Abstract
Worldwide, more than 1 in 10 infants is born prior to 37 weeks gestation. Preterm birth can lead to increased mortality risk and poor life-long health and neurodevelopmental outcomes. Whether environmental risk factors affect preterm birth through epigenetic phenomena is largely unstudied. We sought to determine whether preterm risk factors, such as smoke exposure and education, were associated with cervical DNA methylation in the prostaglandin E receptor 2 gene (PTGER2) and a repetitive element, long interspersed nuclear element-1 Homo sapiens-specific (LINE 1-HS). Second, we aimed to determine whether mid-pregnancy DNA methylation of these regions in cervical samples could predict the length of gestation. We obtained a cervical swab between 16–19 weeks gestation from 80 women participating in a Mexico City birth cohort, used pyrosequencing to analyze DNA methylation of PTGER2 and LINE 1-HS, and examined associations with maternal covariates. We used accelerated failure time models to analyze associations of DNA methylation with the length of gestation. DNA methylation of both sequences was associated with Pap smear inflammation. LINE 1-HS methylation was associated with smoke exposure, BMI and parity. In adjusted models, gestations were 3.3 days longer (95%CI 0.6, 6.0) for each interquartile range of PTGER2 DNA methylation. Higher LINE 1-HS methylation was associated with shorter gestations (-3.3 days, 95%CI -6.5, -0.2). In conclusion, cervical DNA methylation was associated with risk factors for preterm birth and the length of gestation.
Keywords: Cervix, DNA methylation, epigenetics, LINE 1, Preterm birth, PTGER2
Introduction
Preterm birth remains an enormous public health issue affecting over 11% of infants worldwide, contributing to over 1 million deaths annually.1 Spontaneous preterm birth remains particularly enigmatic.2 Mechanisms explaining preterm labor, cervical incompetence, or premature rupture of membranes have eluded investigators. Genetic studies have largely failed to explain preterm birth.3 Instead, investigators discuss unifying themes of inflammation4,5 that can result from a variety of preexisting states, including genital infection or colonization patterns,6 smoking,7 and social stress (i.e., from poverty or racism).8,9 Others postulate a genetic predisposition coupled with the above risk factors could lead to preterm birth in susceptible mothers, suggesting a gene-environment interaction.10
While gene-environment interactions were originally conceived as genetic polymorphisms that either render a host protected from or susceptible to an environmental exposure, epigenetics represents another mechanism by which genes may interact with the environment. Epigenetic processes such as DNA methylation and histone modifications affect gene expression through altering chromatin structure and availability of gene promoter regions to binding with transcription factors and enhancers.11 Non-coding RNAs affect expression at the level of translation by competitively binding mRNA.12 In concert, epigenetic phenomena culminate in alterations in gene expression independent of differences in genetic sequence. Such marks are mitotically heritable and typically tissue specific.
DNA methylation is the most commonly studied epigenetic mark in epidemiologic studies and has been associated with several environmental exposures, some of which overlap with preterm birth risk factors. Smoking,13,14 aging,15 psychosocial stress,16 and air pollution17 all have been shown to be associated with alterations in DNA methylation. Furthermore, we have previously shown that lower first trimester maternal methylation of long interspersed nuclear element-1 Homo sapiens-specific (LINE 1-HS) is associated with preterm birth.18 However, our previous study was limited by the use of white blood cell DNA. Because DNA methylation is tissue specific, blood may not be the most appropriate target tissue for studying preterm delivery, which is often related to cervical dysfunction.19 Other studies of DNA methylation and preterm birth have not only been limited by the use of maternal blood DNA but also by the use of samples obtained from umbilical cord blood. While potentially indicative of fetal health effects, by its nature, cord blood is always collected after prematurity has occurred, limiting its predictive utility. Blood that can only be collected at the time of delivery, does not allow for differentiating natural DNA methylation changes that may occur with advancing gestational age from mechanisms that may precipitate preterm birth. To address these limitations, we studied the cervix, a target tissue relevant to parturition. In addition to their role as a barrier to infection, cervical epithelial cells have been shown to play a role in immunomodulation and labor initiation by releasing metalloproteinases (MMPs) in animal models.20 Because local cervical installation of prostaglandins is used to induce labor, we analyzed a gene plausibly related to labor, prostaglandin E receptor 2 (subtype EP2) (PTGER2), from cervical cells collected between 16–19 wk gestation. Our hypothesis was that higher DNA methylation in the upstream promoter of PTGER2 would be associated with longer gestations, since DNA methylation typically represses gene expression, theoretically making the cervix less sensitive to prostaglandins. We also analyzed LINE 1-HS DNA methylation levels in the cervix with the hypothesis that we would observe an association between hypomethylation and shorter gestations as we had seen in the aforementioned study.
Results
Of the 80 samples collected, 77 were successfully interrogated for PTGER2 DNA methylation and 78 for LINE 1-HS. Mean PTGER2 methylation was 32.5% 5mC (SD 15.9, range 2.7–64.8). Mean LINE 1-HS methylation was 68.8% 5mC (SD 4.3, range 61.8–77.6). Technical replicates for PTGER2 revealed a high Pearson correlation coefficient of 0.99. For LINE 1-HS, between-run correlations for each CpG site ranged from 0.80–0.91. When we averaged the values across each CpG site, the between-run correlation was 0.99.
Four of the women who initially enrolled in the study dropped out before delivery, resulting in 76 live births. Just 7 (9.2%) were born prior to 37 wk gestation (Table 1). Median gestational age was 38 wk (271 d) (range 34–42 wk). More than half (51%) women delivered via Cesarean section (9/39 Cesarean sections were scheduled, and 30/39 done for clinical indications including fetal distress, labor with breech positioning and arrest of descent) and 17% experienced premature rupture of membranes. Most women were between 20 and 35 y of age at the time of enrollment. More than half (51%) of the women had pre-pregnancy BMI < 25 kg/m2. The majority of women (71%) were exposed to household tobacco smoke, but none reported smoking themselves during pregnancy. However, 60% of the women reported smoking prior to pregnancy. Of the 76 women with Papanicolaou (Pap) smear results, 31 (41%) had evidence of inflammation on the Pap smear. None had evidence of dysplasia.
Table 1. Cervical DNA methylation of the prostaglandin E receptor 2 (PTGER2) and long interspersed nuclear element-1 (LINE 1-HS) ELEMENT birth cohort, Mexico City.
| PTGER2 (n = 77) | LINE 1-HS (n = 78) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Participant Characteristics (n = 80)* | n | % | Mean % 5mC | SD | Chi-square P value | Mean % 5mC | SD | Chi-square P value | ||||
| Maternal age (years) | 0.28 | 0.59 | ||||||||||
| <20 | 6 | 7.5 | 35.7 | 10.5 | 68.1 | 3.1 | ||||||
| 20 – <35 | 64 | 80.0 | 31.2 | 15.9 | 69.1 | 4.3 | ||||||
| ≥35 | 10 | 12.5 | 40.3 | 18.3 | 67.6 | 4.7 | 0.585 | |||||
| Parity | 0.46 | 0.17 | ||||||||||
| Nulliparous | 38 | 47.5 | 34.0 | 16.3 | 68.1 | 3.9 | ||||||
| Multiparous | 42 | 52.5 | 31.3 | 15.7 | 69.4 | 4.5 | ||||||
| Smoked before pregnancy | 0.04 | 0.22 | ||||||||||
| Yes | 48 | 60.0 | 29.4 | 16.0 | 69.3 | 4.3 | ||||||
| No | 32 | 40.0 | 37.1 | 15.0 | 68.1 | 4.2 | ||||||
| Smoke exposure in the home | 0.40 | 0.28 | ||||||||||
| Yes | 57 | 71.3 | 30.1 | 15.4 | 68.0 | 4.3 | ||||||
| No | 23 | 28.7 | 33.5 | 16.2 | 69.1 | 4.3 | ||||||
| Education (years) | 0.92 | 0.21 | ||||||||||
| <12 | 22 | 27.5 | 32.8 | 16.3 | 69.8 | 4.1 | ||||||
| ≥12 | 58 | 72.5 | 32.4 | 15.9 | 68.4 | 4.4 | ||||||
| Pre-pregnancy BMI (kg/m2) | 0.12 | 0.17 | ||||||||||
| <25 | 45 | 56.3 | 30.8 | 15.9 | 69.1 | 4.3 | ||||||
| 25 – <30 | 23 | 27.7 | 31.5 | 15.3 | 69.3 | 4.1 | ||||||
| ≥30 | 12 | 15.0 | 41.6 | 15.6 | 66.7 | 4.5 | ||||||
| Pap smear inflammation | <0.001 | <0.001 | ||||||||||
| No | 45 | 59.2 | 38.1 | 13.8 | 67.2 | 3.5 | ||||||
| Yes | 31 | 40.8 | 24.5 | 14.2 | 71.1 | 4.2 | ||||||
| Gestational Age groups | 0.79 | 0.50 | ||||||||||
| <37 | 7 | 9.2 | 31.0 | 15.9 | 70.4 | 4.6 | ||||||
| 37–38 | 33 | 43.4 | 32.0 | 15.7 | 68.8 | 4.4 | ||||||
| ≥39 | 36 | 47.4 | 34.3 | 15.9 | 68.4 | 4.1 | ||||||
| Method of Delivery | 0.13 | 0.79 | ||||||||||
| Vaginal | 37 | 48.7 | 30.1 | 14.8 | 68.9 | 4.3 | ||||||
| Cesarean Section | 39 | 51.3 | 35.6 | 16.0 | 68.6 | 4.2 | ||||||
| Premature rupture of membranes | 0.06 | 0.019 | ||||||||||
| Yes | 13 | 17.1 | 25.1 | 15.1 | 70.2 | 4.5 | ||||||
| No | 63 | 82.9 | 34.5 | 15.4 | 68.5 | 4.1 | ||||||
80 mothers agreed to participate, but 4 mothers dropped out before delivery. 76/80 Pap Smear results, 77/80 successful assays for PTGER2 and 78/80 for LINE 1-HS.
Bivariate associations revealed that evidence of inflammation on the Pap smear was significantly associated with PTGER2 and LINE 1-HS methylation (Table 1). History of smoking prior to pregnancy was associated with higher PTGER2 methylation. Premature rupture of membranes was associated with LINE 1-HS methylation (Table 1). Because we were concerned that cell mixture from the cervical swab might be the primary determinant of DNA methylation patterns, we restricted all multivariable regression models to women with Pap smear results and then adjusted for Pap smear inflammation in a separate model (Table 2). In multivariable models, pre-pregnancy BMI demonstrated a borderline association with PTGER2 DNA methylation that was not statistically significant. Each kg/m2 increment was associated with 0.7% 5mC higher PTGER2 DNA methylation, (95% CI –0.2, 1.6). When further adjusted for Pap smear result, the association between BMI and PTGER2 methylation did not materially change (0.6, 95% CI –0.2, 1.5), even in the presence of a strong association of inflammation with PTGER2 DNA methylation. For LINE 1-HS, multivariable linear regression revealed that pre-pregnancy BMI was inversely associated with LINE 1-HS methylation. For each increment of kg/m2, DNA methylation of LINE 1-HS was 0.2% 5mC lower. When we further adjusted for Pap smear inflammation, we observed that household smoke exposure was associated with lower LINE 1-HS (–2.5% 5mC, 95% CI –4.4, –0.6). However, personal history of smoking prior to pregnancy was not associated with LINE 1-HS DNA methylation (0.8, 95% CI –1.1, 2.6). Primiparous women had higher LINE 1-HS DNA methylation (2.3, 95% CI 0.5, 4.1). We did not observe any association of maternal lead levels with cervical DNA methylation.
Table 2. Associations of maternal characteristics and cervical DNA methylation, ELEMENT birth cohort, Mexico City.
| PTGER2 β^ in % 5mC (95% CI) | LINE 1-HS β^ in % 5mC (95% CI) | ||||
|---|---|---|---|---|---|
| Maternal characteristics | |||||
| Model 1 | Model 2 | Model 1 | Model 2 | ||
| Maternal age (years) | |||||
| < 20 | 5.4 (-8.2, 19.0) | 3.2 (-9.2, 15.6) | -1.6 (-5.3, 2.1) | -0.9 (-4.1, 2.3) | |
| 20- < 35 | ref. | ref. | ref. | ref. | |
| ≥ 35 | 3.8 (-9.1, 16.7) | 6.5 (-5.3, 18.3) | -0.7 (-3.8, 2.4) | -2.1 (-4.8, 0.4) | |
| Maternal pre-pregnancy BMI per kg/m2 | 0.7 (-0.2, 1.6) | 0.6 (-0.2, 1.5) | -0.2 (-0.5, 0.0) | -0.2 (-0.4, 0.0) | |
| Primiparous (vs. multiparous) | -4.1 (-11.8, 3.5) | -6.0 (-13.0, 1.0) | 1.7 (-0.3, 3.7) | 2.3 (0.5, 4.1) | |
| Maternal Education (> = 12 y vs. less) | -3.4 (-11.6, 4.9) | -5.9 (-13.6, 1.7) | -0.5 (-2.7, 1.8) | 0.4 (-1.6, 2.3) | |
| Smoking exposure within the home (vs. not) | -2.2 (-10.3, 6.0) | 0.8 (-6.8, 8.4) | -1.6 (-3.7, 0.9) | -2.5 (-4.4, -0.6) | |
| Smoking prior to pregnancy (vs. never) | -6.8 (-14.6, 1.1) | -4.3 (-11.5, 3.0) | 1.6 (-0.5, 3.7) | 0.8 (-1.1, 2.6) | |
| Maternal second trimester blood lead per μg/dl | 0.5 (-1.6, 2.6) | 0.3 (-1.6, 2.2) | 0.0 (-0.6, 0.5) | 0.1 (-0.4, 0.5) | |
| Pap smear inflammation (vs. not) | -13.5 (-20.6, -6.4) | 4.5 (2.7, 6.3) | |||
PTGER2 prostaglandin E receptor 2 (subtype EP2); LINE 1-HS long interspersed nuclear element-1 Homo sapiens-specific. PTGER2 models include 73 participants; LINE 1-HS models include 75 participants. ^β coefficients obtained from linear regression models inclusive of all variables listed.
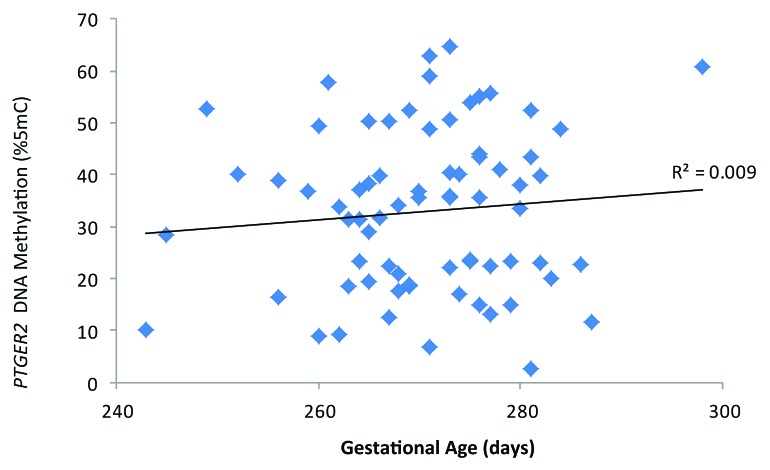
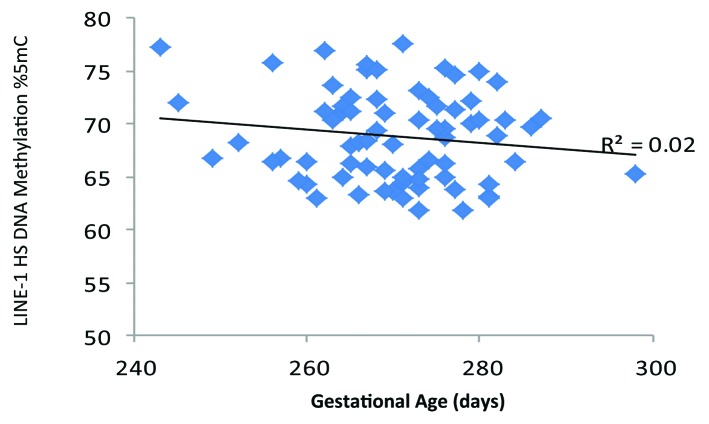
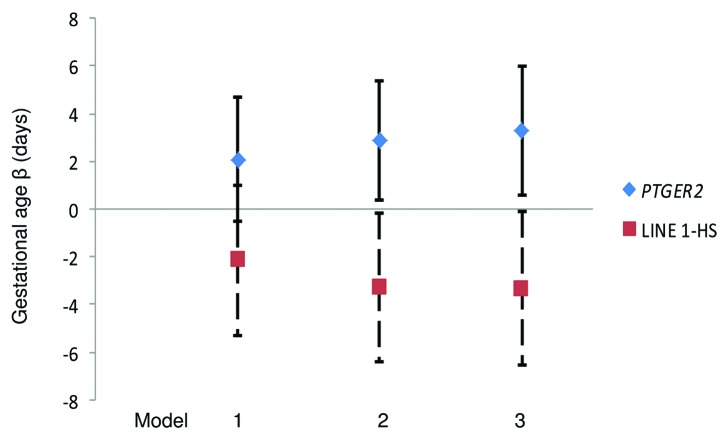
Scatter plots of DNA methylation with gestational age revealed a subtle positive, but non-significant, association between PTGER2 DNA methylation and gestational age (Fig. 1). Conversely, we observed a potentially inverse association between LINE 1-HS DNA methylation and gestational age (Fig. 2). Accelerated failure time models including participants with DNA methylation values, gestational age assessments and interpretable Pap smear results (n = 69 for PTGER2 models and n = 71 for LINE 1-HS models) revealed that the crude associations were not significant between DNA methylation of either PTGER2 or LINE 1-HS and gestational age (Fig. 3). For each interquartile range (IQR) of methylation of PTGER2, gestational age was 2.1 d longer (95% CI–0.5, 4.7). However, adjustment for maternal age resulted in a significant association; for each IQR increment of PTGER2 methylation, gestational age was 2.9 d longer (95% CI 0.4, 5.4). Further adjustment for Pap smear result only strengthened the association (3.3, 95% CI 0.6, 6.0). Counter to our hypothesis, we observed the opposite for LINE 1-HS. For each IQR increment of LINE 1-HS, gestational age was shorter, but not significantly so (–2.2 d, 95% CI –5.3, 1.0). Once we adjusted for maternal age and Pap smear result, this association became significant (–3.3 d, 95% CI –6.5, –0.2). Adjustment for other covariates did not alter the results and thus were excluded from parsimonious models.
Figure 1. Cervical prostaglandin E receptor 2 (subtype EP2) (PTGER2) DNA methylation and gestational age (days), ELEMENT birth cohort, Mexico City (n = 73).
Figure 2. Cervical long interspersed nuclear element-1 Homo sapiens specific (LINE 1-HS) DNA methylation and gestational age (days), ELEMENT birth cohort, Mexico City (n = 74).
Figure 3. Gestational Age Differences per interquartile range of cervical DNA methylation of PTGER2 and LINE 1-HS, ELEMENT birth cohort, Mexico City. Model 1: Unadjusted Model 2: Adjusted for maternal age Model 3: Model 2 additionally adjusted for Pap smear inflammation Abbreviations: PTGER2 prostaglandin E receptor 2 (subtype EP2); LINE 1-HS long interspersed nuclear element-1 Homo sapiens-specific. Footnote: n = 69 for PTGER2 models and n = 71 for LINE 1-HS models; error bars indicate 95% confidence intervals.
Discussion
We found that DNA methylation in the PTGER2 gene and LINE 1-HS repetitive element of cervical cells obtained from swabs during mid-pregnancy predicted the length of gestation. We also found that household tobacco smoke exposure, a known preterm birth risk factor, was associated with mid-pregnancy LINE 1-HS DNA methylation of the cervix. This finding is of particular relevance because the mechanistic link between smoke exposure and spontaneous preterm delivery remains poorly understood.
Most previous epigenetic studies of birth outcomes focus on fetal growth/birth weight.21-27 In a prior prospective, longitudinal US-based birth cohort study, we found that first trimester maternal blood LINE 1-HS DNA methylation was directly associated with the length of gestation and that cord blood LINE 1-HS was inversely associated with the length of gestation.18 However, that study used blood DNA, which may not be the appropriate target tissue for studying preterm birth compared with cervical cells. Another group of investigators studied associations of methylation patterns in 194 full term infants in cord blood with gestational age28 and identified 26 CpG sites associated with gestational age using the Infinium 27K array. Another recent study used the Infinium 450K array and found DNA methylation of 29 CpG sites were associated with preterm birth from cord blood DNA from 22 preterm infants born before 34 wk gestations and 28 full term controls.29 Such cross-sectional studies of cord blood would have limited clinical use in predicting preterm birth during pregnancy.
Our investigation of the cervix as a relevant target tissue for preterm birth is an improvement on prior epigenetic studies. This design strength also makes our findings more biologically plausible than a study that used a surrogate tissue for DNA methylation, as the cervix is a critical component of labor initiation. Women are routinely given prostaglandins to induce labor,30 highlighting the potential functional role of PTGER2 in labor initiation. In a caprine model, investigators found that PTGER2 mRNA levels were higher in laboring mothers than non-pregnant females or during mid-pregnancy, supporting the notion that PTGER2 plays a role in labor.31 Cancer researchers have also found that increased DNA methylation of the CpG islands of PTGER2 is associated with progression of neuroblastomas32 and that aberrant methylation of the PTGER2 gene is frequently observed in non-small cell lung cancer tissue,33 suggesting that DNA methylation of PTGER2 may regulate cellular proliferation. Taken together, these studies support the notion that PTGER2 is biologically relevant to labor initiation and the mechanism may be through cellular proliferation and subsequent sensitivity to prostaglandins.
In addition to biologic plausibility, an additional strength of our study is its prospective study design. We collected cervical cells at 16–19 wk gestation, which is prior to the development of preterm delivery, suggesting that our findings could be useful for a clinically relevant predictive tool to help identify women at risk of preterm labor. Yet, our study has several limitations. First is the lack of ability to study preterm birth as a more relevant health outcome compared with gestational age differences of just a few days. However, if the distribution of gestational age of the entire population were shifted by two days, this would substantially affect the incidence of preterm birth. Nonetheless, future, nested case-control studies may be able to more efficiently determine whether these marks are associated with preterm birth. The second limitation is our inability to accurately identify cell type. The DNA we extracted came from a cell mixture that likely varied across subjects. The clinical pathologist did not provide a detailed assessment of the subtype of leukocytes, nor the different subtypes of epithelial cells noted on the Pap smear thereby limiting our ability to further adjust for cell type. Furthermore, the cells obtained through cervical swabs were too few to perform cell-sorting. In the subset of women without evidence of inflammation, the vast majority of cells are likely epithelial (both squamous and glandular) cells since we sampled internal aspect of the external cervical os. For blood DNA epigenome-wide analyses, statistical approaches to adjust for leukocyte mixture have been developed,34 but for gene-specific or LINE 1-HS analyses, there are no tools to determine which cells were present based on DNA methylation results. It should also be noted that cell type itself is not necessarily a confounder in the relationship between environmental exposures and preterm birth, as cell type may be a mediator. Thus, although our results might be because methylation is a marker of cell type, DNA methylation changes could still be a better predictor of risk than cell type assessments and still deserving of study. We did, however, adjust our analysis using a binary inflammation variable (yes/no). This variable was strongly associated with the methylation of both PTGER2 and LINE 1-HS. We were reassured that, despite these associations, adjustment for the presence of inflammation did not have a large impact on the association between methylation and the length of gestation. As a sensitivity analysis we re-analyzed associations of DNA methylation with covariates in the subset without evidence of Pap smear inflammation and the results did not substantially differ from the combined group.
We defined gestational age as the difference between the birth date and the mother’s report upon enrollment of her last menstrual period. Ultrasound is not routinely performed in Mexico and thus we cannot corroborate the gestational age by that method. Additionally, in a pilot study prior to this cohort, enrolled in an identical way, ultrasound was performed on a subset of 98 women and the correlation between LMP gestational age and ultrasound was 0.89 (P < 0.0001). Nonetheless, we recognize there could be non-differential measurement error with regard to gestational age. Furthermore, there could be pyrosequencing measurement error. Neither of these errors should result in bias, but could add noise to our estimates making associations more difficult to detect. We ascertained smoke exposure using a questionnaire as opposed to potentially more accurate methods such as cotinine measurements and, thus, our study could be limited by exposure misclassification. Because we approached a convenience sample of the final 100 women in the parent cohort, we do not believe there was significant selection bias, but there may have been some. Twenty women declined to participate potentially limiting the generalizability of our findings. Our study was performed solely in Mexico City and, thus, needs replication in other populations to further ensure generalizability and reproducibility.
Our selection of CpG sites was limited by resources and technical methods. We analyzed just one CpG site on the promoter upstream of PTGER2. Although methylation is often highly correlated over short genomic regions (~1000 base pairs),35 one CpG site is insufficient to understand the overall regulation of gene expression of any gene. To assess correlation of DNA methylation across PTGER2, we analyzed data from Farkas and colleagues, who performed a case-control study of cervical cancer and DNA methylation of cervical samples.36 We downloaded their data from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) (data set GSE46306). This data set includes genome-wide DNA methylation from cervical tissue from 20 healthy controls measured with the Illumina HumanMethylation 450 BeadChip. There were 11 methylation sites within +/− 5 kilobases of our selected pyrosequencing site (although our particular site in not included on the BeadChip microarray). The mean spearman correlation of adjacent pairs of these sites was 0.34 (mean distance between these sites was 695 base pairs). We chose our specific CpG site based on its close proximity to a CCAAT box is a sequence known for binding general transcription factors as well as RNA transcription factors.37 In an ideal study, one would interrogate more CpG sites, analyze histone modifications that could also affect transcription and perhaps the non-coding RNAs that affect translation. Ultimately, measurement of mRNA or the resultant protein, combined with measures of the epigenetic and genetic variation in the PTGER2, would provide much greater insight into whether the regulation of this gene plays a causal role in cervical function. The measurement of LINE 1-HS DNA methylation by pyrosequencing is also limited to the representative sequence chosen. Furthermore, sodium-bisulfite conversion, which precedes pyrosequencing, cannot differentiate between CpG to TpG mutation and cytosine methylation. To compensate for both of those limitations, we analyzed 4 CpG sites within LINE 1-HS. LINE 1-HS has been shown to be responsive to several environmental factors, but its role is still not completely understood. Methylation of repetitive elements is thought to silence their expression and promote genomic stability. Our hypothesis was that hypomethylation of LINE 1-HS would be associated with shorter gestations based on our prior study demonstrating lower maternal LINE 1-HS from peripheral blood leukocytes was associated with preterm birth.18 However, we observed the opposite. DNA methylation does not always correlate with less expression and may be associated with higher levels of expression if DNA methylation of LINE 1-HS is a compensatory effect. In other words, the cervix response to inflammation could be to methylate repetitive elements to promote genomic stability. In vitro studies and expression studies analyzing mRNA would be necessary to confirm the occurrence of this compensatory activity.
In conclusion, using a novel approach of studying the cervix, we demonstrated that cervical DNA methylation of PTGER2 and LINE 1-HS obtained mid pregnancy was associated with the length of gestation. We also found that while personal smoking history prior to pregnancy was not associated with LINE 1-HS DNA methylation of the cervix, household smoke exposure was associated with LINE 1-HS, highlighting the notion that environmental health effects may be mediated through epigenetic phenomena. Further work is necessary to determine whether these DNA methylation biomarkers are associated with preterm birth to make them clinically useful as well as to determine whether they are part of the causal pathway linking risk factors for preterm birth with spontaneous early delivery.
Methods
We approached a subset of 100 women being enrolled in the ELEMENT birth cohort in Mexico City. Details of enrollment for the parent cohort are published elsewhere.21 Briefly, women were recruited between 2007 and 2011 during the second trimester through the Mexican social security system (Instituto Mexicano del Seguro Social). The parent cohort consists of 1054 mothers. Eighty of the final 100 parent cohort enrollees provided written, informed consent for an obstetrician to obtain a cervical swab mid-pregnancy (16–19 wk gestation) for epigenetic analysis. Women were offered a free Papanicolaou (Pap) smear in return for their participation, which is the standard of care during pregnancy if a woman is due for a Pap smear in the United States, but is not routinely offered in Mexico. The IRBs of the participating institutions (Brigham and Women’s Hospital and The National Institute of Public Health in Mexico) approved this study.
Participant data collection
Demographics and birth outcomes were collected as part of the parent cohort including maternal age, parity, and years of education. Study staff measured mothers’ height to calculate pre-pregnancy body mass index (BMI) with self-reported pre-pregnancy weight. Staff also conducted in-person surveys which included a question about personal smoking and household smoke exposure. We dichotomized household smoke exposure as yes/no based on whether mothers reported at least one household member smoked during the pregnancy. Gestational age was calculated from the maternally reported last menstrual period. As part of the parent cohort, maternal second trimester blood lead levels were measured using atomic absorption mass spectrometry. Compared with the larger parent cohort, this subcohort did not differ with respect to gestational age or smoke exposure. However, women enrolled in this study had more years of education (P = 0.02) and were more likely to have premature rupture of membranes (P = 0.05).
Cervical specimen collection and analysis
At a mean of 17.9 wk gestation (SD 1.2, range 16.0–19.0 wk) an obstetrician performed a speculum examination and cleared away cervical mucus with a large soft, cotton swab. A narrower cotton swab was used to encircle the cervical os five times. The swab was then placed in 2% n-acetyl cysteine and frozen at –80 °C. The samples were stored in Mexico City and then shipped to Boston on dry ice for DNA methylation analyses. DNA was extracted using Qiagen tissue extraction kits (QIAGEN). One µg of genomic DNA was bisulfite converted using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer’s protocol. Final elution was performed with 30 µl M-Elution Buffer. We performed DNA methylation analyses on bisulfite-treated DNA using a quantitative analysis based on PCR-pyrosequencing. PCR and pyrosequencing primer sequences for LINE1-HS have been previously published.38 We developed a specific assay for PTGER2 measuring % 5mC at one individual CpG dinucleotide located on chromosome 14 (position 52780428) within a CpG island in the gene promoter just 7 nucleotides from a CCAAT box. The CCAAT box is a sequence known for binding general transcription factors and RNA transcription factors thus having a primary function in regulating gene transcription.37 For the gene map, please see Figure S1. Details of the pyrosequencing are provided in supplementary material, Table S1. Briefly, PCR for PTGER2 was performed with 15 μL GoTaqR Hot Start Polymerase (Promega), 1 µl forward primer (PTGER2_F: TGGTTTTTTG GATGGATATA TTTGT), 1 µl biotinylated reverse primer (PTGER2_R [bio]: AATAAAATTA AAAAAACCCA ATCCC), 1 µg bisulfite-treated genomic DNA, and water. PCR cycling conditions were 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s for 40 cycles. PCR products were purified and sequenced by PSQ Q96 MD pyrosequencing System (QIAGEN), as previously described17 using 0.3 μM sequencing primer (PTGER2_Seq: GATATATTTG TTTTA). Each sample was tested two times for each assay to confirm reproducibility of pyrosequencing performance and the average was used for all statistical analyses. As quality controls, we placed duplicate genomic DNA samples and universal PCR products to estimate the internal plate variation, bisulfite conversion efficiency and the pyrosequencing reaction. A second swab was sent to a clinical pathologist for a clinical Pap smear. The pathologist was blinded to both methylation status and the length of gestation. We subsequently dichotomized the Pap smear result as inflammation vs. no inflammation based on the clinically reported result. Inflammation was defined as the presence of leukocytes or cytolysis on the smear.
Statistical approach
We examined univariate descriptive statistics, bivariate associations and performed two types of multivariable modeling. To analyze associations between maternal covariates and DNA methylation we used multivariate linear regression models. To analyze the association of DNA methylation with gestational age we used accelerated failure time models, assuming a Weibull distribution. We used this latter approach because it directly models gestational age, and not hazard ratios. Gestational age has a skewed distribution with an abrupt truncation at ~42 wk gestation, which is reasonably approximated by the Weibull distribution. The result of this model provides a ratio of gestational ages. This ratio can be converted into a percent, which then can be applied to a standard time duration (in this case a normal gestation is 40 wk or 280 d). For instance, if the β coefficient were 0.5%, then for interpretability, the β coefficient would be reported as 0.005*280 = 1.4 d.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported in part by Pilot Project funding from the HSPH-NIEHS Center for Environmental Health (ES000002) and NIH/NIEHS: K23ES022242, P42 ES016454; R01ES013744, R01ES020268; R01ES014930, and the Klarman Scholars Program at Beth Israel Deaconess Medical Center.
Glossary
Abbreviations:
- 5mC
5-methylcytosine
- BMI
body mass index
- CpG
cytosine-phosphate-guanine
- LINE 1-HS
long interspersed nuclear element-1 Homo sapiens-specific
- PTGER2
prostaglandin E receptor 2 (subtype EP2)
References
- 1.Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379:2162–72. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- 2.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med. 2010;362:529–35. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 3.David R, Collins J., Jr. Disparities in infant mortality: what’s genetics got to do with it? Am J Public Health. 2007;97:1191–7. doi: 10.2105/AJPH.2005.068387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecht JL, Fichorova RN, Tang VF, Allred EN, McElrath TF, Leviton A, Elgan Study Investigators Relationship Between Neonatal Blood Protein Concentrations and Placenta Histologic Characteristics in Extremely Low GA Newborns. Pediatr Res. 2011;69:68–73. doi: 10.1203/PDR.0b013e3181fed334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leviton A, Fichorova R, Yamamoto Y, Allred EN, Dammann O, Hecht J, Kuban K, McElrath T, O’Shea TM, Paneth N. Inflammation-related proteins in the blood of extremely low gestational age newborns. The contribution of inflammation to the appearance of developmental regulation. Cytokine. 2011;53:66–73. doi: 10.1016/j.cyto.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldenberg RL, Andrews WW, Guerrant RL, Newman M, Mercer B, Iams J, Meis P, Moawad A, Das A, VanDorsten JP, et al. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network The preterm prediction study: cervical lactoferrin concentration, other markers of lower genital tract infection, and preterm birth. Am J Obstet Gynecol. 2000;182:631–5. doi: 10.1067/mob.2000.104211. [DOI] [PubMed] [Google Scholar]
- 7.Savitz DA, Dole N, Terry JW, Jr., Zhou H, Thorp JM., Jr. Smoking and pregnancy outcome among African-American and white women in central North Carolina. Epidemiology. 2001;12:636–42. doi: 10.1097/00001648-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Kramer MS, Séguin L, Lydon J, Goulet L. Socio-economic disparities in pregnancy outcome: why do the poor fare so poorly? Paediatr Perinat Epidemiol. 2000;14:194–210. doi: 10.1046/j.1365-3016.2000.00266.x. [DOI] [PubMed] [Google Scholar]
- 9.David RJ, Collins JW., Jr. Differing birth weight among infants of U.S.-born blacks, African-born blacks, and U.S.-born whites. N Engl J Med. 1997;337:1209–14. doi: 10.1056/NEJM199710233371706. [DOI] [PubMed] [Google Scholar]
- 10.Dizon-Townson DS. Preterm labour and delivery: a genetic predisposition. Paediatr Perinat Epidemiol. 2001;15(Suppl 2):57–62. doi: 10.1046/j.1365-3016.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- 11.Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb) 2010;105:105–12. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou L, Wang D, Baccarelli A. Environmental chemicals and microRNAs. Mutat Res. 2011;714:105–12. doi: 10.1016/j.mrfmmm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shenker NS, Ueland PM, Polidoro S, van Veldhoven K, Ricceri F, Brown R, Flanagan JM, Vineis P. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology. 2013;24:712–6. doi: 10.1097/EDE.0b013e31829d5cb3. [DOI] [PubMed] [Google Scholar]
- 15.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–9. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulligan CJ, D’Errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7:853–7. doi: 10.4161/epi.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burris HH, Rifas-Shiman SL, Baccarelli A, Tarantini L, Boeke CE, Kleinman K, Litonjua AA, Rich-Edwards JW, Gillman MW. Associations of LINE-1 DNA Methylation with Preterm Birth in a Prospective Cohort Study. J Dev Orig Health Dis. 2012;3:173–81. doi: 10.1017/S2040174412000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iams JD, Goldenberg RL, Meis PJ, Mercer BM, Moawad A, Das A, Thom E, McNellis D, Copper RL, Johnson F, et al. National Institute of Child Health and Human Development Maternal Fetal Medicine Unit Network The length of the cervix and the risk of spontaneous premature delivery. N Engl J Med. 1996;334:567–72. doi: 10.1056/NEJM199602293340904. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez JM, Dong Z, Romero R, Girardi G. Cervical remodeling/ripening at term and preterm delivery: the same mechanism initiated by different mediators and different effector cells. PLoS One. 2011;6:e26877. doi: 10.1371/journal.pone.0026877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burris HH, Braun JM, Byun HM, Tarantini L, Mercado A, Wright RJ, Schnaas L, Baccarelli AA, Wright RO, Tellez-Rojo MM. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu. Epigenomics. 2013;5:271–81. doi: 10.2217/epi.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoyo C, Fortner K, Murtha AP, Schildkraut JM, Soubry A, Demark-Wahnefried W, Jirtle RL, Kurtzberg J, Forman MR, Overcash F, et al. Association of cord blood methylation fractions at imprinted insulin-like growth factor 2 (IGF2), plasma IGF2, and birth weight. Cancer Causes Control. 2012;23:635–45. doi: 10.1007/s10552-012-9932-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilhelm-Benartzi CS, Houseman EA, Maccani MA, Poage GM, Koestler DC, Langevin SM, Gagne LA, Banister CE, Padbury JF, Marsit CJ. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect. 2012;120:296–302. doi: 10.1289/ehp.1103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michels KB, Harris HR, Barault L. Birthweight, maternal weight trajectories and global DNA methylation of LINE-1 repetitive elements. PLoS One. 2011;6:e25254. doi: 10.1371/journal.pone.0025254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabano S, Colapietro P, Cetin I, Grati FR, Zanutto S, Mandò C, Antonazzo P, Pileri P, Rossella F, Larizza L, et al. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics. 2010;5:313–24. doi: 10.4161/epi.5.4.11637. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, Uxa R, Keating S, Kingdom J, Weksberg R. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol. 2008;320:79–91. doi: 10.1016/j.ydbio.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 27.Filiberto AC, Maccani MA, Koestler D, Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, Gagne LA, Marsit CJ. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6:566–72. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder JW, Conneely KN, Cubells JC, Kilaru V, Newport DJ, Knight BT, Stowe ZN, Brennan PA, Krushkal J, Tylavsky FA, et al. Neonatal DNA methylation patterns associate with gestational age. Epigenetics. 2011;6:1498–504. doi: 10.4161/epi.6.12.18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parets SE, Conneely KN, Kilaru V, Fortunato SJ, Syed TA, Saade G, Smith AK, Menon R. Fetal DNA Methylation Associates with Early Spontaneous Preterm Birth and Gestational Age. PLoS One. 2013;8:e67489. doi: 10.1371/journal.pone.0067489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swamy GK. Current methods of labor induction. Semin Perinatol. 2012;36:348–52. doi: 10.1053/j.semperi.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 31.Gu G, Gao Q, Yuan X, Huang L, Ge L. Immunolocalization of adipocytes and prostaglandin E2 and its four receptor proteins EP1, EP2, EP3, and EP4 in the caprine cervix during spontaneous term labor. Biol Reprod. 2012;86:159–, 1-10. doi: 10.1095/biolreprod.111.096040. [DOI] [PubMed] [Google Scholar]
- 32.Sugino Y, Misawa A, Inoue J, Kitagawa M, Hosoi H, Sugimoto T, Imoto I, Inazawa J. Epigenetic silencing of prostaglandin E receptor 2 (PTGER2) is associated with progression of neuroblastomas. Oncogene. 2007;26:7401–13. doi: 10.1038/sj.onc.1210550. [DOI] [PubMed] [Google Scholar]
- 33.Tian L, Suzuki M, Nakajima T, Kubo R, Sekine Y, Shibuya K, Hiroshima K, Nakatani Y, Fujisawa T, Yoshino I, Thoracic Surgery Group Clinical significance of aberrant methylation of prostaglandin E receptor 2 (PTGER2) in nonsmall cell lung cancer: association with prognosis, PTGER2 expression, and epidermal growth factor receptor mutation. Cancer. 2008;113:1396–403. doi: 10.1002/cncr.23694. [DOI] [PubMed] [Google Scholar]
- 34.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farkas SA, Milutin-Gašperov N, Grce M, Nilsson TK. Genome-wide DNA methylation assay reveals novel candidate biomarker genes in cervical cancer. Epigenetics. 2013;8:1213–25. doi: 10.4161/epi.26346. [DOI] [PubMed] [Google Scholar]
- 37.Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29–49. doi: 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]
- 38.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.