Abstract
Bardet–Biedl syndrome (BBS) and autosomal dominant polycystic kidney disease (ADPKD) are two genetically distinct ciliopathies but share common phenotypes such as renal cysts. Seven BBS proteins form a complex called the BBSome which is localized at the basal body or ciliary axoneme and regulates the ciliary entry or flagellar exit of several signaling molecules. Here, we demonstrate that, unlike the seven-span somatostatin receptor 3 or the leptin receptor that interacts with all subunits of the BBSome, the ADPKD protein polycystin-1 (PC1) interacts with BBS1, BBS4, BBS5 and BBS8, four of the seven components of the BBSome. Only depletion or mutation of BBS1, but not depletion of BBS5 and BBS8, or knockout of BBS4, impairs ciliary trafficking of PC1 in kidney epithelial cells. Depletion of these BBS proteins affects neither the ciliary length nor the plasma membrane targeting of PC1. Expression of a pathogenic BBS3/Arl6 mutant (T31R) that locks Arl6 in the GDP form leads to stunted cilia and inhibition of PC1 on primary cilia. We propose that the 11-span membrane protein PC1 is a BBSome cargo and that the components of the BBSome may possess subunit-specific functions. Moreover, physical interactions between the BBS and ADPKD proteins may underline the overlapping renal phenotypes in these two diseases.
INTRODUCTION
Primary cilia are tiny, hair-like sensory organelles projecting from the apical surface of most cells. Bardet–Biedl syndrome (BBS) is a genetically heterogeneous recessive disorder of the primary cilia. BBS is primarily characterized by obesity, retinal degeneration, cognitive impairment, polydactyly, hypogonadism and renal dysfunction including renal cysts (1). Although 19 BBS genes have been identified to date, the precise cellular functions of these BBS proteins are not fully understood. It was reported that some BBS proteins are involved in intracellular transport (2,3) and intraflagellar transport (4). Biochemical studies discovered the BBSome, a protein complex assembled by seven of the BBS proteins including BBS1, 2, 4, 5, 7, 8 and 9 (5). The BBSome localizes and functions at the basal body or ciliary axoneme and interacts with the Rab8 GTPase-exchanging factor, Rabin8, facilitating Rab8 entry into the primary cilia in a BBS3(Arl6)-GTP-dependent manner. This process regulates the ciliary entry of signaling molecules (6,7) and is shown critical for ciliogenesis (8). In addition to protein ciliary entry, BBSome also controls the flagellar exit of signaling proteins such as phospholipase D Type c in Chlamydomonas reinhardtii (9).
Autosomal dominant polycystic kidney disease (ADPKD) (10), affecting over 12 million people worldwide, is characterized by progressive development of epithelial-lined and fluid-filled cysts in various kidney tubular segments (11). Cystic liver and pancreas are also seen. Mutations in two genes PKD1 (12,13) and PKD2 (14), respectively encoding polycystin-1 (PC1) and polycystin-2 (PC2), account for ∼85 and ∼15% of ADPKD cases. To date, PC1 and PC2 have been implicated in modulating a number of cellular events such as Ca2+ signaling (15,16), JAK-STAT (17), mTOR (18), cyclic AMP (19), canonical Wnt (20), Id2 (21), planar cell polarity (22), cMET (23), STAT3 (24), PI3/Akt (25), Jade-1 (26), G protein-coupled receptor (GPCR) (27), epidermal growth factor receptor (28,29), as well as the localization and activity of cystic fibrosis transmembrane conductance regulator (CFTR) (30,31). How the polycystins modulate these pathways remains elusive. Although PC1 and PC2 have been reported to localize to multiple subcellular sites (15,32–34), the primary cilium has been implicated as a key organelle for polycystin function and the pathogenesis of ADPKD (15,35). PC1 and PC2 form a receptor-channel complex at the primary cilium and their role in transducing the extracellular fluid flow shear stress into a Ca2+ signal at this site may be a primary defect in ADPKD (10). A chemosensory role has also been proposed for PC1 and PC2 (36). Elucidation of the polycystin interaction network is one way to pinpoint the proximal events in the complex biochemical networks of polycystins and to facilitate the rational design and evaluation of therapeutics for ADPKD.
Through an unbiased screen using PC1 C-terminal tail as a bait, we identified BBS8, a component of the BBSome to physically interact with ADPKD. By establishing an expression system of full-length PC1 and lentiviral knockdown of individual BBS genes, we further report that PC1 interacts with four of the seven components of the BBSome and that the ciliary localization of PC1 can be regulated by specific components of the BBSome. We found that BBS1, a major BBS gene, is important for the efficient delivery of PC1 to cilia but not to plasma membrane. Re-expression of the wild-type but not a pathogenic mutant BBS1 rescues PC1 ciliary trafficking. Expression of a pathogenic dominant-negative mutant form of BBS3 also affects PC1 ciliary localization. Since both ADPKD and BBS patients develop renal cysts, our data suggest that physical interactions between PC1 and BBS proteins may underline the overlapping renal phenotypes in BBS and ADPKD.
RESULTS
BBS8 and three other subunits of the BBSome interact with PC1
To identify the interaction network of PC1, we screened a human fetal kidney library using a yeast two-hybrid system with the 198-amino acid cytoplasmic tail (4106–4303) of human PC1 as a bait. BBS8, the first BBS protein that was identified to link basal body dysfunction to the cause of BBS (37) became our immediate top candidate binding partner of human PC1. BBS8 encodes a protein with multiple tetratricopeptide repeats (TPRs) and is among the seven BBS proteins that form the BBSome complex at the basal body (8,37). The fragment of BBS8 isolated from the library encodes a peptide from amino acid 281 to the end of the protein, amino acid 593.
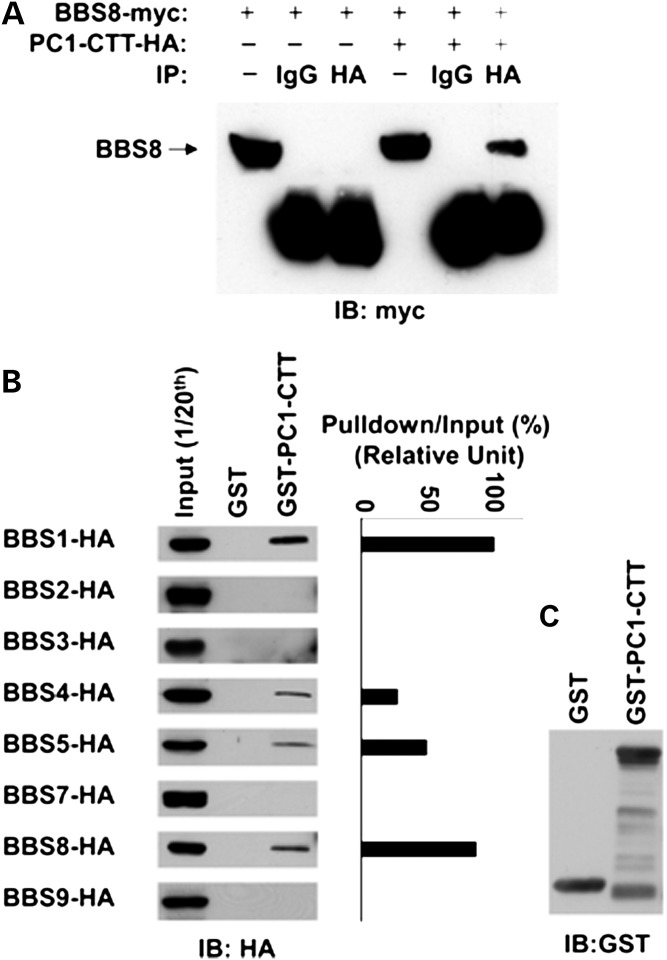
To confirm this interaction in mammalian cells, we performed co-immunoprecipitation in HEK293 cells transfected with both myc-tagged BBS8 and HA-tagged PC1 C-terminal cytoplasmic tail (PC1-CTT). BBS8 was co-immunoprecipitated with PC1-CTT in these cells (Fig. 1A). In contrast, in HEK293 cell lysate transfected with BBS8 alone, no BBS8 was precipitated by the anti-HA antibody, indicating the specificity of the BBS8-PC1-CTT interaction.
Figure 1.
PC1 interacts with BBS8 and several other BBSome subunits. (A) Validation of BBS8 as an interacting partner of PC1 in HEK293 cells. Cells were transfected with BBS8-myc alone or BBS8-myc plus PC1-CTT-HA. Cell lysates were immunoprecipitated with antibody against HA. BBS8-myc (∼63 kDa, arrow) was detected with an antibody against myc. BBS8-myc was immunoprecipitated only in the presence of PC1-CTT-HA. One-twentieth of cell lysates were used as input. (B) PC1-CTT interacts specifically with BBS1, 4, 5 and 8. GST and PC1 C-tail fused with GST (GST-PC1-CTT) beads were used to pull down each of the seven subunits of the BBSome plus BBS3 expressed in HEK293 cells. One-twentieth of cell lysates were used as an input for each pull-down assay. BBS proteins were blotted with an antibody against HA. (C) Blots were also probed with an antibody against GST. The amount of BBS proteins captured by GST-PC1-CTT and inputs were quantified by Image J software and the percentage of captured protein versus input was plot with BBS1 set to 100%.
To determine whether other subunits of the BBSome complex are able to bind PC1, glutathione S-transferase (GST) pull-down experiments were performed using GST-PC1-CTT fusion protein and 293T cell lysates transfected with each of the seven BBS proteins comprising the BBSome (BBS1, 2, 4, 5, 7 and 9) as well as BBS3/Arl6, the small GTPase critical for BBSome function. Interestingly, in addition to BBS8, the PC1-CTT also interacts with BBS1, 4 and 5 (Fig. 1B).
Establishment of a cell model for studying the ciliary trafficking of PC1
The physical association between PC1 and subunits of the BBSome prompted us to explore the functional consequence of this interaction. BBSome complex is implicated in the import of several seven-span GPCRs (6,38) and the single-span leptin receptor into the cilium of mammalian cells and export of specific signaling proteins from the flagella of the Chlamydomonas reinhardtii (9). Therefore, we aim to examine the role of BBSome components in the ciliary localization of PC1 which is known to have 11 transmembrane domains. We made a set of N-terminally and/or C-terminally tagged full-length mouse PC1 constructs (Supplementary Material, Fig. S1A). We inserted enhanced yellow fluorescent protein (YFP) after the first 34 residues downstream from the predicted signal peptide, and/or a triple HA tag, an AviTag or an enhanced green fluorescent protein (eGFP) to the C-terminus of full-length PC1. The expression of YFP-PC1 constructs is readily visualized using fluorescence microscopy in HEK293 cells (Supplementary Material, Fig. S1B) although not in the inner medullary collecting duct 3 (IMCD3) epithelial cells (Supplementary Material, Fig. S1C) owing to a low level of expression (Supplementary Material, Fig. S1D), similar to PC1-eGFP (data not shown). Expression of YFP-PC1-AviTag (YPC1A) promotes tubulogenesis of IMCD3 cells (Supplementary Material, Fig. S1E), consistent with a previously reported role of PC1 in this process (39). Using a GFP or HA tag antibody that recognizes either the YFP/GFP or the HA tag, the expression of recombinant full-length PC1 is readily detectable on the primary cilia of IMCD3 cells (Supplementary Material, Fig. S2A). Since the N-terminus of PC1 is exposed to the extracellular space, cell surface expression of YFP-PC1 can be visualized by direct labeling of live cells with an antibody against GFP without fixation or permeabilization procedures. Ciliary expression of YFP-PC1 is seen in every ciliated IMCD3 cells that stably expressing full-length PC1 (Supplementary Material, Fig. S2B).
The ciliary localization of PC1 is affected by lack of BBS1, but not BBS5, BBS8 or BBS4
We then attempted to deplete BBS1, 4, 5 and 8, the BBSome subunits that were pulled down by PC1-CTT in IMCD3 cells by lentiviral shRNA. With the exception of BBS4, we were able to achieve >70% knockdown efficiency evaluated by quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) (Supplementary Material, Fig. S3A). Knockdown of BBS8 was also confirmed at the protein level by western analysis (Supplementary Material, Fig. S3B and C), as we were able to obtain a working antibody. Depletion of BBS1, 5 or 8 affects neither the ciliary number nor length in IMCD3 cells (Supplementary Material, Fig. S3D), resulting in an average length of cilium at 4.2 µm (Supplementary Material, Fig. S3E) and ciliation rate of 60% (Supplementary Material, Fig. S3F). Because none of the five target sites for BBS4 achieved sufficient knockdown efficiency, we used previously published primary BBS4 knockout mouse kidney tubular epithelial cells (2).
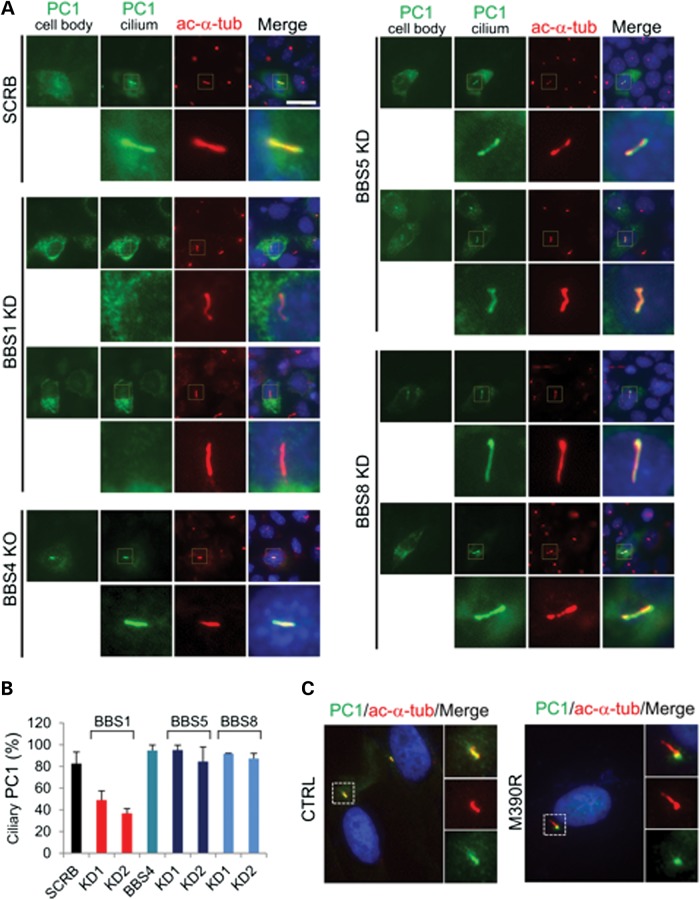
We found that in the scrambled control cells, ∼83% ciliated cells have YFP-PC1 on the cilia (Fig. 2A and B). In the BBS1 knockdown cells, however, the trafficking of YFP-PC1 to cilia was severely diminished with only 40–50% of cells having YFP-PC1 on their cilia. On the contrary, depletion of BBS5 or eight in IMCD3 cells had no effect on the ciliary trafficking of YFP-PC1. The percentage of YFP-PC1-positive ciliated cells (∼90%) is comparable to that of the scrambled control cells. Surprisingly, ciliary localization of YFP-PC1 in the primary BBS4 knockout mouse kidney tubular cells was not affected.
Figure 2.
BBS1, not BBS4, BBS5 or BBS8 is important for the ciliary trafficking of PC1. (A) YFP-PC1 is absent on cilia in BBS1 knockdown cells, although its expression on cell body remains. YFP-PC1 trafficking to cilia in BBS5 or BBS8 knockdown cells and BBS4 knockout renal epithelial cells are normal. SCRB, scrambled control. YFP-PC1 was stained by an antibody against GFP (green). Acetylated α-tubulin (ac-α-tub, red) was used to mark cilia. Scale bar represents 20 µm. The region around the cilium is boxed and enlarged. (B) Percentage of PC1-positive cilia shown in (A). At least 50 YFP-PC1 transfected and ciliated cells were counted for each condition. Error bars represent the SD between microscope fields. (C) The ciliary localization of endogenous PC1 is absent in skin fibroblasts from a human patient with a BBS1 M390R mutation. PC1 was stained by antibody 96521 (green). Acetylated α-tubulin (ac-α-tub, red) was used to mark cilia.
We also examined the ciliary trafficking of PC1 in skin fibroblast cells from a patient with a homozygous BBS1 mutation (M390R). BBS1 (M390R) has been shown to have an impaired function in a mouse model (40). In control fibroblast cells, we observed PC1 at the primary cilia in ∼10% of cells (Fig. 2C). In contrast, in the fibroblast cells derived from a BBS1 patient, PC1 was confined to the basal body area.
Expression of a dominant-negative form of BBS3/Arl6 impairs PC1 localization on cilia
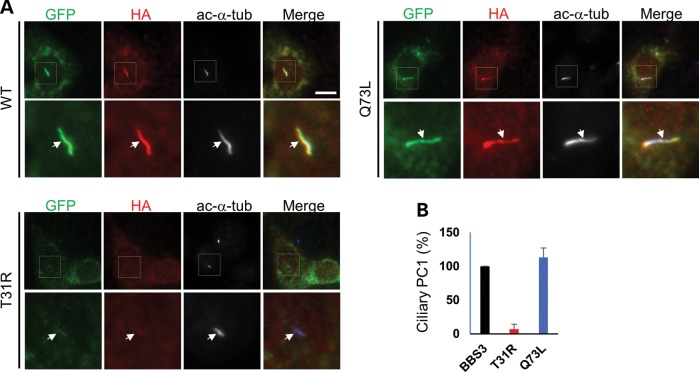
To assess whether BBS3/Arl6, the small GTPase critical for BBSome function, plays a role in the ciliary localization of PC1, we expressed a set of HA-tagged mouse BBS3/Arl6 constructs (wild-type, the pathogenic dominant-negative mutant T31R or constitutively active mutant Q73L) together with YFP-PC1. We found that YFP-PC1 traffics to the primary cilia along with the wild-type and the constitutively active BBS3/Arl6 (Q73L) (Fig. 3 and Supplementary Material, Fig. S4). Overexpression of the dominant-negative mutant of BBS3/Arl6 (T31R) in IMCD3 cells led to stunted cilia, indicating a ciliogenesis defect. Nonetheless, the PC1 signal was absent in the cells with shorter cilia although PC1 expression in the cell body was clearly visible.
Figure 3.
BBS3/Arl6 controls the ciliary trafficking of PC1. (A) YFP-PC1 was co-transfected with either the HA-tagged wild-type (WT), T31R or Q73L BBS3/Arl6 into IMCD3 cells. Cells were stained with antibodies against GFP for YFP-PC1 (green), HA for BBS3/Arl6 (red) and acetylated α-tubulin (ac-α-tub, blue but shown in black and white) to mark the cilia. Scale bar represents 10 µm. (B) Quantification of percentage of YFP-PC1 trafficking to the primary cilia shown in (A). At least 30 YFP-PC1 transfected and ciliated cells were counted for each condition. Error bars represent the SD between microscope fields.
The interaction of PC1 and BBS1 is important for PC1 to traffic to the primary cilia
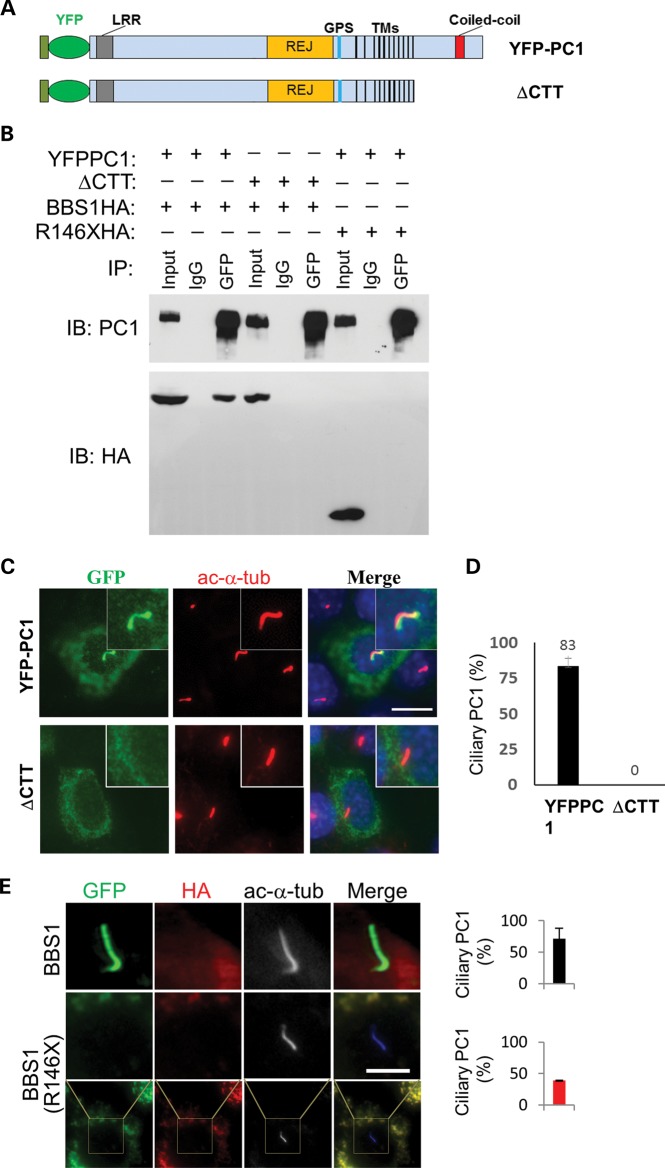
The GST pull-down experiment indicates that BBS1 interacts with PC1 through PC1’s C-terminal tail. Therefore, we sought to investigate the role of PC1-CTT in the ciliary trafficking of PC1. For this purpose, we made a YFP-PC1 construct without the last 198-amino acid of PC1 (YFP-PC1ΔCTT) (Fig. 4A) and tested its ability to interact with BBS1 by a co-immunoprecipitation assay. While YFP-PC1 co-immunoprecipitated HA-tagged BBS1, YFP-PC1ΔCTT failed to do so (Fig. 4B). Additionally, YFP-PC1 also failed to co-immunoprecipitate a common pathogenic mutant of BBS1 in human BBS patients, BBS1 (R146X) (41).
Figure 4.
The interaction of PC1 and BBS1 is important for PC1 to traffic to the primary cilia. (A) Diagram of full-length YFP-PC1 as illustrated in Supplementary Material, Figure S1 and YFP-PC1ΔCTT deletion mutant. The 198-amino acid CTT was deleted from YFP-PC1, generating YFP-PC1ΔCTT. (B) BBS1 interacts with YFP-PC1, but not with YFP-PC1ΔCTT. HEK293 cells were co-transfected with HA-tagged BBS1 and YFP-PC1 or YFP-PC1ΔCTT. Cell lysates were immunoprecipitated with a GFP antibody and blotted for PC1 (7e12) and BBS1 (HA). Full-length BBS1 (BBS1-HA) was co-immunoprecipitated by YFP-PC1 but not YFP-PC1ΔCTT. YFP-PC1 could not co-immunoprecipitate a pathogenic mutant of BBS1 (R146XHA). One-tenth of cell lysates were used as input. (C) Effects of C-terminal deletion on the ciliary trafficking of PC1. YFP-PC1 or YFP-PC1ΔCTT was transfected to IMCD3 as described above, and ciliary expression of these constructs was evaluated by staining with antibodies against GFP (green) and acetylated α-tubulin (ac-α-tub, red). Representative images from each transfection were shown. Scale bar represents 5 µm. (D) Quantification of percentage of ciliary trafficking of the constructs shown in (C). Error bars represent the SD between microscope fields. (E) Expression of WT but not a pathogenic mutant BBS1 (R146X) recues the ciliary trafficking of PC1. Cells were stained with antibodies against GFP for YFP-PC1 (green), HA for BBS1 (red) and acetylated α-tubulin (ac-α-tub, blue but shown in black and white) to mark the cilia. Scale bar represents 5 µm.
Interestingly, YFP-PC1ΔCTT fails to reach primary cilia when transfected to IMCD3 cells (Fig. 4C and D). Furthermore, the defect in PC1 trafficking to the primary cilia in BBS1 knockdown cells can be rescued by reexpressing the normal human BBS1 (Fig. 4E), but not the truncation mutant BBS1 (R146X). Given the inability of YFP-PC1ΔCTT to interact with BBS1 and of YFP-PC1 to interact with BBS1 (R146X), these results suggest that the functional interaction between BBS1 and PC1 through PC1-CTT is important for the ciliary trafficking of PC1.
Trafficking of PC1 to the plasma membrane or basal body is not affected by BBS1 depletion
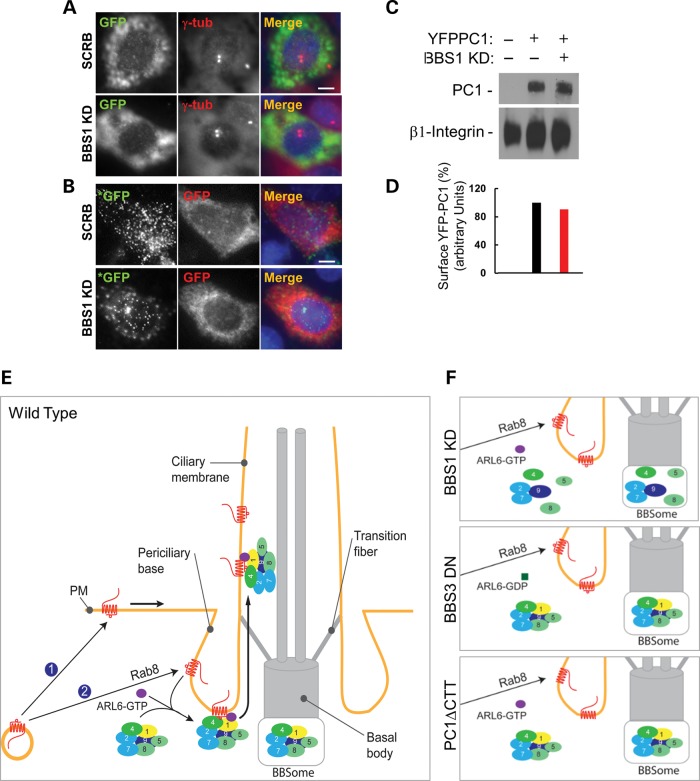
Does PC1 accumulates in certain membrane compartments when it fails to traffic to the primary cilia? Since proteins may traffic to the cilia through either polarized targeting to the basal body or lateral diffusion from the plasma membrane (42), we examined whether BBS1 depletion leads to PC1 accumulation at these sites. Unlike CD8a-SSTR3i3 that accumulates on the plasma membrane of cells depleted of Arl6 or BBS4 (8), there was no apparent accumulation of PC1 on either the basal body or the plasma membrane in BBS1-depleted cells (Fig. 5A and B).
Figure 5.
Effects of BBS1 depletion on the trafficking of PC1 to basal bodies or plasma membrane and a model of BBSome regulation of PC1 trafficking to the primary cilium. (A) YFP-PC1 trafficking to basal bodies in BBS1-depleted IMCD3 cells. YFP-PC1 was detected with an antibody against GFP (green in merged images), and γ-tubulin (denoted by arrows, red in merged images) was used as a basal body marker. Scale bar represents 5 µm. (B) Apical surface trafficking of YFP-PC1 is not affected in BBS1-depleted cells. Live cell staining using a polyclonal rabbit anti-GFP antibody (green, marked with *) was first performed to detect cell surface PC1. After fixation and permeabilization, a second mouse monoclonal antibody against GFP (red) was used to detect all recombinant PC1 including the intracellular pool. Scale bar represents 5 µm. (C) Surface biotinylation assay. Surface biotinylation was performed as described in Materials and Methods. Surface YFP-PC1 was detected with an antibody against GFP, and β1-integrin was used as a control. (D) Quantification of surface PC1 shown in (C). Surface PC1 normalized to β-integrin in scrambled cells was set as 100%. (E) The ciliary trafficking of PC1 is facilitated through BBSome recognition of the PC1-CTT in a BBS3/ARL6-dependent manner. PC1 (marked in red)-containing vesicles target to the plasma membrane (labeled by ①) or the periciliary base (labeled by ②) and enter the cilia from either the plasma membrane via lateral diffusion or from the periciliary base via polarized targeting. (F) Disruption of the PC1-BBSome protein complex function such as depletion of BBS1, dysfunction of BBS3/ARL6 or deletion/mutation of the PC1 C-tail (ΔCTT) impairs the ciliary trafficking of PC1.
The effect of depletion of BBS1 on the plasma membrane targeting of PC1 was also confirmed biochemically by a cell surface biotinylation assay. Cell surface biotinylation of either scrambled control or BBS1-depleted IMCD3 cells transfected with YFP-PC1 revealed that there is no significant reduction of cell surface PC1 in BBS1-depleted cells (Fig. 5C and D), suggesting that trafficking of PC1-containing vesicles destined for the plasma membrane does not rely on the presence of BBS1.
DISCUSSION
The precise mechanism by which PC1 reaches the primary cilia remains elusive. Here, we show, for the first time, through an unbiased screen using PC1 C-terminal tail as a bait, that the ADPKD protein PC1 interacts with proteins encoded by the BBS genes. Moreover, we show that the ciliary trafficking of PC1 is regulated by distinct BBS proteins (Fig. 5E). Given the crucial importance of proper localization and function of the polycystins in normal organ physiology, our findings provide the basis for further understanding of the pleiotropic phenotype and etiology of the BBS.
The relatively large transcript of Pkd1 (∼14 kb) has hampered studies of PC1. Here, we have established an expression system of a both N- and C-terminal-tagged full-length PC1, which is useful for studying the trafficking, biosynthesis and function of full-length PC1 in renal epithelial cells. We found both the N- and C-termini of PC1 on the primary cilia. PC1 was proposed to undergo notch-like cleavage event in which a small C-terminal tail is released (43,44). Together with the critical role of PC1-CTT in its ciliary trafficking, our data suggest that either the large portion of PC1 is not cleaved or the cleavage of PC1 at its C-terminus occurs after reaching the primary cilia.
Data presented here suggest a role of BBSome in the import of PC1 to the primary cilia rather than export of PC1 out of the primary cilia since PC1 is reduced, not accumulated at the primary cilia of IMCD3 cells when BBSome function is lost. Interestingly, however, that the impaired PC1 trafficking to the primary cilia is seen only in the BBS1 knockdown, but not in the BBS5 and BBS8 knockdown or the BBS4 knockout epithelial cells. There are a total of 19 (45) genes whose mutations cause BBS. BBS1 is one of the three major BBS genes mutated in 20–30% of postnatal BBS cases (46). One possibility is that BBS1 may have a distinct role from other BBSome subunits so that the interaction between PC1 and BBS1 is the most critical contributing factor in the ciliary localization of PC1. Unique functions of different BBS genes have been proposed due to the fact that phenotypic differences or variability are often observed among different BBS mutant mice or patients (47). Our data are also consistent with a BBSome assembly model in which depletion of BBS1, 2, 7 and 9 have the most prominent effect on the assembly of the BBSome complex while depletion of BBS4, 5 and 8 had minor or no impact in retinal pigment epithelial cells (48). A recent report suggests that BBS2, 7 and 9 form the core components of the BBSome while BBS1, 4, 5 and 8 are added as the periphery subunits (49). PC1 interacts only with the peripheral subunits as shown in this study. BBS4 and BBS8 are primarily composed of multiple protein–protein interaction motifs, namely the TPRs, which form a solenoid structure (8). Since BBS8 was fished out from a yeast two-hybrid screen using PC1-CTT as a bait, the PC1-BBS8 or the PC1-BBS4 interaction is likely direct, probably mediated by the TPR motifs. BBS1 and BBS5 are structurally distinct from BBS4 and BBS8. In our pull-down assays, PC1 C-tail appears to interact with BBS1 as strongly as with BBS8 (Fig. 1B). The interaction of PC1 with BBS1 was also confirmed by co-immunoprecipitations (Fig. 4B). Given the potential redundant nature of the interaction of PC1 with different BBSome subunits, we believe that in the absence of certain BBSome subunits such as BBS4, 5 or 8, the remaining BBSome complex may preserve sufficient activity for targeting PC1 to the cilia. A recent report showed that in the absence of BBS7, a BBSome subcomplex still forms and the ciliary trafficking of the polycystins appears to be grossly normal and potentially functional (47) also supports our hypothesis.
BBS1 mediates the interaction of the BBSome with Rabin8 (5) and has been proposed to be the cargo recognition subunit of the BBSome for the ciliary entry of the single-span leptin receptor (7). Given only BBS1 depletion affects the ciliary trafficking of PC1, it is also possible that BBS1 plays a crucial role in the recognition of PC1 as a BBSome cargo and therefore the subsequent delivery of PC1 to the cilia. Cargo recognition by BBS1 may be mediated by the C-terminal cytoplasmic tail of PC1 since deletion of this fragment also results in failure of PC1 trafficking to the cilia (Fig. 5F). Similarly, the third intracellular loop of seven-span somatostatin receptor 3 (SSTR3) possesses a cilia-targeting sequence whose recognition by the BBSome is critical for the localization of SSTR3 to cilia (8).
Experiments with BBS3/Arl6 provide additional support for the regulation of the ciliary localization of PC1 by another BBSome protein (Fig. 5F). An elegant study by Jin et al. (8) showed that the GTP-bound form of BBS3/Arl6 is critical for recruiting the polymerized BBSome to lipid vesicles as a coat and for targeting the BBSome to cilia, but not for the assembly of the BBSome. In this study, we showed that overproduction of the GTP-binding competent wild-type BBS3 or the constitutively active form of BBS3 (Q73L) was capable of accompanying PC1 trafficking to cilia efficiently. Overexpression of BBS3 T31R, a GDP-locked, dominant-negative pathogenic mutant found in a human BBS patient, however, results in stunt cilia in IMCD3 cells and failure of PC1 to traffic to the cilia. Notably, BBS3/Arl6 interacts most efficiently with the BBS1 subunit of the BBSome through binding to the N-terminus of BBS1 (8).
The clinical phenotype of BBS patients is highly variable, even between siblings with the same mutations (50). Moreover, the spectrum of BBS phenotype may overlap with other clinically and genetically distinct ciliopathies (50). Multiple allelic interactions between BBS loci or BBS loci with other ciliopathy genes have been shown to contribute to the variability in clinical expression of BBS mutations (41,51). Although here we found that only deficiency of BBS1 and BBS3 impaired PC1 ciliary trafficking, it would not be surprising if multiallelic BBS genes or in combination with other ciliopathic alleles affect the ciliary trafficking and/or function of polycystins and consequently lead to polycystin-related phenotypes such as renal cysts.
Proteins may accumulate at certain location when they fail to traffic to its destination (7,8). In our experiments, however, we could not observe discernable increase of YFP-PC1 signal at the basal body, plasma membrane or other sites examined (data not shown) in BBS1 knockdown cells. Because the ciliary membrane only represents a tiny portion of the massive plasma membrane of IMCD3 cells, the absence of PC1 from cilia may not result in a prominent increase in PC1 signal intensity in the plasma or other organelle membranes. The normal plasma membrane distribution of PC1 in BBS1-depleted cells suggests, that PC1-containing vesicles are able to bud from the trans-Golgi network (TGN), dock and fuse to the plasma membrane under these conditions. It has recently been shown that Smoothened moves through a novel lateral transport pathway from the plasma membrane to the ciliary membrane (52). With our current data, we cannot distinguish the possibilities that the BBSome complex transports PC1-containing cargos residing in the plasma membrane to the primary cilia or directly to from the TGN to the periciliary membrane in renal epithelial cells.
In summary, we demonstrate that the ADPKD protein PC1, an 11-span atypical GPCR, targets to the primary cilia in a BBSome-dependent manner. The interaction of PC1 with BBS1 is a critical step for efficiently transporting PC1-containing vesicles to the primary cilia. This presents an interesting interplay between the critical players underlying the two genotypically distinct but partially phenotypically overlapping genetic diseases. PC1 is also present on the primary cilia of endothelial cells where it mediates mechanosensation and nitric oxide release (53). It is likely that a subset of phenotypes observed in BBS patients such as kidney cysts and hypertension result from mistargeted PC1. Our data also provide novel understanding of the signaling pathways controlled by the BBSome.
MATERIALS AND METHODS
Cell culture and shRNA knockdown
Mouse inner medullary collecting duct cells (mIMCD3) were purchased from ATCC (CRL-2123) and cultured in DMEM/F12 50/50 supplemented with 10% fetal bovine serum (FBS). HEK293 and 293EBNA cells were cultured in Dulbecco's modified Eagle's medium (DMEM) complemented with 10% FBS. Human skin fibroblast control and M390R cells (provided by Dr Beales) were cultured in DMEM supplemented with 10% FBS. Plasmids containing five target sites against each gene were purchased from Sigma. Lentiviral particles were assembled in 293EBNA cells following manufactory's instructions. IMCD3 cells were infected with viruses overnight followed by selection with puromycin at a concentration of 2 µg/ml. Knockdown efficiency in stable cells were tested by quantitative RT-PCR or western blotting. ShRNA target sequences from Sigma found to be effective in this study are: BBS1 (TRCN0000252354 and TRCN0000252355), BBS5 (TRCN0000248514 and TRCN0000248517) and BBS8 (TRCN0000113210 and TRCN0000113212).
For analysis of ciliogenesis, cells were seeded at 70% confluence and starved for 48 h in DMEM/F12 media containing 0.5% FBS after reaching confluence. For analysis of ciliary trafficking of PC1, cells were seeded at 60% confluence and transiently transfected with various constructs overnight. Cells were then serum starved in DMEM/F12 media containing 0.5% FBS for 48 h.
Plasmids and antibodies
Human PC1 C-terminal tail (PC1-CTT) fragment encoding 198 amino acids was PCR amplified from human kidney cDNAs and inserted into pACT2 to create pACT2-hCTT bait plasmid. This fragment was also inserted to pGEX-4T-1 to create pGEX-hCTT construct for production of GST-PC1-CTT fusion protein in bacteria. The myc-tagged BBS8 was a gift provided by Dr Nicholas Katsanis at Duke University. The HA-tagged BBS1, 2, 3, 4, 5, 8 and 9 were generous gifts from Dr Val Sheffield at University of Iowa. The HA-tagged truncated BBS1(R146X) was created from HA-BBS1 by PCR mutagenesis. The mouse BBS3 was amplified from mouse kidney cDNA and cloned into pcDNA3.1-HA vector to create HA-tagged BBS3 (pcDNA3.1-BBS3-HA). T31R and Q73L point mutation was introduced to pcDNA3.1-BBS3-HA by PCR mutagenesis. To generate YFP-PC1, the open reading frame of YFP was inserted into the mouse Pkd1 cDNA construct immediately after the first 34 amino acids by recombinant PCR in a tetracycline-inducible expression plasmid (pcDNA4.1, Invitrogen). YFP-PC1ΔCTT was created from YFP-PC1 by deleting the last 198 amino acids of PC1 C-terminus. The sequences of all constructs generated in this study have been confirmed by DNA sequencing analyses.
Primary antibodies used were: anti-GFP rabbit polyclonal (Abcam) 1:20 000; anti-GFP mouse monoclonal (Covance) 1:1000 dilution, anti-HA rat monoclonal 3F10 (Roche) 1:1000 dilution; anti-HA mouse monoclonal 12CA5 (Roche) 1:100 dilution; anti-myc mouse monoclonal 9e10 (Invitrogen) 1:1000 dilution; anti-γ-tubulin (Sigma) mouse monoclonal 1:2000 dilution; anti-acetylated α-tubulin mouse monoclonal (Sigma) 1:50 000 dilution; anti-PC1 rabbit polyclonal 96521 (15) 1:500 and anti-PC1 mouse monoclonal 7e12 (Santa Cruz) 1:2000, anti-BBS8 rabbit polyclonal (Santa Cruz) 1:500 dilution.
Immunostaining and immunofluorescence microscopy
For microscopic analyses, cultured cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X and blocked with 5% bovine serum albumin. Permeabilized cells were then stained with desired primary antibodies for 1 h at room temperature or overnight at 4°C. After thorough washing, secondary antibodies with appropriate labeling were added for another hour of incubation. Slides were mounted with ProLong® Gold Antifade Reagent with or without 4′,6-diamidino-2-phenylindole (DAPI). For live cell staining, cells were first rinsed twice with phosphate buffered saline (PBS) and incubated with GFP antibody on ice. After 1 h, cells were washed, fixed with 4% paraformaldehyde and continued with regular staining protocol mentioned above.
Fluorescent images were acquired with a Nikon-1000 epifluorescence microscope (Nikon, Tokyo, Japan). All images in the same set of experiment were taken with identical conditions and subjected to brightness or contrast adjustment equally in Photoshop.
Yeast two-hybrid screening
Yeast two-hybrid was performed using Clontech MATCHMAKER III yeast two-hybrid system according to the manufacturer's instructions. The bait plasmid pACT2-hCTT was transformed to yeast strain AH109 first, and a human fetal kidney cDNA library from Invitrogen was transformed to AH109/pACT2-hCTT. Colonies formed were selected on high-stringency plates SD/–Ade/–His/–Leu/–Trp/X-a-Gal. Plasmids were recovered from positive clones and sent for DNA sequencing.
Co-immunoprecipitation
HEK293 cells transfected with various constructs were homogenized with M-PER (Pierce) containing protease inhibitors (Roche) and 1 mm DTT. For co-immunoprecipitation, 2–4 mg of the cell lysates were incubated with either 2–4 µg antibodies or an equal amount of normal IgG with gentle rocking overnight at 4°C. Protein A Sepharose beads (Invitrogen) were added and incubated for another 2 h at 4°C. The washed beads were solubilized in an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer, and proteins were eluted by boiling for 5 min and analyzed by western blotting. One-tenth to 1/20th of lysates were used as inputs, unless otherwise indicated.
GST pull-down assay
GST and the GST fusion proteins were generated as previously described (54). For GST pull-down analysis, 500 µg of 293 T-cell lysates expressing different HA-tagged BBS proteins were incubated with purified GST fusion proteins immobilized on glutathione–Sepharose beads (Amersham Pharmacia Biotech) at 4°C overnight. The beads were centrifuged and were washed three times with ice-cold PBS washing buffer containing 1% NP-40. The washed beads were solubilized in an equal volume of 2× SDS sample buffer, and proteins eluted by boiling for 5 min and analyzed by western blotting. One-twentieth of lysates were used as inputs, unless otherwise indicated.
Surface biotinylation assay
Surface biotinylation assay was performed as previously described with some modifications (55). Cells were transfected overnight and serum starved for 24 h after reaching confluence. Cell surface proteins were then labeled in cell suspension at room temperature with 0.5 mg/ml of NHS-PEG4-Biotin (Thermo Scientific) for 30 min. Cells are lysed in radioimmunoprecipitation buffer, and biotinylated proteins were pulled down by streptavidin beads. Beads were eluted with 2 mm free d-biotin (Pierce), and eluted samples were resolved on 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis for subsequently analysis.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the National Institutes of Health (DK51050, DK099532, DK40703 and DK53357) to J.Z.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Nicholas Katsanis at Duke University, Dr Val C. Sheffield at University of Iowa and Dr Stuart H. Orkin at Dana-Farber Cancer Institute for providing valuable reagents used in this study. We thank Drs Gregory Pazour and George Witman at University of Massachusetts, Dr Jagesh Shah at the Brigham and women's hospital and the members of the Zhou Lab and the Harvard Center for Polycystic Kidney Disease Research for scientific discussions and support.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Beales P.L., Warner A.M., Hitman G.A., Thakker R., Flinter F.A. Bardet–Biedl syndrome: a molecular and phenotypic study of 18 families. J. Med. Genet. 1997;34:92–98. doi: 10.1136/jmg.34.2.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J.C., Badano J.L., Sibold S., Esmail M.A., Hill J., Hoskins B.E., Leitch C.C., Venner K., Ansley S.J., Ross A.J., et al. The Bardet–Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- 3.Yen H.J., Tayeh M.K., Mullins R.F., Stone E.M., Sheffield V.C., Slusarski D.C. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 4.Blacque O.E., Reardon M.J., Li C., McCarthy J., Mahjoub M.R., Ansley S.J., Badano J.L., Mah A.K., Beales P.L., Davidson W.S., et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 6.Berbari N.F., Lewis J.S., Bishop G.A., Askwith C.C., Mykytyn K. Bardet–Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo S., Guo D.F., Bugge K., Morgan D.A., Rahmouni K., Sheffield V.C. Requirement of Bardet–Biedl syndrome proteins for leptin receptor signaling. Hum. Mol. Genet. 2009;18:1323–1331. doi: 10.1093/hmg/ddp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., Nachury M.V. The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lechtreck K.F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., Pazour G.J., Ikebe M., Witman G.B. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell. Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J. Polycystins and primary cilia: primers for cell cycle progression. Annu. Rev. Physiol. 2009;71:83–113. doi: 10.1146/annurev.physiol.70.113006.100621. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J., Pie . The Molecular and Genetic Basis of Kidney Disease. Elsevier; 2008. Autosomal Dominant Polycystic Kidney Disease; p. Y. [Google Scholar]
- 12.The International PKD Consortium, T.I.P.K.D. Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell. 1995;81:289–298. doi: 10.1016/0092-8674(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 13.Hughes J., Ward C.J., Peral B., Aspinwall R., Clark K., San Millan J.L., Gamble V., Harris P.C. The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 1995;10:151–160. doi: 10.1038/ng0695-151. [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki T., Wu G., Hayashi T., Xenophontos S.L., Veldhuisen B., Saris J.J., Reynolds D.M., Cai Y., Gabow P.A., Pierides A., et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 15.Nauli S.M., Alenghat F.J., Luo Y., Williams E., Vassilev P., Li X., Elia A.E., Lu W., Brown E.M., Quinn S.J., et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 16.Nauli S.M., Rossetti S., Kolb R.J., Alenghat F.J., Consugar M.B., Harris P.C., Ingber D.E., Loghman-Adham M., Zhou J. Loss of polycystin-1 in human cyst-lining epithelia leads to ciliary dysfunction. J. Am. Soc. Nephrol. 2006;17:1015–1025. doi: 10.1681/ASN.2005080830. [DOI] [PubMed] [Google Scholar]
- 17.Talbot J.J., Shillingford J.M., Vasanth S., Doerr N., Mukherjee S., Kinter M.T., Watnick T., Weimbs T. Polycystin-1 regulates STAT activity by a dual mechanism. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7985–7990. doi: 10.1073/pnas.1103816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shillingford J.M., Murcia N.S., Larson C.H., Low S.H., Hedgepeth R., Brown N., Flask C.A., Novick A.C., Goldfarb D.A., Kramer-Zucker A., et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi Y.H., Suzuki A., Hajarnis S., Ma Z., Chapin H.C., Caplan M.J., Pontoglio M., Somlo S., Igarashi P. Polycystin-2 and phosphodiesterase 4C are components of a ciliary A-kinase anchoring protein complex that is disrupted in cystic kidney diseases. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10679–10684. doi: 10.1073/pnas.1016214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E., Arnould T., Sellin L.K., Benzing T., Fan M.J., Gruning W., Sokol S.Y., Drummond I., Walz G. The polycystic kidney disease 1 gene product modulates Wnt signaling. J. Biol. Chem. 1999;274:4947–4953. doi: 10.1074/jbc.274.8.4947. [DOI] [PubMed] [Google Scholar]
- 21.Li X., Luo Y., Starremans P.G., McNamara C.A., Pei Y., Zhou J. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat. Cell Biol. 2005;7:1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- 22.Luyten A., Su X., Gondela S., Chen Y., Rompani S., Takakura A., Zhou J. Aberrant regulation of planar cell polarity in polycystic kidney disease. J. Am. Soc. Nephrol. 2010;21:1521–1532. doi: 10.1681/ASN.2010010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin S., Taglienti M., Nauli S.M., Contrino L., Takakura A., Zhou J., Kreidberg J.A. Failure to ubiquitinate c-Met leads to hyperactivation of mTOR signaling in a mouse model of autosomal dominant polycystic kidney disease. J. Clin. Invest. 2010;120:3617–3628. doi: 10.1172/JCI41531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takakura A., Nelson E.A., Haque N., Humphreys B.D., Zandi-Nejad K., Frank D.A., Zhou J. Pyrimethamine inhibits adult polycystic kidney disease by modulating STAT signaling pathways. Hum. Mol. Genet. 2011;20:4143–4154. doi: 10.1093/hmg/ddr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boca M., Distefano G., Qian F., Bhunia A.K., Germino G.G., Boletta A. Polycystin-1 induces resistance to apoptosis through the phosphatidylinositol 3-kinase/Akt signaling pathway. J. Am. Soc. Nephrol. 2006;17:637–647. doi: 10.1681/ASN.2005050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foy R.L., Chitalia V.C., Panchenko M.V., Zeng L., Lopez D., Lee J.W., Rana S.V., Boletta A., Qian F., Tsiokas L., et al. Polycystin-1 regulates the stability and ubiquitination of transcription factor Jade-1. Hum. Mol. Genet. 2012;21:5456–5471. doi: 10.1093/hmg/dds391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delmas P., Nomura H., Li X., Lakkis M., Luo Y., Segal Y., Fernandez-Fernandez J.M., Harris P., Frischauf A.M., Brown D.A., et al. Constitutive activation of G-proteins by polycystin-1 is antagonized by polycystin-2. J. Biol. Chem. 2002;277:11276–11283. doi: 10.1074/jbc.M110483200. [DOI] [PubMed] [Google Scholar]
- 28.Torres V.E., Sweeney W.E., Jr, Wang X., Qian Q., Harris P.C., Frost P., Avner E.D. EGF receptor tyrosine kinase inhibition attenuates the development of PKD in Han:SPRD rats. Kidney Int. 2003;64:1573–1579. doi: 10.1046/j.1523-1755.2003.00256.x. [DOI] [PubMed] [Google Scholar]
- 29.Sweeney W.E., Jr, Hamahira K., Sweeney J., Garcia-Gatrell M., Frost P., Avner E.D. Combination treatment of PKD utilizing dual inhibition of EGF-receptor activity and ligand bioavailability. Kidney Int. 2003;64:1310–1319. doi: 10.1046/j.1523-1755.2003.00232.x. [DOI] [PubMed] [Google Scholar]
- 30.Yang B., Sonawane N.D., Zhao D., Somlo S., Verkman A.S. Small-molecule CFTR inhibitors slow cyst growth in polycystic kidney disease. J. Am. Soc. Nephrol. 2008;19:1300–1310. doi: 10.1681/ASN.2007070828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanaoka K., Devuyst O., Schwiebert E.M., Wilson P.D., Guggino W.B. A role for CFTR in human autosomal dominant polycystic kidney disease. Am. J. Physiol. 1996;270:C389–C399. doi: 10.1152/ajpcell.1996.270.1.C389. [DOI] [PubMed] [Google Scholar]
- 32.Yoder B.K., Hou X., Guay-Woodford L.M. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
- 33.Geng L., Segal Y., Peissel B., Deng N., Pei Y., Carone F., Rennke H.G., Glucksmann-Kuis A.M., Schneider M.C., Ericsson M., et al. Identification and localization of polycystin, the PKD1 gene product. J. Clin. Invest. 1996;98:2674–2682. doi: 10.1172/JCI119090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geng L., Segal Y., Pavlova A., Barros E.J., Lohning C., Lu W., Nigam S.K., Frischauf A.M., Reeders S.T., Zhou J. Distribution and developmentally regulated expression of murine polycystin. Am. J. Physiol. 1997;272:F451–F459. doi: 10.1152/ajprenal.1997.272.4.F451. [DOI] [PubMed] [Google Scholar]
- 35.Ma M., Tian X., Igarashi P., Pazour G.J., Somlo S. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat. Genet. 2013;45:1004–1012. doi: 10.1038/ng.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barr M.M., Sternberg P.W. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 37.Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet–Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura D.Y., Fath M., Mullins R.F., Searby C., Andrews M., Davis R., Andorf J.L., Mykytyn K., Swiderski R.E., Yang B., et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boletta A., Qian F., Onuchic L.F., Bhunia A.K., Phakdeekitcharoen B., Hanaoka K., Guggino W., Monaco L., Germino G.G. Polycystin-1, the gene product of PKD1, induces resistance to apoptosis and spontaneous tubulogenesis in MDCK cells. Mol. Cell. 2000;6:1267–1273. doi: 10.1016/s1097-2765(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 40.Cox K.F., Kerr N.C., Kedrov M., Nishimura D., Jennings B.J., Stone E.M., Sheffield V.C., Iannaccone A. Phenotypic expression of Bardet-Biedl syndrome in patients homozygous for the common M390R mutation in the BBS1 gene. Vision Res. 2012;75:77–87. doi: 10.1016/j.visres.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Beales P.L., Badano J.L., Ross A.J., Ansley S.J., Hoskins B.E., Kirsten B., Mein C.A., Froguel P., Scambler P.J., Lewis R.A., et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet–Biedl syndrome. Am. J. Hum. Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emmer B.T., Maric D., Engman D.M. Molecular mechanisms of protein and lipid targeting to ciliary membranes. J. Cell Sci. 2010;123:529–536. doi: 10.1242/jcs.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chauvet V., Tian X., Husson H., Grimm D.H., Wang T., Hiesberger T., Igarashi P., Bennett A.M., Ibraghimov-Beskrovnaya O., Somlo S., et al. Mechanical stimuli induce cleavage and nuclear translocation of the polycystin-1 C terminus. J. Clin. Invest. 2004;114:1433–1443. doi: 10.1172/JCI21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merrick D., Bertuccio C.A., Chapin H.C., Lal M., Chauvet V., Caplan M.J. Polycystin-1 cleavage and the regulation of transcriptional pathways. Pediatr. Nephrol. 2013;29:505–511. doi: 10.1007/s00467-013-2548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aldahmesh M.A., Li Y., Alhashem A., Anazi S., Alkuraya H., Hashem M., Awaji A.A., Sogaty S., Alkharashi A., Alzahrani S., et al. IFT27, encoding a small GTPase component of IFT particles, is mutated in a consanguineous family with Bardet–Biedl syndrome. Hum. Mol. Genet. 2014;23:3307–3315. doi: 10.1093/hmg/ddu044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Putoux A., Mougou-Zerelli S., Thomas S., Elkhartoufi N., Audollent S., Le Merrer M., Lachmeijer A., Sigaudy S., Buenerd A., Fernandez C., et al. BBS10 mutations are common in ‘Meckel’-type cystic kidneys. J. Med. Genet. 2010;47:848–852. doi: 10.1136/jmg.2010.079392. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q., Nishimura D., Vogel T., Shao J., Swiderski R., Yin T., Searby C., Carter C.S., Kim G., Bugge K., et al. BBS7 is required for BBSome formation and its absence in mice results in Bardet–Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J. Cell Sci. 2013;126:2372–2380. doi: 10.1242/jcs.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo S., Zhang Q., Bugge K., Breslow D.K., Searby C.C., Nachury M.V., Sheffield V.C. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 2011;7:e1002358. doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q., Yu D., Seo S., Stone E.M., Sheffield V.C. Intrinsic protein–protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet–Biedl syndrome protein complex, the BBSome. J. Biol. Chem. 2012;287:20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riise R., Andreasson S., Borgastrom M.K., Wright A.F., Tommerup N., Rosenberg T., Tornqvist K. Intrafamilial variation of the phenotype in Bardet–Biedl syndrome. Br. J. Ophthalmol. 1997;81:378–385. doi: 10.1136/bjo.81.5.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leitch C.C., Zaghloul N.A., Davis E.E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R.A., Eyaid W., Banin E., et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet–Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 52.Milenkovic L., Scott M.P., Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J. Cell. Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nauli S.M., Kawanabe Y., Kaminski J.J., Pearce W.J., Ingber D.E., Zhou J. Endothelial cilia are fluid shear sensors that regulate calcium signaling and nitric oxide production through polycystin-1. Circulation. 2008;117:1161–1171. doi: 10.1161/CIRCULATIONAHA.107.710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parnell S.C., Magenheimer B.S., Maser R.L., Rankin C.A., Smine A., Okamoto T., Calvet J.P. The polycystic kidney disease-1 protein, polycystin-1, binds and activates heterotrimeric G-proteins in vitro. Biochem. Biophys. Res. Commun. 1998;251:625–631. doi: 10.1006/bbrc.1998.9514. [DOI] [PubMed] [Google Scholar]
- 55.Luo Y., Vassilev P.M., Li X., Kawanabe Y., Zhou J. Native polycystin 2 functions as a plasma membrane Ca2+-permeable cation channel in renal epithelia. Mol. Cell. Biol. 2003;23:2600–2607. doi: 10.1128/MCB.23.7.2600-2607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.