Abstract
The Abl interactor 1 (Abi-1) protein has been implicated in the regulation of actin dynamics and localizes to the tips of lamellipodia and filopodia. Here, we show that Abi-1 binds the actin nucleator protein Wave-1 through an amino-terminal Wave-binding (WAB) domain and that disruption of the Abi-1-Wave-1 interaction prevents Abi-1 from reaching the tip of the lamellipodium. Abi-1 binds to the Wave homology domain of Wave-1, a region that is required for translocation of Wave-1 to the lamellipodium. Mouse embryo fibroblasts that lack one allele of Abi-1 and are homozygous null for the related Abi-2 protein exhibit decreased Wave-1 protein levels. This phenotype is rescued by Abi-1 proteins that retain Wave-1 binding but not by Abi-1 mutants that cannot bind to Wave-1. Moreover, we uncovered an overlapping SNARE domain in the amino terminus of Abi-1 that interacts with Syntaxin-1, a SNARE family member. Further, we demonstrated that Abi-1 shuttles in and out of the nucleus in a leptomycin B (LMB)-dependent manner and that complete nuclear translocation of Abi-1 in the absence of LMB requires the combined inactivation of the SNARE, WAB, and SH3 domains of Abi-1. Thus, Abi-1 undergoes nucleocytoplasmic shuttling and functions at the leading edge to regulate Wave-1 localization and protein levels.
Actin polymerization at the leading edge of a moving cell is a highly regulated process that provides the force required for cell movement. Actin polymerization occurs in response to a variety of stimuli at multiple subcellular sites, including the leading edge of the lamellipodium and sites adjacent to moving vesicles (28, 38). Much progress has been achieved over the last few years regarding the identification of signaling complexes that link extracellular stimuli to actin reorganization (38, 60). However, the mechanisms that regulate the spatial and temporal localization of actin-regulatory proteins remain to be defined.
Regulation of actin dynamics in response to extracellular stimuli is mediated in part by the Rac and Cdc42 GTPases (14). Among multiple Rac and Cdc42 effectors are the Wasp/Wave family of proteins that link GTPase activation to Arp2/3 complex-mediated actin nucleation (52). Wave subfamily members (Wave-1, Wave-2, and Wave-3) function to regulate actin dynamics in response to Rac activation (52). Unlike the related Wasp family proteins that are activated directly by binding to activated Cdc42, Wave proteins lack binding sites for activated Rac and are thought to be under tight negative regulation in cells (7, 38). In vitro, purified Wave-1 can stimulate the actin nucleation activity of the Arp2/3 complex (26). The activity of Wave-1 has been shown to be regulated in vitro by a multiprotein complex (12). This complex includes Nap-1, Pir121, HSPC300, and Abl interactor 2 (Abi-2) (12). Studies with purified proteins showed that in response to Rac activation, this complex dissociates, relieving Wave-1 inhibition, which results in Wave-1-dependent actin polymerization through the Arp2/3 complex (12). However, recent studies with cultured cells showed that the association of Nap-1, Pir121, Abi-1, and Wave-2 is insensitive to activated Rac (45).
A role for Wave proteins in the regulation of actin dynamics in vivo has been demonstrated in several organisms. Genetic elimination of Scar/Wave in Dictyostelium discoideum affects macropynocitosis, exocytosis, and endosome traffic (41). Disruption of Wave-2 in mice results in embryonic lethality, cell motility defects, malformation of the ventricles in the developing brain (65), and aberrant lamellipodium formation and sprouting of endothelial cells (64). Genetic elimination of Wave-1 in mice induced sensorimotor and cognitive deficits in one study (44). In a second study, a retroviral gene trap approach produced a disruption of Wave-1 that yielded postnatal lethality, severe limb weakness, a resting tremor, and neuroanatomical malformations (10). Wave-1 deficiency inhibits dorsal ruffle formation in response to platelet-derived growth factor stimulation of mouse embryo fibroblasts (MEFs), while Wave-2 deficiency impairs peripheral ruffle formation under the same conditions (49).
Wave proteins have been reported to localize to the lamellipodium, filopodia, and focal adhesions (20, 32, 61). The carboxy-terminal half of Wave-1 binds to actin, and the Arp2/3 complex and is conserved in the related Wasp/N-Wasp proteins. In contrast, the amino-terminal region contains the Wave homology domain (WHD) that is specific for the Wave subfamily (29, 52). The WHD of Wave-1 was shown to be sufficient for translocation of Wave-1 to the leading edge of the lamellipodium (32). In a similar study, Wave-2 amino acids 1 to 83 were sufficient for translocating this protein to the tip of the filopodia (33). No binding partner for the WHD has been identified to date.
The Abi proteins were originally identified as substrates and binding partners for the Abl tyrosine kinases (11, 42, 57), a family of proteins that bind actin and regulate actin dynamics (36). Subsequently, Abi proteins were shown to interact with the adaptor protein Eps8, Spectrin, the Sos guanine nucleotide exchange factor, and Grb4 (2, 9, 15, 40, 67). Abi proteins are conserved throughout evolution. One Abi gene has been identified in Drosophila melanogaster, Caenorhabditis elegans, and D. discoideum, and three Abi-related genes, Abi-1, Abi-2, and NESH (11, 31, 42), are present in humans and mice. Abi-2 was found in a multiprotein complex regulated by the Rac GTPase; this complex includes the Abi binding protein Nap-1 (63), HSPC300, Pir121, and Wave-1 (12). Abi-1 and Abi-2 were also shown to copurify in a protein complex containing SNIP (SNAP-25 interacting protein), α-tubulin, spectrins αII and βIII, WRP, and Wave-1 (43). Recent studies have shown that downregulation of Abi in Drosophila S2R+ cells by RNA interference (RNAi) treatment induces morphological changes that are similar to those induced by downregulation of Wave/Scar (24). This phenotype is due in part to the reduced Scar/Wave levels in cells treated with RNAi for Abi (24, 39). Although these data link Abi and Wave genetically and biochemically, it is not known whether these proteins interact directly or whether they colocalize in cells. Other studies have shown that Abi-1 regulates Rac in a complex with Eps8 and Sos-1 and that the formation of this complex stimulates the RacGEF activity of Sos-1 (40). Thus, Abi proteins play a role in actin reorganization through regulation of Rac-dependent pathways.
We have previously shown that Abi-1 and Abi-2 localize to the leading edge of the lamellipodium and filopodia in the highly motile B16 melanoma cell line (46). The localization pattern of Abi-1 is different from those of other actin-regulatory proteins such as Vasp or profilin, but it is very similar to that of Wave proteins (20, 46). The localization of Abi-1 to the leading edge of the lamellipodium is dependent on its amino-terminal 145 amino acids (46). The proteins that bind to this conserved Abi region remain to be identified. Analysis of the amino acid sequences of Abi proteins across species revealed that the middle portion of the molecule is highly variable among Abi family members, while the amino- and carboxy-terminal regions are highly conserved. The carboxy terminus of Abi-1 contains an SH3 domain, and the amino terminus contains a homeodomain homologous region (HHR) and a putative SNARE domain (11, 46). SNARE domains were first characterized in SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor) proteins such as Syntaxin-1, SNAP-25, and VAMP-2 (6). SNARE proteins are involved in the fusion of a vesicle with its target membrane (6). Syntaxin-1 plays a role in the fusion between neurotransmitter-filled exocytic vesicles and the presynaptic membrane by entering in a complex with SNAP-25 and VAMP-2 (6). These three molecules form the neuronal SNARE complex (50). Syntaxin-1 may regulate additional cellular processes as it interacts with actin (53), Munc18 (21), Tomosyn (19), ion channels (66), and CDCRel (1).
Here we show that two proteins, Wave-1 and Syntaxin-1, interact with the conserved amino terminus of Abi-1 and bind to different but overlapping regions in the Abi-1 amino terminus. Localization of Abi-1 to the tip of the lamellipodium is dependent on an intact Wave-binding (WAB) domain in the Abi-1 amino terminus. Wave-1 binds to Abi-1 through a region within the Wave-1 WHD, and mutation of this region prevents Wave-1 from reaching the tip of the lamellipodium. Cells deficient in Abi-2 and expressing only one allele of Abi-1 have decreased Wave-1 protein levels that can be rescued by Abi-1 proteins that interact with Wave-1 and localize to the tip of the lamellipodium. We show that Abi-1 not only localizes to sites of actin polymerization but also shuttles in and out of the nucleus in a leptomycin B (LMB)-dependent manner. Nuclear accumulation of Abi-1 is regulated by multiple Abi-1 protein interaction domains that include the SNARE, WAB, and SH3 domains. These results show that the localization and function of Abi-1 are controlled by interactions with distinct proteins that bind to amino- and carboxy-terminal domains conserved among Abi family members.
MATERIALS AND METHODS
Cells and reagents.
293T cells and MEFs were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. MEFs were derived from wild-type mice and mice that were homozygous null for Abi-2 and heterozygous for Abi-1 (M. Grove and A. M. Pendergast, unpublished results). The generation and characterization of the Abi knockout mice will be reported in a separate paper. NIH 3T3 cells were cultured in Dulbecco modified Eagle medium with 10% calf serum. Monoclonal antibodies used were anti-Myc 9E10 (Santa Cruz Biotechnology, Santa Cruz, Calif.), anti-Flag M2 (Sigma, St. Louis, Mo.), anti-green fluorescent protein (GFP) (Roche, Indianapolis, Ind.), β-tubulin (Sigma), and Wave-1 (Transduction Laboratories, Lexington, Ky.). Polyclonal antibodies used were anti-Abi-1 6987 (8) and Sin3A (Santa Cruz Biotechnology). Monoclonal antibodies against Syntaxin-1, SNAP-25, and VAMP-2 were a gift from G. J. Augustine (Duke University Medical Center, Durham, N.C.) (54). Wave-1 polyclonal antibody was a gift from J. D. Scott (Oregon Health Sciences University, Portland, Oreg.) (61). Leptomycin B (kindly provided by M. Yoshida, Chemical Genetics Laboratory, RIKEN, Saitama, Japan) was used at 20 nM for 6 h.
Plasmids.
Flag-tagged Abi-1 wild-type and mutant cDNAs were amplified by PCR and cloned into the pCMV2Flag vector as follows. Human Abi-1 (8) was cloned into NotI-KpnI sites. Abi-1-1-145 and Abi-1ΔSH3 (amino acids 1 to 379) were introduced into the EcoRI site. Abi-1Δ1-145, Abi-1SNARE (amino acids 41 to 117), Abi-1ΔSNARE (lacking amino acids 44 to 111), and Abi-1Δ1-55 were inserted into EcoRI-KpnI. Abi-1Δ56-65 was introduced into EcoRI-KpnI, and the BglII site was introduced into the deleted region, which added one amino acid (serine). Abi-1Δ65-79 was cloned into EcoRI-KpnI, and the BglII site was introduced in the deleted region, which added two amino acids (serine and threonine). Abi-1Δ2-17, Abi-1Δ3-32, and Abi-1Δ18-32 were introduced into BglII-KpnI sites. All Flag-tagged Abi-1 mutants were also subcloned into pEYFP EcoRI-KpnI sites. The constructs pEYFPAbi-1, pEYFPAbi-1Δpro, pEYFPAbi-1ΔHHR, and pEGFPAbi-1ΔSH3 have previously been described (46). pEYFPAbi-1Δ361-383, pEYFPAbi-1Δ275-330, pEYFPAbi-1Δ212-276, pEYFPAbi-1Δ145-210, pEYFPAbi-1Δ277-446, pEYFPAbi-1WP (W421K and P434L point mutations were introduced into the 3′ oligonucleotide), and pEYFPAbi-1Δ65-79WP were cloned into EcoRI-KpnI sites. PEYFPAbi-1Δ1-276 was cloned into SpeI-XbaI. PEYFPAbi-1ΔSNARE/ΔSH3 was cloned into EcoRI-BamHI, and a random tail of amino acids (PDLDN) was added at the C terminus. Flag-Abi-1 was cloned into the MIGR1 EcoRI site. Abi-1, Abi-1ΔSNARE, Abi-1Δ3-32, Abi-1Δ18-32, Abi-1Δ56-65, and Abi-1Δ65-79 were also subcloned into retroviral vector pLEGFPC1 in frame with EGFP with BglII (Abi-1) or XhoI-ApaI. Abi-1-1-276 was subcloned into pLEGFPC1 XhoI-ApaI sites. PCDNA3.1Abi-1 and pCDNA3.1Abi-1ΔSNARE (both under T7 RNA polymerase and cytomegalovirus [CMV] promoters) were cloned into EcoRI and EcoRI-XbaI sites, respectively. The cDNAs for wild-type mouse Wave-1 and mutant forms were amplified by PCR and inserted into the pCMV/Myc vector (Clontech.com) digested with EcoRI-BglII (mutant forms) or EcoRI (wild type). Wave-1ΔV lacks amino acids 498 to 513, and Wave-1ΔProR lacks amino acids 275 to 435. Myc-Wave and Myc-WaveΔ34-92 were cloned into the MIGR1 blunted EcoR1 site. Wave-1NT (amino acids 1 to 277) was subcloned in frame with the glutathione S-transferase (GST) sequence into pGEXKG EcoRI-XhoI sites. Myc-tagged full-length VAMP-2 was cloned into pCMV/Myc EcoRI sites. All PCR products were sequenced to confirm the absence of unwanted mutations. Partial VAMP-2 cDNA was a gift of R. H. Scheller (Stanford University, Stanford, Calif.). PCDNA3.1 Syntaxin-1A was a gift of Z. H. Sheng (National Institutes of Health, Bethesda, Md.) (25). PGEXKG-Syntaxin-1 (lacking the transmembrane region) was a gift of G. Augustine (Duke University Medical Center). PCDNA3 SNAP-25 was a gift of H. Y. Gaisano (University of Toronto, Toronto, Canada) (22).
Infection and transfection of cells.
Retroviral supernatants were used to infect NIH 3T3 cells or MEFs and were generated in 293T cells cotransfected with bicistronic and GFP-expressing MIGR1- or pLEGFPC1-based vectors and pSVψ2 (37). GFP-positive cells were sorted by fluorescence-activated cell sorting. NIH 3T3 cells were transfected with Lipofectamine 2000 (Invitrogene).
Immunoprecipitation and cell fractionation.
Immunoprecipitations were carried out with kinase lysis buffer (KLB) containing 150 mM NaCl, 10 mM sodium phosphate (pH 7), and 1% Triton X-100 plus protease and phosphatase inhibitors. For immunoprecipitation of endogenous proteins from NG108-15 cells, 900 μg of total lysate in KLB was used, and 350 μg of total lysate was used for immunoprecipitation of overexpressed proteins. Immunoprecipitations with mouse brain extract (900 μg) were performed as described previously (34). For nuclear and cytoplasmic fractionation, cells were lysed in CE buffer (10 mM HEPES [pH 7.6], 60 mM KCl, 1 mM EDTA, 0.3% NP-40, 1 mM dithiothreitol) plus protease and phosphatase inhibitors and centrifuged at 400 × g for 5 min. Supernatant was centrifuged at 16,000 × g for 10 min to obtain the cytoplasmic fraction. The nuclei were washed extensively with CE buffer without detergent, lysed by vortexing for 10 min in NE buffer (20 mM Tris-HCl [pH 8], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol) plus protease and phosphatase inhibitors, and centrifuged at 16,000 × g for 10 min.
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, permeabilized in 0.2% Triton X-100 in PBS for 5 min, blocked with 10% normal donkey serum in PBS, and incubated with primary antibodies (overnight at 4°C) and fluorescein isothiocyanate- or rhodamine-conjugated secondary antibodies (1 h at room temperature). Fibronectin was used at 30 μg/ml for wild-type or Abi-2−/− Abi-1−/+ MEFs, and poly-L-lysine at 10 μg/ml for NIH 3T3 cells. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) at 0.0008% (wt/vol) for 2 min, and actin was stained with Alexa fluor 633 Phalloidin as indicated by the manufacturer (Molecular Probes, Eugene, Oreg.). Cells were visualized with an epifluorescence or confocal microscope (Carl Zeiss).
GST pull-down.
pGEXKGWave-1NT was introduced into Escherichia coli BL21-Codonplus (DE3)-RIL (Stratagene.com), and pGEXKGSyntaxin-1 and pGEXKG were introduced into E. coli BL21. Protein induction and purification were performed as described previously (5) except that the cell extracts were solubilized in 1% Triton X-100 and centrifuged once for 12 min. Abi-1 and Abi-1ΔSNARE were synthesized in vitro and labeled with [35S]methionine by using TNT wheat germ extract as indicated by the manufacturer (Promega.com). Abi-1 proteins were diluted 1:4 in incubation buffer (20 mM HEPES [pH 7], 150 mM NaCl, 0.1% [vol/vol] Triton X-100, 10% [vol/vol] glycerol) plus protease and phosphatase inhibitors and incubated for 1.5 h at 4°C with 6 μg of GST fusion proteins. Beads were washed extensively, and bound proteins were resolved with sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
RESULTS
The amino terminus of Abi-1 binds to Syntaxin-1.
We previously identified a region within the amino terminus of Abi proteins that shares significant homology with the SNARE domain (46). The crystal structure of the neuronal SNARE complex reveals that four SNARE domains contributed by Syntaxin-1, SNAP-25 (contributing two domains), and VAMP-2 form a parallel four-helix bundle with conserved hydrophobic amino acids making contacts in the interior of the bundle and an ionic layer in the center of the complex formed by three glutamines and one arginine (17, 50). SNARE proteins can be classified into Q-SNAREs and R-SNAREs. Q-SNAREs are present in the target membrane and contain a glutamine in the central, ionic layer of the SNARE motif, while R-SNAREs contain an arginine and localize to vesicles (17). The Q-SNARE motif of Syntaxin and SNAP-25 family proteins contains conserved layers of interacting amino acid side chains. Interestingly, 13 out of the conserved 15 amino acids are conserved in Abi family proteins (Fig. 1). The glutamine in the central, ionic layer is the most conserved amino acid in the Q-SNARE family and is also conserved in all Abi family members.
FIG. 1.
The Abi amino-terminal region exhibits homology to the SNARE motif. Shown is the alignment of Abi family proteins and Q-SNARE family members in the SNARE motif region. Black and gray backgrounds indicate amino acid identity and similarity, respectively. The SNAP-25 family contains two SNARE motifs and the carboxy-terminal motif is indicated by “-c.” Sx, Syntaxin; hu, human; r, rat; m, mouse; d, D. melanogaster; ce, C. elegans. GenBank accession numbers: Sx1a, P32851; Sx3, Q08849; Sx4, Q08850; Sso1, P32867; Sed5, Q01590; Sx5, Q08851; Vam3, Q12241; SNAP-25B, P13795; SNAP-23, U73143; SNAP-25, U81153; Sec9, L34336. NCBI protein accession numbers for the Abi family: mAbi-1, 16225952; hAbi-1, 7839526; rE3B1, 5882255; huAbi2B, 7839524; HuNESH, 14043609; dAbi, 5051971; ceAbi, 17551826. Numbers at the bottom indicate the amino acid positions in hAbi-1.
To test whether the putative SNARE motif in Abi-1 is functional for binding other SNARE-containing proteins, we examined whether Abi-1 could interact with representative SNARE family members. Abi-1 interacts strongly with Syntaxin-1 and weakly with SNAP-25 and VAMP-2 proteins (Fig. 2A). Interestingly, Syntaxin-4, which is highly similar to Syntaxin-1, and SNAP-23, which displays strong homology to SNAP-25, failed to interact with Abi-1 under these conditions (data not shown). The SNARE proteins SNAP-29, VAMP-4, and Syntaxin-7 did not interact with Abi-1 (data not shown). This finding suggests that Abi-1 has a functional SNARE domain that interacts preferentially with Syntaxin-1.
FIG. 2.
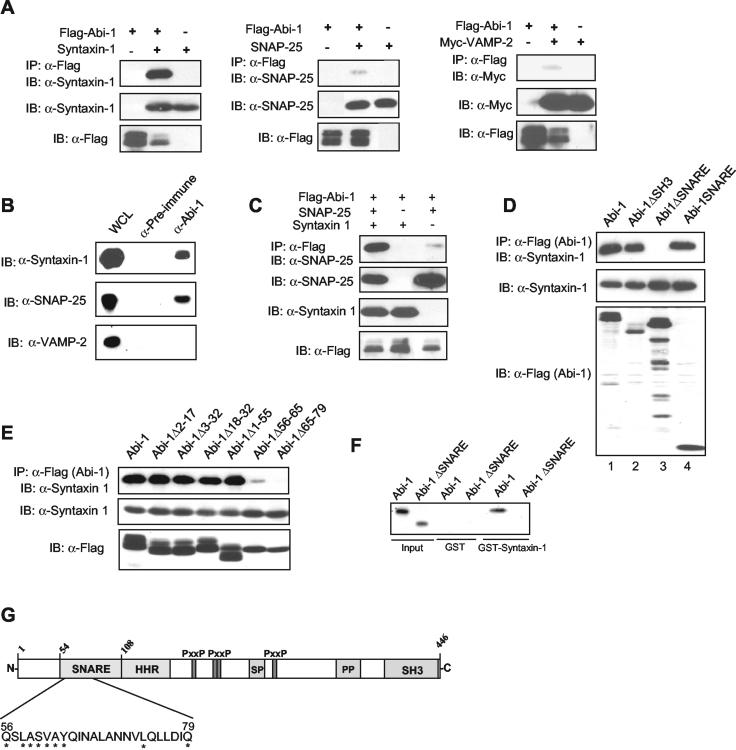
Abi-1 binds Syntaxin-1 through the SNARE motif. (A) Abi-1 specifically interacts with Syntaxin-1. Flag-Abi-1 was transfected into 293T cells with Syntaxin-1 (left panel), SNAP-25 (middle panel), or Myc-VAMP-2 (right panel). Abi-1 was immunoprecipitated (IP) with anti-Flag antibodies, and the coprecipitating SNARE proteins were detected by immunoblotting (IB) with the indicated specific antibodies. A DNA ratio of 6:1 was used for Flag-Abi-1 and SNARE proteins. (B) Endogenous Abi-1 interacts with Syntaxin-1. Endogenous Abi-1 was immunoprecipitated from brain extracts with anti-Abi-1 antibody, and the immunopurified complex was immunoblotted with Syntaxin-1, VAMP-2, and SNAP-25 antibodies. Preimmune serum was used as a negative control. (C) Syntaxin-1 bridges the interaction between Abi-1 and SNAP-25. The interaction between SNAP-25 and Abi-1 was detected as described for panel A in the absence or in the presence of Syntaxin-1. (D) The SNARE motif of Abi-1 is necessary and sufficient for interaction with Syntaxin-1 in cells. Flag-tagged full-length Abi-1, Abi-1ΔSH3, Abi-1ΔSNARE, and Abi-1SNARE were transfected with Syntaxin-1-expressing vector into 293T cells, and Abi-1 was immunoprecipitated with anti-Flag antibody. Syntaxin-1 that copurified with Abi-1 is shown in the upper panel, and the levels of Syntaxin-1 and Abi-1 in the total lysates are shown below. (E) Abi-1 amino acids 56 to 79 are critical for interaction with Syntaxin-1. The indicated Flag-Abi-1 deletion mutants were coexpressed with Syntaxin-1, cell lysates were incubated with anti-Flag as described for panel A, and coprecipitating Syntaxin-1 was detected by immunoblotting with anti-Syntaxin-1 antibody. Total lysates were immunoblotted for Syntaxin-1 and Flag-Abi-1 forms. (F) Abi-1 and Syntaxin-1 interact directly. Abi-1 and Abi-1ΔSNARE were synthesized in vitro by using wheat germ extracts and incubated with GST-Syntaxin-1 or GST alone; ∼1% of the total input was loaded in the first two lanes. (G) Schematic representation of the Syntaxin-1-binding region in Abi-1 and the different Abi-1 domains. SR, serine rich; PP, polyproline; P, proline; x, any amino acid. The amino acids required for Syntaxin-1 binding are shown. Asterisks indicate 100% conservation among all Abi-1 family members (human Abi-1, Abi-2, and NESH, as well as mouse Abi-1, D. melanogaster Abi, Anopheles gambiae Abi, and C. elegans Abi).
Next, we tested whether endogenous Abi-1 and Syntaxin-1 could form a complex. As shown in Fig. 2B, Abi-1 interacts strongly with Syntaxin-1 in cell lysates prepared from mouse brain. Endogenous SNAP-25, but not VAMP-2, also coimmunoprecipitated with Abi-1 under the same conditions (Fig. 2B). Because we did not detect significant interaction between SNAP-25 and Abi-1 following overexpression of the two proteins in transfected 293T cells, we hypothesized that this interaction may be indirect, possibly requiring a bridging protein absent from 293T cells. Syntaxin-1 interacts with SNAP-25 (50) and may serve to bridge the binding of SNAP-25 to Abi-1. In order to test this hypothesis, we examined the interaction between Abi-1 and SNAP-25 following coexpression in 293T cells in the absence or presence of Syntaxin-1. As shown in Fig. 2C, coexpression of Syntaxin-1 significantly enhanced the interaction between Abi-1 and SNAP-25. This finding suggests that Syntaxin-1 binds to Abi-1 and recruits SNAP-25 to the complex. In order to map the Abi-1 region that is involved in the interaction with Syntaxin-1, we made various Abi-1 deletion mutants and tested them for interaction with Syntaxin-1. As expected, the SNARE domain of Abi-1 was necessary for the interaction (Fig. 2D). In contrast, deletion of the SH3 domain (Fig. 2D) or the proline-rich region (data not shown) did not affect Syntaxin-1 binding. Moreover, the Abi-1 SNARE domain alone was sufficient to bind Syntaxin-1 (Fig. 2D, lane 4). Additionally, mutations in the region encompassing Abi-1 amino acids 65 to 79 abolished the interaction between Abi-1 and Syntaxin-1, and the deletion of the region corresponding to amino acids 56 to 65 greatly impaired Syntaxin-1 binding (Fig. 2E). In contrast, the deletion of the region upstream of amino acid 55 or downstream of amino acid 100 in Abi-1 did not affect Syntaxin-1 binding (Fig. 2E and data not shown). In order to determine whether Abi-1 interacts directly with Syntaxin-1, we used a GST pull-down assay. In Fig. 2F, we show that in vitro-synthesized Abi-1, but not Abi-1ΔSNARE, interacted with GST-Syntaxin-1. Taken together, these results indicate that Abi-1 specifically interacts with Syntaxin-1 and that the region spanning amino acids 56 to 79 within the SNARE domain on Abi-1 is critical for this interaction (Fig. 2G).
Abi-1 binds to Wave-1 via a conserved amino-terminal domain.
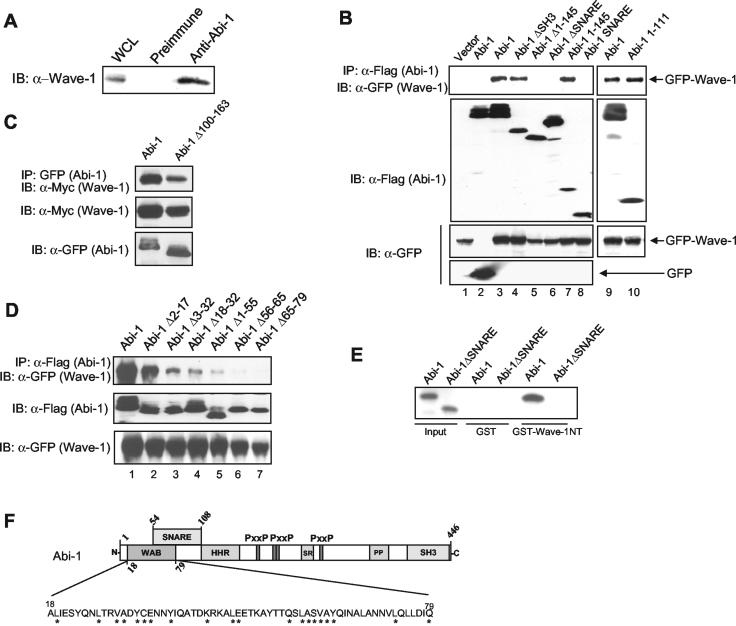
The amino terminus of the Abi family proteins is highly conserved. In addition to the residues involved in binding to Syntaxin-1, residues upstream and downstream of the Abi-1 SNARE domain are also conserved and may be involved in functionally conserved interactions. Recent reports have shown that Abi-1 and Abi-2 are present in a multiprotein complex that includes Wave-1 (12, 43, 45). Moreover, like Abi proteins, Wave-1 localizes to the leading edge of the lamellipodium (20, 33). We observed that Wave-1 coimmunoprecipitated with endogenous Abi-1 from mouse brain lysates when an anti-Abi-1-specific antibody was used (Fig. 3A). Similar results were observed for lysates from NG108-15 cells (data not shown). To identify the Abi-1 sites involved in Wave-1 binding, Flag-tagged Abi-1 full-length proteins or the indicated Abi-1 mutant proteins were coexpressed with GFP-tagged Wave-1 in 293T cells. Abi-1 interacted with GFP-tagged Wave-1 but not with GFP alone (Fig. 3B, lanes 3 and 2). Both Abi-1 and Wave-1 are phosphorylated by the c-Abl tyrosine kinase (42, 61). However, phosphorylation of Abi-1 and Wave-1 by c-Abl did not affect the interaction between the two proteins (data not shown). Wave-1 interacts with other SH3 domain-containing proteins, such as c-Abl and IRSp53, that bind to the proline-rich region of Wave-1 (30, 61). However, deletion of the entire Abi-1 SH3 domain did not affect binding to Wave-1 (Fig. 3B, lane 4). In contrast, deletion of Abi-1 amino acids 1 to 145 or of the entire Abi-1 SNARE domain completely eliminated the interaction with Wave-1 (Fig. 3B, lanes 5 and 6). Therefore, the amino terminus of Abi-1 is necessary for the interaction between Abi-1 and Wave-1. Moreover, Abi-1 amino acids 1 to 145 and 1 to 111 were sufficient for Wave-1 binding (Fig. 3B, lanes 7 and 10). In contrast, the Abi-1 SNARE domain alone did not bind to Wave-1 (Fig. 3B, lane 8). The region comprising Abi-1 amino acids 1 to 145 includes the SNARE motif and a portion of the HHR (11). Deletion of the HHR (amino acids 100 to 163) did not significantly reduce Abi-1 binding to Wave-1 (Fig. 3C). These results indicate that the first 100 amino acids of Abi-1 are important for its interaction with Wave-1. To further define the Wave-1 binding domain, we carried out mutagenesis of this Abi-1 region. The deletion of amino acids 56 to 79 completely abolished the interaction with Wave-1 (Fig. 3D, lanes 6 and 7). The deletion of Abi-1 amino acids 18 to 55 markedly reduced Wave-1 binding (Fig. 3D, lanes 3 to 5), but an Abi-1 mutant lacking amino acids 2 to 17 retained strong Wave-1 binding (Fig. 3D, lane 2). In order to determine whether the interaction between Abi-1 and Wave-1 was direct, we carried out a GST pull-down assay with purified GST-Wave-1NT (amino acids 1 to 277) or a GST control and in vitro-synthesized Abi-1 or Abi-1ΔSNARE produced in wheat germ extracts. GST-Wave-1NT, but not GST, interacted strongly with the Abi-1 wild type but not with Abi-1 lacking the SNARE domain. Taken together, these results show that the Abi-1 region encompassing amino acids 18 to 79 is required for Wave-1 binding and that Wave-1 and Syntaxin-1 have distinct but overlapping binding sites on Abi-1. We propose that the region between Abi-1 amino acids 18 to 79 be designated the WAB domain (Fig. 3F).
FIG. 3.
Abi-1 interacts with Wave-1 through a conserved amino-terminal WAB domain. (A) Endogenous Abi-1 interacts with Wave-1. Endogenous Abi-1 was immunoprecipitated from brain extracts by using anti-Abi-1 antibody (6987), and the immunopurified complex was immunoblotted (IB) with polyclonal Wave-1 antibody. Preimmune serum was used as a negative control. (B) The region comprising amino acids 1 to 111 of Abi-1 is necessary and sufficient to bind Wave-1. Different Flag-tagged Abi-1 forms and GFP-Wave-1 were coexpressed in 293T cells, and the Flag-Abi-1 forms were immunoprecipitated (IP) by using anti-Flag antibody. In lane 2, GFP was coexpressed with Flag-Abi-1 as a negative control. The coprecipitating GFP-Wave-1 was detected with anti-GFP antibody. Due to low expression levels of Flag-Abi-1SNARE, the ratio of Flag-tagged constructs to GFP-Wave-1 vector was 7:1 in all lanes. (C) EYFP-Abi-1 or EYFP-Abi-1Δ100-163 was cotransfected into 293T cells with Myc-Wave-1, and the Abi-1 forms were immunoprecipitated with anti-GFP antibody. Purified immunocomplexes were immunoblotted with Myc antibody to detect Wave-1 (upper panel). Whole-cell lysates were immunoblotted for Myc and GFP. (D) Abi-1 amino acids 18 to 79 are required for interaction with Wave-1. The indicated Flag-Abi-1 deletion mutants were coexpressed with GFP-Wave-1 and immunoprecipitated with anti-Flag. The coprecipitated GFP-Wave-1 protein was detected with anti-GFP antibodies (upper panel). GFP-Wave-1 protein levels in total cell lysates (lower panel) and the immunoprecipitated Flag-Abi-1 forms (middle panel) were detected with the indicated antibodies. (E) Abi-1 and Wave-1 interact directly. Abi-1 and Abi-1ΔSNARE were synthesized in vitro by using wheat germ extracts and incubated with GST-Wave-1NT or GST alone; ∼5% of the input was loaded in the first two lanes. (F) Schematic representation of the WAB domain in Abi-1. The Abi-1 nomenclature is given in the legend to Fig. 2G.
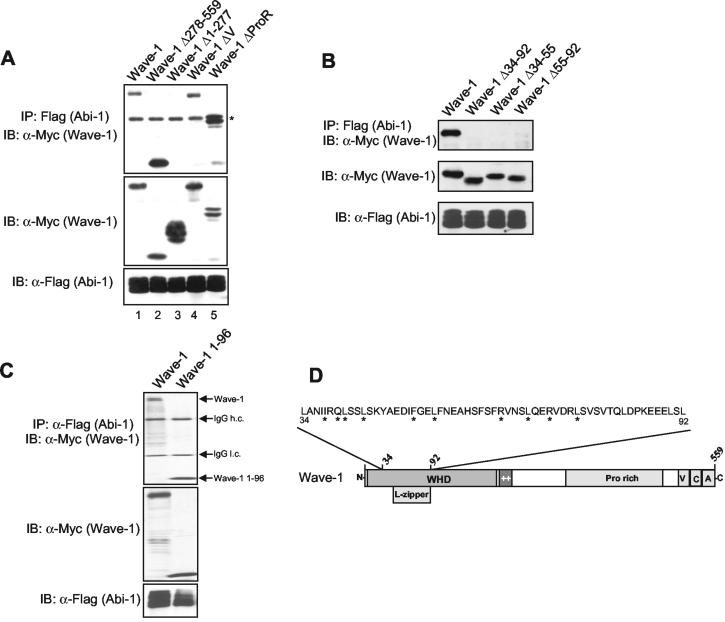
Next, we defined the region of Wave-1 involved in binding to Abi-1. Deletion of the Wave-1 V domain, which is required for G-actin binding, had no effect on Abi-1 binding (Fig. 4A). Similarly, mutation of the Wave-1 proline-rich region or mutation of the entire Wave-1 carboxy-terminal half (amino acids 278 to 559) did not affect Abi-1 binding (Fig. 4A). In contrast, deletion of the Wave-1 amino terminus (amino acids 1 to 277) completely disrupted the interaction with Abi-1 (Fig. 4A, upper panel, lane 3). The region between Wave-1 amino acids 1 to 277 contains the WHD and a basic region (52). This region is conserved among the Wave subfamily proteins, but it is absent in the Wasp subfamily. The WHD of Wave-1 contains a putative leucine-zipper motif that has been implicated in localizing Wave-2 to the leading edge of filopodia, where Abi-1 also localizes (33, 46). As shown in Fig. 4B, deletion of the entire putative leucine zipper (Δ55-92) eliminated binding to Abi-1. Deletion of amino acids 34 to 55 and 34 to 92 in Wave-1 also abolished the interaction with Abi-1 (Fig. 4B). Moreover, Wave-1 amino acids 1 to 96 are sufficient for interaction with Abi-1 (Fig. 4C). Taken together, these results show that Wave-1 and Abi-1 interact inside the cell and that the amino-terminal sequences of both proteins are required for this interaction.
FIG. 4.
Wave-1 interacts with Abi-1 through a region overlapping the putative leucine zipper region. (A) The amino terminus of Wave-1 is necessary for binding to Abi-1. Myc-tagged Wave-1 full-length and Flag-Abi-1 forms were coexpressed in 293T cells, and Flag-Abi-1 forms were immunoprecipitated (IP) with anti-Flag antibodies. Coimmunoprecipitated Wave-1 forms were detected with anti-Myc antibody (upper panel). Expression levels of the Myc-Wave forms are shown in the middle panel, and the levels of Flag-Abi-1 are shown in the lower panel. The asterisk indicates the immunoglobulin G (IgG) heavy-chain band. IB, immunoblot. (B) The region between amino acids 34 to 92 of Wave-1 is necessary to bind Abi-1. Coimmunoprecipitation was performed as described for panel A. The upper panel shows the binding between Flag-Abi-1 and the wild-type and mutant forms of Myc-Wave-1. Wave-1 and Abi-1 levels were visualized by immunoblotting with Myc and Flag antibodies, respectively. (C) Wave-1 amino acids 1 to 96 are sufficient to bind Abi-1. Myc-tagged Wave-1 and Wave-1-1-96 were coexpressed with Flag-Abi-1, and Abi-1 was immunoprecipitated as described for panel A. IgG heavy and light chains are indicated. (D) Schematic representation of the Abi-1-binding region of Wave-1. L-zipper, leucine zipper; ++, basic region; Pro, proline; V, verprolin homology domain; C, cofilin homology domain; A, acidic region. The Wave-1 diagram is not to scale. Asterisks indicate 100% conservation among Wave family members (Wave-1, Wave-2, Wave-3, D. melanogaster Scar, and Dictyostelium discoideum Scar).
The WAB domain of Abi-1 determines its localization to the leading edge of the lamellipodium and regulates Wave-1 protein levels.
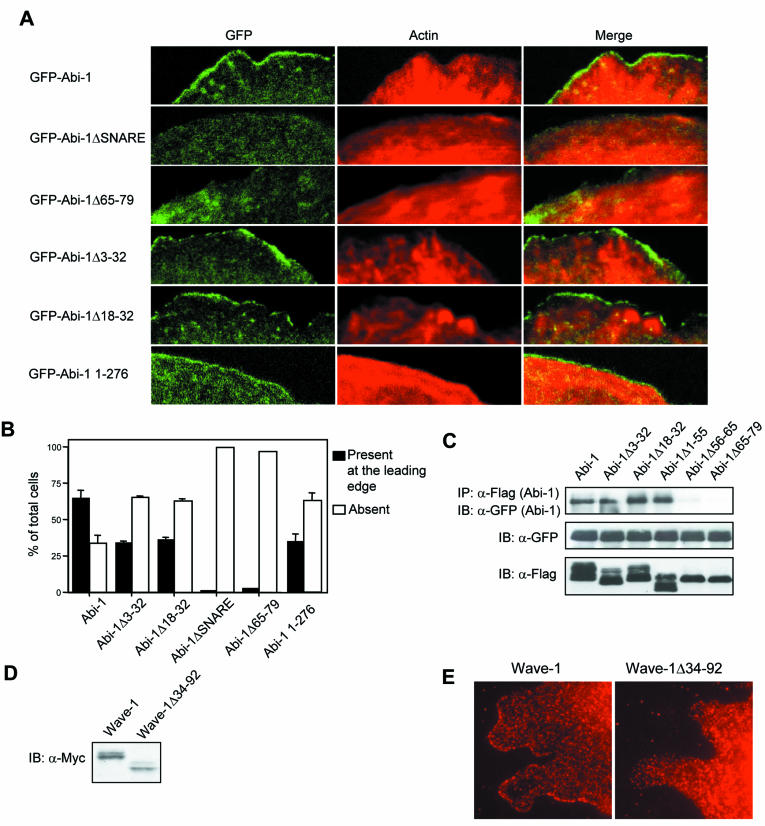
Next, we determined whether Abi-1 binding to Wave-1 or Syntaxin-1 affected its localization to the lamellipodium. It has been previously shown that Abi-1 localization to the leading edge of the lamellipodium requires Abi-1 amino acids 1 to 145 (46). The expression levels of GFP-Abi-1 wild-type and mutant forms were similar to those of endogenous Abi-1 in wild-type MEFs (data not shown). GFP-Abi-1 localized to the leading edge of the lamellipodium (Fig. 5A and B). In contrast, deletion of amino acids 44 to 111 (ΔSNARE) prevented Abi-1 from reaching the leading edge of the lamellipodium (Fig. 5A and B). Similarly, the Abi-1Δ65-79 mutant, which is deficient in Wave-1 binding, did not localize to the leading edge of the lamellipodium (Fig. 5A and B). Two Abi-1 mutants, Abi-1Δ3-32 and Abi-1Δ18-32, which contained small deletions in the Abi-1 WAB domain which reduced Wave-1 binding but did not affect Syntaxin-1 binding, were partially impaired in their ability to reach the tip of the lamellipodium in wild-type MEFs (Fig. 5A and B). While 65% of the cells expressing wild-type Abi-1 showed clear lamellipodium localization, only 34 and 36% of the cells expressing Abi-1Δ3-32 or Abi-1Δ18-32, respectively, showed lamellipodium localization of these Abi-1 mutants (Fig. 5B). The Abi-1 amino terminus alone (amino acids 1 to 276) is sufficient to translocate GFP to the lamellipodium, although it is not as efficient as wild-type Abi-1 (Fig. 5A and B).
FIG. 5.
Localization of Abi-1 and Wave-1 to the leading edge of the lamellipodium is dependent on the WAB domain of Abi-1 and the Abi-binding region of Wave-1, respectively. (A) Wild-type MEFs were infected with retroviruses encoding GFP-tagged Abi-1 wild-type and mutant forms, and the Abi-1 proteins were localized by direct observation of the GFP signal (left panels). Actin staining is shown in red (middle panels), and GFP and actin signals are merged in the right panels. Cells were plated for 30 min on fibronectin-coated coverslips, fixed, and visualized by confocal microscopy. (B) Percentages of cells expressing the indicated GFP-Abi-1 forms that exhibit (black bars) or lack (white bars) GFP signal at the leading edge of the lamellipodium. (C) Abi-1 oligomerization depends on the Syntaxin-1-binding region. The indicated Flag-Abi-1 forms and GFP-Abi-1 were coexpressed in 293T cells, and the Flag-Abi-1 proteins were immunoprecipitated (IP) with anti-Flag antibody. Coimmunoprecipitated GFP-Abi-1 was detected with anti-GFP (upper panel). Whole-cell lysates were immunoblotted (IB) with GPF (middle panel) and Flag antibodies (lower panel). (D) Expression of Myc-tagged wild-type Wave-1 and Wave-1Δ34-92 in MEFs. MEFs were infected with retroviruses encoding the indicated Wave-1 proteins, and cell lysates were immunoblotted with anti-Myc antibody. (E) Localization of Wave-1 to the leading edge of the lamellipodium is dependent on the Abi-1-binding region of Wave-1. Wild-type MEFs were infected with a retrovirus encoding Myc-tagged Wave-1 wild type or Wave-1Δ34-92. Cells were plated for 2 h and 15 min on fibronectin-coated coverslips, fixed, and visualized by using anti-Myc antibody.
We also examined the localization of Abi-1 in MEFs that were null for Abi-2 and heterozygotes for Abi-1 (Grove and Pendergast, unpublished) and observed that GFP-Abi-1Δ3-32 did not accumulate to a significant extent at the leading edges of the lamellipodia in these cells. Similarly, the localization of full-length GFP-Abi-1 to the leading edge of the lamellipodium in the Abi-2−/− Abi-1−/+ MEFs was reduced compared to that observed in wild-type MEFs (data not shown). However, the full-length Abi-1 protein exhibited greater accumulation at the leading edge than did Abi-1Δ3-32 in the Abi-deficient cells. The differential accumulation of the GFP-Abi-1 and GFP-Abi-1Δ3-32 proteins in wild-type and Abi-2−/− Abi-1−/+ MEFs may be explained by oligomerization of the ectopically expressed GFP-Abi-1 and GFP-Abi-1Δ3-32 proteins with the endogenous Abi proteins which may recruit the exogenous Abi-1 proteins to the leading edge. Indeed, overexpressed Abi-1 has been reported to oligomerize through its amino-terminal domain (16). As shown in Fig. 5C, Flag-tagged forms of Abi-1, Abi-1Δ3-32, Abi-1Δ18-32, and Abi-1Δ1-55 interacted with GFP-tagged Abi-1 when overexpressed in 293T cells. In contrast, Abi-1Δ56-65 and Abi-1Δ65-79 did not oligomerize with GFP-Abi-1 (Fig. 5C). The oligomerization region in Abi-1 closely matched the Syntaxin-1-binding SNARE domain. This finding is consistent with the reported ability of SNARE domains to form dimers (62). Thus, Abi-1 proteins that do not interact with Wave-1 cannot reach the leading edge of the lamellipodium, and Abi-1 mutants that weakly interact with Wave-1 exhibit decreased accumulation at this site.
The WHD domain of Wave-1 is required for proper localization to the leading edge of the lamellipodium (32). We observed that a small deletion of amino acids 34 to 92 within the WHD domain disrupted the interaction of Wave-1 with Abi-1 (Fig. 4B). Wild-type Wave-1 localized to the protruding lamellipodia in MEFs spreading on a fibronectin-coated surface (Fig. 5E). In contrast, the Wave-1Δ34-92 mutant protein did not localize to the tips of these protrusive structures (Fig. 5E), even though its expression levels were comparable to those of wild-type Wave-1 (Fig. 5D). This finding suggests that Wave-1 binding to Abi-1 is required for proper Wave-1 localization to the tip of the lamellipodium.
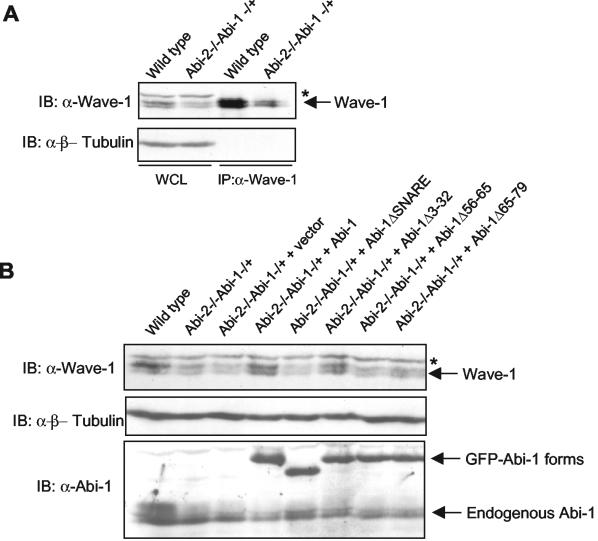
We observed that coexpression of Flag-Abi-1 with GFP-Wave-1 or Myc-Wave-1 in 293T cells resulted in a consistent increase in Wave-1 protein levels (Fig. 3B, third panel, compare lanes 1 and 3; also data not shown). Therefore, we examined whether the levels of endogenous mouse Wave-1 could be affected by genetic elimination of both alleles of Abi-2 and one allele of Abi-1 (Grove and Pendergast, unpublished). As shown in Fig. 6A, the levels of endogenous Wave-1 were reduced in Abi-2−/− Abi-1−/+ MEFs compared to those detected in wild-type MEFs. Most importantly, this phenotype was rescued by the expression of the Abi-1 wild type, Abi-1Δ3-32, and Abi-1Δ18-32, all of which retained Wave-1 binding and localized to the leading edge of the lamellipodia in wild-type MEFs (Fig. 6B and data not shown). In contrast, Abi-1Δ65-79, Abi-1Δ56-65, and Abi-1ΔSNARE did not rescue this phenotype (Fig. 6B). These three Abi-1 mutants cannot bind Wave-1 and are completely absent from the tip of the lamellipodium in wild-type MEFs. Taken together, these results suggest that the Abi-1 proteins that retain Wave-1 binding and localize to the leading edges of lamellipodia stabilize Wave-1 protein levels by binding to the Wave-1 amino terminus.
FIG. 6.
Abi-1 and Abi-2 regulate Wave-1 protein levels. (A) Wild-type MEFs or Abi-2−/− Abi-1−/+ MEFs were lysed in KLB, and the lysate was immunoblotted (IB) for Wave-1 and β-tubulin; 1.25 mg of the same lysate was used to immunoprecipitate Wave-1 with anti-Wave-1 monoclonal antibody. (B) Abi-2−/− Abi-1−/+ MEFs were infected with retroviruses expressing the indicated GFP-Abi-1 forms. Cells were lysed as described for panel A and immunoblotted for Wave-1, β-tubulin, and Abi-1. An asterisk indicates a cross-reacting band not present in Wave-1 immunoprecipitates.
Abi-1 shuttles to the nucleus, and its nucleocytoplasmic equilibrium is dependent on the Abi-1 SNARE, WAB, and SH3 domains.
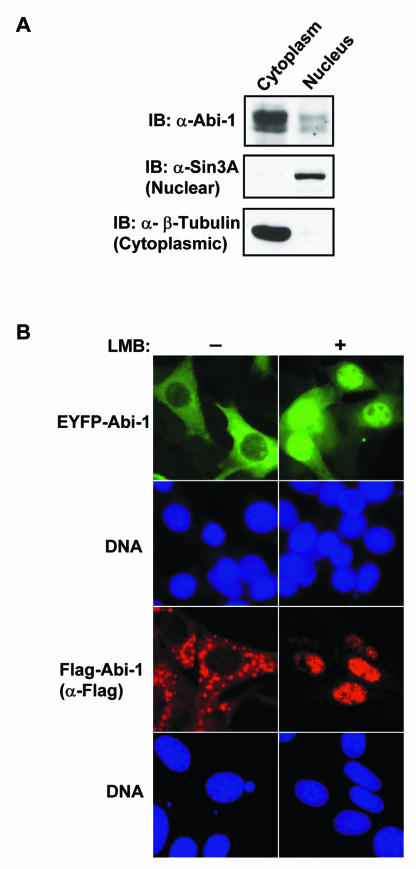
Unexpectedly, we observed that a significant fraction of the cells expressing Abi-1ΔSNARE or Abi-1Δ1-145 displayed Abi-1 nuclear staining, while exogenous full-length Abi-1 did not exhibit significant nuclear accumulation and was largely cytoplasmic (Fig. 7 and 8). In order to determine whether endogenous Abi-1 localized to the nucleus, cytoplasmic and nuclear fractions were prepared by using NIH-3T3 cells and the fractions were immunoblotted for endogenous Abi-1. As shown in Fig. 7A, while the majority of Abi-1 was present in the cytoplasmic fraction, a detectable amount of endogenous Abi-1 was nuclear. This result is consistent with our previous finding that a fraction of the related Abi-2 protein localized to the nucleus (11). The presence of a small fraction of Abi-1 in the nucleus under steady-state conditions suggested that Abi-1 localization may be regulated by limited nuclear import or active nuclear export. To examine these possibilities, we inhibited nuclear export with LMB, an inhibitor of the leucine-rich nuclear export signal (NES)- and Crm-1-dependent pathway (23). As shown in Fig. 7B, full-length EYFP- or Flag-tagged Abi-1 completely localized to the nucleus after LMB treatment. This finding indicates that after import to the nucleus, Abi-1 is actively exported from the nucleus. Approximately 20% of the cells that expressed full-length Abi-1 at high levels displayed large vesicular structures containing Abi-1 that were partially resistant to LMB treatment (data not shown).
FIG. 7.
Abi-1 undergoes nucleocytoplasmic shuttling. (A) Endogenous Abi-1 is present in the nucleus. NIH 3T3 cells were lysed, and the lysates were separated into nuclear and cytoplasmic fractions, followed by immunoblotting (IB) with Abi-1 antibody. The purities of nuclear and cytoplasmic fractions were determined by immunoblotting with Sin3A (a nuclear marker) and β-tubulin (a cytoplasmic marker), respectively. (B) Abi-1 accumulates in the nucleus in an LMB-dependent manner. An immunofluorescence signal (green) of EYFP-tagged Abi-1 was observed in transfected NIH 3T3 cells before (−) and after (+) LMB treatment. Flag-tagged Abi-1 in a bicistronic-GFP-expressing vector was expressed following retroviral transduction in NIH 3T3 cells, and Abi-1 localization was determined before and after LMB treatment by immunostaining with Flag monoclonal antibody (red). DAPI staining (DNA; blue) is also shown.
FIG.8.
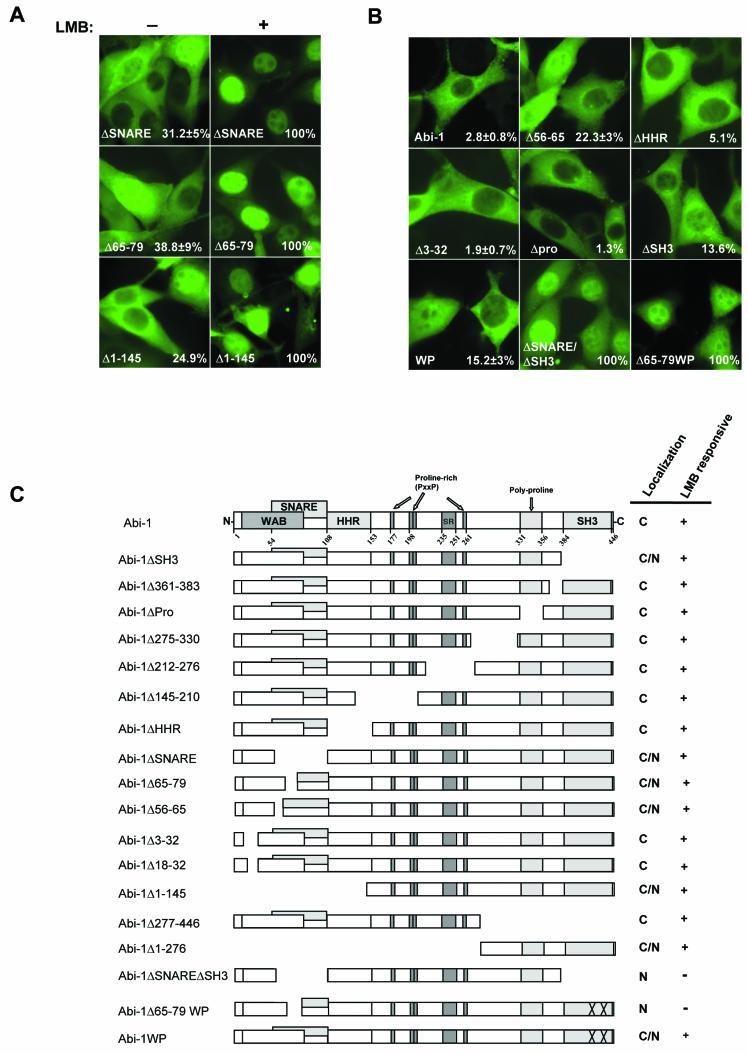
The nucleocytoplasmic distribution of Abi-1 is regulated by the Wave-1- and Syntaxin-1-binding regions and the SH3 domain. (A) Cellular localization of the indicated Abi-1 mutants before (−) and after (+) LMB treatment. The number on the bottom right in each panel is the percentage of cells with Abi-1 nuclear staining. (B) Localization of the indicated EYFP-tagged Abi-1 mutants in NIH 3T3 cells in the absence of LMB. The number on the bottom right in each panel is the percentage of cells with Abi-1 nuclear staining. (C) Schematic representation of the Abi-1 mutants employed in this study; their localization is indicated. C, cytoplasmic localization; N, nuclear localization. The Abi-1 nomenclature is given in the legend to Fig. 2G.
Although there are no obvious nuclear localization signals (NLS) in Abi-1, there is a putative NES within the Abi-1 SNARE domain. Abi-1 amino acids LANNVLQLLDI (amino acids 68 to 78; underlining indicates the NES consensus sequence) conform to the NES consensus (4). In order to determine whether these Abi-1 amino acids contained a functional NES, we analyzed the localization of Abi-1 mutants that lacked the NES in the absence or presence of LMB. Approximately 30 to 40% of cells expressing Abi-1ΔSNARE or Abi-1Δ65-79 displayed nuclear localization of these Abi mutant proteins in the absence of LMB (Fig. 8A). However, in the presence of LMB, virtually 100% of the cells showed Abi-1 nuclear staining (Fig. 8A). The cytoplasmic pool of the Abi-1 mutant proteins lacking the putative NES shuttled to the nucleus in a LMB-dependent manner (Fig. 8A). Thus, this Abi-1 sequence is not likely to be a functional NES. In the absence of LMB, some of the Abi-1Δ65-79-expressing cells (39%) and Abi-1Δ56-65-expressing cells (22%) showed Abi-1 nuclear accumulation. Interestingly, both of these Abi-1 mutants are deficient in binding to Wave-1 and Syntaxin-1. In contrast, Abi-1ΔHHR, which binds to both Wave-1 (Fig. 3C) and Syntaxin-1 (data not shown), showed a localization pattern indistinguishable from that of wild-type Abi-1 (Fig. 8B). The Abi-1Δ3-32 and Abi-1Δ18-32 mutants that exhibited reduced Wave-1 binding but retained binding to Syntaxin-1 displayed cytoplasmic localization similar to that of wild-type Abi-1 (Fig. 8B and C). Thus, there appears to be an inverse correlation between binding to Syntaxin-1 and nuclear accumulation. Taken together, these data suggest that Abi-1 may be anchored in the cytoplasm primarily by interactions mediated by the Syntaxin-1-binding region, but Wave-1 binding may also contribute to cytoplasmic anchoring.
Deletions of Abi-1 amino acids 361 to 383, 330 to 359 (Δpro), 275 to 330, 212 to 276, 145 to 210, and 100 to 163 (ΔHHR) and 277 to 446 had no effect on nuclear localization in the absence of LMB, but these Abi-1 mutant proteins accumulated in the nucleus in response to LMB treatment (Fig. 8C). Abi-1Δ1-276 and Abi-1Δ277-446 mutant proteins containing the carboxy- and amino-terminal halves of the Abi-1 protein, respectively, can shuttle in and out of the nucleus, suggesting that both the amino terminus and the carboxy terminus harbor sequences that induce Abi-1 nucleocytoplasmic shuttling (Fig. 8C).
A significant number (∼60%) of cells expressing the Abi-1Δ65-79 mutant protein that is deficient in binding to both Syntaxin-1 and Wave-1 showed cytoplasmic staining in the absence of LMB (Fig. 8A). Thus, it is likely that additional proteins that bind to Abi-1 sequences distinct from those required for Syntaxin-1 binding may anchor Abi-1 in the cytoplasm. We tested whether the Abi-1 SH3 domain contributed to cytoplasmic retention of Abi-1. Deletion of the entire SH3 domain or mutation of two residues that inactivate SH3 domain binding to proline-rich targets (Abi-1WP) slightly increased the percentages of cells showing Abi-1 nuclear staining in the absence of LMB to 13 and 15%, respectively (Fig. 8B). In contrast, deletion of the polyproline region that binds to Spectrin and Eps8 (40, 67) did not induce nuclear accumulation (Fig. 8B). However, simultaneous inactivation or deletion of both the SH3 and the SNARE domains in Abi-1 (Abi-1ΔSH3/ΔSNARE and Abi-1Δ65-79WP) produced proteins that localized exclusively to the nucleus in all cells (Fig. 8B). Because no single mutation completely affects Abi-1 import and export in the absence of LMB, these results suggest that the factor or factors involved in the nucleocytoplasmic shuttling of Abi-1 bind to different regions of the molecule. Taken together, our data suggest that the putative import factor for Abi-1 does not bind to its SH3 or SNARE domains, that the export factor uses the Crm-1 export pathway, and that dissociation of Abi-1 from proteins that bind to the Abi-1 SNARE, WAB, and SH3 domains is required for Abi-1 to enter the nucleus (Fig. 9).
FIG. 9.
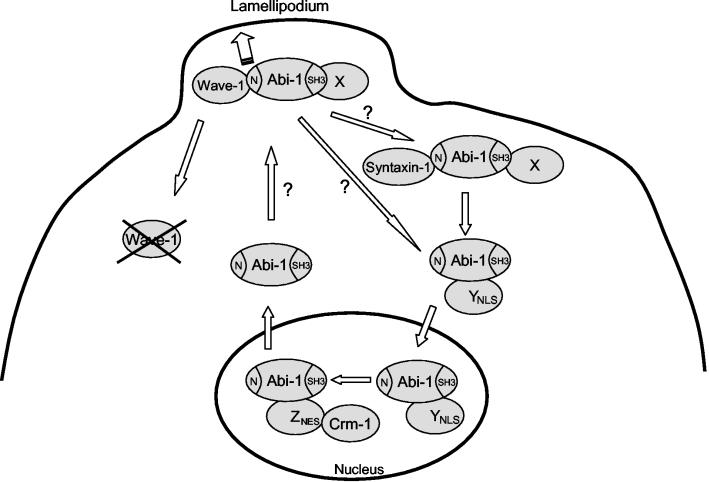
Abi-1 localizes to the leading edge of the lamellipodium and undergoes nucleocytoplasmic shuttling. Distinct pools of Abi-1 bind to Wave-1 or Syntaxin-1 in the cytosol, and localization of Abi-1 to the leading edge of the lamellipodium requires the SNARE and WAB domains in the conserved Abi-1 amino terminus. Disruption of the Abi-1-Wave-1 interaction leads to a reduction of Wave-1 protein levels. Moreover, the binding of Wave-1 to Abi-1 contributes to proper localization of Wave-1 to the leading edge of lamellipodia. Abi-1 undergoes nucleocytoplasmic shuttling and accumulates in the nucleus in response to LMB treatment. Localization of Abi-1 to the nucleus in the absence of LMB requires the simultaneous disruption of the SNARE, WAB, and SH3 domains of Abi-1. These findings suggest that multiple protein-protein interactions are involved in retaining Abi-1 in the cytoplasm. The protein (X) that binds to the Abi-1 SH3 domain and contributes to Abi-1 cytoplasmic retention has not yet been identified. While Wave-1 and Syntaxin-1 bind to the WAB and SNARE domains of Abi-1, respectively, other unknown proteins may also bind to these domains and regulate the nucleocytoplasmic distribution of Abi-1. An unknown factor (Y) is required for the translocation of Abi-1 into the nucleus, and another factor (Z) is required for the nuclear export of Abi-1 through the Crm-1-dependent pathway. YNLS, NLS-containing protein; ZNES, NES-containing protein.
DISCUSSION
Our findings have uncovered an important role for the Abi adaptor proteins in positive regulation of the stability and localization of the actin regulatory protein Wave-1 in mammalian fibroblasts. Abi-1 binds to Wave-1 through an amino-terminal WAB domain that is conserved among Abi family members. Like Wave-1 and Wave-2, Abi-1 and Abi-2 localize to the tips of lamellipodia and filopodia (20, 32, 33, 46). Here, we showed that the disruption of the Abi-1-Wave-1 interaction blocks the proper localization of both proteins to the leading edge of the lamellipodium. Previous studies have shown that a GFP fusion protein containing Wave-2 amino acids 1 to 83 localizes to the tip of the lamellipodium (33). This Wave-1 region is required for Abi-1 binding, and amino acids 1 to 96 are sufficient to bind Abi-1 (Fig. 4). Moreover, the Abi-1-binding-deficient Wave-1Δ34-92 protein cannot reach the leading edge of the lamellipodium (Fig. 5E). These findings suggest that the region between Wave-1 amino acids 1 and 96 is sufficient to bind Abi-1 and translocate Wave-1 to the lamellipodium. In agreement with this finding, downregulation of Drosophila Abi by RNAi treatment prevented Wave-1 from reaching the leading edge of the lamellipodium (24).
Fibroblasts derived from Abi-2−/− Abi-1−/+ mice showed reduced levels of Wave-1 protein that can be restored to normal levels by Abi-1 forms that can interact with Wave-1 and localize to the leading edge of the lamellipodium. Our data suggest that increased, normal Wave-1 protein levels require the physical association of Wave-1 with Abi-1. Wave-1 activity has been reported to be inhibited in vitro by a multiprotein complex that includes HSPC300, Pir121, Nap-1, and Abi-2 (12), and activation of Wave-1 in vitro correlated with the disruption of this complex (12). However, other studies have shown that the expression of several components of the Wave-1 protein complex are required for Wave-1 protein stability. Downregulation of amoeba Pir121 or its fruit fly homolog (Sra-1), Drosophila Kette (Nap-1), and mammalian Nap-1 resulted in unstable Wave/Scar proteins (3, 39, 45). Moreover, downregulation of Abi-1, Nap-1, or Wave/Scar in the Drosophila cells induces similar actin phenotypes, suggesting that Abi-1 and Nap-1 may not antagonize Wave-1 function but, rather, positively regulate Wave-1 stability and downstream signaling. Further studies will be required to determine whether Abi proteins exert both positive and negative regulatory functions on Wave proteins.
The region of Abi-1 critical for Wave-1 binding overlaps the SNARE domain that is conserved among Abi family members (Fig. 3F). Here, we show that the Abi-1 SNARE domain is necessary and sufficient to bind the Q-SNARE protein Syntaxin-1. We detected SNAP-25, but not VAMP-2, in the Abi-1/Syntaxin-1 complex, suggesting that Abi-1 may not be part of the neuronal SNARE complex formed by SNAP-25/Syntaxin-1/VAMP-2. Several studies have suggested that the actin cytoskeleton plays a role in membrane fusion, such as exocytosis (13). Moreover, while Scar/Wave is required for endosome traffic and exocytosis (41), very little is known regarding the molecules that directly link membrane fusion and the actin cytoskeleton. We propose that Abi proteins are good candidates for the cross talk between the fusion machinery and the actin cytoskeleton. We are currently testing this hypothesis.
The secondary structure of the SNARE motif is an α-helix (50), and secondary-structure prediction analysis revealed that Abi-1 amino acids 17 to 27 and 45 to 85 have a high probability of forming an α-helix. Similarly, the majority of the Abi-1-binding region on Wave-1 (amino acids 31 to 77) is predicted to form an α-helix with high probability. Therefore, it is likely that the interaction of Abi-1 with Wave-1 or Syntaxin-1 involves α-helical structures. It was previously reported that Abi-1 could oligomerize through its amino terminus (16). We have confirmed that following overexpression, Abi-1 can oligomerize (Fig. 5C). The interaction between Abi-1 proteins is weaker than the interaction of Abi-1 with either Syntaxin-1 or Wave-1 (A. Echarri and A. M. Pendergast, unpublished observations). However, it is possible that Abi-1 may interact with Syntaxin-1 and/or Wave-1 as a dimer.
Interestingly, while we could detect endogenous Abi-1 in Wave-1 immunoprecipitates from NG108-15 cell lysates, we could not detect Syntaxin-1, which is expressed in these cells. Moreover, increasing amounts of Wave-1 decreased the interaction between Abi-1 and Syntaxin-1 (data not shown). These observations suggest that the Abi-1/Syntaxin-1 complex and the Abi-1/Wave-1 complex may be mutually exclusive and are consistent with our finding that Abi-1 binds to Wave-1 and Syntaxin-1 through distinct but overlapping amino-terminal sequences (Fig. 3F). Our data indicate that the SNARE and WAB regions of Abi-1 are required for binding to Syntaxin-1 and Wave-1, respectively, but these data do not exclude the possibility that Abi-1 binds to other proteins through these domains. This finding is particularly relevant for the Abi-1 SNARE domain, which is likely to bind Syntaxin-1-like proteins in cell types such as fibroblasts, in which Syntaxin-1 is not expressed.
We showed that endogenous Abi-1 localizes not only to the lamellipodium but also to the nucleus. Interestingly, the sequences required for the localization of Abi-1 to the lamellipodium also regulate Abi-1 nuclear accumulation. The Abi-1 SNARE and WAB domains may function to anchor Abi-1 in the cytoplasm, as both domains must be disrupted for Abi-1 to completely enter the nucleus. However, partial disruption of the WAB domain alone (Abi-1Δ3-32) does not influence Abi-1 nuclear accumulation (Fig. 8B and C), suggesting that protein interactions mediated by both the SNARE and the WAB domains must be simultaneously disrupted. Furthermore, the Abi-1 SH3 domain also contributes to the nuclear translocation of Abi-1, as mutation of the SH3, SNARE, and WAB domains is required to induce complete nuclear accumulation of Abi-1 in the absence of LMB.
Abi-1 uses the NES- and Crm-1-dependent pathway to exit the nucleus, suggesting that Abi-1 or an Abi-1-binding partner contains a NES. Although Abi-1 contains a putative NES, it appears to be nonfunctional, because deletion of this region did not result in complete nuclear accumulation of Abi-1 in the absence of LMB and the cytoplasmic pool of this mutant retained sensitivity to LMB treatment. This finding suggests that Abi-1 may use another NES-containing protein to exit the nucleus. In this regard, Myopodin, an actin-bundling protein, undergoes nucleocytoplasmic shuttling in an LMB-dependent manner despite the absence of a leucine-rich NES (59).
The Abi-1 sequence does not contain amino acids with similarity to any known NLS, suggesting that Abi-1 may contain an uncharacterized NLS or binds to an NLS-containing protein to enter the nucleus. Abi-1 binds to c-Abl, a protein that undergoes nucleocytoplasmic shuttling and contains both NLS and NES sequences (51). However, it is unlikely that Abl is responsible for the nuclear entry or exit of Abi-1, because Abi-1 accumulates in the nucleus in c-Abl/Arg null cells after LMB treatment (Echarri and Pendergast, unpublished).
It is intriguing that actin and several actin-binding and actin-regulatory molecules undergo nucleocytoplasmic shuttling (35, 47, 51, 55, 56, 58, 59). The actin-regulatory protein WASP enters the nucleus to regulate transcription (48). Similarly, Wave-1 is found in the nucleus, but its nuclear role is unknown (61). Zyxin, a regulator of actin polymerization (18), also undergoes nucleocytoplasmic shuttling and, like Abi-1, lacks a conventional NLS (58). Moreover, a number of proteins that function at the plasma membrane to regulate signaling cascades also undergo nucleocytoplasmic shuttling. Among these is Sterile 5 (Ste5), a scaffold protein of the yeast mating pathway that must shuttle to the nucleus before it can localize to the plasma membrane and activate the MAPK cascade (27). Nucleocytoplasmic shuttling of Abi-1 may serve to limit Abi-1-dependent regulation of Wave-1 and actin-dependent processes at the leading edge of the lamellipodium in the absence of appropriate extracellular stimuli. Alternatively, nuclear accumulation may be required for yet-to-be-identified Abi-1-regulated processes in the nucleus.
Our work has shown that Abi-1 is one of a growing number of molecules that undergo nucleocytoplasmic shuttling and also function to regulate proteins involved in actin dynamics in the cytosol. Analysis of the phenotypes induced by the loss of Abi-1 and Abi-2 in mice and other species will reveal whether the Abi protein family is required for the regulation of actin dynamics in vivo.
Acknowledgments
This work was supported by NIH grants CA70940 and GM62375 to A.M.P.
We thank J. Scott, R. H. Scheller, G. J. Augustine, H. Y. Gaisano, P. A. Roche, and Z. H. Sheng for kindly providing us with valuable reagents. We also thank M. Grove for construct generation (pCMV2Flag-Abi-1) and generation of the Abi-2−/− Abi-1−/+ MEFs. We thank P. Zipfel for critical reading of the manuscript and construct generation (MIGR1Flag-Abi-1). We thank Heather Weiman for construct generation (pLEGFPC1Abi-1 1-276). We also thank M. Quiroz for excellent technical assistance and M. Cook for fluorescence-activated cell sorting analysis.
REFERENCES
- 1.Beites, C. L., H. Xie, R. Bowser, and W. S. Trimble. 1999. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 5:434-439. [DOI] [PubMed] [Google Scholar]
- 2.Biesova, Z., C. Piccoli, and W. T. Wong. 1997. Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene 14:233-241. [DOI] [PubMed] [Google Scholar]
- 3.Blagg, S. L., M. Stewart, C. Sambles, and R. H. Insall. 2003. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13:1480-1487. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman, E. R., S. An, N. Barton, and R. Jahn. 1994. SNAP-25, a t-SNARE which binds to both syntaxin and synaptobrevin via domains that may form coiled coils. J. Biol. Chem. 269:27427-27432. [PubMed] [Google Scholar]
- 6.Chen, Y. A., and R. H. Scheller. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98-106. [DOI] [PubMed] [Google Scholar]
- 7.Cory, G. O., and A. J. Ridley. 2002. Cell motility: braking WAVEs. Nature 418:732-733. [DOI] [PubMed] [Google Scholar]
- 8.Courtney, K. D., M. Grove, H. Vandongen, A. Vandongen, A. S. LaMantia, and A. M. Pendergast. 2000. Localization and phosphorylation of Abl-interactor proteins, Abi-1 and Abi-2, in the developing nervous system. Mol. Cell. Neurosci. 16:244-257. [DOI] [PubMed] [Google Scholar]
- 9.Cowan, C. A., and M. Henkemeyer. 2001. The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature 413:174-179. [DOI] [PubMed] [Google Scholar]
- 10.Dahl, J. P., J. Wang-Dunlop, C. Gonzales, M. E. Goad, R. J. Mark, and S. P. Kwak. 2003. Characterization of the WAVE1 knock-out mouse: implications for CNS development. J. Neurosci. 23:3343-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai, Z., and A. M. Pendergast. 1995. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 9:2569-2582. [DOI] [PubMed] [Google Scholar]
- 12.Eden, S., R. Rohatgi, A. V. Podtelejnikov, M. Mann, and M. W. Kirschner. 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418:790-793. [DOI] [PubMed] [Google Scholar]
- 13.Eitzen, G. 2003. Actin remodeling to facilitate membrane fusion. Biochim. Biophys. Acta 1641:175-181. [DOI] [PubMed] [Google Scholar]
- 14.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 15.Fan, P.-D., and S. P. Goff. 2000. Abl interactor 1 binds to Sos and inhibits epidermal growth factor- and v-Abl-induced activation of extracellular signal-regulated kinases. Mol. Cell. Biol. 20:7591-7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan, P.-D., F. Cong, and S. P. Goff. 2003. Homo- and hetero-oligomerization of the c-Abl kinase and Abelson-interactor-1. Cancer Res. 63:873-877. [PubMed] [Google Scholar]
- 17.Fasshauer, D., R. B. Sutton, A. T. Brunger, and R. Jahn. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 95:15781-15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fradelizi, J., V. Noireaux, J. Plastino, B. Menichi, D. Louvard, C. Sykes, R. M. Golsteyn, and E. Friederich. 2001. ActA and human zyxin harbour Arp2/3-independent actin-polymerization activity. Nat. Cell Biol. 8:699-707. [DOI] [PubMed] [Google Scholar]
- 19.Fujita, Y., H. Shirataki, T. Sakisaka, T. Asakura, T. Ohya, H. Kotani, S. Yokoyama, H. Nishiok, Y. Matsuura, A. Mizoguchi, R. H. Scheller, and Y. Takai. 1998. Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron 20:905-915. [DOI] [PubMed] [Google Scholar]
- 20.Hahne, P., A. Sechi, S. Benesch, and J. V. Small. 2001. Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 492:215-220. [DOI] [PubMed] [Google Scholar]
- 21.Hata, Y., C. A. Slaughter, and T. C. Sudhof. 1993. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366:347-351. [DOI] [PubMed] [Google Scholar]
- 22.Ji, J., S. Tsuk, A. M. Salapatek, X. Huang, D. Chikvashvili, E. A. Pasyk, Y. Kang, L. Sheu, R. Tsushima, N. Diamant, W. S. Trimble, I. Lotan, and H. Y. Gaisano. 2002. The 25-kDa synaptosome-associated protein (SNAP-25) binds and inhibits delayed rectifier potassium channels in secretory cells. J. Biol. Chem. 277:20195-20204. [DOI] [PubMed] [Google Scholar]
- 23.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 24.Kunda, P., G. Craig, V. Dominguez, and B. Baum. 2003. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13:1867-1875. [DOI] [PubMed] [Google Scholar]
- 25.Lao, G., V. Scheuss, C. M. Gerwin, Q. Su, S. Mochida, J. Rettig, and Z. H. Sheng. 2000. Syntaphilin: a syntaxin-1 clamp that controls SNARE assembly. Neuron 1:191-201. [DOI] [PubMed] [Google Scholar]
- 26.Machesky, L. M., R. D. Mullins, H. N. Higgs, D. A. Kaiser, L. Blanchoin, R. C. May, M. E. Hall, and T. D. Pollard. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 96:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahanty, S. K., Y. Wang, F. W. Farley, and E. A. Elion. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98:501-512. [DOI] [PubMed] [Google Scholar]
- 28.Merrifield, C. J., S. E. Moss, C. Ballestrem, B. A. Imhof, G. Giese, I. Wunderlich, and W. Almers. 1999. Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nat. Cell Biol. 1:72-74. [DOI] [PubMed] [Google Scholar]
- 29.Miki, H., S. Suetsugu, and T. Takenawa. 1998. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17:6932-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miki, H., H. Yamaguchi, S. Suetsugu, and T. Takenawa. 2000. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408:732-735. [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki, K., S. Matsuda, Y. Ichigotani, Y. Takenouchi, K. Hayashi, Y. Fukuda, Y. Nimura, and M. Hamaguchi. 2000. Isolation and characterization of a novel human gene (NESH) which encodes a putative signaling molecule similar to e3B1 protein. Biochim. Biophys. Acta 493:237-241. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa, H., H. Miki, M. Ito, K. Ohashi, T. Takenawa, and S. Miyamoto. 2001. N-WASP, WAVE and Mena play different roles in the organization of actin cytoskeleton in lamellipodia. J. Cell Sci. 114:1555-1565. [DOI] [PubMed] [Google Scholar]
- 33.Nozumi, M., H. Nakagawa, H. Miki, T. Takenawa, and S. Miyamoto. 2003. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 116:239-246. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto, M., S. Schoch, and T. C. Sudhof. 1999. EHSH1/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. A protein connection between exocytosis and endocytosis? J. Biol. Chem. 274:18446-18454. [DOI] [PubMed] [Google Scholar]
- 35.Olave, I. A., S. L. Reck-Peterson, and G. R. Crabtree. 2002. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71:755-781. [DOI] [PubMed] [Google Scholar]
- 36.Pendergast, A. M. 2002. The Abl family kinases: mechanisms of regulation and signaling. Adv. Cancer Res. 85:51-100. [DOI] [PubMed] [Google Scholar]
- 37.Quackenbush, R. C., G. W. Reuther, J. P. Miller, K. D. Courtney, W. S. Pear, and A. M. Pendergast. 2000. Analysis of the biologic properties of p230 Bcr-Abl reveals unique and overlapping properties with the oncogenic p185 and p210 Bcr-Abl tyrosine kinases. Blood 95:2913-2921. [PubMed] [Google Scholar]
- 38.Ridley, A. J., M. A. Schwartz, K. Burridge, R. A. Firtel, M. H. Ginsberg, G. Borisy, J. T. Parsons, and A. R. Horwitz. 2003. Cell migration: integrating signals from front to back. Science 302:1704-1709. [DOI] [PubMed] [Google Scholar]
- 39.Rogers, S. L., U. Wiedemann, N. Stuurman, and R. D. Vale. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scita, G., J. Nordstrom, R. Carbone, P. Tenca, G. Giardina, S. Gutkind, M. Bjarnegard, C. Betsholtz, and P. P. Di Fiore. 1999. EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401:290-293. [DOI] [PubMed] [Google Scholar]
- 41.Seastone, D. J., E. Harris, L. A. Temesvari, J. E. Bear, C. L. Saxe, and J. Cardelli. 2001. The WASp-like protein scar regulates macropinocytosis, phagocytosis and endosomal membrane flow in Dictyostelium. J. Cell Sci. 114:2673-2683. [DOI] [PubMed] [Google Scholar]
- 42.Shi, Y., K. Alin, and S. P. Goff. 1995. Abl-interactor-1, a novel SH3 protein binding to the carboxy-terminal portion of the Abl protein, suppresses v-abl transforming activity. Genes Dev. 21:2583-2597. [DOI] [PubMed] [Google Scholar]
- 43.Soderling, S. H., K. L. Binns, G. A. Wayman, S. M. Davee, S. H. Ong, T. Pawson, and J. D. Scott. 2002. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat. Cell Biol. 4:970-975. [DOI] [PubMed] [Google Scholar]
- 44.Soderling, S. H., L. K. Langeberg, J. A. Soderling, S. M. Davee, R. Simerly, J. Raber, and J. D. Scott. 2003. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. USA 100:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steffen, A., K. Rottner, J. Ehinger, M. Innocenti, G. Scita, J. Wehland, and T. E. Stradal. 2004. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stradal, T., K. D. Courtney, K. Rottner, P. Hahne, J. V. Small, and A. M. Pendergast. 2001. The Abl interactor proteins localize to sites of actin polymerization at the tips of lamellipodia and filopodia. Curr. Biol. 11:891-895. [DOI] [PubMed] [Google Scholar]
- 47.Stüven, T., E. Hartmann, and D. Görlich. 2003. Exportin 6: a novel nuclear export receptor that is specific for profilin-actin complexes. EMBO J. 22:5928-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suetsugu, S., and T. Takenawa. 2003. Translocation of N-WASP by nuclear localization and export signals into the nucleus modulates expression of HSP90. J. Biol. Chem. 278:42515-42523. [DOI] [PubMed] [Google Scholar]
- 49.Suetsugu, S., D. Yamazaki, S. Kurisu, and T. Takenawa. 2003. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell 4:595-609. [DOI] [PubMed] [Google Scholar]
- 50.Sutton, R. B., D. Fasshauer, R. Jahn, and A. T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395:347-353. [DOI] [PubMed] [Google Scholar]
- 51.Taagepera, S., D. McDonald, J. E. Loeb, L. L. Whitaker, A. K. McElroy, J. Y. Wang, and T. J. Hope. 1998. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc. Natl. Acad. Sci. USA 95:7457-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takenawa, T., and H. Miki. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801-1809. [DOI] [PubMed] [Google Scholar]
- 53.Thurmond, D. C., C. Gonelle-Gispert, M. Furukawa, P. A. Halban, and J. E. Pessin. 2003. Glucose-stimulated insulin secretion is coupled to the interaction of actin with the t-SNARE (target membrane soluble N-ethylmaleimide-sensitive factor attachment protein receptor protein) complex. Mol. Endocrinol. 4:732-742. [DOI] [PubMed] [Google Scholar]
- 54.Tokumaru, H., K. Umayahara, L. L. Pellegrini, T. Ishizuka, H. Saisu, H. Betz, G. J. Augustine, and T. Abe. 2001. SNARE complex oligomerization by synaphin/complexin is essential for synaptic vesicle exocytosis. Cell 104:421-432. [DOI] [PubMed] [Google Scholar]
- 55.Van Impe, K., V. De Corte, L. Eichinger, E. Bruyneel, M. Mareel, J. Vandekerckhove, and J. Gettemans. 2003. The nucleo-cytoplasmic actin-binding protein CapG lacks a nuclear export sequence present in structurally related proteins. J. Biol. Chem. 278:17945-17952. [DOI] [PubMed] [Google Scholar]
- 56.Wada, A., M. Fukuda, M. Mishima, and E. Nishida. 1998. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 17:1635-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, B., T. Mysliwiec, D. Krainc, R. A. Jensen, G. Sonoda, J. R. Testa, E. A. Golemis, and G. D. Kruh. 1996. Identification of ArgBP1, an Arg protein tyrosine kinase binding protein that is the human homologue of a CNS-specific Xenopus gene. Oncogene 12:1921-1929. [PubMed] [Google Scholar]
- 58.Wang, Y., and T. D. Gilmore. 2003. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta 1593:115-120. [DOI] [PubMed] [Google Scholar]
- 59.Weins, A., K. Schwarz, C. Faul, L. Barisoni, W. A. Linke, and P. Mundel. 2001. Differentiation- and stress-dependent nuclear cytoplasmic redistribution of myopodin, a novel actin-bundling protein. J. Cell Biol. 155:393-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Welch, M. D., and R. D. Mullins. 2002. Cellular control of actin nucleation. Annu. Rev. Cell. Dev. Biol. 18:247-288. [DOI] [PubMed] [Google Scholar]
- 61.Westphal, R. S., S. H. Soderling, N. M. Alto, L. K. Langeberg, and J. D. Scott. 2000. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 19:4589-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao, W., M. A. Poirier, M. K. Bennett, and Y. K. Shin. 2001. The neuronal t-SNARE complex is a parallel four-helix bundle. Nat. Struct. Biol. 8:308-311. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto, A., T. Suzuki, and Y. Sakaki. 2001. Isolation of hNap1BP which interacts with human Nap1 (NCKAP1) whose expression is down-regulated in Alzheimer's disease. Gene 271:159-169. [DOI] [PubMed] [Google Scholar]
- 64.Yamazaki, D., S. Suetsugu, H. Miki, Y. Kataoka, S. Nishikawa, T. Fujiwara, N. Yoshida, and T. Takenawa. 2003. WAVE2 is required for directed cell migration and cardiovascular development. Nature 424:452-456. [DOI] [PubMed] [Google Scholar]
- 65.Yan, C., N. Martinez-Quiles, S. Eden, T. Shibata, F. Takeshima, R. Shinkura, Y. Fujiwara, R. Bronson, S. B. Snapper, M. W. Kirschner, R. Geha, F. S. Rosen, and F. W. Alt. 2003. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 22:3602-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zamponi, G. W. 2003. Regulation of presynaptic calcium channels by synaptic proteins. J. Pharmacol. Sci. 2:79-83. [DOI] [PubMed] [Google Scholar]
- 67.Ziemnicka-Kotula, D., J. Xu, H. Gu, A. Potempska, K. S. Kim, E. C. Jenkins, E. Trenkner, and L. Kotula. 1998. Identification of a candidate human spectrin Src homology 3 domain-binding protein suggests a general mechanism of association of tyrosine kinases with the spectrin-based membrane skeleton. J. Biol. Chem. 273:13681-13692. [DOI] [PubMed] [Google Scholar]