Abstract
Small proteins, here defined as proteins of 50 amino acids or less in the absence of processing, have traditionally been overlooked due to challenges in their annotation and biochemical detection. In the past several years however, increasing numbers of small proteins have been identified either through the realization that mutations in “intergenic” regions actually are within unannotated small protein genes, or through the discovery that some small, regulatory RNAs (sRNAs) encode small proteins. These insights together with comparative sequence analysis indicate that tens if not hundreds of small proteins are synthesized in a given organism. This review will summarize what has been learned about the functions of several of these bacterial small proteins, most of which act at the membrane, illustrating the astonishing range of processes in which the small proteins act and pointing to several general conclusions. Important questions for future studies of these overlooked proteins also will be discussed.
Keywords: membrane, cell division, sporulation, transport, signal transduction, sprotein
INTRODUCTION
While the activities and structures of hundreds of thousands of proteins have been studied in exquisite detail, one class of proteins has largely been ignored. These are small proteins: polypeptides that are encoded by small open reading frames. As opposed to “peptides”, which may refer to either intrinsically unordered polypeptides regardless of size, smaller polypeptides that arise from proteolytic processing of a larger precursor (such as leader peptides), or are synthesized by a ribosome-independent mechanism (in vitro, for example), we defined small proteins as those proteins that acquire their diminutive size directly by translation of a small ORF. While defining a size limit for what qualifies as “small” may seem arbitrary, in actuality, the genomic age has already, perhaps unwittingly, defined such a limit. For example, soon after the yeast genome was sequenced, an arbitrary minimum cutoff of 100 codons was applied to annotate putative ORFs as such (1). This was more than simply a matter of convenience: had such a cutoff not been applied, annotating every theoretical ORF between 2 and 99 codons would have resulted in 260,000 additional ORFs (2)! Accordingly, a similar minimum cutoff of 100 codons has since been applied in the annotation of most eukaryotic genome sequences. In bacteria, owing to smaller genome size, the arbitrary cutoff generally is shorter, but nonetheless can lead to the exclusion of bona fide small protein-coding genes. Largely in keeping with these practices, GenBank, the genetic sequence database of the National Institutes of Health, presently does not accept submissions of individual sequences with a length of less than 200 nucleotides, corresponding to about 66 codons (3). Surprisingly, this exclusion even extends to newly discovered, previously unannotated small ORFs that demonstrably produce a protein whose function is experimentally elucidated. Thus, proteins arising from small ORFs that are substantially smaller than 100 codons quite literally tend to be ignored.
In addition to the bioinformatic challenges associated with identifying and cataloging small proteins and small ORFs, the functions of small proteins are often difficult to identify, and as such, small proteins, even those that participate in very well studied pathways, may elude discovery for many years. Mutations of genes encoding most small proteins characterized thus far typically do not result in an obvious phenotype on their own; thus, these genes often are not identified in screens for loss-of-function mutants. Additionally, even if such mutations in “intergenic regions” are successfully isolated, genes encoding small proteins run the risk of being ignored by researchers once the mutation is mapped if the small ORF is not annotated; an issue that is compounded if the ORF initiates with either a GTG or TTG start codon. Complicating the matter further, classical biochemical experiments that aim to identify proteins that co-purify with a molecule of interest typically employ methods that miss small proteins. For example, small proteins that may have co-purified are simply run off the gel and consequently are not detected if the gel system employed is not optimized to detect proteins that are less than ~5 kD in size.
Here, we will highlight the diverse functions of several small proteins in bacteria that were discovered either serendipitously by biochemical or genetic methods; or whose functions were elucidated after their corresponding small ORF was identified using bioinformatic approaches. For brevity's sake, we will limit our discussion to polypeptides that are encoded by small ORFs containing 50 or fewer codons that are found either as part of operons or as stand-alone genes. In an effort to focus on these “ignored” proteins, polypeptides that obtain their small size by proteolytic processing of larger precursors (“peptides”, according to our definition), such as signaling molecules and leader peptides, are excluded from this discussion. We also are omitting the increasing number of identified small ribosomal proteins (4), secreted toxins and small proteins encoded by regulatory 5' leader sequences as well as by toxin-antitoxin systems (reviewed in (5)), by phage genomes (6) or by prophage-like regions of bacterial genomes (7). Despite these arbitrary limitations, not only are many small proteins left to discuss in detail here, but even more have been detected and still await characterization to determine their functions.
EXAMPLES OF SMALL PROTEIN FUNCTION
As we begin our discussion of some of the best-characterized small proteins, what is clear is that they participate in diverse cellular functions ranging from morphogenesis and cell division to transport, enzymatic activities, regulatory networks and stress responses (Figure 1). Small proteins may therefore provide insight not only into how biological functions may be carried out with very few amino acids, but also may be used as tools to probe how their interacting proteins of more standard size participate in various cellular processes.
Figure 1.
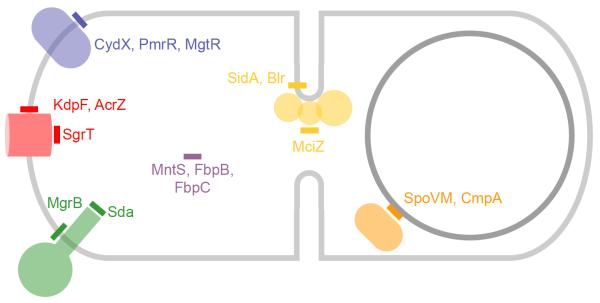
Sites of small protein action. Depicted is the cytosol of a composite bacterium (Gram-positive or negative) bounded by the plasma membrane (light gray). Subcellular locations or proteins associated with various cell functions are colored as follows: kinases, green; transporters, red; membrane-bound enzymes, blue; cell division septum, yellow; forespore during sporulation, orange; soluble chaperones, purple. Small proteins are depicted as rectangles. Transmembrane small proteins are depicted as rectangles that traverse the plasma membrane; amphipathic helical small proteins that are peripherally membrane associated are drawn as rectangles that are parallel to the plane of the membrane; soluble small proteins are shown in the cytosol.
Small proteins that affect spore formation: SpoVM and CmpA
Endospore (hereafter, simply “spore”) formation (“sporulation”) in the gram-positive Bacillus subtilis is triggered when the cell senses the imminent onset of starvation, whereupon the rod-shaped bacterium elaborates a roughly spherical, double membrane-bound organelle called the “forespore” that contains a copy of the genetic material (reviewed in (8)). The outer cell (called the “mother cell”) then deposits a thick protein shell (the “coat”) onto the surface of the forespore and constructs a peptidoglycan shell (the “cortex”) between the two membranes that encapsulates the forespore. Eventually, the forespore core dehydrates and becomes largely metabolically inactive, at which time the mother cell lyses, releasing the now dormant spore into the environment (reviewed in (9)).
SpoVM
Classically, genes in which mutations arrest sporulation at defined morphological stages are named “spo” genes and are readily identified because loss-of-function mutations in them result in a dramatic decrease in the ability of sporulating cells to withstand extreme conditions (high heat or caustic chemicals, for example). This strong phenotype led to the identification of one of the first reported bacterial genes encoding a small protein (10, 11). spoVM is a monocistronic locus that encodes a 26-amino acid protein that is exclusively conserved among endospore-forming species (12). Deletion of the gene results in a six-log decrease in sporulation efficiency, due to massive defects in spore coat and cortex morphogenesis (11). Like many small proteins identified thus far (13), SpoVM is a membrane-associated protein. During sporulation, SpoVM is produced only in the mother cell and localizes on the forespore surface (14). Unlike many other small proteins described below however, SpoVM is not an integral membrane protein. Instead, in the presence of lipid bilayers, SpoVM assumes an α-helical conformation that displays a striking amphipathicity (15), which drives it to orient itself parallel to the plane of the plasma membrane such that its hydrophobic face embeds into the lipid bilayer and its positively charged face is exposed to the mother cell cytosol (16). SpoVM therefore spontaneously inserts into membranes without the need for protein insertion machinery.
What is the basis for the strong sporulation phenotype resulting from the absence of such a small protein? Despite its diminutive size, SpoVM has been reported to perform at least four distinct functions during sporulation. First, SpoVM is among the first coat proteins that localizes to the surface of the developing forespore to mark this membrane as the site for future coat assembly. Recent evidence has suggested that SpoVM preferentially embeds in slightly convex, or positively curved, membranes. Since the surface of the forespore is the only convex membrane surface in the mother cell cytosol (the other surfaces are all concave), this ability to discriminate between degrees of membrane curvature drives the proper localization of SpoVM to its subcellular position (17). Introduction of amino acid substitutions that disrupt the ability of SpoVM to recognize membrane curvature results in the promiscuous localization of SpoVM and, as a consequence, the mis-assembly of the spore coat at incorrect locations in the mother cell cytosol (17).
Second, upon arriving at the forespore surface, SpoVM recruits an ATPase called SpoIVA, which is the structural component of the basement layer of the spore coat (18–20). Interaction with SpoIVA requires at least one amino acid residue on the charged face of SpoVM that faces the mother cell (16). Disruption of this residue abrogates the interaction between SpoVM and SpoIVA and also results in the mis-assembly of the spore coat.
The third function of SpoVM during sporulation is not well understood, but stems from the observation that it is a competitive inhibitor of the membrane-bound protease FtsH (21) and shares a limited amino acid sequence homology with the cIII protein of bacteriophage lambda (22), another small protein that inhibits FtsH in the Gram-negative E. coli (23). While FtsH is required for entry into sporulation (24), the role that SpoVM may play in inhibiting FtsH later during the sporulation program is not known. Nonetheless, the ability of SpoVM to inhibit this protease has been exploited as a tool to understand the role of FtsH during other cellular processes such as biofilm formation (25).
The fourth function of SpoVM concerns a sporulation phenomenon that was first described about forty years ago, wherein it was observed that mutations in B. subtilis which abrogate the initiation of coat (the outer proteinaceous shell surrounding the spore) assembly also abrogate cortex (the inner peptidoglycan shell) assembly, suggesting that the morphogenesis of these two structures separated by a membrane must somehow be linked (26). Of the approximately seventy proteins present in the spore coat of B. subtilis, only SpoVM and SpoIVA were shown to be required for this linkage (11, 18).
CmpA
In an effort to determine how the coordinated assembly of the coat and cortex is achieved, a mutant allele of spoVM was isolated that specifically blocked cortex assembly but allowed normal initiation of coat assembly. A spontaneous suppressor mutation that corrected this sporulation defect was identified in an intergenic region that harbored a previously unannotated small gene subsequently named cmpA (cortex morphogenetic protein A) (27). The mRNA encoding this 26-amino acid small protein was also identified in a global screen for sRNAs that were specifically up-regulated during sporulation (28). Although cmpA is widely conserved in endospore-forming species of the Bacillales order, it is conspicuously absent in members of the Clostridiales order. This observation is consistent with a recent report that coat and cortex assembly likely are not linked in Clostridium difficile (29).
Based on the phenotypes of cmpA single mutants compared to spoVM cmpA double mutants, CmpA is thought to function in a checkpoint that inhibits cortex assembly until coat assembly properly initiates (28). Accordingly, the lack of cmpA results in cells sporulating faster (presumably due to unchecked cortex assembly). Overexpression of the gene impairs cortex assembly and, as a result, reduces sporulation efficiency. Consistent with its cortex-inhibiting function, CmpA-GFP was no longer detected by fluorescence microscopy in those cells that had reached a particular sporulation milestone. This observation suggests that the inhibitory effect of CmpA is eventually overcome in those cells by a post-transcriptional mechanism. Identification of the target of CmpA inhibition (presumably a cortex assembly factor) and factors that participate in the regulatory pathway that ultimately leads to the relief of CmpA inhibition should reveal the mechanism underlying the linked morphogenesis of the coat and cortex during sporulation.
Small proteins that affect cell division: MciZ, SidA and Blr
Cytokinesis in bacteria is dependent on the assembly of the cell division machinery, called the divisome, usually at or near mid-cell. The divisome is composed of approximately ten core proteins that anchor the divisome to the membrane and mediate its constriction (reviewed in (30)). The component of the divisome that actually exerts the force for this constriction is a tubulin homolog called FtsZ that polymerizes into a ring at the division site in a GTP-dependent manner (reviewed in (31)). As the central component of the divisome, FtsZ is a frequent target of positive or negative regulation either by components of the divisome or other species-specific accessory factors, among them small proteins. In this manner, cytokinesis may be carefully regulated both spatially and temporally (reviewed in (32)).
MciZ
At the onset of sporulation in B. subtilis, before the elaboration of an asymmetrically-positioned septum, the FtsZ ring initially assembles at mid-cell, after which time it redeploys to two polar sites at the opposite ends of the cell (33). Only one of the polar FtsZ rings then constricts to form the polar septum (34). Upon elaboration of the polar septum, other cell division events are generally blocked so that the progenitor cell differentiates into two (and only two) dissimilar progeny cells.
In an interaction screen to identify FtsZ-associated proteins, Handler et al. discovered a previously unannotated 40-amino acid small protein they named MciZ (mother cell inhibitor of FtsZ) that was specifically synthesized in the mother cell during sporulation (35). Purified MciZ not only bound directly to FtsZ, but also abrogated polymerization of FtsZ in vitro. Consistent with this observation, overproduction of MciZ in vegetatively growing B. subtilis resulted in cell filamentation that arose from a reduction in the number of FtsZ rings assembled. This cell division defect was suppressed by a single amino acid substitution in FtsZ, near its GTP binding pocket, suggesting that binding of MciZ to FtsZ could directly occlude GTP binding, thereby inhibiting polymerization. A deletion of the mciZ gene revealed the physiological role for this small protein. Whereas vegetative cells harboring a deletion of mciZ did not display an obvious phenotype, sporulating cells continued to form aberrant Z-rings in the mother cell even after the onset of polar septation. Taken together, the data were consistent with a model in which MciZ, by inhibiting FtsZ assembly, prevents additional cell division events after the mother cell commits to the sporulation pathway.
SidA
Although FtsZ, the central component of the divisome, is the most frequently identified target for regulation of cell division, a newly discovered small protein in Caulobacter crescentus inhibits another component of the divisome, FtsW, and prevents membrane constriction after the divisome has already assembled and is poised to constrict. Treatment of C. crescentus with DNA damaging agents results in an arrest in cell division (36), but not in cell elongation. Whole genome microarray analysis revealed that the expression of approximately 5% of the genes in the C. crescentus genome was concomitantly affected, either positively or negatively (37). One of these genes, which the authors renamed sidA (SOS-induced inhibitor of cell division A), was upregulated in response to DNA damage and encoded a putative 29 amino-acid hydrophobic protein containing a predicted transmembrane segment (37). The presence of an upstream LexA binding site suggests that the sidA gene is part of the SOS regulon of C. crescentus.
In a well-studied pathway in E. coli, induction of the SulA protein by DNA damage delays cell division by inhibiting FtsZ polymerization (38). C. crescentus does not harbor a SulA homolog, but the authors demonstrated that SidA could fulfill a similar inhibitory function, as overproduction of SidA during normal growth resulted in cell filamentation. However, unlike SulA, overproduction of SidA did not prevent the localization and assembly of the FtsZ ring at midcell, even though membrane constriction subsequently could not be observed.
A genetic selection revealed that the target of SidA inhibition is likely FtsW, an integral membrane protein that has been implicated to be the lipid II flippase, which transports peptidoglycan precursors from the cytosol into the periplasm (39). Bacterial two-hybrid analysis demonstrated that SidA directly interacts with FtsW (as well as FtsN and indirectly with FtsI, two other late-arriving divisome proteins). In addition, mutations in ftsW that suppressed the SidA overexpression phenotype resulted in FtsW variants that displayed a reduced interaction with the small protein.
One hypothesis that could be drawn from the direct interaction between SidA and FtsW is that SidA disrupts peptidoglycan synthesis at the nascent septum via inhibition of the lipid II flippase activity of FtsW. Surprisingly though, as monitored by fluorescein-labeled vancomycin, the initiation of peptidoglycan synthesis did not appear to be disrupted by SidA overexpression. Moreover, in cells overexpressing SidA, the initiation of membrane invagination at mid-cell also appeared to be normal, even though septum formation did not proceed past this point. Taken together, the data suggest that FtsW may be involved in a previously unappreciated step of cell division that occurs after its peptidoglycan precursor transporting activity and involves the final membrane-remodeling step of cytokinesis. Thus, the discovery of the small protein SidA not only revealed a previously unidentified target for the regulation of cell division, but also implicated that target in a previously undescribed role in cell division.
Blr
A second small protein found to regulate a divisome component other than FtsZ is the 41-amino acid Blr small protein of E. coli. blr was first identified as a stand-alone gene (40) that, when disrupted, caused increased sensitivity to β-lactam antibiotics (β-lactam resistance) (41) and, surprisingly, decreased sensitivity to cell envelope stress (42). Blr was later identified in a bacterial two-hybrid screen (43) for proteins that interacted with FtsL, an integral membrane protein with a very poorly understood function that is nonetheless a core component of the division machinery (30). The two-hybrid assay also suggested that Blr could associate with other components of the cell division machinery, such as FtsI, FtsK, FtsN, FtsQ, and FtsW (43). Consistent with these results, GFP-Blr was found to localize to the division septum of E. coli in a divisome-dependent manner. The authors were unable to identify a significant cell division defect in cells harboring a deletion in the blr gene. However, when combined with a temperature sensitive variant of another cell division protein called FtsQ, which also performs a very poorly understood function during cell division, the cells became elongated when grown in a low osmotic strength medium. This observation suggests that Blr modulates the function of FtsQ during specific growth conditions. Additional characterization of Blr and its interacting partners undoubtedly will provide further insights into small protein action as well as the functions of divisome proteins, similar to the SidA-FtsW example. Future studies of MciZ, SidA and Blr and other divisome regulators also should clarify whether small proteins predominate or whether larger proteins can have similar functions in modulating cell division.
Small protein regulators of transport: KdpF, AcrZ and SgrT
Bacterial cells have a multitude of transporters that allow the import of critical nutrients and the export of detrimental compounds. Several small proteins in E. coli have been found to associate with small molecule transporters with various consequences for activity.
KdpF
The hydrophobic 29-amino acid KdpF protein was one of the first small proteins described to affect a transporter (44). Gaßel et al. noted that there was a small open reading frame initiating with a GTG in the 116 base pair region between the promoter elements of the E. coli kdpABC operon and the first annotated gene in the operon, kdpA (44). The kdpABC mRNA encodes a P-type ATPase high affinity K+ transporter and is induced in response to limiting K+ by the KdpD-KdpE sensor kinase and response regulator encoded downstream (45).
Synthesis of KdpF was confirmed by labeling minicells with 35S methionine, by visualizing the proteins in the purified Kdp complex by silver staining and by purifying the small protein upon extracting membranes with chloroform and methanol. The purified KdpF protein was used to examine its effects on the purified KdpABC complex. Interestingly, the complex purified from a strain lacking kdpF had low activity. A significant increase in activity was observed upon the addition of purified KdpF. The presence of high amounts of E. coli lipids also increased activity though not to the same extent as the addition of KdpF. Together these results led to the conclusion that KdpF helps stabilize the KdpABC complex. Despite the strong effects of KdpF addition in vitro, the lack of kdpF did not obviously affect E. coli growth in low K+, suggesting that KdpF may only be required under specific growth conditions or that there may be redundant mechanisms for stabilizing the KdpABC complex (45).
The kdpFABC genes are also present in Mycobacterium bovis BCG. Bacterial two-hybrid assays conducted with mycobacterial proteins revealed an interaction between KdpF and the sensor kinase KdpD as well as the MmpL7 and MmpL10 proteins involved in lipid synthesis and transport. The significance of these two-hybrid signals is not clear, since high levels of KdpF did not alter the levels of the kdp mRNAs or apolar lipids. However, the overexpression of KdpF led to decreased growth of M. bovis BCG in murine and human macrophages, and cells with high levels of KdpF had altered colony morphology pointing to a physiological role for the small protein (46).
AcrZ
AcrZ, which interacts with the major AcrB-AcrA-TolC efflux pump in E. coli, is another hydrophobic small protein that affects transport (47). This 49 amino acid small protein is encoded by a stand-alone gene near the modEF and modABC operons, which encode proteins involved in molybdenum uptake and in regulating the synthesis of molybdopterin. Despite the synteny with the mod genes across multiple organisms, strains lacking acrZ have not been found to have a phenotype related to molybdenum.
However, expression of acrZ is strongly induced by a range of antibiotics, detergents and oxidizing compounds via the MarA, Rob and SoxS transcription factors (47). Consistent with a role in protecting against deleterious compounds, a functional tagged version of the AcrZ protein (AcrZ-SPA) was found to copurify with AcrB, the inner membrane component of the AcrAB-TolC efflux pump, even in the absence of AcrA and TolC. Bacterial two-hybrid assays further supported an interaction between AcrZ and AcrB as did increased protease sensitivity of AcrB in extracts from cells lacking AcrZ. A dominant negative AcrZ mutant that conferred sensitivity to chloramphenicol was suppressed by a mutation in AcrB, possibly defining the region of interaction between the two proteins. The precise consequences of AcrZ binding to AcrB are not known, but it is noteworthy that ΔacrZ mutants are sensitive to only a subset of compounds to which ΔacrB strains are hypersensitive, both in global phenotypic screens (48) and in individual tests (47). These observations suggested that one function of AcrZ may be to enhance AcrAB-TolC export of certain classes of substrates.
SgrT
The 43-amino acid SgrT protein is encoded on a transcript that was first identified as a sRNA. In fact, the sRNA denoted SgrS binds to the Hfq RNA chaperone protein and has been found to regulate several mRNAs by base pairing, including the ptsG mRNA gene encoding the EIICBGlc glucose transporter of the phosphoenolpyruvate (PEP)-dependent glucose phosphotransferase-system (Glc-PTS) (49). The SgrS transcript is unusually long for an E. coli sRNA and upon further examination of the sequence was found to also encode a small protein whose expression was confirmed by lacZ translational fusions (50). Phenotypic assays showed that SgrT inhibits glucose uptake most likely by inhibiting the activity of the EIICBGlc glucose transporter. Thus the SgrS RNA, whose expression is induced by the SgrR transcription factor in response to glucose-phosphate stress (51), provides an elegant defense against high levels of toxic glucose-6-phosphate: the sRNA basepairs with ptsG to block translation of the EIICBGlc glucose transporter and the encoded small protein SgrT blocks transport by pre-existing EIICBGlc.
Further characterization of the SgrT protein gave results consistent with a direct interaction between EIICBGlc and SgrT (52). The two proteins were found to be crosslinked when cells expressing both C-terminally tagged proteins (EIICBGlu-His5 and SgrT-3HA) from plasmids in a deletion background were treated with paraformaldehyde. These experiments also revealed that SgrT has a preference for the dephosphorylated form of EIICBGlc prominent during glucose uptake. Additional evidence for an interaction between the two proteins came from a bimolecular fluorescence complementation assay in which both proteins were fused to one half of a green fluorescent protein (GFP). This assay was used to delineate the regions of EIICBGlc required for the interaction with SgrT. The results of these assays together with co-purification studies of tagged EIICBGlc carrying different amino acid substitutions suggested that SgrT may interact with a conserved KTPGRED motif present in the glucose transporter but not the transporters of other sugars. The SgrT protein does not have an obvious α-helical transmembrane domain but fluorescence microscopy with SgrT tagged with GFP (SgrT-GFP) indicated that SgrT is localized to the membrane upon the interaction with EIICBGlc.
Small protein regulators of membrane-bound enzymes: CydX, PmrR and MgtR
Some of the very first small proteins to be identified are those associated with photosystem I, photosystem II and cytochrome complexes in cyanobacteria (reviewed in (53–55)). In all cases, the interaction of these single transmembrane domain small proteins with these large complexes has been confirmed by crystallographic studies, although the exact role of each protein is less clear. A survey of the phenotypes associated with the lack of the Psb small protein components of photosystem II proposes that the majority are involved in stabilization, assembly or dimerization of the complex (reviewed in (53)). Similarly, a study of the PetG, PetL and PetN proteins of Synechocystis suggested that PetG and PetN stabilize the cytochrome b6f complex (54). Thus the functions of these large complex associated proteins may be similar to what has been proposed for KdpF.
CydX
In recent studies of the 37-amino acid CydX protein encoded downstream of the CydA and CydB subunits of the cytochrome bd oxidase in E. coli, it was found that a deletion of the cydX gene gave rise to the same phenotypes as cydA or cydB deletions, such as slow growth under aerobic conditions and sensitivity to the reducing agent β-mercaptoethanol (56). Direct measurements of oxidase activity in E. coli also showed that activity is reduced in the absence of CydX. The corresponding protein encoded adjacent to the Brucella abortus cydAB genes is slightly longer than E. coli CydX at 51 amino acids, but again strains lacking this protein have defects consistent with a lack of cytochrome bd oxidase activity as well as impaired growth in macrophages (57).
Co-purification studies with functional tagged derivatives of E. coli CydX (CydX-SPA) confirmed that the small protein tightly associates with tagged CydA (CydA-His6) and CydB (CydB-His6) (56). The AppX homolog encoded by the appABX operon for the anaerobically-induced cytochrome bd oxidase also associated with CydA, albeit to a lower extent than CydX. The presence of a cysteine residue in the predicted transmembrane domain of CydX led to the attractive hypothesis that the cysteine might coordinate a heme in the CydABX complex, but this was not supported by mutational studies. In fact, these studies revealed that substitution of only four residues of the 15 tested prevented full complementation of the β-mercaptoethanol sensitivity phenotype associated with the cydX deletion.
PmrR
The hydrophobic 29-amino acid PmrR protein in Salmonella was discovered by bioinformatics, which revealed a small ORF preceded by a binding site for the PmrA response regulator, both encoded downstream and on the opposite strand of the known PmrA-regulated pmrCAB operon (58, 59). Transcript mapping confirmed the presence of a PmrA-dependent transcript, and a tagged PmrR protein (PmrR-FLAG) that co-fractionated with the inner membrane was detected by immunoblot analysis. Given the roles of the PmrB sensor kinase and PmrA response regulator in controlling genes the mediate the modification of the Salmonella lidopolysaccharide (LPS) and the unexplained observation that a PmrA-regulated gene inhibited LpxT-mediated synthesis of diphosphorylated lipid A (60), Kato et al. tested for an interaction between PmrR and LpxT. The predicted interaction was confirmed in both two-hybrid assays and co-purification of tagged PmrR (FLAG-His6-PmrR) and tagged LpxT (LpxT-LacZ). Overexpression of PmrR and decreased expression of the small protein also had the expected effects on decreasing and increasing diphosphorylated lipid A levels, respectively.
These experiments raised the broader question of the physiological consequences of different levels of diphosphorylated lipid A, which increases the overall negative charge of the LPS (58). Assays of Fe3+ binding revealed that increased PmrR levels (which resulted in decreased diphosphorylated lipid A synthesis and thus an increased positive charge of the LPS) decreased the binding of Fe3+ to the LPS. Since Fe3+ activates the PmrB response regulator, elevated PmrR and decreased Fe3+ binding also had the consequence of decreased activation of PmrB-PmrA target genes. These observations led to the proposal of a negative feedback loop wherein PmrB-PmrA are activated by high external Fe3+ and induce the expression of PmrR, which inhibits LpxT, leading to a change in LPS charge. The resulting decrease in negative charge in turn results in lower Fe3+ binding and finally decreased PmrB activation.
MgtR
The PhoQ sensor kinase and PhoP response regulator have been found to have a critical role in pathogenesis in Salmonella species and control a large regulon in response to low Mg2+, acidic pH and antimicrobial peptides. Expression of at least three small hydrophobic proteins—the 30-amino acid MgtR protein (61), the 47-amino acid MgrB protein (62) and the 31-amino acid YneM protein (63)—is controlled by the PhoQ-PhoP system.
The first of these small proteins, MgtR, was found to modulate the stability of the MgtC virulence factor in Salmonella typhimurium (61). The discovery of MgtR came from studies that followed up the observation that although the levels of the mgtCB transcript are high in Mg2+-depleted medium, the MgtC protein is barely detectable (64). By mapping portions of the mgtCB transcript associated with the instability, Alix and Blanc-Potard found that a region downstream of mgtB is involved and noted that this region could encode a 30-amino acid protein (61). Mutations that correlated MgtR coding potential with MgtC instability provided evidence that the small protein was synthesized. Alix and Blanc-Potard also were able to detect a functional tagged MgtR derivative (His6-MgtR).
Western blot analysis of MgtC levels showed that MgtR affected the stability of MgtC in a manner that was dependent on FtsH, the membrane-bound protease involved in the degradation of membrane proteins. A direct interaction between MgtR and MgtC was supported by bacterial two-hybrid assays. Intriguingly, substitution of only two of 11 residues in plasmid-expressed MgtR had effects on MgtC levels. In MgtC, mutations in the cytoplasmic loop between the third and fourth transmembrane domains confer resistance to MgtR, possibly defining the region of interaction between the two proteins.
Although MgtC was long known to be a virulence determinant, the activity of this protein was uncovered only recently (65). Co-purification experiments after crosslinking revealed that tagged MgtC (MgtC-FLAG) associated with FtsH and MgtR (as expected from the results above), as well as the Foa subunit of the F1Fo ATP synthase. Lee et al. went on to show that the MgtC interaction with the Foa subunit inhibits ATP synthesis and ATP-driven proton translocation and hypothesized that the MgtC-mediated inhibition of the F1Fo ATP synthase could protect the bacteria against acidification that occurs inside macrophages.
In another recent study, MgtR also was shown to post-transcriptionally affect the levels of the MgtA manganese transporter, as well as interact with MgtA in a two-hybrid assay (66). The interplay between the small protein MgtR with MgtA, MgtC, FtsH and the F1Fo ATP synthase is not completely understood but could constitute a type of feedback loop.
Small protein regulators of protein kinases and signal transduction: MgrB and Sda
The frequent localization of small proteins at the inner membrane makes these proteins candidates for regulating membrane-localized sensor kinases and impacting signal transduction.
MgrB
The second of the hydrophobic, PhoQ-PhoP-regulated small proteins, MgrB, was discovered to negatively regulate the PhoQ sensor kinase in E. coli (67). Since PhoQ-PhoP were known to strongly regulate expression of mgrB itself (62), Lippa and Goulian monitored the effects of deleting PhoP target genes in a strain background in which the yfp reporter was fused to the mgrB promoter (67). In this assay the ΔmgrB mutant showed a striking increase in YFP activity as monitored by colony color on plates in low and high magnesium. On the other hand, overexpression of E. coli MgrB as well as homologs from Salmonella and Yersina, led to strong repression of the mgrB-yfp fusion. The hydrophobic nature of the protein prompted the authors to examine subcellular localization. Both cell fractionation followed by immunoblot analysis with antibodies raised against a C-terminal peptide of MgrB as well as localization of a GFP-tagged derivative of MgrB (GFP-MgrB) gave signals consistent with membrane localization.
The mgrB deletion and overexpression phenotypes led to the prediction that the small protein might be interacting with the PhoQ sensor kinase, an expectation that was supported by two-hybrid analysis (67). The binding and repression of PhoQ resulted in a negative feedback loop wherein PhoQ-PhoP activated expression of MgrB, which in turn repressed PhoQ. Since the kinetics of reporter induction were not found to be altered, it was proposed that the MgrB-mediated feedback inhibition could be a mechanism to integrate additional environmental signals into the PhoQ-PhoP circuit. Consistent with this model, MgrB was found to be required for the repressive effects of DsbA (68, 69), a periplasmic disulfide oxidase, on the induction of PhoQ-PhoP regulated genes. Interestingly, MgrB contains three conserved cysteines; C16 predicted to be in the transmembrane domain and C28 and C39 predicted to be in the periplasm. Future studies to determine the redox status of the cysteines under different stress conditions and to examine how oxidation and reduction influence MgrB binding to PhoQ should further elucidate the role of MgrB in modulating the PhoQ-PhoP response.
Sda
The cytosolic 46-amino acid Sda protein, one of the best characterized small proteins, similarly inhibits the first kinase in the histidine kinase phosphorelay that regulates sporulation-specific genes in B. subtilis. When cells encounter starvation and stress, the histidine kinases KinA and KinB are activated and autophosphorylate; phosphates from these kinases are transferred to Spo0F, then to Spo0B, and finally to the transcription regulator Spo0A. An advantage of such a phosphorelay is that the drastic step to initiate sporulation can be modulated at multiple steps in response to a range of environmental signals. A mutant allele of dnaA was previously shown to block replication initiation and, as a result, blocked the entry into sporulation. The sda gene (suppressor of dnaA) was identified in a screen for mutations that bypassed this block in sporulation. (70). The sda promoter has multiple DnaA binding sites and expression of a sda-lacZ fusion was found to be induced by various mutants than affect replication, most likely via DnaA. In vitro assays with purified tagged derivatives of soluble KinA (KinA-His6) and Sda (Sda-His6) revealed that Sda inhibits the kinase activity of KinA. Burkholder et al. also suggested that Sda could inhibit KinB, but were unable to directly test this hypothesis in vitro given difficulty in purifying the integral membrane kinase. They proposed that Sda serves as a checkpoint to inhibit the KinA/KinB-Spo0F-Spo0B-Spo0A phosphorelay and thus sporulation if DNA cannot be replicated properly.
The interaction between Sda and the KinA kinase has been examined in detail. The site of Sda binding to KinA was mapped to the KinA dimerization/phosphotransfer (DHp) domain, first by the determination of the Sda NMR structure and mutational studies (71) and later by small-angle X-ray scattering (SAXS) and neutron contrast variation studies on the Sda-KinA complex (72). It was first proposed that Sda only acts by blocking an interaction between the catalytic and the DHp domains of KinA (71). However, later studies led to the conclusion that Sda inhibits KinA by inducing a conformational change via the DHp domain (72). It is quite possible that the Sda interaction with KinA has multiple consequences. One of the latest studies showed that Sda also interferes with the phosphotransfer from KinA to Spo0F since the Sda and Spo0F binding sites on KinA overlap (73). An X-ray structure of the Geobacillus stearothermophilus Sda protein in complex with the cytoplasmic portion of KinB comprising the catalytic and the DHp domain, supported the suggestion that Sda also inhibits KinB and showed that Sda again blocks the interaction between the catalytic and the DHp domains and the interaction with Spo0F (74). These extensive structural studies together with an X-ray structure of B. subtilis Sda (75), illustrate how a protein of only 46 amino acids and only two α-helices nevertheless can significantly impact interactions between proteins or between domains of a single protein.
Small protein chaperones: MntS, FbpB and FbpC
Several well-characterized protein, nucleic acid, and metal chaperone proteins are of relatively low molecular weight (reviewed in (76, 77)). Thus perhaps it is not surprising that some of the bacterial small proteins of even lower molecular weight also appear to have chaperone function.
MntS
Another example of a small protein discovered as a consequence of characterizing a transcript first annotated as a sRNA in E. coli is MntS. This small ORF, which was recognized on the basis of sequence conservation, was reported to encode a 42 amino acid small protein initiating with an ATG codon (78) but upon further inspection may instead encode a 33-amino acid small protein initiating with an GTG codon (L.S. Waters, personal communication). Clues to MntS function came from the information about its expression. The mntS gene is adjacent to the gene encoding the manganese-responsive MntR transcription repressor and synthesis of a tagged MntS derivative (MntS-SPA) is strongly repressed by high manganese. Consistent with its regulation, overexpression of MntS resulted in severe sensitivity to manganese but not high levels of other metals such as iron, nickel or copper. This observation, together with the finding that another target of MntR is abnormally regulated in an mntS deletion strain, led to the suggestion that MntS functions as a chaperone to deliver manganese to appropriate proteins or locations in the cell. This model is supported by experiments showing that the activities of the manganese-dependent superoxide dismutase and ribonucleotide reductase enzymes are reduced in E. coli mntS mutants (J. Martin, L.S. Waters and J.A. Imlay, personal communication).
FbpB and FbpC
Two other small proteins whose expression is regulated by the presence of metals, in this case iron, are the 48-amino acid FbpB (Fur-regulated basic protein) and the 29-amino acid FbpC proteins of B. subtilis (79). The transcript encoding FbpC was the first identified member of the ferric uptake repressor (Fur) regulon (80). The low-iron induced mRNA encoding FbpB (initially annotated as a 59-amino acid protein) and the slightly larger 54-amino acid FbpA protein was identified in a genome-wide screen for Fur-repressed genes (81). All three of these basic proteins can be detected as FLAG-tagged derivatives. Given that repression of some operons by the regulatory sRNA FsrA requires one or the other of these small proteins, it was proposed that the basic small proteins act as chaperones to facilitate sRNA function (81), but the reasons for the observed differential effects on target operons, binding partners for the three proteins, as well as their exact functions are unknown.
FUNDAMENTAL QUESTIONS
The characterization of an increasing number of small proteins with diverse physiological roles allows some reflection on general principles regarding the discovery and characterization of small proteins, their mechanisms of action, synthesis, and degradation as well as distribution and evolution. At the same time, these studies raise interesting and important general questions that can serve to guide future studies.
Further identification of small proteins and the elucidation of their functions
The full complement of small proteins is not yet known for any organism. A review of how the small proteins described above were found reveals that many were discovered by serendipity: some as mutations in unannotated regions of the chromosome, others by further inspection of potential promoters and short transcripts. There have been a limited number of computational screens based on comparative genomics and the identification of ribosome binding site sequences designed to specifically detect bacterial small proteins (for example (13, 59, 82, 83). However, a general drawback of computational screens is the limited information content of a small protein-coding gene, particularly if it encodes a hydrophobic small protein. This limitation has led to both the under- and the over-annotation of small protein genes in completed genome sequences (84). Thus an essential component of small protein identification is independent data such as direct detection or mutational analysis demonstrating that the small protein is synthesized.
The newly developed technique of ribosome profiling to identify the positions of ribosome occupancy on a transcriptome-wide level is leading to the identification of ribosome binding signatures on small ORFs (85). Whether all of these signals will be correlated with the existence of small proteins is not yet clear. Again, the onus will be to obtain independent data supporting the synthesis of the small proteins. As more small proteins are characterized, it is possible that general features that are uncovered can provide clues that lead to the productive identification of this family of proteins. For example, the hydrophobic nature of the majority of the small proteins studied thus far suggests that membrane extraction, for example by methanol and chloroform as was carried out for KpdF (44), may be a fruitful avenue to pursue. We envision that increased awareness of important roles for small proteins also will increase the scrutiny of mutations in unannotated regions of the chromosome as well as faint bands at the bottom of protein gels. A consequence of the identification of new small proteins is the need to revisit genome annotation of genome sequences, likely by both manual and improved in silico methods, to fill in the missing genes. At this point, we are still left with the exciting question of the true extent of the small proteome.
As the synthesis of new small proteins is confirmed, there will be the challenge of elucidating their functions. This challenge is not substantially different from the problem of characterizing larger proteins of unknown function, but has the added complication that current biochemical assays are biased against small proteins. For example, it is difficult to raise antibodies against a 30 amino acid hydrophobic protein; the two reported antibodies against mature small proteins (α-SgrT and α-MgrB), have only been shown to detect protein overproduced from plasmids (67, 86). With the exception of KdpF and the subunits of the cytochrome bc1 and cytochrome oxidase complexes, all interactions between all of the small proteins described and their interacting protein partners were examined using tagged derivatives of the small proteins. The majority of these tags are larger than the size of the small protein itself. While, in many examples, the tagged protein was confirmed to complement a specific phenotype, this may not be the case for all small proteins and caution regarding unanticipated effects of the tags is warranted. It is worth noting that the identification of interactions in two-hybrid assays was a recurrent theme suggesting that this line of investigation may be productive for the characterization of other small proteins.
The approach of examining the phenotypes of overproducing the small protein and deleting the corresponding gene also has given useful insights for many of the small proteins. None of the small proteins have been found to be essential for viability, and only a subset of the null mutants gave strong phenotypes. Possibly, some small proteins may have partially redundant functions as found for CydX and AppX (56). Alternatively, small proteins may generally be acting in regulatory roles as seen for PmrR (58), MgrB (67) and Sda (70). This would be akin to regulatory sRNAs, which, despite having significant impacts on bacterial cell physiology, also are not essential (reviewed in (87)).
Mechanisms of action
As increasing numbers of small proteins are characterized, we are learning more about their mechanisms of action. By definition, the small size of the proteins limits the number of possible three-dimensional structures into which the proteins can fold and also the number of activities the proteins can have. Accordingly, an α-helical structure appears to be the common inferred structural motif amongst the transmembrane and amphipathic small proteins discussed above. The set of small proteins studied thus far also suggests that it is unlikely that many small proteins will possess enzymatic functions. Instead, as illustrated by the proteins discussed, this class of proteins appears to act in more mechanical ways. They can serve as inhibitors by directly blocking a domain of a target protein as is suggested for MciZ (35) and MgrB (67) or blocking interactions between domains or interactions between proteins as suggested for Sda (73, 74). Alternatively, a small protein might facilitate interactions between domains within or between proteins as well as between proteins and other molecules as suggested for MgtR (61) and MntS (78). An interaction with a small protein also may bring about a conformational change as has been suggested for the binding of Sda to the KinA kinase (72). Finally, small proteins could provide a membrane anchor as found for SpoVM (17).
An important direction for further studies of small proteins will be the elucidation of their structures, particularly in the context of the interacting larger proteins. These types of studies together with mutational analyses to define critical amino acids will help to delineate how small proteins interact with their respective partners. In this context it is interesting that surprisingly few of the mutations isolated thus far have dramatic effects on small protein activity. With only a limited number of amino acids, an initial assumption might be that most of the residues are critical, but this has not been borne out by many of the results obtained thus far. Of course, more systematic scanning mutagenesis of small proteins may reveal residues that are more sensitive to substitutions.
Many of the small toxic proteins synthesized as part of type I toxin-antitoxin systems (reviewed in (5)) or serving as a defense against host cells (reviewed in (88)) are of similar length and hydrophobicity as the transmembrane small proteins we described, yet the small proteins we discussed are not toxic or secreted. It is unclear which residues define the differences in the modes of action. Interestingly, ComI, a member of the TxpA family of toxins (89) encoded on a large plasmid in undomesticated strains of B. subtilis, has recently been found to perform the additional cellular function of inhibiting competence (90) suggesting that the distinctions between the different types of small proteins might in fact be blurred.
A related question is whether there are any lower limits to the size of a functional small protein. The average length of a transmembrane helix is approximately 26 ± 5 residues, but transmembrane segments of as few as 9 residues are possible (91). Other questions that remain to be addressed are whether small proteins form oligomeric complexes, whether one small protein is capable of interacting with multiple larger proteins and whether larger proteins can be the target of a set of small proteins expressed under different conditions. In this context it is worth noting that, in E. coli, the slightly larger 72-amino acid, hydrophobic YmgF protein has been shown to interact with some of the same divisome proteins as Blr (92); the 65-amino acid SafA (B1500), like MgrB, interacts with PhoQ (93); and the 110-amino acid single-transmembrane YajC protein was reported to co-crystallize with AcrB (94).
We assume that small proteins provide advantages not afforded by larger proteins. For example, small proteins may be able to assume functions directly after synthesis without the requirement for folding chaperones. A surprisingly large percentage of the described small proteins have been found localized at or in the membrane. Hydrophobic small proteins that anchor larger proteins to the membrane separate the localization function of the target protein from its activity, thereby providing another level of regulation. Small proteins also can fine-tune an activity absent the synthesis or degradation of an entire enzyme complex. This may be particularly important for membrane complexes given that the membrane may limit the number of ways the complexes can be regulated.
Synthesis, subcellular localization and degradation
The small sizes of small proteins raise interesting fundamental questions about the synthesis, subcellular localization, and degradation of this class of proteins. The first question is whether the small size of the ORF exerts any unusual demands on translation, particularly since many of the small proteins are hydrophobic. Do RNA secondary structures, alternative start codons, stop codons and frameshift mutations have the same effects on small proteins as on larger proteins? Are there any impediments to releasing small proteins that might not be much longer than the ribosome exit channel? In this context it is notable that the hydrophobic sequences of several of the small proteins are not very different from the hydrophobic sequences of small ORFs found in the 5' leaders of antibiotic resistance operons. These 5' leader-encoded peptides regulate the expression of the downstream operons by interacting with the exit channel to lead to the formation of a stalled ribosome in the presence of antibiotics (reviewed in (95)).
Similarly, how are small transmembrane proteins, themselves approximately the size of a signal sequence, inserted into the membrane? Studies to examine the subcellular localization of small proteins are still in their infancy, but initial experiments indicate that small proteins display diversity in topology and membrane insertion pathways (96). For example, the tagging of single transmembrane domain small proteins in E. coli with proteins which are only active in either the cytoplasm (GFP) or the periplasm (alkaline phosphatase), revealed some small proteins have an Nin-Cout and others an Nout-Cin orientation, while a few even have dual topology. Mutational analysis of fusions to one of the small proteins showed that positive residues adjacent to the transmembrane domain impact topology, similar to what has been found for polytopic membrane proteins. A potential problem with these experiments though is that the tags employed could impact the orientation of small proteins more than they do for larger proteins. CydX was reported to have an Nout-Cin orientation in the E. coli study but an Nin-Cout orientation in a B. abortus study using a different tag (β-lactamase) (57). Since several relatively small bacteriophage proteins require the YidC protein for membrane insertion (reviewed in (97)), it was assumed that insertion of the E. coli small proteins might also be dependent on YidC. However, the depletion of the essential YidC protein as well as the essential SecE component of the Sec translocase again revealed diversity; some small proteins were affected by the depletion of both proteins, others by the depletion of only one or the other and some were not affected by either depletion (96). Finally, higher resolution imaging is needed to determine if small proteins generally are inserted into membranes uniformly or in punctate patterns that may indicate a preference for particular lipid microdomains or rafts. Overall the initial studies revealed that much remains to be learned about small protein insertion into membranes.
Given the hypothesis that small proteins most frequently act as regulators that modulate processes or act under specific conditions, one must also assume that there are pathways to specifically degrade the small proteins, but this has not been explored. In addition, the possibility of post-translational modifications to small proteins needs to be considered.
Evolution
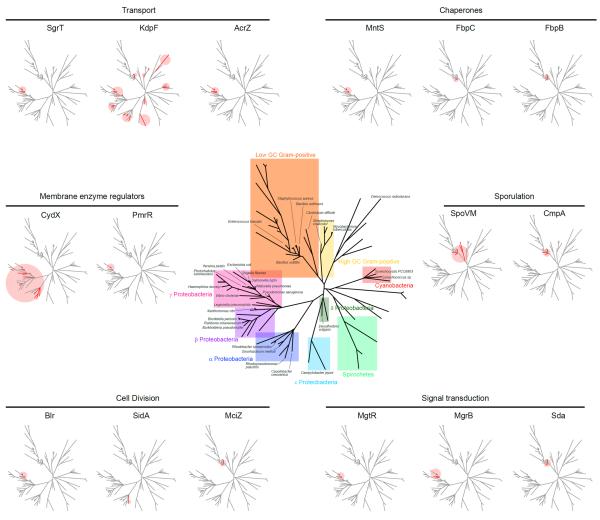
Other fundamental questions relate to the distribution and evolution of small proteins. The short length of the coding sequences and the fact that many of the small proteins are comprised of hydrophobic amino acids such as leucine, isoleucine and valine coupled with the uneven annotation of small protein genes in completed genome sequences, provide formidable challenges to identifying small protein orthologs. In one study, linkage to the adjacent SgrR protein coding gene allowed the recognition of SgrT orthologs in other enteric bacteria (98), but conservation of synteny has not been established for many of the small protein genes. Despite the caveats associated with the identification of orthologs, the phylogenetic distribution of the small proteins described in this study is shown in Figure 2. While a few of the small proteins, particularly those encoded in operons with larger proteins, spans more than one phylogenetic class such as the α, β and γ proteobacteria, most are conserved in only a limited number of organisms.
Figure 2.
Phylogenetic distribution of small proteins. Unrooted prokaryotic phylogenetic tree based on 16S rRNA sequences, adapted from Nelson et al. (106). Center: selected major phylogenetic groupings are colored and selected organisms are indicated. Phylogenetic distribution of small proteins discussed in this review are grouped according to biological function and highlighted in red on individual trees shown at lower magnification. Homologs were identified by a PSI_BLAST search of 2262 completely sequenced genomes (as of February 2013). A table summarizing the presence of absence of particular small proteins is given as Supplemental Table 1. This analysis illustrated the disparate nature of small protein annotation; for example, the pmrR gene is annotated in only six out of the 95 genomes in which homologs were found. The alignments of annotated small proteins generated by MUSCLE are given in Supplemental Table 2 and are available in FASTA format at ftp://ftp.ncbi.nih.gov/pub/wolf/_suppl/small.
As for all proteins, the small protein distribution has implications for evolution. Did these small genes evolve independently or are they fragments of genes that originally encoded larger proteins; for example, larger proteins with which the small proteins interact? Conversely, do small protein genes serve as building blocks for the evolution of genes encoding larger proteins? Again, due to the low information content of small proteins, it may be difficult to establish clear evolutionary relationships between small proteins and the rest of the proteome. In one study in which translation, conservation and other features of small ORFs in “non-genic” sequences of Saccharomyces cerevisiae were examined, Carvunis et al. noted that less conserved ORFs in S. cerevisiae had higher hydropathicity and a higher tendency to form transmembrane regions and concluded that many of these small ORFs are “proto-genes” that serve as a reservoir for the “birth” of new genes (99). Larger proteins are less affected by thermal noise due to the energy of interactions between the residues in the polypeptide chain; as a result, small proteins may be unordered on their own and therefore may depend on an external factor (another protein or a phospholipid bilayer, for example) to obtain a stable conformation. Such a dependency may explain the skewed subcellular distribution of small proteins, either as components of larger complexes or as membrane-associated protein. Determination of whether these are general evolutionary constraints will require further structural studies and larger genomic data sets.
BROADER VIEW
The focus of this review has been on the small proteins encoded by specific genes on bacterial chromosomes, but it is likely that what has been learned can inform our understanding of the many potential small proteins encoded by bacteriophage and viruses as well as by archaeal and eukaryotic genomes. In addition, the small size of the gene products makes them attractive candidates for biotechnological applications.
Small proteins encoded by bacteriophage
We suggest that bacteriophage may be an ideal system in which to study the presence and functions of small proteins given the much smaller viral genome sizes and the enormous evolutionary timescale that allowed the streamlining of phage proteins to maximize efficiency. Recently, a ribosome profiling study of bacteriophage lambda revealed that 55 potential ORFs of at least 5 codons in length, all previously unannotated, show evidence of translation (6). Although many of these ORFs had ribosome binding at levels comparable to, or higher than, well-studied lambda genes, they probably remained undiscovered because they did not display obvious deletion phenotypes under routine laboratory conditions.
One well-characterized small protein of bacteriophage lambda is the 22 amino acid-long product of the cIII gene, which harbors some sequence similarity to the SpoVM small protein of B. subtilis discussed above. Like SpoVM, cIII is a membrane-associated amphipathic α-helix and is both a substrate and inhibitor of the membrane bound protease FtsH (22). Kobiler et al. demonstrated that cIII oligomerizes and competitively inhibits FtsH from degrading lambda cII, the transcription factor that mediates lambda lysogeny. We speculate that many more small bacteriophage proteins that interact with and modulate larger bacterial proteins will be found.
Small proteins synthesized by eukaryotes
Similar to efforts to study small proteins in bacteria, efforts directed at identifying small ORFs in eukaryotes are also beginning to reveal potentially thousands of small proteins in these more complex organisms (reviewed in (100)). Indeed, there is a growing realization that previously discovered regulatory or “noncoding” RNAs actually harbor small ORFs that are translated. For example, Hanada et al. employed a bioinformatic approach to identify small ORFs with a high probability of encoding a protein by exploiting differences in codon usage between coding and non-coding DNA sequences in the Arabidopsis thaliana genome (101). This analysis led to 7,901 candidate small ORFs, all previously unannotated, located in intergenic regions that the authors deemed highly likely to produce a small protein. An array analysis led to the observation that over 2,000 of these candidates were highly expressed at the mRNA level during at least one of many experimental conditions. In separate experiments, overexpression of 49 of these genes that were highly conserved across other plant species resulted in obvious, discernable phenotypes in A. thaliana. However, a systematic analysis to confirm that the observed overexpression phenotypes are specifically related to the over-production of an encoded small protein, rather than simply the putative mRNA remains to be done.
More detailed studies of small proteins have recently been carried out in Drosophila, when it was realized that some transcripts annotated as “non-coding” RNAs actually encode small proteins (102–104). For example, an RNA denoted pri (or tal) was found to encoded four small proteins ranging in size from 11 to 32 amino acids (102, 103). Deletion of pri resulted in defects in epidermal differentiation, and reduction in pri levels led to defects in leg morphogenesis. Flies lacking a gene called svb, which encodes a transcription factor, showed similar defects. Although deletion of pri did not reduce the expression of svb, it did prevent a normal switch in the localization pattern of Svb protein in the nucleus, which correlated with a switch in the activity of Svb from a transcriptional repressor to a transcriptional activator (105). Induced expression of pri was sufficient to elicit this switch in Svb localization. In addition, the expression of pri resulted in a shift in the electrophoretic mobility of Svb that corresponded to the removal of its repressor domain, but not its activator domain. Taken together, Kondo et al. concluded that the Pri small proteins mediate the processing of the Svb transcription factor to provide temporal regulation of Svb activity, first as a repressor, then as an activator. It will be interesting to see how the Pri small proteins mediate Svb processing since it is unlikely the small proteins harbor proteolytic activity. If the precedents from bacterial studies apply, the Pri proteins could be promoting interactions with factors that are responsible for the processing.
Potential for exploiting small proteins
Given the low molecular weight of small proteins and their propensity to modulate the activities of proteins with which they interact, one intriguing possibility is that small proteins may be used as externally added agents, analogous to the way that small molecules are currently employed, either as probes or to affect specific processes. SpoVM, when added in culture media, was able to inhibit the FtsH protease and prevent biofilm formation by B. subtilis (25) providing preliminary evidence that this approach could be fruitful. Thus, for example, it might be possible to exploit the interactions of small proteins with efflux pumps and components of the cell division machinery as genetic tools to probe the active sites of these targets in an effort to design antimicrobial agents that block either drug efflux or cell division.
CONCLUSIONS
We suggest that the study of small proteins is a field that is poised to explode. The characterization of a subset of small proteins in bacteria has shown that they are present, particularly in membranes. Furthermore, their synthesis is regulated and they impact diverse processes ranging from spore formation and cell division to the movement of molecules across the membrane, enzymatic activites and signal transduction. Thus, small proteins should not be overlooked in any organism. Most small proteins probably will act mechanically to block protein domains or block or facilitate interactions between domains or membranes, but it is likely there are hundreds of these proteins. In addition to the identification and characterization of small proteins, there are numerous fundamental questions about the nature of small protein interactions with other molecules, their synthesis and degradation as well as their evolution that remain to be answered not only in bacteria but in viruses and eukaryotes as well. We look forward to the future exploration and exploitation of the ignored proteome.
Supplementary Material
SUMMARY POINTS.
Small proteins in bacteria, here defined as polypeptides of 50 or fewer amino acids, have been ignored because they are difficult to detect using routine biochemical methods and the corresponding genes historically are not annotated in sequenced genomes and are overlooked in classical genetic screens.
Several small proteins discovered in bacteria are beginning to be characterized. These small proteins participate in morphogenesis, cell division, DNA uptake, ion transport and efflux of macromolecules, regulation of enzyme activity, signal transduction, and can function as chaperones.
Most bacterial small proteins characterized thus far are membrane-associated, and none to date have been reported to harbor an enzymatic activity. Rather, the small proteins either modulate the enzymatic activity of larger proteins, positively or negatively, via a direct interaction; or play a structural role in stabilizing large complexes or anchoring larger soluble proteins to the membrane.
Multi-pronged strategies, including enhanced bioinformatics, twists on classical biochemical techniques like ribosome profiling, and other directed global screens should lead to the discovery of previously unappreciated small proteins in bacterial, eukaryotic, archaeal and viral genomes.
FUTURE ISSUES.
What is the full complement of small proteins for a single organism?
What other common features and mechanisms of action will be discerned for small proteins?
How are small proteins folded and how do they interact with other molecules?
What is the smallest size for a functional small protein?
How do small proteins evolve?
ACKNOWLEDGMENTS
We thank current and former members of the Storz lab, S. Gottesman, K. Makarova, M. Laub, M. Goulian, E. Groisman, C. Vanderpool and A. Blanc-Potard for helpful discussions and comments on the review. This work was funded by the Intramural Research Program of the NIH, Eunice Kennedy Shriver National Institute of Child Health and Human Development (G.S.), National Center for Biotechnology Information, National Library of Medicine (Y.I.W.), and National Cancer Institute, Center for Cancer Research (K.S.R.).
Mini-glossary
- Small protein
experimentally-detected short polypeptide, here defined as less than 50 amino acids in length.
- Small ORF
DNA sequence with the potential to encode a small protein
- Sporulation
a developmental program in some Gram-positive bacteria in which a cell differentiates into a hardy, dormant cell called an endospore (“spore”).
- Spore coat
complex proteinaceous shell surrounding bacterial endospores.
- Spore cortex
an inner shell, made of peptidoglycan, surrounding bacterial endospores. Along with the coat, it confers resistance to the spore against environmental insults.
- FtsZ
a tubulin homolog found in most bacteria that is responsible for membrane constriction during cell division.
- Divisome
the name given to the bacterial cell division machinery, composed of approximately ten core proteins.
- SOS response
bacterial response to DNA damage in which genes required for cell cycle arrest and DNA repair are induced.
- Competence
the ability to uptake exogenous DNA by bacteria.
- Sensor kinase
in bacterial two component signal transduction systems, the protein that serves as a phosphor-donor to a response regulator.
- Response regulator
in bacterial two component signal transduction systems, the protein (usually a transcription factor) that receives the phosphoryl group from the sensor kinase.
- Ribosome profiling
Global method to determine mRNA sites bound by a ribosome at any given time.
Acronyms
- sRNA
small, regulatory RNA.
- GFP
green fluorescent protein.
- ORF
open reading frame
- LPS
lipopolysaccharide
LITERATURE CITED
- 1.Harrison PM, Kumar A, Lang N, Snyder M, Gerstein M. A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 2002;30:1083–90. doi: 10.1093/nar/30.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7:768–71. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- 3.National Center for Biotechnology . The GenBank Submissions Handbook. Bethesda, MD: 2011. What Kind of Data Can be Submitted to GenBank? http://www.ncbi.nlm.nih.gov/books/NBK53707/ [Google Scholar]
- 4.Yutin N, Puigbò P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–89. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Jiang H, Gu Z, Roberts JW. High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Proc. Natl. Acad. Sci. USA. 2013;110:11928–33. doi: 10.1073/pnas.1309739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansaldi M, Théraulaz L, Méjean V. TorI, a response regulator inhibitor of phage origin in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2004;101:9423–8. doi: 10.1073/pnas.0401927101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 9.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–80. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Sandman K, Losick R, Youngman P. Genetic analysis of Bacillus subtilis spo mutations generated by Tn917-mediated insertional mutagenesis. Genetics. 1987;117:603–17. doi: 10.1093/genetics/117.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin PA, Fan N, Ricca E, Driks A, Losick R, Cutting S. An unusually small gene required for sporulation by Bacillus subtilis. Mol. Microbiol. 1993;9:761–71. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 12.Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ. Microbiol. 2012;14:2870–90. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 2008;70:1487–501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 2003;185:1391–8. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prajapati RS, Ogura T, Cutting SM. Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim. Biophys. Acta. 2000;1475:353–9. doi: 10.1016/s0304-4165(00)00089-1. [DOI] [PubMed] [Google Scholar]
- 16.Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol. Microbiol. 2006;62:1547–57. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 17.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–7. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 1992;174:575–85. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol. Cell. 2008;31:406–14. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castaing JP, Nagy A, Anantharaman V, Aravind L, Ramamurthi KS. ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc. Natl. Acad. Sci. USA. 2013;110:E151–60. doi: 10.1073/pnas.1210554110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutting S, Anderson M, Lysenko E, Page A, Tomoyasu T, et al. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J. Bacteriol. 1997;179:5534–42. doi: 10.1128/jb.179.17.5534-5542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobiler O, Rokney A, Oppenheim AB. Phage lambda CIII: a protease inhibitor regulating the lysis-lysogeny decision. PLoS One. 2007;2:e363. doi: 10.1371/journal.pone.0000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halder S, Banerjee S, Parrack P. Direct CIII-HflB interaction is responsible for the inhibition of the HflB (FtsH)-mediated proteolysis of Escherichia coli σ32 by λCIII. FEBS J. 2008;275:4767–72. doi: 10.1111/j.1742-4658.2008.06610.x. [DOI] [PubMed] [Google Scholar]
- 24.Le AT, Schumann W. The Spo0E phosphatase of Bacillus subtilis is a substrate of the FtsH metalloprotease. Microbiology. 2009;155:1122–32. doi: 10.1099/mic.0.024182-0. [DOI] [PubMed] [Google Scholar]
- 25.Yepes A, Schneider J, Mielich B, Koch G, García-Betancur JC, et al. The biofilm formation defect of a Bacillus subtilis flotillin-defective mutant involves the protease FtsH. Mol. Microbiol. 2012;86:457–71. doi: 10.1111/j.1365-2958.2012.08205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coote JG. Sporulation in Bacillus subtilis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J. Gen. Microbiol. 1972;71:1–15. doi: 10.1099/00221287-71-1-1. [DOI] [PubMed] [Google Scholar]
- 27.Ebmeier SE, Tan IS, Clapham KR, Ramamurthi KS. Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol. Microbiol. 2012;84:682–96. doi: 10.1111/j.1365-2958.2012.08052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmalisch M, Maiques E, Nikolov L, Camp AH, Chevreux B, et al. Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol. 2010;192:5402–12. doi: 10.1128/JB.00534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putnam EE, Nock AM, Lawley TD, Shen A. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J. Bacteriol. 2013;195:1214–25. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 2012;69:778–90. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 2010;74:504–28. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkpatrick CL, Viollier PH. New(s) to the (Z-)ring. Curr. Opin. Microbiol. 2011;14:691–7. doi: 10.1016/j.mib.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–66. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 34.Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–88. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 35.Handler AA, Lim JE, Losick R. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol. Microbiol. 2008;68:588–99. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wortinger M, Sackett MJ, Brun YV. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 2000;19:4503–12. doi: 10.1093/emboj/19.17.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25:1328–43. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Milam SL, Erickson HP. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry. 2012;51:3100–9. doi: 10.1021/bi201669d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–32. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–8. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong RS, McMurry LM, Levy SB. `Intergenic' blr gene in Escherichia coli encodes a 41-residue membrane protein affecting intrinsic susceptibility to certain inhibitors of peptidoglycan synthesis. Mol. Microbiol. 2000;37:364–70. doi: 10.1046/j.1365-2958.2000.01998.x. [DOI] [PubMed] [Google Scholar]
- 42.Hobbs EC, Astarita JL, Storz G. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a barcoded mutant collection. J. Bacteriol. 2010;192:59–67. doi: 10.1128/JB.00873-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karimova G, Davi M, Ladant D. The β-lactam resistance protein Blr, a small membrane polypeptide, is a component of the Escherichia coli cell division machinery. J. Bacteriol. 2012;194:5576–88. doi: 10.1128/JB.00774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaßel M, Möllenkamp T, Puppe W, Altendorf K. The KdpF subunit is part of the K+-translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 1999;274:37901–7. doi: 10.1074/jbc.274.53.37901. [DOI] [PubMed] [Google Scholar]
- 45.Hamann K, Zimmann P, Altendorf K. Reduction of turgor is not the stimulus for the sensor kinase KdpD of Escherichia coli. J. Bacteriol. 2008;190:2360–7. doi: 10.1128/JB.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gannoun-Zaki L, Alibaud L, Carrère-Kremer S, Kremer L, Blanc-Potard AB. Overexpression of the KdpF membrane peptide in Mycobacterium bovis BCG results in reduced intramacrophage growth and altered coding morphology. PLoS One. 2013;8:e60379. doi: 10.1371/journal.pone.0060379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc. Natl. Acad. Sci. USA. 2012;109:16696–701. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–56. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phophostransferase system. Mol. Microbiol. 2004;54:1076–89. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 50.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA. 2007;104:20454–9. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanderpool CK, Gottesman S. The novel transcripton factor SgrR coordinates the response to glucose-phosphate stress. J. Bacteriol. 2007;189:2238–48. doi: 10.1128/JB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosfeld A, Jahreis K. Characterization of the interaction between the small regulatory peptide SgrT and the EIICBGlu of the glucose-phosphotransferase system of E. coli K-12. Metabolites. 2012;2:756–74. doi: 10.3390/metabo2040756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi LX, Schröder WP. The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim. Biophys. Acta. 2004;1608:75–96. doi: 10.1016/j.bbabio.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Schneider D, Volkmer T, Rögner M. PetG and PetN, but not PetL, are essential subunits of the cytochrome b6f complex from Synechocystis PCC 6803. Res. Microbiol. 2007;158:45–50. doi: 10.1016/j.resmic.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Nelson N, Yocum CF. Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 2006;57:521–65. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 56.Vanorsdel CE, Bhatt S, Allen RJ, Brenner EP, Hobson JJ, et al. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J. Bacteriol. 2013 doi: 10.1128/JB.00324-13. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun YH, de Jong MF, den Hartigh AB, Roux CM, Rolán HG, Tsolis RM. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front. Cell. Infect. Microbiol. 2012;2 doi: 10.3389/fcimb.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kato A, Chen HD, Latifi T, Groisman EA. Reciprocal control between a bacterium's regulatory system and the modification status of Its lipopolysaccharide. Mol. Cell. 2012;47:897–908. doi: 10.1016/j.molcel.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alix E, Blanc-Potard AB. Hydrophobic peptides: novel regulators within bacterial membrane. Mol Microbiol. 2009;72:5–11. doi: 10.1111/j.1365-2958.2009.06626.x. [DOI] [PubMed] [Google Scholar]
- 60.Herrera CM, Hankins JV, Trent MS. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 2010;76:1444–60. doi: 10.1111/j.1365-2958.2010.07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alix E, Blanc-Potard AB. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008;27:546–57. doi: 10.1038/sj.emboj.7601983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyashiro T, Goulian M. Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc. Natl. Acad. Sci. USA. 2007;104:16305–10. doi: 10.1073/pnas.0700025104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moon K, Gottesman S. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 2009;74:1314–30. doi: 10.1111/j.1365-2958.2009.06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moncrief MB, Maguire ME. Magnesium and the role of MgtC in growth of Salmonella typhimurium. Infect. Immun. 1998;66:3802–9. doi: 10.1128/iai.66.8.3802-3809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee EJ, Pontes MH, Groisman EA. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium's own F1Fo ATP synthase. Cell. 2013;154:146–56. doi: 10.1016/j.cell.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi E, Lee KY, Shin D. The MgtR regulatory peptide negatively controls expression of the MgtA Mg2+ transporter in Salmonella enterica serovar Typhimurium. Biochem. Biophys. Res. Commun. 2012;417:318–23. doi: 10.1016/j.bbrc.2011.11.107. [DOI] [PubMed] [Google Scholar]
- 67.Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lippa AM, Goulian M. Perturbation of the oxidizing environment of the periplasm stimulates the PhoQ/PhoP system in Escherichia coli. J. Bacteriol. 2012;194:1457–63. doi: 10.1128/JB.06055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cardenal-Muñoz E, Ramos-Morales F. DsbA and MgrB regulate steA expression through the two-component system PhoQ/PhoP in Salmonella enterica. J. Bacteriol. 2013;195:2368–78. doi: 10.1128/JB.00110-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–79. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 71.Rowland SL, Burkholder WF, Cunningham KA, Maciejewski MW, Grossman AD, King GF. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell. 2004;13:689–701. doi: 10.1016/s1097-2765(04)00084-x. [DOI] [PubMed] [Google Scholar]
- 72.Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, et al. The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J. Mol. Biol. 2007;368:407–20. doi: 10.1016/j.jmb.2007.01.064. [DOI] [PubMed] [Google Scholar]
- 73.Cunningham KA, Burkholder WF. The histidine kinase inhibitor Sda binds near the site of autophosphorylation and may sterically hinder autophosphorylation and phosphotransfer to Spo0F. Mol. Microbiol. 2009;71:659–77. doi: 10.1111/j.1365-2958.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- 74.Bick MJ, Lamour V, Rajashankar KR, Gordiyenko Y, Robinson CV, Darst SA. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J. Mol. Biol. 2009;386:163–77. doi: 10.1016/j.jmb.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacques DA, Streamer M, Rowland SL, King GF, Guss JM, et al. Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state. Acta Crystallogr. D. Biol. Crystallogr. 2009;65:574–81. doi: 10.1107/S090744490901169X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005;12:842–6. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 77.Robinson NJ, Winge DR. Copper metallochaperones. Annu. Rev. Biochem. 2010;79:537–62. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waters LS, Sandoval M, Storz G. The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J. Bacteriol. 2011;193:5887–97. doi: 10.1128/JB.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, et al. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. USA. 2008;105:11927–32. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen L, James LP, Helmann JD. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 1993;175:5428–37. doi: 10.1128/jb.175.17.5428-5437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smaldone GT, Antelmann H, Gaballa A, Helmann JD. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. J. Bacteriol. 2012;194:2586–93. doi: 10.1128/JB.05567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Samayoa J, Yildiz FH, Karplus K. Identification of prokaryotic small proteins using a comparative genomic approach. Bioinformatics. 2011;27:1756–71. doi: 10.1093/bioinformatics/btr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ibrahim M, Nicolas P, Bessières P, Bolotin A, Monnet V, Gardan R. A genome-wide survey of short coding sequences in streptococci. Microbiology. 2007;153:3631–44. doi: 10.1099/mic.0.2007/006205-0. [DOI] [PubMed] [Google Scholar]
- 84.Skovgaard M, Jensen LJ, Brunak S, Ussery D, Krogh A. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 2001;17:425–8. doi: 10.1016/s0168-9525(01)02372-1. [DOI] [PubMed] [Google Scholar]
- 85.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–41. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Balasubramanian D, Vanderpool CK. Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J. Bacteriol. 2013 doi: 10.1128/JB.00586-13. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 2010;64:143–62. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 89.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–59. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Konkol MA, Blair KM, Kearns DB. Plasmid-encoded ComI inhibits competence in the ancestral strain of Bacillus subtilis. J. Bacteriol. 2013;195:4085–93. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jaud S, Fernández-Vidal M, Nilsson I, Meindl-Beinker NM, Hübner NC, et al. Insertion of short transmembrane helices by the Sec61 translocon. Proc. Natl. Acad. Sci. USA. 2009;106:11588–93. doi: 10.1073/pnas.0900638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karimova G, Robichon C, Ladant D. Characterization of YmgF, a 72-residue inner membrane protein that associates with the Escherichia coli cell division machinery. J. Bacteriol. 2009;191:333–46. doi: 10.1128/JB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, et al. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2007;104:18712–7. doi: 10.1073/pnas.0705768104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Törnroth-Horsefield S, Gourdon P, Horsefield R, Brive L, Yamamoto N, et al. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure. 2007;15:1663–73. doi: 10.1016/j.str.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 95.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009;71:811–24. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 96.Fontaine F, Fuchs RT, Storz G. Membrane localization of small proteins in Escherichia coli. J. Biol. Chem. 2011;286:32464–74. doi: 10.1074/jbc.M111.245696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 2008;6:234–44. doi: 10.1038/nrmicro3595. [DOI] [PubMed] [Google Scholar]
- 98.Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37:5465–76. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, et al. Protogenes and de novo gene birth. Nature. 2012;487:370–4. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by small open reading frames. Nat. Rev. Genet. 2013 doi: 10.1038/nrg3520. In press. [DOI] [PubMed] [Google Scholar]
- 101.Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc. Natl. Acad. Sci. USA. 2013;110:2395–400. doi: 10.1073/pnas.1213958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 2007;9:660–5. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 103.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013 doi: 10.1126/science.1238802. In press. [DOI] [PubMed] [Google Scholar]
- 105.Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–9. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 106.Nelson KE, Paulsen IT, Heidelberg JF, Fraser CM. Status of genome projects for nonpathogenic bacteria and archaea. Nat. Biotechnol. 2000;18:1049–54. doi: 10.1038/80235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.