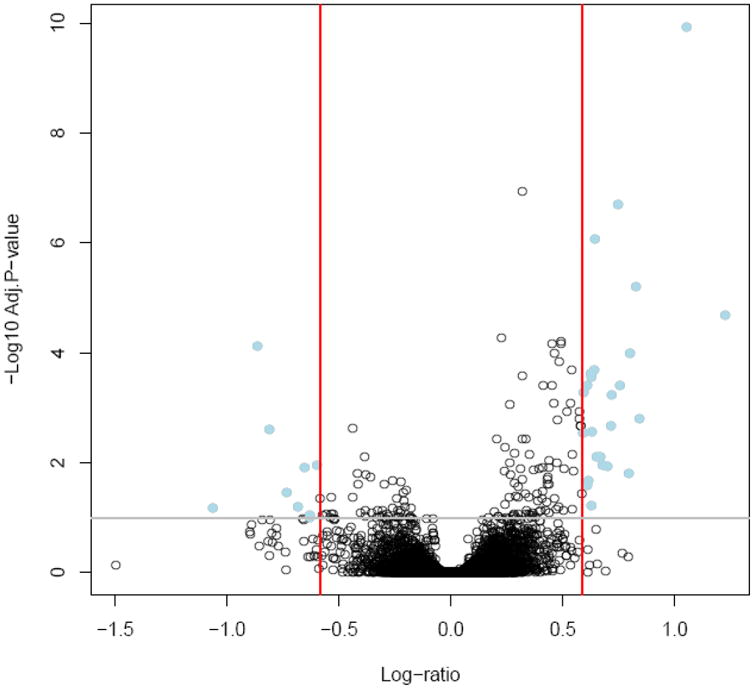
A “volcano plot” shows the differential expression of all the annotated probe sets between early-onset preeclampsia and control groups on the Affymetrix GeneChip HG-U133 PLUS 2.0 array with the log (base10) of the FDR-adjusted probability values (y-axis) plotted against the log (base 2) fold changes (x-axis).
Gene ontology analysis of the differentially expressed genes was performed and 41 biological processes were enriched (pFDR <0.05 and at least two significant genes per GO term); the top 20 significant processes (ranking based on odds ratio) are presented in Table 3 (see full list at supplementary Table 3) and include negative regulation of T cell activation, defense response to bacterium, acute inflammatory response and positive regulation of I-kappaB kinase/NF-kappaB cascade.
Table 3.
Top 20 biological processes (ranking by odds ratio) significantly enriched in comparison between early-onset preeclampsia and controls.
| GOBPID | Biological Process | Genes in Differentially Expressed List, n | Genes in GOBPID List | Odds Ratio | Adjusted p-value |
|---|---|---|---|---|---|
| GO:0031424 | keratinization | 2 | 8 | 101.47 | 0.0080 |
| GO:0030225 | macrophage differentiation | 2 | 12 | 60.86 | 0.0133 |
| GO:0018149 | peptide cross-linking | 2 | 16 | 43.45 | 0.0179 |
| GO:0042742 | defense response to bacterium | 4 | 39 | 39.11 | 0.0005 |
| GO:0032945 | negative regulation of mononuclear cell proliferation | 2 | 27 | 24.31 | 0.0192 |
| GO:0009913 | epidermal cell differentiation | 2 | 35 | 18.40 | 0.0241 |
| GO:0050868 | negative regulation of T cell activation | 2 | 37 | 17.35 | 0.0241 |
| GO:0030855 | epithelial cell differentiation | 4 | 83 | 16.23 | 0.0057 |
| GO:0002695 | negative regulation of leukocyte activation | 2 | 44 | 14.87 | 0.0305 |
| GO:0051707 | response to other organism | 8 | 227 | 13.15 | 0.0001 |
| GO:0042129 | regulation of T cell proliferation | 2 | 52 | 12.13 | 0.0329 |
| GO:0007398 | ectoderm development | 3 | 100 | 9.61 | 0.0233 |
| GO:0043627 | response to estrogen stimulus | 2 | 69 | 9.03 | 0.0386 |
| GO:0002526 | acute inflammatory response | 2 | 70 | 8.90 | 0.0386 |
| GO:0070663 | regulation of leukocyte proliferation | 2 | 71 | 8.77 | 0.0391 |
| GO:0007204 | elevation of cytosolic calcium ion concentration | 2 | 77 | 8.06 | 0.0405 |
| GO:0050865 | regulation of cell activation | 3 | 147 | 6.44 | 0.0329 |
| GO:0008624 | induction of apoptosis by extracellular signals | 2 | 97 | 6.35 | 0.0476 |
| GO:0045664 | regulation of neuron differentiation | 2 | 97 | 6.35 | 0.0476 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | 2 | 98 | 6.29 | 0.0477 |
Late-onset PE vs controls
When comparing the late-onset PE group and controls, 32 probe sets that represented 28 genes were differentially expressed in maternal whole blood after adjustment for WBC count and gestational age (Table 4; without adjustment, there were 22 genes differentially expressed). A total of six genes had higher and 22 genes had lower expression in women with late-onset PE compared to those with uncomplicated pregnancy. A “volcano plot” for this comparison is displayed in Figure 2. Thirty-one biological processes were enriched; the top 20 significant biological processes (ranking based on odds ratio) are presented in Table 5 (see full list at supplementary Table 4) and include defense response to bacterium, leukocyte cell-cell adhesion, negative regulation of immune system process and negative regulation of cell activation.
Table 4.
Differentially expressed genes between late-onset preeclampsia and controls based on microarray data.
| Entrez Gene | Symbol | Gene Name | Fold Change | Adjusted p-value |
|---|---|---|---|---|
| Higher Expression | ||||
| 728577 | CNTNAP3B | contactin associated protein-like 3B | 1.78 | 0.05735 |
| 2012 | EMP1 | epithelial membrane protein 1 | 1.63 | 0.02499 |
| 79937 | CNTNAP3 | contactin associated protein-like 3 | 1.63 | 0.09549 |
| 79937 | CNTNAP3 | contactin associated protein-like 3 | 1.61 | 0.03953 |
| 23243 | ANKRD28 | ankyrin repeat domain 28 | 1.57 | 0.00291 |
| 1894 | ECT2 | epithelial cell transforming sequence 2 oncogene | 1.53 | 0.02362 |
| 7252 | TSHB | thyroid stimulating hormone, beta | 1.5 | 0.00116 |
| Lower Expression | ||||
| 1669 | DEFA4 | defensin, alpha 4, corticostatin | 2.49 | 0.00116 |
| 1088 | CEACAM8 | carcinoembryonic antigen-related cell adhesion molecule 8 | 2.09 | 0.00388 |
| 4057 | LTF | lactotransferrin | 2.03 | 0.00099 |
| 671 | BPI | bactericidal/permeability-increasing protein | 2.01 | 0.00519 |
| 4317 | MMP8 | matrix metallopeptidase 8 (neutrophil collagenase) | 1.99 | 0.03667 |
| 4680 | CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | 1.99 | 0.02288 |
| 4317 | MMP8 | matrix metallopeptidase 8 (neutrophil collagenase) | 1.98 | 0.02162 |
| 6037 | RNASE3 | ribonuclease, RNase A family, 3 (eosinophil cationic protein) | 1.89 | 0.02677 |
| 4070 | TACSTD2 | tumor-associated calcium signal transducer 2 | 1.89 | 0.02091 |
| 10562 | OLFM4 | olfactomedin 4 | 1.88 | 0.07735 |
| 4680 | CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | 1.87 | 0.03082 |
| 3934 | LCN2 | lipocalin 2 | 1.85 | 0.00126 |
| 10321 | CRISP3 | cysteine-rich secretory protein 3 | 1.83 | 0.05188 |
| 1991 | ELANE | elastase, neutrophil expressed | 1.78 | 0.02841 |
| 260429 | PRSS33 | protease, serine, 33 | 1.73 | 0.08714 |
| 5806 | PTX3 | pentraxin-related gene, rapidly induced by IL-1 beta | 1.69 | 0.00206 |
| 1511 | CTSG | cathepsin G | 1.69 | 0.09089 |
| 6947 | TCN1 | transcobalamin I (vitamin B12 binding protein, R binder family) | 1.68 | 0.00847 |
| 820 | CAMP | cathelicidin antimicrobial peptide | 1.66 | 0.00178 |
| 1232 | CCR3 | chemokine (C-C motif) receptor 3 | 1.64 | 0.001 |
| 932 | MS4A3 | membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | 1.63 | 0.08534 |
| 260429 | PRSS33 | protease, serine, 33 | 1.57 | 0.08534 |
| 4973 | OLR1 | oxidized low density lipoprotein (lectin-like) receptor 1 | 1.54 | 0.0919 |
| 56729 | RETN | resistin | 1.53 | 0.05967 |
| 100133941 | CD24 | CD24 molecule | 1.52 | 0.03254 |
Figure 2.
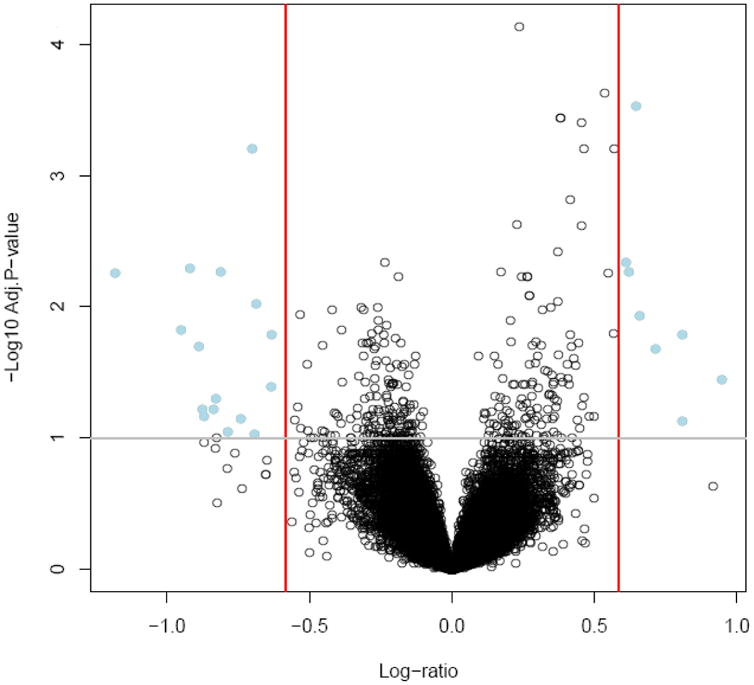
A “volcano plot” shows the differential expression of all the annotated probe sets between late-onset preeclampsia and control groups on the Affymetrix GeneChip HG-U133 PLUS 2.0 array with the log (base10) of the FDR-adjusted probability values (y-axis) plotted against the log (base 2) fold changes (x-axis).
Table 5.
Top 20 biological processes (ranking by odds ratio) significantly enriched in the comparison between late-onset preeclampsia and controls.
| GOBPID | Biological Process | # Differentially Expressed Genes in GO term | # Genes in GO term | Odds Ratio | Adjusted p-value |
|---|---|---|---|---|---|
| GO:0042742 | defense response to bacterium | 5 | 51 | 60.76 | 0.0000 |
| GO:0007159 | leukocyte cell-cell adhesion | 2 | 25 | 40.26 | 0.0129 |
| GO:0051707 | response to other organism | 8 | 227 | 22.46 | 0.0000 |
| GO:0050866 | negative regulation of cell activation | 2 | 48 | 20.09 | 0.0163 |
| GO:0002683 | negative regulation of immune system process | 2 | 68 | 13.97 | 0.0255 |
| GO:0043627 | response to estrogen stimulus | 2 | 69 | 13.76 | 0.0256 |
| GO:0006874 | cellular calcium ion homeostasis | 3 | 117 | 12.63 | 0.0129 |
| GO:0055066 | di-, tri-valent inorganic cation homeostasis | 4 | 165 | 12.44 | 0.0101 |
| GO:0007204 | elevation of cytosolic calcium ion concentration | 2 | 77 | 12.29 | 0.0273 |
| GO:0043406 | positive regulation of MAP kinase activity | 2 | 79 | 11.96 | 0.0278 |
| GO:0030003 | cellular cation homeostasis | 4 | 175 | 11.70 | 0.0105 |
| GO:0055065 | metal ion homeostasis | 3 | 131 | 11.23 | 0.0142 |
| GO:0008624 | induction of apoptosis by extracellular signals | 2 | 97 | 9.68 | 0.0323 |
| GO:0032101 | regulation of response to external stimulus | 2 | 105 | 8.92 | 0.0351 |
| GO:0006955 | immune response | 8 | 581 | 8.29 | 0.0008 |
| GO:0031347 | regulation of defense response | 2 | 114 | 8.20 | 0.0371 |
| GO:0055082 | cellular chemical homeostasis | 4 | 261 | 7.72 | 0.0142 |
| GO:0050801 | ion homeostasis | 4 | 278 | 7.23 | 0.0142 |
| GO:0042330 | taxis | 2 | 131 | 7.11 | 0.0442 |
| GO:0010324 | membrane invagination | 3 | 211 | 6.86 | 0.0273 |
Pathway analysis of the differentially expressed genes was undertaken with an overrepresentation method and the Signaling Pathway Impact Analysis method. No pathways emerged as significantly enriched or impacted based on either method in both comparisons (between both early-onset vs. controls and between late-onset vs. controls).
qRT-PCR analysis
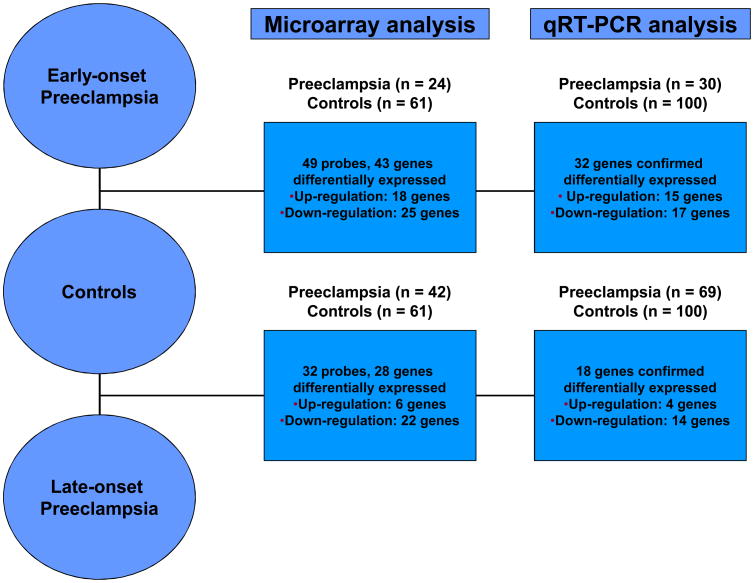
Figure 3 summarizes the numbers of genes differentially expressed in microarray and qRT-PCR experiments. For the comparison between early-onset PE and controls, qRT-PCR confirmed the microarray results for both the direction of fold change and the significance in 77% (33/43) of genes. Specifically, qRT-PCR confirmed increased expression of 15 genes and decreased expression of 18 genes in early-onset PE (see supplementary Table 5) when compared to uncomplicated pregnancies. Similarly, qRT-PCR validated the microarray results for the comparison between late-onset PE and controls in 71% (20/28) of genes. qRT-PCR confirmed up-regulation of 4 genes and down-regulation of 16 genes (see supplementary Table 6).
Figure 3.
The numbers of genes differentially expressed in microarray and qRT-PCR experiments between early-onset, late-onset preeclampsia and uncomplicated pregnancies. For the comparison between early-onset preeclampsia and controls, qRT-PCR confirmed the microarray results for both the direction of fold change and the significance in 77% (33/43) of genes. Similarly, qRT-PCR confirmed the microarray results for the comparison between late-onset PE and controls in 71% (20/28) of genes.
Genes differentially expressed by both microarray and qRT-PCR analysis are summarized in Table 6. Twenty genes (12 higher expressions and 8 lower expression) were differentially expressed in early-onset PE alone. Among these genes, H19 - an imprinted maternally expressed gene, which is involved in developmental processes, had the highest fold change (5.7). Genes involved in coagulation/fibrinolytic system (SERPINI2), immune regulation (VSIG4, CD24), and inflammation (S100A10) were detected as differentially regulated. In contrast, only seven genes (1 higher expression and 5 lower expressions) that are involved in innate immunity (LTF, ELANE) and cell-to-cell recognition in the nervous system (CNTNAP3) were differentially expressed in late-onset PE alone. Thirteen genes (3 higher expressions and 10 lower expression) that are essential to host defense (DEFA4, BPI, CTSG, LCN2), tight junctions in blood-brain barrier (EMP1) and liver regeneration (ECT2) were differentially expressed in late-onset PE alone compared to uncomplicated pregnancies.
Table 6. Summary of genes which are differentially expressed in both microarray analysis and by qRT-PCR.
| Number | Entrez Gene | Symbol | Gene Name | Microarray | qRT-PCR | ||
|---|---|---|---|---|---|---|---|
| Fold Change | Adjusted P-value | Fold Change | Adjusted P-value | ||||
| Early-onset preeclampsia alone | |||||||
| Higher expression | |||||||
| 1 | 224997_x_at | H19 | H19, imprinted maternally expressed transcript (non-protein coding) | 1.66 | 0.0011 | 5.74 | 0.0000 |
| 2 | 204787_at | VSIG4 | V-set and immunoglobulin domain containing 4 | 1.63 | 0.0026 | 2.76 | 0.0000 |
| 3 | 207636_at | SERPINI2 | serpin peptidase inhibitor, clade I (pancpin), member 2 | 1.53 | 0.0077 | 2.74 | 0.0000 |
| 4 | 202768_at | FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 2.06 | 0.0000 | 2.66 | 0.0095 |
| 5 | 209191_at | TUBB6 | tubulin, beta 6 | 1.62 | 0.0106 | 2.26 | 0.0000 |
| 6 | 205081_at | CRIP1 | cysteine-rich protein 1 (intestinal) | 1.59 | 0.0000 | 2.21 | 0.0001 |
| 7 | 212089_at | LMNA | lamin A/C | 1.52 | 0.0011 | 2.05 | 0.0003 |
| 8 | 227862_at | TRNP1 | TMF1-regulated nuclear protein 1 | 1.55 | 0.0508 | 1.96 | 0.0011 |
| 9 | 230422_at | FPR3 | formyl peptide receptor 3 | 1.63 | 0.0139 | 1.86 | 0.0009 |
| 10 | 219833_s_at | EFHC1 | EF-hand domain (C-terminal) containing 1 | 1.52 | 0.0047 | 1.74 | 0.0000 |
| 11 | 231077_at | C1orf192 | chromosome 1 open reading frame 192 | 1.54 | 0.0008 | 1.52 | 0.0057 |
| 12 | 238909_at | S100A10 | S100 calcium binding protein A10 | 1.69 | 0.0000 | 1.44 | 0.0003 |
| Lower expression | |||||||
| 1 | 1553605_a_at | ABCA13 | ATP-binding cassette, sub-family A (ABC1), member 13 | 1.56 | 0.0855 | 2.03 | 0.0045 |
| 2 | 208650_s_at | CD24 | CD24 molecule | 1.63 | 0.0387 | 2.00 | 0.0465 |
| 3 | 229144_at | RP1-21O18.1 | kazrin | 1.82 | 0.0051 | 1.95 | 0.0014 |
| 4 | 214523_at | CEBPE | CCAAT/enhancer binding protein (C/EBP), epsilon | 1.59 | 0.0036 | 1.77 | 0.0208 |
| 5 | 205513_at | TCN1 | transcobalamin I (vitamin B12 binding protein, R binder family) | 1.75 | 0.0120 | 1.75 | 0.0007 |
| 6 | 207384_at | PGLYRP1 | peptidoglycan recognition protein 1 | 1.59 | 0.0065 | 1.61 | 0.0005 |
| 7 | 206209_s_at | CA4 | carbonic anhydrase IV | 1.52 | 0.0398 | 1.52 | 0.0094 |
| 8 | 206464_at | BMX | BMX non-receptor tyrosine kinase | 1.50 | 0.0332 | 1.32 | 0.0165 |
| Late-onset preeclampsia alone | |||||||
| Higher expression | |||||||
| 1 | 233202_at | CNTNAP3 | contactin associated protein-like 3 | 1.61 | 0.0395 | 1.87 | 0.0016 |
| Lower expression | |||||||
| 1 | 202018_s_at | LTF | lactotransferrin | 2.03 | 0.0010 | 2.27 | 0.0001 |
| 2 | 212768_s_at | OLFM4 | olfactomedin 4 | 1.88 | 0.0774 | 1.97 | 0.0023 |
| 3 | 206871_at | ELANE | elastase, neutrophil expressed | 1.78 | 0.0284 | 1.70 | 0.0159 |
| 4 | 207802_at | CRISP3 | cysteine-rich secretory protein 3 | 1.83 | 0.0519 | 1.69 | 0.0027 |
| 5 | 210004_at | OLR1 | oxidized low density lipoprotein (lectin-like) receptor 1 | 1.54 | 0.0919 | 1.47 | 0.0180 |
| 6 | 220570_at | RETN | resistin | 1.53 | 0.0597 | 1.46 | 0.0462 |
| Both early- and late-onset preeclampsia | |||||||
| Higher expression in early-onset | |||||||
| 1 | 201324_at | EMP1 | epithelial membrane protein 1 | 2.33 | 0.0001 | 3.01 | 0.0000 |
| 2 | 237241_at | ECT2 | epithelial cell transforming sequence 2 oncogene | 1.71 | 0.0077 | 2.05 | 0.0001 |
| 3 | 229307_at | ANKRD28 | ankyrin repeat domain 28 | 1.72 | 0.0233 | 1.66 | 0.0000 |
| Higher expression in late-onset | |||||||
| 1 | 201324_at | EMP1 | epithelial membrane protein 1 | 1.63 | 0.0250 | 2.34 | 0.0000 |
| 2 | 237241_at | ECT2 | epithelial cell transforming sequence 2 oncogene | 1.53 | 0.0236 | 1.58 | 0.0007 |
| 3 | 1561079_at | ANKRD28 | ankyrin repeat domain 28 | 1.57 | 0.0029 | 1.45 | 0.0000 |
| Lower expression in early-onset | |||||||
| 1 | 207269_at | DEFA4 | defensin, alpha 4, corticostatin | 2.36 | 0.0110 | 3.22 | 0.0000 |
| 2 | 211657_at | CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | 2.06 | 0.0334 | 2.69 | 0.0001 |
| 3 | 210244_at | CAMP | cathelicidin antimicrobial peptide | 1.93 | 0.0002 | 2.51 | 0.0000 |
| 4 | 206676_at | CEACAM8 | carcinoembryonic antigen-related cell adhesion molecule 8 | 1.90 | 0.0508 | 2.36 | 0.0005 |
| 5 | 205653_at | CTSG | cathepsin G | 1.93 | 0.0993 | 2.27 | 0.0126 |
| 6 | 212531_at | LCN2 | lipocalin 2 | 1.58 | 0.0984 | 2.03 | 0.0002 |
| 7 | 205557_at | BPI | bactericidal/permeability-increasing protein | 2.00 | 0.0214 | 1.98 | 0.0006 |
| 8 | 1554892_a_at | MS4A3 | membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | 1.83 | 0.0962 | 1.95 | 0.0033 |
| 9 | 208304_at | CCR3 | chemokine (C-C motif) receptor 3 | 1.83 | 0.0002 | 1.39 | 0.0343 |
| 10 | 206157_at | PTX3 | pentraxin-related gene, rapidly induced by IL-1 beta | 1.57 | 0.0448 | 1.37 | 0.0236 |
| Lower expression in late-onset | |||||||
| 1 | 207269_at | DEFA4 | defensin, alpha 4, corticostatin | 2.49 | 0.0012 | 2.04 | 0.0006 |
| 2 | 205653_at | CTSG | cathepsin G | 1.69 | 0.0909 | 1.93 | 0.0151 |
| 3 | 206676_at | CEACAM8 | carcinoembryonic antigen-related cell adhesion molecule 8 | 2.09 | 0.0039 | 1.91 | 0.0011 |
| 4 | 203757_s_at | CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | 1.99 | 0.0229 | 1.83 | 0.0034 |
| 5 | 210244_at | CAMP | cathelicidin antimicrobial peptide | 1.66 | 0.0018 | 1.82 | 0.0006 |
| 6 | 205557_at | BPI | bactericidal/permeability-increasing protein | 2.01 | 0.0052 | 1.56 | 0.0059 |
| 7 | 212531_at | LCN2 | lipocalin 2 | 1.85 | 0.0013 | 1.52 | 0.0108 |
| 8 | 1554892_a_at | MS4A3 | membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | 1.63 | 0.0853 | 1.46 | 0.0321 |
| 9 | 208304_at | CCR3 | chemokine (C-C motif) receptor 3 | 1.64 | 0.0010 | 1.38 | 0.0113 |
| 10 | 206157_at | PTX3 | pentraxin-related gene, rapidly induced by IL-1 beta | 1.69 | 0.0021 | 1.33 | 0.0193 |
Discussion
Principal findings of the study
1) microarray analysis in maternal whole blood revealed 43 and 28 differentially expressed genes in early-onset and late-onset PE, respectively, compared to uncomplicated pregnancy; 2) qRT-PCR confirmed microarray results for early- and late-onset PE in 77% (33/43) and 71% (20/28) of genes, respectively; and 3) twenty genes that are involved in coagulation, immune regulation, developmental process and inflammation were differentially expressed in early-onset PE alone. In contrast, only seven genes implicated in innate immunity and cell-to-cell recognition in the nervous system were differentially expressed in late-onset PE alone. Thirteen genes that are essential to host defense, tight junctions in blood-brain barrier and liver regeneration were differentially expressed in both early- and late-onset PE compared to uncomplicated pregnancy.
The results of our study were reported after adjustment for gestational age and WBC count, since variation in the proportion of peripheral blood cell types could be responsible for differences of gene expression profiles observed among groups [99,135]. Among uncomplicated pregnant women, the expression of 56 genes was correlated with WBC count, whereas the expression of only two genes changed as a function of gestational age after adjustment for the WBC count. After adjustment for WBC count and gestational age, the differentially expressed genes between PE and controls increased from 31 to 43 genes and from 22 to 28 genes for early- and late-onset PE, respectively.
In the current study, we did not compare transcriptomic profiles of early- and late-onset PE directly, because we could not reasonably adjust for gestational age differences (by definition, these two conditions have completely different ranges of gestational age at blood sampling). It is noteworthy that several differentially expressed genes in early- and late-onset PE change in the same direction. One interpretation of these findings is that early- and late-onset PE are characterized by a common signature in the transcriptional profile of whole blood. However, a small set of genes were differentially expressed in early- or late-onset PE alone when compared to uncomplicated pregnancies. Selected differentially expressed genes are discussed below.
H19, imprinted maternally expressed transcript (non-protein coding) (H-19)
The H-19 gene produces a non-coding RNA which may have growth suppressive function. This gene is located in an imprinted region of chromosome 11 near the insulin-like growth factor-2 (IGF2) gene. Expression of H-19 and IGF2 are imprinted so that H-19 is only expressed from the maternal allele, and IGF2 is only expressed from the paternal allele [136-138].Experiments in mice have demonstrated that placental IGF2 knockout leads to fetal growth restriction, while H-19 silencing results in fetal overgrowth [139]. These phenotypes also observed in humans -Silver Russell Syndrome [116,140-142]and Beckwith-Wiedemann Syndrome [143-145], respectively.
H-19 is highly expressed in cytotrophoblasts [136]. Hypomethylation, along with increased expression of H-19, has been observed in the placenta of pregnancies complicated by fetal growth restriction [146], while hypermethylation with decreased expression of H-19 has been reported in the placenta of patients with early-onset PE [147]. Furthermore, H-19 could inhibit trophoblast proliferation through miR-675 and the lower expression of H-19 in the placenta may result in an excessive proliferation of trophoblasts in early-onset PE [148].
Another study also reports biallelic expression (loss of imprinting: LOI) of the H-19 gene in 46% (6/13) of the placentas from patients with PE whereas there is no bialleic expression in 26 cases of normal pregnancy [138]. The clinical symptoms of patients with LOI on the H-19 gene are more severe than those of patients without LOI [138]. Moreover, H-19 expression may be related to hypoxia since over-expression of HIF-1α and suppression of p53, a tumor suppressor gene, induces H-19 expression in response to hypoxia in several tumor cell lines [149]. We found a 5.7 fold higher mRNA expression of H-19 in the whole blood of patients with early-onset, but not late-onset PE, compared to uncomplicated pregnancies. The biological consequence of increased expression of H-19 genes on leukocytes deserves further investigation.
V-set and immunoglobulin domain containing 4 (VSIG4)
This gene encodes a v-set and immunoglobulin-domain containing protein, also referred to as complement receptor of the immunoglobulin superfamily (CRIg). This protein is a receptor for the complement component 3 (C3), fragments C3b, iC3b and implicated in the clearance of systemic pathogens and autologous cells [150]. The activation of complement system resulting in the generation of split products with proinflammatory properties has been observed in PE [151,152]. In the current study, a 2.7 fold higher expression of VSIG4 gene was observed in early-onset PE, but not in the late-onset disease. However, VSIG4 is believed to play a role in tissue homeostasis and resolution rather than initiation of inflammatory response [153]. In mice, this protein inhibits both cytotoxic T and B cell responses to viral antigen, and thus, may also be a negative regulator of T-cell responses [154].
CD24 molecule (CD24)
CD24, a cell-surface sialoglycoprotein, is expressed on multiple cell types and disappears after these cells have reached their final stage of differentiation [155,156]. Recently, CD24 has been proposed to be a genetic check point in T cell homeostasis and autoimmune disease since CD24 expression in T cells is necessary for optimal T cell homeostatic proliferation in the lymhopenic host, while CD24 expressed by non-T cells acts as a negative regulator that control the pace of T cell proliferation [157]. CD24 polymorphisms are also associated with multiple sclerosis [158] and systemic lupus erythematosus [159].
CD 24 also plays a role in the innate immune response. In a mouse model of acetaminophen-induced liver necrosis, CD24 interacts with sialic acid binding immunoglobulin-like lectins (Siglecs) and decreases the host response to danger- associated molecular patterns (i.e.: high mobility group box 1, heat shock protein 70 and heat shock protein 90), but not to pathogen associated molecular pattern (eg: Toll-like receptor), by selectively repression of NF-κB activation [160]. Furthermore, a recent study reported that cytotrophoblasts and the syncytiotrophoblasts expressed CD24 and treatment placental explants with adiponectin (an adipokine which has been reported to be elevated in circulation of patients with PE [161]) increased expression of Siglec10 (both mRNA and protein) [162] suggesting that CD24/Siglecs interaction may be one mechanism whereby trophoblasts protect themselves against a danger signal [160,162]. However, the consequences of decreased CD24 expression in the peripheral blood of patients with early-onset PE reported herein remains to be determined.
ATP-binding cassette, sub-family A (ABC1), member 13 (ABCA13)
ABCA 13 is a new member of the ATP-binding cassette (ABC) gene subfamily A (ABCA). The 12 members of this subfamily share a high degree of sequence conservation and have been mostly related to lipid trafficking [133,160,163-170]. For example; ABCA 2, which is predominantly expressed in the brain [171], plays an important role in neuronal lipid transport [168,170] and is associated with Alzheimer's disease [172,173]. ABCA 3 is predominantly expressed in alveolar type II cells [174], plays a role in the excretion of the lipid fraction of the pulmonary surfactant [166,175]. However, the function of ABCA 13, which is expressed in trachea, testis and bone marrow [176], remains to be determined. The murine ABCA13 promoter region contains transcription factor-binding sites associated with myeloid and lymphoid cell types, and that its ubiquitous expression in blood derived cells suggests a role for ABCA 13 in hematopoiesis [177].In the current study, ABCA13 gene expression in peripheral whole blood was lower in early-onset PE than in uncomplicated pregnancies.
Contactin associated protein-like 3 (CNTNAP3)
The protein encoded by this gene belongs to the Neuroxin-IV/CNTNAP/Paranodin (NCP) family, the biological function of which is thought to be the mediation of neuron–glia cells interactions [178]. Thus, this protein may play a role in cell recognition within the nervous system. At least five CNTNAP genes, with expression restricted primarily to the brain and peripheral nerves, have been cloned so far and are presumed to play a role in cell to cell interactions [179-182]. CNTNAP3 was the only gene that up-regulated significantly in late-onset PE alone.
Epithelial membrane protein 1 (EMP1)
This gene encodes a protein that is integral to the formation of tight junctions in the blood-brain barrier [183]. This protein is a liver cell-junction protein [184] and is expressed in tissues that have significant epidermal growth factor (EGF) receptor expression [185,186]. EMP1 gene expression was higher in the peripheral blood of patients with early- and late-onset PE than in uncomplicated pregnancies.
Preeclampsia: a condition with down-regulation of anti-inflammatory genes in peripheral whole blood
An interesting finding of this study is that several genes involved in the innate immune response of neutrophils and monocytes/macrophages, such as neutrophil elastase (ELANE), cathepsin-G (CTSG), bactericidial/permeability increasing protein (BPI), defensin alpha (DEFA)-4, and lactotransferrin (LTF), were down-regulated either in both subtypes of PE (DEFA4, BPI, CTSG) or in late-onset PE (ELANE, LTF). Since the nature of systemic inflammation in PE is chronic [1,187,188], it is not surprising that these genes (which are involved in the acute inflammatory response and participated in the first line of defense against pathogens) are not up-regulated in whole blood. However, the fact that their expression was down-regulated in PE may seem paradoxical. Nonetheless, recent evidence may provide a possible explanation for this phenomenon.
Previous studies indicate that neutrophil elastase and cathepsin G have not only pro-inflammatory, but anti-inflammatory properties, as well [189-191].A study by Gardiner et al. demonstrated that when both proteins were applied to neutrophils, they were capable of cleaving P-selectin glycoprotein ligand-1 (PSGL-1) on neutrophils, which led to the loss of PSGL-1 protein from neutrophil surfaces, and rapidly abolished their capacity to bind P-selectin, a PSGL-1 ligand on activated endothelium [189]. This study suggested that the cathepsin G and neutrophil elastase-mediated PSGL-1 proteolysis is part of an autocrine mechanism for the down-regulation of neutrophil adhesion to activated endothelium. Similarly, another study reported that elastase and cathepsin G containing fractions (derived from neutrophil lysates and degranulation supernatants), as well as purified elastase and cathepsin G, inhibited C5a-dependent neutrophil functions [191]. These findings indicate that elastase and cathepsin G play an important role in the down-regulation of acute inflammation.
Neutrophil cathepsin G may also exert its anti-inflammatory functions on monocytes since it is capable of cleaving CD14 (endotoxin receptor) from monocyte surfaces, thus limiting inflammation [190]. Since the activation of the endothelium and complement system is part of the exaggerated systemic maternal inflammation in PE [28,31,151,152,192], the down-regulation of neutrophil elastase and cathepsin G may indicate defective up-regulation of important anti-inflammatory molecules in peripheral leukocytes that is intended to limit inflammation.
Of interest, a microarray study by Williams at el. found that genes encoding for alpha-defensin A3, cathelicidin, cathepsin G, and lactotransferrin were down-regulated in monocytes that had a transmigratory phenotype [193]. It is possible that our findings showing down-regulation of three out of four of these genes, as well as of DEFA-4, may reflect a phenotypic change in monocytes and/or neutrophils, in which this phenomenon may be related to their altered migratory potential in PE.
Previous microarray studies of the transcriptome of peripheral blood of patients with preeclampsia
Four studies have used microarray to examine transcriptomic signatures in peripheral blood of patients with PE. Sun et al. reported 72 differentially regulated genes between six patients with early-onset PE and five uncomplicated pregnant women [194]. The authors reported the up-regulation of the VSIG4 gene, which is the only finding that is consistent with the current study. However, this gene was found to be down regulated in their qRT-PCR validation. In contrast to our study, Sun et al. used Trizol and isolated lymphocytes prior to conducting microarray analysis. Rajakumar et al. compared the transcriptomic profiles of peripheral blood mononuclear cells isolated from five patients with PE and women with uncomplicated pregnancies (n=5) [70]. Matrix metalloproteinase (MMP)-9 was found to be differentially expressed in that study, but was not confirmed in our qRT-PCR analysis. Dahlstrom et al., using the PAX gene Blood RNA system, reported 19 differentially expressed genes between 8 early-onset PE and 8 normal pregnant women [195]. Only lower expression of CCR3 (CC-chemokine receptor 3) gene in late-onset PE was confirmed in our study. Although this study was the only one that evaluated gene expression at the time of diagnosis in whole blood of PE using the PAX gene Blood RNA system (as in our study), the difference in analytic approach (principal component analysis) and the inclusion of patients who received medications may explain the discrepancy between their results and our findings. A recent study by Enquobahrie et al. examined gene expression profiles in peripheral whole blood with the PAX gene Blood RNA system at 16 weeks of gestation in 16 patients destined to develop PE [196]. Differential expressed genes included COL1A1, IKBKB and RB1, none of which was significantly differentially expressed between uncomplicated pregnancies and PE at the time of diagnosis in our study. However, Enquobahrie et al. did not stratify patients into early- or late- onset PE.
Advantage and disadvantage of whole blood transcriptome
The current study used microarray technology, which allows unbiased genome-wide examination of the transcriptome of peripheral blood in patients with PE. In order to minimize alteration in RNA expression after blood sampling, the PAX gene blood RNA system, which contains a stabilizing additive in a blood collection tube to immediately capture the transcriptional profile, was utilized. This approach allows analysis of intracellular RNA-expression with reduced laboratory complexities and less artificial activation compared to other approaches, which isolated each cellular subpopulation prior to microarray experiments. Yet, results of microarray analysis with the use of the PAX gene blood RNA system have been reported to provide useful information in several studies [175,197,198].However, the microarray hybridization rates reported in whole blood are usually lower than those using individual cell types [87,199]. The Encore™ Biotin Module and Ovation® RNA Amplification System V2 were used in this study for amplification of whole blood total RNA to increase hybridization rates. We did not use globin reduction procedures in order to avoid the interference with the observed transcriptional profiles caused by these methods [200].
Since PAXgene blood system stabilizes intra-cellular RNA, the trancriptomic profiles of whole blood observed in this study mainly originated from maternal leukocytes (especially neutrophils) and other cellular components, including reticulocytes and immature platelets. However, we also observed transcripts of placental specific or pregnancy specific genes such as PLAC4, PLAC1, Placental lactogen (CSH1), chorionic gonadotropin, beta popypeptide (CGB) genes in some samples (data not shown). This observation is consistent with a study reported by Okazaki et al. that the mRNA expression of trophoblasts can be observed through analysis of the cellular component of maternal blood [201,202], although PLAC4, and PLAC1 may not be specific to placenta [203] since they could be detectable in non-pregnant whole blood.
Strengths and limitations
This is the largest study to date that used microarray for the transcriptomic profiling of whole blood in patients with PE. Patients with early- and late-onset PE were included in the study. Moreover, all differentially expressed genes from microarray analysis were validated by qRT-PCR. The limitations of the study include: 1) the differentially expressed genes reflect the changes in total intra-cellular mRNA components of whole blood and cannot be traced to the subpopulation of leukocytes or reticulocytes that are responsible for the observed differences; and 2) since blood samples were obtained at the time of diagnosis, it remains to be determined whether the differentially expressed genes reported herein are causally related to PE.
Conclusion
Microarray analysis in maternal whole blood revealed several differentially expressed genes in early- and late-onset PE compared to uncomplicated pregnancy. Differentially expressed genes are involved in coagulation, immune regulation, growth/developmental process, host defense and tight junctions in blood-brain barrier. However, early- and late-onset PE are characterized by a common signature in the transcriptional profile of whole blood. Future studies of the biological function, expression timetable and protein expression of differentially expressed genes may provide insight into the pathophysiology of PE.
Supplementary Material
Acknowledgments
This research was supported, in part, by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services (NICHD/NIH); and, in part, with Federal funds from NICHD, NIH under Contract No. HHSN275201300006C.
Footnotes
Presented at the 58th annual meeting of the Society for Gynecologic Investigation, March 16-19, 2011, Miami, FL
Reference List
- 1.Romero R. Prenatal medicine: the child is the father of the man. 1996. J Matern Fetal Neonatal Med. 2009;22:636–9. doi: 10.1080/14767050902784171. [DOI] [PubMed] [Google Scholar]
- 2.Di Renzo GC. The great obstetrical syndromes. J Matern Fetal Neonatal Med. 2009;22:633–5. doi: 10.1080/14767050902866804. [DOI] [PubMed] [Google Scholar]
- 3.Roberts JM, Taylor RN, Goldfien A. Clinical and biochemical evidence of endothelial cell dysfunction in the pregnancy syndrome preeclampsia. Am J Hypertens. 1991;4:700–8. doi: 10.1093/ajh/4.8.700. [DOI] [PubMed] [Google Scholar]
- 4.Brosens IA. Morphological changes in the utero-placental bed in pregnancy hypertension. Clin Obstet Gynaecol. 1977;4:573–93. [PubMed] [Google Scholar]
- 5.Brosens I, Pijnenborg R, Vercruysse L, Romero R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am J Obstet Gynecol. 2011;204:193–201. doi: 10.1016/j.ajog.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman SA, Schiff E, Emeis JJ, Dekker GA, Sibai BM. Biochemical corroboration of endothelial involvement in severe preeclampsia. Am J Obstet Gynecol. 1995;172:202–3. doi: 10.1016/0002-9378(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 7.Romero R, Lockwood C, Oyarzun E, Hobbins JC. Toxemia: new concepts in an old disease. Semin Perinatol. 1988;12:302–23. [PubMed] [Google Scholar]
- 8.Pijnenborg R, Anthony J, Davey DA, Rees A, Tiltman A, Vercruysse L, et al. Placental bed spiral arteries in the hypertensive disorders of pregnancy. Br J Obstet Gynaecol. 1991;98:648–55. doi: 10.1111/j.1471-0528.1991.tb13450.x. [DOI] [PubMed] [Google Scholar]
- 9.Myatt L, Kossenjans W, Sahay R, Eis A, Brockman D. Oxidative stress causes vascular dysfunction in the placenta. J Matern Fetal Med. 2000;9:79–82. doi: 10.1002/(SICI)1520-6661(200001/02)9:1<79::AID-MFM16>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 10.Hubel CA. Oxidative stress in the pathogenesis of preeclampsia. Proc Soc Exp Biol Med. 1999;222:222–35. doi: 10.1177/153537029922200305. [DOI] [PubMed] [Google Scholar]
- 11.Gervasi MT, Chaiworapongsa T, Naccasha N, Blackwell S, Yoon BH, Maymon E, et al. Phenotypic and metabolic characteristics of maternal monocytes and granulocytes in preterm labor with intact membranes. Am J Obstet Gynecol. 2001;185:1124–9. doi: 10.1067/mob.2001.117681. [DOI] [PubMed] [Google Scholar]
- 12.Brosens JJ, Pijnenborg R, Brosens IA. The myometrial junctional zone spiral arteries in normal and abnormal pregnancies: a review of the literature. Am J Obstet Gynecol. 2002;187:1416–23. doi: 10.1067/mob.2002.127305. [DOI] [PubMed] [Google Scholar]
- 13.Chaiworapongsa T, Romero R, Yoshimatsu J, Espinoza J, Kim YM, Park K, et al. Soluble adhesion molecule profile in normal pregnancy and pre-eclampsia. J Matern Fetal Neonatal Med. 2002;12:19–27. doi: 10.1080/jmf.12.1.19.27. [DOI] [PubMed] [Google Scholar]
- 14.Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003;111:649–58. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koga K, Osuga Y, Yoshino O, Hirota Y, Ruimeng X, Hirata T, et al. Elevated serum soluble vascular endothelial growth factor receptor 1 (sVEGFR-1) levels in women with preeclampsia. J Clin Endocrinol Metab. 2003;88:2348–51. doi: 10.1210/jc.2002-021942. [DOI] [PubMed] [Google Scholar]
- 16.Chaiworapongsa T, Romero R, Espinoza J, Bujold E, Mee KY, Goncalves LF, et al. Evidence supporting a role for blockade of the vascular endothelial growth factor system in the pathophysiology of preeclampsia. Young Investigator Award. Am J Obstet Gynecol. 2004;190:1541–7. doi: 10.1016/j.ajog.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 17.Bujold E, Romero R, Chaiworapongsa T, Kim YM, Kim GJ, Kim MR, et al. Evidence supporting that the excess of the sVEGFR-1 concentration in maternal plasma in preeclampsia has a uterine origin. J Matern Fetal Neonatal Med. 2005;18:9–16. doi: 10.1080/14767050500202493. [DOI] [PubMed] [Google Scholar]
- 18.Chaiworapongsa T, Romero R, Gotsch F, Espinoza J, Nien JK, Goncalves L, et al. Low maternal concentrations of soluble vascular endothelial growth factor receptor-2 in preeclampsia and small for gestational age. J Matern Fetal Neonatal Med. 2008;21:41–52. doi: 10.1080/14767050701831397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torry DS, Wang HS, Wang TH, Caudle MR, Torry RJ. Preeclampsia is associated with reduced serum levels of placenta growth factor. Am J Obstet Gynecol. 1998;179:1539–44. doi: 10.1016/s0002-9378(98)70021-3. [DOI] [PubMed] [Google Scholar]
- 20.Burton GJ, Yung HW, Cindrova-Davies T, Charnock-Jones DS. Placental endoplasmic reticulum stress and oxidative stress in the pathophysiology of unexplained intrauterine growth restriction and early onset preeclampsia. Placenta. 2009;30(Suppl A):S43–S48. doi: 10.1016/j.placenta.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaiworapongsa T, Romero R, Tarca AL, Kusanovic JP, Gotsch F, Mittal P, et al. A decrease in maternal plasma concentrations of sVEGFR-2 precedes the clinical diagnosis of preeclampsia. Am J Obstet Gynecol. 2010;202:550–10. doi: 10.1016/j.ajog.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cindrova-Davies T. Gabor Than Award Lecture 2008: pre-eclampsia - from placental oxidative stress to maternal endothelial dysfunction. Placenta. 2009;30(Suppl A):S55–S65. doi: 10.1016/j.placenta.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert JS, Ryan MJ, LaMarca BB, Sedeek M, Murphy SR, Granger JP. Pathophysiology of hypertension during preeclampsia: linking placental ischemia with endothelial dysfunction. Am J Physiol Heart Circ Physiol. 2008;294:H541–H550. doi: 10.1152/ajpheart.01113.2007. [DOI] [PubMed] [Google Scholar]
- 24.George EM, Granger JP. Linking placental ischemia and hypertension in preeclampsia: role of endothelin 1. Hypertension. 2012;60:507–11. doi: 10.1161/HYPERTENSIONAHA.112.194845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molvarec A, Szarka A, Walentin S, Szucs E, Nagy B, Rigo J., Jr Circulating angiogenic factors determined by electrochemiluminescence immunoassay in relation to the clinical features and laboratory parameters in women with pre-eclampsia. Hypertens Res. 2010;33:892–8. doi: 10.1038/hr.2010.92. [DOI] [PubMed] [Google Scholar]
- 26.Ogge G, Chaiworapongsa T, Romero R, Hussein Y, Kusanovic JP, Yeo L, et al. Placental lesions associated with maternal underperfusion are more frequent in early-onset than in late-onset preeclampsia. J Perinat Med. 2011;39:641–52. doi: 10.1515/JPM.2011.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrozella L, Mahendroo M, Timmons B, Roberts S, McIntire D, Alexander JM. Endothelial microparticles and the antiangiogenic state in preeclampsia and the postpartum period. Am J Obstet Gynecol. 2012;207:140–6. doi: 10.1016/j.ajog.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redman CW, Sacks GP, Sargent IL. Preeclampsia: an excessive maternal inflammatory response to pregnancy. Am J Obstet Gynecol. 1999;180:499–506. doi: 10.1016/s0002-9378(99)70239-5. [DOI] [PubMed] [Google Scholar]
- 29.Roberts JM, Hubel CA. The two stage model of preeclampsia: variations on the theme. Placenta. 2009;30(Suppl A):S32–S37. doi: 10.1016/j.placenta.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson WB, Brosens I, Dixon G. Maternal uterine vascular lesions in the hypertensive complications of pregnancy. Perspect Nephrol Hypertens. 1976;5:115–27. [PubMed] [Google Scholar]
- 31.Sacks GP, Studena K, Sargent K, Redman CW. Normal pregnancy and preeclampsia both produce inflammatory changes in peripheral blood leukocytes akin to those of sepsis. Am J Obstet Gynecol. 1998;179:80–6. doi: 10.1016/s0002-9378(98)70254-6. [DOI] [PubMed] [Google Scholar]
- 32.Tidwell SC, Ho HN, Chiu WH, Torry RJ, Torry DS. Low maternal serum levels of placenta growth factor as an antecedent of clinical preeclampsia. Am J Obstet Gynecol. 2001;184:1267–72. doi: 10.1067/mob.2001.113129. [DOI] [PubMed] [Google Scholar]
- 33.Tsatsaris V, Goffin F, Munaut C, Brichant JF, Pignon MR, Noel A, et al. Overexpression of the soluble vascular endothelial growth factor receptor in preeclamptic patients: pathophysiological consequences. J Clin Endocrinol Metab. 2003;88:5555–63. doi: 10.1210/jc.2003-030528. [DOI] [PubMed] [Google Scholar]
- 34.Vaughan JE, Walsh SW. Oxidative stress reproduces placental abnormalities of preeclampsia. Hypertens Pregnancy. 2002;21:205–23. doi: 10.1081/PRG-120015848. [DOI] [PubMed] [Google Scholar]
- 35.Verlohren S, Stepan H, Dechend R. Angiogenic growth factors in the diagnosis and prediction of pre-eclampsia. Clin Sci (Lond) 2012;122:43–52. doi: 10.1042/CS20110097. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Zhang GY, Wang J, Lu SL, Cao J, Sun LZ. A novel bridge between oxidative stress and immunity: the interaction between hydrogen peroxide and human leukocyte antigen G in placental trophoblasts during preeclampsia. Am J Obstet Gynecol. 2012;206:447–16. doi: 10.1016/j.ajog.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Duan DM, Niu JM, Lei Q, Lin XH, Chen X. Serum levels of the adipokine chemerin in preeclampsia. J Perinat Med. 2012;40:121–7. doi: 10.1515/JPM.2011.127. [DOI] [PubMed] [Google Scholar]
- 38.Gandevani SB, Banaem LM, Mohamadi B, Moghadam NA, Asghari M. Association of high-sensitivity C-reactive protein serum levels in early pregnancy with the severity of preeclampsia and fetal birth weight. J Perinat Med. 2012;0:1–5. doi: 10.1515/jpm-2011-0190. [DOI] [PubMed] [Google Scholar]
- 39.Kwon JH, Kim YH, Kwon JY, Park YW. Clinical significance of serum sRAGE and esRAGE in women with normal pregnancy and preeclampsia. J Perinat Med. 2011;39:507–13. doi: 10.1515/jpm.2011.055. [DOI] [PubMed] [Google Scholar]
- 40.Vaisbuch E, Romero R, Mazaki-Tovi S, Erez O, Kim SK, Chaiworapongsa T, et al. Retinol binding protein 4--a novel association with early-onset preeclampsia. J Perinat Med. 2010;38:129–39. doi: 10.1515/JPM.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazaki-Tovi S, Romero R, Vaisbuch E, Kusanovic JP, Erez O, Gotsch F, et al. Maternal serum adiponectin multimers in preeclampsia. J Perinat Med. 2009;37:349–63. doi: 10.1515/JPM.2009.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng YC, Lin QD, Lin JH, Ding CW, Zuo Y. Coagulation and fibrinolysis related cytokine imbalance in preeclampsia: the role of placental trophoblasts. J Perinat Med. 2009;37:343–8. doi: 10.1515/JPM.2009.060. [DOI] [PubMed] [Google Scholar]
- 43.Chaiworapongsa T, Romero R, Gotsch F, Kusanovic JP, Mittal P, Kim SK, et al. Acute pyelonephritis during pregnancy changes the balance of angiogenic and anti-angiogenic factors in maternal plasma. J Matern Fetal Neonatal Med. 2010;23:167–78. doi: 10.3109/14767050903067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conde-Agudelo A, Villar J, Lindheimer M. Maternal infection and risk of preeclampsia: systematic review and metaanalysis. Am J Obstet Gynecol. 2008;198:7–22. doi: 10.1016/j.ajog.2007.07.040. [DOI] [PubMed] [Google Scholar]
- 45.Stampalija T, Chaiworapongsa T, Chaemsaithong P, Korzeniewski SJ, Schwartz AG, Ferrazzi EM, et al. Maternal Plasma Concentrations of sST2 and Angiogenic/Anti-angiogenic Factors in Preeclampsia. J Matern Fetal Neonatal Med. 2013 doi: 10.3109/14767058.2013.784256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaiworapongsa T, Romero R, Korzeniewski SJ, Kusanovic JP, Soto E, Lam J, et al. Maternal plasma concentrations of angiogenic/antiangiogenic factors in the third trimester of pregnancy to identify the patient at risk for stillbirth at or near term and severe late preeclampsia. Am J Obstet Gynecol. 2013;208:287. doi: 10.1016/j.ajog.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SM, Romero R, Lee YJ, Park IS, Park CW, Yoon BH. Systemic inflammatory stimulation by microparticles derived from hypoxic trophoblast as a model for inflammatory response in preeclampsia. Am J Obstet Gynecol. 2012;207:337–8. doi: 10.1016/j.ajog.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loisel DA, Billstrand C, Murray K, Patterson K, Chaiworapongsa T, Romero R, et al. The maternal HLA-G 1597DeltaC null mutation is associated with increased risk of pre-eclampsia and reduced HLA-G expression during pregnancy in African-American women. Mol Hum Reprod. 2013;19:144–52. doi: 10.1093/molehr/gas041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding D, Scott NM, Thompson EE, Chaiworapongsa T, Torres R, Billstrand C, et al. Increased protein-coding mutations in the mitochondrial genome of African American women with preeclampsia. Reprod Sci. 2012;19:1343–51. doi: 10.1177/1933719112450337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mousa AA, Cappello RE, Estrada-Gutierrez G, Shukla J, Romero R, Strauss JF, III, et al. Preeclampsia is associated with alterations in DNA methylation of genes involved in collagen metabolism. Am J Pathol. 2012;181:1455–63. doi: 10.1016/j.ajpath.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J, Romero R, Dong Z, Lee DC, Dong Y, Mittal P, et al. Glycogen phosphorylase isoenzyme BB plasma concentration is elevated in pregnancy and preterm preeclampsia. Hypertension. 2012;59:274–82. doi: 10.1161/HYPERTENSIONAHA.111.177444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaiworapongsa T, Romero R, Savasan ZA, Kusanovic JP, Ogge G, Soto E, et al. Maternal plasma concentrations of angiogenic/anti-angiogenic factors are of prognostic value in patients presenting to the obstetrical triage area with the suspicion of preeclampsia. J Matern Fetal Neonatal Med. 2011;24:1187–207. doi: 10.3109/14767058.2011.589932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gotsch F, Romero R, Kusanovic JP, Chaiworapongsa T, Dombrowski M, Erez O, et al. Preeclampsia and small-for-gestational age are associated with decreased concentrations of a factor involved in angiogenesis: soluble Tie-2. J Matern Fetal Neonatal Med. 2008;21:389–402. doi: 10.1080/14767050802046069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berg CJ, Mackay AP, Qin C, Callaghan WM. Overview of maternal morbidity during hospitalization for labor and delivery in the United States: 1993-1997 and 2001-2005. Obstet Gynecol. 2009;113:1075–81. doi: 10.1097/AOG.0b013e3181a09fc0. [DOI] [PubMed] [Google Scholar]
- 55.Khan KS, Wojdyla D, Say L, Gulmezoglu AM, Van Look PF. WHO analysis of causes of maternal death: a systematic review. Lancet. 2006;367:1066–74. doi: 10.1016/S0140-6736(06)68397-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Meikle S, Trumble A. Severe maternal morbidity associated with hypertensive disorders in pregnancy in the United States. Hypertens Pregnancy. 2003;22:203–12. doi: 10.1081/PRG-120021066. [DOI] [PubMed] [Google Scholar]
- 57.Studd J. Pre-eclampsia. Br J Hosp Med. 1977;18:52–62. [PubMed] [Google Scholar]
- 58.Sibai BM. Hypertensive disorders of pregnancy: the United States perspective. Curr Opin Obstet Gynecol. 2008;20:102–6. doi: 10.1097/GCO.0b013e3282f73380. [DOI] [PubMed] [Google Scholar]
- 59.Ozkan H, Cetinkaya M, Koksal N, Ozmen A, Yildiz M. Maternal preeclampsia is associated with an increased risk of retinopathy of prematurity. J Perinat Med. 2011;39:523–7. doi: 10.1515/jpm.2011.071. [DOI] [PubMed] [Google Scholar]
- 60.Schneider S, Freerksen N, Maul H, Roehrig S, Fischer B, Hoeft B. Risk groups and maternal-neonatal complications of preeclampsia--current results from the national German Perinatal Quality Registry. J Perinat Med. 2011;39:257–65. doi: 10.1515/jpm.2011.010. [DOI] [PubMed] [Google Scholar]
- 61.Report of the National High Blood Pressure Education Program Working Group on High Blood Pressure in Pregnancy. Am J Obstet Gynecol. 2000;183:S1–S22. [PubMed] [Google Scholar]
- 62.Funai EF, Friedlander Y, Paltiel O, Tiram E, Xue X, Deutsch L, et al. Long-term mortality after preeclampsia. Epidemiology. 2005;16:206–15. doi: 10.1097/01.ede.0000152912.02042.cd. [DOI] [PubMed] [Google Scholar]
- 63.Ray JG, Vermeulen MJ, Schull MJ, Redelmeier DA. Cardiovascular health after maternal placental syndromes (CHAMPS): population-based retrospective cohort study. Lancet. 2005;366:1797–803. doi: 10.1016/S0140-6736(05)67726-4. [DOI] [PubMed] [Google Scholar]
- 64.Smith GC, Pell JP, Walsh D. Pregnancy complications and maternal risk of ischaemic heart disease: a retrospective cohort study of 129,290 births. Lancet. 2001;357:2002–6. doi: 10.1016/S0140-6736(00)05112-6. [DOI] [PubMed] [Google Scholar]
- 65.Irgens HU, Reisaeter L, Irgens LM, Lie RT. Long term mortality of mothers and fathers after pre-eclampsia: population based cohort study. BMJ. 2001;323:1213–7. doi: 10.1136/bmj.323.7323.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kralj M, Kraljevic S, Sedic M, Kurjak A, Pavelic K. Global approach to perinatal medicine: functional genomics and proteomics. J Perinat Med. 2005;33:5–16. doi: 10.1515/JPM.2005.001. [DOI] [PubMed] [Google Scholar]
- 67.Mohaupt M. Molecular aspects of preeclampsia. Mol Aspects Med. 2007;28:169–91. doi: 10.1016/j.mam.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 68.Nejatizadeh A, Stobdan T, Malhotra N, Pasha MA. The genetic aspects of pre-eclampsia: achievements and limitations. Biochem Genet. 2008;46:451–79. doi: 10.1007/s10528-008-9163-9. [DOI] [PubMed] [Google Scholar]
- 69.Ward K. Searching for genetic factors underlying pre-eclampsia: recent progress and persistent challenges. Minerva Ginecol. 2008;60:399–419. [PubMed] [Google Scholar]
- 70.Rajakumar A, Chu T, Handley DE, Bunce KD, Burke B, Hubel CA, et al. Maternal gene expression profiling during pregnancy and preeclampsia in human peripheral blood mononuclear cells. Placenta. 2011;32:70–8. doi: 10.1016/j.placenta.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toft JH, Lian IA, Tarca AL, Erez O, Espinoza J, Eide IP, et al. Whole-genome microarray and targeted analysis of angiogenesis-regulating gene expression (ENG, FLT1, VEGF, PlGF) in placentas from pre-eclamptic and small-for-gestational-age pregnancies. J Matern Fetal Neonatal Med. 2008;21:267–73. doi: 10.1080/14767050801924118. [DOI] [PubMed] [Google Scholar]
- 72.Uuskula L, Mannik J, Rull K, Minajeva A, Koks S, Vaas P, et al. Mid-gestational gene expression profile in placenta and link to pregnancy complications. PLoS One. 2012;7:e49248. doi: 10.1371/journal.pone.0049248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varkonyi T, Nagy B, Fule T, Tarca AL, Karaszi K, Schonleber J, et al. Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta. 2011;32(Suppl):S21–S29. doi: 10.1016/j.placenta.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baker PN, Myers JE. Preeclamptic toxemia: a disease ripe for proteomic discovery. Expert Rev Proteomics. 2009;6:107–10. doi: 10.1586/epr.09.5. [DOI] [PubMed] [Google Scholar]
- 75.Blumenstein M, McMaster MT, Black MA, Wu S, Prakash R, Cooney J, et al. A proteomic approach identifies early pregnancy biomarkers for preeclampsia: novel linkages between a predisposition to preeclampsia and cardiovascular disease. Proteomics. 2009;9:2929–45. doi: 10.1002/pmic.200800625. [DOI] [PubMed] [Google Scholar]
- 76.Carty DM, Siwy J, Brennand JE, Zurbig P, Mullen W, Franke J, et al. Urinary proteomics for prediction of preeclampsia. Hypertension. 2011;57:561–9. doi: 10.1161/HYPERTENSIONAHA.110.164285. [DOI] [PubMed] [Google Scholar]
- 77.Feng YL, Zhou CJ, Li XM, Liang XQ. Alpha-1-antitrypsin acts as a preeclampsia-related protein: a proteomic study. Gynecol Obstet Invest. 2012;73:252–9. doi: 10.1159/000334820. [DOI] [PubMed] [Google Scholar]
- 78.Gharesi-Fard B, Zolghadri J, Kamali-Sarvestani E. Proteome differences of placenta between pre-eclampsia and normal pregnancy. Placenta. 2010;31:121–5. doi: 10.1016/j.placenta.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 79.Jin H, Ma KD, Hu R, Chen Y, Yang F, Yao J, et al. Analysis of expression and comparative profile of normal placental tissue proteins and those in preeclampsia patients using proteomic approaches. Anal Chim Acta. 2008;629:158–64. doi: 10.1016/j.aca.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 80.Lee SM, Park JS, Norwitz ER, Kim SM, Kim BJ, Park CW, et al. Characterization of discriminatory urinary proteomic biomarkers for severe preeclampsia using SELDI-TOF mass spectrometry. J Perinat Med. 2011;39:391–6. doi: 10.1515/jpm.2011.028. [DOI] [PubMed] [Google Scholar]
- 81.Liu C, Zhang N, Yu H, Chen Y, Liang Y, Deng H, et al. Proteomic analysis of human serum for finding pathogenic factors and potential biomarkers in preeclampsia. Placenta. 2011;32:168–74. doi: 10.1016/j.placenta.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mary S, Patil GV, Kulkarni AV, Kulkarni MJ, Joshi SR, Mehendale SS, et al. Dynamic proteome in enigmatic preeclampsia: an account of molecular mechanisms and biomarker discovery. Proteomics Clin Appl. 2012;6:79–90. doi: 10.1002/prca.201100089. [DOI] [PubMed] [Google Scholar]
- 83.Myers J, Macleod M, Reed B, Harris N, Mires G, Baker P. Use of proteomic patterns as a novel screening tool in pre-eclampsia. J Obstet Gynaecol. 2004;24:873–4. doi: 10.1080/01443610400018791. [DOI] [PubMed] [Google Scholar]
- 84.Rasanen J, Girsen A, Lu X, Lapidus JA, Standley M, Reddy A, et al. Comprehensive maternal serum proteomic profiles of preclinical and clinical preeclampsia. J Proteome Res. 2010;9:4274–81. doi: 10.1021/pr100198m. [DOI] [PubMed] [Google Scholar]
- 85.Shin JK, Baek JC, Kang MY, Park JK, Lee SA, Lee JH, et al. Proteomic analysis reveals an elevated expression of heat shock protein 27 in preeclamptic placentas. Gynecol Obstet Invest. 2011;71:151–7. doi: 10.1159/000315162. [DOI] [PubMed] [Google Scholar]
- 86.Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, et al. Metabolomics and first-trimester prediction of early-onset preeclampsia. J Matern Fetal Neonatal Med. 2012;25:1840–7. doi: 10.3109/14767058.2012.680254. [DOI] [PubMed] [Google Scholar]
- 87.Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, et al. First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2013;208:58–7. doi: 10.1016/j.ajog.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 88.Heazell AE, Brown M, Worton SA, Dunn WB. Review: The effects of oxygen on normal and pre-eclamptic placental tissue--insights from metabolomics. Placenta. 2011;32(Suppl 2):S119–S124. doi: 10.1016/j.placenta.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 89.Kenny LC, Broadhurst DI, Dunn W, Brown M, North RA, McCowan L, et al. Robust early pregnancy prediction of later preeclampsia using metabolomic biomarkers. Hypertension. 2010;56:741–9. doi: 10.1161/HYPERTENSIONAHA.110.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Odibo AO, Zhong Y, Longtine M, Tuuli M, Odibo L, Cahill AG, et al. First-trimester serum analytes, biophysical tests and the association with pathological morphometry in the placenta of pregnancies with preeclampsia and fetal growth restriction. Placenta. 2011;32:333–8. doi: 10.1016/j.placenta.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esplin MS, Merrell K, Goldenberg R, Lai Y, Iams JD, Mercer B, et al. Proteomic identification of serum peptides predicting subsequent spontaneous preterm birth. Am J Obstet Gynecol. 2011;204:391–8. doi: 10.1016/j.ajog.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gercel-Taylor C, Taylor DD, Rai SN. Proteomic identification of serum peptides predicting subsequent spontaneous preterm birth. Am J Obstet Gynecol. 2012;206:e3–e4. doi: 10.1016/j.ajog.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 93.Gracie S, Pennell C, Ekman-Ordeberg G, Lye S, McManaman J, Williams S, et al. An integrated systems biology approach to the study of preterm birth using “-omic” technology--a guideline for research. BMC Pregnancy Childbirth. 2011;11:71. doi: 10.1186/1471-2393-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hitti J, Lapidus JA, Lu X, Reddy AP, Jacob T, Dasari S, et al. Noninvasive diagnosis of intraamniotic infection: proteomic biomarkers in vaginal fluid. Am J Obstet Gynecol. 2010;203:32–8. doi: 10.1016/j.ajog.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mittal P, Romero R, Tarca AL, Gonzalez J, Draghici S, Xu Y, et al. Characterization of the myometrial transcriptome and biological pathways of spontaneous human labor at term. J Perinat Med. 2010;38:617–43. doi: 10.1515/JPM.2010.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pereira L, Reddy AP, Alexander AL, Lu X, Lapidus JA, Gravett MG, et al. Insights into the multifactorial nature of preterm birth: proteomic profiling of the maternal serum glycoproteome and maternal serum peptidome among women in preterm labor. Am J Obstet Gynecol. 2010;202:555–10. doi: 10.1016/j.ajog.2010.02.048. [DOI] [PubMed] [Google Scholar]
- 97.Romero R, Mazaki-Tovi S, Vaisbuch E, Kusanovic JP, Chaiworapongsa T, Gomez R, et al. Metabolomics in premature labor: a novel approach to identify patients at risk for preterm delivery. J Matern Fetal Neonatal Med. 2010;23:1344–59. doi: 10.3109/14767058.2010.482618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stella CL, Bennett MR, Devarajan P, Greis K, Wyder M, Macha S, et al. Preterm labor biomarker discovery in serum using 3 proteomic profiling methodologies. Am J Obstet Gynecol. 2009;201:387–13. doi: 10.1016/j.ajog.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 99.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–7. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 100.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 101.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–40. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 103.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 105.Hsu FD, Nielsen TO, Alkushi A, Dupuis B, Huntsman D, Liu CL, et al. Tissue microarrays are an effective quality assurance tool for diagnostic immunohistochemistry. Mod Pathol. 2002;15:1374–80. doi: 10.1097/01.MP.0000039571.02827.CE. [DOI] [PubMed] [Google Scholar]
- 106.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 107.Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O'Connell JX, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–7. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 108.Perou CM, Jeffrey SS, van de RM, Rees CA, Eisen MB, Ross DT, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96:9212–7. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perou CM, Sorlie T, Eisen MB, van de RM, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 110.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–42. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 111.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 113.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2001;98:1176–81. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goyal R, Yellon SM, Longo LD, Mata-Greenwood E. Placental gene expression in a rat ‘model’ of placental insufficiency. Placenta. 2010;31:568–75. doi: 10.1016/j.placenta.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 115.Hoegh AM, Borup R, Nielsen FC, Sorensen S, Hviid TV. Gene expression profiling of placentas affected by pre-eclampsia. J Biomed Biotechnol. 2010;2010:787545. doi: 10.1155/2010/787545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta. 2007;28:487–97. doi: 10.1016/j.placenta.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 117.Reimer T, Koczan D, Gerber B, Richter D, Thiesen HJ, Friese K. Microarray analysis of differentially expressed genes in placental tissue of pre-eclampsia: up-regulation of obesity-related genes. Mol Hum Reprod. 2002;8:674–80. doi: 10.1093/molehr/8.7.674. [DOI] [PubMed] [Google Scholar]
- 118.Sitras V, Paulssen RH, Gronaas H, Leirvik J, Hanssen TA, Vartun A, et al. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30:424–33. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 119.Zhou R, Zhu Q, Wang Y, Ren Y, Zhang L, Zhou Y. Genomewide oligonucleotide microarray analysis on placentae of pre-eclamptic pregnancies. Gynecol Obstet Invest. 2006;62:108–14. doi: 10.1159/000092857. [DOI] [PubMed] [Google Scholar]
- 120.Achiron A, Grotto I, Balicer R, Magalashvili D, Feldman A, Gurevich M. Microarray analysis identifies altered regulation of nuclear receptor family members in the pre-disease state of multiple sclerosis. Neurobiol Dis. 2010;38:201–9. doi: 10.1016/j.nbd.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 121.Bansard C, Lequerre T, Derambure C, Vittecoq O, Hiron M, Daragon A, et al. Gene profiling predicts rheumatoid arthritis responsiveness to IL-1Ra (anakinra) Rheumatology (Oxford) 2011;50:283–92. doi: 10.1093/rheumatology/keq344. [DOI] [PubMed] [Google Scholar]
- 122.Moore DF, Li H, Jeffries N, Wright V, Cooper RA, Jr, Elkahloun A, et al. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation. 2005;111:212–21. doi: 10.1161/01.CIR.0000152105.79665.C6. [DOI] [PubMed] [Google Scholar]
- 123.Riveros C, Mellor D, Gandhi KS, McKay FC, Cox MB, Berretta R, et al. A transcription factor map as revealed by a genome-wide gene expression analysis of whole-blood mRNA transcriptome in multiple sclerosis. PLoS One. 2010;5:e14176. doi: 10.1371/journal.pone.0014176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tang BM, McLean AS, Dawes IW, Huang SJ, Cowley MJ, Lin RC. Gene-expression profiling of gram-positive and gram-negative sepsis in critically ill patients. Crit Care Med. 2008;36:1125–8. doi: 10.1097/CCM.0b013e3181692c0b. [DOI] [PubMed] [Google Scholar]
- 125.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med. 2009;37:882–8. doi: 10.1097/CCM.0b013e31819b52fd. [DOI] [PubMed] [Google Scholar]
- 126.Tang Y, Xu H, Du X, Lit L, Walker W, Lu A, et al. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab. 2006;26:1089–102. doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
- 127.van Baarsen LG, Bos WH, Rustenburg F, van der Pouw Kraan TC, Wolbink GJ, Dijkmans BA, et al. Gene expression profiling in autoantibody-positive patients with arthralgia predicts development of arthritis. Arthritis Rheum. 2010;62:694–704. doi: 10.1002/art.27294. [DOI] [PubMed] [Google Scholar]
- 128.ACOG practice bulletin. Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. Obstet Gynecol. 2002;99:159–67. doi: 10.1016/s0029-7844(01)01747-1. [DOI] [PubMed] [Google Scholar]
- 129.Vatten LJ, Skjaerven R. Is pre-eclampsia more than one disease? BJOG. 2004;111:298–302. doi: 10.1111/j.1471-0528.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 130.von Dadelszen P, Magee LA, Roberts JM. Subclassification of preeclampsia. Hypertens Pregnancy. 2003;22:143–8. doi: 10.1081/PRG-120021060. [DOI] [PubMed] [Google Scholar]
- 131.Alexander GR, Himes JH, Kaufman RB, Mor J, Kogan M. A United States national reference for fetal growth. Obstet Gynecol. 1996;87:163–8. doi: 10.1016/0029-7844(95)00386-X. [DOI] [PubMed] [Google Scholar]
- 132.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Albrecht C, Viturro E. The ABCA subfamily--gene and protein structures, functions and associated hereditary diseases. Pflugers Arch. 2007;453:581–9. doi: 10.1007/s00424-006-0047-8. [DOI] [PubMed] [Google Scholar]
- 134.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 135.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Goshen R, Rachmilewitz J, Schneider T, de Groot N, Ariel I, Palti Z, et al. The expression of the H-19 and IGF-2 genes during human embryogenesis and placental development. Mol Reprod Dev. 1993;34:374–9. doi: 10.1002/mrd.1080340405. [DOI] [PubMed] [Google Scholar]
- 137.Jinno Y, Ikeda Y, Yun K, Maw M, Masuzaki H, Fukuda H, et al. Establishment of functional imprinting of the H19 gene in human developing placentae. Nat Genet. 1995;10:318–24. doi: 10.1038/ng0795-318. [DOI] [PubMed] [Google Scholar]
- 138.Yu L, Chen M, Zhao D, Yi P, Lu L, Han J, et al. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta. 2009;30:443–7. doi: 10.1016/j.placenta.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 139.Esquiliano DR, Guo W, Liang L, Dikkes P, Lopez MF. Placental glycogen stores are increased in mice with H19 null mutations but not in those with insulin or IGF type 1 receptor mutations. Placenta. 2009;30:693–9. doi: 10.1016/j.placenta.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bliek J, Terhal P, van den Bogaard MJ, Maas S, Hamel B, Salieb-Beugelaar G, et al. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am J Hum Genet. 2006;78:604–14. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Eggermann T, Begemann M, Spengler S, Schroder C, Kordass U, Binder G. Genetic and epigenetic findings in Silver-Russell syndrome. Pediatr Endocrinol Rev. 2010;8:86–93. [PubMed] [Google Scholar]
- 142.Lin SY, Lee CN, Hung CC, Tsai WY, Lin SP, Li NC, et al. Epigenetic profiling of the H19 differentially methylated region and comprehensive whole genome array-based analysis in Silver-Russell syndrome. Am J Med Genet A. 2010;152A:2521–8. doi: 10.1002/ajmg.a.33629. [DOI] [PubMed] [Google Scholar]
- 143.Choufani S, Shuman C, Weksberg R. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:343–54. doi: 10.1002/ajmg.c.30267. [DOI] [PubMed] [Google Scholar]
- 144.Cohen MM., Jr Beckwith-Wiedemann syndrome: historical, clinicopathological, and etiopathogenetic perspectives. Pediatr Dev Pathol. 2005;8:287–304. doi: 10.1007/s10024-005-1154-9. [DOI] [PubMed] [Google Scholar]
- 145.Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, et al. Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2005;13:1025–32. doi: 10.1038/sj.ejhg.5201463. [DOI] [PubMed] [Google Scholar]
- 146.Koukoura O, Sifakis S, Zaravinos A, Apostolidou S, Jones A, Hajiioannou J, et al. Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction. Placenta. 2011;32:51–7. doi: 10.1016/j.placenta.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 147.Gao WL, Li D, Xiao ZX, Liao QP, Yang HX, Li YX, et al. Detection of global DNA methylation and paternally imprinted H19 gene methylation in preeclamptic placentas. Hypertens Res. 2011;34:655–61. doi: 10.1038/hr.2011.9. [DOI] [PubMed] [Google Scholar]
- 148.Gao WL, Liu M, Yang Y, Yang H, Liao Q, Bai Y, et al. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1) RNA Biol. 2012;9 doi: 10.4161/rna.20807. [DOI] [PubMed] [Google Scholar]
- 149.Matouk IJ, Mezan S, Mizrahi A, Ohana P, bu-Lail R, Fellig Y, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–51. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 150.Helmy KY, Katschke KJ, Jr, Gorgani NN, Kljavin NM, Elliott JM, Diehl L, et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124:915–27. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 151.Haeger M, Unander M, Bengtsson A. Complement activation in relation to development of preeclampsia. Obstet Gynecol. 1991;78:46–9. [PubMed] [Google Scholar]
- 152.Soto E, Romero R, Richani K, Espinoza J, Chaiworapongsa T, Nien JK, et al. Preeclampsia and pregnancies with small-for-gestational age neonates have different profiles of complement split products. J Matern Fetal Neonatal Med. 2010;23:646–57. doi: 10.3109/14767050903301009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.He JQ, Wiesmann C, van Lookeren CM. A role of macrophage complement receptor CRIg in immune clearance and inflammation. Mol Immunol. 2008;45:4041–7. doi: 10.1016/j.molimm.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 154.Vogt L, Schmitz N, Kurrer MO, Bauer M, Hinton HI, Behnke S, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116:2817–26. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fang X, Zheng P, Tang J, Liu Y. CD24: from A to Z. Cell Mol Immunol. 2010;7:100–3. doi: 10.1038/cmi.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hunte BE, Capone M, Zlotnik A, Rennick D, Moore TA. Acquisition of CD24 expression by Lin-CD43+B220(low)ckit(hi) cells coincides with commitment to the B cell lineage. Eur J Immunol. 1998;28:3850–6. doi: 10.1002/(SICI)1521-4141(199811)28:11<3850::AID-IMMU3850>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 157.Liu Y, Zheng P. CD24: a genetic checkpoint in T cell homeostasis and autoimmune diseases. Trends Immunol. 2007;28:315–20. doi: 10.1016/j.it.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 158.Zhou Q, Rammohan K, Lin S, Robinson N, Li O, Liu X, et al. CD24 is a genetic modifier for risk and progression of multiple sclerosis. Proc Natl Acad Sci U S A. 2003;100:15041–6. doi: 10.1073/pnas.2533866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sanchez E, Abelson AK, Sabio JM, Gonzalez-Gay MA, Ortego-Centeno N, Jimenez-Alonso J, et al. Association of a CD24 gene polymorphism with susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2007;56:3080–6. doi: 10.1002/art.22871. [DOI] [PubMed] [Google Scholar]
- 160.Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 2009;323:1722–5. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nien JK, Mazaki-Tovi S, Romero R, Erez O, Kusanovic JP, Gotsch F, et al. Adiponectin in severe preeclampsia. J Perinat Med. 2007;35:503–12. doi: 10.1515/JPM.2007.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.McDonald EA, Wolfe MW. The Pro-Inflammatory Role of Adiponectin at the Maternal-Fetal Interface. Am J Reprod Immunol. 2011 doi: 10.1111/j.1600-0897.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 163.Hayashi M, Abe-Dohmae S, Okazaki M, Ueda K, Yokoyama S. Heterogeneity of high density lipoprotein generated by ABCA1 and ABCA7. J Lipid Res. 2005;46:1703–11. doi: 10.1194/jlr.M500092-JLR200. [DOI] [PubMed] [Google Scholar]
- 164.Kaminski WE, Orso E, Diederich W, Klucken J, Drobnik W, Schmitz G. Identification of a novel human sterol-sensitive ATP-binding cassette transporter (ABCA7) Biochem Biophys Res Commun. 2000;273:532–8. doi: 10.1006/bbrc.2000.2954. [DOI] [PubMed] [Google Scholar]
- 165.Linsel-Nitschke P, Jehle AW, Shan J, Cao G, Bacic D, Lan D, et al. Potential role of ABCA7 in cellular lipid efflux to apoA-I. J Lipid Res. 2005;46:86–92. doi: 10.1194/jlr.M400247-JLR200. [DOI] [PubMed] [Google Scholar]
- 166.Nagata K, Yamamoto A, Ban N, Tanaka AR, Matsuo M, Kioka N, et al. Human ABCA3, a product of a responsible gene for abca3 for fatal surfactant deficiency in newborns, exhibits unique ATP hydrolysis activity and generates intracellular multilamellar vesicles. Biochem Biophys Res Commun. 2004;324:262–8. doi: 10.1016/j.bbrc.2004.09.043. [DOI] [PubMed] [Google Scholar]
- 167.Takahashi K, Kimura Y, Nagata K, Yamamoto A, Matsuo M, Ueda K. ABC proteins: key molecules for lipid homeostasis. Med Mol Morphol. 2005;38:2–12. doi: 10.1007/s00795-004-0278-8. [DOI] [PubMed] [Google Scholar]
- 168.Tanaka Y, Yamada K, Zhou CJ, Ban N, Shioda S, Inagaki N. Temporal and spatial profiles of ABCA2-expressing oligodendrocytes in the developing rat brain. J Comp Neurol. 2003;455:353–67. doi: 10.1002/cne.10493. [DOI] [PubMed] [Google Scholar]
- 169.Wang N, Lan D, Gerbod-Giannone M, Linsel-Nitschke P, Jehle AW, Chen W, et al. ATP-binding cassette transporter A7 (ABCA7) binds apolipoprotein A-I and mediates cellular phospholipid but not cholesterol efflux. J Biol Chem. 2003;278:42906–12. doi: 10.1074/jbc.M307831200. [DOI] [PubMed] [Google Scholar]
- 170.Zhou C, Zhao L, Inagaki N, Guan J, Nakajo S, Hirabayashi T, et al. Atp-binding cassette transporter ABC2/ABCA2 in the rat brain: a novel mammalian lysosome-associated membrane protein and a specific marker for oligodendrocytes but not for myelin sheaths. J Neurosci. 2001;21:849–57. doi: 10.1523/JNEUROSCI.21-03-00849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vulevic B, Chen Z, Boyd JT, Davis W, Jr, Walsh ES, Belinsky MG, et al. Cloning and characterization of human adenosine 5′-triphosphate-binding cassette, sub-family A, transporter 2 (ABCA2) Cancer Res. 2001;61:3339–47. [PubMed] [Google Scholar]
- 172.Chen ZJ, Vulevic B, Ile KE, Soulika A, Davis W, Jr, Reiner PB, et al. Association of ABCA2 expression with determinants of Alzheimer's disease. FASEB J. 2004;18:1129–31. doi: 10.1096/fj.03-1490fje. [DOI] [PubMed] [Google Scholar]
- 173.Mace S, Cousin E, Ricard S, Genin E, Spanakis E, Lafargue-Soubigou C, et al. ABCA2 is a strong genetic risk factor for early-onset Alzheimer's disease. Neurobiol Dis. 2005;18:119–25. doi: 10.1016/j.nbd.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 174.Yamano G, Funahashi H, Kawanami O, Zhao LX, Ban N, Uchida Y, et al. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett. 2001;508:221–5. doi: 10.1016/s0014-5793(01)03056-3. [DOI] [PubMed] [Google Scholar]
- 175.Asare AL, Kolchinsky SA, Gao Z, Wang R, Raddassi K, Bourcier K, et al. Differential gene expression profiles are dependent upon method of peripheral blood collection and RNA isolation. BMC Genomics. 2008;9:474. doi: 10.1186/1471-2164-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prades C, Arnould I, Annilo T, Shulenin S, Chen ZQ, Orosco L, et al. The human ATP binding cassette gene ABCA13, located on chromosome 7p12.3, encodes a 5058 amino acid protein with an extracellular domain encoded in part by a 4.8-kb conserved exon. Cytogenet Genome Res. 2002;98:160–8. doi: 10.1159/000069852. [DOI] [PubMed] [Google Scholar]
- 177.Barros SA, Tennant RW, Cannon RE. Molecular structure and characterization of a novel murine ABC transporter, Abca13. Gene. 2003;307:191–200. doi: 10.1016/s0378-1119(03)00465-7. [DOI] [PubMed] [Google Scholar]
- 178.Bhat MA, Rios JC, Lu Y, Garcia-Fresco GP, Ching W, St Martin M, et al. Axon-glia interactions and the domain organization of myelinated axons requires neurexin IV/Caspr/Paranodin. Neuron. 2001;30:369–83. doi: 10.1016/s0896-6273(01)00294-x. [DOI] [PubMed] [Google Scholar]
- 179.Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, et al. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24:1037–47. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 180.Rios JC, Melendez-Vasquez CV, Einheber S, Lustig M, Grumet M, Hemperly J, et al. Contactin-associated protein (Caspr) and contactin form a complex that is targeted to the paranodal junctions during myelination. J Neurosci. 2000;20:8354–64. doi: 10.1523/JNEUROSCI.20-22-08354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Spiegel I, Salomon D, Erne B, Schaeren-Wiemers N, Peles E. Caspr3 and caspr4, two novel members of the caspr family are expressed in the nervous system and interact with PDZ domains. Mol Cell Neurosci. 2002;20:283–97. doi: 10.1006/mcne.2002.1110. [DOI] [PubMed] [Google Scholar]
- 182.Traut W, Weichenhan D, Himmelbauer H, Winking H. New members of the neurexin superfamily: multiple rodent homologues of the human CASPR5 gene. Mamm Genome. 2006;17:723–31. doi: 10.1007/s00335-005-0157-1. [DOI] [PubMed] [Google Scholar]
- 183.Bangsow T, Baumann E, Bangsow C, Jaeger MH, Pelzer B, Gruhn P, et al. The epithelial membrane protein 1 is a novel tight junction protein of the blood-brain barrier. J Cereb Blood Flow Metab. 2008;28:1249–60. doi: 10.1038/jcbfm.2008.19. [DOI] [PubMed] [Google Scholar]
- 184.Lee HS, Sherley JL, Chen JJ, Chiu CC, Chiou LL, Liang JD, et al. EMP-1 is a junctional protein in a liver stem cell line and in the liver. Biochem Biophys Res Commun. 2005;334:996–1003. doi: 10.1016/j.bbrc.2005.06.194. [DOI] [PubMed] [Google Scholar]
- 185.Jain A, Tindell CA, Laux I, Hunter JB, Curran J, Galkin A, et al. Epithelial membrane protein-1 is a biomarker of gefitinib resistance. Proc Natl Acad Sci U S A. 2005;102:11858–63. doi: 10.1073/pnas.0502113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li YQ, Xue T, Wang L, Xu ZC, Xi ZQ, Yuan J, et al. Up-regulation of epithelial membrane protein-1 in the temporal neocortex of patients with intractable epilepsy. Neurochem Res. 2009;34:1594–602. doi: 10.1007/s11064-009-9948-1. [DOI] [PubMed] [Google Scholar]
- 187.Gant NF, Daley GL, Chand S, Whalley PJ, MacDonald PC. A study of angiotensin II pressor response throughout primigravid pregnancy. J Clin Invest. 1973;52:2682–9. doi: 10.1172/JCI107462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gant NF, Chand S, Worley RJ, Whalley PJ, Crosby UD, MacDonald PC. A clinical test useful for predicting the development of acute hypertension in pregnancy. Am J Obstet Gynecol. 1974;120:1–7. doi: 10.1016/0002-9378(74)90170-7. [DOI] [PubMed] [Google Scholar]
- 189.Gardiner EE, De Luca M, McNally T, Michelson AD, Andrews RK, Berndt MC. Regulation of P-selectin binding to the neutrophil P-selectin counter-receptor P-selectin glycoprotein ligand-1 by neutrophil elastase and cathepsin G. Blood. 2001;98:1440–7. doi: 10.1182/blood.v98.5.1440. [DOI] [PubMed] [Google Scholar]
- 190.Le-Barillec K, Pidard D, Balloy V, Chignard M. Human neutrophil cathepsin G down-regulates LPS-mediated monocyte activation through CD14 proteolysis. J Leukoc Biol. 2000;68:209–15. [PubMed] [Google Scholar]
- 191.Tralau T, Meyer-Hoffert U, Schroder JM, Wiedow O. Human leukocyte elastase and cathepsin G are specific inhibitors of C5a-dependent neutrophil enzyme release and chemotaxis. Exp Dermatol. 2004;13:316–25. doi: 10.1111/j.0906-6705.2004.00145.x. [DOI] [PubMed] [Google Scholar]
- 192.Roberts JM. Endothelial dysfunction in preeclampsia. Semin Reprod Endocrinol. 1998;16:5–15. doi: 10.1055/s-2007-1016248. [DOI] [PubMed] [Google Scholar]
- 193.Williams MR, Sakurai Y, Zughaier SM, Eskin SG, McIntire LV. Transmigration across activated endothelium induces transcriptional changes, inhibits apoptosis, and decreases antimicrobial protein expression in human monocytes. J Leukoc Biol. 2009;86:1331–43. doi: 10.1189/jlb.0209062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sun CJ, Zhang L, Zhang WY. Gene expression profiling of maternal blood in early onset severe preeclampsia: identification of novel biomarkers. J Perinat Med. 2009;37:609–16. doi: 10.1515/JPM.2009.103. [DOI] [PubMed] [Google Scholar]
- 195.Dahlstrom B, Esbensen Y, Vollan H, Oian P, Bukholm G. Genome profiles in maternal blood during early onset preeclampsia and towards term. J Perinat Med. 2010;38:601–8. doi: 10.1515/jpm.2010.095. [DOI] [PubMed] [Google Scholar]
- 196.Enquobahrie DA, Qiu C, Muhie SY, Williams MA. Maternal peripheral blood gene expression in early pregnancy and preeclampsia. Int J Mol Epidemiol Genet. 2011;2:78–94. [PMC free article] [PubMed] [Google Scholar]
- 197.Kruhoffer M, Dyrskjot L, Voss T, Lindberg RL, Wyrich R, Thykjaer T, et al. Isolation of microarray-grade total RNA, microRNA, and DNA from a single PAXgene blood RNA tube. J Mol Diagn. 2007;9:452–8. doi: 10.2353/jmoldx.2007.060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vartanian K, Slottke R, Johnstone T, Casale A, Planck SR, Choi D, et al. Gene expression profiling of whole blood: comparison of target preparation methods for accurate and reproducible microarray analysis. BMC Genomics. 2009;10:2. doi: 10.1186/1471-2164-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Feezor RJ, Baker HV, Mindrinos M, Hayden D, Tannahill CL, Brownstein BH, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–54. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 200.Liu J, Walter E, Stenger D, Thach D. Effects of globin mRNA reduction methods on gene expression profiles from whole blood. J Mol Diagn. 2006;8:551–8. doi: 10.2353/jmoldx.2006.060021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Okazaki S, Sekizawa A, Purwosunu Y, Iwasaki M, Farina A, Okai T. Measurement of mRNA of trophoblast-specific genes in cellular and plasma components of maternal blood. J Med Genet. 2006;43:e47. doi: 10.1136/jmg.2005.040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Okazaki S, Sekizawa A, Purwosunu Y, Farina A, Wibowo N, Okai T. Placenta-derived, cellular messenger RNA expression in the maternal blood of preeclamptic women. Obstet Gynecol. 2007;110:1130–6. doi: 10.1097/01.AOG.0000286761.11436.67. [DOI] [PubMed] [Google Scholar]
- 203.Heung MM, Jin S, Tsui NB, Ding C, Leung TY, Lau TK, et al. Placenta-derived fetal specific mRNA is more readily detectable in maternal plasma than in whole blood. PLoS One. 2009;4:e5858. doi: 10.1371/journal.pone.0005858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.