Abstract
Based solely on in vitro results, two contrasting models have been proposed for the recognition of the brome mosaic virus (BMV) subgenomic core promoter by the replicase. The first posits that the replicase recognizes at least four key nucleotides in the core promoter, followed by an induced fit, wherein some of the nucleotides base pair prior to the initiation of RNA synthesis (S. Adkins and C. C. Kao, Virology 252:1-8, 1998). The second model posits that a short RNA hairpin in the core promoter serves as a landing pad for the replicase and that at least some of the key nucleotides help form a stable hairpin (P. C. J. Haasnoot, F. Brederode, R. C. L. Olsthoorn, and J. Bol, RNA 6:708-716, 2000; P. C. J. Haasnoot, R. C. L. Olsthoorn, and J. Bol, RNA 8:110-122, 2002). We used transfected barley protoplasts to examine the recognition of the subgenomic core promoter by the BMV replicase. Key nucleotides required for subgenomic initiation in vitro were found to be important for RNA4 levels in protoplasts. In addition, additional residues not required in vitro and the formation of an RNA hairpin within the core promoter were correlated with wild-type RNA4 levels in cells. Using a template competition assay, the core promoter of ca. 20 nucleotides was found to be sufficient for replicase binding. Mutations of the key residues in the core promoter reduced replicase binding, but deletions that disrupt the predicted base pairing in the proposed stem retained binding at wild-type levels. Together, these results indicate that key nucleotides in the BMV subgenomic core promoter direct replicase recognition but that the formation of a stem-loop is required at a step after binding. Additional functional characterization of the subgenomic core promoter was performed. A portion of the promoter for BMV minus-strand RNA synthesis could substitute for the subgenomic core promoter in transfected cells. The comparable sequence from Cowpea Chlorotic Mottle Virus (CCMV) could also substitute for the BMV subgenomic core promoter. However, nucleotides in the CCMV core required for RNA synthesis are not identical to those in BMV, suggesting that the subgenomic core promoter can induce the BMV replicase in interactions needed for subgenomic RNA transcription in vivo.
Viral RNA-dependent RNA synthesis requires the specific interaction of the replication enzymes and the viral RNA (6, 28). Specificity can be achieved in a number of ways. For example, RNA synthesis can take place within the virion, or the replicase can use the RNA template from which it is translated. Brome mosaic virus (BMV) is a positive-strand tripartite RNA virus whose replication proteins are encoded by RNA1 and RNA2 while the proteins for encapsidation and cell-to-cell spread are in a third RNA, RNA3. Hence, the replicase must locate the specificity elements in RNA3 in trans (4, 15). In addition to the synthesis of genome-length RNAs, minus-strand BMV RNA3 directs the transcription of a subgenomic RNA4 (20). The subgenomic promoter consists of an A/U-rich sequence, a poly(U) tract, and a 20-nucleotide (nt) core subgenomic promoter (Fig. 1A) (2, 3, 8, 19, 20). The core promoter is of functional interest, since it positions the replicase for accurate initiation and resembles simple DNA promoters.
FIG. 1.
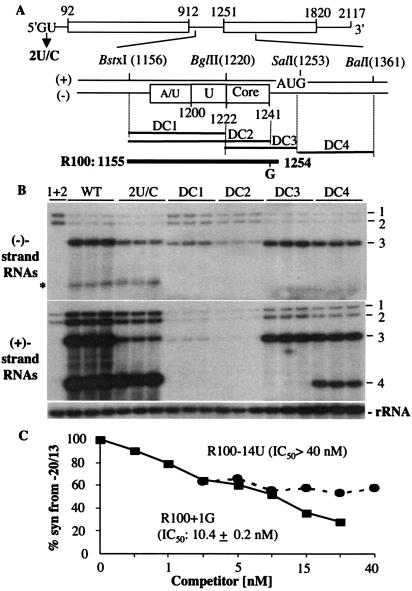
Analysis of the regions in the BMV RNA3 intercistronic region required for RNA synthesis. (A) Schematic diagram of the intercistronic region in plus- and minus-strand RNA3. Deletions designed to test the roles of the A/U-rich and poly(U) sequences, the core promoter in the intercistronic sequence, and a portion of the capsid-encoding sequence in BMV RNA accumulation. The deletions, marked by dark lines, were made by use of the restriction sites shown. (B) Autoradiogram of a Northern blot showing the effect of mutations on genomic minus-strand and genomic plus-strand accumulation. The identities of the RNA bands in the autoradiograms are listed to the sides of the autoradiogram. All reactions tested were performed with three independent samples to allow assessment of the reproducibility of the reactions. Except for the leftmost lane, which has barley protoplasts transfected with only BMV RNA1 and RNA2, the other reactions were transfected with the RNA3 indicated above the autoradiogram and BMV RNA1 and RNA2. The faint band that corresponds to the length of minus-strand RNA4 (identified by an asterisk) is minus-strand RNA4 that has a template of subgenomic RNA4 (11). The bottom slice of the autoradiogram containing the18S rRNAs is intended as an internal loading control to assess the amount of RNA in each lane. (C) Results of a representative competition assay with competitor RNA, R100 + 1G containing the BMV intercistronic sequence spanning nt 1155 to 1254 and a mutation of the initiation cytidylate to prevent RNA synthesis, and a second competitor with an identical sequence except for a uridylate substitution at −14A. The graphs represent the syntheses from the reference template, −20/13, in response to the concentration of the competitors. WT, wild type.
The RNA recognition elements in the BMV subgenomic core promoter have been characterized by using minimal-length promoter templates called proscripts and enriched viral replicase extracted from infected plants (2, 9, 10, 27, 28). Analyses of the specificity elements for other modes of BMV RNA synthesis have provided results that are generally in excellent agreement with those from cells transfected with viral RNA, thus validating the use of this biochemical approach as one useful for dissecting the requirements for RNA synthesis (11, 29).
Previously, it was determined that RNA synthesis in vitro from proscripts requires at least 4 nt at positions −17G, −14A, −13C, and −11G relative to the initiation cytidylate (+1C) (2, 27, 28). RNA synthesis also involves mutual adjustment, or induced fit, between the replicase and the RNA core promoter. This conclusion came from observations that the Cowpea chlorotic mottle virus (CCMV) core promoter can direct the BMV replicase to recognize different nucleotides for the initiation of RNA synthesis in vitro than the usual BMV ones (1). Furthermore, recognition of the promoter for minus-strand RNA synthesis has features consistent with an induced fit mechanism (16).
Contrary to a mechanism of sequence-specific recognition of the BMV subgenomic core promoter, Jaspars (13) identified through sequence analysis a short RNA hairpin in the subgenomic core promoters of plant RNA viruses. Haasnoot et al. (9) characterized the sequence forming this hairpin in BMV and proposed that it has a trinucleotide loop and is required to direct BMV subgenomic RNA synthesis in vitro. Furthermore, Hassnoot et al. (10) proposed that the recognition of the subgenomic promoter occurs in a manner identical to the recognition of the genomic minus-strand core promoter. In this work, we examine the regulation of BMV RNA4 levels in transfected barley protoplasts and the replicase-core promoter interactions in vitro by using a number of mutations that may affect the sequence and/or structure of the core promoter RNA. We also examine RNA4 levels when the subgenomic core promoter is replaced with the core promoter for minus-strand initiation and the subgenomic core promoters from other bromoviruses.
MATERIALS AND METHODS
BMV replication in barley protoplasts.
Infectious transcripts used for transfection of barley protoplasts were made from cDNA copies of wild-type BMV RNA1, RNA2, and RNA3 contained in pB1TP3, pB2TP5, and pB3TP8, respectively (12). Deletions within the subgenomic promoter of pB3TP8 were made by use of appropriately located restriction sites, and point mutations were constructed by using a site-directed mutagenesis kit (Stratagene, San Diego, Calif.) with desired DNA oligonucleotides. The presence of the introduced mutations and the absence of spurious mutations were confirmed by sequencing the region encompassing the engineered mutation. Capped full-length transcripts were made by using a Message Machine kit as described by the manufacturer (Epicentre Inc., Madison, Wis.), with EcoRI-linearized plasmids.
Protoplasts were generated from 5-day-old primary barley leaves as described by Kroner et al. (18). Protoplasts were transfected with a mixture of capped full-length transcripts of RNA1, RNA2, and either RNA3 or a mutant derivative of RNA3. Transfected protoplasts were incubated at a constant 23°C temperature and with illumination for 14 h, unless stated otherwise. Following the incubation period, total RNA was extracted with phenol and chloroform, and Northern blot hybridization was done with probes specific to the 3′-terminal 200 nt of the plus- or minus-strand BMV RNA3. Most blots were first probed to detect minus-strand RNA, then stripped in a low-salt buffer at 95°C to remove the probe, confirmed to have no remaining radiolabel, and probed with an RNA that recognizes plus-strand BMV RNAs. Lastly, the membranes were stripped and probed with a transcript that recognizes the 18S rRNA. Hybridizations and washing of the membranes used conditions that do not allow cross-recognition of the plus- and minus-strand RNAs. Quantification used a PhosphorImager and Molecular Dynamics software. Each value listed in the figures represents a minimum of two independent assays. Where standard deviations are shown, the values are based on a minimum of four independent assays.
Transcripts used in RNA-dependent RNA synthesis assays were prepared with T7 RNA polymerase or were chemically synthesized (Dharmacon Inc., Boulder, Colo.). The transcripts were electrophoresed on denaturing gels, separating RNAs that differed in length by 1 nt. Fragments of the correct length were excised with a razor blade, and the gel slices were crushed to elute the RNAs with 0.3 M ammonium acetate overnight. The eluted RNAs were extracted with phenol-chloroform and precipitated with ethanol. Recovered RNAs were quantified by spectrophotometry and checked for quality in denaturing gels stained with Toluidine blue.
RNA replicase assay.
BMV replicase was prepared from infected barley as previously described (30). Standard replicase assays were carried out as described by Adkins et al. (2). Template competition assays measured the synthesis from a chemically synthesized proscript, −20/13, as affected by an increasing concentration of competitor RNAs, as described by Siegel et al. (27). Each assay consisted of a 40-μl reaction mixture containing the desired amount of template, 7 μl of BMV replicase, 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12 mM dithiothreitol, 0.5% (vol/vol) Triton X-100, 1 mM MnCl2, 200 μM ATP, 200 μM UTP, 500 μM GTP, and 242 nM [α-32P]CTP (400 Ci/mmol, 10 mC/ml; ICN). After incubation for 60 min at 25°C, reactions were terminated by phenol-chloroform extraction followed by ethanol precipitation in the presence of 10 μg of glycogen and 0.4 M ammonium acetate. Products were suspended in a denaturing loading buffer (45% [vol/vol] deionized formamide, 1.5% [vol/vol] glycerol, 0.04% [vol/vol] bromophenol blue, 0.04% [wt/vol] xylene cyanol), heated for 3 min at 90°C, and separated by 20% denaturing polyacrylamide gel electrophoresis. Gels were wrapped in plastic and exposed to film at −80°C. RNA products were quantified with a PhosphorImager (Amersham, Inc., San Diego, Calif.). Each value represents a mean of the results from at least three independent experiments with at least two replicates for each proscript.
RESULTS
Requirements for subgenomic RNA4 synthesis in cells.
To examine the boundaries of the BMV RNA3 intercistronic sequence required for subgenomic RNA synthesis in a cell-based assay, we made three deletion mutants, DC1 to DC3, that removed the A/U-rich, the poly(U), and/or the subgenomic core promoter in the intercistronic sequence of RNA3 (Fig. 1A) for transfection into barley protoplasts. Since these deletions could affect multiple classes of BMV RNA (i.e., genomic plus, genomic minus, and subgenomic), a number of controls were included in the experiment. One control was 2U/C, which has been characterized previously to preferentially affect the accumulation of RNA3 but not RNA4 (11). In addition, the different classes of RNAs were detected by Northern blot analyses. Each mutant RNA3 was transfected into barley protoplasts along with BMV RNA1 and RNA2. As controls, protoplasts were transfected with (i) wild-type BMV transcripts, (ii) only RNA1 and RNA2, and (iii) 2U/C, which has a single nucleotide change in the 5′ noncoding region of RNA3 that affects RNA3, but not RNA4, synthesis (11). Consistent with the previous results of French and Ahlquist (8), we observed that the A/U-rich, poly(U), and core promoter regions were all required for RNA4 synthesis in cells (Fig. 1B). Furthermore, the A/U-rich and poly(U) sequences deleted in DC1 and DC2 also affected both minus- and plus-strand RNA3 synthesis. The deletion of the core promoter in DC3, which removes the core promoter and the untranslated region (12-nt) up to and including the translation initiation site, had a more modest effect on both minus- and plus-strand RNA3, while severely reducing RNA4 (Fig. 1B, lanes DC3). Deletion of a portion of the capsid-encoding sequence called DC4 had a less pronounced effect on both minus-strand RNA3 and subgenomic RNA4 accumulation. Thus, the intercistronic sequence has multiple roles in minus-, plus-, and subgenomic RNA synthesis while the core promoter sequence (nt 1222 to 1241 in plus-strand RNA3) has a more specific role in subgenomic RNA synthesis.
Requirements for replicase-subgenomic promoter interaction in vitro.
We examined whether the core promoter is the primary determinant that binds the BMV replicase in vitro by using a template competition assay (28). This approach has fewer requirements than analysis of RNA synthesis in vivo or in vitro. Briefly, the assay determines the level of synthesis in vitro from a reference promoter template in the presence of increasing concentrations of a competitor RNA. The competitor concentration needed to reduce synthesis from the reference template to 50% is defined as the 50% inhibitory concentration (IC50). Proscript RNA −20/13 was capable of robust synthesis when present at a final concentration of 2 nM (S.-K. Choi, data not shown); hence, this concentration of −20/13 was used in all of the template competition assays.
An RNA named R100 + 1G that contains the intercistronic sequence from nt 1155 to 1254 was made to determine the relative contributions of the different motifs for replicase binding (Fig. 1A). We wanted to separate the requirements for RNA binding from those for RNA synthesis in the competitor. Hence, R100 + 1G has its initiation cytidylate changed to a guanylate; it does not direct RNA synthesis by the BMV replicase in vitro (Choi, data not shown). An identical change of the +1C made in the context of −20/13 abolished RNA synthesis but did not affect binding to the replicase (28). Also, a version of R100 + 1G named R100 − 14U, with a transversion at the −14U of the core promoter, was made for use as a control in the template competition assay. R100 + 1G had IC50s lower than 10.4 nM while R100 − 14U had an IC50 of >40 nM (Fig. 1C), suggesting that the core promoter is primarily responsible for binding the BMV replicase. The core promoter sequence will be the focus of the remainder of this analysis.
Recognition of the BMV core promoter in barley protoplasts.
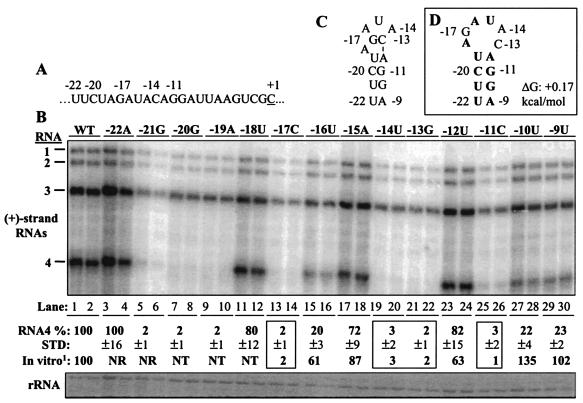
To examine whether the key nucleotides important for subgenomic RNA synthesis in vitro are also important for BMV replication in cells, RNAs containing substitutions at every position from −22 to −9 of the core promoter were transfected along with wild-type BMV RNA1 and RNA2 into barley protoplasts (Fig. 2A). Except as noted, mutant RNAs were named by their positions relative to the + 1C and the final identity of the nucleotide. In comparison to the levels of wild-type RNAs, mutations in the four key nucleotides identified in vitro (−17, −14, −13, and −11) all had RNA4 levels near the background (Fig. 2B). Furthermore, several mutations that had less severe effects for RNA synthesis in vitro, including changes at the −18, −16, −15, −12, −10, and −9 positions, produced detectable levels of RNA synthesis in cells. In vitro, the nucleotides 3′ of position −20 were not required for synthesis (2). In protoplasts, a change of −22U to A transcribed RNA4 at near wild-type levels (Fig. 2B, lanes 3 to 4). However, when −21U was changed to G, RNA4 accumulation was reduced to near background (Fig. 2B, lanes 5 to 6). Similar effects were seen for nucleotide substitutions at the −19 and −20 positions (Fig. 2B, lanes 7 to 10). These results indicate that the in vitro RNA synthesis assay did identify some of the crucial residues required in vivo but would miss other requirements.
FIG. 2.
Effects of mutations in key residues of the BMV subgenomic core promoter in transfected barley protoplasts. (A) The locations of the key residues in the core promoter are identified by their position relative to the initiation cytidylate (position +1 is underlined). (B) An autoradiogram of a Northern blot showing the effects of the mutations on BMV plus-strand RNA accumulation. The names of mutant RNAs used in transfection are shown above the lanes, and the identities of the RNAs shown are to the left of the autoradiogram. RNA4 is the RNA that should be most directly affected by the mutations in the core promoter. Quantifications of the amounts of RNA4, after normalization to the wild-type (WT) transfection, are shown under the autoradiogram. The amount of RNA synthesis in vitro by the BMV replicase is also shown to allow comparison of the effects of the mutations in vitro and in vivo. The boxes identify the four key nucleotides of the core promoter (27). STD, one standard deviation; NR, not required in vitro; NT, not tested. (C) The secondary structure of the BMV core promoter element required for replicase recognition, as reported by Haasnoot et al. (9, 10). (D) The most stable structure predicted by the computer program MFOLD (35) to exist in the BMV core promoter sequence.
Results from mutations of BMV RNA4 synthesis in protoplasts could be used to examine the two models for the recognition of the BMV core promoter. Haasnoot et al. (10) proposed that nt −22 to −9 of the subgenomic core promoter form a hairpin with a 3-nt loop and a 5-bp stem, with positions −17 and −13 forming the loop-closing base pair (Fig. 2C). However, the bulged −18 should destabilize this hairpin, and the structure predicted by MFOLD (35) contained a 6-nt loop and a 4-bp stem (Fig. 2D). Since a base substitution of the bulged −18A did not significantly affect RNA synthesis (Fig. 2B, lanes 11 to 12), it is unlikely to be specifically involved in stabilizing tertiary interactions and retaining the identical conformation. Furthermore, in the one-dimensional nuclear magnetic resonance (NMR) analysis performed by Haasnoot et al. (10), none of the identities of the imino peaks were assigned, but the one ascribed to −17G was quite broad, indicating that the loop nucleotides lack a stable closing base pair (even at 4°C, the temperature of the measurements). The change in entropy for this structure was predicted by MFOLD (35) to be −1.8 kcal/mol, which suggests that it is likely to be quite a dynamic structure in solution. Whether these nucleotides form more-complex structures remains to be determined. We will consider the loop to be 6 nt long until there is additional information to indicate otherwise (Fig. 2D).
Two changes in the loop that do not affect the key nucleotides (−18A to U and −15U to A) had only minor effects on RNA4 accumulation in protoplasts and no negative effects on RNA synthesis in vitro (27) (Fig. 2B, lanes 11 to 12 and 17 to 18). At the top of the putative stem, a change of −12A to U resulted in RNA4 levels at 82% of the wild type. Therefore, the U-A base pair formed by nt −19 and −12 is either not essential for RNA4 accumulation in vivo, or an alternative base pairing(s) within the loop is acceptable for subgenomic transcription. A change of −19U to A reduced RNA4 levels to background, suggesting that base pairing at this position was needed. Formation of the U-A base pair at the bottom of the stem is not required, since a change of −22U to A retained wild-type levels of RNA synthesis (Fig. 2B, lanes 3 and 4). However, a change of the complementary −9A to U reduced synthesis to 15% of the wild type (Fig. 2B, lanes 29 to 30). These results indicate that while the formation of some base pairs in the core promoter is required in the middle and bottom of the stem, there is some flexibility at the top of the hairpin in Fig. 2D.
Effects of select mutations on the stabilities of the RNAs.
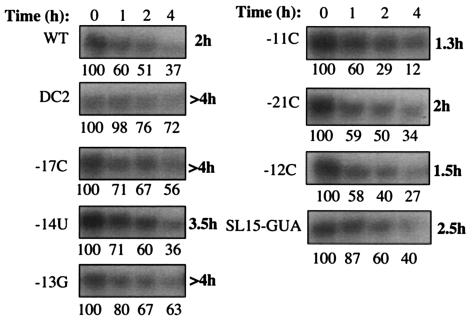
In vivo results could be affected by a combination of factors, including the stability of the transfected RNAs and the production of the capsid protein. Most of the base substitutions in the BMV core promoter tend to preferentially affect subgenomic RNA4 synthesis rather than RNA3, suggesting that there is no major defect in the stabilities of the mutant RNAs (Fig. 2B). It was confirmed that minus-strand RNA3 accumulated to normal levels from 6 to 12 h posttransfection (M. Hema, data not shown). Nonetheless, we tested the stabilities of the transfected mutant RNAs directly. Radiolabeled transcripts of the wild-type and mutant RNA3s were prepared and transfected into barley protoplasts along with unlabeled RNA1 and RNA2. Total RNAs were then harvested at 0, 1, 2, and 4 h posttransfection, electrophoresed on a denaturing gel, and autoradiographed. Radiolabeling of the RNAs varied somewhat during in vitro transcription. Therefore, the stability of each transcript was measured relative to the sample from 0 h, which was extracted from cells immediately after transfection. The results show that 4 h after transfection, wild-type BMV RNA3 was present at 37% of the initial inoculum and that the half-life is approximately 2 h (Fig. 3). Of the nine mutant RNAs tested, including the deletion DC2 that lacks much of the intercistronic region, only −11C and −12C had lower half-lives than wild-type RNA3. Even these two RNAs are easily detectable at 4 h after transfection. All indications are that there is no rapid turnover of our transfected transcripts in barley protoplasts, although we cannot rule out minor effects on RNA stability.
FIG. 3.
Assessment of the stability of the various wild-type and mutant RNA3s in barley protoplasts. Radiolabeled RNAs were transfected into barley protoplasts along with wild-type (WT) RNA1 and RNA2. The protoplasts were then harvested after incubation for the number of hours above the two top autoradiograms, electrophoresed onto a denaturing gel, dried, and exposed to X-ray film. The names of the mutant RNAs are to the left of the autoradiograms, and the estimated half-lives are on the right. Only a select number of mutants that are generally affected in RNA4 accumulation were tested. Numbers under the autoradiograms represent the amount of signal in each band relative to time zero.
Requirements for a stem-loop structure in the BMV subgenomic core promoter.
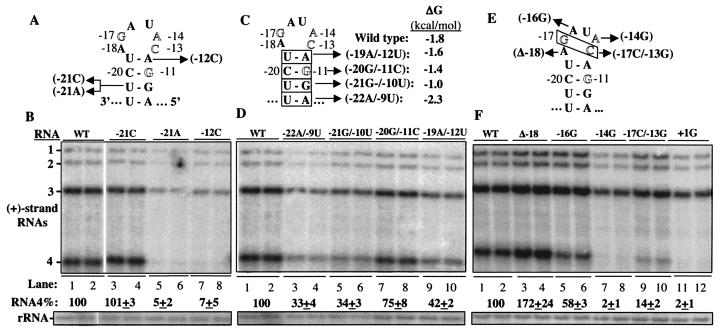
To examine further the requirements for base pairing in the putative hairpin, we made additional mutations at the −12 and −21 positions (Fig. 4A). With the change of −12A to C, which would result in a U and a C being the two loop-closing nucleotides, RNA synthesis was reduced to 7% of that of the wild type. Since a change of −12A to U retained 80% of the wild-type level of RNA synthesis (Fig. 2B, lanes 23 to 24), we also changed −21U to C and to A, resulting in a C-G pair and an A-G pair, respectively. The C-G pair was capable of efficient synthesis while the A-G pair was not (Fig. 4B, lanes 3 to 6). These results confirm that the middle and bottom portions of the stem must be maintained for wild-type RNA4 levels.
FIG. 4.
Features in the BMV subgenomic core promoter that may affect RNA4 accumulation. (A) Schematic of mutations in the subgenomic core promoter. The RNA shown is the minus sense of BMV RNA3, with the polarities shown. (B) Autoradiograms of a Northern blot of positive-strand BMV RNAs accumulated by the mutant RNA3s and the positive control. In some of the autoradiograms, portions of the original image were removed to facilitate presentation of the results. However, all RNAs shown within a panel were originally from one autoradiogram. Quantifications shown below the autoradiogram were derived from the experiments shown and two other independently performed experiments. The slice containing the rRNA signal was obtained from the same blot probed after the analysis of BMV RNAs. (C) Changes in the BMV subgenomic core promoter hairpin designed to examine whether stability of the stem was correlated to RNA4 levels. (D) Autoradiogram of the RNA accumulation by RNAs with the mutations shown in panel C. (E) Mutations within the loop portion of the core promoter hairpin. The box indicates that both boxed nucleotides were mutated. (F) Autoradiogram of the effects of the mutations on RNA4 accumulation. WT, wild type.
To examine whether the predicted stability of the subgenomic hairpin in the subgenomic core promoter is correlated with RNA4 levels, we made four mutant RNAs that reversed the bases within the 4 bp of the stem (Fig. 4C). ΔG values of the resultant RNA hairpins were calculated by using the nearest-neighbor rules under conditions of 30°C, 125 mM Na+, and 1 mM Mg2+ (to mimic intracellular concentrations) and were found to differ only slightly from that of the wild-type hairpin (Fig. 4C). In protoplasts, however, all four mutant RNAs accumulated at levels lower than the wild type. Taken with results from mutants −12U and −18U (Fig. 2B), we posit that the stability of the stem in the hairpin is not directly correlated with the level of BMV RNA4. Notably, the switch of the base pair at the −20 and −11 positions resulted in 75% of the wild-type RNA synthesis, indicating that base-specific recognition at the key −11G position is not essential in vivo.
Next, we tested the effects of nucleotide changes within the loop of the putative RNA hairpin. The loop contains three of the key nucleotides found to be important for RNA synthesis in vitro and in protoplasts (Fig. 4E) (27). A deletion of −18A increased RNA4 levels to 172% of the wild type (Fig. 4F, lanes 3 to 4). This deletion could allow −17G and −13C to base pair without possible steric interference of −18A. To examine whether the pairing of the 2 nt or their base identities are more important for RNA4 levels, we reversed the bases of the −17 and −13 positions from the normal G-C to a C-G base pair. This switch resulted in an RNA that produced RNA4 at only 14% of the wild type, indicating that the identities of the bases at positions −17 and −13 positions are more important than their ability to pair (Fig. 4F, lanes 9 to 10). Similarly, −14A and +1C are required for RNA4 accumulation, since substitutions at these positions resulted in RNA4 at background levels (Fig. 4E and F, lanes 7 to 8 and 11 to 12). These results indicate that the key nucleotides in the putative loop of the hairpin are important for RNA4 transcription. Whether these nucleotides form more complex structures remains to be determined.
Minimal length of the BMV subgenomic core promoter required to interact with the BMV replicase in vitro.
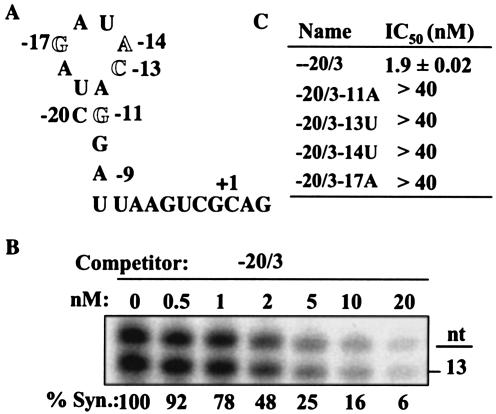
The complex requirements for subgenomic RNA4 synthesis and accumulation in cells prompted us to dissect in vitro the requirements for the interactions between the BMV replicase and the core promoter. We used the template competition assay to elucidate the sequences and structures needed for replicase binding. The first competitor tested is named −20/3, an RNA known to retain interaction with the replicase but which can direct RNA synthesis in vitro at less than 5% of the amount made by −20/13 (28; Choi, data not shown), thus allowing the separation of the requirements for RNA synthesis from replicase binding. An MFOLD prediction performed under the conditions of RNA synthesis in vitro revealed no stable structure. For the sake of examining the requirements of the hairpin in vitro, however, the base pairs that would remain in the predicted hairpin are shown in Fig. 5A. RNA −20/3 had an IC50 of 1.9 nM and was at least as capable of interacting with the BMV replicase as the intercistronic sequence within R100 + 1G (Fig. 5B). This value is lower than the 25 nM previously reported by Siegel et al. (28), likely because we now use a lower concentration of the reference template (2 versus 25 nM). To confirm that −20/3 could be a prototype competitor RNA for meaningful analysis of the requirements of replicase binding, we made Watson-Crick transversions at each of the four key residues that are required for RNA synthesis (Fig. 5A). All four mutant RNAs had IC50s greater than 40 nM (Fig. 5C). These results suggest that these four key nucleotides are important for initial recognition by the BMV replicase in the context of −20/3.
FIG. 5.
Nucleotides in the subgenomic core promoter that confer higher affinity binding to the BMV RNA replicase in vitro. (A) Sequence of −20/3, modified according to the secondary structure proposed by Haasnoot et al. (10). (B) Representative result from a template competition assay with the reference template −20/13 and the competitor −20/3. The final concentration of the competitor RNA is shown above the autoradiogram. (C) Summary of the concentrations of the competitor RNAs derived from −20/3 needed to reduce RNA synthesis (Syn.) from the reference template to 50%.
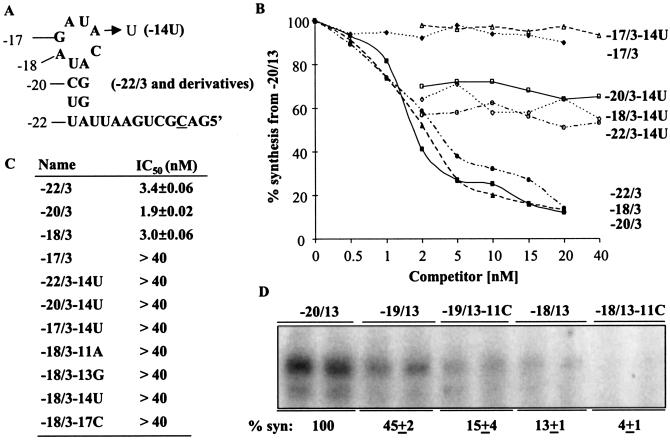
Next, we determined the minimal length of the core promoter required for replicase binding. A series of RNAs with their 3′ ends terminating at nt −22, −20, −18, and −17 was tested. Each RNA was made in a version that also had a change of −14A to U, thus providing a control for RNAs of different lengths (Fig. 6A). Templates −22/3, −20/3, and −18/3 all had IC50s lower than 3.4 nM (Fig. 6B). However, −17/3 had an IC50 of >40 nM, suggesting that a minimal core promoter length of 18 nt is sufficient to effectively bind the BMV replicase. Changes of the −14A to U in the contexts of −22/3, −20/3, and −18/3 all resulted in IC50s greater than 40 nM. We note that the predicted ΔG for −22/3 was −1.8 kcal/mol and that the other truncated RNAs, including −18/3, are not predicted by MFOLD to have a stable structure. RNA −17/3 (20 nt) is of a sufficient length to bind to the BMV replicase, since RNAs as short as 13 nt can bind to the BMV replicase with IC50s similar to that of −20/3 and can direct RNA synthesis in vitro (34).
FIG. 6.
Minimal length of the subgenomic core promoter needed to interact with the BMV replicase in vitro. (A) Sequence and predicted structures of −22/3. The locations of nt −20, −18, and −17 are shown. Each of these four RNAs was also made with a change of −14A to U to serve as a parallel negative control in the template competition assay. The initiation cytidylate is underlined, and a mutation of the −14 residue is indicated by an arrow. (B) Results from competition assays in which products from 2 nM −20/13 are plotted against the concentration of the competitor RNAs. (C) Summary of the results from the template competition assays shown in panel B. The IC50s of −22/3, −20/3, and −18/3 were from six independent assays, with the values for one standard deviation shown after the means. Other values were derived from the results of two independent assays that yielded consistent results. (D) Synthesis (syn) from RNAs with different deletions from the 3′ end of the BMV core promoter. The names of the RNAs tested denote the 3′ and 5′ nucleotides that are present at the termini of the RNA. The quantitative values are from four independent assays.
Since mutating −17G to C in the context of −18/3 affected replicase binding, we changed the other three key recognition nucleotides in −18/3 to determine whether their specificities for the core promoter would be changed. All three mutant RNAs were reduced in binding to the BMV replicase (Fig. 6C), suggesting that specific replicase binding required the key nucleotides but not the formation of a stable hairpin.
As nt −19 and −20 are required for BMV RNA4 accumulation in transfected protoplasts (Fig. 2B, lanes 7 to 10) but not for replicase binding, we wanted to address whether the hairpin in the BMV subgenomic core promoter is needed for RNA synthesis. Previously, a 5′ truncation of the subgenomic proscript to position −17 directed RNA synthesis at 6% of the level of an RNA with a 20-nt core promoter. RNAs −19/13 and −18/13 were tested for RNA synthesis by the BMV replicase in vitro. RNA −19/13 was able to direct RNA synthesis at 45% of the wild-type level (Fig. 6D). To address whether this level of RNA synthesis depended on specific recognition of the core promoter, we mutated −11C in the context of −19/13 and found that this change reduced synthesis to 15%. We also observed that −18/13 directed RNA synthesis at 13% of the level of −20/13 and that this synthesis was reduced to 4% upon mutation of the −11G residue. These results are consistent with our previous report and indicate that the −11G residue can contribute to RNA synthesis in a manner independent of the formation of a base pair with the −20 residue. Furthermore, we note that since a 3′ deletion to position −18 can retain replicase binding but not RNA synthesis, the two activities have overlapping but nonidentical requirements.
Analysis of chimeric subgenomic promoters in vivo.
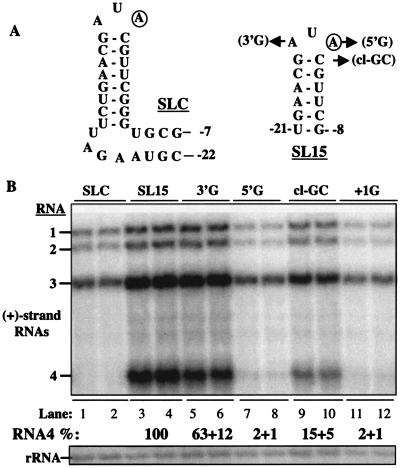
Haasnoot et al. (10) proposed that the BMV core promoter is identical to the core promoter for genomic minus-strand RNA synthesis. The basis for this claim is that the subgenomic hairpin could potentially form an AUA triloop that mimics the specificity element for genomic minus-strand RNA initiation (10, 17). However, the assumption of an AUA triloop requires that −18A be bulged from the stem at a position adjacent to the closing base pair, but as discussed previously, evidence for this is lacking. We sought to examine the effects of replacing the subgenomic hairpin with the promoter for genomic minus-strand RNA synthesis. The wild-type core promoter for genomic minus-strand RNA synthesis is in a structure named stem-loop C (SLC), which is composed of a short stem, a bulge of 4 nt, and a longer terminal stem-triloop (Fig. 7A). In protoplasts, replacement of the subgenomic sequence from position −22 to −9 with SLC resulted in an RNA that was incapable of directing RNA4 accumulation (Fig. 7B, lanes 1 to 2). This result is consistent with an analysis of a similar replacement in vitro (24).
FIG. 7.
Effects of replacing a portion of the subgenomic core promoter with SLC or the terminal hairpin within SLC. (A) Sequences of SLC and SL15 that are used to replace the subgenomic core promoter hairpin. In both structures, the clamped adenine is circled to facilitate its identification. The numbers at the two ends of SLC and SL15 denote the positions of the subgenomic core promoter to which the foreign sequence was fused. In SL15, several mutations were made to examine the requirement for a clamped adenine motif in subgenomic RNA synthesis. The names of the mutant RNAs are in parentheses.(B) An autoradiogram of the effects of replacing the subgenomic core promoter hairpin with SLC, SL15, or mutants that are derived from SL15. Names of the mutant RNAs are above the lanes in the autoradiogram. The quantification of the amount of RNA4 produced is normalized to SL15.
SLC may fail to replace the subgenomic core promoter because it is more than twice the length of the subgenomic hairpin. Therefore, we made a construct wherein the subgenomic hairpin from position −21 to −9 was replaced with the 15-nt terminal hairpin in SLC. Given that deletion of −18A resulted in an RNA that could direct better than wild-type levels of RNA4, such a construct is more likely to replace the normal subgenomic hairpin. The resulting RNA, SL15, was able to direct RNA4 accumulation in protoplasts at levels comparable to that of the wild type (Fig. 7B, lanes 3 to 4). This result is in agreement with the predictions of Haasnoot et al. (10) and raises the interesting possibility that an identical factor(s) recognizes the promoters for minus-strand and subgenomic RNA synthesis.
Since the requirements for the minus-strand core promoter are known (16, 17, 29), we tested whether mutations known to affect BMV minus-strand RNA synthesis will have the same effects on subgenomic RNA synthesis. First, a crucial recognition element in SLC is the 5′ adenine of the triloop, which forms a clamped adenine motif (CAM) (17). This adenine is comparable to −14A, a key nucleotide in the BMV subgenomic promoter. Second, the loop-closing C-G base pair is required to maintain a stable CAM, and a reversal of the bases in mutant cl-GC decreased RNA synthesis in vitro to a third of the wild type (16). Third, the 3′ adenine of the triloop could be changed to a guanine without significantly affecting RNA synthesis in vitro and in vivo (16, 29). In the context of SL15, mutations of the 5′, 3′, and closing base pairs all yielded results consistent with the requirements in SLC (Fig. 7B, lanes 5 to 10). Therefore, it is likely that SL15 is recognized by the BMV replicase in the same way as the core promoter for genomic minus-strand RNA synthesis.
Bromovirus chimeras.
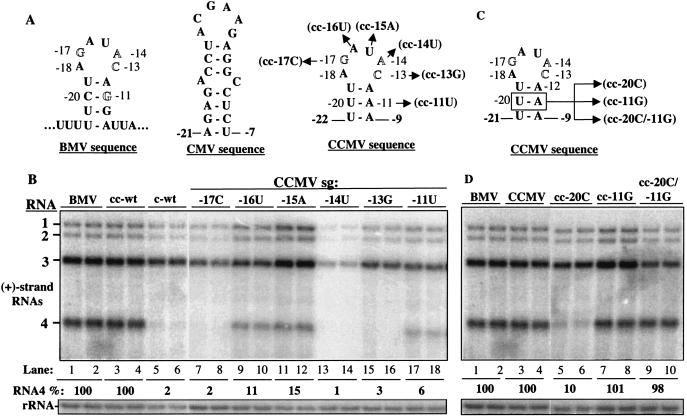
To probe further the function of the BMV subgenomic core promoter, we made chimeric subgenomic core promoters by using sequences from Cucumber Mosaic Virus (CMV) and CCMV and tested these in barley protoplasts. The minimal functional core promoter for CMV RNA4 synthesis, consisting of a 30-nt sequence, was used to replace the BMV core promoter in an RNA named c-wt (7) (Fig. 8A). RNA c-wt was unable to direct BMV RNA4 synthesis in protoplasts (Fig. 8B, lanes 5 to 6).
FIG. 8.
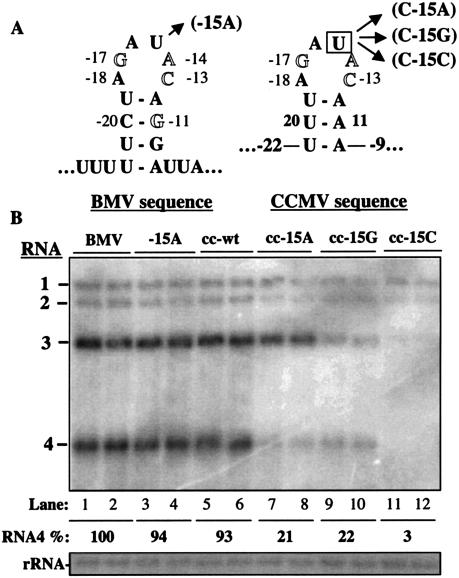
Effects of replacing a portion of the subgenomic core promoter with a core promoter of CMV or the comparable sequence from CCMV. (A) Schematics of the BMV subgenomic core promoter hairpin, the putative hairpin in the CCMV core promoter, and the nucleotide substitutions in the CCMV subgenomic hairpin. The numbers next to the nucleotide sequences denote the positions of the BMV core promoter that were fused to the foreign sequence. The numbers next to the foreign sequence are the positions from the BMV core promoter adjacent to the foreign sequence. (B) Effects of chimeric promoters on plus-strand BMV RNA4 accumulation. Identities of the most relevant RNA transfected into protoplasts are listed above autoradiogram. The lanes labeled with BMV denote products from transfection with wild-type BMV transcripts. Names beginning with “cc-” denote a chimeric RNA3 where the subgenomic hairpin sequence comes from CCMV. c-wt lanes denote transfections performed with a chimeric RNA3 that contains the CMV core promoter that was characterized by Chen et al. (7). (C) Mutations in the CCMV subgenomic core promoter hairpin used to demonstrate base pairing requirements in the stem of the hairpin. (D) Autoradiogram of the effects of mutations shown in panel C on BMV RNA accumulation.
Recognition of the CCMV core promoter by the BMV replicase in vitro was generally similar to that of the BMV subgenomic promoter, with two significant differences: (i) the −11G in BMV is an A in CCMV and recognition required base pairing with the U at the −20 position (Fig. 8A) (1) and (ii) additional residues are needed for the CCMV sequence to direct RNA synthesis by the BMV replicase, including −15, a position in the BMV core promoter which is not required in vitro or in barley protoplasts (Fig. 2B, lanes 17 to 18). The sequence within and including positions −20 and −9 of the BMV core promoter was replaced with the comparable sequence from CCMV in a chimeric RNA named cc-wt. The resulting RNA directed efficient accumulation of BMV RNA4 (Fig. 8B, lanes 1 to 4). One implication of this result is that the BMV and CCMV subgenomic promoters are recognized by the same factor in vivo. Further, this result confirms that one property of the −11 base is to form a base pair, likely with the −20 base. To examine the recognition of cc-wt further, we changed either nt −20U or −11A, or both, in the context of cc-wt. RNAs cc−20C, cc−11G, and cc−20C/−11G will have −20 and −11 bases that are, respectively, C-A, U-G, or C-G (Fig. 8C and D). A U-G or C-G pair was found to functionally substitute for the U-A base pair in cc-wt while a C-A pair was not (Fig. 3D, lanes 5 to 10), confirming that RNA4 levels require the base pairing of nt −11 and −20.
Adkins and Kao (1) previously found that the key residues in the CCMV core promoter were at positions −10, −11, −13, −14, −15, −16, −17, and −20. We examined the requirement for most of these key residues by testing the effects of single nucleotide substitutions (Fig. 9). Changes of the nucleotides at positions from −17 to −13 and at −11 all decreased RNA4 levels to less than 15% of that of cc-wt (Fig. 9B, lanes 7 to 12). These results are consistent with those in vitro (1) and are notable in that, in protoplasts, the replicase, encoded by BMV, will change its recognition of the subgenomic core promoter with a change in the promoter sequence.
FIG. 9.
Mutational analysis of position −15 in the CCMV-BMV chimera. (A) Schematics of the relevant BMV and CCMV sequences and the names of the mutant RNAs. (B) Autoradiogram of a Northern blot showing the effects of mutations at position −15 on RNA4s produced from normal BMV and chimeric CCMV-BMV subgenomic promoters. RNA cc-wt is a version of BMV RNA3 in which nt −9 to −22 were replaced with the wild-type CCMV sequence. RNAs cc-15A, cc-15G, and cc-15C were derivations of cc-wt in which the −15 positions were changed. Quantifications were from four independently transfected protoplast preparations.
Recognition of −15U in cc-wt was examined further, since this base is not essential in the BMV core promoter either in vitro or in protoplasts (Fig. 2B, lanes 17 to 18, and 9B, lanes 11 to 12). Changes of the U to any of the other three residues decreased RNA synthesis to less than 22% in this experiment. Thus, the −15 position of the CCMV core promoter is recognized in a base-specific manner by the BMV replicase. This position in cc-wt may also have a concomitant effect on RNA3 levels but not as much as on RNA1 and RNA2.
DISCUSSION
The goal of this study was to elucidate the mechanism of BMV replicase-subgenomic promoter recognition. We analyzed the effects of mutations in the BMV subgenomic core promoter on RNA4 levels in barley protoplasts by using chimeras and mutated versions of BMV RNA3 and in vitro by using the enriched BMV replicase. In protoplasts, most mutations had more of an effect on RNA4 than RNA3, indicating that their primary effects were on subgenomic RNA synthesis. Wild-type RNA4 levels required the four key nucleotides identified in vitro and an RNA secondary structure that is formed by nt −21 to −9 of the core promoter. However, the stability of the RNA hairpin is not correlated with RNA4 levels. In examining replicase-core promoter interaction in vitro, we found that the four key nucleotides are required for binding at wild-type levels but that the hairpin is largely dispensable. Based on these and previously published results suggesting an induced fit mode of recognition (2), we propose that subgenomic transcription requires at least two steps: binding to the core promoter sequence and the subsequent formation of a hairpin structure in the presence of the replicase. We found that the subgenomic core promoter can be functionally replaced with the terminal stem-loop within SLC named SL15 and with the comparable sequence from CCMV. The recognition of the CCMV and BMV subgenomic hairpins by the BMV replicase have overlapping but nonidentical requirements (Fig. 9), indicating that the core promoter sequence can direct the BMV replicase to form alternative contacts.
Results from Haasnoot et al. and Jaspars (9, 10, 13) and those from our lab (1-3, 27, 28) were all based on similar in vitro RNA synthesis assays, thus raising the question of why one group observed a requirement for an unstable stem-loop while the other did not. One difference was that Haasnoot et al. (10) used a 44-nt proscript that extends from −25 to +19 relative to the initiation site, whereas our laboratory's studies primarily used a 33-nt proscript that spans nt −20 to +13. Our lab had demonstrated, through a series of deletions, that a proscript of this length was not compromised for the level or the accuracy of subgenomic RNA initiation in comparison to longer templates (2). In this study, we demonstrate that even shorter RNAs will retain similar levels of replicase binding (Fig. 6B and C) but not necessarily RNA synthesis (Fig. 6D). The longer RNA used by Haasnoot et al. (10) for RNA synthesis reactions lengthened the stem to 4 bp (not counting the −17G/−13C base pair at the base of the loop nucleotides) (Fig. 2A) and would contribute to the detection of a stable RNA structure (Fig. 6B and C). Also, the RNase T1 digestion and one-dimensional NMR analyses used to demonstrate the existence of a stable structure were performed on ice and at 4°C, respectively (10). Based on the rather broad imino peaks detected in the NMR analysis under these conditions, the RNAs are likely to be quite dynamic at the temperatures required for BMV replication.
Multistep mechanism for the initiation of BMV subgenomic RNA synthesis.
We propose that of the four key nucleotides, −11G participates in base pairing with the −20 residue while −13C, −14A, and −17G are recognized in a base-specific manner. The latter 3 nt cannot be replaced by other bases or base analogs and result in RNAs that retained RNA synthesis in vitro (27, 28). Substitutions tested in transfected protoplasts are also debilitated for RNA4 levels (Fig. 2 and 4). Lastly, the bases at the −17 and −13 positions that are proposed to form a base pair (10) cannot be reversed and retain wild-type RNA4 levels in transfected cells (Fig. 4F, lanes 9 to 10).
While there is strong evidence for the base-specific recognition of the key residues, there is also strong evidence that the core promoter sequences in bromoviruses can form a short hairpin (13) and that its stem is required for RNA4 transcription. An important interaction is the G-C base pair formed by nt −11 and −20, which is not as important for interacting with the replicase or RNA synthesis in vitro (Fig. 6C and D). Consistent with recognition of the stem in infected cells, the CCMV subgenomic promoter has an A-U base pair at this position and is recognized by the BMV replicase (Fig. 8D). A notable feature of the structures of the subgenomic core promoters for BMV and CCMV is that they are both relatively unstable, even when compared to the structures found in other plus-strand RNA viruses (7, 21). Hence, BMV and CCMV may have different requirements even when compared to related RNA viruses. In template competition assays, the hairpin structure is not essential for specific recognition by the BMV replicase in vitro. We cannot presently rule out unusual structures formed in the loop nucleotides that would help stabilize this structure. However, we did not find evidence for additional stabilizing interactions in UV melt analyses (14). Instead, we propose that the structure may be stabilized by its interaction with the replicase, which requires the key nucleotides but not the hairpin. This would make BMV subgenomic transcription a multistep process that involves (i) binding of the minus-strand RNA3 by the BMV replicase, (ii) formation of the hairpin as a result of the interaction, and (iii) initiation of subgenomic RNA synthesis. DNA-dependent RNA polymerases are known to undergo multiple steps involving conformational changes during the initiation of RNA synthesis (for an example, see reference 25). There is also evidence that binding by the T7 polymerase will cause changes in the promoter that go beyond the simple unwinding of the initiation site (31). Multistep recognition of the core promoter could allow for increased specificity and potential regulation in subgenomic RNA synthesis.
There are currently three general models for viral subgenomic RNA synthesis: (i) initiation that takes place from a fully formed minus-strand RNA (21), (ii) discontinuous minus-strand RNA synthesis, as is seen with coronavirus transcription (26), (iii) premature termination during minus-strand RNA synthesis that allows either the same replicase complex or a different one to use the nascent RNA as the template for subgenomic RNA synthesis (reference 32 and references therein). For BMV subgenomic RNA initiation, we propose that the replicase binding could be coupled to premature termination. French and Ahlquist (8) observed that when a series of BMV subgenomic promoters was placed in RNA3, the one closest to the 5′ end of minus-strand RNA produced the most subgenomic transcript. Therefore, subgenomic synthesis may not require the completion of minus-strand RNA3 synthesis. It is possible that an interaction of the nascent minus-strand RNA with either the transcribing replicase or another replicase that acts in trans is part of the promoter recognition mechanism. In fact, the sequence of nascent minus-strand RNA3 containing the core promoter resembles an intrinsic, or Rho-independent, termination signal in bacteria (33), which is a hairpin followed by a stretch of uridylates. Alternatively, DNA-dependent RNA polymerase III can terminate RNA synthesis after it synthesizes four or more uridylates (22, 23). A poly(U) sequence is 3′ of the core promoter sequence in the minus-strand BMV and CCMV intercistronic sequences.
The complexity associated with the formation of a hairpin in the core promoter during nascent minus-strand RNA synthesis could provide additional opportunities for the regulation of transcription. Furthermore, the demonstration of the CCMV core promoter requiring additional contacts with the BMV replicase (Fig. 7) indicates that there is intimate communication and likely induced fit between the core promoter and the replicase.
A common element for subgenomic and genomic minus-strand RNA synthesis?
Recognition of SL15 occurs in a manner consistent with requirements for genomic minus-strand initiation (Fig. 5). This was a result predicted by Haasnoot et al. (10) based on similarities in the sequences of the terminal loop of SLC and a presumed loop in the subgenomic hairpin. One implication of this result is that the core promoters for genomic minus-strand and subgenomic RNAs are initiated by a similar mechanism. Should this be the case, then the highly regulated levels and timing of BMV genomic minus-strand and subgenomic RNA must be due to factors other than the core promoter. There is ample precedence for this in transcription from DNA templates, where basal transcription uses the same core polymerase but the frequency and timing of initiation are influenced by other trans and cis-acting factors (reviewed in reference 5). This result also raises the question of whether the initiation of genomic plus-strand RNA is specified by the same factor.
While it is appealing to simplify the mechanisms for the different modes of BMV RNA synthesis, we do caution, however, that the functional replacement of one promoter with another does not necessarily mean that they are recognized by the same mechanism. Use of foreign promoters to direct transcription in vitro and in vivo is common but does not indicate that the foreign and natural promoters are identical. Furthermore, the sequences of the BMV and CCMV core promoters are not optimal for the formation of the CAM, which directs minus-strand RNA synthesis in vitro and in vivo (16, 17). The CAM is formed in a large part due to the base stacking along the terminal stem of SLC, which stabilizes the loop-closing base pairs and forces the displaced adenine to form other interactions with moieties in the stem (16). The presence of −18A in the subgenomic core promoter should negatively affect the interactions necessary to form a stable stem and the CAM unless −18A forms an unusual interaction with the loop nucleotides. It is interesting that −18A is found in all BMV isolates and in CCMV isolates (Fig. 8C), and its retention suggests a relevant role in BMV infection. Lastly, the formation of a CAM requires a stable stem, and the lower stem for SLC is significantly more stable (ΔG of −6.8 kcal/mol) than the subgenomic sequences (ΔG values of −1.8 and +1.8 kcal/mol for the BMV and CCMV subgenomic hairpins, respectively). Whether the subgenomic core hairpin resembles the CAM in the promoter for genomic minus-strand RNA synthesis will require the elucidation of the structure of the BMV subgenomic core promoter.
Acknowledgments
We acknowledge the helpful discussions and editing of the BMV group at Texas A&M University, the Cereal Killers, and C. H. Kim of the CSU—Hayward.
S.-K.C. acknowledges a fellowship by the Korean Science & Engineering Foundation (KOSEF). Funding was provided by the National Science Foundation.
REFERENCES
- 1.Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, S., R. W. Siegel, J.-H. Sun, and C. C. Kao. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3:634-647. [PMC free article] [PubMed] [Google Scholar]
- 3.Adkins, S., S. Stawicki, G. Faurote, R. Siegel, and C. C. Kao. 1998. Mechanistic analysis of RNA synthesis RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 4.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:271-276. [DOI] [PubMed] [Google Scholar]
- 5.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 25:1-13. [DOI] [PubMed] [Google Scholar]
- 6.Buck, K. W. 1996. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M. H., M. J. Roossinck, and C. C. Kao. 2000. Efficient and specific initiation of subgenomic RNA synthesis by cucumber mosaic virus replicase in vitro requires an upstream RNA stem-loop. J. Virol. 74:11201-11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haasnoot, P. C. J., F. Brederode, R. C. L. Olsthoorn, and J. Bol. 2000. A conserved hairpin structure in alfamovirus and bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haasnoot, P. C. J., R. C. L. Olsthoorn, and J. Bol. 2002. The brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 8:110-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hema, M., and C. C. Kao. 2004. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 78:1169-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 13.Jaspars, E. M. J. 1998. A core promoter hairpin is essential for subgenomic RNA synthesis in alfalfa mosaic alfamovirus and is conserved in other bromoviridae. Virus Genes 17:233-242. [DOI] [PubMed] [Google Scholar]
- 14.Kao, C. C. 2002. Lessons learned from the core RNA promoters of Brome mosaic virus and Cucumber mosaic virus. Mol. Plant Pathol. 3:53-59. [DOI] [PubMed] [Google Scholar]
- 15.Kao, C. C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1:91-98. [DOI] [PubMed] [Google Scholar]
- 16.Kim, C. H., and C. C. Kao. 2001. A mutant viral RNA promoter with an altered conformation retains efficient recognition by a viral RNA replicase through a solution-exposed adenine. RNA 27:1476-1485. [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, C.-H., C. C. Kao, and I. Tinoco. 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Biol. 7:415-423. [DOI] [PubMed] [Google Scholar]
- 18.Kroner, P. A., B. M. Young, and P. Ahlquist. 1990. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J. Virol. 64:6110-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 21.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 22.Myslinski, E., J. C. Ame, A. Krol, and P. Carbon. 2001. An unusually compact external promoter for RNA polymerase III transcription of the human H1 RNA gene. Nucleic Acids Res. 29:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paule, M. R., and R. J. White. 2000. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28:1283-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranjith-Kumar, R., X. Zhang, and C. C. Kao. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saecher, R. M., O. V. Tsodikov, K. L. McQuade, P. E. Schlax, M. W. Capp, and M. T. Record. 2003. Kinetic studies and structural models of the association of E. coli sigma(70) RNA polymerase with the lambda PR promoter: large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 319:649-671. [DOI] [PubMed] [Google Scholar]
- 26.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 44:215-219. [DOI] [PubMed] [Google Scholar]
- 27.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel, R. W., L. Bellon, L. Beigelman, and C. C. Kao. 1998. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 95:11613-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivakumaran, K., M. Hema, and C. C. Kao. 2003. Brome mosaic virus RNA syntheses in vitro and in barley protoplasts. J. Virol. 77:5703-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, J. H., S. Adkins, G. Faurote, and C. Kao. 1996. Initiation of (-)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 31.Ujvari, A., and C. T. Martin. 1996. Thermodynamic measurements of promoter binding by T7 RNA polymerase. Biochemistry 35:14574-14582. [DOI] [PubMed] [Google Scholar]
- 32.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304:147-154. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, K. S., and P. H. Von Hippel. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl. Acad. Sci. USA 92:8793-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, X., C.-H. Kim, and C. Kao. 2003. RNA secondary structure inhibits the initiation of RNA synthesis by a viral RNA replicase. RNA 9:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. In J. Barciszewski and B. F. C. Clark (ed.), Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide in RNA biochemistry and biotechnology, p. 11-43. NATO ASI Series. Kluwer Academic Publishers, Dordrecht, The Netherlands.