Abstract
Imaging mass spectrometry (IMS) has become a prime tool for studying the distribution of biomolecules in tissue. Although IMS data sets can become very large, computational methods have made it practically feasible to search these experiments for relevant findings. However, these methods lack access to an important source of information that many human interpretations rely upon: anatomical insight. In this work, we address this need by (1) integrating a curated anatomical data source with an empirically acquired IMS data source, establishing an algorithm-accessible link between them and (2) demonstrating the potential of such an IMS-anatomical atlas link by applying it toward automated anatomical interpretation of ion distributions in tissue. The concept is demonstrated in mouse brain tissue, using the Allen Mouse Brain Atlas as the curated anatomical data source that is linked to MALDI-based IMS experiments. We first develop a method to spatially map the anatomical atlas to the IMS data sets using nonrigid registration techniques. Once a mapping is established, a second computational method, called correlation-based querying, gives an elementary demonstration of the link by delivering basic insight into relationships between ion images and anatomical structures. Finally, a third algorithm moves further beyond both registration and correlation by providing automated anatomical interpretation of ion images. This task is approached as an optimization problem that deconstructs ion distributions as combinations of known anatomical structures. We demonstrate that establishing a link between an IMS experiment and an anatomical atlas enables automated anatomical annotation, which can serve as an important accelerator both for human and machine-guided exploration of IMS experiments.
Understanding the spatial context in which molecular interactions take place is becoming increasingly important in the study of biological and pathological processes in living organisms. The spatial distribution of biomolecules and the localization of biochemical interactions throughout tissue often hold crucial clues toward determining the biological functions of these biomolecules. Imaging mass spectrometry (IMS)1,2 is a molecular imaging technology that can deliver such spatial information with high chemical specificity for various classes of biomolecules, including metabolites, lipids, peptides, and proteins. IMS has been gaining considerable momentum in recent years, primarily in the field of tissue biomarkers3,4 and drug delivery,5,6 and has been successfully applied to tissues of various origin, including insect,7 mammalian,8 and human tissue.9−11 IMS makes it possible to monitor many hundreds of biomolecules simultaneously, making it a prime technology for exploratory studies. However, this exploratory advantage is hampered by the large amount of data that a single IMS experiment can deliver, making interpretation and analysis difficult.
Previous work has employed both supervised and unsupervised computational methods,12−15 such as hierarchical clustering, principal component analysis,16,17 and probabilistic latent semantic analysis,18 to perform comprehensive analysis of these large data sets. While these methods aid human interpretation by reducing the data size and complexity, they often operate in a blind fashion in the sense that they lack access to an important source of information that many human interpretations rely upon: anatomical information on the tissue in question. This information is available in textbooks and through publically accessible anatomical atlases for various organ types and organisms. However, the use of such information in IMS studies remains largely restricted to manual comparison,19−21 which poses a practical challenge for larger multiexperiment studies and brings with it a risk of introducing human bias into the analysis. In order to fully utilize this body of anatomical insight for the interpretation of ion distributions in IMS data, a computer-traversable bridge between IMS and curated anatomical info is essential. Recent work by Abdelmoula et al.22 has taken a first step toward this integration by developing a workflow that performs automated spatial registration of IMS data to anatomical information through microscopy. We extend this line of research further by focusing on the applications that become possible once a registration is available. In order to do so, we first (1) establish and demonstrate our own algorithm-accessible link between curated anatomical data and empirically acquired IMS data and then (2) move beyond registration by applying the established IMS-anatomical atlas link toward automated anatomical interpretation of the ion images obtained through IMS.
Since a substantial amount of IMS research focuses on the rodent brain,23 our case studies use MALDI-TOF IMS data from mouse brain tissue as the empirical data source and the Allen Mouse Brain Atlas24 as the curated anatomical data source. Both data types have been used in studies of neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and healthy mouse brain.25,26 However, the methods developed in this paper are not specific to these case studies, a particular species, disease model, or atlas. They can be readily applied to any IMS-atlas combination that makes sense within the context of a particular study.
Methods introduces the two data sources and describes the three computational methods that implement the anatomy-aware analysis approach we developed: (i) registration, (ii) correlation-based querying, and (iii) automated anatomical interpretation. Results & Discussion applies the developed methods to both a protein-focused and a lipid-focused case study, with complementary details in the Supporting Information.
Methods
The first objective, the integration of the two data sources, entails development of two computational methods. The first method spatially registers the IMS data to the anatomical atlas. Registration is a necessary step that precedes anatomical interpretation and makes direct mapping of findings across data sources possible by establishing a common coordinate system.27−29 Since the methods that follow are independent of how this spatial mapping is established and are quite robust against registration errors, we provide a relatively basic registration implementation. A more advanced and automated registration procedure is available in Abdelmoula et al.22 Since the registration needs to account for cutting artifacts, tissue deformations due to extraction and freezing of the brain, and other spatial perturbations, nonrigid registration techniques27,28 play a central role here. The second method utilizes the established link to interrogate the combined data sources for correlations. Correlation-based queries deliver insight into the spatial correlations between ion images in IMS and anatomical areas in the atlas. In previous studies the authors, as well as several other groups, have successfully demonstrated the potential of correlation-based approaches to guide the user toward relevant findings.22,30−34 When the integration objective is complete, anatomical regions are implicitly annotated with biochemical findings from mass spectrometry, and biomolecular distributions are inherently mapped to a set of anatomical definitions.
The second objective moves beyond registration and simple correlation and uses the established IMS-anatomical atlas link to develop an automated anatomical interpretation method for IMS data. We define the anatomical interpretation of an ion image as decomposing the ion distribution into a combination of known anatomical areas that are tentatively tied to that specific ion. Anatomical interpretation becomes possible once a registration of IMS data to an atlas is available. Since it is independent of the particular method that was utilized to attain such registration, and it is assumed that registration errors are always present to some extent, automated anatomical interpretation methods can be developed largely orthogonal to but still benefit from any registration advancements. The interpretation method we develop can therefore be used in any setting where IMS is coupled to an atlas. Although the correlation-based queries demonstrate the functioning of the IMS-to-atlas mapping and provide a first step toward exploration of the combined data, they are insufficient to power automated anatomical interpretation of ion images. The main reason is their inadequate handling of ions that are present in several anatomical areas simultaneously. An automated anatomical interpretation of ion images therefore needs to be able to handle membership of an ion to multiple anatomical structures and preferably should include a measure of abundance. To this end, the third computational method of this work develops an algorithm that uses the IMS-atlas link to automatically interpret any ion image in the IMS data set as a combination of atlas-provided anatomical structures, without the need for human intervention.
Anatomy Data
The publicly accessible Allen Mouse Brain Atlas (AMBA) is used as the anatomical data source. This atlas is based on the brain of a 56-day-old C57BL/6J mouse, and has a user base that exceeds 10000 users per month.25 The brain is dissected into 528 coronal tissue sections at 25 μm separation, which are Nissl-stained, registered to each other, and assembled into a reconstructed brain volume. A low resolution (25 μm voxel width) gray scale version of this reconstructed brain volume is accessible through the application programming interface provided by the AMBA Web site35 and is imported into MATLAB 2012b (The Mathworks Inc., Natick, MA) to establish a local copy for further computation (Figure 1).
Figure 1.
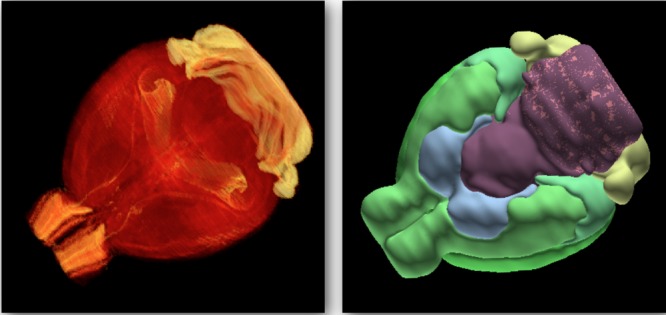
Registered microscopy and anatomy in the Allen Mouse Brain Atlas. The microscopy volume contains 528 coronal Nissl-stained tissue sections (left). These data are imported into and visualized using MATLAB, color-coding intensity from red to yellow. The 3D anatomical reference atlas contains over 800 anatomical structures (right). These data are visualized using Brain Explorer 2.
Of the 528 coronal tissue sections, 132 are hand annotated and combined to create a three-dimensional (3D) anatomical reference atlas (Figure 1). This atlas is registered to the brain volume constructed from the Nissl stains and contains over 800 anatomical structures. It can be consulted using Brain Explorer 2 (AMBA Web site).
IMS Data
Coronal tissue sections of a 12 μm thickness were acquired from a healthy adult mouse brain that had been frozen in liquid nitrogen. Two neighboring sections from this brain were selected for IMS measurement and mounted on ITO-coated glass slides. A third neighboring section was mounted on a glass slide and Nissl-stained for matching against the Nissl stains of the AMBA. We give an overview of the staining and IMS measurements below and refer to the Supporting Information for full details. One IMS measurement focuses on protein imaging, acquiring ions between m/z 3000 and 22000. The other IMS measurement focuses on lipids with a m/z range from 400 to 1000. The tissue sections were sublimated with sinapinic acid (protein-oriented) and 1,5-diaminonaphthalene (lipid-oriented). The measurements were acquired on a Bruker Autoflex Speed MALDI-TOF mass spectrometer in the positive linear mode with a laser spot size of 80 μm on target and at a pitch of 100 μm (protein-oriented) and in the negative reflector mode with a laser spot size of 30 μm on target and at a pitch of 80 μm (lipid-oriented), using FlexControl 3.3. Approximately 100 shots/spot were acquired at a 1 kHz repetition rate using a Smartbeam II Nd:YAG laser. Image acquisition was carried out using FlexImaging 2.1, and further processing took place in MATLAB. The spectra were normalized on the basis of their common ion current, disregarding differential peaks.36 They were baseline-corrected using a spline approximation of the baseline at the 10%-quantile of ion intensities and employing window sizes of 500 and 50 and step sizes of 250 and 25 for the protein and lipid-focused spectra, respectively. The spectra were also optimally aligned along the m/z axis to reduce peak drift, allowing a maximum m/z shift of 12 and 0.5 for protein and lipid-focused spectra, respectively. Both steps were performed using the Bioinformatics Toolbox of MATLAB (The Mathworks Inc., Natick, MA).
Registration of Data Sources
Coupling IMS data to the atlas requires the two data sources to be registered to each other, thus transforming them to a common coordinate system, in which their pixel locations describe the same space and can be directly compared.27,28 The IMS-atlas registration process requires multiple steps and uses both rigid and nonrigid registration to handle the complexities that are commonly encountered in tissue (e.g., deformation during extraction and freezing of the brain, differences between individual mouse brains, cutting artifacts, etc.). To deal with these complex deformation cases, nonrigid registration techniques37,38 are essential.
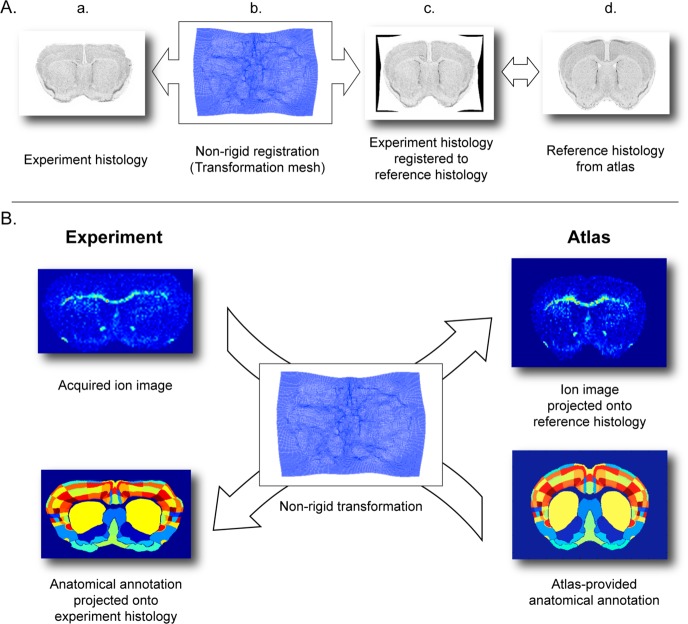
The IMS data is registered to the anatomical atlas via a modality common to both data sources: stained microscopy (Figure 2). The registration process entails: (1) rigid registration of IMS data to the experiment histology, (2) rigid registration of atlas data to the reference histology, and (3) nonrigid registration of the experiment histology to the reference histology. Although it is technically possible to register IMS data directly to the atlas, it is preferable to go through histology since the IMS data and the atlas data are of a substantially different nature with very differing spatial resolutions (80–100 and 0.3 μm, respectively). Using microscopy as an intermediate offers several advantages: (i) registration is more straightforward within a single modality, certainly given the complexity involved in nonrigid registration, (ii) the resolutions are high and comparable, and (iii) histological microscopy is readily available in most state-of-the-art IMS experiments.
Figure 2.
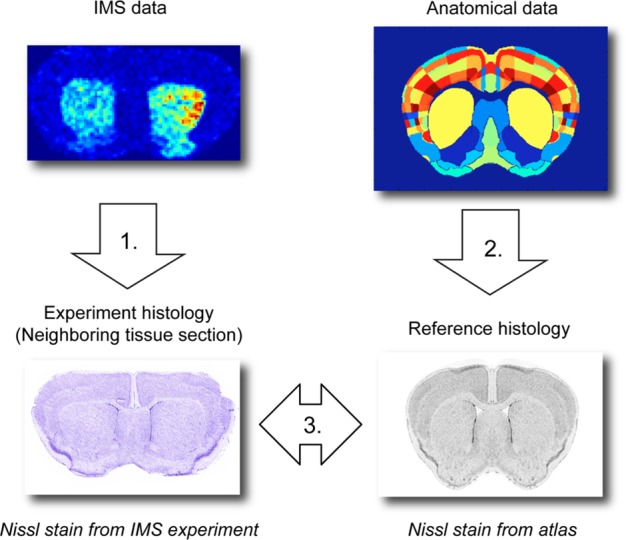
Workflow of the registration process. Spatially registering IMS data to anatomical data consists of 3 individual registration steps: (1) rigid registration of IMS data to experiment histology, (2) rigid registration of anatomical data to reference histology, and (3) nonrigid registration of experiment histology to reference histology.
Details of the registration process can be found in the Supporting Information. To summarize, (1) the rigid registration between IMS data and experiment histology is performed in MATLAB through manual selection of fiducial markers, (2) the rigid registration from atlas to reference histology is provided by the AMBA, and (3) the nonrigid registration between experiment histology and reference histology is performed using the Medical Image Registration Toolbox (MIRT) by Myronenko,29,39 making use of a free form deformation (FFD) model.
Correlation-Based Querying
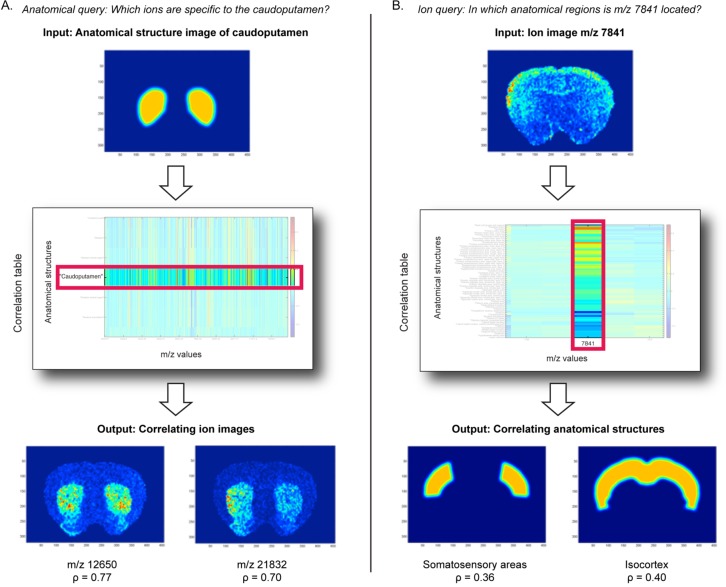
Once registration is complete, it becomes possible to find out which anatomical structures correlate with a measured ion distribution or which ions correlate with a certain anatomical zone of interest. To enable this simple form of correlation-based querying, we create an anatomical structure image for each individual anatomical structure that is present in the tissue slice. Such an anatomical structure image (caudoputamen example in Figure 4A, top) represents the spatial location of a single anatomical annotation and contains ones in locations where the anatomical structure is present, and zeros elsewhere. To soften the binary assignments somewhat at the edges of the structure, we apply a Gaussian filter, which eliminates crisp borders.
Figure 4.
(A) Example of an anatomical query, finding ions specific to the caudoputamen. The anatomical structure image of the caudoputamen is given as an input and the correlation table returns the spatial correlation to this structure for each ion image. Two examples of ion images that positively correlate with the target anatomical structure are displayed. (B) Example of an ion query, finding anatomical regions in which m/z 7841 is highly expressed. The ion image of m/z 7841 is given as an input, and the correlation table returns the spatial correlation to this ion image for all the anatomical regions. Two examples of anatomical structure images that positively correlate with the target ion image are displayed.
Next, the spatial correlation between an ion image and an anatomical structure image is obtained by calculating the Pearson correlation coefficient between the intensities of both images over all IMS measurement locations. To prevent bias, only pixels for which both types of data are available are part of the correlation analysis. The analysis is performed by reshaping both 2D images to 1D vectors, removing any rows for which only one type of information is available, and then calculating the correlations between the resulting vectors. The correlation coefficient for each possible anatomical structure/ion image combination is calculated and collected into a correlation table for easy visualization and querying.
Automated Anatomical Interpretation
The goal of the anatomical interpretation method is to examine the pattern in an ion image, and, without human intervention, determine which anatomical structures are involved and what their ion intensity contribution is. In other words, once an ion image is mapped to the atlas (using the proposed registration pipeline or an automated variant thereof22), the interpretation method takes that ion distribution pattern as an input and then automatically decomposes it into a combination of atlas-provided anatomical structures.
At its core, anatomical interpretation is a problem of approximating the spatial pattern of an ion with a combination of patterns selected from a provided vocabulary of anatomical patterns. The model we employ in our algorithm considers an ion image to be a sum of products, each product multiplying a pattern from the finite set of anatomical patterns with its contribution coefficient. Since we know both the ion image and the anatomical patterns and their relationship is established by the model, the search for the optimal anatomical contribution coefficients (and thus the optimal anatomical interpretation) can be approached as a multivariate optimization problem. The mathematical details of this approach are provided in the Supporting Information.
Our implementation uses CVX, a package for specifying and solving convex programs,40,41 to solve the optimization problem for each ion image we want interpreted. The anatomical patterns are used as building blocks to construct an approximation of each ion image, and the coefficients specify how each anatomical structure contributes to the overall approximation. A nice feature of the method is that an anatomical contribution coefficient tends to be proportional to the ion intensity in that anatomical structure, inherently assigning a notion of importance or weight to each anatomical zone involved. Anatomical images, and thus structures, corresponding to high absolute coefficients are important for approximating the ion distribution of interest and are therefore considered part of the anatomical interpretation of that ion image. Also, note that in our examples the coefficients are not constrained to positive values. This allows the anatomical interpretation to say things like “the ion seems to be present in zone A plus zone B minus zone C.”
Results and Discussion
We demonstrate the developed methods both in a protein-focused and a lipid-focused case study of coronal mouse brain sections, illustrating the potential of incorporating anatomical information into IMS analysis.
Results on Registration of the Data Sources
A nonrigid registration algorithm is used to register the experiment histology (Figure 3A.a) to the atlas reference histology (Figure 3A.d). Although the experiment histology is strongly deformed compared to the reference histology, the registration results (Figure 3A.c) demonstrate that the nonrigid registration can deal well with these soft-tissue deformations. The FFD (free form deformation) mesh of control points (Figure 3A.b) that constitutes the transformation between the two images shows how the experiment histology image is “warped” to register to the atlas histology. By using a FFD transformation mesh, we ensure that neighboring IMS pixels remain neighbors and prevent excessive distortion of the original data.
Figure 3.
Overview of registration results. (A) Nonrigid registration of the experiment histology onto the reference histology from the atlas. (a) Despite the large initial deformation of the experiment histology, (b) the nonrigid transformation successfully compensates and (c) registers onto the (d) reference histology. (B) The nonrigid registration enables traversal from the coordinate system of one data source to the other: the acquired ion image can be projected onto the reference histology for direct comparison to the anatomical annotations. Inversely, the anatomical annotations, provided by the atlas, can be projected onto the experiment histology, to provide anatomical guidance within the sectioned tissue.
Figure 3B shows how the transformation established by the nonrigid registration process is used to transform and project data from one data source to the other and vice versa. The nonrigid transformation is reversible (up to a rounding error), and there is a one-to-one relationship between locations in “IMS space” and locations in “atlas space”. This means that we are able to project IMS data onto the reference histology (Figure 3B, top right). Such a projection effectively brings empirical MS measurements acquired from imperfectly sectioned or deformed tissue into the reference shape of the mouse brain, and it directly links these observations to reference atlas annotations. Similarly, we can project the atlas annotations onto the experiment histology (Figure 3B lower left). This direction of projection draws annotations that are typically defined in an ideally shaped version of the mouse brain into the practical tissue sample that was sectioned. Casting the anatomy information to the experiment histology also enables quick visual verification of the correctness of the projection since the neighborhood structure of the different subareas should be retained regardless of tissue deformation.
Results on Querying the Correlation between Anatomy and Ions
Once registration of the two data sources is complete, we construct a correlation table by calculating the spatial correlation between the anatomical structure images and the ion images of the peak-picked IMS protein data. The correlation table (available in the Supporting Information) allows two types of queries: anatomical queries and ion queries.
Anatomical Query
The anatomical query provides an answer to the question “Which ions are specific to anatomical region X?”. To demonstrate, we use the caudoputamen as an example anatomical structure. Figure 4A shows the input, the anatomical structure image of the caudoputamen, and displays the part of the correlation table that pertains to this structure, highlighting its spatial correlation to the various ion images. Ions m/z 12650 (ρ = 0.77) and m/z 21832 (ρ = 0.70) are two examples of ion images that exhibit high positive correlation in this case. These ions show a clear spatial overlap with the anatomical structure image of the caudoputamen and are almost exclusively located therein.
Ion Query
The ion query provides an answer to the question “In which anatomical regions is ion Y located?”. To demonstrate this query, we use the ion image for m/z 7841 as an example. Figure 4B shows the ion image as input and, analogous to the anatomical query, it displays the part of the correlation table that pertains to this ion. The isocortex (ρ = 0.40) and the somatosensory areas (ρ = 0.36), which are substructures of the isocortex, are two examples of anatomical structures that exhibit high positive correlation with m/z 7841. Additional examples are provided in the Supporting Information.
Correlation-based queries can deliver fast insight into relationships between ions and anatomical structures but have several disadvantages. First, it is difficult to define a generic threshold to determine when these correlations become significant. Second, since several thousands of correlations are being calculated in parallel, the multiple testing problem needs to be considered when drawing conclusions from these results. However, the most important roadblock for using correlation toward automated anatomical interpretation is the concept of “multimembership”. Correlation considers only the relationship between a single anatomical structure and a single ion. An ion that appears in several anatomical structures simultaneously will exhibit a relatively low correlation to each of the individual anatomical structures that it is a member of. Such an ion will not give a strong signal in the correlation table and could go undetected as a result. In these multimembership situations, which are quite common in most biological tissue types, correlation-based querying falls short and is not capable of dealing with the complexities of the biology. In fact, in such complex cases, any univariate querying strategy will provide skewed results.
Results on Automated Anatomical Interpretation
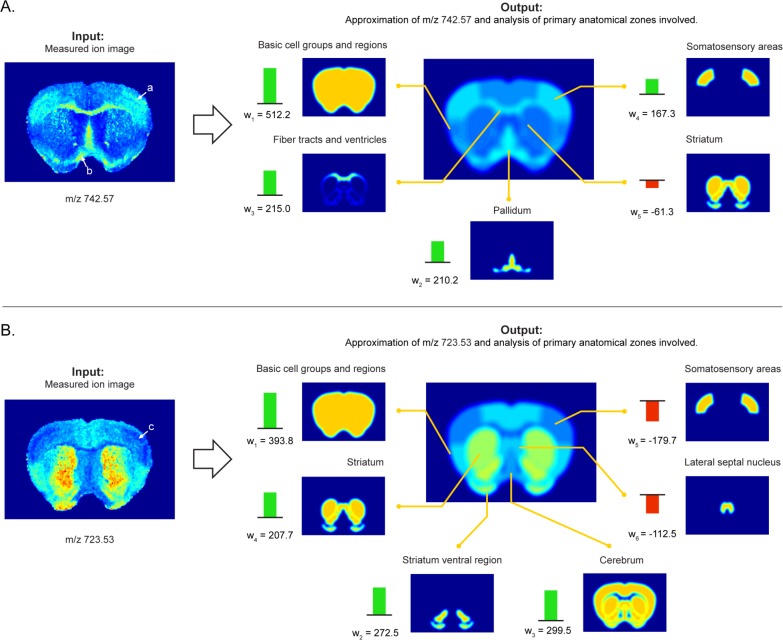
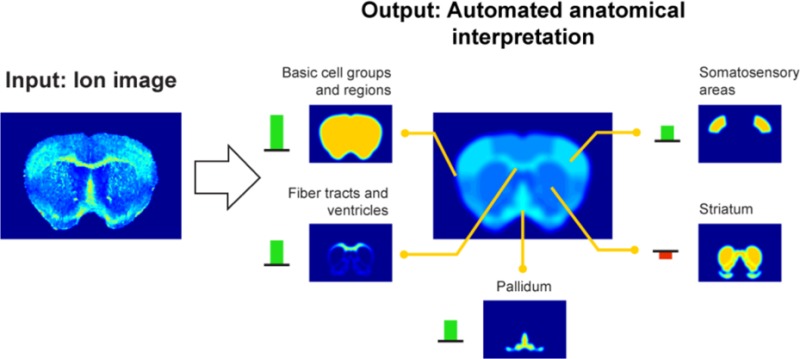
Since the membership of an ion to multiple anatomical structures cannot be clearly captured by a univariate strategy, the use of correlation to drive automated anatomical interpretation of ion images is limited. Instead, a more advanced approach, using multivariate models to account for multimembership, is necessary. Methods introduces a linear model, capable of capturing the multimembership aspect effectively, to tie ion image patterns to anatomical structure patterns. By applying convex optimization to this model and the given ion and anatomy patterns, it is possible to obtain an optimal anatomical explanation for each ion image. Figure 5 shows the automated anatomical interpretation of several ion images from the lipid case study, using our method. Each ion image gets interpreted, automatically and without user-intervention, as an optimal combination of anatomical zones from the AMBA. The interpretation is optimal in the sense that it selects the combination of atlas patterns that gives the closest approximation of the measured ion image. If there are multiple combinations that come equally close, the combination with the least amount of anatomical structures (and thus the simplest explanation) is selected. In short, the automated anatomical interpretation method provides (i) the closest approximation of the measured ion image using atlas structures (Figure 5, right, middle) and (ii) an overview of the contributing anatomical structures, specifying name, reference location, and contributing intensity or weight in the interpretation (Figure 5, right, outer ring). This visualization delivers quick insight into the major anatomical zones associated with an ion image, while also providing a notion of the relative contributions of each underlying structure involved.
Figure 5.
Examples of automated anatomical interpretation. When an ion image is given as input (left), the interpretation method provides an optimal anatomical explanation for the observed ion pattern (right), using the library of provided anatomical structures. Specifically, the ion intensity pattern is decomposed without user intervention into an optimal combination of contributing anatomical structures from the atlas. The interpretation method provides (i) the closest approximation of the measured ion image using atlas structures (right, middle) and (ii) an overview of the contributing anatomical structures, specifying name, reference location, and contributing intensity or weight in the interpretation (right, outer ring). This visualization delivers quick insight into the major anatomical zones associated with an ion image, while also providing a notion of the relative contributions of each underlying structure involved. Negative weights indicate a relative decrease of the ion in those areas. (A) Ion m/z 742.57 is highly expressed in the somatosensory areas, the fiber tracts, and the pallidum. (B) Ion m/z 723.53 shows a decrease specifically in the somatosensory areas, as indicated by the negative weight. The empirical ion distributions show good congruence with the boundaries of the anatomical structures defined in the atlas, indicating good spatial mapping between the data sources and strong biological signals in the IMS measurements.
The example in panel A shows a clearly defined expression of the ion m/z 742.57 in the fiber tracts and ventricles (more specifically the corpus callosum, which is not annotated as a separate region in the AMBA), the pallidum, and the somatosensory areas. Using negative contribution coefficients, the algorithm also reports areas where the ion exhibits reduced presence, such as in the striatum. Panel B shows the ion m/z 723.53, which is specifically absent from the somatosensory areas. The strong congruence between empirically observed patterns in the ion distributions and anatomical structures extracted from the atlas indicate good spatial mapping between these data sources and strong biological signals in the IMS measurements. Additional examples of the in total 1405 ion images that we applied the automated anatomical interpretation methodology to are provided in the Supporting Information.
It should be noted that the heads-up-display type visualization of Figure 5 is meant for human consumption and is but one of many possible representations of the interpretation results. In essence, the interpretation algorithm provides for each ion image a set of contribution coefficients, one for each structure in the atlas. Each contribution coefficient can be considered to report a degree of membership of an ion to a particular structure. Custom visualizations of these coefficients can be developed as demanded by the application. In the case of a machine-based follow-up, no visualization will be necessary at all and the anatomical membership coefficients for each ion image in an IMS experiment can be passed on directly to the next computational step in the analysis.
The automated breakdown of an ion image into contributing anatomical structures is a powerful tool, particularly for the histological nonexpert. It supplies the researcher directly with the relationships between an ion and the anatomical structures in which it is expressed. The interpretation algorithm can substantially aid in unraveling the function of biomolecular ions. It can incorporate the body of pathological research that is currently publicly available into the analysis of an individual IMS experiment and this without much additional effort. Essentially, the results of the analysis constitute a table of anatomical membership coefficients, linking each ion image to each anatomical structure. This table can be queried in the same way as the correlation table from the correlation-based queries but does away with the disadvantages of a univariate approach. Most importantly, the multimembership of ions to different anatomical zones is now taken into account, providing the user with a much more complete and reliable list of anatomical-structure-to-ion relationships.
The greatest benefit of an automated anatomical interpretation method is the potential for parallelization. A computational interpretation method can deliver anatomical interpretations for every ion image in an experiment, even if the number of ion images runs into the hundreds or thousands. It can provide these interpretations concurrently, without human intervention, and in a single calculation. This avoids the labor and time-intensive step of having a histological expert manually interpret hundreds of ion images and the potential risk for human bias that comes with it. It also enables the researcher to pursue broad exploratory measurements, after which the focus can be narrowed to only the most promising ions that have demonstrated a relationship to a particular anatomical structure of interest.
A possible downside to using an anatomical atlas for the interpretation of IMS data is that zones that have not been previously discovered in pathological and anatomical research and thus are not present in the atlas cannot be found in the ion images and cannot be part of their interpretation. However, examining those ions that cannot be adequately approximated with an atlas could provide a route to the automated discovery of new physiological or chemical subdivisions within tissue areas that are considered to be homogeneous by anatomy. We elaborate on this in the Supporting Information.
Conclusions
Establishing a link between an IMS experiment and an anatomical atlas can serve as an important accelerator both for human and machine-guided exploration of IMS experiments.
As the number and complexity of IMS experiments increases and studies aim to compare a growing number of IMS experiments to each other, the importance of automated approaches to filter the massive data sets for patterns of interest will become increasingly important. The automated anatomical interpretation of ion images can serve as a formidable catalyst for IMS analysis, due to its speed and ability to interpret thousands of ion images concurrently without human supervision.
On a less application-specific scale, a mapping between curated anatomy and IMS data can make a body of anatomical research available to any IMS-related computational method, to integrate into its analysis and improve its performance. In the case of multiple IMS experiments and multiple mappings to the same atlas, one can compare the anatomical interpretations of multiple experiments to each other by using a common reference (atlas) space. This would allow, for example, the automated detection of differences in activities of anatomical structures between healthy versus diseased tissues. Mapping multiple IMS data sets to a common reference coordinate system would also enable the creation of an IMS-based chemical reference atlas for lipidomics, proteomics, and metabolomics. In the specific case of the Allen Mouse Brain Atlas, this offers several promising perspectives for the future, since IMS-based data could then even be combined with non-IMS data sources, such as gene expression and neuron connectivity measurements, which are currently being linked to the Allen Brain Atlas by other research initiatives. The combination of these data sources can serve as a valuable multimodal resource for systems biology research, bringing together insights from many different technologies and fields.
Acknowledgments
We acknowledge Andriy Myronenko for his work on the MIRT toolbox and making this tool available to us. This research was funded by a PhD grant of the Agency for Innovation by Science and Technology (IWT), the Research Council KU Leuven: GOA/10/09 MaNet, GOA/12/24, Industrial Research fund (IOF): IOF/HB/13/027 Logic Insulin, the Flemish Government: FWO: projects: G.0871.12N (Neural circuits), IWT: TBM-Logic Insulin (100793), TBM Rectal Cancer (100783), TBM IETA (130256), PhD grants, iMinds Medical Information Technologies SBO 2014, VLK Stichting E. van der Schueren: rectal cancer, Federal Government: FOD: Cancer Plan 2012-2015 KPC-29-023 (prostate), IAP P7/13, COST: Action BM1104: Mass Spectrometry Imaging. This work was supported by the U.S. National Institutes of Health Grants NIH/NIGMS R01 GM058008-14 and NIH/NIGMS P41 GM103391-03. The scientific responsibility is assumed by its authors.
Supporting Information Available
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions from all authors. All authors have given approval for the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Caprioli R. M.; Farmer T. B.; Gile J. Anal. Chem. 1997, 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- Mcdonnell L. A.; Heeren R. M. A. Mass Spectrom. Rev. 2007, 26, 606–643. [DOI] [PubMed] [Google Scholar]
- Chughtai K.; Heeren R. M. A. Chem. Rev. 2010, 110, 3237–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch A.; Rauser S.; Deininger S.-O.; Höfler H. Histochem. Cell Biol. 2008, 130, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N.; Tanaka Y.; Inazawa K.; Saji H. Rapid Commun. Mass Spectrom. 2012, 26, 1549–1556. [DOI] [PubMed] [Google Scholar]
- Trim P. J.; Henson C. M.; Avery J. L.; McEwen A.; Snel M. F.; Claude E.; Marshall P. S.; West A.; Princivalle A. P.; Clench M. R. Anal. Chem. 2008, 80, 8628–8634. [DOI] [PubMed] [Google Scholar]
- Sugiura Y.; Konishi Y.; Zaima N.; Kajihara S.; Nakanishi H.; Taguchi R.; Setou M. J. Lipid Res. 2009, 50, 1776–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckli M.; Chaurand P.; Hallahan D. E.; Caprioli R. M. Nat. Med. 2001, 7, 493–496. [DOI] [PubMed] [Google Scholar]
- Khatib-Shahidi S.; Andersson M.; Herman J. L.; Gillespie T. A.; Caprioli R. M. Anal. Chem. 2006, 78, 6448–6456. [DOI] [PubMed] [Google Scholar]
- Eberlin L. S.; Norton I.; Dill A. L.; Golby A. J.; Ligon K. L.; Santagata S.; Cooks R. G.; Agar N. Y. R. Cancer Res. 2012, 72, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazares L. H.; Troyer D.; Mendrinos S.; Lance R. A.; Nyalwidhe J. O.; Beydoun H. A.; Clements M. A.; Drake R. R.; Semmes O. J. Clin. Cancer Res. 2009, 15, 5541–5551. [DOI] [PubMed] [Google Scholar]
- Jones E. A.; Deininger S.-O.; Hogendoorn P. C. W.; Deelder A. M.; Mcdonnell L. A. J. Proteomics 2012, 75, 4962–4989. [DOI] [PubMed] [Google Scholar]
- Bruand J.; Alexandrov T.; Sistla S.; Wisztorski M.; Meriaux C.; Becker M.; Salzet M.; Fournier I.; Macagno E.; Bafna V. J. Proteome Res. 2011, 10, 4734–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville J. M.; Carter C. L.; Pizarro L.; Steven R. T.; Palmer A. D.; Griffiths R. L.; Lalor P. F.; Lindon J. C.; Nicholson J. K.; Holmes E.; Bunch J. Anal. Chem. 2013, 85, 1415–1423. [DOI] [PubMed] [Google Scholar]
- Alexandrov T. BMC Bioinf. 2012, 13Suppl 1S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Plas R.; Ojeda F.; Dewil M.; Van Den Bosch L.; De Moor B.; Waelkens E. In Proceedings of the Pacific Symposium on Biocomputing (PSB), Maui, Hawaii, 2007; pp 458–469. [PubMed]
- McCombie G.; Staab D.; Stoeckli M.; Knochenmuss R. Anal. Chem. 2005, 77, 6118–6124. [DOI] [PubMed] [Google Scholar]
- Hanselmann M.; Kirchner M.; Renard B. Y.; Amstalden E. R.; Glunde K.; Heeren R. M. A.; Hamprecht F. A. Anal. Chem. 2008, 80, 9649–9658. [DOI] [PubMed] [Google Scholar]
- Boggio K. J.; Obasuyi E.; Sugino K.; Nelson S. B.; Agar N. Y. R.; Agar J. N. Expert Rev. Proteomics 2011, 8, 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead S. N.; Chan K. H. N.; Gangaraju S.; Slinn J.; Li J.; Hou S. T. PLoS One 2011, 6, e20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov T.; Kobarg J. H. Bioinformatics 2011, 27, i230–i238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmoula W. M.; Carreira R. J.; Shyti R.; Balluff B.; van Zeijl R. J. M.; Tolner E. A.; Lelieveldt B. F. P.; van den Maagdenberg A. M. J. M.; McDonnell L. A.; Dijkstra J. Anal. Chem. 2014, 86, 3947–3954. [DOI] [PubMed] [Google Scholar]
- Sköld K.; Svensson M.; Nilsson A.; Zhang X.; Nydahl K.; Caprioli R. M.; Svenningsson P.; Andrén P. E. J. Proteome Res. 2006, 5, 262–269. [DOI] [PubMed] [Google Scholar]
- Lein E. S.; Hawrylycz M. J.; Ao N.; Ayres M.; Bensinger A.; Bernard A.; Boe A. F.; Boguski M. S.; Brockway K. S.; Byrnes E. J.; Chen L.; et al. Nature 2007, 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Jones A. R.; Overly C. C.; Sunkin S. M. Nat. Rev. Neurosci. 2009, 10, 821–828. [DOI] [PubMed] [Google Scholar]
- Hanrieder J.; Phan N. T. N.; Kurczy M. E.; Ewing A. G. ACS Chem. Neurosci. 2013, 4, 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L. G. ACM Comput. Surv. 1992, 24, 325–376. [Google Scholar]
- Crum W.; Hartkens T.; Hill D. L. Br. J. Radiol. 2004, 77, S140–S153. [DOI] [PubMed] [Google Scholar]
- Myronenko A.Non-rigid Image Registration: Regularization, Algorithms and Applications; Oregon Health & Science University, Department of Science & Engineering School of Medicine: Portland, OR, 2010; p 316. [Google Scholar]
- Bruand J.; Sistla S.; Mériaux C.; Dorrestein P. C.; Gaasterland T.; Ghassemian M.; Wisztorski M.; Fournier I.; Salzet M.; Macagno E.; Bafna V. J. Proteome Res. 2011, 10, 1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suits F.; Fehniger T. E.; Végvári A.; Marko-Varga G.; Horvatovich P. Anal. Chem. 2013, 85, 4398–4404. [DOI] [PubMed] [Google Scholar]
- Fehniger T. E.; Suits F.; Végvári Á.; Horvatovich P.; Foster M.; Marko-Varga G. Proteomics 2014, 14, 862–871. [DOI] [PubMed] [Google Scholar]
- Van de Plas R.; Pelckmans K.; De Moor B.; Waelkens E. In The NIPS workshop on Machine Learning in Computational Biology (NIPS MLCB), Whistler, B.C., 2007; pp 1–4.
- McDonnell L. A.; van Remoortere A.; van Zeijl R. J. M.; Deelder A. M. J. Proteome Res. 2008, 7, 3619–3627. [DOI] [PubMed] [Google Scholar]
- Allen Institute for Brain Science Allen Brain Atlas. http://mouse.brain-map.org/ (accessed Oct 7, 2013).
- Van de Plas R.Tissue Based Proteomics and Biomarker Discovery: Multivariate Data Mining Strategies for Mass Spectral Imaging; KU Leuven, Faculty of Engineering: Leuven, Belgium, 2010; p 245. [Google Scholar]
- Eastman R. D.; Le Moigne J.; Netanyahu N. S. In 2007 IEEE Conference on Computer Vision and Pattern Recognition; IEEE: Piscataway, NJ, 2007; pp 1–8. [Google Scholar]
- Rueckert D.; Sonoda L. I.; Hayes C.; Hill D. L. G.; Leach M. O.; Hawkes D. J. IEEE Transactions on Medical Imaging 1999, 18, 712–721. [DOI] [PubMed] [Google Scholar]
- Myronenko A. Medical Image Registration Toolbox: Andriy Myronenko https://sites.google.com/site/myronenko/research/mirt (accessed Oct 7, 2013).
- Grant M.; Boyd S. In Recent Advances in Learning and Control; Blondel V., Boyd S., Kimura H., Eds.; Springer-Verlag Limited: Heidelberg, Germany, 2008; pp 95–110. [Google Scholar]
- Grant M.; Boyd S.. CVX: Matlab Software for Disciplined Convex Programming, version 2.0 beta (http://cvxr.com/cvx); CVX Research, Inc.: Stanford, CA, 2012.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.