Abstract
Rapamycin, a drug that has been shown to increase lifespan in mice, inhibits the target of rapamycin (TOR) pathway, a major pathway that regulates cell growth and energy status. It has been hypothesized that rapamycin and dietary restriction (DR) extend lifespan through similar mechanisms/pathways. Using microarray analysis, we compared the transcriptome of white adipose tissue from mice fed rapamycin or DR-diet for six months. Multidimensional scaling and heatmap analyses showed that rapamycin had essentially no effect on the transcriptome as compared to DR. For example, only six transcripts were significantly altered by rapamycin while mice fed DR showed a significant change in over 1,000 transcripts. Using ingenuity pathway analysis, we found that stearate biosynthesis and circadian rhythm signaling were significantly changed by DR. Our findings showing that DR, but not rapamycin, have an effect on the transcriptome of the adipose tissue, suggesting that these two manipulations increase lifespan through different mechanisms/pathways.
Keywords: Adipose, Fat, Dietary Restriction, Rapamycin, Transcriptome, Microarrays
Introduction
Dietary restriction (DR) is one of the most studied interventions known to extend lifespan across multiple organisms, e.g., it has been shown to increase the lifespan of animals ranging from invertebrates such as yeast, nematodes, and fruit flies to rodents and dogs (Fontana et al., 2010; Lawler et al., 2008; Weindruch et al., 1986). DR is a dietary manipulation in which animals are fed 30-50% less food than that consumed by animals fed ad libitum (AL). Recently, it has been shown that rapamycin increased the lifespan of mice (Anisimov et al., 2011; Fok et al., 2014; Harrison et al., 2009; Miller et al., 2011; Zhang et al., 2013) as well as Saccharomyces cerevisiae (Powers et al., 2006), Caenorhabditis elegans (Robida-Stubbs et al., 2012), and Drosophila melanogaster (Bjedov et al., 2010). Rapamycin was initially discovered by its ability to inhibit the Target of Rapamycin (TOR) pathway, which is a nutrient sensing pathway involved in regulation of multiple functions in the cell from growth to energy metabolism (Foster and Fingar, 2010). In mammals, mTOR forms two complexes, mTORC1 and mTORC2, and rapamycin was shown to inhibit mTORC1 signaling through the specific binding to FKBP12 which inhibits the interaction of mTOR and Raptor (Hay and Sonenberg, 2004). Although rapamycin was initially thought to inhibit only mTORC1 signaling, more recent studies suggest that long term rapamycin treatment may also affect mTORC2 signaling (Thomson et al., 2009).
While it is unclear as to the mechanism responsible for the increased lifespan by DR, it has been hypothesized that DR and rapamycin increase lifespan through similar mechanisms/pathways. For example, Kaeberlein's group reported that a mutation in TOR in S. cerevisiae increased replicative lifespan similar to DR and that DR does not extend the lifespan of TOR mutants (Kaeberlein et al., 2005). TOR inhibition by RNAi in C. elegans also does not show further increased lifespan in the eat-2 mutant, a DR mimetic in C. elegans, compared to the wild type N2 background (Hansen et al 2008). However, Drosophila fed rapamycin showed an increase in lifespan beyond the extension shown with flies on DR alone (Bjedov et al., 2010), suggesting that rapamycin and DR increased lifespan in part with pathways independent of those used by DR to extend lifespan.
In this study, we focused on the effect of DR and rapamycin on the transcriptome of epididymal fat, a white adipose tissue. The adipose tissue is one of the largest organs in mammals, and plays an important role in inflammation and insulin sensitivity, which have been proposed to be important factors in aging (Tchkonia et al., 2010). In addition, an increase in obesity is associated with increase in age-associated diseases (Fontaine et al., 2003), and mice fed a high fat diet to increase obesity have been shown to have a decreased lifespan (Minor et al., 2011). In addition, removal of epididymal and perirenal visceral adipose tissue has been shown to increase the lifespan of rats (Muzumdar et al., 2008), while also improving insulin sensitivity (Gabriely et al., 2002). Clearly, the role of the adipose tissue is important in aging and longevity. Because of the role of adipose in the modulation of lifespan, we studied the transcriptome of mice fed DR or rapamycin and found major differences in the expression of transcripts in white adipose tissue. DR significantly altered the expression of over 1,000 transcripts, while the expression of only six transcripts were altered significantly by rapamycin.
Material and methods
Animals and feeding regiment
Male C57BL/6 mice (N=8 per group) were purchased from The Jackson Labs (Bar Harbor, ME) and placed on a commercial mouse chow, 7012 Teklad LM-450 (Harlan Laboratories, Madison, WI), until 2 months of age. At 2 months of age, the mice were separated into three dietary regimens: ad libitium (AL), 40% diet restriction (DR), and AL diet plus 14 ppm of rapamycin in the food. The AL group was fed without restriction a commercial mouse chow, Purina Mills Test Diet Control #1810306 (Purina Mills, St. Louis, MO). The DR group was fed 40% less food than eaten by the AL mice. The rapamycin group was fed the AL diet with 14 ppm of encapsulated rapamycin in the food as described Harrison et al (2009). Mice were maintained on these dietary conditions until 8 months of age (6 months of treatment). Mice were then euthanized by carbon dioxide and adipose tissues were collected, snap frozen in liquid nitrogen, and stored at -80°C until used. All procedures followed the guidelines approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio.
RNA processing
Total RNA from frozen epididymal fat (100 mg) was extracted using RNeasy kit (Qiagen, Valencia, CA) following manufacturer's protocols. RNA quality was assessed by agarose gel and Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA), and RNA quantity was determined using the Nanodrop (Thermo Scientific, Wilmington, DE). RNA was then processed into cRNA probes for hybridization to arrays using Illumina Total RNA prep kit from Ambion (Life Technologies, Grand Island, NY) following manufacturer's protocols. Adipose cRNA probes were then hybridized to Illumina Mouse Ref8 microarrays (V2.0, Illumina, San Diego, CA) following manufacturer's protocol. Hybridized arrays were scanned by iSCAN system (Illumina, San Diego, CA). Hybridization of probes and scanning of the arrays were done at the Genomics Resources Core, University of Texas Health Science Center at San Antonio.
Microarray Analysis
Data from iDAT files was extracted by Genome Studios software (v 1.6, Illumina, San Diego, CA). Data was then transformed and normalized using log2 transformation and the z-normalization method as described by Cheadle et al (2003). For statistical analysis, one way ANOVA with pairwise comparisons was done on the microarray data using R (v 2.15.1, R Development Core Team). False discovery analysis was then done to the dataset using R package “qvalue” (v 1.30.0, Dabney, A and Storey, J). Multidimensional scaling analysis was done using Matlab (2011a, The Mathworks, Natick, MA). For gene analysis, we analyzed microarray data with filtering criteria of q<0.05 and median fold change greater than 15% in DR vs. AL and Rapa vs. AL. Significantly altered genes were then plotted in a heatmap using Matlab (2011a, The Mathworks, Natick, MA). Pathway analysis was done using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) which provides association of significantly changed genes into pathways.
Quantitative Real-Time PCR
Using the total RNA extracted from frozen adipose tissues described above, one μg of RNA was used to make cDNA using the Ambion retroscript kit following manufacturer's instructions (Life Technologies, Grand Island, NY). Primers were designed using the NCBI primer blast and was obtained from Sigma-Aldrich (St. Louis, MO). The sequences of the primers are listed in Supplementary Table 1. For qRT-PCR, three dilution of cDNA (200 ng/μl, 20 ng/μl, and 2 ng/μl) in triplicate was used in the analysis of each gene using the EvaGreen master mix (Biotium, Hayward, CA) with 1 μl of cDNA at the indicated dilutions in a 10 μl reaction following manufacturer's protocol inABI 7900HT qPCR system (Applied Biosystem, Inc., Foster City, CA). The ΔΔCT method was used to quantify the qRT-PCR results and gene products were assayed using agarose gels and dissociation curves. The qRT-PCR data were analyzed using one-way ANOVA with Tukey's post-hoc test.
Measurement of NEFAs, glycerol and triglycerides in serum samples
Serum levels of non-esterified fatty acids (NEFAs), triglycerides, and glycerol, were measured in serum from un-fasted mice treated for 6 months with DR or rapamycin using a colorimetric assay, using a kit purchased from Wako Chemicals (NEFAs and triglycerides; Richmond, VA), and Sigma (glycerol; St. Louis, MO).
Statistical Analysis
Unless specifically indicated, all statistical analysis was done using one-way ANOVA with Tukey's post-hoc test.
Database linking
Microarray data for this study has been deposited in Gene Expression Omnibus with accession number GSE52825 and can be viewed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=gporsicovnqvrgh&acc=GSE52825
Results
The effect of DR and rapamycin on body fat and specific fat depots was characterized by quantitative magnetic resonance imaging for percent body fat and determined by fat depots weight vs. total body weight. No significant differences were observed in overall fat mass and lean mass between AL and rapamycin groups (Figure1A). In contrast, DR mice show a major decrease in both lean and fat mass, i.e., fat mass decreased 63% and lean body mass 20% compared to AL. Moreover, our data showed that the decrease in fat mass by DR is mainly due to a decrease in the weight of all fat depots: eipdidymal, perirenal, subcutaneous (SQ), and mesenteric fat depots (Figure 1B).
Figure 1. Effect of DR and rapamycin on body fat and fat depots.
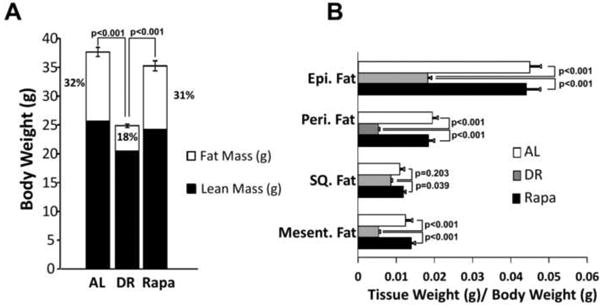
Weight (gr) of the distribution of lean mass (solid bars) and fat mass to body weight (open bars), respect to total body weight. (A), determined in AL (open bars), DR (grey bars), and Rapa (solid bars) in fed mice at 8 months of age (after 6 months of treatment). Fat depot weights are shown relative to body weight (B) at 8 months of age. The data were obtained from 11-12 mice per group and expressed as mean ± standard error of the mean (SEM). Data were analyzed using one-way ANOVA with the Tukey's post-hoc test; the p-values are marked for the values that are significant.
Using Illumina microarrays, we measured the levels of transcripts in eipdidymal white adipose tissue from mice fed AL, DR, or rapamycin. We first analyzed the data obtained from all probes detected with a p-detection value of less than 0.02 (24,114 transcripts) using an unbiased multidimentional scaling analysis (MDS). MDS uses data from principle components analysis, which allows one to determine which samples are similar based on variance. The data in Figure 2 shows that the DR mice group separately compared to mice in the AL and rapamycin groups, while mice fed AL and rapamycin group together. Using heatmap analysis with hierarchical clustering, we compared the similarities and differences in the expression of individual genes in each of the animals studied. The heatmap in Figure 3 shows that all of the DR mice showed a similar expression pattern that was distinct from the expression patterns of the mice fed AL or rapamycin. Thus, compared to DR, rapamycin has essentially no effect on transcriptome of epididymal fat of mice after 6 months of feeding.
Figure 2. Multidimensional Scaling (MDS) analysis shows that DR is different from AL and Rapa.
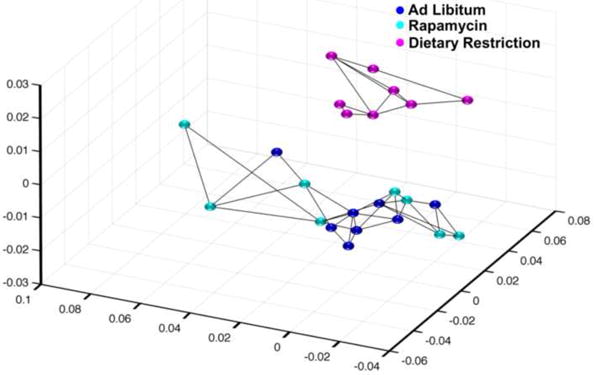
Using all the probes detected (24,114 probes) with a p-detection value of 0.02, the MDS analysis is shown in a 3D plot. Each point represents one sample, with AL groups in blue, DR groups in magenta, and Rapa groups in cyan color. The lines between points indicate the nearest neighbors.
Figure 3. Transcriptome analysis indicate that DR is different than AL but that Rapa is highly similar to AL.
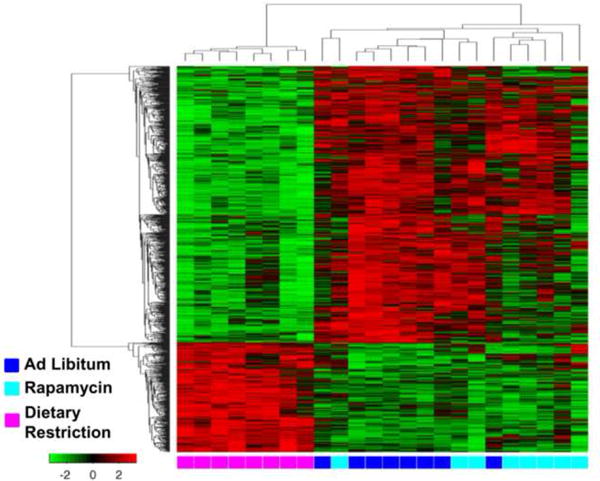
Genes that are significantly different to AL are shown for DR and Rapa using filtering criteria of a false discovery rate of less than 0.05 and a median change of greater than 15%.The heatmap shows all the probes significantly changed by DR or Rapa (1023 probes) using average linkage hierarchical cluster with Euclidean distance for both the ranking of genes and samples. Gene expression levels are shown by red for high and green for low expressing genes. The source for each RNA sample is shown at the bottom of the heat map: blue for AL, magenta for DR, and cyan for Rapa are shown at the bottom of the heat map. A list of the genes that changed is shown in supplementary file 1 tab 1 in the same order as the heatmap from top to bottom.
To identify the specific gene transcripts that change with DR or rapamycin compared to mice fed AL, we used a filtering criteria of a false discovery rate q<0.05 and >15% median change from that observed in the AL mice. For DR mice, we found that 1,020 transcripts are altered compared to AL mice; the levels of 287 transcripts were up-regulated and 733 transcripts down-regulated. A list of the transcripts that significantly changed is given in supplementary file 1 tab1. The top 3 genes that showed the greatest increase (5-6 fold) in expression with DR were Acsm3, Elovl6, and Acacb and the top 3 genes that showed the greatest decrease (82-83%) in expression with DR were Sfrp5, Rbm28, and Trp53inp2. Interestingly, we found that that the levels of only six transcripts changed significantly in the rapamycin-fed mice compared to AL-fed mice; all transcripts were reduced. The six genes down-regulated by rapamycin were Cox8b, Gpc1, Aldh5a1, Mogat2, Serpina3, and Zfp91. Interestingly, three of the six genes down-regulated by rapamycin were also significantly down-regulated by DR. Those 3 genes were Gpc1, Mogat2, and Serpina3.
To confirm the microarray data, we measured the levels of transcripts using quantitative real-time PCR. We picked three genes that were highly changed by DR or rapamycin from each of the four possible categories: genes that were significantly down-regulated by DR and rapamycin, genes up-regulated by DR and did not change in rapamycin, genes down-regulated by DR and did not change with rapamycin and genes down-regulated by rapamycin and did not change with DR. As shown in Figure 4A, qRT-PCR analysis showed that the mRNA levels of Gpc1, Mogat2, and Serpina3b were down-regulated by both DR and rapamycin as we had found by microarray analysis. The levels of Gpc1, Mogat2, and Serpina3b mRNA transcripts were reduced 48%, 48%, and 56% in DR and 56%, 68%, 88% in mice treated with rapamycin, respectively compared to AL. Acsm3, Elovl6, and Acacb were genes that were up-regulated by DR and did not change significantly with rapamycin (Figure 4B). We found that DR increased the mRNA levels of these genes 7- to 17-fold over the mRNA levels found in the AL group, and rapamycin did not significantly alter the levels of these transcripts. Sfrp5, Rbm28, and Trp53inp2 were genes that were down-regulated by DR and did not change with rapamycin (Figure 4C). For these genes, we found that DR down-regulated the mRNA levels significantly (60 to 85%) compared to the AL group while rapamycin had no effect on the mRNA levels of these genes. The genes that were significantly down-regulated by rapamycin, and did not change with DR were Aldh5a1, Cox8b, and Zfp91 (Figure 4D). For these three genes, we found that the mRNA levels for both Aldh5a1 and Cox8b were significantly reduced (31 and 77%, respectively) for the rapamycin mice compared to AL mice. Although the mRNA levels of Zfp91 were reduced for the rapamycin mice compared to AL mice, this decrease (15%) was not statistically significantly, however, the mRNA levels of Zfp91 were significantly lower than the DR mice. One exception we found with our real-time PCR analysis was the expression of Cox8b by DR mice. The data in Figure 3D show a 110% increase in this transcript in DR mice compared to AL mice while the microarray data show no increase. Thus, the qRT-PCR analysis confirmed our microarray results with exception of Zfp91 and Cox8b.
Figure 4. qRT-PCR analysis of genes found to be significantly changed with DR or Rapa.
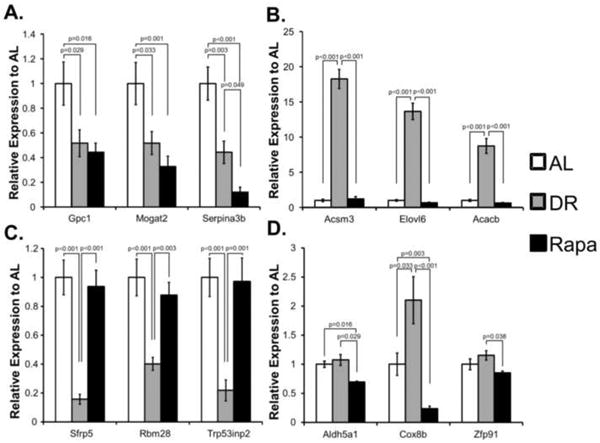
To confirm the microarray data, the levels of 12 mRNA transcripts that were observed to change with either DR or Rapa were measured by qRT-PCR analysis using the mRNA samples used for microarray analysis. (A) Genes down-regulated in DR and Rapa groups: Gpc1, Mogat2, and Serpina3b. (B) Genes down-regulated in Rapa but not significantly changed in DR: Aldh5a1, Cox8b, and Zfp91. (C) Genes down-regulated in DR only but not significantly changed in Rapa: Sfp5, Rbm28, and Trp53inp2. (D) Genes up-regulated by DR but not significantly changed in Rapa Acsm3, Elvol6, and Acacb. The mean ± standard error of the mean is shown. The data were analyzed using one-way ANOVA with Tukey's post-hoc test and the p-values are shown.
We analyzed the transcriptome further by conducting a pathway analysis on the data using Ingenuity pathway analysis, a program that places the transcripts that change into pathways using a database constructed from the literature. This analysis allows one to determine if genes in a pathway are altered, which will give one insight into potential mechanisms whereby the changes in the transcriptome could physiologically impact the organism. Because only six genes were significantly changed by rapamycin, we were unable to perform this analysis on the mice treated with rapamycin. The ingenuity pathway analysis identified 41 pathways that were significantly (p<0.05 determined by Fisher's exact test) altered in the adipose tissue of DR mice. However, when we correct the pathway analysis for false discovery, only 2 pathways, the stearate biosynthesis (I) and circadian rhythm signaling, were found to be significantly altered by DR using a FDR significance cutoff of adjusted p<0.05 (Figure 5A). Because our data indicate that DR and rapamycin have different effects on lipid metabolism, we measured the lipid profile in the serum of DR and rapamycin treated mice. While our data showed that both, DR and rapamycin, have a similar effect on serum glycerol and non-esterified fatty acids (Figure 5B and 5C), the amount of serum triglycerides were significantly reduced by DR, but not by rapamycin treatment (Figure 5D).
Figure 5. Pathway analysis of genes found significantly changed with DR and serum lipid profile analysis.
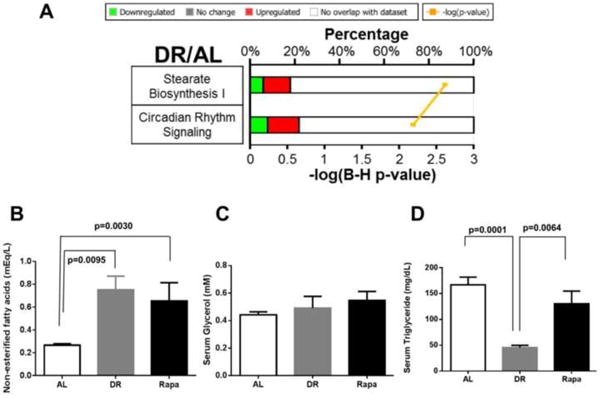
Pathway analysis using Ingenuity Pathway analysis program showed 2 pathways were found to be significantly changed by DR with B-H p<0.05 (A). The bold number on the left of each pathway indicates the possible number of genes for that particular pathway while the color bars indicate percentage of genes matching from the significant gene list in DR. The color red indicates up-regulated while green indicates down-regulated genes. The yellow line indicates the –log(B-H p-value) with the smallest p-value at the top. A list of the pathways that is significantly changed by p-value is shown in supplementary file 1 tab2 and by B-H p-value in supplementary file 1 tab 3. Serum levels of non-esterified fatty acids (NEFAs) (B), glycerol (C), and triglycerides (D) were determined by a colorimetric assay. The data were obtained from 6-7 mice per group [AL (solid bars), DR (gray bars), and Rapa (open bars)]. The data were expressed as mean ± SEM and analyzed using one-way ANOVA with the Holm-Sidek post-hoc test.
Discussion
DR is the most studied manipulation to increase lifespan. Although the mechanism(s) responsible for the life-extending action of DR is unknown, knockdown of autophagy genes in C. elegans has been shown to negate the lifespan extension effects of the DR mimetic mutant, eat-2 (Hansen et al., 2008), and the extension of lifespan in invertebrate models of decreased TOR signaling is not increased further by DR in S. cerevisiae or the eat-2 mutant in C. elegans (Hansen et al., 2007). These studies suggest that autophagy and TOR are key pathways in the extension of lifespan seen in DR. Because the inhibition of TOR signaling is well known to be key to rapamycin's mechanism of action; it was logical to assume when Harrison et al. (2009) showed that rapamycin increased the lifespan of mice that rapamycin was a mimetic of DR. However, there are no studies comparing the actions of rapamycin and DR in mice and a study in Drosophila suggest that DR and rapamycin may have differential effects on lifespan, e.g., a combination of rapamycin and DR treatment extended the lifespan of flies further than either treatment alone (Bjedov et al., 2010).
To compare the effects of rampamycin and DR on the physiology/biology of mice, our group has studied the effect of these two aging interventions on gene expression. Initially, we compared the effect of DR and rapamycin on the liver transcriptome (Fok et al., 2013a) because the liver is one the first tissue to be exposed to rapamycin and it expresses a diverse number of metabolomics pathways. We found that less than 20% of 2724 transcripts that changed significantly by either DR or rapamycin were shared by both treatments. Fok et al. (2013a) also compared the liver metabolome of mice fed rapamycin and a DR-diet. An even greater difference between DR and rapamycin was observed in the 546 metabolites detected in liver. No metabolites were significantly changed by rapamycin while 173 known and unknown metabolites were significantly altered by DR. Thus, our data on the liver transcriptome and metabolome suggest that DR and rapamycin have different effects in mice. However, these data are limited to only one tissue.
In this study, we extended our research to epididymal fat, a white adipose tissue, to determine the similarity and differences in the effect of DR and rapamycin on mice. We chose the epididymal depot because is the largest depot of visceral fat in the mice and it is the site where most of the lipids are stored (Sackmann-Sala et al., 2012). In addition, it has been argued that the decrease in fat mass arising from DR has several beneficial effects that could be important in DR's anti-aging action, e.g., insulin sensitivity and inflammation (Barzilai and Gabriely, 2001). For example Barzilai's group (Muzumdar et al., 2008) showed that the removal of epydidmyal and perirenal visceral adipose tissue increases the lifespan of rats, while also improving insulin sensitivity (Gabriely et al., 2002). In addition, an increase in obesity is associated with increase in age-associated diseases and decrease with life expectancy (Fontaine et al., 2003), e.g., mice fed a high fat diet to increase obesity have been shown to have a decreased lifespan (Minor et al., 2011). Adipose tissue is an important regulator of inflammation because it can release and regulate adipocytokines that have important roles in inflammation and immunity. Adipocytokines such as adiponectin, which decreases with age and obesity, can suppress inflammation through the inhibition of macrophage activity, (Tilg and Moschen, 2006). Chronic inflammation is detrimental in healthy aging and can increase risks of age-associated diseases. For example, studies done by Masternak et al. (2012) with long-the lived growth hormone receptor knockout (GHRKO) mice demonstrated that the secretory activity of the visceral fat (epididymal and perirenal fat) is necessary and sufficient to promote the pro-longevity phenotype in these mice, i.e., high insulin sensitivity and lower levels of inflammatory cytokines. Clearly, the role of the adipose tissue is important in aging and longevity.
Adipose tissue has been reported to be affected by rapamycin as shown by a decrease in mTOR signaling, i.e., phosphorylation of ribosomal protein S6 (Blanchard et al., 2012; Harrison et al., 2009; Zhang et al., 2013). In addition, 15 days of rapamycin injection in Sprague-Dawley rats reduces adiposity, decrease fat cell numbers, and down-regulate genes involved in lipid uptake, and storage (Houde et al., 2010). However using C57BL/6 mice, we showed that rapamycin had no effect on total fat mass or fat depots (Figure 1) in contrast to DR treatment, which resulted in ∼30% reduction in body weight that largely arose from a loss of adipose weight across all fat deposits (Fok et al., 2013b). Our group showed that male or female mice fed rapamycin for 6 months or 21 months had body weight and fat mass similar to mice fed a control diet (Fok et al., 2013a; Fok et al., 2014; Fok et al., 2013b; Zhang et al., 2013).
In this study, we found that over 1,000 genes in white adipose tissue were altered by 6 months of DR, but no major changes were observed in rapamycin treated mice. The changes in expression of transcripts by DR were similar to that reported by two previous studies. Higami et al. (2004) reported that DR altered the expression of 622 transcripts in white adipose tissue of male C57BL/6 mice fed 41% DR from 6 weeks of age to 11 months of age using Affymetrix microarrays. Genes in the following pathways were observed to change with DR in our study and the study by Higami et al. (2004) using epididymal fat tissue: glycolysis (Pdha1 expression was up-regulated and Pfkp was down-regulated) gluconeogenesis (Pdk1 was up-regulated), amino acid metabolism (Bcat2 was up-regulated), lipogenesis (an up-regulation of Slc27a, Enpep, and Acly) and steroid biosynthesis (up-regulation of Hsd11b1 and Hsd17b10). Wheatley et al. (2011) measured the expression of transcripts in visceral fat from 4-month-old C57Bl/6 mice fed a 30% DR diet for 8 weeks after 8 weeks of a high fat diet using Affymetrix microarrays. They found 705 genes were significantly changed by DR. Comparing our study and the study by Wheatley et al. (2011), we found a similar up-regulation of Gsta4, Cidea, Acss2, Pdk1, Aacs, Sorl1, Acly, Enpep, and Pdha1 and down-regulation of Prps1, Mmp3, Lgmn, Capg, and Cyba in the DR mice. All three microarray studies reported similar effect of DR in the adipose tissue on several families of genes, e.g, the glutathione-S-transferases, histone associated genes, and collagen genes. Subunits for glutathione S-transferase subunits, such as Gstt2, Gsta3, and Gsta4, are up-regulated in DR fed mice, suggesting an increase glutathione biosynthesis, which is consistent with the report by Fok et al. (2013b) showing an increase in the reduced form glutathione in mice fed DR. Histone associated genes, that code for histone cluster 1 and 2, and collagen genes, that code for extra-cellular collagen fibers, are down-regulated by DR, which suggest a decrease in lipid cell growth and expansion.
We identified two pathways from our transcriptome data that changed with DR using IPA analysis: circadian rhythm controls the sleep-wake period for animals and there is also evidence that adiposity and circadian rhythm are correlated in mice. Multiple genetic knockout mouse models of genes that regulate energy in the white adipose tissue have shown that a decrease in adiposity is correlated with alterations is circadian rhythm as well as activity during the light cycle (Gimble et al., 2011). In addition, mice fed a high fat diet show altered the expression of circadian rhythm genes (Kohsaka et al., 2007), and rats that have a decreased light cycle or sleep cycle, have increased adiposity and loss of circadian rhythm (Wideman and Murphy, 2009). Interestingly, mice null for Bmal1, one of the genes involved in generating circadian rhythm, show a significant decrease lifespan. Thus the up-regulation of circadian rhythm signaling in white adipose tissue of the DR mice would be consistent with improved healthspan and increased lifespan, which are observed in mice and rats fed DR-diets.
Stearate biosynthesis I and circadian rhythm signaling. Both IPA pathways appear from our transcriptome data to be up-regulated by DR. Stearate biosynthesis (I) is a pathway that produces stearic acid, which is one of the most abundant saturated fatty acids, from the precursor palmitate. Elongation of palmitate to stearate involves elongation factors, of which there are 6 in mice. Using qRT-PCR analysis, we showed Elovl6, one of the elongation factors involved in stearate biosynthesis (I), to be highly up-regulated. The increase stearate biosynthesis suggests that mice fed DR, which show a major loss in fat mass, are compensating by increasing the production in stearic acid that can then be converted to oleic acid to produce triglycerides that can be used by the other tissues (Miyazaki et al., 2001). For example, the increase in stearate biosynthesis pathway could stimulate the increase in brown fat to regulate thermogenesis (Westerberg et al., 2006) to compensate for changes in the core body temperature of mice on DR (Rikke et al., 2003). Our data on serum lipid profile indicate that DR and rapamycin have similar effects on the levels of glycerol and non-esterified fatty acids, suggesting that these two treatments have a similar effect on lipid metabolism in the liver, which serves as the hub for fatty acid synthesis and lipid transport in circulation through lipoprotein synthesis (Nguyen et al., 2008). However, the serum levels of triglycerides (TG) is significantly reduced by DR (Figure 5D), which may be due to a significant decrease in fat depots mass, which are the main store for TG (Coleman and Mashek, 2011).
The major observation of our study was that rapamycin had a negligible effect of the transcriptome of white adipose tissue compared to DR. Only six transcripts were altered by rapamycin: Aldh5a1, Cox8b, Zfp91, Gpc1, Mogat2, and Serpina3b. Gpc1, Mogat2, and Serpina3b were also down-regulated by DR. Thus, the response of white adipose tissue is quite different for DR and rapamycin, however these data are only in adipose tissue and might not reflect what is occurring in other tissues. Based on the transcriptome date in adipose tissue it appears that DR and rapamycin have quite different effects on gene expression and are acting through different mechanisms, at least with respect to this tissue. Our studies in the liver and fat suggest that lifespan extension by DR and rapamycin most-likely occurs through different mechanisms and pathways, suggesting that a combination of DR and rapamycin could have a further extension of lifespan than either manipulation alone.
Supplementary Material
Highlights.
First study to describe the transcriptome of white adipose tissue from mice fed rapamycin.
Rapamycin changed few transcripts compared to mice fed dietary restriction.
Dietary restriction significantly altered the sterate biosynthesis I and circadian rhythm signaling in white adipose tissue.
Acknowledgments
We would like to thank UTHSCSA LAR for mouse husbandry and the Genomics Core Shared Resource, which is supported by UTHSCSA, NIH-NCI P30CA054174 (CTRC of UTHSCSA) for microarray hybridization and scanning, and Sherry Dodds for help in RNA isolation.
Funding: Financial support was provided by The San Antonio Nathan Shock Aging Center (1p30-AG-13319, AR), National Institutes of Health (NIH) RC2 Grand Opportunity grant (AG036613, AR), NIH T32 Training Grant (AG021890, WF), the Ellison Medical Foundation (VP), and the Intramural Research Program of the NIH, National Institute on Aging, and VA Merit grant (AR) from the Department of Veteran Affairs. Computation Support was provided by the Computational System Biology Core funded by the National Institute on Minority Health and Health Disparities (G12MD007591) from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anisimov VN, Zabezhinski MA, Popovich IG, et al. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10:4230–4236. doi: 10.4161/cc.10.24.18486. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Gabriely I. The role of fat depletion in the biological benefits of caloric restriction. J Nutr. 2001;131:903S–906S. doi: 10.1093/jn/131.3.903S. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard PG, Festuccia WT, Houde VP, et al. Major involvement of mTOR in the PPARgamma-induced stimulation of adipose tissue lipid uptake and fat accretion. J Lipid Res. 2012;53:1117–1125. doi: 10.1194/jlr.M021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, et al. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Mashek DG. Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem Rev. 2011;111:6359–6386. doi: 10.1021/cr100404w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok WC, Bokov A, Gelfond J, et al. Combined treatment of rapamycin and dietary restriction has a larger effect on the transcriptome and metabolome of liver. Aging Cell. 2013a doi: 10.1111/acel.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok WC, Chen Y, Bokov A, et al. Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PLoS ONE. 2014;9:e83988. doi: 10.1371/journal.pone.0083988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok WC, Zhang Y, Salmon AB, et al. Short-term treatment with rapamycin and dietary restriction have overlapping and distinctive effects in young mice. J Gerontol A Biol Sci Med Sci. 2013b;68:108–116. doi: 10.1093/gerona/gls127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine KR, Redden DT, Wang C, et al. Years of life lost due to obesity. JAMA. 2003;289:187–193. doi: 10.1001/jama.289.2.187. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KG, Fingar DC. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem. 2010;285:14071–14077. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely I, Ma XH, Yang XM, et al. Removal of visceral fat prevents insulin resistance and glucose intolerance of aging: an adipokine-mediated process? Diabetes. 2002;51:2951–2958. doi: 10.2337/diabetes.51.10.2951. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Sutton GM, Ptitsyn AA, et al. Circadian rhythms in adipose tissue: an update. Curr Opin Clin Nutr Metab Care. 2011;14:554–561. doi: 10.1097/MCO.0b013e32834ad94b. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Higami Y, Pugh TD, Page GP, et al. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- Houde VP, Brule S, Festuccia WT, et al. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 2010;59:1338–1348. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Lawler DF, Larson BT, Ballam JM, et al. Diet restriction and ageing in the dog: major observations over two decades. Br J Nutr. 2008;99:793–805. doi: 10.1017/S0007114507871686. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor RK, Baur JA, Gomes AP, et al. SRT1720 improves survival and healthspan of obese mice. Scientific reports. 2011;1:70. doi: 10.1038/srep00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Man WC, Ntambi JM. Targeted disruption of stearoyl-CoA desaturase1 gene in mice causes atrophy of sebaceous and meibomian glands and depletion of wax esters in the eyelid. J Nutr. 2001;131:2260–2268. doi: 10.1093/jn/131.9.2260. [DOI] [PubMed] [Google Scholar]
- Muzumdar R, Allison DB, Huffman DM, et al. Visceral adipose tissue modulates mammalian longevity. Aging Cell. 2008;7:438–440. doi: 10.1111/j.1474-9726.2008.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Leray V, Diez M, et al. Liver lipid metabolism. J Anim Physiol Anim Nutr (Berl) 2008;92:272–283. doi: 10.1111/j.1439-0396.2007.00752.x. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, et al. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikke BA, Yerg JE, 3rd, Battaglia ME, et al. Strain variation in the response of body temperature to dietary restriction. Mech Ageing Dev. 2003;124:663–678. doi: 10.1016/s0047-6374(03)00003-4. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Morbeck DE, Von Zglinicki T, et al. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9:667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol. 2006;6:772–783. doi: 10.1038/nri1937. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, et al. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Westerberg R, Mansson JE, Golozoubova V, et al. ELOVL3 is an important component for early onset of lipid recruitment in brown adipose tissue. J Biol Chem. 2006;281:4958–4968. doi: 10.1074/jbc.M511588200. [DOI] [PubMed] [Google Scholar]
- Wheatley KE, Nogueira LM, Perkins SN, et al. Differential effects of calorie restriction and exercise on the adipose transcriptome in diet-induced obese mice. Journal of obesity 2011. 2011:265417. doi: 10.1155/2011/265417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman CH, Murphy HM. Constant light induces alterations in melatonin levels, food intake, feed efficiency, visceral adiposity, and circadian rhythms in rats. Nutr Neurosci. 2009;12:233–240. doi: 10.1179/147683009X423436. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bokov A, Gelfond J, et al. Rapamycin Extends Life and Health in C57BL/6 Mice. J Gerontol A Biol Sci Med Sci. 2013 doi: 10.1093/gerona/glt056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


