Summary
Background
Autophagy as a conserved lysosomal/vacuolar degradation and recycling pathway is important in normal development and physiology, and defects in this process are linked to many kinds of disease. Because too much or too little autophagy can be detrimental, the process must be tightly regulated both temporally and in magnitude. Two parameters that affect this regulation are the size and the number of autophagosomes; however, although we know that the amount of Atg8 affects the size of autophagosomes, the mechanism for regulating their number has not been elucidated. The transcriptional induction and repression of the autophagy-related (ATG) genes is one crucial aspect of autophagy regulation, but the transcriptional regulators that modulate autophagy are not well characterized.
Results
We detected increased expression levels of ATG genes, and elevated autophagy activity, in cells lacking the transcriptional regulator Pho23. Using transmission electron microscopy, we found that PHO23 null mutant cells contain significantly more autophagosomes than the wild-type. By RNA sequencing transcriptome profiling, we identified ATG9 as one of the key targets of Pho23, and our studies with strains expressing modulated levels of Atg9 show that the amount of this protein directly correlates with the frequency of autophagosome formation and the level of autophagy activity.
Conclusions
Our results identified Pho23 as a master transcriptional repressor for autophagy that regulates the frequency of autophagosome formation through its negative regulation of ATG9.
Introduction
Macroautophagy, hereafter referred to as autophagy, is a highly conserved, intracellular degradation and recycling process that is tightly regulated by nutrient depletion and many other kinds of stress. In yeast, autophagy is essential for survival in nutrient-poor conditions and for adaptive responses to other changing environmental conditions through cellular remodeling [1]. In higher eukaryotes, autophagy is required for normal development and physiology, whereas autophagic dysfunction is associated with many types of human disease, including cancer, neurodegeneration, and metabolic disorders [2, 3]. Under stress conditions, portions of the cytoplasm are sequestered within a transient double-membrane structure, termed the phagophore. The phagophore is initiated and expanded from the phagophore assembly site [4], a perivacuolar punctum nucleated by most of the essential molecular machinery of autophagy. After elongation and completion, the phagophore forms a double-membrane vesicle, called the autophagosome. The autophagosome is then delivered to the vacuole (functionally analogous to mammalian lysosomes) and its outer membrane fuses with the vacuole limiting membrane, releasing its inner vesicle together with the enclosed cargo into the vacuole lumen. The cargo, including cytosolic proteins and even entire organelles, is broken down into macromolecular constituents such as amino acids by vacuolar hydrolases and recycled into the cytosol, to allow cells to survive during stress conditions [5].
Our understanding of the molecular mechanism of autophagy has been greatly advanced by yeast genetic studies in the past two decades, and more than 30 autophagy-related (ATG) genes have been identified as important players that are needed to initiate, complete, and regulate this complex process [1, 6–8]. Although an early study examining genomewide changes in gene expression under different environmental conditions provided the first indication that many ATG genes were transcriptionally regulated upon autophagy induction [9], little is known about the specific transcriptional regulators of autophagy. Furthermore, how differential induction of ATG gene expression leads to different levels of Atg proteins and contributes to each step of the autophagy pathway is not well understood.
Altered expression of many ATG genes has been observed in various human diseases [10–13], suggesting a connection between autophagy regulation and disease progression. At present, ATG8 is one of the best-characterized genes encoding a component of the core autophagy machinery, and it represents the only example in which studies have shown how protein level directly contributes to function in autophagosome formation: expression of ATG8 is negatively regulated by the transcription factor Ume6, and the Atg8 protein controls phagophore expansion during autophagosome formation; the amount of Atg8 correlates with autophagosome size [14, 15]. However, how the induction of expression of other ATG genes contributes to autophagy is still unknown, and which transcriptional regulators participate in this event remains to be determined.
Here, we identified Pho23 as a transcriptional repressor for autophagy, which negatively controls the mRNA levels, and subsequent protein levels, of many components of the autophagy core machinery, including ATG1, ATG7, ATG8, ATG9, and ATG14. We also detected elevated autophagy activity and increased frequency of autophagosome formation in pho23Δ cells. We subsequently focused on the regulation of ATG9, the gene for which transcription was most strongly affected by Pho23. Atg9 is the only transmembrane protein required for autophagosome formation, and it displays a relatively unique subcellular distribution and trafficking pattern [16–18]. Therefore, we wanted to determine how the amount of Atg9 affects autophagosome function and morphology separate from other Atg proteins such as Atg8, which mainly regulates the size of autophagosomes [15]. We found that Atg9 has a role in regulating the frequency of autophagosome formation and that Atg9 protein levels correlate with the number of autophagosomes. These findings advance our understanding of the molecular events occurring during autophagy induction, of how gene expression is regulated, and of how this regulation modulates different aspects of autophagosome formation.
Results
Pho23 Represses the Transcription of ATG Genes
ATG8 is an essential component of the autophagy machinery, and both its mRNA and protein levels are significantly elevated after a short time of autophagy induction [19]. Furthermore, among the ATG genes, ATG8 is the most well-studied example of how altered expression levels affect the autophagy outcome; reduced ATG8 expression results in a decrease in the average size of autophagosomes and a corresponding reduction in autophagy activity [15]. These features make the Atg8 protein level a good indicator for transcriptional induction of autophagy. Accordingly, we screened more than 200 yeast null mutant strains in the BY4742 background focusing on transcriptional regulators that could potentially control ATG gene expression, using the Atg8 protein level as detected by western blot for the initial readout. From the screen, a consistently increased Atg8 protein level in growing conditions (SD-N, t = 0), but not after nitrogen starvation, was detected in a PHO23 deletion mutant strain relative to the wild-type control, and this phenotype was further confirmed in two additional yeast backgrounds (Figure 1A; Table S1 available online).
Figure 1. Pho23 Represses the Transcription of Several ATG Genes When Autophagy Is Suppressed.
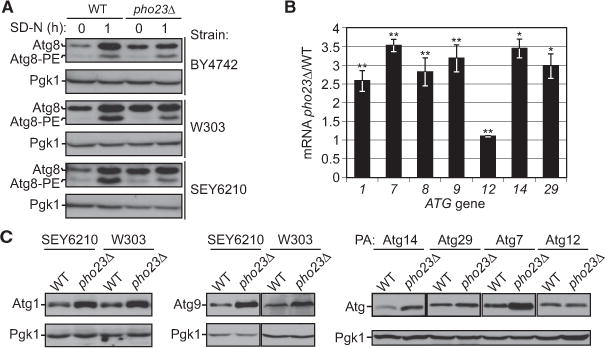
(A) Protein extracts were generated from wild-type and pho23Δ strains in the indicated backgrounds after growth in YPD to mid-log phase (growing conditions) and then shifted to SD-N medium (nitrogen starvation). Proteins were resolved by SDS-PAGE, then detected by western blot with anti-Atg8 and anti-Pgk1 (loading control) antisera. The Atg8 protein level was increased in growing conditions in pho23Δ cells relative to the wildtype in all three strain backgrounds.
(B) The ratio of pho23Δ to wild-type mRNA levels of the indicated ATG genes was measured by qRT-PCR. RNA extracts were prepared from wild-type (SEY6210) and pho23Δ (JMY047) cells after growth in YPD to mid-log phase. The error bars represent the SEM of at least three independent experiments. Two-tailed t test was used for statistical significance; *p < 0.05, **p < 0.01.
(C) Protein extracts were prepared as in (A) from wild-type and pho23Δ strains in growing conditions. The indicated proteins were detected by western blot using antisera to the endogenous proteins or an antibody that detects the protein A (PA) tag. Pgk1 was used as a loading control.
To determine whether Pho23 also regulates the expression of other ATG genes, we also examined the mRNA levels of several genes encoding the core machinery of autophagy, including ATG1, ATG7, ATG8, ATG9, ATG12, ATG14, and ATG29, in both wild-type and pho23Δ cells, by real-time quantitative RT-PCR (qRT-PCR; Figure 1B). For most of these genes, the mRNA levels during vegetative growth clearly increased in pho23Δ cells compared to the wild-type, with the exception being ATG12, which showed less than a 10% increase. We further tested whether the transcriptional increase in these genes seen in the pho23Δ cells resulted in a correlative change in protein levels, similar to the result with Atg8 (Figure 1A). Protein extracts were prepared from cells in growing conditions (i.e., when Pho23 is predicted to act as a negative regulator) and analyzed by western blot. We found elevated levels of Atg1, Atg7, Atg9, and Atg14, and to a lesser extent Atg29, in the pho23Δ strain relative to the wild-type (Figure 1C). In contrast, there was no clear change in the level of Atg12. Together, these results identified Pho23 as a potential transcriptional repressor of autophagy.
Cells Lacking Pho23 Have Increased Autophagy Activity and Generate More Autophagosomes
To determine whether Pho23 can regulate autophagy activity through its transcriptional regulation of ATG genes, different well-established assays were performed to measure autophagy activity in pho23Δ versus wild-type cells. A plasmid that expresses GFP-Atg8 driven by the CUP1 promoter was transformed into both wild-type and PHO23 deletion strains to examine GFP-Atg8 processing; we used the CUP1 promoter to eliminate Pho23-dependent effects on GFP-Atg8 expression. The GFP-Atg8 processing assay is based on the fact that a population of GFP-Atg8 is attached on the inner membrane of the autophagosome and released into the vacuole upon autophagy; this chimera is processed through the action of vacuolar hydrolases to generate free GFP in the vacuole lumen. Thus, the conversion of GFP-Atg8 into GFP is used as a readout for nonselective autophagic degradation [20]. An increasing amount of GFP-Atg8 processing was observed in the wild-type and pho23Δ strains following autophagy induction (Figure 2A). A reduced amount of the full-length chimera was seen in pho23Δ cells compared with the wildtype after 1 hr starvation, and this difference became more significant following 2 hr of nitrogen starvation, suggesting increased autophagy flux in pho23Δ cells.
Figure 2. Pho23 Negatively Regulates Autophagy Activity.
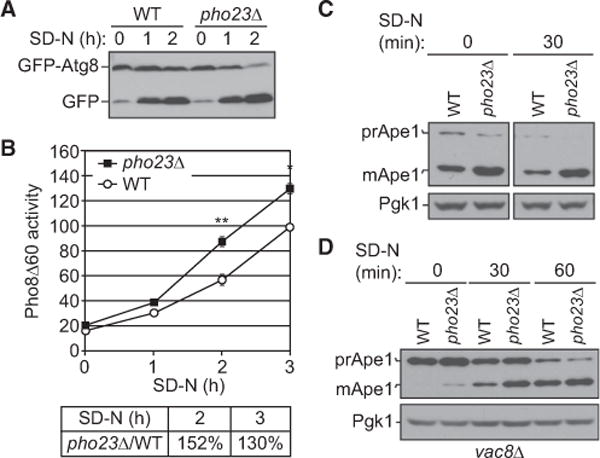
(A) Wild-type (BY4742) and pho23Δ (JMY018) cells with a centromeric plasmid expressing CUP1 promoter-driven GFP-ATG8 were grown to mid-log phase inSMD-Ura and shifted to SD-N for the indicated times. Autophagy activity was measured by the GFP-Atg8 processing assay.
(B) Wild-type (WLY176) and pho23Δ (JMY048) cells were grown to mid-log phase in YPD and shifted to SD-N for the indicated times of nitrogen starvation, and autophagy activity was monitored by the Pho8Δ60 assay. Pho8Δ60 activity was normalized to the wild-type strain (set to 100%) after 3 hr of nitrogen starvation. The graph shows the average activity from three different experiments. Error bars represent the SEM. Two-tailed paired t test was used for statistical significance; *p < 0.05, **p < 0.01.
(C) Wild-type (SEY6210) and pho23Δ (JMY047) cells were grown overnight, diluted to 0.1 optical density 600 (OD600), grown to mid-log phase (0.6 OD600) in YPD, and shifted to SD-N for 30 min of nitrogen starvation. The precursor (pr) and mature (m) forms of Ape1 were separated by SDS-PAGE and detected with anti-Ape1 antiserum by western blotting. Pgk1 was detected with anti-Pgk1 antiserum as a loading control.
(D) Precursor Ape1 processing in wild-type (vac8Δ; CWY230) and pho23Δ (pho23Δ vac8Δ; JMY146) cells at 0, 30, and 60 min after nitrogen starvation was detected by western blotting as in (C).
To extend our analysis, we took advantage of the quantitative pho8Δ60 assay. The Pho8Δ60 protein is an altered form of the vacuolar alkaline phosphatase that cannot be delivered to the vacuole via the secretory pathway; the cytosolic zymogen can be sequestered within an autophagosome and activated within the vacuole lumen [21]. Measuring the Pho8Δ60-dependent alkaline phosphatase activity can therefore be used to monitor nonselective autophagy. The Pho8Δ60 activity was measured for samples prepared from wild-type and pho23Δ cells after 0, 1, 2, and 3 hr of nitrogen starvation. In agreement with the GFP-Atg8 processing assay, pho23Δ cells showed increased autophagy activity compared with the wild-type cells after 2 and 3 hr of nitrogen starvation (Figure 2B).
We further tested whether Pho23 may affect selective autophagy by looking at the maturation of the precursor form of aminopeptidase I (prApe1). The mature form of this enzyme (Ape1) is a vacuole-resident hydrolase, and prApe1 is constitutively delivered to this organelle through the cytoplasm-to-vacuole targeting (Cvt) pathway. Following delivery, prApe1 is processed to a mature, active form that migrates as a lower-molecular-weight form during SDS-PAGE; vacuolar delivery can thus be monitored by western blot. The Cvt pathway is considered to be a selective type of autophagy, which is dependent on Atg19, the receptor for prApe1 [22]. When we compared prApe1 maturation between wild-type and pho23Δ cells, the majority of the protein was present as the mature form even in growing conditions, although pho23Δ cells appeared to have even less of the precursor form remaining; however, under these conditions it was difficult to determine whether there was a significant difference in prApe1 processing between the two strains (Figure 2C). To solve this technical problem, we took advantage of the phenotype of the vac8Δ mutant; in these cells, prApe1 processing is essentially blocked in growing conditions but is restored by nitrogen starvation [23]. Importantly, vacuolar delivery of prApe1 in these cells requires the specificity components Atg19 and Atg11, indicating that the import is still a selective process. In the vac8Δ pho23Δ strain, we observed a low level of prApe1 processing even in growing conditions and an increased level of processing relative to the control vac8Δ (WT) strain after a short time of nitrogen starvation (Figure 2D), indicating increased vacuolar delivery of prApe1 by selective autophagy in the absence of Pho23.
The elevated autophagy activity seen in the pho23Δ cells could be accounted for by an increased size or number of autophagosomes. Accordingly, we used transmission electron microscopy (TEM) to examine autophagic bodies, the single-membrane vesicles that result from the fusion of autophagosomes with the vacuole. We generated strains deleted for the PEP4 gene, which codes for a key hydrolase that is needed to break down autophagic bodies, and the VPS4 gene, which is necessary for the multivesicular body pathway. The wild-type (pep4Δ vsp4Δ) and pep4Δ pho23Δ vsp4Δ strains were grown to mid-log phase and switched to starvation conditions. Samples were prepared and analyzed by TEM (Figure 3A). Quantification of the TEM images indicated that the average sizes of autophagosome in the pho23Δ and wild-type strains were similar (Table S2); in contrast, the number of autophagic bodies per cell was nearly doubled in the pho23Δ cells relative to the wild-type after 3 hr of starvation (Figure 3B). These results further support the hypothesis that Pho23 negatively regulates autophagy and that this regulation may operate by controlling the frequency of autophagosome formation rather than the extent of autophagosome expansion (i.e., regulating the number rather than the size of autophagosomes).
Figure 3. pho23Δ Cells Have an Increased Frequency of Autophagosome Formation.
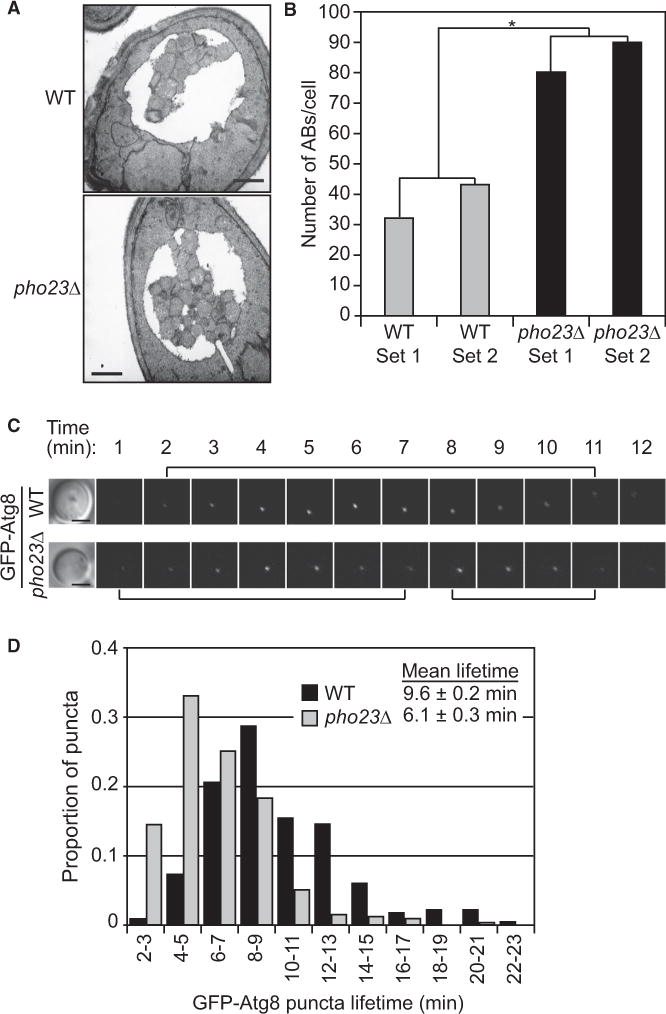
(A) Representative TEM images of wild-type (pep4Δ vps4Δ; FRY143) and pho23Δ (pep4Δ pho23Δ vps4Δ; JMY050) cells after 3 hr of nitrogen starvation. More autophagic bodies accumulated in the vacuole of pho23Δ cells. Scale bar, 500 nm.
(B) Estimated average number of autophagic body numbers per cell in wild-type and pho23Δ strains after 3 hr of nitrogen starvation. Estimation was based on the number of autophagic body cross sections observed by TEM in two independent experimental sets of more than 100 cells each for each strain. Two-tailed t test was used for statistical significance; *p < 0.05.
(C) Representative images of GFP-Atg8 in wildtype (MZY089) and pho23Δ (SKB233) cells. Brackets indicate the lifetime of each punctum. Scale bar, 2 μm.
(D) Distribution of GFP-Atg8 puncta lifetimes. Cells were imaged for 45 min beginning 40 min after a shift to nitrogen starvation. The lifetime of each individual punctum was determined as the time from when the punctum first appeared and began brightening to the time when it either disappeared or began a second round of brightening (indicating a second round of autophagosome formation from the same phagophore assembly site). N > 300 puncta from >65 cells per condition. Mean lifetime numbers show the SEM.
See also Table S2.
The increased number of autophagosomes generated in the pho23Δ cells indicated an increased frequency of autophagosome formation. To test whether this difference in frequency of formation was due to the increased rate of generating single autophagosomes, or to shortened gaps between the formation of two autophagosomes, we used fluorescence live imaging of cells expressing GFP-Atg8, which labels the forming autophagosome. The lifetime of each individual GFP-Atg8 punctum reflects the time needed to complete the formation of the represented autophagosome [24]. The average lifetime of the GFP-Atg8 puncta was 6.1 min in pho23Δ, cells versus 9.6 min in the wild-type (Figures 3C and 3D), indicating that in the Pho23-depleted cells each single autophagosome could be generated in a shorter time than in the wild-type cells. Overall, our results suggest an accelerated process of autophagosome formation when PHO23 is deleted; therefore, more autophagosomes can be generated in a given time of autophagy induction in the mutant cells.
Pho23 Regulates Autophagy in an Rpd3-Dependent Manner
To further understand the mechanism of how Pho23 regulates the transcription of ATG genes, we looked at Rpd3, a histone deacetylase (HDAC), which is associated with Pho23 and is required for the function of the Rpd3 large (Rpd3L) complex in gene repression [25]. Rpd3 occupies the promoter of many genes, and it is brought to different clusters of gene promoters by alternative recruiters [26]. Although Pho23 is not required for the enzyme activity of the Rpd3L complex, it functions as one of the recruiters essential for the specific localization of the Rpd3L complex to its target promoters [27, 28].
To understand whether the Pho23-dependent repression of ATG genes is related to Rpd3, we analyzed available data from a previous genomic study in which the transcriptome of 165 histone-modifying genes, including PHO23 and RPD3, was established by two-channel microarray [4]. We compared the genes that showed more than a 1.5-fold increase in the pho23Δ and rpd3Δ strains from their microarray study and found that Rpd3 regulates most of the genes targeted by Pho23 (122 of 143), including ATG1, ATG7, and ATG9, indicating that Pho23 may suppress transcription in an Rpd3-dependent manner (Figure S1A). To test this hypothesis, we compared the mRNA level of these ATG genes in wild-type, pho23Δ, rpd3Δ, and pho23Δ rpd3Δ deletion strains (Figure S1B). All of the single-and double-deletion mutants showed increased mRNA levels of ATG1, ATG7, ATG8, ATG9, and ATG14 compared with the wild-type strain. Furthermore, an additive increase of ATG1, ATG7, ATG9, and ATG14 mRNA levels was not detected in the pho23Δ rpd3Δ double-deletion strain relative to the rpd3Δ mutant, suggesting that in rpd3Δ cells, Pho23 no longer represses the transcription of these ATG genes.
Of the ATG genes we tested, ATG8, was an exception in that the pho23Δ rpd3Δ double mutant displayed a higher level of expression relative to either single mutant (Figure S1B). Previously, we showed that Ume6, a DNA-binding protein that belongs to the Rpd3L complex, suppresses autophagy (at least in part) through its direct repression of ATG8 transcription [14]. We further tested the possibility that Pho23 regulates ATG gene transcription through Ume6 by monitoring the ATG gene mRNA levels in a ume6Δ strain compared to the pho23Δ ume6Δ double-deletion mutant (Figure S1B). Whereas deletion of PHO23 in the ume6Δ background did not result in an additional increase of ATG8 mRNA, other ATG genes, including ATG1, ATG7, ATG9, and ATG14, showed a higher mRNA level in the double-deletion mutant. These data suggest that the role of Pho23 in the repression of ATG8 transcription is regulated less by Rpd3, and rather is Ume6 dependent, whereas for the other ATG genes tested, the deletion of UME6 does not release the Pho23-dependent transcriptional repression.
Finally, we examined the protein level of Atg1, Atg8, and Atg9 by western blot in cells lacking RPD3, UME6, and/or PHO23 (Figure S1C) and found that the results were consistent with the respective mRNA patterns. That is, in the rpd3Δ background, deletion of PHO23 did not further increase the Atg protein levels, whereas in the ume6Δ background Atg1 and Atg9, but not Atg8, were more highly expressed when PHO23 was also deleted. These results support the model that there are different forms of Rpd3L that target various groups of genes through different subunits of the complex; Pho23 and Ume6 belong to different, but partly overlapping, Rpd3 complexes, which have different effects on autophagy gene expression. This model may explain why in pho23Δ cells we observed more, but not enlarged, autophagosomes (Figure 3A and 3B), which is different from the phenotype of larger autophagosomes in ume6D cells observed in the previous study [14].
Pho23 Is Required for Extended Repression of ATG9 Transcription
The Atg proteins participate in different steps of autophagy; therefore, the change of expression of different ATG genes may cause different effects on autophagy. At present, Atg8 is the only protein reported to affect the size of autophagosomes [15]; however, there are no clear data as to whether any of the Atg proteins play a role in regulating the frequency of autophagosome formation.
To better understand how Pho23 regulates the autophagosome number per cell, we next examined the Pho23-dependent regulation of the known ATG genes relative to the entire genome. Polyadenylated RNA was isolated from wild-type and pho23Δ yeast cells in growing and starvation conditions and analyzed by RNA sequencing (RNA-seq) using the Illumina platform. In growing conditions, 12 out of 28 (41%) of the ATG genes showed a more than 2-fold increase in mRNA levels in pho23Δ cells compared to the wildtype, while only 322 out of all 6,650 (5%) genes from the whole genome showed a similar increase (Figure 4A; Tables S3 and S4). We performed a gene ontology (GO) enrichment analysis for these 322 transcripts and identified the autophagy pathway as one of the top hits (Table S1). After 2 hr of nitrogen starvation, none of the ATG genes showed a more than 2-fold increase in transcript levels in pho23Δ cells relative to the wild-type (Figure 4B). These results suggest that Pho23 acts as a potential autophagy-specific transcriptional repressor, which keeps autophagic transcription at a basal level when the process is suppressed, and its negative effect on most target genes is released by the starvation-dependent induction of autophagy.
Figure 4. pho23Δ Cells Maintain Higher ATG9 Expression Levels Relative to the Wild-Type after Autophagy Is Activated.
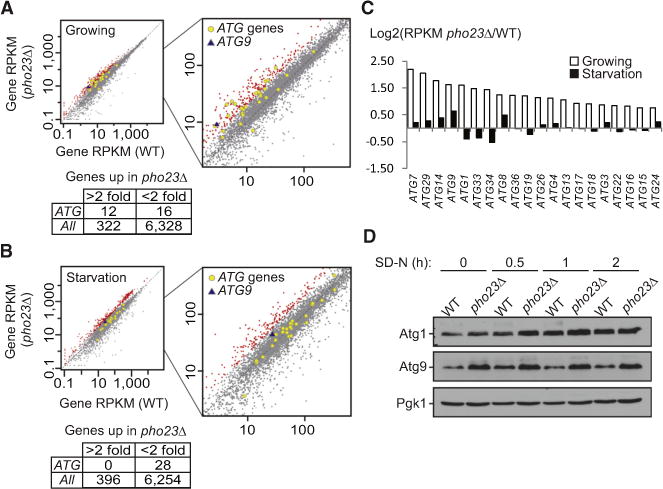
(A and B) Gene RPKM (reads per kilobase per million mapped reads) values under growing (A) or 2 hr nitrogen starvation (B) conditions are shown for the wild-type (SEY6210) versus the pho23Δ (JMY047) strains. Genes with expression changes more than 2-fold in pho23Δ cells are highlighted in red, while ATG genes are indicated by gray-outlined yellow circles and ATG9 is indicated by a blue triangle.
(C) The ratio of the gene RPKM of the pho23Δ strain to the wild-type is calculated for the top hits among the ATG genes.
(D) Atg1 and Atg9 protein levels in wild-type (TVY1) and pho23Δ (JMY020) cells in the pep4Δ background after 0, 0.5, 1, and 2 hr nitrogen starvation were analyzed by western blot. Pgk1 was used as a loading control.
See also Tables S3 and S4.
From the RNA-seq data, we further compared mRNA levels of the ATG genes that showed the greatest increase in the absence of Pho23 (Figure 4C). After nitrogen starvation, when the role of Pho23 in repressing the transcription of most of its ATG targets is largely diminished, we still observed a moderate repression of Pho23 on a limited numbers of its genes including ATG8 and ATG9. Because Atg8 regulates autophagosome size rather than number [15], ATG9 seemed a likely candidate to be a key target gene whose regulation by Pho23 controls the frequency of autophagosome formation.
To determine whether the extended repression of ATG9 mRNA transcription by Pho23 correlated with an altered level of protein synthesis, we examined the protein levels of Atg9 at various time points (0, 0.5, 1, and 2 hr) after autophagy induction by nitrogen starvation. To prevent the potential degradation of Atg proteins by autophagy, we again utilized the pep4Δ mutation. The expression pattern of the Atg9 protein was consistent with that of the mRNA; the pho23Δ strain displayed a higher level of Atg9 relative to the wildtype at time zero, and this was maintained throughout the time course (Figure 4D). In contrast, the Atg1 protein level, which we used as a control because ATG1 expression was not dependent on Pho23 after starvation (Figure 4C), showed a clear difference between the wild-type and pho23Δ strains only at the early time points, and this difference essentially disappeared by 1 hr of starvation.
These results highlighted ATG9 as the ATG gene that is most strongly affected by Pho23 repression; Pho23 regulates ATG9 transcription even after starvation, when the transcriptional repression by Pho23 on most of the other ATG genes is released.
The Level of ATG9 Expression Regulates the Frequency of Autophagosome Formation
Atg9 is the only characterized transmembrane protein among the autophagy core machinery, and it displays a dynamic trafficking pattern between the phagophore assembly site and the tubulovesicular clusters that are proposed to represent membrane reservoirs for phagophore expansion [17, 18, 29, 30]. These characteristics support a model wherein Atg9 functions in directing membrane to the phagophore from other organelles and the movement of Atg9 is required for autophagosome formation. Single-membrane vesicles containing Atg9 are proposed to fuse to initiate phagophore formation, indicating that Atg9 may also have roles in early steps of autophagosome biogenesis [31]. The apparent extended repression of ATG9 transcription by Pho23 and the increase in autophagic bodies/autophagosomes seen in the pho23Δ strain led us to hypothesize that the Atg9 protein level correlates with the frequency of autophagosome formation and thus determines in part the number of autophagosomes per cell.
To test this hypothesis, we generated four strains that express different levels of ATG9. HDY001 (referred to here as A9Δ) is the atg9Δ negative control, HDY003 (A9Lo) expresses ATG9-GFP under the control of the ATG23 promoter and thus synthesizes a level of Atg9 lower than that of the wild-type, HDY002 (A9WT) expresses ATG9-GFP driven by the endogenous ATG9 promoter, and HDY007 (A9OE) expresses ATG9-GFP driven by the ATG8 promoter integrated into a wild-type strain in addition to the endogenous ATG9 gene and expresses a higher level of Atg9 than the wild-type. The Atg9 or Atg9-GFP protein levels in these strains were confirmed by western blotting using anti-Atg9 antiserum (Figure 5A). Next, we quantified the autophagy activity of these strains using the Pho8Δ60 assay. All four strains showed similar levels of basal Pho8Δ60 activity in growing conditions, but the Pho8Δ60 activities after 3 hr of nitrogen starvation varied (Figure 5B). As expected, the A9WT strain showed a clear induction of Pho8Δ60 activity after starvation (this value was set to 100% and used for the normalization of the other activities), while the activity of the A9Δ strain remained at the background level. The A9Lo strain showed approximately 60% of the Pho8Δ60 activity of A9WT, and the A9OE strain displayed approximately 130% of the activity of A9WT, being significantly higher than the wild-type level. These results indicated that the Atg9 protein level correlates with/regulates autophagy activity.
Figure 5. The Atg9 Protein Level Correlates with Autophagosome Formation Frequency and Autophagy Activity.
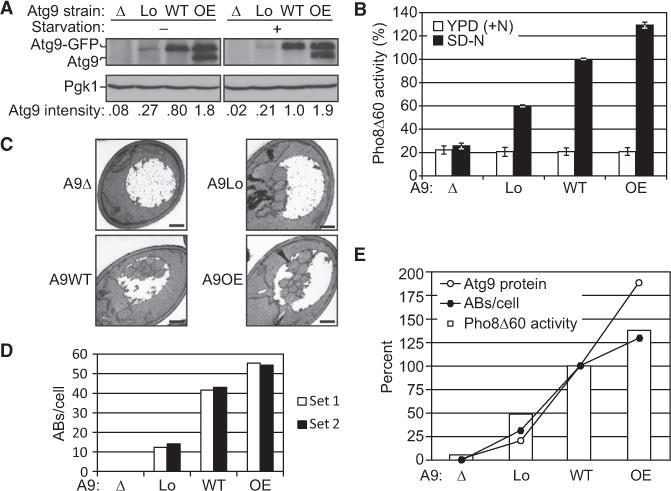
(A) Four strains expressing different levels of Atg9-GFP (A9Δ, the null control strain; A9Lo, expression controlled by the ATG23 promoter and thus lower than WT; A9WT, controlled by the endogenous promoter; and A9OE, expression controlled by the ATG8 promoter in addition to the endogenous copy of ATG9 and thus higher than WT) were grown in YPD to mid-log phase and shifted to SD-N for 3 hr. Proteins were resolved by SDS-PAGE and detected by western blot with anti-Atg9 and anti-Pgk1 (loading control) antisera. Atg9 intensity was measured in ImageJ and represents an average of three independent experiments.
(B) Cell lysates were generated as in (A) and autophagy activity was monitored by the Pho8Δ60 assay. Pho8Δ60 activity was normalized to the A9WT strain (set to 100%) after 3 hr starvation. The graph shows the average activity from at least four different experiments. Error bars represent the SEM.
(C) Representative TEM images of the four A9 strains after 3 hr starvation. Scale bar, 500 nm.
(D) Estimated average number of autophagic bodies per cell in the four A9 strains after 3 hr of nitrogen starvation. Estimation was based on the number of autophagic body cross sections observed by TEM in two independent experimental sets of more than 190 cells each for each strain.
(E) Atg9 protein level detected by western blot, average autophagic body number per cell estimated by TEM, and starvation-induced Pho8Δ60 activity (the average Pho8Δ60 activity in growing condition was subtracted) of the four different A9 strains after 3 hr starvation were plotted on one graph for comparison.
See also Table S5.
Finally, as with the pho23Δ strain, the changes in autophagy activity could be due to differences in autophagosome size and/or number. Therefore, we deleted PEP4 in each of these strains and measured the size and counted the number of autophagic bodies after 3 hr of nitrogen starvation using TEM. As expected, no autophagic bodies were observed in the A9Δ strain. For the other strains, the estimated average number of autophagic bodies per cell was increased in the order of increased ATG9 expression (Figures 5C and 5D), which again strongly supports our hypothesis that the Atg9 protein level regulates the frequency of autophagosome formation and thus controls the autophagic body number in each cell. At higher levels of Atg9 expression, the highest correlation was seen between autophagic body number and Pho8Δ60 activity, whereas Atg9 protein expression appeared to be saturating near that of the wild-type (Figure 5E). These results also provide a partial explanation for the increased autophagic body numbers we observed in the Pho23Δ cells (Figures 3A and 3B), since ATG9 expression was significantly increased in these cells before, and continued after, autophagy induction.
Discussion
Here, we identified Pho23 as a transcriptional repressor, the absence of which causes increased transcription of several autophagy genes, increased frequency of autophagosome formation, and elevated autophagy activity. Pho23, together with Yng1 and Yng2, compose the inhibitor of growing (ING) family in yeast [32]. The ING family proteins share a significant sequence identity on their conserved C-terminal plant homeodomain (PHD) finger, and each of them has been identified as a subunit of distinct chromatin modification complexes [28]. Pho23 has two human homologs, ING1 and ING2, which associate with a HDAC complex, and are candidate tumor suppressors [33]. In yeast, Pho23 is associated with Rpd3, an HDAC, and is required for the gene-repressive function of the Rpd3L complex [25]. Although Pho23 is not required for the HDAC activity of Rpd3, it is essential for the specific recruitment of Rpd3 to the promoters of its target genes [27, 28]. In this study, we found that Pho23 represses the transcription of certain ATG genes in an Rpd3-dependent manner, which raises the possibility that Rpd3 controls ATG gene transcription specifically through Pho23. However, there is no evidence that Pho23 can directly bind to DNA, and therefore, the recruitment of the Pho23-Rpd3 complex may further depend on additional DNA binding molecules. One of the DNA-binding components of the Rpd3L complex, Ume6, represses the transcription of ATG8 and autophagy activity [27, 28], but our data suggest that Pho23-dependent transcriptional repression of ATG genes other than ATG8 is independent of Ume6. Thus, the DNA-binding partner of Pho23 that may target the Pho23-Rpd3 complex to the promoters of ATG genes remains to be identified.
Recently, HDACs have become potential targets for various disease therapies, including cancer and metabolic disorders [34]. Our findings reveal a possible mechanism for how HDACs may play a role in these diseases through the regulation of autophagy. HDAC inhibitors have been studied as candidate drugs for various diseases including cancer, and our finding that Pho23 may target autophagy genes in a more specific manner than Rpd3 suggests a potential therapeutic strategy; targeting the ING proteins rather than the histone deacetylase itself may increase the efficiency, while decreasing the side effects, of particular drugs.
In cells lacking PHO23, several ATG genes displayed an increase in expression. Thus, it was not immediately clear which gene product(s) accounted for the observed increase in autophagosome number. In order to specifically examine the role of Atg9, we constructed strains in which only the expression of the ATG9 gene was altered. In this report, we show, by comparing Pho8Δ60 activity and autophagic body numbers, that the amount of Atg9 regulates autophagy activity by modulating the number of autophagosomes (Figure 5). Generally, higher levels of Atg9 result in more autophagic bodies and higher autophagy activity; however, with a more careful comparison, we found that the Atg9 level is not correlated in an absolutely linear manner with autophagic body number or autophagy activity (Figure 5E). The A9Lo cells, which express only 21% of the Atg9 of the A9WT cells and form only one-third the number of autophagosomes, showed as high as 48% of the induction of the autophagy activity relative to the A9WT cells. This observation can be explained by the increased average size of the autophagic bodies observed in the A9Lo cells (Table S5). We do not know the reason for this increase in size; however, we suggest that this may be due to the delay of autophagosome formation caused by the reduced amount of Atg9, which could provide additional time for the phagophore to expand. Overall, the reduced number of autophagosomes seen with lower levels of Atg9 reflects a role for this protein in the rate of autophagosome expansion or closure, but further studies will be needed to make this determination. Conversely, the A9OE cells, whose Atg9 expression level is almost twice that of the A9WT cells, showed only a moderate increase (~25%) of autophagic bodies and autophagy activity. Most likely, only increasing the Atg9 level beyond that of the wild-type is not sufficient to dramatically increase autophagosome formation, as other Atg proteins may become limiting.
In cells lacking PHO23, a significantly increased number of autophagic bodies per cell was detected (Figure 3B), even higher than in the A9OE cells. This increase is likely primarily due to the elevated Atg9 protein levels in the pho23Δ cells, but its magnitude suggests that the increased expression level of other Atg proteins such as Atg7, Atg14, and Atg29 whose expression is also increased in pho23Δ mutants may be involved as well. Carefully designed experiments will be needed to separate the functions of these other proteins from Atg9 in order to test their roles in regulating autophagosome number, because some of them affect the trafficking of this protein.
In summary, our results suggest a relatively direct connection between Atg9 level and autophagosome formation frequency. Additionally, the data show that Pho23 represses ATG gene expression levels, especially in growing conditions, and that this repression—in particular, the repression of ATG9—decreases the frequency of autophagosome formation and limits autophagic activity.
Experimental Procedures
Yeast Strains, Media, and Culture
Yeast strains used are listed in Table S1. Gene deletions or integrations were performed using a standard method [35]. Samples for growing conditions are collected from mid-log phase yeast cells grown in rich medium (YPD: 1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] glucose; or SMD: 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed). Autophagy was induced through nitrogen starvation by shifting cells in mid-log phase from YPD to SD-N (0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% [w/v] glucose) for the indicated times.
Plasmids
Plasmid pRS406-ATG9p-ATG9-GFP contains 1,000 bp of ATG9 5′ sequence in front of the ATG9-GFP open reading frame, plasmid pRS406-ATG23p-ATG9-GFP contains 750 bp of the ATG23 5′ sequence instead of the ATG9 promoter, and plasmid pRS406-ATG8p-ATG9-GFP replaces the ATG9 promoter with 1,000 bp of the ATG8 5′ sequence. These ATG9-GFP-expressing plasmids or the corresponding empty vector (pRS406) were linearized and integrated into the wild-type or atg9Δ strain to generate the A9 strains indicated in Table S1.
RNA-Seq
Yeast cells were grown in YPD to mid-log phase and then shifted to SD-N. Total RNAs were extracted using the Master-Pure yeast RNA purification Kit (Epicenter Biotechnologies). cDNA library preparation and Illumina high-throughput sequencing was performed by the DNA sequencing core at the University of Michigan. Reads from multiplexed libraries were scanned for a perfect match to each unique 6 nt barcode. If more than one barcode was matched perfectly, the 3′-most barcode was chosen. Remaining reads were scanned for one mismatch to each 6 nt barcode. If more than one barcode was matched with one mismatch, the 3′ -most barcode was chosen. The mean sequencing error rate for each library was estimated from the mean quality score. Reads were mapped using Bowtie2 [36] (parameters: -f -v 3 -k 500–best–strata) to the yeast transcriptome derived from yeast genome version S228C. Alignments with quality score less than 88 were discarded. A majority of mapped reads mapped to a unique locus in the transcriptome; these reads were used to calculate reads per kilobase per million mapped reads (RPKM) for each yeast gene (for details, see Tables S4 and S5).
qRT-PCR
Yeast cells were grown in YPD to mid-log phase and then shifted to SD-N. Total RNAs were extracted using the RNeasy Mini kit (QIAGEN), and reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems).
GFP-Atg8 Live Imaging
Cells were immobilized for live imaging as described previously [15]. Images were collected on a Deltavision Elite deconvolution microscope (GE Healthcare/Applied Precision) with a 100× objective and a CCD camera (CoolSnap HQ, Photometrics). Twelve-image stacks (0.4 μm spacing, to cover the entire cell) were taken each minute for 45 min, deconvolved, projected, and manually analyzed to determine GFP-Atg8 puncta lifetime.
Other Methods
Western blot, the GFP-Atg8 processing and Pho8D60 assays, and TEM were performed as described previously [20, 37, 38]. Antisera to Atg8 [39], Atg1 [40], Atg9 [16], Ape1 [41], Pgk1 (a generous gift from Dr. Jeremy Thorner, University of California, Berkeley), and a commercial antibody that reacts with protein A (no longer available) were used as described previously.
Statistical Analysis
Two-tailed Student’s t test and two-tailed paired Student’s t test was used to determine statistical significance.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM053396 (to D.J.K.) and GM088565 (to J.K.K.) and by the National Science Foundation Open Data IGERT grant 0903629 (to M.A.F.) and was funded in part through the Protein Folding Diseases Fast Forward Initiative, University of Michigan.
Footnotes
Accession Numbers
The Gene Expression Omnibus accession number for the pho23Δ sequencing data reported in this paper is GSE57031.
Supplemental Information
Supplemental Information includes one figure and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2014.04.048.
References
- 1.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 6.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 8.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 9.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, Pang S, Feng X, Huang W, Hawley RG, Yan B. Genetic analysis of the ATG7 gene promoter in sporadic Parkinson’s disease. Neurosci Lett. 2013;534:193–198. doi: 10.1016/j.neulet.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, He Z, von Rutte T, Yousefi S, Hunger RE, Simon HU. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med. 2013;5:202ra123. doi: 10.1126/scitranslmed.3005864. [DOI] [PubMed] [Google Scholar]
- 12.Jo YK, Kim SC, Park IJ, Park SJ, Jin DH, Hong SW, Cho DH, Kim JC. Increased expression of ATG10 in colorectal cancer is associated with lymphovascular invasion and lymph node metastasis. PLoS ONE. 2012;7:e52705. doi: 10.1371/journal.pone.0052705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Pan XL, Ding LJ, Liu DY, Da-Peng Lei, Jin T. Aberrant expression of Beclin-1 and LC3 correlates with poor prognosis of human hypopharyngeal squamous cell carcinoma. PLoS ONE. 2013;8:e69038. doi: 10.1371/journal.pone.0069038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci USA. 2012;109:11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 18.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondriaand the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 22.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott SV, Nice DC, 3rd, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 24.Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008;182:129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loewith R, Smith JS, Meijer M, Williams TJ, Bachman N, Boeke JD, Young D. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J Biol Chem. 2001;276:24068–24074. doi: 10.1074/jbc.M102176200. [DOI] [PubMed] [Google Scholar]
- 26.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 27.Wang SS, Zhou BO, Zhou JQ. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol Cell Biol. 2011;31:3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avvakumov N, Côté J. The MYST family of histone acetyl-transferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 29.Yen WL, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guérillon C, Larrieu D, Pedeux R. ING1 and ING2: multifaceted tumor suppressor genes. Cell Mol Life Sci. 2013;70:3753–3772. doi: 10.1007/s00018-013-1270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang J, Yan H, Zhuang S. Histone deacetylases as targets for treatment of multiple diseases. Clin Sci. 2013;124:651–662. doi: 10.1042/CS20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 38.Backues SK, Chen D, Ruan J, Xie Z, Klionsky DJ. Estimating the size and number of autophagic bodies by electron microscopy. Autophagy. 2014;10:155–164. doi: 10.4161/auto.26856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang WP, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 40.Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


