Abstract
Nrf2, a central regulator of the cellular defense against oxidative stress and inflammation, participates in modulating hepatocyte proliferation during liver regeneration. It is not clear, however, whether Nrf2 regulates hepatocyte growth, an important cellular mechanism to regain the lost liver mass after partial hepatectomy (PH). To determine this, various analyses were performed in wild-type and Nrf2-null mice following PH. We found that, at 60 h post-PH, the vast majority of hepatocytes lacking Nrf2 reduced their sizes, activated hepatic progenitor markers (CD133, TWEAK receptor, and trefoil factor family 3), depleted HNF4α protein, and downregulated the expression of a group of genes critical for their functions. Thus, the identity of hepatocytes deficient in Nrf2 was transiently but massively impaired in response to liver mass loss. This event was associated with the coupling of protein depletion of hepatic HNF4α, a master regulator of hepatocyte differentiation, and concomitant inactivation of hepatic Akt1 and p70S6K, critical hepatocyte growth signaling molecules. We conclude that Nrf2 participates in maintaining newly regenerated hepatocytes in a fully differentiated state by ensuring proper regulation of HNF4α, Akt1, and p70S6K during liver regeneration.
Introduction
Nuclear factor erythroid 2-related factor 2 (Nrf2), a basic leucine zipper region-containing transcription factor, is ubiquitously expressed all over the body, but more predominantly in metabolic organs, including the liver [1]. The various beneficial effects exerted by the activation of Nrf2 in detoxification, antioxidation, and anti-inflammation have attracted numerous investigators to study its regulation, modes of action, target genes, and functions in health and disease, especially over the past decade [2]–[5]. Upon activation by actin-binding Kelch-like ECH-associated protein 1 (Keap1)-dependent and Keap1-independent mechanisms, Nrf2 translocates into the nucleus and regulates the transcription of a network of genes involved in various cellular activities, including redox balance, metabolism, proliferation, and apoptosis. Thereby, Nrf2 is required for maintaining tissue homeostasis. Moreover, several lines of evidence support its important roles in modulating the injury and repair of tissues including the liver [6]–[10]. We are interested in how Nrf2 regulates liver regeneration.
Liver regeneration triggered by partial hepatectomy (PH) is considered to be fundamentally driven by the replication of the remaining mature hepatocytes without the complications of massive cellular injury and inflammation [11], [12]. A recent report highlights the importance of hepatocyte hypertrophy in liver regeneration. Proliferation and hypertrophy of remnant hepatocytes almost equally contribute to the restoration of lost liver mass after 70% liver resection [13]. It has been demonstrated that Nrf2 regulates hepatocyte proliferation by ensuring normal insulin/IGF-1 and Notch1 signaling during liver regeneration [6], [7]. However, it remains unclear whether Nrf2 participates in modulating hepatocyte growth following liver resection. Using the mouse PH model with the presence or absence of Nrf2, we demonstrate here that Nrf2 deficiency results in transient but extensive impairment of hepatocyte growth and identity during liver regeneration.
Results
Nrf2 deficiency causes transient but massive reduction in hepatocyte size after PH
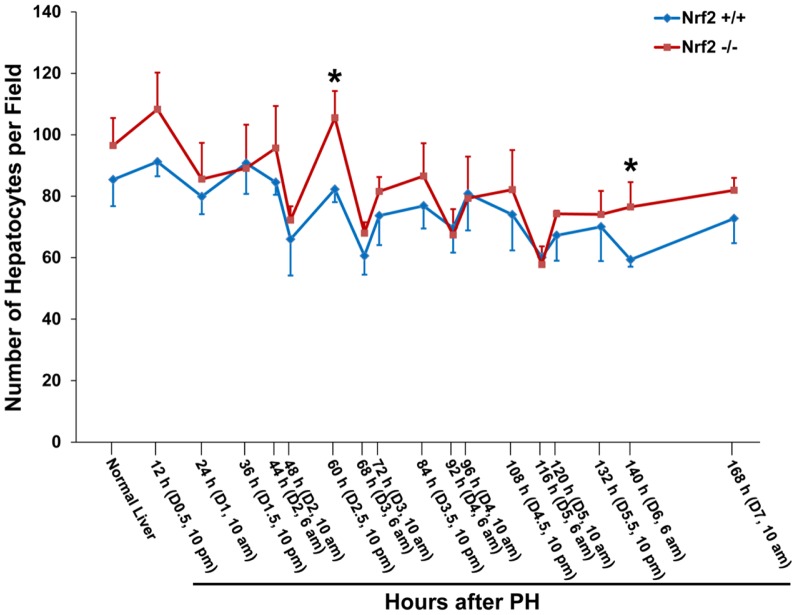
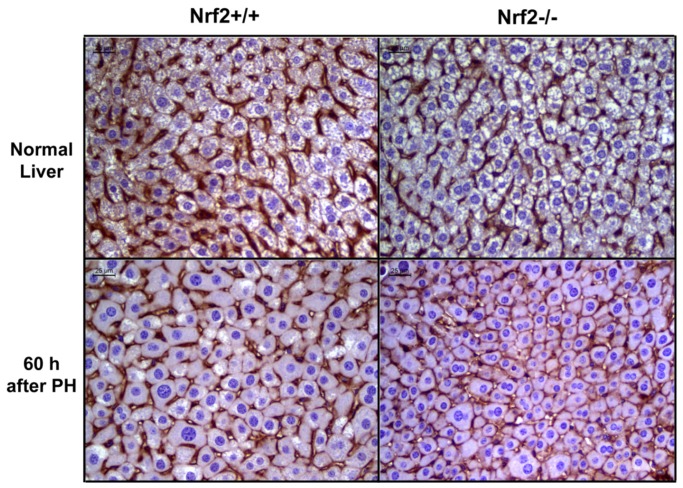
Hepatocyte hypertrophy constitutes an important but often neglected cellular mechanism in PH-induced liver regeneration [13]–[16]. To determine whether Nrf2 is associated with hepatocyte size regulation, we assessed hepatocyte density during the course of liver regrowth in wild-type and Nrf2-null mice (Fig. 1). Although the average number of hepatocytes per microscope field in normal wild-type mice was lower than that in Nrf2-null mice, statistical significance was not achieved. After PH, the hepatocyte density showed dynamic changes with an overall trend of a decrease in both genotypes of mice. The data indicate that hepatocytes undergo progressive growth (hypertrophy) as liver regeneration advances. Our observation is in line with a report [13]. However, at 60 h and 140 h post-PH, Nrf2-null mice exhibited significantly higher hepatocyte densities than wild-type controls. To clearly visualize hepatocyte sizes, β-catenin immunostaining was performed on tissue sections of the livers collected from both genotypes of mice at 60 h following PH. Obviously, hepatocytes deficient in Nrf2 were smaller than wild-type controls at the time point (Fig. 2). These data suggest that hepatocyte growth was transiently inhibited due to Nrf2 null mutation. This finding prompted us to focus on the temporary event taking place earlier (60 h after PH) to gain insight at the cellular and molecular levels. In addition, we counted binuclear hepatocytes in regenerating livers at 60 h after PH. As a result, there is no significant difference in the percentage of binuclear hepatocytes at this time point between wild-type mice and Nrf2-null mice (wild-types: 13.86±1.2 vs. Nrf2 knockouts: 16.17±3.63, p = 0.23). The data suggest that Nrf2 may not play a role in hepatocyte cytokinesis during liver regeneration.
Figure 1. Changes in hepatocyte density after partial hepatectomy (PH) in Nrf2+/+ and Nrf2−/− mice.
Livers were collected from the two genotype groups of mice at the indicated time points post-PH. Liver sections were prepared and subjected to hematoxylin and eosin staining. Hepatocytes were counted in five randomly chosen fields per liver section (400x magnification) with Image-Pro Plus software. The results are shown as the means per field ± SD (n = 3 to 5 mice/time point/genotype; *, p<0.05 between Nrf2+/+ and Nrf2−/− mice).
Figure 2. β-catenin immunostaining.
Liver sections were prepared from the resting livers and the livers at 60 h after PH in Nrf2+/+ and Nrf2−/− mice and subjected to immunostaining with β-catenin primary antibody. Hepatocyte membrane was stained dark brown.
Nrf2 deficiency results in transient but extensive activation of liver progenitor markers and depletion of HNF4α in regenerating livers
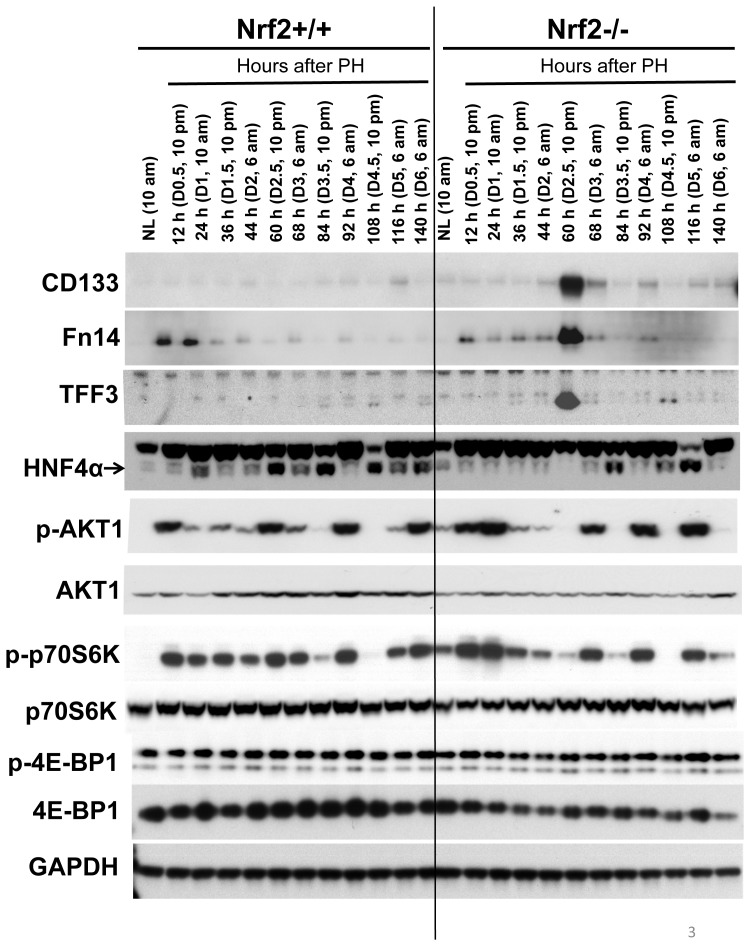
Reduction in size is characteristic of cells undergoing dedifferentiation including hepatocytes [17]. Thus, we hypothesized that hepatocytes lacking Nrf2 may transiently lose their identity during liver regeneration. To test this, we searched for dedifferentiation makers by comparing the gene expression profiles between wild-type and Nrf2-null regenerating livers at the specific time point (60 h after PH). We found that CD133, tumor necrosis factor-like weak inducer of apoptosis (TWEAK) receptor (Fn14), and trefoil factor family 3 (TFF3) were highly upregulated due to the lack of Nrf2. CD133 is widely used as a progenitor cell marker in a number of tissues including the liver [18], [19]. Fn14 has been shown to be expressed in hepatic progenitor cells [20]–[23]. TFF3 is a stable secretory protein predominantly produced by gastrointestinal mucosa and is associated with epithelial restitution [24]–[26]. Very recently, TFF3 was found to be expressed by hepatocellular carcinoma progenitors [27]. We analyzed the protein expression of the three genes throughout the course of liver regeneration in wild-type and Nrf2-null mice. In addition, hepatocyte nuclear factor 4α (HNF4α) was included in the assay because it is essential to the maintenance of hepatocyte differentiation [28]. As shown in Figure 3, the expression of hepatic CD133, Fn14, and TFF3 proteins were drastically activated at 60 h after PH in Nrf2-null, but not wild-type, mice. Wild-type regenerating livers showed marked upregulation in HNF4α protein expression at most timepoints measured. In contrast, the dynamic increases in hepatic HNF4α protein level were prevented prior to 84 h following PH due to Nrf2 absence. Especially, at 60 h after PH, HNF4α protein completely vanished in Nrf2-deficient regenerating livers. Thus, Nrf2 null mutation caused severe dysregulation in HNF4α protein expression post-PH.
Figure 3. Protein expression of CD133, Fn14, TFF3, HNF4α, p-Akt1 (T308), Akt1, p-p70S6K (T389), p70S6K, p-4E-BP1 (T37/46), and 4E-BP1 in regenerating livers of Nrf2+/+ and Nrf2−/− mice.
Livers were collected from normal mice and the mice subjected to partial hepatectomy (PH) at the indicated time points following surgery. Liver lysates prepared from three mice per time point per genotype were pooled with equal amount of protein from each preparation. Western blotting was performed with antibodies against the proteins indicated. Glyceraldehyde 3-phosphate dehydrogenase (GADPH) was used as a loading control. NL, normal liver.
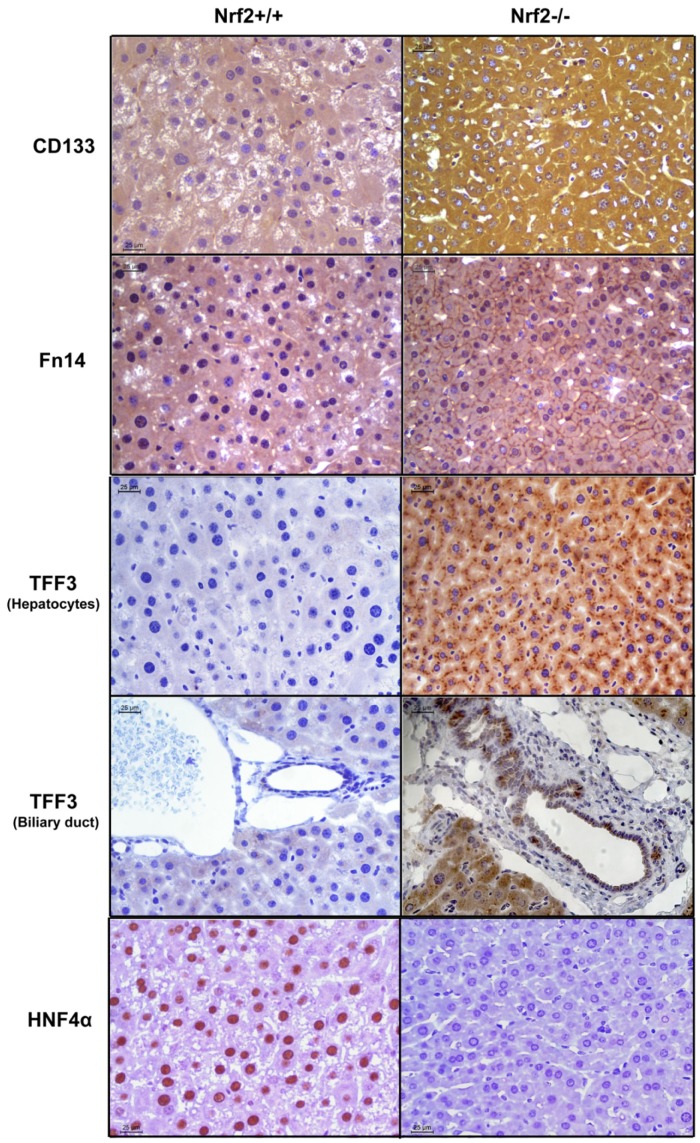
Immunohistochemical analysis revealed the distribution of the four proteins in the liver tissue and hepatocytes (Fig. 4). At 60 h following PH, almost all hepatocytes lacking Nrf2 expressed CD133 evenly in their cytosol, Fn14 on their surface, and TFF3 cytosolically concentrated as aggregates. Simultaneously, HNF4α was abundantly expressed in the nuclei of wild-type hepatocytes, but was not detectable in Nrf2-null hepatocytes. Thus, the vast majority of hepatocytes deficient in Nrf2 exhibited a phenotype of CD133+/Fn14+/TFF3+/HNF4α−. In addition, TFF3+ biliary epithelial cells were also seen exclusively in Nrf2-null livers at the same time point after PH.
Figure 4. Immunohistochemical analysis of CD133, Fn14, TFF3, and HNF4α in regenerating livers of Nrf2+/+ and Nrf2−/− mice.
Liver sections were prepared from the livers isolated at 60 h after PH from three mice per genotype. All liver sections were subjected to immunostaining with primary antibodies against CD133, Fn14, TFF3, or HNF4α. Representative immunohistochemically stained liver sections are shown.
Nrf2 deficiency leads to downregulation of a group of genes critical for liver function post-PH
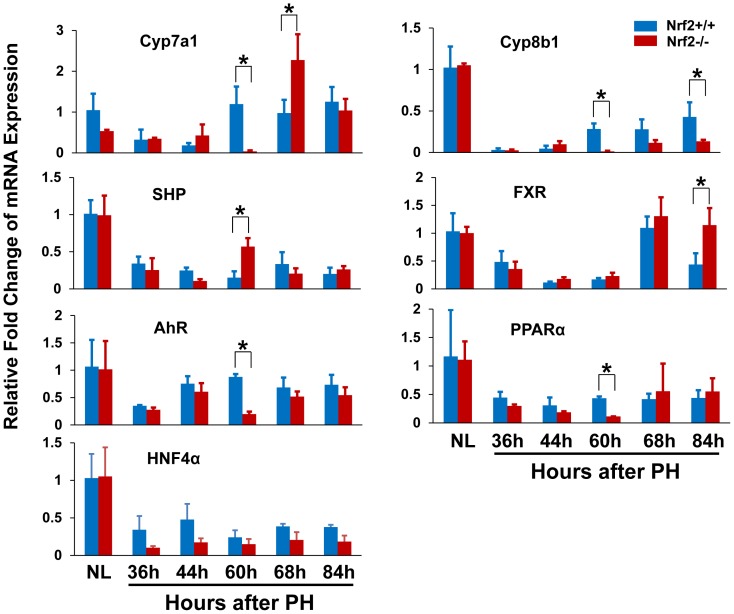
Furthermore, we found that a group of genes governing critical hepatocyte functions were markedly downregulated at 60 h following PH due to Nrf2 absence, including bile acid synthesis enzymes Cyp7a1 and Cyp8b1, xenobiotic metabolism-regulating transcription factor Aryl hydrocarbon receptor (AhR), and fat metabolism-regulating nuclear receptor peroxisome proliferator-activated receptor alpha (PPARα) (Fig. 5). At this time point in Nrf2-null regenerating livers, almost diminished expression of Cyp7a1 gene was companied by increased mRNA level of its negative regulator small heterodimer partner (SHP), whereas SHP-positive regulator Farnesoid X receptor (FXR) exhibited PH-dependent downregulation irrelevant to Nrf2. This observation suggests that SHP-mediated prevention of Cyp7a1 expression could be one of the most sensitive responses to the impairment of hepatocyte identity. In addition, we found that hepatic HNF4α mRNA expression was downregulated after PH, irrespective of genotypes. Nrf2 deficiency did not result in significant differences in hepatic HNF4α transcript levels prior to and following PH (Fig. 5). The data are not correlated with PH- and Nrf2-dependent HNF4α protein expression in regenerating liver (Fig. 3). Thus, hepatic HNF4α is regulated at both mRNA and protein levels during liver regeneration and Nrf2-dependent regulation in HNF4α protein expression occurs post transcriptionally.
Figure 5. The mRNA expression of a group of genes associated with liver functions in regenerating livers of Nrf2+/+ and Nrf2−/− mice.
Total RNA was prepared from the livers at the indicated time points after partial hepatectomy (PH). Hepatic expression levels of the genes indicated were measured by qRT-PCR and are expressed as the means of fold changes relative to the mRNA level in normal livers in Nrf2+/+ mice ± SD (n = 3 mice/time point/genotype; *, p<0.05 between Nrf2+/+ and Nrf2−/− mice). NL, normal liver.
Nrf2 deficiency causes dysregulation of Akt and p70S6K activities in regenerating livers
It is known that Akt/mTOR signaling controls the growth and thus sizes of cells, including hepatocytes, via its downstream targets p70S6K and 4E-BP1 [16], [29]. We found that, in contrast to wild-type controls, Nrf2 null mutation led to the inactivation of both Akt1 and p70S6K at 60 h and 140 h after PH (Fig. 3), consistent with the marked reduction in hepatocyte volume at the same time points in Nrf2-null mice (Figures 1 & 2). Following PH, Nrf2 absence resulted in decreases in total hepatic 4E-BP1 protein, but did not severely affect the phosphorylation of the protein, at most time points measured (Fig. 3).
Collectively, we identified a temporal event of massive impairment in hepatocyte identity caused by Nrf2 deficiency during liver regeneration. This event is characterized by the reduction in hepatocyte size, the activation of hepatic progenitor markers, the loss of HNF4α protein, and the downregulation of a subset of genes associated with important hepatocyte functions. Mechanistically, we linked the Nrf2-dependent phenomenon to the depletion of HNF4α protein and the inactivation of Akt1 and p70S6K in regenerating liver.
Discussion
In this study, we revealed a transient phenomenon of massive damage in hepatocyte identity in a pathological condition (Nrf2 genetic deletion and liver mass loss). This event is most likely indicative of hepatocyte dedifferentiation. We have provided three lines of evidence to support our assumption. Firstly, hepatocytes lacking Nrf2 markedly reduced their sizes at 60 h after PH. Sahin's group reported that mature hepatocytes are able to directly dedifferentiate into liver progenitor cells in vitro. Cell condensation or shrinkage is proposed to be one potential unique cellular mechanism underlying mature hepatocyte dedifferentiation [17]. Secondly, simultaneous with the cell volume decrease, hepatocytes deficient in Nrf2 exhibited a dedifferentiation phenotype of CD133+/Fn14+/TFF3+/HNF4α−. CD133, Fn14, and TFF3 are known to be coexpressed by hepatic progenitor cells [20], [27]. HNF4α is not only a marker of hepatocyte lineage, but also a master regulator of hepatocyte differentiation [28], [30]. Thirdly, the expression of a subgroup of genes conferring critical liver functions was downregulated and even diminished. It is remarkable that almost all hepatocytes synchronously shrank their sizes, activated hepatic progenitor markers, and reduced the expression of hepatocyte signature genes at this particular stage of live regeneration (60 h after PH) due to the absence of a single molecule (Nrf2). The data reveal an important role for Nrf2 in maintaining newly regenerated hepatocytes in a fully differentiated state during liver repair.
Mammalian cell dedifferentiation in vivo has been observed in the pancreas in adult mice [31], [32]. In light of those findings in pancreas, our study strongly suggests that hepatocytes could undergo dedifferentiation in response to a pathological stimulus. A previous study in gene expression profiling performed within 48 hours after PH revealed that replicating hepatocytes reduce the expression of a large set of genes involved in liver metabolic function [33], [34]. This phenomenon is considered as transient transcriptional dedifferentiation of normal mature hepatocytes undergoing proliferation, although the activation of the signature genes characteristic of fetal hepatic cells or HCC was not observed [34]. Here, we found that a subset of hepatic progenitor markers (CD133, Fn14, and TFF3) were activated, following the completion of the first wave of hepatocyte replication (60 h after PH), when Nrf2 was lacking. Our findings implicate that Nrf2 is required to prevent the progression of hepatocyte dedifferentiation during liver regeneration.
Recent reports have highlighted the plasticity exhibited by mature hepatocytes. They are able to dedifferentiate into liver progenitor cells in vitro and to transdifferentiate into biliary epithelial cells during chronic liver injury [17], [35]. Notably, Nrf2-null mice exhibit high susceptibility to carcinogenesis induced by several carcinogens or inflammation in multiple organ systems [36]–[39]. Hence, it is very likely that, during the regenerative response to chemical-induced tissue injury, the cells deficient in Nrf2 undergo temporal dedifferentiation, thus promoting tumor formation. Furthermore, TFF3 is associated with the transformation, growth, and migration of cancer cells [40]. Therefore, our findings provide a potential new explanation for the high incidence of carcinogenesis caused by Nrf2 ablation.
It is intriguing how Nrf2 null mutation triggers the loss of mature hepatocyte identity and even hepatocyte dedifferentiation during the process of liver repair. Our study suggests that Nrf2-dependent regulation of the expression of HNF4α protein and the activities of Akt1 and p70S6K may contribute to the event. It has been well established that HNF4α is required for liver development, hepatic specific expression of many genes associated with metabolism, and maintaining hepatocyte differentiation [30]. Among all the time points studied, 60 h post-PH was the only one when the deletion of HNF4α protein and the inactivation of Akt1 and p70S6K occurred simultaneously (Fig. 3). Presumably, at this time point after PH in Nrf2-null regenerating liver, complete loss of HNF4α protein coupled with the inactivation of Akt1 and p70S6K may synergistically exert detrimental effects. These effects trigger hepatocytes to reduce their sizes, activate progenitor markers, and decrease the expression of liver functional genes at the same time. As a result, hepatocytes display severe impairment in their identity and may go dedifferentiation. Notably, at 140 h after PH, Nrf2-null hepatocytes inactivated Akt1 and p70S6K and thereby reduced their sizes compared with wild-type controls (Figures 1 & 3). However, Nrf2-null hepatocytes decreased, but not fully prohibited, the expression of HNF4α protein relative to wild-type hepatocytes at the same time point. This may explain why liver progenitor markers (CD133, Fn14, and TFF3) were not activated at 140 h post-PH in Nrf2-null regenerating livers. Several lines of evidence support a role of insulin/IGF signaling in hepatocyte proliferation and liver growth [41]–[43]. It has been shown that Nrf2 absence results in reduced hepatic insulin/IGF1 signaling at 3 h after PH [6]. Further studies are needed to determine whether the disrupted activities of Akt1 and p70S6K are caused by dysregulated insulin/IGF signaling in Nrf2-null regenerating liver.
A recent report demonstrates that Nrf2 activation exerts negative effects on liver regeneration by delaying proliferation and inducing apoptosis of hepatocytes [8]. However, another recent report shows that Nrf2 displays a beneficial effect by promoting compensatory liver hypertrophy after portal vein ligation [44]. We believe that Nrf2 activity needs to be tightly controlled during liver regeneration, because both overly activated and deficient Nrf2 activity impairs hepatic regenerative response.
In summary, we identified a Nrf2-depedent phenomenon of hepatocyte identity impairment during liver repair. We presume that, as liver regeneration proceeds, Nrf2 presence enables HNF4α to keep newly regenerated hepatocytes in a differentiated state and allows Akt1 and p70S6K to exert appropriate growth signaling, thus maintaining hepatocyte identity in regenerating livers.
Materials and Methods
Animal care and use
The mice were housed in plastic cages at 22±1°C on a 12-hour light/12-hour dark cycle with lights on from 6:00 am to 6:00 pm. Standard rodent chow and water were provided ad libitum throughout the entire feeding period. Wild-type and Nrf2-deficient male mice (3 months old) of a C57BL6/129SV mixed background were used for the study [1]. Standard two-thirds liver resections were performed following the procedure described previously [45], [46]. The gall bladders were kept intact. The surgery was performed between 10:00 am and 12:00 pm to minimize the potential variability in the progression of liver regeneration associated with the surgical time and the circadian clock [47]. All of the animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Protocols for the care and use of animals were approved by the Indiana University-Purdue University Indianapolis Animal Care and Use Committee.
Immunohistochemistry
Formalin-fixed and paraffin-embedded liver sections were subjected to a standard procedure of immunohistochemistry. Primary antibodies against β-catenin (BD Biosciences, San Jose, CA), CD133 (orb18124, Biorbyt, San Francisco, CA), Fn14 (ab109365, Abcam, Cambridge, MA), HNF4α (SC-6556, Santa Cruz Biotechnology, Santa Cruz, CA), and TFF3 (PAB19439, Abnova, Walnut, CA) were used.
Western blot analysis
Liver homogenates (10 or 30 µg) were separated by polyacrylamide gel electrophoresis under reducing conditions. Proteins from the gels were electrophoretically transferred to polyvinylidene difluoride membranes. Primary antibodies used include p-p70S6K (T389) (#9234), p70S6K (#2708), p-4E-BP1 (T37/46) (#2855), and 4E-BP1 (#9644) (Cell Signaling Technology, Danvers, MA); Fn14 (ab109365, Abcam, Cambridge, MA); TFF3 (SC-18273), HNF4α (SC-6556), and glyceraldehyde 3-phosphate dehydrogenase (GADPH) (SC-25778) (Santa Cruz Biotechnology, Santa Cruz, CA); CD133 (PAB12663, Abnova, Walnut, CA); and Akt1 (#1081-1) and p-Akt1 (T308) (#2214-1) (Epitomics). Immune complexes were detected using an enhanced chemiluminescence system (Pierce, Rockford, IL).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from frozen liver tissue using TRIzol reagent according to the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). cDNAs were synthesized from the total RNA (1 µg) of each sample using a Verso cDNA Kit (Thermo Scientific, Rockford), diluted four times with water, and subjected to qRT-PCR to quantify mRNA levels. TaqMan Universal PCR Master Mix and the primers and TaqMan MGB probes of mouse Cyp7a1 (mm00484150_m1), Cyp8b1 (Mm00501637_s1), SHP (Mm00442278_m1), FXR (Mm00436425_m1), AhR (Mm00478932_m1), PPARα (Mm00440939_m1), HNF4α (Mm01247712_m1) and albumin (Mm00802090_m1) were purchased from Applied Biosystems (Foster City, CA). The amplification reactions were carried out with the ABI Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA) with initial hold steps (50°C for 2 min followed by 95°C for 10 min) and 40 cycles of a 2-step PCR (92°C for 15 seconds and 60°C for 1 min). The comparative CT method was used for the relative quantification of the amount of mRNA in each sample normalized to the albumin transcript levels.
Hepatocyte density measurement
Formalin-fixed and paraffin-embedded liver sections were stained with hematoxylin and eosin. Hepatocytes were counted with Image-Pro Plus software (Media Cybernetics, MD, USA) in five randomly chosen microscope fields at 400x magnification for each sample.
Statistical analysis
Data are shown as the means ± standard deviation (SD). Statistical analysis was performed using a one-way analysis of variance. Comparisons of means were determined by post-hoc analysis. Significant differences were defined when P<0.05.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (7RO1DK07596). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chan K, Lu R, Chang JC, Kan YW (1996) NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A 93: 13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes JD, Dinkova-Kostova AT (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. [DOI] [PubMed]
- 3. Jaramillo MC, Zhang DD (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27: 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitsuishi Y, Motohashi H, Yamamoto M (2012) The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Front Oncol 2: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bryan HK, Olayanju A, Goldring CE, Park BK (2013) The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85: 705–717. [DOI] [PubMed] [Google Scholar]
- 6. Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, et al. (2008) Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J 27: 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, et al. (2010) Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal 3: ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler UA, Kurinna S, Schwitter D, Marti A, Schafer M, et al. (2013) Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell cycle control and apoptosis. Hepatology. [DOI] [PubMed]
- 9. Dayoub R, Vogel A, Schuett J, Lupke M, Spieker SM, et al. (2013) Nrf2 activates augmenter of liver regeneration (ALR) via antioxidant response element and links oxidative stress to liver regeneration. Mol Med 19: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buitrago-Molina LE, Marhenke S, Longerich T, Sharma AD, Boukouris AE, et al. (2013) The degree of liver injury determines the role of p21 in liver regeneration and hepatocarcinogenesis in mice. Hepatology 58: 1143–1152. [DOI] [PubMed] [Google Scholar]
- 11. Michalopoulos GK (2010) Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol 176: 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mitchell C, Willenbring H (2008) A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 3: 1167–1170. [DOI] [PubMed] [Google Scholar]
- 13. Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, et al. (2012) Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 22: 1166–1175. [DOI] [PubMed] [Google Scholar]
- 14. Minamishima YA, Nakayama K (2002) Recovery of liver mass without proliferation of hepatocytes after partial hepatectomy in Skp2-deficient mice. Cancer Res 62: 995–999. [PubMed] [Google Scholar]
- 15. Haga S, Ogawa W, Inoue H, Terui K, Ogino T, et al. (2005) Compensatory recovery of liver mass by Akt-mediated hepatocellular hypertrophy in liver-specific STAT3-deficient mice. J Hepatol 43: 799–807. [DOI] [PubMed] [Google Scholar]
- 16. Haga S, Ozaki M, Inoue H, Okamoto Y, Ogawa W, et al. (2009) The survival pathways phosphatidylinositol-3 kinase (PI3-K)/phosphoinositide-dependent protein kinase 1 (PDK1)/Akt modulate liver regeneration through hepatocyte size rather than proliferation. Hepatology 49: 204–214. [DOI] [PubMed] [Google Scholar]
- 17. Chen Y, Wong PP, Sjeklocha L, Steer CJ, Sahin MB (2012) Mature hepatocytes exhibit unexpected plasticity by direct dedifferentiation into liver progenitor cells in culture. Hepatology 55: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suzuki A, Sekiya S, Onishi M, Oshima N, Kiyonari H, et al. (2008) Flow cytometric isolation and clonal identification of self-renewing bipotent hepatic progenitor cells in adult mouse liver. Hepatology 48: 1964–1978. [DOI] [PubMed] [Google Scholar]
- 19. Grosse-Gehling P, Fargeas CA, Dittfeld C, Garbe Y, Alison MR, et al. (2013) CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol 229: 355–378. [DOI] [PubMed] [Google Scholar]
- 20. Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, et al. (2008) Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology 47: 636–647. [DOI] [PubMed] [Google Scholar]
- 21. Tirnitz-Parker JE, Viebahn CS, Jakubowski A, Klopcic BR, Olynyk JK, et al. (2010) Tumor necrosis factor-like weak inducer of apoptosis is a mitogen for liver progenitor cells. Hepatology 52: 291–302. [DOI] [PubMed] [Google Scholar]
- 22. Jakubowski A, Ambrose C, Parr M, Lincecum JM, Wang MZ, et al. (2005) TWEAK induces liver progenitor cell proliferation. J Clin Invest 115: 2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karaca G, Swiderska-Syn M, Xie G, Syn WK, Kruger L, et al. (2014) TWEAK/Fn14 signaling is required for liver regeneration after partial hepatectomy in mice. PLoS One 9: e83987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suemori S, Lynch-Devaney K, Podolsky DK (1991) Identification and characterization of rat intestinal trefoil factor: tissue- and cell-specific member of the trefoil protein family. Proc Natl Acad Sci U S A 88: 11017–11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Podolsky DK, Lynch-Devaney K, Stow JL, Oates P, Murgue B, et al. (1993) Identification of human intestinal trefoil factor. Goblet cell-specific expression of a peptide targeted for apical secretion. J Biol Chem 268: 6694–6702. [PubMed] [Google Scholar]
- 26. Kanai M, Mullen C, Podolsky DK (1998) Intestinal trefoil factor induces inactivation of extracellular signal-regulated protein kinase in intestinal epithelial cells. Proc Natl Acad Sci U S A 95: 178–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, et al. (2013) Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell 155: 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ (2001) Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21: 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fingar DC, Salama S, Tsou C, Harlow E, Blenis J (2002) Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 16: 1472–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gonzalez FJ (2008) Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 23: 2–7. [DOI] [PubMed] [Google Scholar]
- 31. Talchai C, Xuan S, Lin HV, Sussel L, Accili D (2012) Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150: 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Landsman L, Parent A, Hebrok M (2011) Elevated Hedgehog/Gli signaling causes beta-cell dedifferentiation in mice. Proc Natl Acad Sci U S A 108: 17010–17015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. White P, Brestelli JE, Kaestner KH, Greenbaum LE (2005) Identification of transcriptional networks during liver regeneration. J Biol Chem 280: 3715–3722. [DOI] [PubMed] [Google Scholar]
- 34. Klochendler A, Weinberg-Corem N, Moran M, Swisa A, Pochet N, et al. (2012) A transgenic mouse marking live replicating cells reveals in vivo transcriptional program of proliferation. Dev Cell 23: 681–690. [DOI] [PubMed] [Google Scholar]
- 35. Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, et al. (2013) Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev 27: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, et al. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A 98: 3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, et al. (2004) Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res 64: 6424–6431. [DOI] [PubMed] [Google Scholar]
- 38. Xu C, Huang MT, Shen G, Yuan X, Lin W, et al. (2006) Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res 66: 8293–8296. [DOI] [PubMed] [Google Scholar]
- 39. Khor TO, Huang MT, Prawan A, Liu Y, Hao X, et al. (2008) Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res (Phila) 1: 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perry JK, Kannan N, Grandison PM, Mitchell MD, Lobie PE (2008) Are trefoil factors oncogenic? Trends Endocrinol Metab 19: 74–81. [DOI] [PubMed] [Google Scholar]
- 41. Starzl TE, Porter KA, Putnam CW (1976) Insulin, glucagon, and the control of hepatic structure, function, and capacity for regeneration. Metabolism 25: 1429–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jirtle RL, Carr BI, Scott CD (1991) Modulation of insulin-like growth factor-II/mannose 6-phosphate receptors and transforming growth factor-beta 1 during liver regeneration. J Biol Chem 266: 22444–22450. [PubMed] [Google Scholar]
- 43. Amaya MJ, Oliveira AG, Guimaraes ES, Casteluber MC, Carvalho SM, et al. (2014) The insulin receptor translocates to the nucleus to regulate cell proliferation in liver. Hepatology 59: 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirasaki K, Taguchi K, Unno M, Motohashi H, Yamamoto M (2014) Nrf2 promotes compensatory liver hypertrophy after portal vein branch ligation in mice. Hepatology. [DOI] [PubMed]
- 45. Greene AK, Puder M (2003) Partial hepatectomy in the mouse: technique and perioperative management. J Invest Surg 16: 99–102. [PubMed] [Google Scholar]
- 46. Dai G, He L, Bu P, Wan YJ (2008) Pregnane X receptor is essential for normal progression of liver regeneration. Hepatology 47: 1277–1287. [DOI] [PubMed] [Google Scholar]
- 47. Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, et al. (2003) Control mechanism of the circadian clock for timing of cell division in vivo. Science 302: 255–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.