Abstract
The ability to discriminate between closely related contexts is a specific form of hippocampal-dependent learning that may be impaired in certain neurodegenerative disorders such as Alzheimer's and Down Syndrome. However, signaling pathways regulating this form of learning are poorly understood. Previous studies have shown that the calcium-dependent exchange factor Ras-GRF1, an activator of Rac, Ras and R-Ras GTPases, is important for this form of learning and memory. Moreover, the ability to discriminate contexts was linked to the ability of Ras-GRF1 to promote high-frequency stimulation (HFS)-LTP via the activation of p38 Map kinase. Here, we show that R-Ras is involved in this form of learning by using virally-delivered miRNAs targeting R-Ras into the CA1 region of dorsal hippocampus and observing impaired contextual discrimination. Like the loss of GRF1, knockdown of R-Ras in the CA1 also impairs the induction of HFS-LTP and p38 Map kinase. Nevertheless, experiments indicate that this involvement of R-Ras in HFS-LTP that is required for contextual discrimination is independent of Ras-GRF1. Thus, R-Ras is a novel regulator of a form of hippocampal-dependent LTP as well as learning and memory that is affected in certain forms of neurodegenerative diseases.
Keywords: Ras-GRF1, synaptic plasticity, hippocampus
Background
The hippocampus is a critical processor of contextual information from the outside world and essential contributor to the formation of long-term memories. One specific form of hippocampus dependent memory is contextual discrimination, the specialized ability to distinguish between closely related contexts (Frankland et al., 1998, Giese et al., 2001, Jeffery et al., 2004). Aging results in progressive weakening of the ability to process contextual information for the purpose of new memories. This behavior can be mimicked in mouse models of neurodegenerative disorders such as Down syndrome(Hyde and Crnic, 2001) and Alzheimer's disease and thus may be useful as a biomarker for these hallmark disorders (Tronche et al., 2010). However, the mechanisms underlying this form of learning are poorly understood.
In mice, contextual discrimination ability requires Ras-GRF1(Giese et al., 2001), which is expressed in the post-synaptic space of CNS neurons (Shou et al., 1992, Sturani et al., 1997, Zippel et al., 1997). Along with Ras-GRF2 (Fam et al., 1997), they form a family of calcium activated exchange factors for Ras and Rac GTPases (Feig, 2011). Both Ras-GRF1 and Ras-GRF2 have Dbl homology exchange factor domains for Rac and CDC25 exchange factor domains for Ras. Interestingly, the CDC25 domain of RasGRF1, but not Ras-GRF2, also has the capacity to activate the Ras-related R-Ras GTPase (Gotoh et al., 1997, Gotoh et al., 2001, Tian and Feig, 2001).
R-Ras is similar to Ras proteins, but clear differences exist in that they share only a subset of both downstream targets and upstream regulators and have distinct negative regulators. While GTP-bound Ras proteins can activate the Raf/Erk Map kinase, Ral GTPase, and PI3 kinase signaling cascades, GTP-bound R-Ras only activates PI3 kinase . Moreover, only R-Ras activates inside-out integrin signaling (Reuther and Der, 2000). While both Ras and R-Ras can be activated by GRF1 only R-Ras is activated by the exchange factor C3G (Overbeck et al., 1995). In contrast, negative regulators of these proteins are distinct in that Ras-GAPs only promote hydrolysis of GTP to GDP bound to Ras proteins (Iwashita and Song, 2008) and R-Ras GAPs such as pleixin-B1 only suppress R-Ras proteins (Negishi et al., 2005).
R-Ras has been implicated in cell adhesion and neurite outgrowth associated with developing hippocampal or cortical neurons (Ivins et al., 2000, Negishi et al., 2005). R-Ras localizes to active zones to regulate axonal morphogenesis and branching (Oinuma et al., 2007, Iwasawa et al., 2012). This involves downregulation of R-Ras by Plexin-B1 receptor activation in response to its ligand Semaphorin 4D to control growth cone collapse (Oinuma et al., 2004a, Oinuma et al., 2004b, Oinuma et al., 2010). While these studies identified a role for R-Ras in development of the presynaptic nerve terminal, a role for R-Ras in mature neurons has not been explored.
We previously reported that RasGRF1 contributes to contextual discrimination by regulating a specific form of postsynaptic plasticity called high frequency long-term potentiation (HFS-LTP, (Jin et al., 2013) in the CA1 region of the hippocampus. Additionally, this signaling was dependent on p38 activation. Because RasGRF1 can potentially activate R-Ras through the same CDC25 exchange domain that activates R-Ras, in this paper we tested the possibility that R-Ras contributes to HFS-LTP and/or contextual discrimination. Using viral delivered miRNAs specific for knockdown of R-Ras in vitro and in vivo, we found that R-Ras is necessary for HFS-LTP and contextual discrimination, but it is not functioning as an effector of Ras-GRF1 in this context.
EXPERIMENTAL PROCEDURES
Animals
2-3 month old male C57Bl/6J animals were used for all experiments. All procedures were carried out in accordance with the Institutional Animal Care and Use Committee guidelines of Tufts University.
Generation of R-Ras miRNA constructs
R-Ras miRNA sequences were purchased from Open Biosystems as an shRNAmir set (TRCN0000077618, TRCN0000077619, TRCN0000077620, TRCN0000077622) cloned into the pGIPZ expression vector. Unrelated miRNA sequence was used as a control. Each clone was individually transfected into 3T3 cells to test for endogenous knockdown efficacy of R-Ras. 1.5mL Opti-MEM media containing DNA for the pGIPZ expression vector (3μg), VSVG (3μg), and delta 8.91 plasmids (6μg) were combined with 36μl Lipofectamine 2000 (Invitrogen) and 1.5mL Opti-MEM, allowed to mix for 20 minutes and then added to 3T3 cells. After overnight incubation, the cells were washed with PBS and lysed in ice cold RIPA buffer containing protease inhibitors. Cell lysates were spun at 13,000g for 15 min, SDS-loading buffer was added to the supernatant, and SDS-PAGE was run to assay for total R-Ras using anti-R-ras antibody (sc-523, Santa Cruz) and anti-ERK antibody (sc-94, Santa Cruz) for loading control. Positive sequences (miRNA #1: CCGGCCACCATTGAAGATTCCTACACTCGAGTGTAGGAATCTTCAATGGTGGTTTTTG, miRNA #2: CCGGGTCTGAATGTCGATGAGGCATCTCGAGATGCCTCATCGACATTCAGACTTTTTG, sense in bold) and an unrelated control sequence were sent to Virovek (Hayward, CA) for cloning into the full AAV9-CMV-RRasmiRNA-mCherry vector to use for in vivo experiments.
Stereotaxic surgery
2-3 month old male C57Bl/6J animals were used for injection of AAV9-RRasmiRNAs into the CA1 region of the hippocampus. 1μl of virus (titered at 1.45e13 vg/mL) was injected on each side of the hippocampus at a rate of 0.06 μl/min using the following coordinates: Dorsal CA1: anterioposterior: −2.0mm from bregma, lateral: ±1.6mm, ventral: −1.7mm and/or Ventral CA1: anterioposterior: −2.5mm from bregma, lateral: ± 2.5mm, ventral: −1.75mm using a stereotaxic frame (BENCHmark). Animals were allowed 2 weeks for recovery and expression of the virus within the hippocampus prior to being used for electrophysiology or behavioral experiments.
Electrophysiology
Recordings using theta-burst stimulation (TBS-LTP), high frequency stimulation (HFS-LTP) or low frequency stimulation (LTD) were performed as described previously (Jin et al., 2010; Jin et al., 2013). Briefly, transverse acute hippocampal slices (350 μm) were cut in ice-cold oxygenated sucrose-enhanced artificial cerebrospinal fluid (ACSF) containing (in mM): 124 NaCl, 2 KCl, 2 MgSO4, 1.25 NaH2PO4, 2 CaCl2, 26 NaHCO3, 10 D-glucose, , saturated with 95% O2 and 5% CO2 (pH 7.4). in which they were allowed to recover for at least 90 min before recording. Schaffer collaterals were stimulated with a unipolar stimulating electrode (World Precision Instruments) placed in the lateral CA1 subfield. A borosilicate glass recording electrode filled with ACSF was positioned in the stratum radiatum of CA1. For HFS-induced LTP : two consecutive trains (1 s) of stimuli at 100 Hz separated by 20 s were applied to the CA1. 2) For TBS stimulation: 15 bursts of four pulses at 100 Hz delivered at an interburst interval of 200 ms.
p-p38 activation
For experiments testing the effect of HFS-LTP on p38 activation, slices were given the stimulation but with 2X the intensity to quantify differences between control and experimental samples and then fixed 10 minutes later and processed for immunostaining as described below.
Immunohistochemistry
All mice were deeply anesthetized with ketamine/xylazine and transcardially perfused with 0.1 M phosphate buffer (PB) followed by 4% paraformaldehyde (4% PFA) dissolved in 0.1M PB. Brains were extracted and post-fixed in 4% PFA for 24 h. Brains were transferred to 30% sucrose for 48–72 h before slicing 30 μm coronal sections through the extent of the hippocampus using a cryostat. Sections were stored in cryoprotectant at −20°C until use. For post-processing of hippocampal slices used in electrophysiology experiments, slices were bath fixed in 4% PFA overnight at 4°C prior to incubation in 30% sucrose. 18 μm coronal sections through the extent of the hippocampus were cut and directly mounted onto charged slides and processed for staining. For staining, slices were washed extensively in PBS + 0.1% Triton-X (PBST) before blocking in 5% normal goat serum (NGS) + PBST for 1 hour to reduce non-specific background. Slices were incubated with p-p38 antibodies (9211S, Cell Signaling) overnight at room temperature in 5% NGS + PBST. The next day, slices were rinsed extensively in PBST and incubated in AlexaFluor 488 secondary antibodies (A11008, Invitrogen) for 1.5 h at room temperature. The slices were washed with PBST and counterstained with DAPI to observe nuclei before mounting on slides with Vectashield (H-1400, Vector Labs) to prevent fading.
Quantification of images and statistical analysis
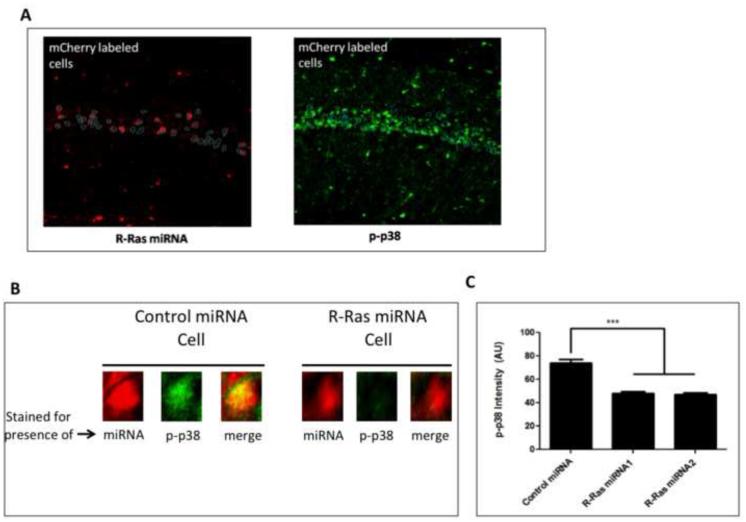
Images to assess the levels of p-p38 were captured with a 20x objective from a Nikon 80i epifluorescent microscope. Cell bodies in area CA1 positive for viral infection of R-Ras miRNA (mCherry+) were identified, traced, and saved as an ROI using ImageJ. This ROI was collected blind to the images taken for positive p-p38 staining. The ROI for virally infected cells was then overlayed onto the corresponding image for p-p38 stained sections and the intensity of the signal was measured for each corresponding cell body using ImageJ (See Figure 3A). The remaining cells positive for p-p38 were then traced and saved as a separate ROI (mCherry −). The intensity of this signal was again measured using ImageJ. An average of 187 cells for each (mCherry + vs. mCherry −) condition were collected and analyzed. Student's two-tailed t-test was performed for statistical significance set at p <0.05.
Figure 3. Knockdown of R-Ras in CA1 attenuates p38 MAPK activation after HFS-LTP stimulation.
Hippocampal slices prepared from mice injected with viruses expressing R-Ras miRNA1 or R-Ras miRNA2 into CA1 were stimulated for HFS-LTP and processed for immunostaining of phosphorylated p38. A. Representative image of CA1 pyramidal neurons positive for miRNA (mCherry) (left) traced and overlayed onto the CA1 layer of p-p38-labeled cells. Fluorescent intensity of p-p38 was quantified for each R-Ras miRNA positive cell and compared to the population of non-infected cells. B. Magnified region of A showing examples of p-p38 in both uninfected cells and cells infected with R-Ras miRNA expressing virus in the same brain slice. C. Quantification of p-p38 fluorescent intensity in R-Ras miRNA1− or R-Ras miRNA2− positive cells compared to virus-negative cells (One-Way ANOVA, F 2,732 = 199, p < 0.0001, Bonferroni post-hoc: R-Ras miRNA1+ vs R-Ras miRNA−, t= 15.93; p < 0.05, R-Ras miRNA2+ vs R-Ras miRNA−, t = 18.04, p < 0.05.
Contextual Discrimination
Contextual discrimination behavior was performed as described previously (Giese et al., 2001; (Jin et al., 2013). The mice were trained to discriminate between two contexts, one being the chamber itself in which they were shocked and a similar context in which the walls of the chamber were changed to alternating white and black bars but retained the metal grid floor, in which they were not. The first day, mice were pre-exposed to both contexts for 10 minutes. On days 2 and 3, the mice were shocked in one context (paired); after 148 s, a 2-s shock (0.75mA) was delivered, and the mice remained for another 30s in the context. After 3 hrs., the mice were exposed to the other context (unpaired) and received no shock over the 180s time period. On day 4, the mice were tested for their freezing behavior over the duration of 3 minutes in each of the two contexts.
Quantification of freezing
Freezing behavior was measured using a digital camera connected to a computer with Actimetrics FreezeFrame software. The bout length was 1 s and the threshold for freezing behavior was determined by an experimenter blind to experimental conditions and animal group. Two-Way ANOVA followed by Bonferroni's correction was performed to test for statistical significance set at p < 0.05.
RESULTS
R-Ras knockdown in the CA1 hippocampus suppresses HFS-LTP but not TBS-LTP
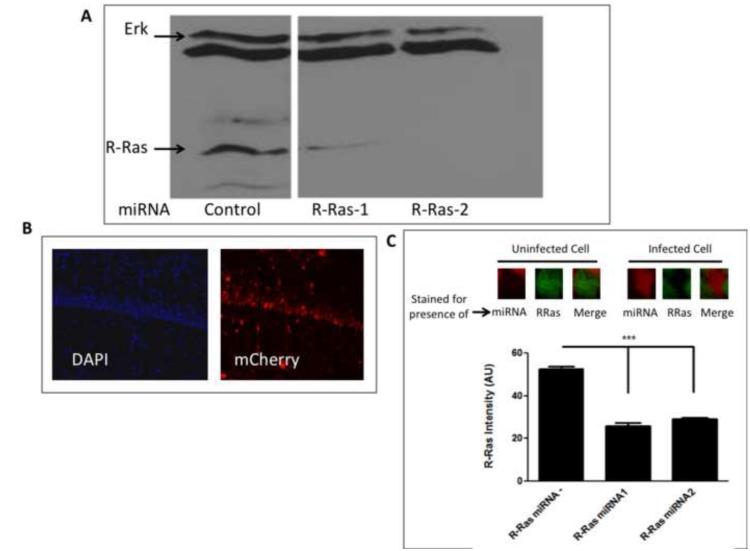
GRF1 is known to be capable of activating R-Ras, as well as Ras. Thus, we began to test whether R-Ras may function as an effector for GRF1 in synaptic plasticity by testing the consequence of knocking down the expression of R-Ras in vivo on a synaptic plasticity paradigm known to be mediated by Ras-GRF1, high frequency stimulated LTP (HFS-LTP), and one that is not, theta-burst induced LTP (TBS-LTP). To this end, we generated adeno-associated viruses (AAV) that express miRNA against R-Ras. First, two previously validated miRNAs against different segments of the R-Ras sequence were tested upon transfection of DNA encoding them into NIH 3T3 cells and compared to a control unrelated miRNA sequence for the ability to block endogenous R-Ras expression. Fig.1A shows that both miRNAs were effective in suppressing expression of R-Ras but not a control protein, Erk Map kinase, with R-Ras 2 being more effective than R-Ras1. Next, both miRNAs and a control unrelated miRNA were cloned into vectors for AAV9 co-expressing mCherry and high titer virus were generated for infection into the CA1 hippocampus by stereotactic injection. After 14 days, the CA1 hippocampus in brain slices were inspected for DAPI staining in all cells and mCherry staining in infected cells. Figure 1B shows that a significant proportion of cells of the CA1 hippocampus were infected. To confirm efficacy of the R-Ras miRNAs in vivo, immunostaining of R-Ras was performed on hippocampal tissue infected with either R-Ras miRNA in area CA1. As expected, we observed a significant reduction in R-Ras immunostaining in cells containing either miRNA compared to neighboring cells lacking viral expression (..compared to slices infected with the control miRNA) (Fig. 1C). AAV9 is known to preferentially infect neurons over non-neuronal cells (Aschauer et al., 2013). Nevertheless, we co-stained infected cells with antibodies to either NeuN or S100, which specifically stain neurons or glia, respectively. As expected, viral expression shows that over 95% of mCherry expressing cells co-expressed NeuN, while less than 5% of mCherry cells co-expressed S100 (data not shown).
Figure 1. miRNA knockdown of R-Ras in cells and in animals.
A. Selection of R-Ras miRNAs for in vivo use. Lysates of NIH 3T3 cells transfected with two R-Ras specific miRNAs or a control unrelated miRNA sequence were immunoblotted to detect endogenous R-Ras expression. B. Expression of AAV encoding R-Ras miRNA and mCherry in the CA1 hippocampus. Hippocampal brain slices from mice infected with AAV ncoding R-Ras miRNA were stained with DAPI to detect cell nuclei and mCherry fluorescence to detect expression of virus. C. R-Ras miRNAs reduce endogenous R-Ras expression in area CA1 hippocampus. Top: Representative images of cells uninfected or infected with R-Ras miRNAs and immunostained for endogenous R-Ras. Bottom: Cells infected with either R-Ras miRNA showed significantly lower staining intensity compared to neighboring cells that were uninfected (One-way ANOVA, F 2, 512 = 112.6, p < 0.0001; Bonferroni post hoc, R-Ras miRNA - vs. R-Ras miRNA1, t = 8.40, p < 0.05, R-Ras miRNA - vs. R-Ras miRNA2, t = 13.73, p < 0.05, R-Ras miRNA1 vs. R-Ras miRNA2, t = 0.98, p > 0.05).
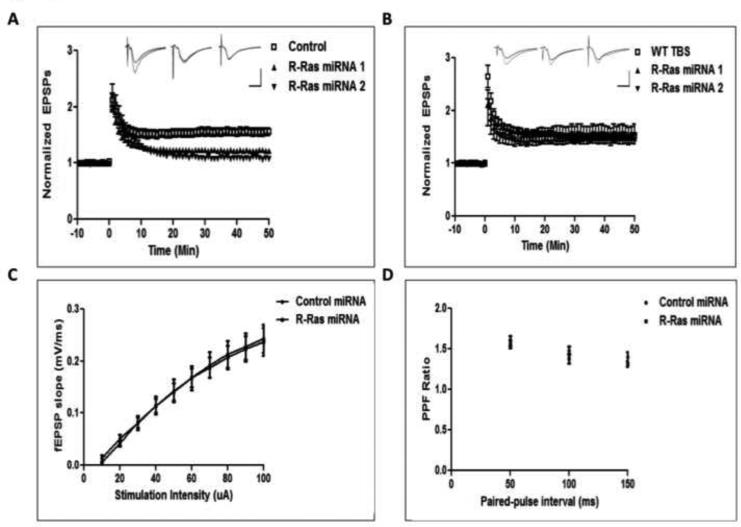
To test whether brain slices with R-Ras expression knocked down in CA1 neurons displayed defective HFS-LTP like brain slices from GRF1 knockout mice, 2-month old WT mice were stereotactically injected with either R-Ras miRNA1, R-Ras miRNA2 or control miRNAs and 14 days later HFS-LTP was measured in the area of the CA1 expressing mCherry. Figure 2A shows brain slices expressing either R-Ras miRNA , but not control miRNA, treated brain slices did, in fact, display suppressed HFS-LTP. Next, we tested TBS-LTP where GRF1 does not to play a role (Jin et al., 2013). Strikingly, R-Ras knockdown by both R-Ras miRNAs failed to suppress TBS-LTP (Fig. 2B). In addition, we injected mice with control or R-Ras miRNA into area CA1 and tested for presynaptic function to further characterize the localized effect of R-Ras knockdown in area CA1. R-Ras knockdown had no effect on the input-output relationship when compared to control miRNA , suggesting no impairment in presynaptic transmission (Fig. 1C). We also tested paired-pulse facilitation and again observed no differences between R-Ras miRNA and control miRNA (Fig. 1D). Together, these results show that both R-Ras miRNAs are quite specific in that they block HFS-LTP but not TBS-LTP, and that this effect is, as expected from viral infection conditions, postsynaptic in nature. They also suggest that R-Ras functions as an effector for GRF1 in promoting HFS-LTP in 2-month old mice that has been shown to involve a GRF1/p38 Map kinase signaling cascade (Jin et al., 2013).
Figure 2. R-Ras knockdown impairs HFS- but not TBS-LTP.
A. Viruses expressing either control miRNA, R-Ras miRNA1 (R-Ras1) or R-Ras miRNA2 (R-Ras2) were injected into CA1 and tested for HFS-LTP induction. (Control-HFS 157 ± 7.4%; R-Ras miRNA1-HFS, 127 ± 3.6%; R-Ras miRNA2-HFS, 122 ± 2.6%; Two-Way ANOVA, miRNA effect, F2,1298 = 13.89, p = 0.0001). Inset shows representative traces for control, R-Ras1, and R-Ras2 miRNAs, respectively. Calibration: Horizontal = 10ms, Vertical = 1mV. B. A separate group of mice injected with viruses expressing R-Ras miRNA1 or R-Ras miRNA2 were tested for TBS-LTP induction in area CA1. (WT-TBS, 159 ± 4.1%; R-Ras miRNA1-TBS, 149 ± 2.7%; R-Ras miRNA2-TBS, 154 ± 16.3%; Two-Way ANOVA, miRNA effect, F 2,767 = 0.64, p = 0.64). Inset shows representative traces for control, R-Ras1, and R-Ras2 miRNAs, respectively. Calibration: Horizontal = 10ms, Vertical = 1mV. C. Input-output relationship is normal in cells infected with R-Ras miRNA compared to control miRNA in area CA1 (Two-Way ANOVA, miRNA × intensity, F 9,108 = 0.05, p = 1.00). D. Paired-pulse facilitation is normal in cells infected with R-Ras miRNA compared to control miRNA in area CA1 (Two-Way ANOVA, miRNA × interval, F 2,36 = 0.64, p = 0.53).
R-Ras knockdown suppresses HFS-induced p38 Map kinase activation
HFS-LTP in mice at least 2-months of age involves GRF1-mediated activation of p38 Map kinase activation (Jin et al., 2013). Thus, in order to further test whether R-Ras may be an effector of GRF1 in HFS-LTP induction, we tested whether the ability of suppressed R-Ras expression to specifically block this form of LTP involved inhibition of p38 Map kinase activation. In these experiments virus expressing R-Ras miRNA(1) or R-Ras miRNA2 were injected into the CA1 hippocampus and then 14 days later brain slices were stimulated with the HFS induction protocol. 10 minutes later, slices were fixed and stained for active p-p38 Map kinase. Cells positive for mCherry (i.e. expressing miRNA) were selected and overlaid onto the corresponding images for p-p38 staining (Fig. 3A). A comparison between p-p38 fluorescent intensity in mCherry+ cells to neighboring cells that were uninfected with virus showed p-p38 activation to be significantly reduced (Fig. 3B). These results suggest R-Ras expression correlates with p-p38 activation and is consistent with being an effector for the GRF1/p38 signaling cascade.
R-Ras knockdown suppresses contextual discrimination
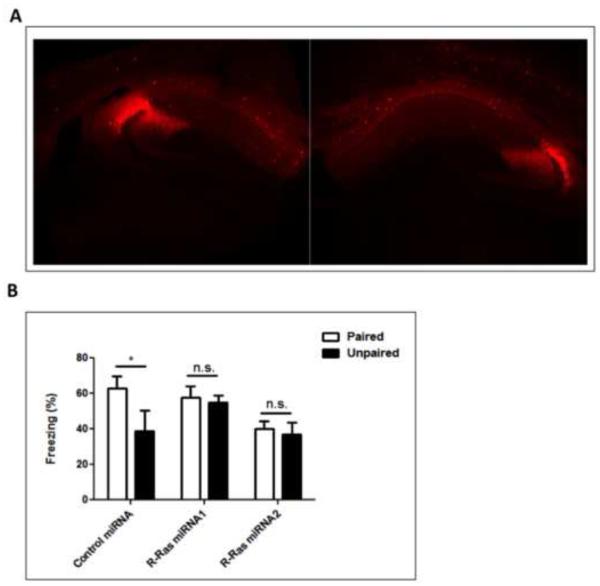
RasGRF1 is known to contribute to the hippocampal-dependent learning task contextual discrimination (Giese) through a HFS-LTP/GRF1/p38 signaling cascade (Jin et al., 2013). To determine whether R-Ras knockdown also leads to behavior changes consistent with its effect on HFS-LTP and p38 activation, mice infected bilaterally in the dorsal CA1 hippocampus with virus expressing control, R-RasmiRNA1 or R-RasmiRNA2 were tested for their ability to display contextual discrimination. In this assay, mice were exposed to one context in which they were given a brief foot shock (paired) and another closely related one in which they were not (unpaired). Mice that display contextual discrimination freeze for a longer duration when placed back in the context within which they were shocked than in the closely related one in which they were not. Figure 4 shows that mice expressing control miRNA displayed proper contextual discrimination by freezing for a longer total duration in the paired context compared to the unpaired context. Mice infected with virus expressing either R-Ras miRNA1 or R-RasmiRNA2 froze equally in the paired and unpaired contexts. Overall, mice infected with either of the R-Ras miRNA viruses froze the same in the unpaired context as in the paired context indicating that like GRF1 knockout mice both sets of mice expressing R-Ras miRNA lost contextual discrimination learning.
Figure 4. Knockdown of R-Ras in the dorsal CA1 impairs contextual discrimination.
Animals injected with either control miRNA, R-Ras miRNA1 or R-RasmiRNA2 into dorsal CA1 were tested for their ability to discriminate between two similar contexts. (Two-Way ANOVA, miRNA effect, F 2,42 = 5.35, p = 0.009; Control, t = 2.89, p < 0.05; R-Ras miRNA1, t = 0.35, p > 0.05; R-Ras miRNA2, t = 0.35, p > 0.05).
Together, these findings show that knocking down expression of R-Ras in the CA1 hippocampus gives the same phenotype in these assays of synaptic plasticity, signal transduction and behavior defects as GRF1 knockout mice, suggesting that R-Ras may mediate at least part of the effects of GRF1 in promoting these processes.
R-Ras functions independently from GRF1 in promoting HFS-LTP
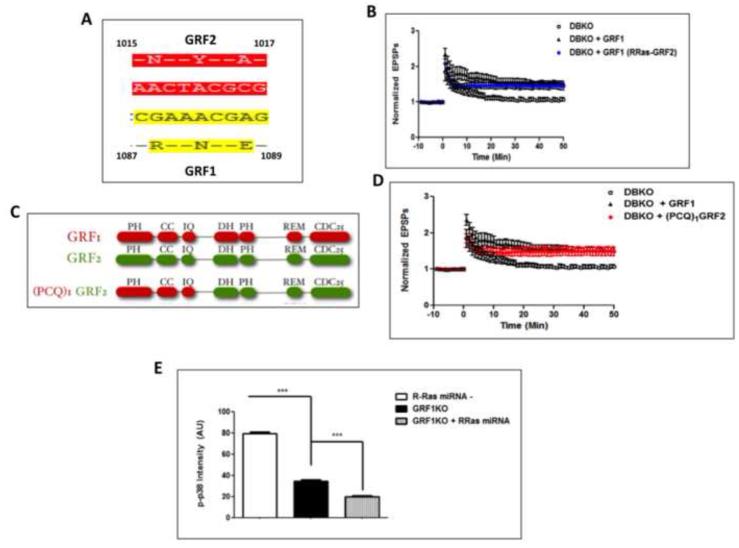
Despite the fact that the loss of GRF1 expression in GRF1 knockout mice and knockdown of R-Ras expression in the CA1 led to similar reductions in HFS-LTP and p38 activation that is necessary for it, additional experiments did not support the hypothesis that R-Ras functions as a downstream effector of GRF1 in these contexts. First, we showed previously that the ability of GRF1, but not GRF2, to activate R-Ras was due to a 3 amino acid differences in their CDC25 Ras-activating domains (Fig. 5A) (Gotoh et al., 2001). A chimera of GRF1, which contained just those amino acids of GRF2 failed to activate R-Ras, while maintaining its ability to activate Ras (Gotoh et al., 2001). Thus, this chimera was tested to determine whether it lost the ability promote HFS-LTP. To this end, we used an assay system we previously generated that involves restoring GRF1 expression into the CA1 of GRF1 knockout mice via stereotactic injection of adenovirus expressing GRF1 under the control of the neuron-specific synapsin promoter (Jin et al., 2013). In that system reconstituted GRF1 behaved like endogenous GRF1 and restored HFS-LTP, but not TBS-LTP. Thus, we used this system to test the chimera described above. Strikingly, Figure 5B shows that expression of this chimera reconstituted HFS-LTP as efficiently as WT GRF1. To confirm this finding, another chimera that cannot activate R-Ras was tested. It had the entire C-terminus of GRF1, including the DH/PH and CDC25 domains, replaced with those of GRF2. It also reconstituted HFS-LTP like GRF1 when expressed in GRF knockout mice (Fig. 5C). Thus, GRF1 does not induce HFS-LTP through R-Ras activation.
Figure 5. GRF1 induces HFS-LTP in an R-Ras-independent manner and R-Ras acts independently of GRF1 in promoting HFS-induced p38 activation.
A. The amino acid differences in the CDC25 domains between GRF1 and GRF2 responsible for activation of R-Ras by GRF1 (Gotoh et al., 2001) B. HFS-LTP in brain slices from double GRF1/GRF2 knockout (DBKO) mice and DBKO mice injected with adenovirus expressing wild-type GRF1 or the GRF1/(R)GRF2 chimera that can no longer activate R-Ras into the CA1 hippocampus (DBKO 111 ± 4.3% ; WT-GRF1, 165 ± 9.4%; GRF1/(R-Ras-GRF2, 148 ± 8.5%, Two-Way ANOVA, Virus effect, F 2,944 = 7.76, p = 0.0044; Bonferroni post-hoc, WT-GRF1 vs. GRF1/(R-Ras-GRF2), p > 0.05 at all time points). C. Functional domains in chimera that replaces the whole C-terminus of GRF1 with that of GRF2, including the CDC25 domain of GRF1 that is required for R-Ras activation. (PH- pleckstrin homology, CC- coiled-coil, IQ- calmodulin binding domain, DH- Dbl homology, REM- Ras exchange motif, CDC25- Ras and R-Ras activating domain) D. HFS-LTP in brain slices of DBKO mice is reconstituted in the CA1 with (PCQ)1GRF2. (DBKO, 111.63 ± 2.23%; WT-GRF1, 165 ± 9.4%; (PCQ)1GRF2, 150 ± 10.7%, Two-Way ANOVA, Virus effect, F2,944 = 6.06, p = 0.011; Bonferroni post-hoc, WT-GRF1 vs. (PCQ)1GRF2, p > 0.05 at all time points, also see data as reported in (Jin et al., 2014, Fig. 3A) E. Hippocampal slices from mice previously injected with virus expressing R-Ras miRNA1 or 2 into the CA1 area of RasGRF1 KO mice were stimulated with HFS-LTP and processed for immunostaining of phosphorylated p38. A comparison of levels of phosphorylated p38 immunostaining between wild-type cells (WT), GRF1 KO cells and GRF1 KO cells containing R-Ras miRNAs revealed an additive effect on p-p38 inhibition by R-Ras in cells already lacking RasGRF1 (One-Way ANOVA, F 2,535 = 817.5, p < 0.0001, Bonferroni post-hoc: R-Ras miRNA− vs. GRF1-KO, t = 19.23, p < 0.05; GRF1-KO vs. GRF1-KO+R-Ras miRNA, t = 5.95, p < 0.05).
Second, we showed previously that GRF1 knockout mice display decreased HFS-induced p-38 activation, but that it was not completely eliminated. Thus, we tested here whether R-Ras independently regulates p38 activity in response to HFS stimulation by injecting R-Ras miRNA-1 virus into GRF1 knockout mice to determine whether it could further suppress HFS-induced p38 activation. Figure 5E shows that it did, arguing that R-Ras does not mediate this function of GRF1, but rather independently regulates p38 Map kinase.
Overall, these findings argue that R-Ras, through its ability to regulate p38 Map kinase and HFS-LTP in a GRF1-independent manner plays a unique role in promoting the ability of mice to display contextual discrimination
DISCUSSION
This study reveals a novel role for the R-Ras GTPase in the brain by demonstrating its requirement for a distinct form of LTP in the CA1 hippocampus, HFS-LTP in adult mice, and a form of learning, contextual discrimination, that depends upon it. These conclusions are based on the findings that two independent mechanisms of suppressing R-Ras expression in neurons of the CA1 hippocampus blocked HFS-LTP, but not other forms of synaptic plasticity such as theta-burst LTP or LTD. The mechanism involved suppressing p38 Map kinase activation by HFS stimulation, which has been shown previously to be required for the induction of HFS-LTP in the CA1 hippocampus of mice at least 2 months of age (Jin et al., 2013). Consistent with our previous finding that HFS-LTP induction via p38 Map kinase activation is required for contextual discrimination learning, we show here that R-Ras knockdown also suppresses this form of learning.
The phenotypes associated with R-Ras knockdown in the CA1 were similar to those found in GRF1 knockout mice that have been tied to its role in promoting contextual discrimination, including suppression of both p38 activation and LTP induction by HFS stimulation. Nevertheless, despite the fact that R-Ras is a potential GTPase effector of GRF1, we found that R-Ras regulates HFS-LTP and p38 activation by HFS-stimulation independently of GRF1. That is because mutants of GRF1 that can no longer activate R-Ras still reconstitutes HFS-LTP when expressed in a GRF knockout mice and suppression of R-Ras in GRF1-knockout mice further suppresses the residual p38 Map kinase activation present in these knockout mice.
The best-documented effectors of R-Ras are PI3-kinase and integrins, not p38 Map kinase. In fact, in one study expression of activated R-Ras in tissue culture cells did not lead to enhanced p38 activity (Self et al., 2001), however in another it did (Pozzi et al., 2006). In yet another study activated R-Ras activated p38 indirectly by promoting TGFb signaling (Erdogan et al., 2008). Finally, immune cells lacking R-Ras were shown to display decreased p38 activity (Singh et al., 2012). The mechanism underlying how R-Ras influences p38 in neurons is not known at present. Our attempt to document that HFS-stimulation leads to R-Ras activation has been unsuccessful because using standard pulldown assays that have been used by others to quantify the GTP bound state we could not detect active GTP-bound R-Ras in lysates from brain slices used in these studies. Thus, it is possible that basal R-Ras activity may not be altered by HFS-stimulation in the CA1 hippocampus to directly mediate p38 activation. Instead, R-Ras may play a permissive role in p38 activation, like that found in epithelial cells described above (Erdogan et al., 2008) by regulating the expression of a component of the activation mechanism used by HFS stimulation to activate this kinase, a possibility presently under investigation.
These findings also add to a growing list of examples where p38 Map kinase can contribute to LTP induction, which has been more tightly linked in the past to Erk Map kinase induction. For example, in addition to HFS-LTP induced by GRF1 described already, p38 Map kinase activity has also been associated with enhanced theta-burst LTP associated with exposure to an enriched environment (Li et al., 2006). Here we show a third example, via R-Ras.
R-Ras has been studied in neurons mainly in the context of the discovery that Plexin-B, a receptor for semaphorins, is a GTPase activating protein that inhibits R-Ras (Ivins et al., 2000, Negishi et al., 2005). Semaphorins are best known for their role in promoting the collapse of growth cones of axons during development of synapses, and suppression of R-Ras activity is a required step in this process (Oinuma et al., 2004a, Oinuma et al., 2004b, Oinuma et al., 2010). R-Ras has also been detected in an shRNA screen for genes that contribute to neuronal toxicity associated with Huntington’s disease (Miller et al., 2012). The results presented here are the first to identify a normal role for R-Ras in a specific form of animal behavior, contextual discrimination, that may be impaired in certain neurodegenerative disorders, and a form of synaptic plasticity that contributes to it.
Highlights.
Presents data implicating a role for the GTPase, R-Ras in a specific form of synaptic plasticity and hippocampus-derived learning.
Adds support to previous papers implicating p38 Map kinase in promoting LTP and not just LTD.
R-Ras and Ras-GRF1 have similar abilities to promote LTP in the CA1 hippocampus and contextual discrimination, but they are most likely regulated by different upstream signals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aschauer DF, Kreuz S, Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan M, Pozzi A, Bhowmick N, Moses HL, Zent R. Transforming growth factor-beta (TGF-beta) and TGF-beta-associated kinase 1 are required for R-Ras-mediated transformation of mammary epithelial cells. Cancer Res. 2008;68:6224–6231. doi: 10.1158/0008-5472.CAN-08-0513. [DOI] [PubMed] [Google Scholar]
- Fam NP, Fan W-T, Wang Z, Zhang L-J, Chen Z, Moran MF. Cloning and characterization of Ras-GRF2, a novel guanine nucleotide exchange factor for Ras. Mol Cell Biol. 1997;17:1396–1406. doi: 10.1128/mcb.17.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Regulation of Neuronal Function by Ras-GRF Exchange Factors. Genes Cancer. 2011;2:306–319. doi: 10.1177/1947601911408077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PW, Cestari V, Filipkowski RK, McDonald RJ, Silva AJ. The dorsal hippocampus is essential for context discrimination but not for contextual conditioning. Behav Neurosci. 1998;112:863–874. doi: 10.1037//0735-7044.112.4.863. [DOI] [PubMed] [Google Scholar]
- Giese KP, Friedman E, Telliez JB, Fedorov NB, Wines M, Feig LA, Silva AJ. Hippocampus- dependent learning and memory is impaired in mice lacking the Ras-guanine-nucleotide releasing factor 1 (Ras-GRF1) Neuropharmacology. 2001;41:791–800. doi: 10.1016/s0028-3908(01)00096-x. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Niino Y, Tokuda M, Hatase O, Nakamura S, Matsuda M, Hattori S. Activation of R-Ras by Ras-guanine nucleotide-releasing factor. J Biol Chem. 1997;272:18602–18607. doi: 10.1074/jbc.272.30.18602. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Tian X, Feig LA. Prenylation of target GTPases contributes to signaling specificity of Ras-guanine nucleotide exchange factors. J Biol Chem. 2001;276:38029–38035. doi: 10.1074/jbc.M104658200. [DOI] [PubMed] [Google Scholar]
- Hyde LA, Crnic LS. Age-related deficits in context discrimination learning in Ts65Dn mice that model Down syndrome and Alzheimer's disease. Behav Neurosci. 2001;115:1239–1246. [PubMed] [Google Scholar]
- Ivins JK, Yurchenco PD, Lander AD. Regulation of neurite outgrowth by integrin activation. J Neurosci. 2000;20:6551–6560. doi: 10.1523/JNEUROSCI.20-17-06551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasawa N, Negishi M, Oinuma I. R-Ras controls axon branching through afadin in cortical neurons. Mol Biol Cell. 2012;23:2793–2804. doi: 10.1091/mbc.E12-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashita S, Song SY. RasGAPs: a crucial regulator of extracellular stimuli for homeostasis of cellular functions. Mol Biosyst. 2008;4:213–222. doi: 10.1039/b716357f. [DOI] [PubMed] [Google Scholar]
- Jeffery KJ, Anderson MI, Hayman R, Chakraborty S. A proposed architecture for the neural representation of spatial context. Neurosci Biobehav Rev. 2004;28:201–218. doi: 10.1016/j.neubiorev.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Jin SX, Arai J, Tian X, Kumar-Singh R, Feig LA. Acquisition of Contextual Discrimination Involves the Appearance of a Ras-GRF1/p38 Map Kinase-Mediated Signaling Pathway that Promotes LTP. J Biol Chem. 2013 doi: 10.1074/jbc.M113.471904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SX, Bartolome C, Arai JA, Hoffman L, Uzturk BG, Kumar-Singh R, Waxham MN, Feig LA. J Biol Chem. 2014;289:16551–16564. doi: 10.1074/jbc.M114.557959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian X, Hartley DM, Feig LA. The environment versus genetics in controlling the contribution of MAP kinases to synaptic plasticity. Curr Biol. 2006;16:2303–2313. doi: 10.1016/j.cub.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Miller JP, Yates BE, Al-Ramahi I, Berman AE, Sanhueza M, Kim E, de Haro M, DeGiacomo F, Torcassi C, Holcomb J, Gafni J, Mooney SD, Botas J, Ellerby LM, Hughes RE. A genome-scale RNA-interference screen identifies RRAS signaling as a pathologic feature of Huntington's disease. PLoS Genet. 2012;8:e1003042. doi: 10.1371/journal.pgen.1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M, Oinuma I, Katoh H. R-ras as a key player for signaling pathway of plexins. Mol Neurobiol. 2005;32:217–222. doi: 10.1385/MN:32:3:217. [DOI] [PubMed] [Google Scholar]
- Oinuma I, Ishikawa Y, Katoh H, Negishi M. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004a;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- Oinuma I, Ito Y, Katoh H, Negishi M. Semaphorin 4D/Plexin-B1 stimulates PTEN activity through R-Ras GTPase-activating protein activity, inducing growth cone collapse in hippocampal neurons. J Biol Chem. 2010;285:28200–28209. doi: 10.1074/jbc.M110.147546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I, Katoh H, Negishi M. Molecular dissection of the semaphorin 4D receptor plexin- B1-stimulated R-Ras GTPase-activating protein activity and neurite remodeling in hippocampal neurons. J Neurosci. 2004b;24:11473–11480. doi: 10.1523/JNEUROSCI.3257-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I, Katoh H, Negishi M. R-Ras controls axon specification upstream of glycogen synthase kinase-3beta through integrin-linked kinase. J Biol Chem. 2007;282:303–318. doi: 10.1074/jbc.M607979200. [DOI] [PubMed] [Google Scholar]
- Overbeck AF, Brtva TR, Cox AD, Graham SM, Huff SY, Khosravi-Far R, Quilliam LA, Solski PA, Der CJ. Guanine nucleotide exchange factors: activators of Ras superfamily proteins. Mol Reprod Dev. 1995;42:468–476. doi: 10.1002/mrd.1080420415. [DOI] [PubMed] [Google Scholar]
- Pozzi A, Coffa S, Bulus N, Zhu W, Chen D, Chen X, Mernaugh G, Su Y, Cai S, Singh A, Brissova M, Zent R. H-Ras, R-Ras, and TC21 differentially regulate ureteric bud cell branching morphogenesis. Mol Biol Cell. 2006;17:2046–2056. doi: 10.1091/mbc.E05-08-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- Self AJ, Caron E, Paterson HF, Hall A. Analysis of R-Ras signalling pathways. J Cell Sci. 2001;114:1357–1366. doi: 10.1242/jcs.114.7.1357. [DOI] [PubMed] [Google Scholar]
- Shou C, Farnsworth CL, Neel BG, Feig LA. Molecular cloning of cDNAs encoding a guanine- nucleotide releasing factor for Ras p21. Nature. 1992;358:351–354. doi: 10.1038/358351a0. [DOI] [PubMed] [Google Scholar]
- Singh G, Hashimoto D, Yan X, Helft J, Park PJ, Ma G, Qiao RF, Kennedy CR, Chen SH, Merad M, Chan AM. R-Ras is required for murine dendritic cell maturation and CD4+ T-cell priming. Blood. 2012;119:1693–1701. doi: 10.1182/blood-2011-05-357319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturani E, Abbondio A, Branduardi P, Ferrari C, Zippel R, Martegani E, Vanoni M, Denis-Donini S. The Ras Guanine nucleotide Exchange Factor CDC25Mm is present at the synaptic junction. Exp Cell Res. 1997;235:117–123. doi: 10.1006/excr.1997.3660. [DOI] [PubMed] [Google Scholar]
- Tian X, Feig LA. Basis for Signaling Specificity Difference between Sos and Ras-GRF Guanine Nucleotide Exchange Factors. J Biol Chem. 2001;276:47248–47256. doi: 10.1074/jbc.M107407200. [DOI] [PubMed] [Google Scholar]
- Zippel R, Gnesutta N, Matus-Leibovitch N, Mancinelli E, Saya D, Vogel Z, Sturani E. Ras-GRF, the activator of Ras, is expressed preferentially in mature neurons of the central nervous system. Brain Res Mol Brain Res. 1997;48:140–144. doi: 10.1016/s0169-328x(97)00120-4. [DOI] [PubMed] [Google Scholar]