Abstract
Like other herpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV, also designated human herpesvirus 8) can establish a latent infection in the infected host. During latency a small number of genes are expressed. One of those genes encodes latency-associated nuclear antigen (LANA), which is constitutively expressed in cells during latent as well as lytic infection. LANA has previously been shown to be important for the establishment of latent episome maintenance through tethering of the viral genome to the host chromosomes. Under specific conditions, KSHV can undergo lytic replication, with the production of viral progeny. The immediate-early Rta, encoded by open reading frame 50 of KSHV, has been shown to play a critical role in switching from viral latent replication to lytic replication. Overexpression of Rta from a heterologous promoter is sufficient for driving KSHV lytic replication and the production of viral progeny. In the present study, we show that LANA down-modulates Rta's promoter activity in transient reporter assays, thus repressing Rta-mediated transactivation. This results in a decrease in the production of KSHV progeny virions. We also found that LANA interacts physically with Rta both in vivo and in vitro. Taken together, our results demonstrate that LANA can inhibit viral lytic replication by inhibiting expression as well as antagonizing the function of Rta. This suggests that LANA may play a critical role in maintaining latency by controlling the switch between viral latency and lytic replication.
Kaposi's sarcoma-associated herpesvirus (KSHV), also designated human herpesvirus 8 (HHV-8), is a recently identified human gammaherpesvirus that is closely associated with several malignancies, including Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (5, 6, 8-10, 45, 47, 48). KSHV is a large, double-stranded DNA virus with a genome which contains a 140-kb unique coding region flanked with multiple GC-rich terminal repeats (45). Herpesviruses are characterized by an initial lytic infection in hosts, followed by establishment of a lifelong latent infection in which the viral genome is maintained episomally in different tissues and can occasionally reactivate from latency to lytic replication (57).
Like infection by other herpesviruses, KSHV infection also displays two modes in its life cycle: latent and lytic replication (34). KSHV can maintain a tightly latent infection in infected cells but is able to undergo lytic infection in a percentage of cells in the population. In KS, for example, KSHV can latently persist in more than 90% of tumor cells, with a small number of tumor cells also undergoing spontaneous lytic viral replication. This is thought to be essential for sustaining KS lesions through a paracrine mechanism (40, 50, 54). Although more than 90 genes or open reading frames have been identified in the KSHV genome, only a small subset of these viral genes are typically expressed during latency. This pattern of viral gene expression allows the virus to escape host immune surveillance and to establish a persistent latent infection (45, 46).
Among the limited number of latent genes, the ORF73 gene, which encodes latency-associated nuclear antigen (LANA), is believed to be essential for the establishment of latent KSHV infection (3, 4, 41). LANA is a large nuclear protein (222 to 234 kDa based on analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) and has three distinct domains (Fig. 1A): a proline-rich N-terminal region with a putative nuclear localization signal (NLS), a long glutamic acid-rich internal repeat domain, and a carboxy-terminal domain including a putative NLS (13, 28, 43, 55). LANA functions in maintaining the viral episome in cells latently infected with KSHV by tethering the viral episome to cellular mitotic chromosomes via interaction with histone H1 and possibly other cellular proteins which include MeCP and DEK (14, 30). LANA has also been shown to modulate the transcriptional activity of the human immunodeficiency virus long terminal repeat promoter and to transactivate the LMP1 and Cp promoters of Epstein-Barr virus (EBV) (22, 27, 44). Importantly, LANA may also contribute to oncogenesis by promoting cell survival through targeting and alteration of p53 function, interaction with the retinoblastoma protein, and activation of the telomerase promoter (18, 21). Most recently, it was demonstrated that the ORF73 gene product of herpesvirus saimiri (HVS) can regulate viral gene expression by acting as a transcriptional modulator of latent and lytic viral promoters. ORF73 of HVS down-regulates the viral ORF50A and ORF50B promoters in permissive OMK cells, and it can inhibit the ORF50-mediated expression of viral early replication genes (49).
FIG. 1.
Scheme showing structurally important domains of LANA, Rta, and the Rta promoter. (A) As shown, LANA is a 1,162-amino-acid protein (strain BC-1). Numbers indicate amino acids (aa). Putative domains include the following: an N-terminal proline-rich domain (P-rich); an aspartic acid, glutamic acid repeat region (DE); glutamine-, glutamic acid-, and proline-rich repeats (QEP); glutamine-, arginine-, glutamic acid-, and proline-rich repeats (QFRP); glutamine-, aspartic acid-, and glutamic acid-rich repeats (QDE); and a leucine zipper (LZ). LANA also appears to have NLSs in both the amino- and carboxy-terminal regions, and a DNA binding domain (DBD) has been mapped to the distal carboxy terminus (13, 45). (B) The various known functional domains of the Rta protein. Three domains—the DNA binding and dimerization domain, located at the amino terminus (530 aa), the proline-rich domain, located in the middle, and the acidic activation domain, located in the carboxy terminus—are shown. Two NLSs are indicated: one at the amino terminus and one located at aa 514 to 528 of the Rta protein. AD1 through AD4 represent the conserved motifs that are essential for the Rta transactivation function (57). (C) Scheme showing the Rta promoter used in the present study. Putative transcriptional factor binding sites are shown. Numbers indicate nucleotides according to the KSHV genome of BC-1 (15, 25, 26, 45).
The specific role of lytic replication in pathogenesis is not fully understood; however, lytic replication is likely to be important in the spread of HHV-8 from the lymphoid compartment to endothelial cells, where KS tumors are most frequently observed (35, 40, 50, 54). This is likely to contribute to the development of KS. The molecular events and genes that control the switch from latency to the lytic phase have been studied in more detail for EBV-infected lymphoblastoid cell lines. Reactivation of EBV from latency was found to be regulated primarily by two immediate-early genes, BZLF1 and BRLF1, encoding Zta and Rta (replication and transcription activator), respectively. These molecules are both transcriptional activators that trigger the lytic cascade of viral replication. Expression of Zta in lymphocytes is sufficient to trigger the lytic cycle (16). However, it has recently been shown that Rta is also able independently to disrupt latency in epithelial cells and certain B-cell lines (58). In contrast to EBV, the switch between latency and lytic-cycle gene expression in KSHV is initiated by the EBV Rta homologue encoded by the KSHV ORF50 gene. KSHV-encoded Rta is a replication and transcription activator which was initially identified on the basis of positional homology and sequence homology to the EBV and HVS genomes; it is one of the earliest immediate-early genes induced upon viral reactivation and has been shown to be sufficient for the ectopic reactivation of lytic replication in PEL cell lines (33, 53, 54). Therefore, Rta plays a critical role as the master regulator of the switch between latency and lytic replication of KSHV (33, 53, 59).
Previous studies have addressed the mechanism by which Rta regulates downstream genes such as ORF57 and K8 to initiate lytic replication; however, the regulation of Rta is poorly understood (32). Deng and colleagues showed that Rta can autoactivate its own promoter (15). This suggests that with specific stimuli, Rta can autoactivate its own expression to initiate the lytic cycle. The Rta promoter region is heavily methylated in latently infected cells (11). Therefore, Rta gene expression is tightly controlled in these cells and may provide an explanation for a potential mechanism by which KSHV can latently persist in more than 90% of tumor cells. During KSHV latency, LANA is consistently expressed in all infected cells, and studies have shown that LANA is critical for the maintenance of latency (3, 4, 14, 23). Additionally, recent reports indicate that LANA's homologue in HVS can repress the expression of ORF50 encoded by HVS (49). Therefore, we hypothesized that LANA may inhibit viral lytic replication by antagonizing the function of Rta in KSHV.
In this study, we show that LANA can repress the Rta promoter and inhibit Rta-mediated transactivation. We also show that LANA associates with Rta both in vitro and in vivo. This interaction results in a reduction in the production of viral progeny. Taken together, our data suggest that LANA can inhibit the switch from latency to lytic replication by repressing the expression of ORF50 through interaction with the Rta protein, negatively regulating the positive feedback of Rta on its native promoter.
MATERIALS AND METHODS
Antibodies, cell lines, and viruses.
A rabbit polyclonal antibody against KSHV Rta was provided by Gary S. Hayward (Johns Hopkins University School of Medicine). A mouse monoclonal antibody against KSHV Rta was a kind gift from Koichi Yamanishi (Osaka University, Osaka, Japan). Human polyclonal serum which recognizes LANA is designated HS and was provided by Gary Nabel (Vaccine Institute, National Institutes of Health, Bethesda, Md.).
BJAB cells are EBV-negative B cells isolated from Burkitt's lymphoma and were provided by Elliott Kieff. Human embryonic kidney fibroblast 293 and 293T cells, transformed with E1A and T antigens, respectively, were obtained from Jon Aster (Brigham and Women's Hospital, Boston, Mass.). BC3 and BCBL1 are KSHV-positive body cavity-based, lymphoma-derived cell lines obtained from the American Type Culture Collection. JSC-1 cells are also latently KSHV infected B-lymphoma cells, provided by Richard F. Ambinder. KSHV was induced from the BCBL-1, BC-3, and JSC-1 cell lines by using 20 ng of tetradecanoyl phorbol acetate (TPA)/ml and 1.5 mM butyrate for 4 to 5 days. Induced cells were then freeze-thawed four times, after which the debris pelleted. The supernatants were collected and filtered through a 0.45-μm-pore-size filter. The filtered virus supernatant can then be frozen at −80°C for storage and thawed for infections with minimal loss in stability.
Transfection.
BJAB, 293, and 293T cells were transfected by electroporation using a Bio-Rad Gene Pulser II electroporator. Ten million cells harvested in exponential phase were collected, washed in phosphate-buffered saline (PBS), and then resuspended in 400 μl of RPMI medium or Dulbecco's modified Eagle medium with DNA for transfection. Resuspended cells were transferred to a 0.4-cm cuvette and electroporated at 975 μF and 220 V. The electroporated cells were then transferred to 10 ml of complete medium, followed by incubation at 37°C under 5% CO2. Transfected cells were harvested after 24 h and assayed for activity. BC3, BCBL1, and JSC cells were transfected by electroporation at 975 μF and 260 V.
Luciferase assay.
HEK 293 cells were collected for transfections at 70% confluency. Ten million cells were resuspended, along with plasmid DNA, in 400 μl of medium. The pRpluc reporter plasmid was a generous gift from Ren Sun (University of California, Los Angeles) (15). The cells were transfected by electroporation with the Bio-Rad Gene Pulser II at 210 V and 975 μF. Transfected cells were transferred to 100-mm-diameter plates in 10 ml of Dulbecco's modified Eagle medium with 10% fetal bovine serum. The plates were incubated at 37°C in a humidified environment supplemented with 5% CO2 for 20 h. BJAB cells were collected at 5 × 105/ml, and 10 million cells were used for transfection at 220 V and 975 μF. At 20 h, cells were harvested and counted to normalize the total number of surviving cells from each transfection. The cells were subsequently washed once with PBS (Invitrogen, Inc.), followed by lysis with 200 μl of reporter lysis buffer (Promega, Inc.). A 40-μl volume of the lysate was mixed with 100 μl of luciferase assay reagent. Luminescence was measured for 10 s by the OpticompI Luminometer (MGM Instruments, Inc.). The lysates were also diluted to ensure that luciferase activity was within the linear range of the assay. The results shown represent experiments performed in triplicate.
Immunoprecipitation and Western blotting.
Thirty million 293T or BJAB cells were transfected with pA3M-LANA and/or pCR3.1-Rta and incubated for 24 h. The cells were then lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, aprotinin [1 μg/ml], and pepstatin [1 μg/ml]) for 1 h on ice with brief vortexing every 15 min. A portion of the lysate was removed for use as a control. The lysates were precleared by a 1-h incubation with protein A-Sepharose beads. Anti-Myc or anti-Rta antibodies were incubated with the lysates overnight at 4°C. Immunoprecipitates were collected by rotating with protein A-Sepharose beads for 1 h and were washed four times in radioimmunoprecipitation assay buffer. The protein was then heated in SDS-β-mercaptoethanol lysis buffer and analyzed by SDS-PAGE. Western blot analyses were performed by using antibodies specific for the detection of LANA or Rta. For BC3 or BCBL1 cells, a total of 108 cells were used and the protocol described above was followed.
Preparation of GST fusion proteins and in vitro binding assays.
Briefly, DH5α cells were transformed with the plasmid constructs for each fusion protein, and following selection with ampicillin, single colonies were selected. Five hundred milliliters of Luria-Bertani medium was inoculated at a dilution of 1:200 and allowed to shake at 37°C until mid-exponential-growth phase. Cells were then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h with continuous shaking at 37°C. Cells were harvested and sonicated, and the proteins were solubilized in the presence of protease inhibitors. Solubilized proteins were incubated with glutathione S-transferase (GST)-Sepharose beads for 6 h or overnight at 4°C with rotation and were then collected by centrifugation and washed three times in NETN (0.5% NP-40, 20 mM Tris, 1 mM EDTA, 100 mM NaCl) buffer with protease inhibitors. GST fusion proteins bound to beads were used for binding assays and were stored in NETN buffer with protease inhibitors and 1 mM phenylmethylsulfonyl fluoride at 4°C. To determine the binding of the GST-Rta fusion protein to full-length LANA and its various truncations, proteins were translated in vitro with [35S]Met-Cys (Tran35S-label; NEN-Dupont) by using the TNT system (Promega Inc.) in accordance with the manufacturer's instructions. Labeled LANA proteins were incubated with equivalent amounts of the GST-Rta fusion protein bound to beads and rotated at 4°C for 4 h, followed by four washes in 1 ml of NETN buffer with protease inhibitors. Bound proteins were eluted from the beads in SDS lysis buffer by heating at 95°C for 10 min and were then fractionated by SDS-10% PAGE. Dried gels were analyzed with a PhosphorImager (Molecular Dynamics Inc.), and signals were quantified with ImageQuant software.
Immunofluorescence.
Immunofluorescence analyses were performed essentially as described previously (27). At 24 h posttransfection or postinduction, the cells were briefly fixed, blocked in the appropriate serum, and then incubated with the specific primary antibody for LANA, Myc, or Rta for 1 h. Cells were washed and then further incubated with the appropriate secondary antibody conjugated to fluorescein isothiocyanate or Texas red at 1:1,000 dilutions in PBS for 1 h. Slides were washed and visualized with an XI70 inverted fluorescence microscope (Olympus Inc.) and photographed by using a digital PixelFly camera and software (Cooke, Inc.).
Induction and PCR analysis of virus progeny.
To determine whether LANA can repress KSHV replication and reduce the production of viral progeny, we collected 10 million exponentially growing BC3, BCBL1, and JSC-1 cells, which were centrifuged and resuspended in 400 μl of RPMI 1640 medium along with increasing amounts of LANA plasmid and then transfected under the conditions described above. At 12 h posttransfection, cells were induced by using TPA at a concentration of 20 ng/ml of medium and butyrate at a concentration of 1.5 mM. Cells were further incubated for 2 days at 37°C under 5% CO2. The supernatant was then collected, and viral particles were spun down at 15,000 rpm for 20 min. Intact cells were discarded to prevent possible lysis and contamination from cellular viral DNA. The pellet was resuspended in 50 μl of 0.2× PBS, heated to 95°C for 15 min, and then switched to 56°C for 1 h with proteinase K treatment (10 mg/ml). The enzyme was then killed by treatment at 95°C for 30 min. A 5-μl portion of virus lysate was used for PCR amplification of the KSHV specific region located between ORF18 and ORF19, coordinates 33167 to 33473 (KSHV genome sequence accession number, NC_003409), with forward primer 5′-CCTTGGTGCGTTTAACAACA-3′ and reverse primer 5′-ACAACCCGGACCATTTGTAA-3′ for 25 cycles. The amplicon was fractionated on a 3% NuSieve agarose gel to determine the production of viral progeny.
RESULTS
LANA represses the ORF50 promoter in HEK 293 and B cells.
We sought to investigate if LANA operates as a transcriptional modulator of the ORF50 promoter encoded by KSHV. The pRpluc reporter plasmid, containing the Rta upstream promoter sequence cloned into the pGL2-basic vector, which has the coding sequence for firefly luciferase, was transfected into HEK 293 cells (Fig. 2A) or BJAB cells (a KSHV-negative lymphoma cell line) (Fig. 2B). The pA3 M-LANA expression construct, a Myc-tagged LANA expression vector, was transfected along with the reporter construct. Cotransfection of the LANA expression vector with the pRpluc reporter construct consistently resulted in repression of the reporter activity relative to that with the reporter construct alone (Fig. 2). Increasing the concentration of the LANA expression construct repressed the ORF50 Rta promoter in a dose-dependent manner. These data indicate that the repression of the Rta promoter in the HEK 293 and BJAB cell lines is directly proportional to the quantity of LANA expressed, as demonstrated by the observed dose-response relationship. Further increasing the amounts of LANA resulted in elimination of this activity and increased cell death (data not shown), suggesting that the levels of LANA in the cell are stringently maintained to prevent cell death and to control the switch to lytic replication. The Rta promoter contains a number of sites for transcription factors known to repress promoters, including the RBP-Jκ cellular repressor (see Fig. 1C). Additional factors that can be targeted include the Ap1, Sp1, and NF-κB sites, known to have roles in transcription regulation. It is possible that LANA can target these factors for down-regulating the Rta promoter. We next wanted to determine if LANA can affect the ability of Rta, the known transactivator of the promoter, to autoactivate its own promoter.
FIG. 2.
LANA represses the transcriptional activity of the Rta gene promoter in human cells. The pRpluc reporter plasmid contains a 3-kb sequence upstream of the translation initiation site of the rta gene that drives the expression of firefly luciferase (15). Five micrograms of the pRpluc reporter plasmid was cotransfected into HEK 293 cells (A) or Burkitt lymphoma BJAB cells (B) with either 2.5, 5, 10, or 15 μg of pA3M-LANA. Total transfected DNA was normalized with the pA3 M vector. Promoter activity was expressed as the fold activation relative to activity with pRpluc alone (control). Means and standard deviations from three independent transfections are shown. Protein lysates were analyzed by Western blotting for levels of expression of transfected protein with the Myc monoclonal antibody to detect LANA and the RTA monoclonal antibody to detect Rta protein. Protein levels were shown by Ponceau S staining.
LANA inhibits Rta-mediated transactivation of the ORF50 promoter.
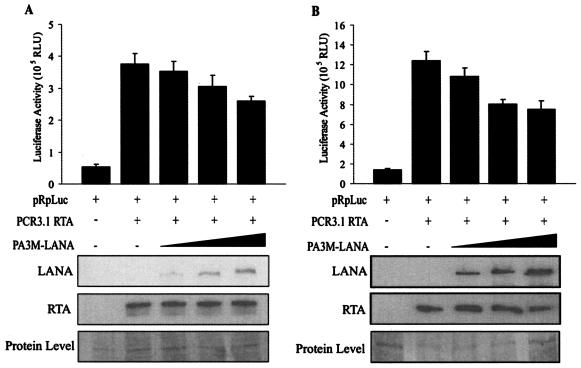
Previous studies showed that Rta can autoactivate its own viral promoter. Therefore, we wanted to test whether LANA expressed exogenously from a heterologous promoter can affect the ability of Rta to activate its own promoter. HEK 293 (Fig. 3A) or BJAB (Fig. 3B) cells were transiently transfected with expression vectors containing Rta, the pRpluc reporter plasmid, and LANA in incremental amounts. As has previously been shown (15), Rta greatly augmented the activation of the reporter, as seen by luciferase activity, from its native promoter in pRpluc in both HEK 293 and BJAB cells. Coexpression of LANA in increasing amounts, however, resulted in inhibition of the transactivation activity of Rta in a dose-dependent manner. As expected, this inhibition was not as dramatic as that seen in the absence of Rta expressed from a heterologous promoter. Increasing amounts of LANA and Rta resulted in cell death; therefore, the levels of plasmid used in these assays were closely monitored and normalized with a plasmid containing the vector alone. This equalized the total DNA contents in all transfections. We also saw greater inhibition with LANA in BJAB cells (Fig. 3B), suggesting the involvement of an additional factor that may work together with LANA in repressing the effects of Rta and that could be LANA dependent or independent. Nevertheless, this inhibition was convincingly consistent in multiple experiments (Fig. 3).
FIG. 3.
LANA represses Rta-mediated autoactivation. Ten million HEK 293 cells (A) or Burkitt lymphoma BJAB cells (B) were transfected with 5 μg of the pRpluc luciferase reporter construct, 10 μg of pCR3.1-Rta, and 2.5, 5, 10, or 15 μg of the pA3M-LANA expression construct as indicated. Total transfected DNA was normalized with the pA3M-vector. Promoter activity was expressed as the fold activation relative to activity with pRpluc alone (control). Means and standard deviations from three independent transfections are shown. Protein lysates were analyzed by Western blotting for levels of expression of transfected protein with the Myc monoclonal antibody to detect LANA and the Rta monoclonal antibody to detect Rta protein. The protein lysate was used as a loading control and was stained with Ponceau S.
LANA inhibits production of KSHV progeny virions through repression of Rta expression from its native promoter.
The experiments described above suggest that LANA is capable of inhibiting the Rta promoter. To address the functional significance of this activity in the context of KSHV, we employed BCBL1, BC3, and JSC1 cells, known to be positive for KSHV but negative for EBV. These latently KSHV infected cells can be induced by chemical inducers such as TPA or n-butyrate to undergo lytic reactivation, producing new viral particles. We wanted to determine if increased expression of LANA from a heterologous promoter can result in decreased production of virion particles.
Ten million cells from each of the KSHV-positive cell lines were grown in culture for transfection and harvested when the cultures reached a density of 5 × 105 cells per milliliter. These exponentially growing cells were resuspended in 400 μl of RPMI 1640 medium along with increasing amounts of LANA. The cells were then transfected at 260 V and 975 μF and were incubated at 37°C under 5% CO2. At 12 h posttransfection, cells were induced by using TPA at a concentration of 20 ng/ml of medium. Cells were further incubated for another 72 h. The supernatant was collected at this time point and then prepared for collection of virus as described above (31). Once the virus was collected by centrifugation, it was resuspended in PBS buffer and lysed. An aliquot of the lysate was used for PCR analysis of virus DNA. The virus lysate was used for PCR amplification of the region of the KSHV genome located between ORF18 and ORF19, which amplified a 260-bp amplicon. This assay provided a semiquantitative look at the viral progeny produced with increasing amounts of LANA. The results obtained indicate that increasing amounts of LANA transfected from a heterologous promoter can reduce the production of viral particles. The effect was a decrease in the total levels of viral DNA collected from viral particles, as shown by PCR analysis for both BC3 (Fig. 4) and BCBL1 (data not shown) cells. Interestingly, Western blotting showed that the expression of Rta protein from its native promoter was reduced (Fig. 4, lower panels). This relative reduction in Rta levels, observed by using this semiquantitative assay, was associated with increasing amounts of exogenous LANA transfected in these KSHV-infected cells (Fig. 4, lower panels, LANA). To determine if this effect was limited to only a couple of cell lines, we performed the assay with another transfectable KSHV-positive cell line, JSC-1. Similar results were obtained from the JSC-1 cell line, indicating that LANA can repress the Rta promoter, resulting in reduced virus replication and virion progeny production (data not shown). Collectively, these data with three independent KSHV-positive cell lines suggest that LANA may directly down-regulate Rta expression through targeting of the native Rta promoter. This results in inhibition of the downstream cascade of lytic gene activation which is dependent on Rta and thus in the eventual reduction in the production of viral progeny.
FIG. 4.
LANA down-regulates KSHV replication and virion production through repression of Rta. Ten million BC3 cells were transfected with 2.5, 5, or 10 μg of the LANA-Myc expression vector. At 12 h posttransfection, cells were induced with TPA and butyrate. At 48 h (A) or 72 h (B) postinduction, supernatants of transfected cells were harvested for PCR to check the virion production level of KSHV. The relative densities of the bands from the PCR products were measured with ImageQuant software (Molecular Dynamics). Cell lysates were analyzed by Western blotting (WB) for levels of expression of transfected protein with the Myc monoclonal antibody to detect LANA and the Rta monoclonal antibody to detect Rta protein. The protein lysate was used as a loading control and was stained with Ponceau S. Lanes 1, mock transfection; lanes 2, 10.0 μg of the pA3M-vector; lanes 3, 2.5 μg of pA3M-LANA and 7.5 μg of the pA3M-vector; lanes 4, 5.0 μg of pA3M-LANA and 5.0 μg of the pA3M-vector; lanes 5, 10.0 μg of pA3M-LANA.
LANA associates with Rta in the epithelial cell line 293T and the B-cell line BJAB when transiently expressed from heterologous promoters.
The experiments described above strongly suggest that LANA can modulate the autoactivation of Rta on its own promoter (15). We therefore wanted to determine if LANA's activity occurred through direct binding to Rta. The results from our immunoprecipitation experiments show that LANA associates with Rta in human cells (Fig. 5). Myc-tagged LANA and Rta expression plasmids (pA3 M-LANA and pCR3.1-Rta, respectively) were cotransfected into HEK 293T cells or BJAB cells by electroporation. Twenty-four hours posttransfection, cells were harvested and lysed. LANA-Myc was immunoprecipitated by using the mouse monoclonal antibody 9E10, and the complexes were resolved by SDS-PAGE. The blot was probed for the presence of LANA-Myc-bound Rta by using a mouse monoclonal anti-Rta antibody. As anticipated, Rta coimmunoprecipitated with LANA-Myc in BJAB and HEK 293T cells (Fig. 5A and B, respectively), while control immunoprecipitates of BJAB and HEK 293T cells transfected with pA3 M-LANA or pCR3.1-Rta alone did not contain Rta. In the reverse assay using an anti-Rta antibody, the Western blots indicated that LANA-Myc coimmunoprecipitated with Rta but was not seen in the control immunoprecipitation lanes (Fig. 5).
FIG. 5.
Immunoprecipitation analysis with a mouse anti-Myc antibody or a mouse anti-Rta antibody showed that Rta was directly immunoprecipitated with LANA in BJAB (A) and 293T (B) cells. For each experiment, 30 million cells were either cotransfected with Rta and LANA-Myc expression vectors or were transfected with one of these vectors. At 24 h posttransfection, cells were harvested and lysed; lysates were used for immunoprecipitation analysis. L, lysate; PC, precleared lysate; IP, immunoprecipitate; WB, Western blotting..
LANA associates with Rta in TPA-induced, KSHV-infected BCBL1 and BC3 cells.
To further corroborate the interaction between endogenous LANA and Rta, we induced 100 million BCBL1 and BC3 cells latently infected with KSHV by using TPA and sodium butyrate, respectively. Forty-eight hours postinduction, when the levels of Rta protein expression were clearly detected by Western blotting, cells were harvested and lysed. The lysate was collected, and Rta was immunoprecipitated with a polyclonal rabbit antiserum. Complexes were resolved by SDS-PAGE and probed for the presence of Rta-bound LANA by using a human polyclonal serum which recognizes LANA. As expected, LANA coimmunoprecipitated with Rta in BC3 and BCBL1 cells (Fig. 6A and B, respectively). However, control immunoprecipitates of KSHV-negative BJAB cells did not show complexes of Rta and LANA, and uninduced cells showed no signals in the immunoprecipitated lanes (Fig. 6).
FIG. 6.
LANA interacts with endogenously induced Rta in KSHV-infected pleural effusion lymphoma cells. Immunoprecipitation analysis with a polyclonal rabbit antiserum against Rta showed that LANA was directly immunoprecipitated with Rta in BC3 (A) and BCBL1 (B) cells. For each experiment, 100 million cells were induced with 20 ng of TPA/ml and 1.5 mM sodium butyrate. At 48 h postinduction, cell lysates were used for immunoprecipitation analysis. L, lysate; PC, precleared lysate; IP, immunoprecipitate; WB, Western blotting.
Rta interacts directly with the C-terminal domain of LANA.
The immunoprecipitation experiments discussed above provide data corroborating the hypothesis that LANA and Rta associate in human cells. We therefore wanted to further identify the domain in LANA that is responsible for direct binding of Rta. Full-length LANA as well as carboxy-terminal and amino-terminal clones of LANA were in vitro translated with [35S]Met labeling. The labeled proteins were then incubated with either GST alone or GST-Rta. The results of the binding, shown in Fig. 7A, indicate that full-length LANA bound to GST-Rta, as was expected from the immunoprecipitation experiments. In addition, the carboxy-terminal truncation of LANA, consisting of amino acids 751 to 1162, also bound to Rta (Fig. 7A). The two amino-terminal truncations, however, showed no binding affinity toward LANA (Fig. 7A). This result therefore shows that the carboxy terminus of LANA is responsible for binding to Rta.
FIG. 7.
The carboxy-terminal region of LANA interacts with Rta in vitro. Full-length LANA and Rta as well as LANA clones with truncations in the carboxy- and amino-terminal regions were in vitro transcribed and translated. 35S-labeled products were incubated with GST as well as either GST-LANA or GST-Rta. Pulldown products were electrophoresed on 8% SDS-PAGE gels, dried, and exposed to a PhosphorImager. Input controls of 10% for LANA and 5% for Rta were run as well. A schematic for the LANA clones used is shown below the gels. AD, acidic domain; LZ, leucine zipper; DBD, DNA binding domain.
In order to further verify the site of Rta binding to LANA, we performed a reverse binding experiment using two LANA-GST truncation constructs, one consisting of amino acids 341 to 939 of LANA and the other consisting of the carboxy-terminal amino acids 990 to 1162. Full-length Rta was in vitro translated with [35S]Met labeling and incubated with GST and one of the GST-LANA constructs. The results demonstrated that Rta binds to the carboxy terminus of LANA (Fig. 7) and that the extreme C terminus is required for binding. These 172 carboxy-terminal-most amino acids (amino acids 990 to 1162) of LANA are responsible for direct interaction with Rta (Fig. 7B). The construct comprising the central region of LANA showed no binding (data not shown).
LANA colocalizes with Rta when transiently transfected in human cells and in KSHV-positive cells induced for lytic replication.
To support the binding results from immunoprecipitation and in vitro binding assays, we performed immunofluorescence analysis to determine whether LANA and Rta were in the same nuclear compartment. First, we transiently cotransfected 293T and BJAB cells with LANA-Myc and pCR3.1-Rta expression constructs. At 24 h posttransfection, cells were harvested, fixed, and probed with a mouse monoclonal antibody against Myc and a rabbit antiserum against Rta. Fluorescein isothiocyanate and Texas red conjugated to the appropriate secondary antibody were used for detection of the signals. The results of this study showed that LANA localized to the same nuclear compartments in 293T and BJAB cells (Fig. 8A), suggesting that exogeneously transfected LANA and Rta protein can colocalize in the nucleus. Second, we used TPA and butyrate to induce KSHV-positive BCBL1 and BC3 cells. Twenty-four hours postinduction, when Rta was expressed, cells were fixed for immunofluoresence analysis and stained as described above. The results showed quite strikingly that endogenous LANA and Rta were colocalized in the same nuclear compartments in BCBL1 as well as BC3 cells. However, in control uninduced cells, LANA localized to punctate signals as expected, but no Rta signals were observed (Fig. 8B and C).
FIG. 8.
Immunofluorescence analysis showed that Rta was localized to the same nuclear compartment as LANA in different cells. (A) BJAB or 293T cells were cotransfected with 10 μg each of the pCR3.1-Rta and pA3 M-LANA expression vectors. At 24 h posttransfection, cells were harvested for immunofluorescence analysis. (B and C) Uninduced (Un) or induced (In) BC3 and BCBL1 cells were also used for immunofluorescence analysis. For induction, BC3 and BCBL1 cells were treated with 10 ng of TPA/ml and 1.5 mM butyrate. After 24 or 48 h of treatment, cells were harvested. LANA is expressed in almost every cell, and Rta is expressed in some induced cells.
DISCUSSION
KSHV-encoded LANA is expressed during latent infection and is detected in all forms of KS-associated malignancies, including PELs and MCD (6, 7, 10, 37, 52). LANA has been shown to have a multifunctional role in the establishment of latent infection and the control of cell cycle progression by modulating various cellular pathways (17-20, 42, 44, 51). LANA has also been shown to be important for episomal maintenance of the viral genome by tethering the viral genome to the host chromosome (3, 4, 12, 23). Tethering of the viral genome to the host chromosome helps in efficient maintenance and equal segregation of the viral genome to the daughter nuclei during cell division (3). Besides maintenance of the episomal DNA, LANA has been shown to interact with various cellular molecules, down-modulating their activity, and to interact with the tumor suppressors p53 and pRb, down-modulating their activity and thus blocking the apoptosis they mediate (18, 42). Additionally, LANA has also been shown to regulate various other cellular pathways, including the Wnt signaling pathway, by stabilizing beta-catenin as a result of binding to the negative regulator GSK-3β, causing a cell cycle-dependent nuclear accumulation of GSK-3β (19, 20). These roles of LANA strongly suggest that it is an important player in the maintenance of latent infection, like the EBNA-1 protein of the lymphocryptovirus EBV, which has been shown to be an important viral molecule involved in transcriptional regulation of various cellular pathways via the family of repeat sites (29).
Although only a small subset of tumor cells undergo spontaneous viral lytic replication, this event is thought to be essential for sustaining KS lesions through a paracrine mechanism (36, 38, 39). However, only a small number of viral genes are actually expressed during latency, and this pattern of gene expression during latency allows escape from host immune surveillance, establishing persistent infection (41, 42).
Within the Rhadinovirus subfamily, all viruses encode an ORF73 protein with homology to KSHV LANA. The sizes of these different ORF73 proteins differ due to a highly repetitive central repeat sequence, but their roles have been shown to be similar in terms of tethering the viral episome to the host chromosome (24, 49, 56). Recently, HVS ORF73 was shown to repress its own ORF50 promoter, supporting our initial hypothesis that KSHV LANA could also down-regulate Rta expression and thus prevent KSHV from undergoing lytic replication. These studies show for the first time that LANA is important for the down-regulation of Rta, which plays an essential role in the induction of the KSHV lytic cycle. Thus, LANA contributes to the maintenance of viral latency by supporting episomal maintenance as well as transcriptional regulation of the “master switch,” Rta, which is essential for inducing lytic infection.
In this study, we showed that LANA can independently repress the Rta promoter and can inhibit the ability of Rta to autoactivate its own promoter. We also showed that this activity occurs through direct binding with Rta in a dose-dependent manner. Therefore, in latently KSHV infected cells, LANA, which is constitutively expressed during latent infection, can directly down-regulate the transcription of Rta by targeting its native promoter, thus stringently controlling the expression of Rta and keeping lytic replication in check. Rta can autoactivate its own native promoter, providing a probable explanation of why lytic replication is triggered in a small subset of KSHV-infected cells and why under certain stress conditions KSHV automatically enters the lytic stage (15). It is therefore interesting to demonstrate through these studies that in addition to its ability to modulate the Rta promoter, LANA can also regulate Rta's autoactivation. Interestingly, the increased expression of LANA reduced Rta-mediated transcription in a dose-dependent manner. This LANA-mediated down-modulation of Rta-mediated transcription could be an important mechanism by which KSHV prevents expression of Rta, suppressing the level of lytic replication (Fig. 9) This down-regulation of Rta results in inhibition of the Rta-initiated lytic gene expression cascade.
FIG. 9.
Hypothetical model for LANA inhibition of lytic replication by targeting of Rta. Rp, Rta promoter; M, methylation.
The results of the transient studies using reporter assays were strongly supported by the data for latently KSHV infected BC3 and BCBL1 cells. These cells are not coinfected with EBV, which would complicate the results due to the expression and possible conservation of the same EBV genes that may have a role in activating KSHV lytic infection (1, 2). The fact that increasing amounts of exogenously expressed LANA can down-regulate virion production suggests that LANA plays a pivotal role in regulating lytic reactivation and may be a primary control switch required for the maintenance of KSHV latent infection.
Taken together, the data presented here are consistent with the model in which LANA plays a critical role in the control of lytic reactivation (see Fig. 9). Rta can initiate lytic viral replication by transactivation of early viral replication genes. Additionally, LANA is also capable of repressing the ORF50 promoter. Rta-mediated transactivation of the delayed-early genes can be blocked by LANA through direct interaction of LANA with Rta, and this blockage inhibits the induction of lytic replication.
Our studies revealed an important function of LANA in the control of the lytic replication of KSHV in the standard cell culture system. Further studies will be necessary to identify additional molecules involved in this complex containing LANA and Rta which leads to the down-regulation of Rta. It is possible that LANA can also mediate modifications of Rta including phosphorylation, ribosylation, or glycosylation. These modifications may also be essential for Rta function. This said, LANA may also be capable of regulating Rta by multiple mechanisms.
Acknowledgments
We thank R. Sun (Department of Molecular and Medical Pharmacology, University of California) for the pRpluc reporter plasmid, J. U. Jung (New England Primate Research Center, Harvard Medical School) for Rta cDNA, Gary S. Hayward (Johns Hopkins University School of Medicine) for the rabbit polyclonal antibody against KSHV Rta, and Koichi Yamanishi (Osaka University) for the mouse monoclonal antibody against KSHV Rta. We also thank Paul M. Lieberman (The Wistar Institute) and Yan Yuan (Dental School of University of Pennsylvania) for suggestions.
This work was supported by grants from the Leukemia and Lymphoma Society of America and by Public Health Service grants from the NCI (CA072510 and CA091792) and from NIDCR (DE01436) (to E.S.R.). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.Aguirre, A. J., and E. S. Robertson. 1999. Characterization of intertypic recombinants of the Epstein-Barr virus from the body-cavity-based lymphomas cell lines BC-1 and BC-2. Virology 264:359-369. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, A. J., and E. S. Robertson. 2000. Epstein-Barr virus recombinants from BC-1 and BC-2 can immortalize human primary B lymphocytes with different levels of efficiency and in the absence of coinfection by Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boshoff, C., and R. A. Weiss. 2001. Epidemiology and pathogenesis of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B 356:517-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 8.Cesarman, E., R. G. Nador, K. Aozasa, G. Delsol, J. W. Said, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus in non-AIDS-related lymphomas occurring in body cavities. Am. J. Pathol. 149:53-57. [PMC free article] [PubMed] [Google Scholar]
- 9.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, C. Parravicini, M. Corbellino, and K. Yamanishi. 2001. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc. Natl. Acad. Sci. USA 98:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 13.Cotter, M. A., II, and E. S. Robertson. 2002. Molecular biology of Kaposi's sarcoma-associated herpesvirus. Front. Biosci. 7:d358-d375. [DOI] [PubMed] [Google Scholar]
- 14.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 15.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 16.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-Acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flore, O., S. Rafii, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 18.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 19.Fujimuro, M., and S. D. Hayward. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus manipulates the activity of glycogen synthase kinase-3β. J. Virol. 77:8019-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 21.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 75:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, A. F. Markham, and A. Whitehouse. 2000. Characterization of the herpesvirus saimiri ORF73 gene product. J. Gen. Virol. 81:2653-2658. [DOI] [PubMed] [Google Scholar]
- 25.Heinemeyer, T., X. Chen, H. Karas, A. E. Kel, O. V. Kel, I. Liebich, T. Meinhardt, I. Reuter, F. Schacherer, and E. Wingender. 1999. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny, and N. A. Kolchanov. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyun, T. S., C. Subramanian, M. A. Cotter II, R. A. Thomas, and E. S. Robertson. 2001. Latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus interacts with Tat and activates the long terminal repeat of human immunodeficiency virus type 1 in human cells. J. Virol. 75:8761-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy, G., and B. Sugden. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 23:6901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, W., Y. H. Hwang, S. K. Lee, C. Subramanian, and E. S. Robertson. 2001. An Epstein-Barr virus isolated from a lymphoblastoid cell line has a 16-kilobase-pair deletion which includes gp350 and the Epstein-Barr virus nuclear antigen 3A. J. Virol. 75:8556-8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monini, P., S. Colombini, M. Sturzl, D. Goletti, A. Cafaro, C. Sgadari, S. Butto, M. Franco, P. Leone, S. Fais, G. Melucci-Vigo, C. Chiozzini, F. Carlini, G. Ascherl, E. Cornali, C. Zietz, E. Ramazzotti, F. Ensoli, M. Andreoni, P. Pezzotti, G. Rezza, R. Yarchoan, R. C. Gallo, and B. Ensoli. 1999. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi's sarcoma. Blood 93:4044-4058. [PubMed] [Google Scholar]
- 36.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 37.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 38.Moore, P. S., and Y. Chang. 1998. Kaposi's sarcoma-associated herpesvirus-encoded oncogenes and oncogenesis. J. Natl. Cancer Inst. Monogr. 1998:65-71. [DOI] [PubMed] [Google Scholar]
- 39.Moore, P. S., and Y. Chang. 2001. Molecular virology of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B 356:499-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neipel, F., and B. Fleckenstein. 1999. The role of HHV-8 in Kaposi's sarcoma. Semin. Cancer Biol. 9:151-164. [DOI] [PubMed] [Google Scholar]
- 41.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 43.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarid, R., A. Klepfish, and A. Schattner. 2002. Virology, pathogenetic mechanisms, and associated diseases of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8). Mayo Clin. Proc. 77:941-949. [DOI] [PubMed] [Google Scholar]
- 48.Sarid, R., S. J. Olsen, and P. S. Moore. 1999. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv. Virus Res. 52:139-232. [DOI] [PubMed] [Google Scholar]
- 49.Schafer, A., D. Lengenfelder, C. Grillhosl, C. Wieser, B. Fleckenstein, and A. Ensser. 2003. The latency-associated nuclear antigen homolog of herpesvirus saimiri inhibits lytic virus replication. J. Virol. 77:5911-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulz, T. F., and P. S. Moore. 1999. Kaposi's sarcoma-associated herpesvirus: a new human tumor virus, but how? Trends Microbiol. 7:196-200. [DOI] [PubMed] [Google Scholar]
- 51.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 53.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma, S. C., and E. S. Robertson. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 222:155-163. [DOI] [PubMed] [Google Scholar]
- 56.Verma, S. C., and E. S. Robertson. 2003. ORF73 of herpesvirus saimiri strain C488 tethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J. Virol. 77:12494-12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West, J. T., and C. Wood. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150-5163. [DOI] [PubMed] [Google Scholar]
- 58.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]