Abstract
The effect of reduced dielectric environments on the conformational sampling of DNA was examined through molecular dynamics simulations. Different dielectric environments were used to model one aspect of cellular environments. Implicit solvent based on the Generalized Born methodology was used to reflect different dielectric environments in the simulations. The simulation results show a tendency of DNA structures to favor noncanonical A-like conformations rather than canonical A- and B-forms as a result of the reduced dielectric environments. The results suggest that the reduced dielectric response in cellular environments may be sufficient to enhance the sampling of A-like DNA structures compared to dilute solvent conditions.
DNA is an essential biomolecule due to its role in numerous biological processes. Its structure, flexible and sensitive to environmental conditions, is one of the most crucial physical determinants of its biological roles. The major conformations of DNA, B-form and A-form, are well-known from in vitro studies; the B-form is the major conformation in solution1 while the A-form is seen for GC-rich sequences under low-humidity environments and, sometimes, when bound to other biomolecules.2 The structural preferences and transitions of DNA between these two forms have been studied extensively for many years not only to gain insight into the structure–function relationship of DNA, but also because DNA is a fascinating biophysical model system that exquisitely balances electrostatic and solvent interactions.3−27 Experimental studies on DNA structure have shown that B- to A-form transitions occur under conditions of “low water activity”, for example, when ethanol is introduced as a cosolvent4,9,12,14 and also when salt is added to solution which bridges the phosphate groups in the major groove of A-DNA.6,11,15 These findings have also been reproduced by computational approaches, especially by molecular dynamics (MD) simulation techniques for the exploration of these conformational preferences in atomistic detail.16−27 These MD studies showed that DNA maintained its B-DNA form in aqueous solvent, while A-DNA was stabilized in solutions containing explicit salt ions or cosolvents such as ethanol as expected from the experiments.16,18,21−25 The effect of salt on DNA structure is easily rationalized by increased electrostatic screening that allows subsequent phosphate groups along the backbone to come closer to each other so that A-DNA can be formed. It is less clear, however, whether cosolvents affect DNA structure through specific molecular interactions or via a more general physical effect such as an altered dielectric response of the environment.
Reduced dielectric environments are also interesting in the context of understanding DNA structure in the cellular context. Cellular environments are highly crowded systems with high concentrations of proteins, nucleic acids, and numerous cosolvents. It has been reported that 20–30% of the cellular volume is occupied by macromolecules.28 Previous studies have shown that the effect of such crowding on protein structure and dynamics can be significant.29 Crowded environments alter the balance between enthalpic and entropic contributions to the conformational free energy either directly through macromolecular interactions or indirectly by modifying solvent properties.29−42 Essentially, the effect of crowding is threefold: (1) volume exclusion by surrounding macromolecules resulting in an entropic penalty for assuming more expanded states;30,39 (2) reduced dielectric response of the environment due to the displacement of 20–30% of the water with less polar macromolecules43−48 and due to slowed dynamics of the water itself;33,49 (3) specific interactions between different macromolecules. Therefore, studying the effect of reduced dielectric environments on DNA structure addresses one major consequence of cellular crowding. Such an approach neglects the effect of specific interactions between DNA and protein crowder molecules as well as the volume-exclusion effect, but it allows for more fundamental insight about how DNA structure is altered by different environments.
In this study, we rely on MD simulations using an implicit continuum dielectric model based on the Generalized Born (GB) methodology50−52 to directly observe the effect of reduced dielectric environments on the conformational sampling of DNA. The GB methodology approximates the solvation free energies obtained from Poisson(-Boltzmann) theory in a numerically convenient and computationally efficient manner that is suitable for the application in MD simulations. This methodology has been applied previously to study peptides in reduced dielectric environments.40,53 We are focusing here on reduced dielectric constants of 20 and 40 that are compared with ε = 80. The reduced values cover the effective dielectric response of ethanol–water mixtures as well as that of cellular environments. As mentioned above, 20–30% of water in the cell is replaced by macromolecules with an internal dielectric constant of 2–2043−47 and cosolvents.48 Furthermore, water itself was found to have a reduced dielectric constant in crowded environments.33,49 As a result, the effective total dielectric constant of cellular environments is assumed to be in the range of 20–60 depending on the degree of crowding.40,53,54 We considered only scalar, static dielectric constants here as the dielectric response of aqueous solvent is isotropic and largely independent of frequency for the range of dynamics considered here (ps−μs). Implicit solvent simulations of DNA using GB models with ε = 80 were previously shown to closely approximate the conformational sampling of DNA seen in explicit solvent.55−58 Therefore, we have confidence in applying the implicit solvent methodology to nucleic acids.
As test systems, we considered the Drew-Dickerson dodecamer d(CGCGAATTCGCG)259 known to be exceptionally stable in B-form and the GC-rich dodecamer d(CGCCCGCGGGCG)260 which tends toward A-form conformations under conditions of high salt, low humidity, or in the presence of cosolvents. Simulations of both systems were started from the standard B-form experimental structure of the Drew-Dickerson dodecamer and we hypothesized that a reduced dielectric response may favor A-like structures akin to the effect of cosolvents shifting the A/B balance toward A. This is indeed what we observe as described in more detail below, although the A-like conformations we observe here appear to be distinct from salt-induced A-DNA. In the following, results are described and discussed.
DNA conformations are distinguished by the orientation of their bases, captured by helical parameters, and by backbone torsional angles. Both were analyzed from our simulations and will be described in the following. The most distinctive helical parameters to characterize A- and B-type conformations are the base inclination relative to the helical axis, the twist between subsequent base pairs, and the x-displacement of base pairs from the helical axis. Secondary parameters of common interest are the relative displacement between subsequent base pairs along the base pair axis (slide) and along the helical axis (rise) as well as the propeller twist between bases forming a base pair. The average values from the simulation are given in Tables 1 and 2. Distributions are shown as Supporting Information Figures S1 and S2. The simulation results are compared with averaged helical parameters for ten A-form and ten B-form DNA duplexes and for the X-ray structures of the exact sequences that were studied here. In all analyses, only the eight inner base pairs were taken into consideration, because of structural distortions at the duplex ends due to occasional base fraying.
Table 1. Helicoidal Parameters for the GC-Rich Dodecamer Compared to Experimental Resultsa.
| canonicalb |
MD simulationsc |
|||||
|---|---|---|---|---|---|---|
| X-rayd | A-DNA | B-DNA | ε = 20 | ε = 40 | ε = 80 | |
| propeller (deg) | –12.49 (2.0) | –9.00 (7.1) | –11.72 (5.8) | 2.16 (15.5) | –2.85 (12.0) | –7.26 (14.1) |
| slide (Å) | –1.71 (0.4) | –1.67 (0.4) | 0.17 (0.7) | –1.48 (0.8) | –1.23 (0.9) | –0.52 (1.4) |
| twist (deg) | 29.59 (3.5) | 30.24 (4.0) | 34.81 (5.7) | 27.50 (10.0) | 29.70 (7.3) | 36.27 (17.7) |
| X-displacement (Å) | –5.01 (1.1) | –4.54 (1.3) | –0.13 (1.1) | –2.68 (3.6) | –2.80 (2.9) | –1.19 (2.5) |
| helical rise (Å) | 2.66 (0.6) | 2.77 (0.5) | 3.25 (0.2) | 3.42 (1.0) | 3.36 (0.8) | 3.36 (0.8) |
| inclination (deg) | 20.71 (11.4) | 16.00 (10.7) | 4.01 (6.7) | 0.45 (20.7) | 3.73 (15.1) | 5.43 (16.4) |
All values are averaged over nonterminal base pairs with standard deviations given in parentheses.
Averages over the A-form structures 3V9D, 3QK4, 2B1B, 1ZEX, 1ZEY, 1ZF1,1ZF6, 1ZF8, 1ZF9, 1ZFA and the B-form structures 2M2C, 4AGZ, 4AH0, 4AH1, 3U05, 3U08, 1VTJ, 3U2N, 3OIE, 3BSE.
Averages over snapshots at 300 K from replica exchange MD simulations.
PDB ID code: 399D.
Table 2. Helicoidal Parameters for the Drew-Dickerson Dodecamer Compared to Experimental Resultsa.
| canonicalb |
MD simulationsc |
|||||
|---|---|---|---|---|---|---|
| X-rayd | A-DNA | B-DNA | ε = 20 | ε = 40 | ε = 80 | |
| propeller (deg) | –13.34 (5.9) | –9.00 (7.1) | –11.72 (5.8) | –6.93 (12.5) | –10.67 (12.7) | –11.32 (13.7) |
| slide (Å) | 0.07 (0.5) | –1.67 (0.4) | 0.17 (0.7) | –0.86 (0.8) | –0.50 (0.7) | –0.32 (0.7) |
| twist (deg) | 34.22 (5.7) | 30.24 (4.0) | 34.81 (5.7) | 30.62 (6.4) | 32.36 (5.9) | 33.12 (6.7) |
| X-displacement (Å) | –0.23 (0.5) | –4.54 (1.3) | –0.13 (1.1) | –2.33 (2.8) | –1.65 (2.3) | –1.23 (2.4) |
| helical rise (Å) | 3.29 (0.1) | 2.77 (0.5) | 3.25 (0.2) | 3.24 (0.7) | 3.25 (0.6) | 3.18 (0.7) |
| inclination (deg) | 4.02 (7.2) | 16.00 (10.7) | 4.01 (6.7) | 7.26 (19.1) | 8.00 (16.7) | 6.30 (19.8) |
All values are averaged over nonterminal base pairs with standard deviations given in parentheses.
Averages over the A-form structures 3V9D, 3QK4, 2B1B, 1ZEX, 1ZEY, 1ZF1,1ZF6, 1ZF8, 1ZF9, 1ZFA and the B-form structures 2M2C, 4AGZ, 4AH0, 4AH1, 3U05, 3U08, 1VTJ, 3U2N, 3OIE, 3BSE.
Averages over snapshots at 300 K from replica exchange MD simulations.
PDB ID code: 1BNA.
X-displacement is one of the clearest indicators of A- vs B-DNA conformations. It is expected to have near zero values for B-DNA and significantly negative values for A-DNA, where bases are displaced from the helical axis. We fnd that, for both the GC-rich and the Drew-Dickerson dodecamers, reduced dielectric environments resulted in a clear shift toward more negative x-displacement values that are about halfway between canonical B- and A-DNA. The shift was slightly greater for the GC-rich sequence and was equally large at ε = 40 and ε = 20. On the other hand, the Drew-Dickerson dodecamer values changed more gradually as the dielectric constant was reduced. A similar observation was made for the twist angle that changed from values of 33–36°, typical for B-DNA, to 28–31°, typical for A-DNA, as the dielectric constant was changed from ε = 80 to ε = 20. Again, the change was already complete at ε = 40 for the GC-rich sequence but occurred more gradually for the Drew-Dickerson dodecamer. The inclination of base pairs relative to the helical axis in A-type conformations is typically quite pronounced compared to B-type conformations, which are mostly perpendicular to the helical axis. Interestingly, we find that the average for the GC-rich dodecamer shifted to more B-like values as the dielectric constant was reduced. This appears to be in part due to increased sampling of negative inclination angles at ε = 20 (see Supporting Information Figure S1). On the other hand, the Drew-Dickerson dodecamer showed a slightly increased inclination angle upon reduction of the dielectric response of the environment. In contrast to what x-displacement and twist values suggest, the changes in base inclination angles are not fully consistent with a classical transition from B-DNA to A-DNA. However, as commented by Dickerson and Ng,61 the inclination angle can be problematic in short helical segments for distinguishing A- from B-form due to the difficulties in separating inclination from local helix bending.
The slide of a base pair along its long axis relative to its neighboring base pair is another measure that can discriminate between A- and B-form DNA structures. While a significant slide is not observed for base pairs in typical B-DNA, A-DNA base pairs have a more pronounced propensity to slide along their long axes. We observed a significant increase in negative slide values for the GC-rich dodecamer at reduced dielectric constants and to a lesser extent for the Drew-Dickerson dodecamers. Another discriminative helical property is helical rise, which is smaller for typical A-DNA, around 2.7–2.8 Å, compared to B-DNA, where it is typically around 3.3–3.4 Å. Interestingly, we found the helical rise to remain unchanged upon reduction of the dielectric constants. Finally, the propeller twist of bases in a base pair with respect to each other is known to be highly sequence dependent with more negative values for A/T base pairs than for C/G base pairs, but it is also reduced in A-DNA vs B-DNA. As expected, the propeller twist was less pronounced for the GC-rich dodecamers than for the AT-base pair containing Drew-Dickerson dodecamer. There was a trend, however, toward less negative propeller twist values upon a decrease of the dielectric constant. This is again indicative of a transition toward A-like structural features. Overall, the analysis of the helical parameters suggests a tendency toward A-type conformations upon a reduction in the dielectric constant. The tendency toward A-DNA was more pronounced for the GC-rich dodecamer and it manifested itself at higher dielectric constants compared to the Drew-Dickerson dodecamer. The helical parameters in the X-ray structure of the B-form Drew-Dickerson dodecamer agree closely with the simulation results at ε = 80 while the helical parameters of the A-form X-ray structure of the GC-rich dodecamer are reproduced best by the simulation results at ε = 20 and ε = 40 rather than ε = 80.
Among backbone torsion angles, the ribose sugar pseudorotation angle is the most significant indicator of A- vs B-DNA conformations. While A-DNA structures commonly have C3′-endo or C2′-exo sugar conformations, sugars in B-DNA structures are more often found in C3′-exo or C2′-endo conformations. The preferred sugar conformations for each base in the GC-rich and the Drew-Dickerson dodecamers in different dielectric environments are depicted in Figure 1. As expected from DNA simulations with the CHARMM force field,62 there was extensive sampling of both C2′-endo and C3′-endo conformations at ε = 80 but with an overall preference for C2′-endo conformers. With a decreasing dielectric constant of the environment, sugar conformations in the GC-rich dodecamer were switched largely to C3′-endo or C2′-exo conformations, further indicating a B to A transition for this sequence. In contrast, the ribose sugars in the Drew-Dickerson dodecamer were largely unaffected by a change in the dielectric environment, suggesting that the backbone retained B-like features.
Figure 1.
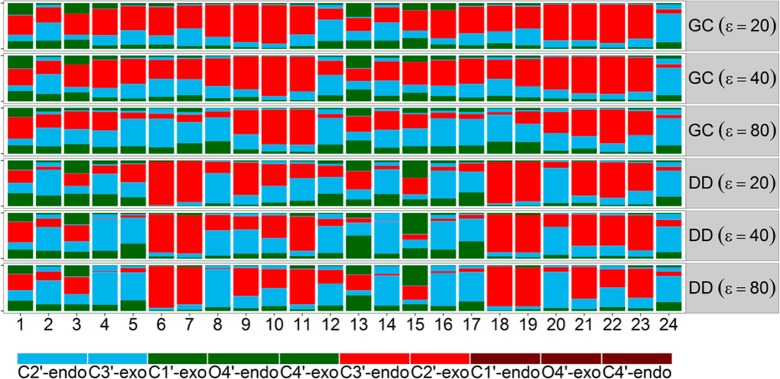
Sugar pucker conformations of each base from simulations of the GC-rich dodecamer (GC), the Drew-Dickerson dodecamer (DD) in different dielectric environments with ε = 20, 40, and 80.
We analyzed the ε and ζ backbone torsion angles as an indicator of the relative sampling of BI and BII conformations. A value of ε – ζ near −90° corresponds to the BI conformation whereas +90° characterizes BII conformations. The potential of mean force as a function of ε/ζ is given in Figure 2. Frequent BI/BII transitions were observed in both dodecamers for all dielectric constants as expected from the most recent version of the CHARMM nucleic acid force field62 used in this study. Closer inspection shows that the sampling of BII conformations diminished at ε = 20 and ε = 40 for the GC-rich dodecamer and at ε = 20 for the Drew-Dickerson dodecamer. This suggests that excursions to the less-populated BI conformer may be further suppressed in reduced dielectric environments. We also found two minima at ε = 80 corresponding to noncanonical conformations of the GC-rich dodecamers that appear to be missing at the lower dielectric constants. However, it is likely that this observation is due to limited sampling in our simulations and/or structural distortions due to the implicit solvent model used here (see Figure 2).
Figure 2.
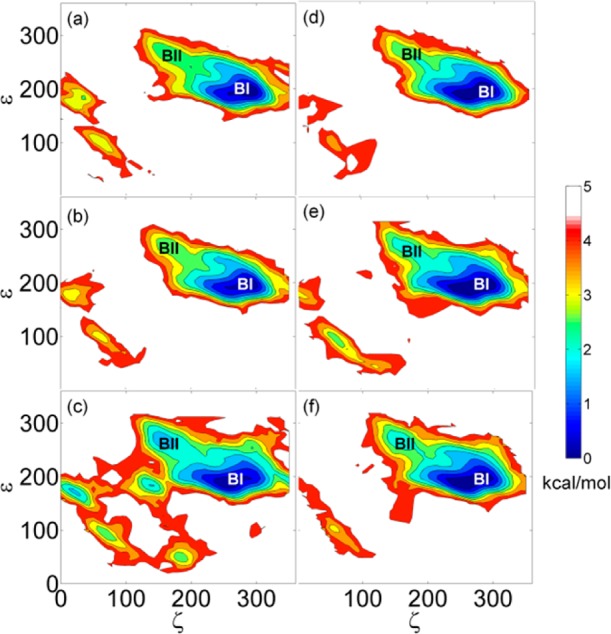
Potential of mean force (kcal/mol) from simulations as a function of ε and ζ torsion angles for the GC-rich at ε = 20 (a), ε = 40 (b), and ε = 80 (c) and for the Drew-Dickerson dodecamer at ε = 20 (d), ε = 40 (e), and ε = 80 (f).
Next, we clustered snapshots from each of the simulations to obtain representative structures at different dielectric environments. The resulting structures for both studied dodecamers are shown in Figure 3. Using a clustering radius of 3 Å resulted in five major conformations for the GC-rich dodecamer and three major conformations for the Drew-Dickerson dodecamer. The average helical parameters of these clusters were calculated and compared with canonical A-, B-, and C-DNA forms (Supporting Information). According to this analysis, GC1 and GC2, GC5 and DD3, and DD1 and DD2 have A-like, B-like, and mixed A- and B-type features, respectively, further confirming the prevalence of B-form conformations (GC5 and DD3) at ε = 80 and a shift toward A-like structures at lower dielectric constants. We also observed a minor population of a distorted conformation (GC4) at ε = 80 that may be a simulation artifact. At the lowest dielectric, a new conformation appears (GC3) that has mixed characteristics of A- and B- type structures. As for the Drew-Dickerson dodecamer, the B-form DD3 conformation persists at ε = 40, but disappears at ε = 20. Instead, A-like structures (DD1 and DD2) appear with a small percentage at ε = 40 while becoming dominant at ε = 20. None of the structures observed here resemble other previously characterized conformations of DNA, such as C-DNA.
Figure 3.
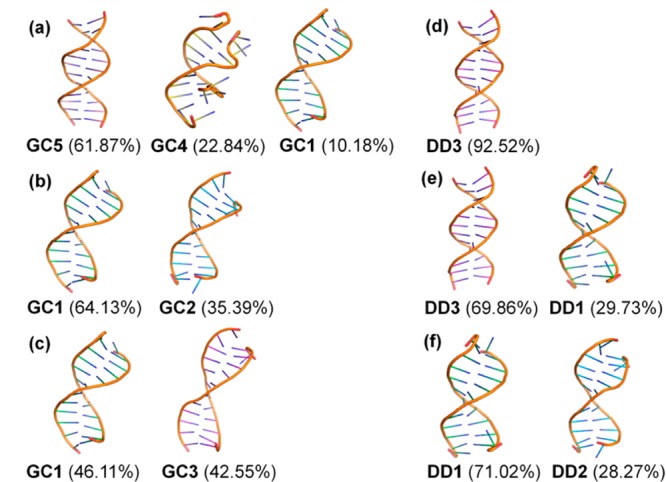
Representative conformations from clustering analysis in different dielectric environment for GC-rich dodecamer with ε = 80 (a), 40 (b), and 20 (c) and for the Drew-Dickerson dodecamer with ε = 80 (d), 40 (e), and 20 (f). Cluster populations are given in the parentheses.
Finally, we estimated relative conformational free energies using the MMPB/SA approach25 and compared the effect of reduced dielectric environments to previously described effects of increased salt concentrations on the A- vs B-DNA equilibrium.6,11,15,18,25 Calculated free energy differences between A- and B-form structures are given in Tables 3 and 4 with positive values indicating a preference for the A-form while negative values indicate a preference for the B-form.
Table 3. Conformational Free Energies from MMPB/SA Analysis for the GC-Rich Dodecamera.
| Ab | Bc | canonical Ad | (B – A) | (B – Can.A) | |
|---|---|---|---|---|---|
| ⟨E(gas)⟩e | 2181.2 (4.1) | 2331.1 (4.1) | 2371.2 (4.2) | 149.9 | –40.1 |
| ⟨E(nonpolar)⟩f | 26.9 (0.0) | 26.9 (0.0) | 26.3 (0.0) | 0.0 | 0.6 |
| ⟨E(PB), ε = 80⟩g | –5837.5 (3.2) | –5965.5 (3.5) | –6069.4 (3.7) | –128.0 | 103.9 |
| ⟨E(total), ε = 80⟩ | –3629.4 (4.0) | –3607.5 (3.9) | –3672.0 (2.3) | 21.9 | 64.5 |
| ⟨ΔE(total), salt: 0.1 M⟩ | –40.6 (0.0) | –41.1 (0.0) | –42.9 (0.0) | –0.5 | 1.8 |
| ⟨ΔE(total), salt: 1.0 M⟩ | –53.4 (0.1) | –54.5 (0.1) | –58.5 (0.1) | –1.1 | 4.0 |
| ⟨ΔE(total), ε = 20⟩ | 242.6 (0.2) | 247.3 (0.2) | 261.1 (0.3) | 4.7 | –13.8 |
Standard errors are given in the parentheses.
100 snapshots selected from GC1 and GC2 clusters closest to cluster centers and with minimal distortions at terminal base pairs.
100 snapshots selected from GC5 cluster as in b.
Snapshots from 1 ns restrained implicit solvent simulation of canonical A-DNA structure taken at 5 ps intervals starting from 0.5 ns.
CHARMM force field energy in vacuum.
Solvent-accessible surface area term: γ*SASA+β with γ = 0.00542 kcal/mol/Å2 and β = 0.92 kcal/mol.63,64
Solution of Poisson–Boltzmann equation using PBEQ module65 in CHARMM program with a grid spacing of 0.25 Å.
Table 4. Conformational Free Energies from MMPB/SA Analysis for the Drew-Dickerson Dodecamera.
| Ab | Bc | canonical Ad | (B – A) | (B – Can.A) | |
|---|---|---|---|---|---|
| ⟨E(gas)⟩e | 2742.6 (5.8) | 2812.8 (7.3) | 2944.2 (5.3) | 70.2 | –131.4 |
| ⟨E(nonpolar)⟩f | 26.9 (0.0) | 26.7 (0.0) | 26.2 (0.0) | –0.2 | 0.5 |
| ⟨E(PB), ε = 80⟩g | –5818.1 (3.9) | –5931.7 (6.1) | –6090.8 (4.6) | –113.6 | 159.1 |
| ⟨E(total), ε = 80⟩ | –3048.6 (5.5) | –3092.3 (3.5) | –3120.4 (2.0) | –43.7 | 28.1 |
| ⟨ΔE(total), salt: 0.1 M⟩ | –40.5 (0.0) | –41.1 (0.1) | –43.1 (0.0) | –0.6 | 2.0 |
| ⟨ΔE(total), salt: 1.0 M⟩ | –53.1 (0.1) | –54.2 (0.1) | –59.1 (0.1) | –1.1 | 4.9 |
| ⟨ΔE(total), ε = 20⟩ | 239.2(0.2) | 245.0 (0.3) | 260.4 (0.3) | 5.8 | –15.4 |
Standard errors are given in the parentheses.
100 snapshots selected from DD1 and DD1 clusters as in Table 3.
100 snapshots selected from DD3 cluster as in Table 3.
Snapshots from 1 ns restrained implicit solvent simulation of canonical A-DNA structure taken at 5 ps intervals starting from 0.5 ns.
CHARMM force field energy in vacuum.
Solvent-accessible surface area term: γ*SASA+β with γ = 0.00542 kcal/mol/Å2 and β = 0.92 kcal/mol.63,64
Solution of Poisson–Boltzmann equation using PBEQ module65 in CHARMM program with a grid spacing of 0.25 Å.
The total free energy differences (⟨E(total)⟩) at ε = 80 between simulated B- and A-forms and between simulated B- and canonical A-structures were more positive for the GC-rich dodecamer (Table 3) than for the Drew-Dickerson dodecamer (Table 4). This indicates that the A-form is relatively more favorable in aqueous medium for the GC-rich dodecamer as we would expect. Furthermore, at ε = 80, both dodecamers appear to be more favorable in canonical A-DNA than in the A-form structures observed in our simulations. This is also consistent with expectations since past in vitro studies have established the canonical A-DNA structure as the relevant A-form in noncellular environments instead of the somewhat different A-form reported here. We note that the energetic differences are relatively large (tens of kcal/mol instead of physically probably more realistic values of less than 10 kcal/mol). This is likely a result of the approximate nature of the MMPB/SA methodology and/or limitations in sampling. Overall, we estimate that the errors of the MMPB/SA estimates are on the order of tens of kcal/mol, consistent with previous studies.21,25,66 Nevertheless, the MMPB/SA estimates appear to give at least qualitatively the correct picture.
Addition of salt to the environment is known to favor A-DNA over B-DNA structures.6,11,15,18,25 Within the MMPB/SA framework, salt can be considered by solving the Poisson–Boltzmann equation with added salt.25 While the overall MMPB/SA estimate is prone to uncertainties because, in part, the vacuum force field term is highly sensitive to small conformational changes, and because entropic effects are not fully taken into account, the relative change in the electrostatic solvation term as a result of added salt or a changed dielectric constant can be estimated with much higher accuracy.67 We find that canonical A-DNA was favored with increasing salt as expected. However, the trend was reversed when the A-like structures from our simulations were considered instead of the canonical forms. Increasing salt concentration appeared to stabilize the B-form over our A-form structures. On the other hand, a reduced dielectric resulted in a stabilization of our A-like structures while canonical A-DNA structures were destabilized for both dodecamers when compared to ε = 80. This suggests that the effects of increased salt and reduced dielectric environment are different and that, although both changes in the environment appear to lead to A-like structures, the resulting conformational ensembles appear to have distinct features.
The main goal of this study has been to examine the conformational sampling of DNA in reduced dielectric environments to both understand the origin of cosolvent induced A-DNA stabilization as well as explore the possible effects of cellular environments. At ε = 80 we observe stable sampling of mostly B-DNA structures with the implicit solvent methodology used here in agreement with experimental expectations. This suggests that the implicit solvent methodology can reasonably describe the conformational sampling of DNA in aqueous solutions. Additional simulations were then carried out at lower dielectric constants. This approach emphasizes mean field solvation properties while neglecting specific molecular interactions with the environment. %85 (v/v) Ethanol/water solutions have dielectric constants of 30.68 At the same time, while ε = 20 may be at the low end of what is found in crowded environments, an effective dielectric near ε = 40 is assumed to be a typical value for crowded biological cells.40,53,54
Our results indicate an overall shift toward A-like conformations that was moderate at ε = 40 and became more pronounced at ε = 20 for both studied dodecamers. The tendency to form A-like conformations was greater for the GC-rich dodecamer, but even the classical B-DNA Drew-Dickerson dodecamer exhibited A-like features in low-dielectric environments. The transition to canonical A-form DNA was nearly complete for the GC-rich dodecamer, but the Drew-Dickerson dodecamers retained many B-like features even at ε = 20. However, even for the GC-rich dodecamer at ε = 20, the resulting structures differ somewhat from canonical A-form. MMPB/SA analytical results further show that increased salt appears to destabilize the A-like structures seen in our simulations, while it stabilizes canonical A-DNA. This suggests the important conclusion that salt effects and reduced dielectric environments, for example, by reducing “water activity” through cosolvents or crowding, affect DNA structure in similar but different ways. The results are not fully consistent with fiber diffractions studies where DNA remains in canonical B-DNA form with its two hydration layers.69,70 However, the fiber environment is likely not as packed as crowded environments, and hence, the effective dielectric environment may not actually be reduced dramatically. Furthermore, DNA–DNA interactions and the presence of salt in fibers may further explain differences between the fiber experiments and our findings. Specific interactions of DNA with proteins in crowded cellular environments as well as volume exclusion effects were also not considered here and may significantly modulate the results reported here. Nevertheless, any tendency toward A-DNA features under crowded cellular conditions could have significant energetic consequences for protein–DNA interactions since most DNA bound to proteins is in B-form.
It is clear that further studies are needed to fully understand DNA structure in crowded environments and in the presence of cosolvents. One specific question to be addressed is whether protein–DNA interactions could counteract the apparent tendency toward A-form DNA and modulate these findings. Furthermore, the studies performed here involved short oligomers vs the very long polymeric DNA found in vivo. Further investigations are also required to see whether longer DNA molecules would exhibit the same tendencies. It is our hope that the results reported will also stimulate new experimental studies of DNA structure under crowded conditions—none of which are available to our knowledge.
Computational Details
Replica exchange molecular dynamics simulations71 of d(CGCGAATTCGCG)2 and d(CGCCCGCGGGCG)2 dodecamers were performed to enhance the sampling by accelerating barrier crossings at elevated temperatures. The Generalized Born with molecular volume (GBMV) implicit solvation model50−52 was used with dielectric constants 20, 40, and 80. The initial d(CGCGAATTCGCG)2 structure was obtained from X-ray analysis (PDB ID code: 1BNA)59 and the initial d(CGCCCGCGGGCG)2 structure was obtained by mutating the base sequence in the X-ray structure of 1BNA. Structures were minimized and equilibrated before the replica exchange simulations. Replica exchange simulations were performed using eight replicas between temperatures 300 and 400 K. Although the replicas visit higher temperatures than 300 K, only the sampling at 300 K is considered and reported here. Exchange probabilities of the simulations were around 25–30%. Langevin dynamics72 was performed using a friction coefficient of 50 ps–1 to control the temperature of the system. Simulations were carried out for 50 ns for each replica with a total simulation time of 400 ns and replica exchange was attempted every 10 ps. The first 6 ns of each replica was excluded during the analysis of the simulations. Nonbonded interactions were cut off at 18 Å with a switching function becoming effective at 16 Å and a cutoff at 20 Å was used for the nonbonded list.
All simulations were performed using the CHARMM program package (v c37a2)65 with CHARMM36 force field62,73 for nucleic acids. Replica exchange simulations were carried out using the Multiscale Modeling Tools for Structural Biology (MMTSB)74 in combination with CHARMM. Cluster analysis of the conformations was carried out with the kclust program in MMTSB. Helicoidal and backbone parameters of dodecamers were analyzed by using the 3DNA program package.75 VMD76 and PyMOL77 were used for the visualization of structures.
Acknowledgments
Funding from NSF (MCB 1330560) and NIH (R01 GM092949) is acknowledged.
Supporting Information Available
Distribution diagrams for helicoidal parameters. Supplemental figures S1–S2 and tables S1–S2. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Watson J. D.; Crick F. H. Molecular Structure of Nucleic Acids; a Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [DOI] [PubMed] [Google Scholar]
- Franklin R. E.; Gosling R. G. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [DOI] [PubMed] [Google Scholar]
- Erfurth S. C.; Bond P. J.; Peticolas W. L. Characterization of a Reversible B Transition of DNA in Fibers and Gels by Laser Raman-Spectroscopy. Biopolymers 1975, 14, 1245–1257. [DOI] [PubMed] [Google Scholar]
- Fang Y.; Spisz T. S.; Hoh J. H. Ethanol-Induced Structural Transitions of DNA on Mica. Nucleic Acids Res. 1999, 27, 1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W.; Wilkins M. H.; Wilson H. R.; Hamilton L. D. The Molecular Configuration of Deoxyribonucleic Acid. Iv. X-Ray Diffraction Study of the a Form. J. Mol. Biol. 1965, 12, 60–76. [DOI] [PubMed] [Google Scholar]
- Gao Y. G.; Robinson H.; Vanboom J. H.; Wang A. H. J. Influence of Counterions on the Crystal-Structures of DNA Decamers - Binding of [Co(NH3)6]3+ and Ba2+ to a-DNA. Biophys. J. 1995, 69, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov V. I.; Krylov D. A-DNA in Solution as Studied by Diverse Approaches. Methods Enzymol. 1992, 211, 111–127. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I.; Minchenkova L. E.; Burckhardt G.; Birch-Hirschfeld E.; Fritzsche H.; Zimmer C. The detection of B-form/A-form Junction in a Deoxyribonucleotide Duplex. Biophys. J. 1996, 71, 3344–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov V. I.; Minchenkova L. E.; Minyat E. E.; Frank-Kamenetskii M. D.; Schyolkina A. K. The B to A Transition of DNA in Solution. J. Mol. Biol. 1974, 87, 817–833. [DOI] [PubMed] [Google Scholar]
- Jose D.; Porschke D. Dynamics of the B-A Transition of DNA Double Helices. Nucleic Acids Res. 2004, 32, 2251–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H.; Wang A. H. J. Neomycin, Spermine and Hexaamminecobalt(III) Share Common Structural Motifs in Converting B- to A-DNA. Nucleic Acids Res. 1996, 24, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht A.; Piskur J.; Schultz J.; Nordenskiold L.; Song Z. Y.; Lahajnar G. Mechanochemical Study of Conformational Transitions and Melting of Li-, Na-, K-, and CSDNA Fibers in Ethanol-Water Solutions. Biopolymers 1994, 34, 897–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason J. M.; Henderson K.; Ho P. S. A Crystallographic Map of the Transition from B-DNA to A-DNA. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 7265–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B.; Pheiffer B. H. Direct Demonstration That the Ethanol-Induced Transition of DNA Is Between the A-Forms and B-Forms - X-Ray-Diffraction Study. J. Mol. Biol. 1979, 135, 1023–1027. [DOI] [PubMed] [Google Scholar]
- Xu Q. W.; Shoemaker R. K.; Braunlin W. H. Induction of B-A Transitions of Deoxyoligonucleotides by Multivalent Cations in Dilute Aqueous-Solution. Biophys. J. 1993, 65, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham T. E.; Crowley M. F.; Fox T.; Kollman P. A. A Molecular Level Picture of the Stabilization of A-DNA in Mixed Ethanol-Water Solutions. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 9626–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham T. E. 3rd; Kollman P. A. Observation of the A-DNA to B-DNA Transition during Unrestrained Molecular Dynamics in Aqueous Solution. J. Mol. Biol. 1996, 259, 434–444. [DOI] [PubMed] [Google Scholar]
- Cheatham T. E.; Kollman P. A. Insight into the Stabilization of A-DNA by Specific Ion Association: Spontaneous B-DNA to A-DNA Transitions Observed in Molecular Dynamics Simulations of d[ACCCGCGGGT](2) in the Presence of Hexaamminecobalt(III). Structure 1997, 5, 1297–1311. [DOI] [PubMed] [Google Scholar]
- Cheatham T. E.; Srinivasan J.; Case D. A.; Kollman P. A. Molecular Dynamics and Continuum Solvent Studies of the Stability of PolyG-PolyC and PolyA-PolyT DNA Duplexes in Solution. J. Biomol. Struct. Dyn. 1998, 16, 265–280. [DOI] [PubMed] [Google Scholar]
- Cieplak P.; Cheatham T. E.; Kollman P. A. Molecular Dynamics Simulations Find That 3′ Phosphoramidate Modified DNA Duplexes Undergo a B to A Transition and Normal DNA Duplexes an A to B Transition. J. Am. Chem. Soc. 1997, 119, 6722–6730. [Google Scholar]
- Jayaram B.; Sprous D.; Young M. A.; Beveridge D. L. Free Energy Analysis of the Conformational Preferences of A and B Forms of DNA in Solution. J. Am. Chem. Soc. 1998, 120, 10629. [Google Scholar]
- Noy A.; Perez A.; Laughton C. A.; Orozco M. Theoretical Study of Large Conformational Transitions in DNA: the B↔A Conformational Change in Water and Ethanol/Water. Nucleic Acids Res. 2007, 35, 3330–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor N. The B- to A-DNA Transition and the Reorganization of Solvent at the DNA Surface. Biophys. J. 2005, 88, 3262–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprous D.; Young M. A.; Beveridge D. L. Molecular Dynamics Studies of the Conformational Preferences of a DNA Double Helix in Water and an Ethanol/Water Mixture: Theoretical Considerations of the A ↔ B Transition. J. Phys. Chem. B 1998, 102, 4658–4667. [Google Scholar]
- Srinavasan J.; Cheatham T. E.; Cieplak P.; Kollman P. A.; Case D. A. Continuum Solvent Studies of the Stability of DNA, RNA, and Phosphoramidate-DNA Helices. J. Am. Chem. Soc. 1998, 120, 9401–9409. [Google Scholar]
- Gu B.; Zhang F. S.; Wang Z. P.; Zhou H. Y. Solvent-Induced DNA Conformational Transition. Phys. Rev. Lett. 2008, 100, 088104. [DOI] [PubMed] [Google Scholar]
- Arscott P. G.; Ma C.; Wenner J. R.; Bloomfield V. A. DNA Condensation by Cobalt Hexaammine (III) in Alcohol-Water Mixtures: Dielectric Constant and Other Solvent Effects. Biopolymers 1995, 36, 345–364. [DOI] [PubMed] [Google Scholar]
- Ellis R. J. Macromolecular Crowding: an Important but Neglected Aspect of the Intracellular Environment. Curr. Opin Struct. Biol. 2001, 11, 114–119. [DOI] [PubMed] [Google Scholar]
- Zhou H. X.; Rivas G.; Minton A. P. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu. Rev. Biophys 2008, 37, 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A. Theory for the Folding and Stability of Globular Proteins. Biochemistry 1985, 24, 1501–1509. [DOI] [PubMed] [Google Scholar]
- Elcock A. H. Models of Macromolecular Crowding Effects and the Need for Quantitative Comparisons with Experiment. Curr. Opin. Struct. Biol. 2010, 20, 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig M.; Sugita Y. Variable Interactions Between Protein Crowders and Biomolecular Solutes are Important in Understanding Cellular Crowding. J. Phys. Chem. B 2012, 116, 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada R.; Sugita Y.; Feig M. Protein Crowding Affects Hydration Structure and Dynamics. J. Am. Chem. Soc. 2012, 134, 4842–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada R.; Tochio N.; Kigawa T.; Sugita Y.; Feig M. Reduced Native State Stability in Crowded Cellular Environment Due to Protein-Protein Interactions. J. Am. Chem. Soc. 2013, 135, 3696–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.; Gierasch L. M. Macromolecular Crowding Remodels the Energy Landscape of a Protein by Favoring a More Compact Unfolded State. J. Am. Chem. Soc. 2010, 132, 10445–10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K.; Ohno A.; Tochio H.; Isogai S.; Tenno T.; Nakase I.; Takeuchi T.; Futaki S.; Ito Y.; Hiroaki H.; Shirakawa M. High-Resolution Multi-Dimensional NMR Spectroscopy of Proteins in Human Cells. Nature 2009, 458, 106–109. [DOI] [PubMed] [Google Scholar]
- Miklos A. C.; Sarkar M.; Wang Y.; Pielak G. J. Protein Crowding Tunes Protein Stability. J. Am. Chem. Soc. 2011, 133, 7116–7120. [DOI] [PubMed] [Google Scholar]
- Minton A. P.; Wilf J. Effect of Macromolecular Crowding upon the Structure and Function of an Enzyme: Glyceraldehyde-3-Phosphate Dehydrogenase. Biochemistry 1981, 20, 4821–4826. [DOI] [PubMed] [Google Scholar]
- Onuchic J. N.; Wolynes P. G. Theory of Protein Folding. Curr. Opin. Struct. Biol. 2004, 14, 70–75. [DOI] [PubMed] [Google Scholar]
- Predeus A. V.; Gul S.; Gopal S. M.; Feig M. Conformational Sampling of Peptides in the Presence of Protein Crowders from AA/CG-Multiscale Simulations. J. Phys. Chem. B 2012, 116, 8610–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger A. P.; Wang Y.; Tadeo X.; Millet O.; Pielak G. J. Macromolecular Crowding Fails to Fold a Globular Protein in Cells. J. Am. Chem. Soc. 2011, 133, 8082–8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Sarkar M.; Smith A. E.; Krois A. S.; Pielak G. J. Macromolecular Crowding and Protein Stability. J. Am. Chem. Soc. 2012, 134, 16614–16618. [DOI] [PubMed] [Google Scholar]
- Schutz C. N.; Warshel A. What Are the Dielectric ″Constants″ of Proteins and How to Validate Electrostatic Models?. Proteins 2001, 44, 400–417. [DOI] [PubMed] [Google Scholar]
- Gilson M. K.; Honig B. H. The Dielectric Constant of a Folded Protein. Biopolymers 1986, 25, 2097–2119. [DOI] [PubMed] [Google Scholar]
- Antosiewicz J.; McCammon J. A.; Gilson M. K. The Determinants of pKas in Proteins. Biochemistry 1996, 35, 7819–7833. [DOI] [PubMed] [Google Scholar]
- Dwyer J. J.; Gittis A. G.; Karp D. A.; Lattman E. E.; Spencer D. S.; Stites W. E.; Garcia-Moreno B. High Apparent Dielectric Constants in the Interior of a Protein Reflect Water Penetration. Biophys. J. 2000, 79, 1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A.; Papazyan A. Electrostatic Effects in Macromolecules: Fundamental Concepts and Practical Modeling. Curr. Opin Struct Biol. 1998, 8, 211–217. [DOI] [PubMed] [Google Scholar]
- Akhadov Y. Y.Dielectric Properties of Binary Solutions; Pergamon Press: Oxford, U.K., 1981. [Google Scholar]
- Despa F.; Fernandez A.; Berry R. S. Dielectric Modulation of Biological Water. Phys. Rev. Lett. 2004, 93, 228104. [DOI] [PubMed] [Google Scholar]
- Lee M. S.; Feig M.; Salsbury F. R. Jr.; Brooks C. L. III New Analytic Approximation to the Standard Molecular Volume Definition and Its Application to Generalized Born Calculations. J. Comput. Chem. 2003, 24, 1348–1356. [DOI] [PubMed] [Google Scholar]
- Lee M. S.; S. J F. R.; Brooks C. L. III Novel Generalized Born Methods. J. Chem. Phys. 2002, 116, 10606–10614. [Google Scholar]
- Feig M.; Im W.; Brooks C. L. III Implicit Solvation Based on Generalized Born Theory in Different Dielectric Environments. J. Chem. Phys. 2004, 120, 903–911. [DOI] [PubMed] [Google Scholar]
- Tanizaki S.; Clifford J.; Connelly B. D.; Feig M. Conformational Sampling of Peptides in Cellular Environments. Biophys. J. 2008, 94, 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H.; Zhou H. X. Prediction of Protein Solubility from Calculation of Transfer Free Energy. Biophys. J. 2008, 95, 2601–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chocholousova J.; Feig M. Implicit Solvent Simulations of DNA and DNA-Protein Complexes: Agreement with Explicit Solvent vs Experiment. J. Phys. Chem. B 2006, 110, 17240–17251. [DOI] [PubMed] [Google Scholar]
- Ruscio J. Z.; Onufriev A. A Computational Study of Nucleosomal DNA Flexibility. Biophys. J. 2006, 91, 4121–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui V.; Case D. A. Theory and Applications of the Generalized Born Solvation Model in Macromolecular Simulations. Biopolymers 2000, 56, 275–291. [DOI] [PubMed] [Google Scholar]
- Tsui V. C.; D A. Molecular Dynamics Simulations of Nucleic Acids with a Generalized Born Solvation Model. J. Am. Chem. Soc. 2000, 122, 2489–2498. [Google Scholar]
- Drew H. R.; Wing R. M.; Takano T.; Broka C.; Tanaka S.; Itakura K.; Dickerson R. E. Structure of a B-DNA Dodecamer: Conformation and Dynamics. Proc. Natl. Acad. Sci. U. S. A. 1981, 78, 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinina L.; Fernandez L. G.; Huynh-Dinh T.; Subirana J. A. Structure of the d(CGCCCGCGGGCG) Dodecamer: a Kinked A-DNA Molecule Showing Some B-DNA Features. J. Mol. Biol. 1999, 285, 1679–1690. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E.; Ng H. L. DNA Structure from A to B. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 6986–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart K.; Foloppe N.; Baker C. M.; Denning E. J.; Nilsson L.; MacKerell A. D. Optimization of the CHARMM Additive Force Field for DNA: Improved Treatment of the BI/BII Conformational Equilibrium. J. Chem. Theory Comput. 2012, 8, 348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B.; Sharp K.; Yang A. S. Macroscopic Models of Aqueous Solutions: Biological and Chemical Applications. J. Phys. Chem. 1993, 97, 1101. [Google Scholar]
- Sitkoff D.; Sharp K. A.; Honig B. Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models. J. Phys. Chem. 1994, 98, 1978. [Google Scholar]
- Brooks B. R.; Brooks C. L. 3rd; Mackerell A. D. Jr.; Nilsson L.; Petrella R. J.; Roux B.; Won Y.; Archontis G.; Bartels C.; Boresch S.; et al. CHARMM: the Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman P. A.; Massova I.; Reyes C.; Kuhn B.; Huo S. H.; Chong L.; Lee M.; Lee T.; Duan Y.; Wang W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. [DOI] [PubMed] [Google Scholar]
- Brice A. R.; Dominy B. N. Analyzing the Robustness of the MM/PBSA Free Energy Calculation Method: Application to DNA Conformational Transitions. J. Comput. Chem. 2011, 32, 1431–1440. [DOI] [PubMed] [Google Scholar]
- Akerlof G. Dielectric Constants of Some Organic Solvent-Water Mixtures at Various Temperatures. J. Am. Chem. Soc. 1932, 54, 4125–4139. [Google Scholar]
- Zimmerman S. B.; Pheiffer B. H. Helical Parameters of DNA Do Not Change When DNA Fibers Are Wetted - X-Ray-Diffraction Study. Proc. Natl. Acad. Sci. U. S. A. 1979, 76, 2703–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B.; Pheiffer B. H. Does DNA Adopt the C-Form in Concentrated Salt-Solutions or in Organic-Solvent Water Mixtures - an X-Ray-Diffraction Study of DNA Fibers Immersed in Various Media. J. Mol. Biol. 1980, 142, 315–330. [DOI] [PubMed] [Google Scholar]
- Sugita Y. O. Yuko Replica-Exchange Molecular Dynamics Method for Protein Folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar]
- Brooks C. L. III; Berkowitz M.; Adelman S. A. Generalized Langevin Theory for Many-Body Problems in Chemical-Dynamics-Gas-Surface Collisions, Vibrational-Energy Relaxation in Solids, and Recombination Reaction in Liquids. J. Chem. Phys. 1980, 73, 4353–4364. [Google Scholar]
- Best R. B.; Zhu X.; Shim J.; Lopes P. E.; Mittal J.; Feig M.; Mackerell A. D. Jr. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone Phi, Psi and Side-Chain Chi(1) and Chi(2) Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig M.; Karanicolas J.; Brooks C. L. III MMTSB Tool Set: Enhanced Sampling and Multiscale Modeling Methods for Applications in Structural Biology. J. Mol. Graphics Modeling 2004, 22, 377–395. [DOI] [PubMed] [Google Scholar]
- Lu X. J.; Olson W. K. 3DNA: a Software Package for the Analysis, Rebuilding and Visualization of Three-Dimensional Nucleic Acid Structures. Nucleic Acids Res. 2003, 31, 5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 1433–3827–38. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System 1.5.0.4; Schrödinger, LLC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


