Abstract
Xanthomonas phytopathogenic bacteria produce unique transcription activator-like effector (TALE) proteins that recognize and activate specific plant promoters through a set of tandem repeats. A unique TALE-DNA-binding code uses two polymorphic amino acids in each repeat to mediate recognition of specific nucleotides. The order of repeats determines effector’s specificity toward the cognate nucleotide sequence of the sense DNA strand. Artificially designed TALE-DNA-binding domains fused to nuclease or activation and repressor domains provide an outstanding toolbox for targeted gene editing and gene regulation in research, biotechnology and gene therapy. Gene editing with custom-designed TALE nucleases (TALENs) extends the repertoire of targeted genome modifications across a broad spectrum of organisms ranging from plants and insect to mammals.
Keywords: TALE, TALEN, ZFN, FokI, DNA editing
INTRODUCTION
Engineered DNA-binding domains (DBDs) fused with different catalytic or effector domains allow researchers to edit DNA sequences or regulate gene expression at specific DNA loci within complex eukaryotic genomes. There are two main classes of engineered site-specific DBDs: zinc finger-based DBDs and transcription activator-like effector (TALE)-based DBDs. Site-specific zinc finger nucleases (ZFN) for genome editing (reviewed in [1]) pawed the road for the TALE Nuclease (TALEN) technology, which is based on a unique modular DBD of TALEs from plant-pathogenic bacterial genus Xanthomonas. A commonly used nuclease domain in ZNFs and TALENs is the dimerizing FokI endonuclease cleavage domain, which introduces a double-strand break (DSB) [2, 3]. DSBs at targeted loci rapidly increase local frequencies of homologous recombination. This enables the extension of genetic manipulations to virtually any model organisms and cell line.
In this review, we first recapitulate discovery of TALEs and deciphering of their binding code. Next, we describe the structure of TALE DBD and its implications for biotechnology. Finally, we discuss TALE-based nucleases and genome regulators as distinct categories of engineered site-specific proteins that share a common DBD but differ in their effector domains, hence in their mode of action.
TALEs—VIRULENCE FACTORS OF XANTHOMONAS
Gram-negative γ-proteobacteria of the genus Xanthomonas are important plant pathogens affecting worldwide yields of crop plants such as wheat, rice, cassava or cotton. Xanthomonas enter host plants through surface wounds or natural openings and multiply inside plant tissues (reviewed in detail in [4]). To facilitate a productive bacterial infection in plants, Xanthomonas secrete a cocktail of effector proteins into host cells, including the TALE family proteins (originally denoted AvrBs3-family effectors) that function as eukaryotic-like transcription factors. TALEs are secreted directly into the plant cell cytoplasm [5] and transported into the nucleus via importin-α [6]. Recognition of specific promoters and subsequent interaction with the basal transcriptional machinery induce transcription of specific host plant genes.
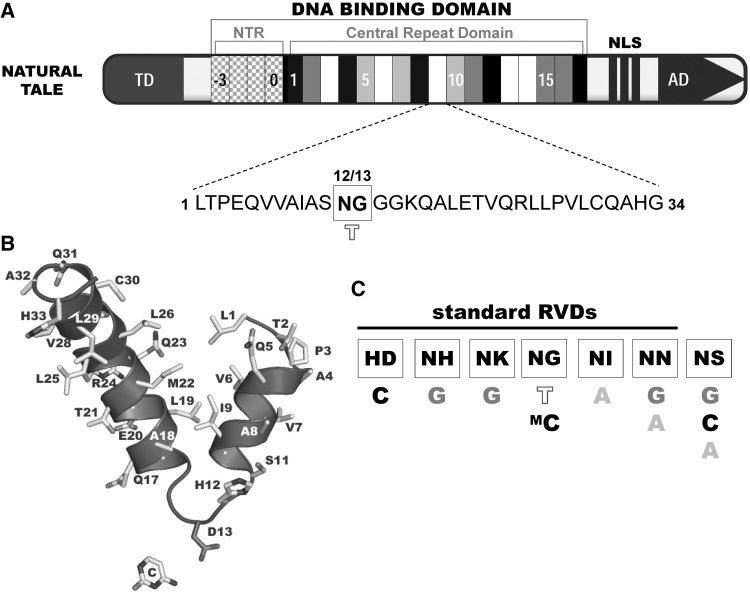
TALEs exhibit exceptional DNA-binding specificity stemming from a unique domain organization [7, 8] (Figure 1A). The common feature of natural TALEs is their DBD composed of 7–34 highly homologous direct repeats in the central part of the protein [9]. Typically, each repeat module (Figure 1B) has 34 amino acids (aa) in length; the last C-terminal truncated repeat, so-called half-repeat, consists of 20 aa. Two polymorphic aa residues at positions 12 and 13 form the repeat-variable diresidue (RVD), where the residue 13 is responsible for preferential binding of the repeat module to a single specific nucleotide in the major groove of target DNA sequence (summarized in [10]). The binding code was deciphered independently in 2009 by two groups who found a simple cipher, where common RVDs HD, NG/HG and NI recognize almost exclusively cytosine, thymine and adenine, respectively; whereas NN or NS has more degenerated specificity [7, 8] (Figure 1C). The order of repeat modules from N to C-terminus within TALE DBD then corresponds to the recognized DNA sequence in 5′ to 3′ direction such that each repeat contacts one specific DNA base pair via the RVD.
Figure 1:
TALE domain composition and DNA-binding code. (A) TALEs contain nuclear localization signals (NLS) and an activation domain (AD) to function as transcriptional activators. A central tandem repeat domain confers specific DNA-binding and host specificity. Translocation signal (TD) and four cryptic repeats required for initiation of DNA binding and for the recognition of 5′-T0 are located at the N-terminus (chequered rectangles). Each 34 amino acid (aa) long repeat in the CRD binds to one nucleotide with specificity determined mainly by aa at position 13. One sample repeat is shown below the protein scheme. Numbers 12/13 refer to aa positions within the repeat. (B) Structure of an individual TALE repeat module. The repeat has 34 amino acids in length and takes a loop–helix secondary structure where two α-helices are linked by short ‘RVD loop’. The residue 13 is responsible for preferential binding of the repeat module to a single specific nucleotide in the major groove of target DNA sequence (C, in this case). (C) Repeat types have specificity for one or several nucleotides. Only bases of the DNA leading strand are shown. Adapted from [7, 9, 10, 11].
While the TALE central repeat domain (CRD) determines the specificity, the DBD is further extended ∼150 aa into the N-terminal region (NTR), immediately preceding the first canonical repeat [12–14]. This region is composed of four cryptic repeats and substantially contributes to the overall basic charge of TALE proteins [12, 15]. The NTR is necessary for binding of TALEs to DNA and mediates interaction with a conserved thymine at position 0 (discussed in more detail later). The N-terminus of natural TALE proteins also contains secretion and translocation signals required for delivery into host cells [16]. The C-terminal region carries conserved three monopartite nuclear localization signals and a conserved eukaryotic-like acidic transcriptional activation domain [6, 17–19]. Notably, TALE-like proteins were also identified in the plant pathogenic bacterium Ralstonia solanacearum [20, 21] offering additional options for engineering DBDs.
SPECIFICITY OF DNA BINDING BY TALEs
Crystallographic studies of TALEs bound to their target sequences unraveled that TALE DBD forms a right-handed superhelical assembly wrapped around B-form DNA duplex (Figure 2) and explained specific repeat-nucleotide interactions [15, 23, 24]. Individual TALE repeats have helix–loop–helix secondary structure where two α-helices are linked by short ‘RVD loop’ (Figure 1B). The first short α-helix spans residues 3–11 and the longer bended second α-helix spans residues 15–33. The RVD loop of each TALE repeat reaches into the major groove of the DNA duplex and contacts a single nucleotide in the sense strand with the residue at position 13 [15, 23]. Interestingly, the residue at position 12 (mainly histidine or asparagine) points away from the major groove and does not contribute to the specific base recognition but, rather stabilizes the position of the RVD loop [15, 23]. Within each repeat, lysine and glutamine residues at positions 16 and 17, respectively, contribute to non-specific interactions with negatively charged DNA backbone [15, 23]. The characteristic angle between inter-repeat helices distinguishes the TALE repeat domain from other known α-helical repeat domains [23].
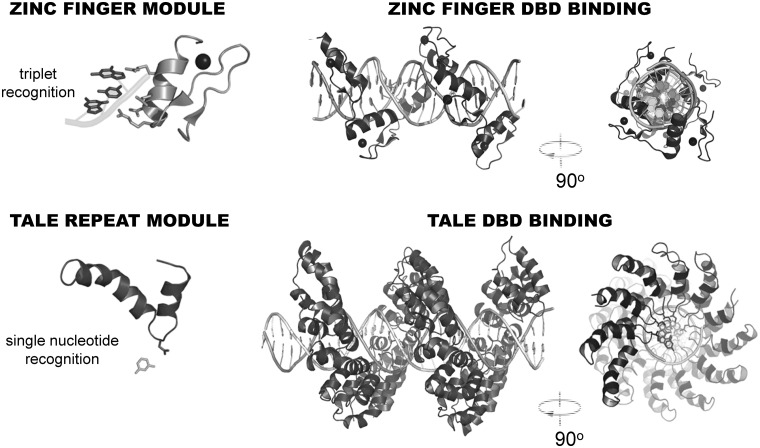
Figure 2:
Comparison of zinc finger and TALE DNA-binding domains. A single zinc finger module recognizes three nucleotides of DNA while a TALE repeat module recognizes a single nucleotide of DNA. Next are shown front and lateral views of zinc finger and TALE DBDs. Shown is a six-finger zinc finger protein that consists of six tandem repeats of C2H2 zinc finger motifs, each consisting of approximately 30 amino acids and a TALE DBD consisting of 2 cryptic repeats and 22 canonical repeat modules. Structures were rendered using available structural information deposited in the Protein Data Bank [15, 22].
Recognition of nucleotides in the cognate sequence
Different types of interactions are responsible for recognition of different nucleotides. This is important for designing custom TALE domains. Direct H- bonds are involved in base selectivity for C, G, G/A and A/G/C mediated by RVDs HD, NH, NN and NS, respectively. Weaker van der Waals contacts are responsible for base selectivity of NI and NG for A and T, respectively [15,, 23 24]. Nucleotide-binding specificity is determined not only by possible contacts with nucleotides but also by steric exclusion of interactions with alternative nucleotides (reviewed in [25]). Notably, the use of HD and NG enables partial discrimination of targets with unmethylated or methylated cytosines with custom TALEs because NG can accommodate a methylated cytosine, whereas HD does not [15, 23]. In addition, a 33 aa long N* repeat (missing the residue at position 13) exhibits complete recognition promiscuity explained by absenting physical contact with nucleotides [15]. Therefore, N* also allows for accommodating methylated cytosines and for designing TALE domains with highly degenerated target specificity [26].
RVDs NI, HD, NH/NK and NG are highly specific, recognizing A, C, G and T nucleotides, respectively [7, 8]. NG and HD bind cognate bases with high, NH with ‘intermediate’ and NI and NK with weaker affinity [15, 27–29]. NN and NS have degenerated specificity; NN repeat selects both for G and A (with a preference for G) and binds them with high affinity [7, 27, 29–32]. NS can bind A, C and G; interaction with T is probably sterically excluded [7, 8, 15, 25]. Guanine is exclusively recognized by NK and NH [27, 29, 32, 33]. NH recognizes guanine with ‘intermediate’ affinity, whereas NK was classified as ‘weak’ and also performed poorly in reporter assays compared with both NN and NH [27, 29, 30, 32, 34, 35]. Thus, NH seems to be a good choice for G targeting, especially if flanked by a few strong RVDs (NG, NN and HD) [29, 32]. Repeats included in available TALE assembly kits (HD, NG, NI, NN, NH and NK) are further referred as standard RVDs, all other RVDs are referred as ‘non-standard’ (Figure 1C).
TALE-DNA-binding mechanism is apparently asymmetric across the protein–DNA interface [27]. NTR ensures 5′-T0 recognition and probably serves as a binding-anchor from which the protein wraps around a DNA helix and probes a nucleotides sequence [12]. Therefore, mutations at the 5′ end of a corresponding TALE target site impair activity more than mutations at the 3′ end [27, 31]. Furthermore, too many strong RVDs at the N-terminal part of CRD may pose a risk of multiple off-target effects. At the same time, weak RVDs at the C-terminal part of CRD may also impair TALE activity [27].
It seems that evolutionary optimal length of TALE arrays is between 17 and 20 RVDs, as most of natural TALEs fall within this range [9]. This possibly reflects a critical TALE size above which deformations in superhelical assembly could lead to registration errors. Thus, adding more repeats to an array may have no positive effect to overall binding affinity [9, 15, 27]. Moreover, a systematic study of TALEN specificity revealed that excess non-specific DNA-binding energy (which is increasing with an array length) results in tolerating more mismatches and, therefore, in greater off-target cleavage [31]. Accordingly, TALENs mutated at the C-terminal domain to reduce non-specific DNA-binding energy still retain high activity and exhibit improved specificity [31].
The invariant 5′-thymine base
Interestingly, well-conserved thymine is present at the position 0 (T0) of most of natural TALE target sites [8] and is necessary for full target gene activation [7, 36] and activity of TALE fusion proteins [12, 37, 38]. Although structural data can explain the 5′-T0 preference [15], TALE fusion proteins functioning on 5′-T0-deficient target sites were also reported [27, 28, 39]. The significance of 5′-T0 differs for wild-type TALEs and artificial TALEs created with standard RVDs suggesting that the latter bind DNA with higher affinity and may not require the invariant 5’-T0 [27]. Recently, redesigned scaffolds allowing non-constrained target site selection were reported [38]. However, it is advisable to design artificial TALEs with 5’-T0, as this natural TALE’s feature does not seriously constrain target site selection in eukaryotic genomes.
USE OF TALE DBD FOR GENE EDITING AND REGULATION
In their pioneering work, Boch et al. [7] demonstrated that artificial TALEs could be synthesized, hence allowing for exploitation of the TALE-binding code for targeting almost any DNA sequence with artificial TALE DBDs. Properties of the TALE DBD offer a great potential for research, biotechnology and gene therapy. Repeat modules can be arranged in a desired order to produce a DBD with high sequence specificity. Such a DBD can be combined with a catalytic or effector domain, e.g. a nuclease to obtain an exceptional tool for DNA editing [40]. High specificity, reliable activity and low cytotoxicity are desired features of an ideal customized nuclease.
TALE fusion proteins use the C-terminal region downstream of CRD as a linker between TALE DBD and the effector domain. The optimal length of the linker may vary for different effector domains, e.g. a short 17–65 aa linker is used for the dimerizing FokI nuclease domain [13, 28], whereas a longer linker (∼65 aa) was used for activation domains [14, 28]. This difference likely reflects different steric requirements of particular effector domains.
Gene editing with TALE nucleases
Organisms repair DSBs through two major pathways: non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ is an error-prone process, which often leads to small insertions or deletions (indels) at the break site, and thus can cause a frameshift mutation in the coding sequence of targeted gene. HR is generally an error-free process, which can use a sister chromatid or exogenous homologous template to repair the damage. Traditional gene targeting relies on DSB-independent HR to replace (knock-in) or disrupt (knock-out) gene sequences in a pre-determined locus (reviewed in [41]). Low frequency of DSB-independent HR limits this approach to just a few model organisms (e.g. Mus musculus, or Saccharomyces cerevisiae) and cell types (e.g. embryonic stem cells). Even in suitable cells, the frequency of HR with the donor sequence is low (1/104–7), requiring some selection system to identify cells where HR occurred. A remedy for this problem represents nuclease-induced DSBs, which stimulate HR [42, 43]. This nuclease-mediated approach is referred to as gene editing.
One of the first tools for gene editing was synthetic ZFN (Figure 2). A ZFN is created by linking the FokI nuclease domain [44] to a Cis2His2 zinc-finger array, which provides the sequence specificity [3]. The FokI nuclease domain functions as a dimer [2]; therefore, two zinc-finger arrays, each carrying a FokI monomer, are targeted to neighboring sites between which FokI dimerization occurs [1]. ZFN technology yielded substantial achievements in a variety of model organisms and cell types, which were previously inaccessible by the classical gene targeting methods. In contrast to traditional gene targeting, gene editing with custom nucleases yields high mutation frequencies; therefore, selectable markers are not necessary. Principles established during more than a decade of ZFNs research were subsequently adapted to TALENs once the TALE-DNA-binding code was deciphered. In TALENs, the FokI nuclease (or its heterodimeric variants [45, 46], Figure 3A) is recruited to two adjacent target sites separated with a short spacer (12–20 nt) (reviewed in [48]). In contrast to a zinc-finger DBD, where one finger predominantly recognizes a nucleotide triplet [49], each module of TALE DBD recognizes a single nucleotide within the target sequence (Figure 2). The initial TALEN fusions with the homodimeric FokI demonstrated successful TALEN-mediated alterations [28, 37, 40, 50].
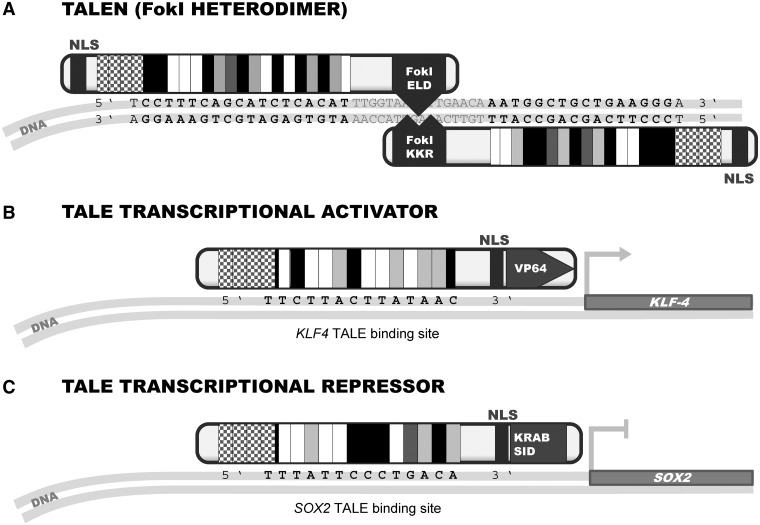
Figure 3:
TALE-based gene editors and regulators. (A) A pair of TALENs with a heterodimerizing FokI domain [47]. (B) A TALE-based transcriptional activator [14]. (C) A TALE-based transcriptional repressor [32].
TALEN technology was successfully used for targeted genome editing in yeasts [50], Drosophila melanogaster and other insect species [51–53], Danio rerio [34, 54], Caenorhabditis elegans [55], Xenopus laevis [56, 57], mouse [58, 59], rat [60] and livestock, including pig and cow [61]. Plants are also accessible for TALEN-mediated gene editing, including not only model organisms such as Arabidopsis [62, 63] but also crop plants such as rice [64] and tobacco [65]. Current efficiency varies usually from 10 to >50% with an average around 22% cells mutated [66, 67]. We achieved TALEN cleavage efficiency of 18% when inducing a ∼0.7 kb deletion with two TALEN pairs in the mouse genome [47] (Figure 4).
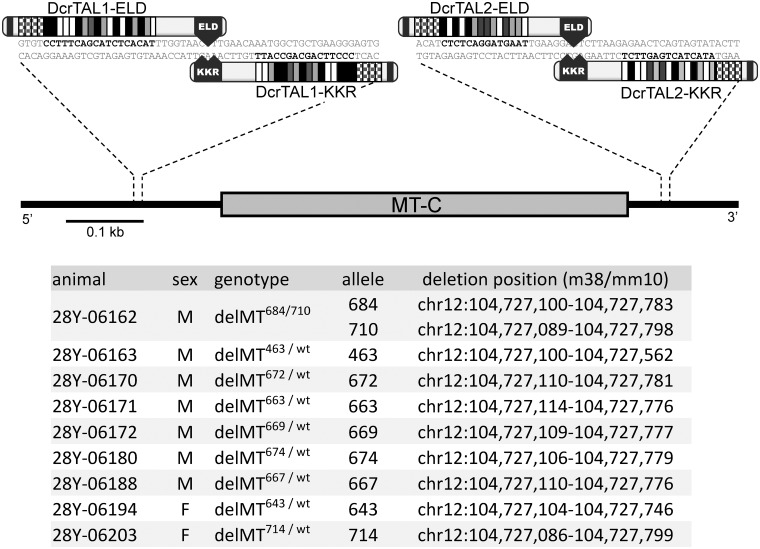
Figure 4:
An example of genomic deletion achieved with two TALEN pairs [47]. Shown are relative positions of DSBs introduced by TALENs. Individual TALEN recognition sites are shown in bold black letters. Nine founder mice carrying 10 deletion alleles were found among 51 animals originating from TALEN-injected zygotes. Deletion positions are listed in the extreme right column.
Early studies typically used NHEJ-mediated mutagenesis. DSB-driven HR with dsDNA donor templates was subsequently used as well, e.g. in human cells [28, 68] and zebrafish zygotes [69]. Single strand oligonucleotides with ∼50 nt long arms of homology were used as a donor template for precise modifications in zebrafish and mouse models [59,70]. Furthermore, introduction of two DSBs simultaneously allows for additional genome alterations [47, 52, 61]. A widely applied and generally successful approach is microinjection of in vitro synthesized mRNAs encoding a custom TALEN pair into the zygote [47, 59–61, 69, 70]. This allows for fast and effective preparation of knock-out models [71]. Heterozygous mutant mice can be prepared within 18 weeks [47, 59]. Biallelic mutations may also occur [47, 56, 61, 70], which strongly reduce time necessary for preparation of homozygous animals. We have produced and analyzed a knock-out mouse model within a year with frequency of genomic deletion of ∼20%; 1/51 founder animals carried the desired deletion on both chromosomes [47].
TALENs are highly specific and can distinguish sites, which differ only in two mismatched bases [13, 54]. Mussolino et al. [13] compared cytotoxicity and specificity of a CCR5-specific TALEN pair with a well-established ZFN pair. Off-target site in highly homologous CCR2 gene differed from CCR5 only in one base and 5′-T0. The TALEN pair induced only 1% mutation in the CCR2 off-target, whereas the ZFN pair induced 11%. Moreover, 2-fold higher cell survival was reported for the TALEN pair. Numerous other results suggest that TALENs are more mutagenic and less cytotoxic than ZFN [34, 39, 66, 72, 73].
Enhanced TALEN-mediated gene disruption in rat zygotes was achieved by co-injection of engineered TALENs with Exonuclease 1 [74] or Trex exonuclease [75], which degrade one DNA strand in DSB site and therefore promote alternative mutagenic correction pathway [74]. Mutagenicity can be further improved by adoption of the more effective FokI nuclease such as Sharkey [76] or by transient hypothermia [28].
Superior TALEN specificity can be achieved by adopting a heterodimeric FokI architecture, by mutating cationic residues in TALE C-terminal domain [31], or via fusion with other cleavage domains with intrinsic sequence specificity such as meganucleases (MegaTALs) or TevI nuclease. Recently reported MegaTALs are compact, active and hyper-specific endonucleases valuable for future widespread, safe and reliable therapeutic use [75, 77]. TevI may work either as a monomeric nuclease (fused to N-terminus of TALE array over a TevI native linker) or as a nicking enzyme (fused to C-terminus of TALE array over shorter artificial linker), cleaving only one DNA strand [78]. The TevI cleavage domain (only ∼200 aa) has degenerated site specificity (CN↑NN↓GN), which limits possible target site selection, but substantially reduces the TALEN size [78]. Targeted nickases could be used to promote gene correction via HR in selected loci, with reduced cytotoxicity, because no DSBs are created [79, 80].
Gene regulation with TALEs-DBDs
TALE DBDs were used not only for gene editing but also for targeted endogenous gene regulation in a form of artificial TALE transcription factors (Figure 3). The first study demonstrated activation of plant genes in Arabidopsis using a native AvrBs3 scaffold with designed CRD matching their promoters [33]. Zhang et al. [14] developed an artificial TALE activator (Figure 3B) using a truncated scaffold fused to the VP64 activation domain (tetrameric version of VP16 activation domain from Herpes simplex virus) and successfully induced expression of SOX2 and KLF4 in human cells but failed to activate OCT4 and c-MYC genes [14]. Similarly, two other groups used different TALE architectures for activation of human genes with the VP16 domain [28, 81].
Activation of Oct4 gene was achieved with a TALE-VP16 activator in murine embryonic stem cells and derived neural stem cells [82]. TALE-mediated gene activation seemed to depend on the binding-site position in a target promoter and consequent interactions with basal transcription factors. Authors also demonstrated that methylation of target promoters impairs TALE activity and that specific activation of silenced genes is possible once cells are treated with low concentration of histone deacetylases and/or DNA methyltransferases inhibitors [82]. Negative effects of DNA methylation on TALE binding can be solved by using NG and N* RVDs, which allow for accommodating 5′-methylcytosine [26, 83]. A set of human genes including non-coding microRNA cluster miR-302/367 was activated in another study, which also showed that using multiple TAL Effector based transcriptiont factors (TALE-TFs) targeting a single gene has a synergistic effect on target expression [84].
TALE fusions with effector domains offer a broad range of applications, ranging from simple locus-specific transcriptional activation and repression [82, 85], through direct induction of epigenetic changes on DNA [86] or on histones [87], to using them for visualization and pull-down of specific genomic loci [88–91].
Design and assembly of TALE repeat domain
Several rules for rational design of TALE-CRD (and inherently for the selection of target site in DNA) could be inferred from known properties of particular repeat types and from the TALE-DNA-binding mode:
Select target sites with 5′-T0 base preceding the CRD-specified sequence. If that is not possible, one can use reengineered scaffold with unrestricted specificity for 5′-N0 [38].
Confirm that your selected target site is truly unique (e.g. not representing a unique polymorphism within a highly repetitive element).
Although optimal repeat lengths likely vary for individual cognate sequences [31] as a rule of thumb [9, 48, 59], we recommend at least 14 repeats for each TALEN in a pair and 18–20 repeats for TALE transcription factors.
Include at least four evenly positioned strong RVDs (e.g. HD > C, NG > T or NN > G/A), especially at termini of CRD to stabilize TALE-DNA interaction [27, 32]
Avoid stretches of more than three identical RVDs, especially of NG, which was shown to adopt a deformed fold even with three repeats in a row [29].
Use NH for targeting G instead of NN, if discrimination between A and G is necessary [29].
Use NI for specific recognition of A along with sufficiently strong RVDs [29].
Use validated TALE scaffold, which includes whole NTR (∼150 aa) and suitable C-terminal linker to the effector domain. One of the most common scaffolds established in multiple organisms is Miller’s [28]. Also Mussolino’s [13] and Zhang’s [14] architectures are reliable and were used repeatedly.
Finally, we highly recommend to search for online tools for TALEN design and off-targeting analysis, which become increasingly available. Several of them are listed in Table 1.
Table 1:
Selected TALEN design web tools
| Tool | URL | References |
|---|---|---|
| E-TALEN | http://www.e-talen.org/E-TALEN/ | [92] |
| tDnA | http://baolab.bme.gatech.edu/Research/BioinformaticTools/assembleTALSequences.html | [93, 94] |
| TALE-NT | https://tale-nt.cac.cornell.edu/node/add/talen | [95] |
| TALeffectors | http://taleffectors.genome-engineering.org/tools/ | [96] |
| Mojo Hand | http://www.talendesign.org/ | [97] |
Because the assembly of designed TALE DBDs from nearly identical repeats was challenging for classical cloning techniques, several platforms have emerged for efficient and rapid (less than a week) construction of expression plasmids containing a TALE scaffold with a designed DBD (reviewed in [48]). A widely used platform is the ‘Golden Gate Cloning’, which allows for highly efficient assembly of designed TALEs in a single reaction [14, 62, 81, 98–100]. Recent advances in TALEN assembly methods include ligation-independent cloning [101] and solid-phase cloning such as Fast Ligation-based Automatable Solid-phase High-throughput platform for large scale assembly of TALENs (FLASH) [67] and Iterative Capped Assembly (ICA) [102], the latter allowing for a rapid automatized robotic assembly with a high-throughput capability. Needless to say, custom TALE nucleases are also available from numerous commercial sources.
SUMMARY AND OUTLOOK
Simple design, fast and low-cost assembly, high specificity combined with low cytotoxicity and a practically unlimited target site selection make TALE DBDs an excellent choice for DNA targeting. The TALEN technology has superior mutagenic potential associated with lower cytotoxicity and higher target specificity compared with ZFNs. Simple design and publicly available assembly toolkits allow for adoption of this technology by laboratories worldwide. Modular nature of TALE-DNA recognition, no significant inter-repeat context effects in contrast to zinc fingers and a possibility to target practically any sequence in are other important features. Although TALENs currently face competition from recently developed RNA-guided clustered regularly interspaced short palindromic repeats (CRISPR) approach [103], their outstanding potential for research and therapy remains undisputed.
Key points.
TALEs interact with cognate sequences via tandem repeats, which bind individual nucleotides.
A selected locus can be targeted with a designed TALE fused with an effector domain
TALENs allow for genetic alterations in virtually any model system.
Acknowledgement
The authors thank Radek Malik for help with preparation of the manuscript.
Biographies
Radek Jankele is an undergraduate student at the Charles University in Prague. He is currently pursuing his MSc degree in the laboratory of Petr Svoboda.
Petr Svoboda received his PhD degree at the University of Pennsylvania in 2002. After a postdoctoral stay at the Friedrich Miescher Institute (Basel, Switzerland), he established his own group at the Institute of Molecular Genetics of the Czech Academy of Sciences in Prague in 2007.
FUNDING
This research was funded by the Czech Science Foundation. The Czech Science Foundation grant P305/12/G034 and the Czech Ministry of Education Youth and Sports grant KONTAKT II (LH13084) (to P.S.). Institutional support was provided by RVO 68378050.
References
- 1.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–82. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitinaite J, Wah DA, Aggarwal AK, et al. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA. 1998;95:10570–5. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc Natl Acad Sci USA. 1994;91:883–7. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan RP, Vorhölter F-J, Potnis N, et al. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol. 2011;9:344–55. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 5.Rossier O, Wengelnik K, Hahn K, et al. The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc Natl Acad Sci USA. 1999;96:9368–73. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szurek B, Marois E, Bonas U, et al. Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 2001;26:523–34. doi: 10.1046/j.0960-7412.2001.01046.x. [DOI] [PubMed] [Google Scholar]
- 7.Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–12. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 8.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 9.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Ann Rev Phytopathol. 2010;48:419–36. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 10.Mak AN-S, Bradley P, Bogdanove AJ, et al. TAL effectors: function, structure, engineering and applications. Curr Opin Struct Biol. 2013;23:93–9. doi: 10.1016/j.sbi.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaj T, Gersbach CA, Barbas Iii CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao H, Wu X, Chai J, et al. Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell Res. 2012;22:1716–20. doi: 10.1038/cr.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mussolino C, Morbitzer R, Lutge F, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–93. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, Cong L, Lodato S, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–53. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mak AN-S, Bradley P, Cernadas RA, et al. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–9. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casper-Lindley C, Dahlbeck D, Clark ET, et al. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc Natl Acad Sci USA. 2002;99:8336–41. doi: 10.1073/pnas.122220299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Gabriel DW. Xanthomonas avirulence/pathogenicity gene family encodes functional plant nuclear targeting signals. Mol Plant Microbe Interact. 1995;8:627–31. doi: 10.1094/mpmi-8-0627. [DOI] [PubMed] [Google Scholar]
- 18.Zhu W, Yang B, Chittoor JM, et al. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Mol Plant Microbe Interact. 1998;11:824–32. doi: 10.1094/MPMI.1998.11.8.824. [DOI] [PubMed] [Google Scholar]
- 19.Zhu WG, Yang B, Wills N, et al. The C terminus of AvrXa10 can be replaced by the transcriptional activation domain of VP16 from the herpes simplex virus. Plant Cell. 1999;11:1665–74. doi: 10.1105/tpc.11.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lange O, Schreiber T, Schandry N, et al. Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 2013;199:773–86. doi: 10.1111/nph.12324. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Atef A, Piatek A, et al. Characterization and DNA-binding specificities of Ralstonia TAL-like effectors. Mol Plant. 2013;6:1318–30. doi: 10.1093/mp/sst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peisach E, Pabo CO. Constraints for zinc finger linker design as inferred from X-ray crystal structure of tandem Zif268-DNA complexes. J Mol Biol. 2003;330:1–7. doi: 10.1016/s0022-2836(03)00572-2. [DOI] [PubMed] [Google Scholar]
- 23.Deng D, Yan C, Pan X, et al. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–3. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng D, Yan C, Wu J, et al. Revisiting the TALE repeat. Protein Cell. 2014;5:297–306. doi: 10.1007/s13238-014-0035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochtler M. Structural basis of the TAL effector-DNA interaction. Biol. Chem. 2012;393:1055–66. doi: 10.1515/hsz-2012-0164. [DOI] [PubMed] [Google Scholar]
- 26.Valton J, Dupuy A, Daboussi F, et al. Overcoming Transcription Activator-like Effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem. 2012;287:38427–32. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meckler JF, Bhakta MS, Kim M-S, et al. Quantitative analysis of TALE–DNA interactions suggests polarity effects. Nucleic Acids Res. 2013;41:4118–28. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 29.Streubel J, Blücher C, Landgraf A, et al. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–5. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 30.Christian ML, Demorest ZL, Starker CG, et al. Targeting G with TAL Effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable DI-residues. PLoS One. 2012;7:e45383. doi: 10.1371/journal.pone.0045383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilinger JP, Pattanayak V, Reyon D, et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Methods. 2014;11:429–35. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cong L, Zhou R, Kuo Y-C, et al. Comprehensive interrogation of natural TALE DNA binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morbitzer R, Romer P, Boch J, et al. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci USA. 2010;107:21617–22. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang P, Xiao A, Zhou M, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Zhang Y, Yuan P, et al. Complete decoding of TAL effectors for DNA recognition. Cell Res. 2014;24:628–31. doi: 10.1038/cr.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Römer P, Recht S, Strauss T, et al. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. Oryzae. New Phytologist. 2010;187:1048–57. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- 37.Mahfouz MM, Li L, Shamimuzzaman M, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA. 2011;108:2623–8. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb BM, Mercer AC, Barbas CF., III Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res. 2013;41:9779–85. doi: 10.1093/nar/gkt754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun N, Liang J, Abil Z, et al. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol BioSyst. 2012;8:1255–63. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 40.Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–61. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capecchi MR. Essay: gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–12. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 42.Brenner DA, Smigocki AC, Camerini-Otero RD. Effect of insertions, deletions, and double-strand breaks on homologous recombination in mouse L cells. Mol Cell Biol. 1985;5:684–91. doi: 10.1128/mcb.5.4.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song KY, Chekuri L, Rauth S, et al. Effect of double-strand breaks on homologous recombination in mammalian cells and extracts. Mol Cell Biol. 1985;5:3331–6. doi: 10.1128/mcb.5.12.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc Natl Acad Sci USA. 1992;89:4275–9. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doyon Y, Vo TD, Mendel MC, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2010;8:74–9. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 46.Szczepek M, Brondani V, Buchel J, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–93. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 47.Flemr M, Malik R, Franke V, et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–16. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–17. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 50.Li T, Huang S, Jiang WZ, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–72. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J. Efficient and specific modifications of the Drosophila genome by means of an easy talen strategy. J Genet Genomics. 2012;39(5):209–15. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Ma S. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One. 2012;7(9):e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun. 2012;3:1017. doi: 10.1038/ncomms2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cade L, Reyon D, Hwang WY, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–10. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood AJ, Lo T-W, Zeitler B, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishibashi S, Cliffe R, Amaya E. Highly efficient bi-allelic mutation rates using TALENs in Xenopus tropicalis. Biol Open. 2012;1:1273–6. doi: 10.1242/bio.20123228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei Y, Guo X, Liu Y, et al. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci USA. 2012;109:17484–9. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sung YH, Baek I-J, Kim DH, et al. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31:23–4. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 59.Wefers B, Meyer M, Ortiz O, et al. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci USA. 2013;110:3782–7. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tesson L, Usal C, Ménoret S, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–6. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 61.Carlson DF. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109:17382–7. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christian M, Qi Y, Zhang Y, et al. Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3. 2013;3:1697–705. doi: 10.1534/g3.113.007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li T, Liu B, Spalding MH, et al. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–2. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Zhang F, Li X, et al. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013;161:20–7. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen S, Oikonomou G, Chiu CN, et al. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41:2769–78. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reyon D, Tsai SQ, Khayter C, et al. FLASH assembly of TALENs enables high-throughput genome editing. Nat Biotechnol. 2012;30:460–5. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zu Y, Tong X, Wang Z, et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods. 2013;10:329–31. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 70.Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–8. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wefers B, Panda SK, Ortiz O, et al. Generation of targeted mouse mutants by embryo microinjection of TALEN mRNA. Nat Protoc. 2013;8:2355–79. doi: 10.1038/nprot.2013.142. [DOI] [PubMed] [Google Scholar]
- 72.Garg A, Lohmueller JJ, Silver PA, et al. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–95. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore FE, Reyon D, Sander JD, et al. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mashimo T, Kaneko T, Sakuma T, et al. Efficient gene targeting by TAL effector nucleases coinjected with exonucleases in zygotes. Sci Rep. 2013;3:1253. doi: 10.1038/srep01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boissel S, Jarjour J, Astrakhan A, et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42:2591–601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo J, Gaj T, Barbas CF. Directed evolution of an enhanced and highly efficient FokI cleavage domain for Zinc Finger Nucleases. J Mol Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeuchi R, Choi M, Stoddard BL. Redesign of extensive protein-DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc Natl Acad Sci USA. 2014;111:4061–6. doi: 10.1073/pnas.1321030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beurdeley M, Bietz F, Li J, et al. Compact designer TALENs for efficient genome engineering. Nat Commun. 2013;4:1762. doi: 10.1038/ncomms2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim E, Kim S, Kim DH, et al. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22:1327–33. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramirez CL, Certo MT, Mussolino C, et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–68. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Geissler R, Scholze H, Hahn S, et al. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bultmann S, Morbitzer R, Schmidt CS, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–77. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deng D, Yin P, Yan C, et al. Recognition of methylated DNA by TAL effectors. Cell Res. 2012;22:1502–4. doi: 10.1038/cr.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maeder ML, Linder SJ, Reyon D, et al. Robust, synergistic regulation of human gene expression using TALE activators. Nat Methods. 2013;10:243–5. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Konermann S, Brigham MD, Trevino AE, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–6. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maeder ML, Angstman JF, Richardson ME, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31:1137–42. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mendenhall EM, Williamson KE, Reyon D, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013;31:1133–6. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Byrum SD, Taverna SD, Tackett AJ. Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res. 2013;41:e195. doi: 10.1093/nar/gkt822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thanisch K, Schneider K, Morbitzer R, et al. Targeting and tracing of specific DNA sequences with dTALEs in living cells. Nucleic Acids Res. 2014;42:e38. doi: 10.1093/nar/gkt1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma H, Reyes-Gutierrez P, Pederson T. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc Natl Acad Sci USA. 2013;110:21048–53. doi: 10.1073/pnas.1319097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol. 2013;20:1321–4. doi: 10.1038/nsmb.2680. [DOI] [PubMed] [Google Scholar]
- 92.Heigwer F, Kerr G, Walther N, et al. E-TALEN: a web tool to design TALENs for genome engineering. Nucleic Acids Res. 2013;41:e190. doi: 10.1093/nar/gkt789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fine EJ, Cradick TJ, Zhao CL, et al. An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res. 2014;42:e42. doi: 10.1093/nar/gkt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin Y, Fine EJ, Zheng Z, et al. SAPTA: a new design tool for improving TALE nuclease activity. Nucleic Acids Res. 2014;42:e47. doi: 10.1093/nar/gkt1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.D.oyle EL, Booher NJ, Standage DS, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–22. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sanjana NE, Cong L, Zhou Y, et al. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–92. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Neff KL, Argue DP, Ma AC, et al. Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinformatics. 2013;14:1. doi: 10.1186/1471-2105-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li T, Huang S, Zhao X, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–25. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morbitzer R, Elsaesser J, Hausner J, et al. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–9. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weber E, Gruetzner R, Werner S, et al. Assembly of designer TAL Effectors by golden gate cloning. PLoS One. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schmid-Burgk JL, Schmidt T, Kaiser V, et al. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat Biotechnol. 2012;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Briggs AW, Rios X, Chari R, et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]