Abstract
Circadian clocks regulate numerous physiological processes that vary across the day-night (diurnal) cycle, but if and how the circadian clock regulates the adaptive immune system is mostly unclear. Interleukin-17-producing CD4+ T helper (Th17) cells are proinflammatory immune cells that protect against bacterial and fungal infections at mucosal surfaces. Their lineage specification is regulated by the orphan nuclear receptor RORγt. We show that the transcription factor NFIL3 suppresses Th17 cell development by directly binding and repressing the Rorγt promoter. NFIL3 links Th17 cell development to the circadian clock network through the transcription factor REV-ERBα. Accordingly Th17 lineage specification varies diurnally and is altered in Rev-erbα−/− mice. Light cycle disruption elevated intestinal Th17 cell frequencies and increased susceptibility to inflammatory disease. Thus, lineage specification of a key immune cell is under direct circadian control.
The development and function of the immune system is profoundly affected by environmental factors such as microorganisms (1, 2), nutrients (3), and light cues (4). Interleukin-17 (IL-17A/F)-producing CD4+ T helper (Th17) cells are a key immune cell lineage that protects against bacterial and fungal infection (5) and is associated with inflammatory disease (6). Th17 cell frequencies in the intestine are influenced by microbiota composition (2, 7), but few other environmental cues are known to regulate Th17 cell development.
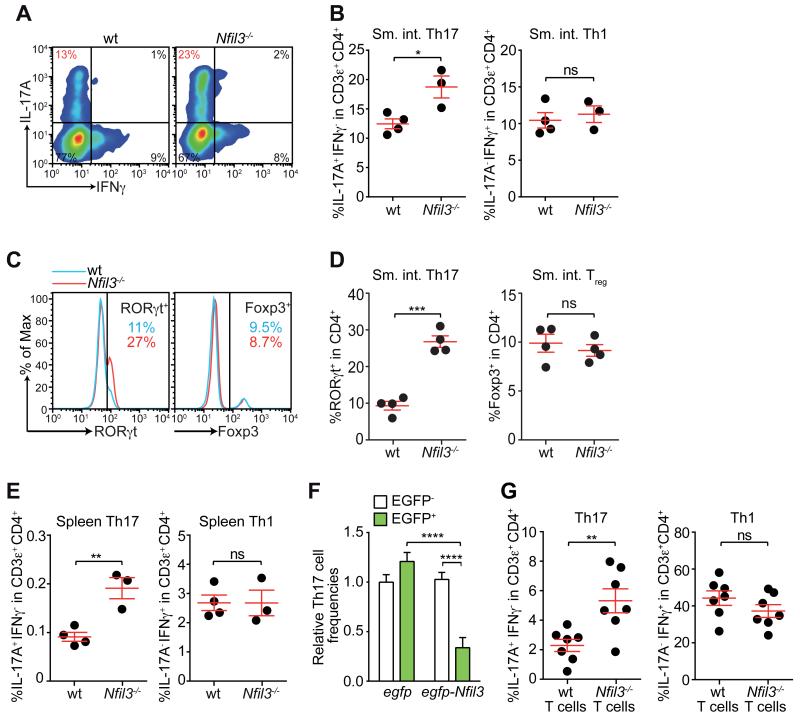
NFIL3, also known as E4BP4, is a basic leucine zipper (bZIP) transcription factor that regulates a number of immune processes (8). Nfil3 polymorphisms are associated with human inflammatory bowel disease (IBD) (9). In agreement with this finding, approximately 10% of Nfil3−/− mice but none of the wild-type mice housed in our specified pathogen-free (SPF) barrier facility exhibited rectal prolapse and immune cell infiltration into the intestine at 6-9 months of age (fig. S1A,B). These abnormalities prompted us to examine CD4+ T cells, which are critical for intestinal immune homeostasis (10). Nfil3−/− mice had higher IL-17A+ and RORγt+ Th17 cell frequencies than wild-type mice in both small intestine (Fig. 1A-D) and colon (fig. S1C,D)). In contrast, there were no significant differences in IFNγ+ Th1 (Fig. 1A,B) or Foxp3+ regulatory T (Treg) cell frequencies (Fig. 1C,D), in agreement with prior findings (11). Thus, NFIL3 deficiency impacts intestinal Th17 but not Th1 or Treg cell frequencies.
Figure 1. NFIL3 suppresses Th17 cell development in a T cell-intrinsic manner.
Intestinal Th17 cell frequencies were analyzed in wild-type (wt) and Nfil3−/− mice by intracellular staining of IL-17A and IFNγ (A,B) and nuclear staining of RORγt and Foxp3 (C,D). Representative flow cytometry plots are shown in (A) and (C) and combined data from multiple mice are shown in (B) and (D). (E) Splenic Th17 and Th1 cell frequencies in wild-type and Nfil3−/− mice. (F) Naïve wild-type CD4+ T cells were transduced by lentivirus encoding EGFP only or EGFP-tagged NFIL3, followed by culture under Th17-polarizing conditions. Th17 cell frequencies were compared between transduced (EGFP+) and non-transduced (EGFP−) cell populations in the same well. (G) Naïve wild-type and Nfil3−/− CD4+ T cells were transferred intravenously into Rag1−/− mice and LPLs were analyzed 4 weeks later. Data are from two independent experiments. Groups were plotted as mean ± SEM and compared by two-tailed student’s t-test (B, D, E, H) or one-way ANOVA (F). *,p<0.05; **,p<0.01; ***, p<0.001; ****,p<0.0001; ns, not significant.
Because intestinal Th17 cell development is sensitive to microflora composition (2, 7), we considered whether the higher Th17 frequencies in Nfil3−/− mice were due to an altered microflora. Microbiota transfer from conventionally-raised wild-type and Nfil3−/− mice into germ-free wild-type mice yielded similar intestinal Th17 cell frequencies in the two groups of recipient mice (fig. S2A,B), indicating that intestinal Th17 cell expansion in Nfil3−/− mice was not due to altered microbiota composition. This is consistent with the elevated Th17 cell frequencies in the spleens of Nfil3−/− mice (Fig. 1E), indicating that loss of NFIL3 leads to a systemic defect in suppression of Th17 cell development. However, because microbiota composition and age are known to impact Th17 cell frequencies (2, 7), we used age- and sex-matched littermates that were co-caged to minimize microbiota differences in all experiments.
To assess whether NFIL3 suppresses Th17 development, we overexpressed EGFP-tagged NFIL3 in naïve CD4+ T cells by lentiviral transduction and grew cells under Th17-polarizing conditions. Since only a fraction of the T cells became transduced, we were able to analyze both transduced (EGFP+) and non-transduced (EGFP−) cells in each sample. CD4+ T cells transduced with lentivirus encoding NFIL3 yielded lower Th17 cell frequencies than non-transduced T cells (Fig. 1F, fig. S3). In contrast, transduced and non-transduced T cells yielded similar Th17 cell frequencies when control lentivirus (egfp only) was used. Thus, NFIL3 suppresses Th17 cell development in a T cell-intrinsic manner in vitro. Although a prior study found that retroviral transduction of Nfil3 did not significantly impact Th17 cell development (12), differences in the transduction protocol likely account for the different experimental outcomes.
To test whether NFIL3 has a T cell-intrinsic role in Th17 development in vivo, we adoptively transferred naïve wild-type and Nfil3−/− CD4+ T cells into lymphopenic Rag1−/− mice (13). More Nfil3−/− T cells differentiated into Th17 cells than did wild-type T cells, indicating that NFIL3 suppresses Th17 cell development in a T cell-intrinsic manner (Fig. 1G, fig. S4A). In accordance with the pathogenic role of Th17 cells in this model, Rag1−/− mice receiving Nfil3−/− T cells exhibited greater weight loss than mice receiving wild-type T cells (fig. S4B), as shown previously (11). IFNγ+ Th1 cell frequencies between the two groups of recipient mice were similar, confirming that NFIL3 preferentially impacts Th17 cell development (Fig. 1G).
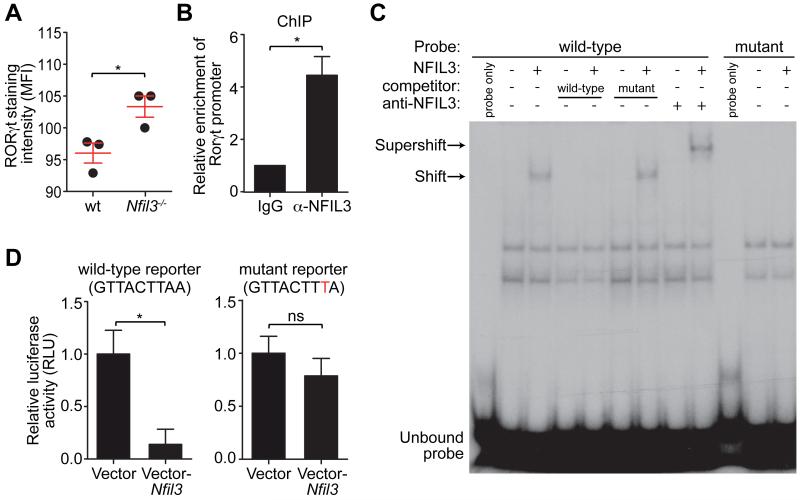
Th17 cell specification requires the orphan nuclear receptor RORγt (12, 14). Analysis of the Rorγt promoter sequence revealed a putative NFIL3 binding site (GTTACTTAA) that was conserved between human and mouse (fig. S5). Accordingly, Rorγt expression was higher in Nfil3−/− Th17 cells than in wild-type cells (Fig. 2A). A chromatin immunoprecipitation (ChIP) assay with an NFIL3-specific antibody (15) indicated that NFIL3 bound to the Rorγt promoter in mouse CD4+ T cells (Fig. 2B). Binding of NFIL3 to the conserved GTTACTTAA motif was demonstrated by electrophoretic mobility-shift assay (EMSA) and binding specificity of NFIL3 was further established by competition with unlabeled probes and supershift with the anti-NFIL3 antibody (Fig. 2C). Finally, overexpression of NFIL3 suppressed Rorγt promoter activity in Jurkat T cells as measured by a luciferase reporter assay (Fig. 2D). Repression was dependent on the GTTACTTAA motif, as introduction of a point mutation (GTTACTTTA) abolished the repressive effect. Thus, NFIL3 binds to the GTTACTTAA motif in the Rorγt promoter and represses promoter activity.
Figure 2. NFIL3 represses Rorγt transcription by binding directly to its promoter.
(A) LPLs from wild-type and Nfil3−/− mice were analyzed by nuclear staining of RORγt and mean fluorescence intensities (MFI) were plotted. (B) ChIP analysis of CD4+ T cells using IgG or anti-NFIL3 antibody. Enrichment of the Rorγt promoter was calculated as the ratio of the anti-NFIL3 to the IgG control pull-down. (C) EMSA with nuclear extracts of HEK293T cells transfected with an empty vector or an NFIL3-encoding vector. A 30-bp DNA fragment encompassing the NFIL3-binding site from the Rorγt promoter was synthesized as a wild-type probe. The mutant probe has the same sequence except that the NFIL3 binding site was mutated. NFIL3 binding specificity was demonstrated by competition with non-radioactively labeled probes and supershift with the anti-NFIL3 antibody. (D) Luciferase reporter assay. A 1018 bp (from −1013 to +5) fragment of the Rorγt promoter was fused with firefly luciferase and position 8 of the NFIL3 binding site was mutated from A to T in the mutant reporter. Jurkat T cells were transfected with reporters and an empty vector or an NFIL3-encoding vector. Luciferase activity was normalized to cells transfected with vector-only controls. Groups were plotted as mean ± SEM and compared by two-tailed student’s t-test. *,p<0.05; ns, not significant.
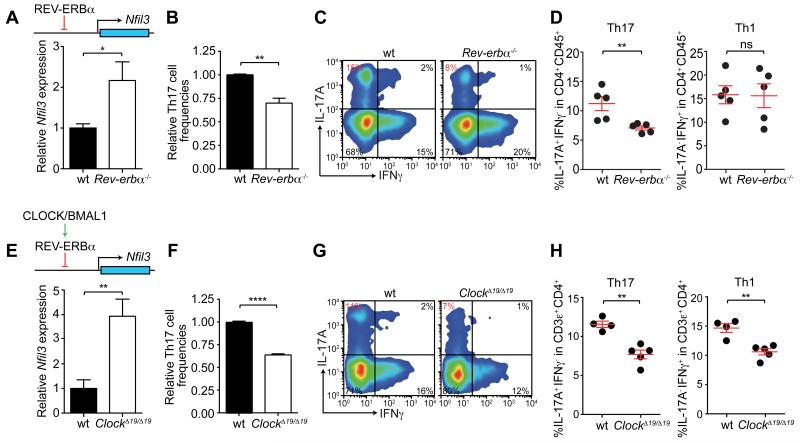
NFIL3 coordinates inputs from multiple regulatory pathways, including the circadian clock (16, 17). The circadian clock is an autoregulatory transcriptional network driven by the primary activators BMAL1 and CLOCK. It is negatively regulated by two feedback arms, one of which comprises the nuclear receptor REV-ERBα and its close homolog REV-ERBβ (18). The circadian clock circuitry has been shown to function in CD4+ T cells (19). REV-ERBα directly represses Nfil3 transcription by binding to a consensus sequence in the Nfil3 gene locus (Fig. 3A, fig. S6) (20). Accordingly, Nfil3 expression was higher in activated Rev-erbα−/− CD4+ T cells than in WT cells (Fig. 3A) (21). Activated CD4+ T cells were used in this experiment because Nfil3 mRNAs are more abundant in activated than in naïve CD4+ T cells (fig. S7A), consistent with the global increase in transcription in activated lymphocytes (22). Additionally, naïve Rev-erbα−/− CD4+ T cells showed a decreased capacity to differentiate into Th17 cells when cultured under Th17 polarizing conditions (Fig. 3B), and intestinal Th17 cell frequencies were reduced in Rev-erbα−/− mice (Fig. 3C,D). In contrast, no differences were observed in Th1 cell frequencies (Fig. 3C,D). Thus, Th17 cell lineage specification is linked to the clock regulatory network through NFIL3 and REV-ERBα.
Figure 3. Th17 cell development is regulated by the circadian clock transcriptional network.
(A) Nfil3 expression levels were quantified in activated CD4+ T cells by real-time PCR. (B) Naïve wild-type and Rev-erbα−/− CD4+ T cells were polarized to Th17 cells in vitro. (C,D) Small intestinal Th17 cell frequencies in wild-type and Rev-erbα−/− mice. Representative flow cytometry plots are shown in (C) and combined data are shown in (D). (E) Nfil3 expression in activated wild-type and ClockΔ19/Δ19 CD4+ T cells. (F) Naïve wild-type and ClockΔ19/Δ19 CD4+ T cells were cultured under Th17-polarizing conditions. (G,H) Small intestinal Th17 cell frequencies of wild-type and ClockΔ19/Δ19 mice. Representative flow cytometry plots are shown in (G) and combined data are shown in (H). Data are plotted as mean ± SEM and statistics were performed with the two-tailed student’s t-test. *,p<0.05; **,p<0.01; ***,p<0.001; ns, not significant.
To confirm the role of the circadian clock in Th17 development, we assessed ClockΔ19/Δ19 mice, which produce a dominant-negative CLOCK that inhibits the function of the BMAL1/CLOCK complex (23). BMAL1/CLOCK is required for REV-ERBα expression (Fig. 3E) (24), and accordingly ClockΔ19/Δ19 mice exhibited higher Nfil3 expression in activated CD4+ T cells (Fig. 3E), lowered capacity for Th17 cell differentiation in naïve T cells (Fig. 3F), and reduced intestinal Th17 cell frequencies when compared to wild-type mice (Fig. 3G,H). Interestingly, unlike Rev-erbα−/− mice, ClockΔ19/Δ19 mice also exhibited lower intestinal Th1 cell frequencies (Fig. 3G,H), suggesting that the circadian clock also impacts other intestinal CD4+ T cell subsets.
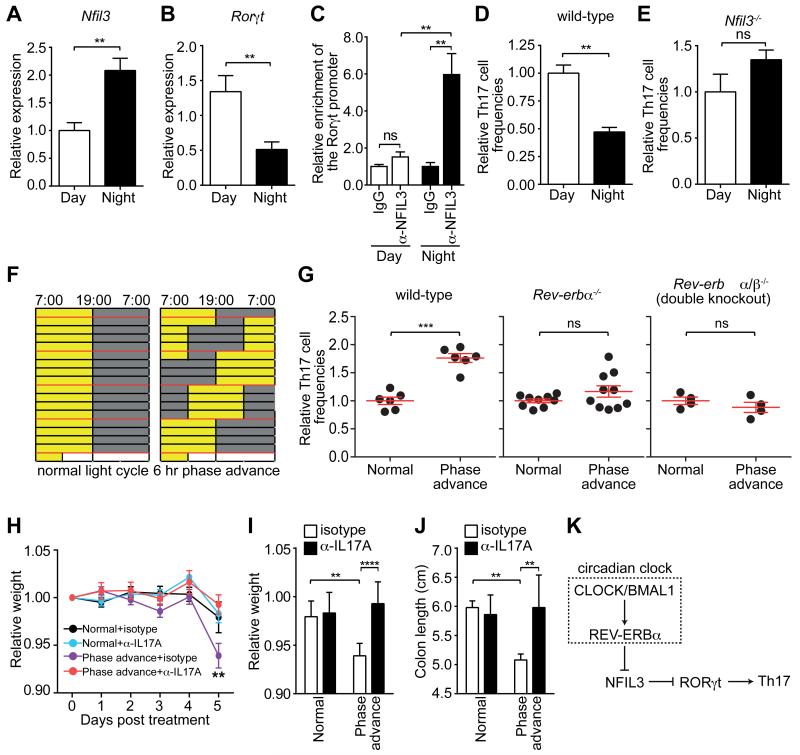
To examine Th17 development during the circadian cycle, we housed age- and sex-matched mice under either normal light cycles (LD, 12hr light: 12hr dark) or reversed light cycles (DL, 12hr dark: 12hr light) (fig. S8A). We found that Nfil3 expression was lower during the day and higher at night, while Rorγt expression was higher during the day and lower at night (Fig. 4A,B). There were no significant differences in CD4+ T cell composition at these timepoints (fig. S8B,C). The expression of Nfil3 and Rorγt in opposite phases of the circadian cycle was consistent with diurnal variation in binding of NFIL3 to the Rorγt promoter (Fig. 4C). Accordingly, naïve CD4+ T cells isolated during the day were more likely to differentiate into Th17 cells after in vitro polarization than those isolated at night (Fig. 4D). This difference was abolished in Nfil3−/− cells (Fig. 4E), showing that the diurnal variation in Th17 lineage specification is NFIL3-dependent. Thus, Th17 lineage specification is regulated in a diurnal manner and is synchronized across the T cell population by the circadian clock.
Figure 4. Diurnal regulation of Th17 cell development.
(A) Nfil3 and (B) Rorγt expression in activated CD4+ T cells during the circadian cycle. (C) Diurnal NFIL3 binding to the Rorγt promoter as determined by ChIP assay. (D,E) Relative percentage of IL-17A-producing CD4+ T cells after in vitro polarization of naïve wild-type (D) or Nfil3−/− (E) CD4+ T cells. (F) Wild-type, Rev-erbα−/− or Rev-erbα/β−/− double knockout mice were maintained under a normal light cycle (left) or were subjected to perturbed light cycles (6 hour phase advance every 4 days). Age-matched mice were co-housed prior to the experiment to minimize microbiota differences between mice in each group. (G) Intestinal Th17 cell frequencies in wild-type, Rev-erbα−/−, and Rev-erbα/β−/− mice subjected to light cycle perturbation. Th17 cell frequencies were calculated relative to the age-matched, co-caged controls in each experiment. (H-J) Mouse weight loss during DSS treatment of mice previously subjected to normal or perturbed light cycles. Mice were treated with anti-IL17A or an isotype control along with DSS challenge. Disease severity on Day 5 was assessed by weight loss (H,I) and colon shortening (J). (K) Schematic diagram summarizing how NFIL3 links the circadian clock circuitry to Th17 cell development. Data are plotted as mean ± SEM and statistical analysis was performed with two-tailed student’s t-test (A-E,G) or two-way ANOVA (H-J). *,p<0.05; **,p<0.01; ***,p<0.001; ns, not significant.
Consistent with the relatively long half-life (25) and week-long differentiation process of Th17 cells, we found that intestinal Th17 cell frequencies were unaltered during a single 24 hour cycle (fig. S9). We therefore tested whether circadian disruption by chronic light cycle perturbations altered Th17 cell frequencies (Fig. 4F). Th17 cell frequencies were higher in the intestines and spleens of mice subjected to perturbed light cycles as compared to mice maintained under a normal light cycle (Fig. 4G, fig. S10A), with no significant impact on cell proliferation or cell survival (fig. S11). This was coincident with a decrease in intestinal Th1 cell frequencies (fig. S10B), but no decrease in spleen Th1 cell frequencies (fig. S10A). Microbiota from mice under normal and perturbed light cycles yielded similar intestinal Th17 cell frequencies when transplanted into germ-free recipients (fig. S12A,B), suggesting that Th17 cell expansion was not due to altered microbiota composition. The increased Th17 cell frequencies was suppressed in Rev-erbα−/− and, to a larger extent, in Rev-erbα−/−β−/− (double knockout) mice (Fig. 4G), indicating that the increased Th17 cell frequencies required REV-ERBα/β and were unlikely to arise from non-specific effects of light cycle perturbation. Mice subjected to perturbed light cycles were more susceptible to dextran sulfate sodium (DSS)-induced colitis, as measured by weight loss and colon shortening (Fig. 4H-K). The enhanced pathology could be ameliorated by neutralizing IL17A (Fig. 4H-K), suggesting that the increased susceptibility to DSS treatment was due in part to the elevated Th17 cell frequencies in the mice subjected to perturbed light cycles. Thus, chronic light cycle perturbation leads to elevated Th17 cell frequencies in the intestine and enhanced susceptibility to inflammatory disease. This suggests that diurnal regulation of Th17 cell differentiation is important for maintaining homeostatic Th17 cell frequencies and restraining inflammation.
Together, our results demonstrate that NFIL3 suppresses Th17 cell development by directly repressing Rorγt transcription, and links Th17 cell development to the circadian clock (Fig. 4K). This ensures that Th17 lineage specification preferentially occurs at a specific stage of the circadian cycle and is thus synchronized across the entire T cell population. We suggest that overaccumulation of Th17 cells may be limited by ensuring that all T cells within a population traverse this critical developmental checkpoint in synchrony rather than at random times during the day-night cycle.
Modern life often involves chronic circadian disruptions, such as night shift work or jet lag, that are linked to human inflammatory diseases (26, 27). Our findings suggest that the pathologic consequences of circadian disruption may be due in part to direct interactions between the circadian clock and the pathways that regulate pro-inflammatory immune cell development.
Supplementary Material
Acknowledgements
We thank C. Behrendt, C. Clements, and S. Murray for assistance with mouse experiments. M. Izumo provided advice and assistance with the circadian cycle experiments. We also thank F. Yarovinsky for critical reading of the manuscript. The data presented in this manuscript are tabulated in the main paper and the supplementary materials. This work was supported by NIH R01 DK070855 (LVH), a Burroughs Wellcome Foundation New Investigators in the Pathogenesis of Infectious Diseases Award (LVH), and the Howard Hughes Medical Institute (JST and LVH).
Footnotes
References
- 1.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 4.Arjona A, Silver AC, Walker WE, Fikrig E. Immunity’s fourth dimension: approaching the circadian-immune connection. Trends Immunol. 2012;33:607. doi: 10.1016/j.it.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009;27:485. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 6.Maloy KJ, Kullberg MC. IL-23 and Th17 cytokines in intestinal homeostasis. Mucosal Immunol. 2008;1:339. doi: 10.1038/mi.2008.28. [DOI] [PubMed] [Google Scholar]
- 7.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Male V, Nisoli I, Gascoyne DM, Brady HJ. E4BP4: an unexpected player in the immune response. Trends Immunol. 2012;33:98. doi: 10.1016/j.it.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 11.Motomura Y, et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat. Immunol. 2011;12:450. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostanin DV, et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G135. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov II, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 15.Kashiwada M, et al. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc. Natl. Acad. Sci. U. S. A. 2010;107:821. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 18.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012;35:445. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bollinger T, et al. Circadian clocks in mouse and human CD4+ T cells. PLoS One. 2011;6:e29801. doi: 10.1371/journal.pone.0029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duez H, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology. 2008;135:689. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 21.Preitner N, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 22.Kouzine F, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Triqueneaux G, et al. The orphan receptor Rev-erbα gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 2004;33:585. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pepper M, et al. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat. Immunol. 2010;11:83. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bechtold DA, Gibbs JE, Loudon AS. Circadian dysfunction in disease. Trends Pharmacol. Sci. 2010;31:191. doi: 10.1016/j.tips.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4453. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitaterna MH, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W, Troutman TD, Edukulla R, Pasare C. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity. 2011;35:1010. doi: 10.1016/j.immuni.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huh JW, et al. Multivalent di-nucleosome recognition enables the Rpd3S histone deacetylase complex to tolerate decreased H3K36 methylation levels. EMBO J. 2012;31:3564. doi: 10.1038/emboj.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.