Abstract
The adult human gut presents a complicated ecosystem where host-bacterium symbiosis plays an important role. Bacteroides thetaiotaomicron is a predominant member of the gut microflora, providing the human digestive tract with a large number of glycolytic enzymes. Expression of many of these enzymes appears to be controlled by histidine kinase receptors that are fused into unusual hybrid two-component systems that share homologous periplasmic sensor domains. These sensor domains belong to the third most populated (HK3) family based on a previous bioinformatics analysis of predicted histidine kinase sensors. Here, we present crystal structures of two sensor domains representative of the HK3 family. Each sensor is folded into three domains: two seven-bladed β-propeller domains and one β-sandwich domain. Both sensors form dimers in crystals and one sensor appears to be physiologically relevant. The folding characteristics in the individual domains, the domain organization, and the oligomeric architecture are all unique to the HK3 sensors. The sequence analysis of the HK3 sensors indicates that these sensors are shared among other signaling molecules, implying a combinatorial molecular evolution.
Keywords: beta-propeller domain, beta-sandwich domain, hybrid two-component system, saccharide sensing, signal transduction, dimerization
Introduction
The adult human gut is colonized with 10–100 trillion microbes, which provide an extra 2–4 million genes that provide beneficial metabolic capacities that are not encoded in our genome [1–3]. It has been estimated that 10% of our daily harvest of calories comes from intestinal absorption of microbial polysaccharide fermentation [4]. Two divisions of bacteria, the Bacteroidetes and the Firmicutes account for >99% of phylotypes in this intestinal microbial community, and Bacteroides thetaiotaomicron is a most prevalent human gut symbiont, accounting for 6% of all human gut microbes based on 16 S rDNA analysis [4]. The B. thetaiotaomicron genome contains a large number of two-component systems (TCSs), many of which are involved in the adaptation to the host diet [5, 6]. The microbial flora constitutes a unique ecosystem with important implications for human health and disease.
TCS signaling predominates in bacterial adaptation to environmental changes [7, 8]. Although TCS signaling is also prevalent in plants, fungi and other protists, it is absent in metazoan animals, which renders systems are potential antibiotic targets. The classical TCS components are a histidine kinase (HK) receptor and its cognate response regulator (RR). Typically, signaling initiates upon ligand binding to an extracellular sensor domain of the HK receptor, which is usually flanked by two transmembrane helices (TM1 and TM2), and the signal arising from this binding propagates across the cell membrane to a dimeric cytoplasmic unit.
The cytoplasmic portion of an HK receptor minimally comprises a dimerization and histidine-containing phosphotransfer domain (DHp) and a characteristic catalytic kinase domain (CA) [9]; other domains may be present. Depending on the receptor, ligand binding or its absence may induce the kinase activity for histidine autophosphorylation. Some receptors autophosphorylate in trans [10, 11] and others do so in cis [10, 11]. Subsequent phosphotransfer to a conserved aspartate residue in the cognate RR receiver domain (REC) causes a conformational change that activates the RR effector domain, which is typically a transcription factor [7]. Many HK receptors also possess a phosphatase activity by which the phospho-RR is dephosphorylated when the kinase-inducing signal is absent [12, 13].
The nature of signal transduction across the lipid bilayer remains controversial; however, based on biochemical analyses and atomic structures, mechanisms have been suggested including piston-like movement [14–19], asymmetric rotation [20], and scissor-like closure [21]. Irrespective of ligand state, HK receptor kinase activity correlates with symmetric sensor-domain dimers and phosphatase activity correlates with asymmetric sensor-domain dimers [22].
In contrast to highly conserved sequence and tertiary structure of CA and REC domains [7, 23–26], HK sensor domains and RR effector domains are modular, having varied organizations reflecting the diversity of signals that are detected and outputs that are generated in TCS signaling. An earlier bioinformatics analysis identified HK sensor domains as segments located between two transmembrane helices and followed by a CA domain (J. Cheung, J. Liu, B. Rost and W.A. Hendrickson, unpublished), finding a few hundred families of different sensor domains based on PSI-BLAST E cut-off at 0.001. Despite considerable effort to fill the structural landscape of HK sensor domains [27–29], many families remained without structural representatives.
Among HK sensor domains without previously known structures, the third most populated family, HK3, is unique in that all of its original 38 members have unusually long sequences (>700 amino acids) and include many sequence repeats. Secondary structure prediction indicates these sensors have an all-β structure and may form a β-propeller type fold. Interestingly, 23 of these 38 sensors are from B. thetaiotaomicron and they are all from a unique class of hybrid TCS (HTCS) having both components fused into one single polypeptide [30]. An analysis of the B. thetaiotaomicron genome found 32 such hybrid TCSs [31], 31 of which contain a sensor homologous to HK3 family members. Our previous bioinformatic study did not catch all these members due to the restrictions on sequence selection.
Here we describe crystal structures for the sensor domains from two B. thetaiotaomicron HK3 receptors, namely BT3049 and BT4673. We previously described an alternative structure determination for one of these [32], and a structure of the BT4663 sensor domain was reported subsequently [21]. The folded chains from these HK3 sensors comprise three domains each, including two seven-bladed β-propeller domains and one C-terminal β-sandwich domain. Both sensor proteins dimerize in solution, and one of the crystal structures captures dimers that appear to have functional significance. Our bioinformatics analyses found 36 of these sensors in B. thetaiotaomicron and more than 1500 in the non-redundant protein database. More than 1000 HK3-like sequences also contain putative functional domains, including >800 histidine kinase CA domains and domains from other signaling pathways. Thus, our structural characterization of this unique family of sensor proteins provides insights into understanding of symbiosis in the human gut.
Results
Based on our previous bioinformatic analyses [29], which classified various HK sensor families at the PSI-BLAST level of E ≤ 1×10−3, we identified as HK3 the family with the third most members. Family HK3 sensor domains are distinguished as being exceptionally large and in being all-β structures by sequence prediction. The original, restricted analysis had 38 sequences; here a fuller PSI-BLAST expansion based solely on the sensor domains found 1658 homologs at the same 10−3 level, including 36 in the genome of B. thetaiotaomicron. We extended our sequence analysis seeking to understand the distribution and evolution of these sensors. We also cloned and expressed several of these domains, we have determined crystal structures for two HK3 sensors from B. thetaiotaomicron, and we have further analyzed these proteins.
Sequence analyses
PSI-BLAST searches through the B. thetaiotaomicron genome, with any single Family 3 member as the template, all converge to the same set of 36 sequences including 23 among our original HK3 members. A multiple sequence alignment was performed to evaluate the relationship of these sequences (Figure S1). All 36 HK3 proteins from B. thetaiotaomicron have a predicted transmembrane helix following the sensor domain, and all have a C-terminal domain homologous with AraC, a transcription factor first described for controlling the uptake and catabolism of L-arabinose in Escherichia coli [33]. Thirty of the 36 proteins appear to complete hybrid systems [30], in which a histidine kinase receptor is fused directly to a response regulator (sensor-DHp-HKase-REC-AraC; Figure 1A). Thereby, AraC appears to be an effector coupled directly to its sensor. The remaining six proteins relate in varying degrees to the hybrid model: BT4236 has all the domains, but it lacks the conserved DHp histidine residue; BT4673 lacks both the REC and DHp domains; BT3951 and BT3957 do not have DHp and HKase domains; and BT0138 and BT3660 lack all intracellular domains except AraC. Of the genes corresponding to these HK3 proteins, 28 are located within previously described polysaccharide utilization loci (PULs) [34], thus implying a likely connection to polysaccharide metabolism.
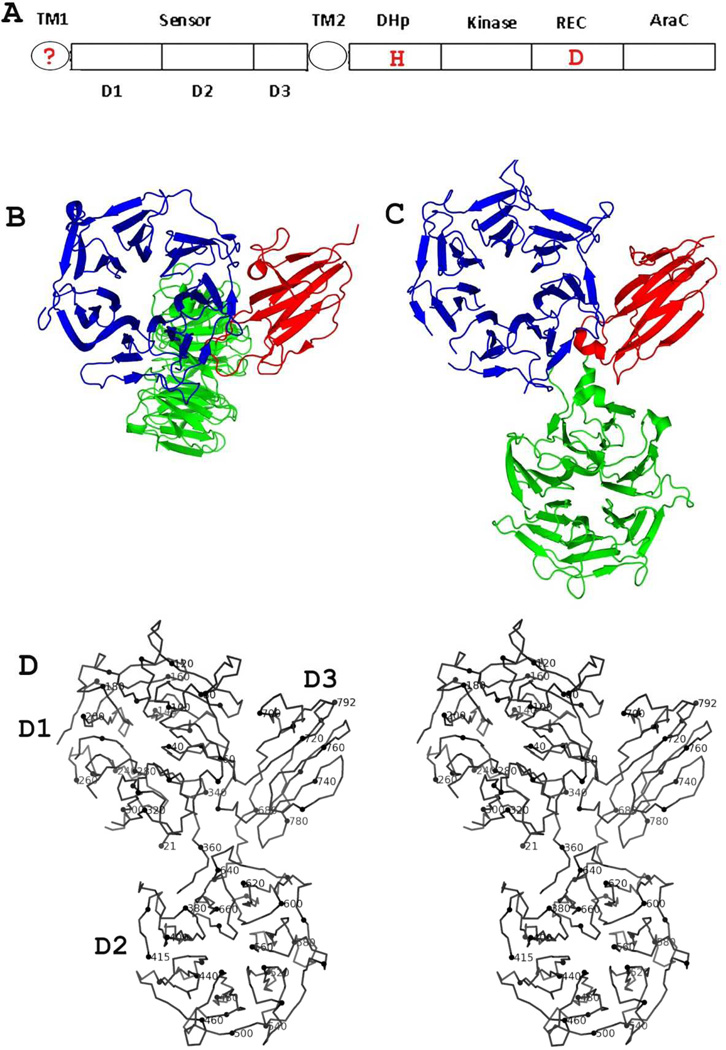
Figure 1. Structure of family 3 (HK3) histidine kinase receptors.
(A) Prototypical domain organization of HK3-type receptors. The question mark in TM1 indicates that it may not be present as a TM helix (see Supplemental Figure S1). Positions of signature two-component phosphorylation sites at histidine (H) and aspartic acid (D) residues are marked in red.
(B) Ribbon diagram of HK3 sensor domain BT4673S. The orientation views domain D1 into the top view [38] of the β-propeller domain. The coloring is done by domain: D1 (blue), D2 (green) and D3 (red).
(C) Ribbon diagram of HK3 sensor domain BT3049S. The orientation of the D1/D3 unit is as in Fig. 1B. The coloring is as in (B). Figs. 1C and 1D were generated using PyMol [69].
(D) Stereodiagram of the α-carbon backbone trace of HK3 BT3049S. Every 10th residue (modulo 10) is highlighted and every 20th residue is labeled.
A broader PSI-BLAST search for full-length HK3 sensor domains found 1658 such sequences in the non-redundant protein database, and 1071 of them contain at least one other functional domain in addition to the sensor domain. The majority of these HK3 sensor homologs (613/1071) have only one predicted transmembrane helix each. Among the others, 136 (12.7%) have no predicted transmembrane helices, 309 (28.9%) are predicted to have two, and only 13 (1.2%) have more than two. We further characterized the 1071 putatively functional proteins based on the nature of cytoplasmic domains. The main group (822, 76.8%) are putative histidine kinase receptors. These include 676 that contain a REC domain and 620 that contain a DNA-binding domain; thus, as in B. thetaiotaomicron, HTCS systems predominate. Each in a second group (112; 10.5%) contains a diguanylate cyclase (DGC) domain, each in a third group (25) contains a type 2C protein phosphatase (PP2C) domain, and the other variants include one methyl-accepting chemotaxis protein (MCP). As noted previously [15, 31, 35], the swapping of common extracellular sensor domains among various cytoplasmic functional domains seems to be commonplace in molecular evolution.
Structure determinations
We crystallized and determined crystal structures for the sensor domains from two HK3 receptors from B. thetaiotaomicron, BT4673S and BT3049S. Each of these sensor proteins is a three-domain structure comprising two N-terminal seven-bladed β-propeller domains (D1 and D2) followed by a C-terminal β-sandwich domain (D3). The structures are obviously similar to one another, but inter-domain orientations differ (Figures 1B and 1C). At the time, the top hits from Dali searches [36] were other β-propeller proteins and β-sandwich proteins, notably tenascin [37], with the best Z scores of ~26. Now, of course, the top hit is for the BT4663 sensor [21]. Searches with individual domains returned the same hits as when combined; however, except now for BT4663S, none of these hits has the same three-domain organization as in HK3 sensors.
HK3 BT4673S structure
The structure of HK3 BT4673S was solved by single-wavelength anomalous diffraction (SAD) phasing from a Ta6Br12 derivative and refined at 2.30 Å resolution (Table 2, Figure 1B). These crystals are in space group P41212 and have two BT4673S molecules, two tantalum clusters, and 280 ordered water molecules in the asymmetric unit. The two BT4673S molecules are related by approximate non-crystallographic two-fold symmetry. D1 and D3 in one molecule are closely packed together and D2 sticks out to make contacts with D1/D3 from the other molecule and vice versa. Several of the intra-propeller loops in D1 and D2 as well as the D2-D3 connector are disordered in the crystal structure; however, the domain connectivity is unambiguous. The tantalum clusters are well ordered and may contribute to the stabilization of derivatized protein for better diffraction. Without tantalum derivatization, BT4673S crystals diffracted to about 2.8 Å. For the selenomethionyl (SeMet) version, most crystals diffracted weakly to only about 3.5 Å; and we needed multiple crystals to enhance signal-to-noise ratio for structure determination by SAD phasing [32].
Table 2.
Refinement statistics
| Proteins | HK3BT4673s | HK3BT3049s | |
|---|---|---|---|
| Derivative label | Tantalum cluster | SeMet | |
| Resolution (Å) | 2.3 | 3.3 | |
| Space Group | P41212 | P212121 | |
|
Unit cell |
a (Å) | 88.2 | 79.9 |
| b (Å) | 88.2 | 114.1 | |
| c (Å) | 433.0 | 487.5 | |
| Zaa | 2 | 4 | |
| Solvent content | 50% | 61% | |
| Unique reflections | 81,707 | 59,376 | |
| Total atoms | 12,329 | 23,560 | |
| Protein atoms | 12,099 | 23,560 | |
| Water molecules | 280 | 0 | |
| Tantalum cluster (Ta6Br12) | 2 | ||
| Rworkb | 19.5 | 22.5 | |
| Rfreec | 25.3 | 28.0 | |
| RMS bond (Å) | 0.020 | 0.012 | |
| RMS angle (Å) | 1.761 | 1.504 | |
| Bfactor (Å2) | 22.8 | 101 | |
|
Ramachandran analysisd Favored/allowed (%) |
94.6/99.3 | 87.7/97.6 | |
| PDB code | 3OTT | 3V9F | |
Za stands for number of molecules per asymmetric unit.
Rwork = (Σ | |Fo| − |Fc| |) / Σ|Fo|, where Fo and Fc denote observed and calculated structure factors, respectively.
Rfree was calculated using 5% of data excluded from refinement.
Molprobity [68].
HK3 BT3049S structure
The crystal structure of HK3 BT3049S was solved by SeMet SAD phasing at 3.3 Å resolution (Table 2, Figure 1C). There are four protein molecules in each asymmetric unit of a P212121 lattice. The four BT3049S molecules form two similar dimers wherein the protomers of each dimer are diad related. There is no rational symmetry between dimers. Based on analyses of the dimeric associations, we concluded that the conformation observed for BT3049S has the two C-termini poised appropriately for transmembrane signaling, whereas the conformation captured for BT4673S does not. The BT3049S conformation is shown in stereographic detail in Figure 1D. Due to the low resolution and intrinsic flexibility, many loops are disordered and hence were not built in the model of BT3049S.
Structural analyses
Propeller packing
Comparing to known β-propeller domains [38], to our best knowledge, the HK3 propellers present a novel sequence conservation and structural packing pattern (Figure 2A and 2B). The repeat units of each propeller are four-stranded anti-parallel β-sheets (comprising strands A to D named from N-terminus to C-terminus); these sheets serve as blades of the propeller with strand A being innermost. The blades circulate counterclockwise around the central pore (Figure 2C) when viewed from the conventional ‘top’ side [37] in Figures 1 and 2. All of the blades are similar in structure (Figure 2D). The circular configuration is such that sequence repeats are overlapping in the structural blades. In particular, strands A, B and C of blade 7 in propeller D1 are from the seventh sequence repeat, but strand D of this blade is from the beginning of the first sequence repeat, just ahead of strand A of blade 1 in D1; similarly, in propeller D2, strand D of blade 7 is from the segment that starts the eighth sequence repeat (just ahead of strand A in its blade 1 and just after strand C of blade 7 in D1) whereas strands A, B and C of this blade are from the 14th repeat (Figure 2F). The hydrogen-bonded inter-digitizations of these circular permutations further stabilize each propeller by adding a covalent linkage to the non-bonded inter-blade interactions (Figure 2C and 2E).
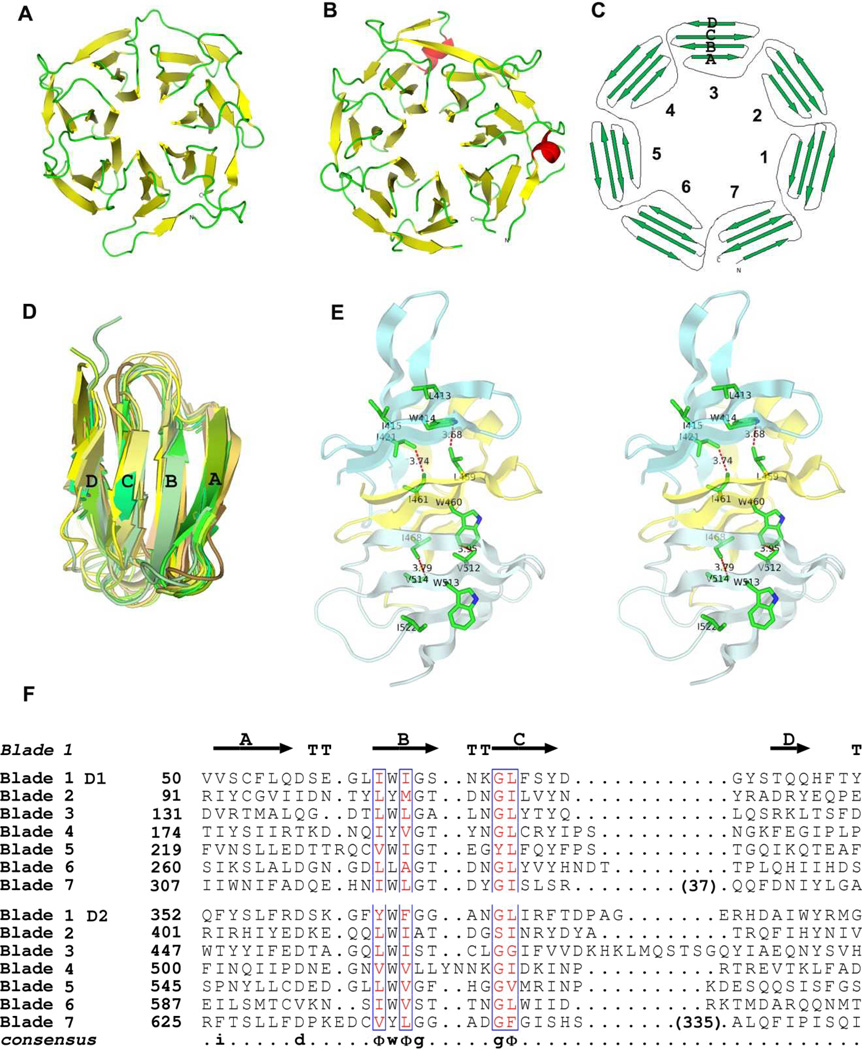
Figure 2. Structure and sequence characteristics of HK3 β-propeller domains.
(A) Ribbon diagram of the D1 domain of HK3 BT4673S viewing the top side [38]. Strands are colored yellow and loops are green.
(B) Ribbon diagram of the D2 domain of HK3 BT4673S oriented and colored as in A except that two short helices are in red.
(C) Topology diagram of the β-propeller domains.
(D) Superimposition of ribbon diagrams of all 14 propeller blades from HK3 BT4673S.
(E) Stereodiagram showing the interactions between three adjacent propeller blades. Blades 9–11 from D2 of HK3 BT4673S are shown as ribbon diagrams with contact residues drawn in stick representation. Hydrophobic contacts are shown as red dotted lines with the shortest distance labeled.
(F) The structure-based sequence alignment of all 14 blades of HK3 BT4673S.
The ribbon figures were generated using PyMol [69], and the sequence alignment figure was drawn using ESPript [70–72]. Strand and turn residues in blade 1 of D1 are shown as arrows or by T, respectively.
Each repeat unit shares the consensus sequence Dx3–5Φ1WΦ2Gx3–5GΦ3, where Φ stands for a hydrophobic residue (Figure 2E and 2F). The ΦWΦ triplet in strand B forms the core for propeller packing. The tryptophan in each blade forms hydrophobic interactions with Φ1 in the next blade until the seventh blade where the tryptophan packs with Φ1 in the first blade. In addition, Φ2 each blade makes hydrophobic interactions with Φ3 from the previous blade, which is most often the first residue in the strand C of each blade (Figure 2E). The interactions appear to be two circular strings linking the seven blades of the propeller. The aspartate residue (D) at the start of the consensus sequence follows right after strand A, in many cases making contacts with a residue in the loop between strand C and strand D in the next blade. The aspartate interaction is less conservative and less energetically favorable than the interactions involving the hydrophobic core triplet. The two glycine residues flank a type I turn between strands B and C; however, the conformation does not require both positions to be glycine. There are many interactions between the innermost strand A from neighboring blades while there are few contacts from strand C and D due to the further distance; moreover, these outer interactions are variable.
β-Sandwich domain
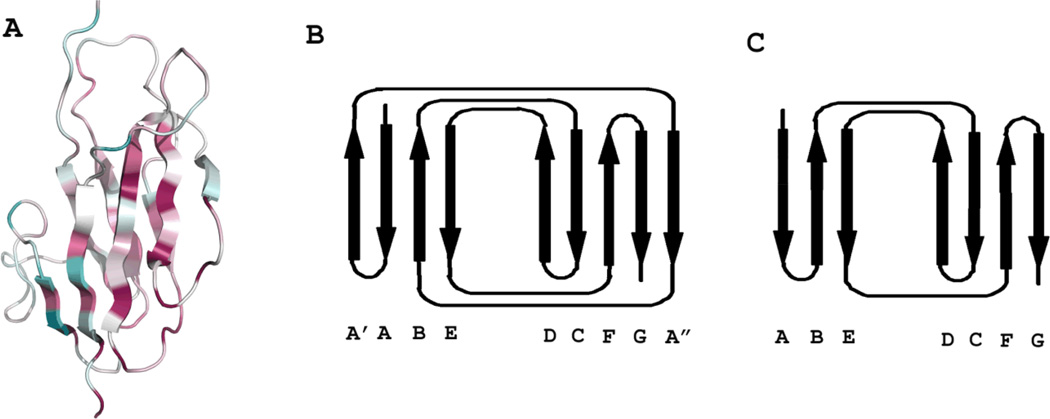
The β-sandwich domain D3 has two layers of slightly tilted β-sheets, with one layer containing four anti-parallel strands and the other having four strands in anti-parallel and the fifth in parallel (Figure 3A). Following the strand nomenclature of the closely related tenascin structure [37], the D3 topology can be described as A′ABE(DCFGA″), where strands in bracket are in the front layer and the italic A″ is the strand parallel to G. In the topology diagrams (Figure 3B for D3 and Figure 3C for tenascin), the front layers have been rotated open like a book about an imaginary axis next to strands D to view the sheets from within the sandwich. All side chains pointing into the space between the two sheets are hydrophobic residues, which form a tightly packed hydrophobic core to stabilize the structure. This core comprises the most conservative residues in all B. thetaiotaomicron HK3 sensors (Figure 3A). The loop between A′ and A″ covers the left side of the hydrophobic core, protecting it from the solvent. The front face has a mixture of hydrophobic and hydrophilic residues, while the back face is purely hydrophilic.
Figure 3. Overall fold of HK3 β-sandwich domains.
(A) Stereodiagram of the β-sandwich domain D3 of HK3 BT4673S. Coloring is based on the conservation of residues through all HK3 homologs in B. thetaiotaomicron using the ConSurf scheme [73] whereby degree of conservation are reflected in increasing saturations of purple and degree of variability is reflected in increase saturations of cyan. The figure was generated using PyMol [69].
(B) Topology diagram of HK3 domain D3, opened as a book to view the two sheets from inside the β-sandwich.
(C) Topology diagram of tenascin (modified from [37]).
Structural comparison and superimposition
There are 12 β-propeller domains and six β-sandwich domains in the asymmetric units of the two crystals and all these individual domains from each fold are very similar to one another. All β-propeller domains can be superimposed with an RMSD less than 2.00 Å for more than two thirds of all Cα atoms superimposed. The RMSD between D3 domains of both proteins is 1.08 Å with 84 Cα atoms superimposed. In each structure, D1 and D3 form similarly extensive interfaces while D2 makes only a few contacts with D1 and D3. With all six D3 domains from our two structures superimposed, their associated D1 domains can be superimposed by rotations that average χ = 6.9° (4.3° when comparing chains of the same protein; 9.2° when comparing BT3049S-BT4673S pairs). There is substantial flexibility at the D2-D3 juncture, however. With the D1/D3 units superimposed, the D2 domains from the two copies of BT4673S differ by 6.7°, the four D2 domains from BT3049S vary on average by 12.1°, and rotations of χ = 145.5° ± 3.5° are required to bring D2 domains of BT4673S and BT3049S into alignment. Thus, while the copies within each crystal structure are similar to one another, BT4673S and BT3049S are very different overall (Figures 1B and 1C).
In contrast to other HK sensors with known structures, which have their N- and C-termini close to one another as appropriate for connection to TM helices in a dimeric, two-helix transmembrane domain [27, 39], the N-terminus in domain D1 and the C-terminus in domain D3 are almost 60 Å apart. It appears impossible for HK3 receptors to be organized with TM domains as found previously for the sensory rhodopsin receptor [39].
Dimerization analysis
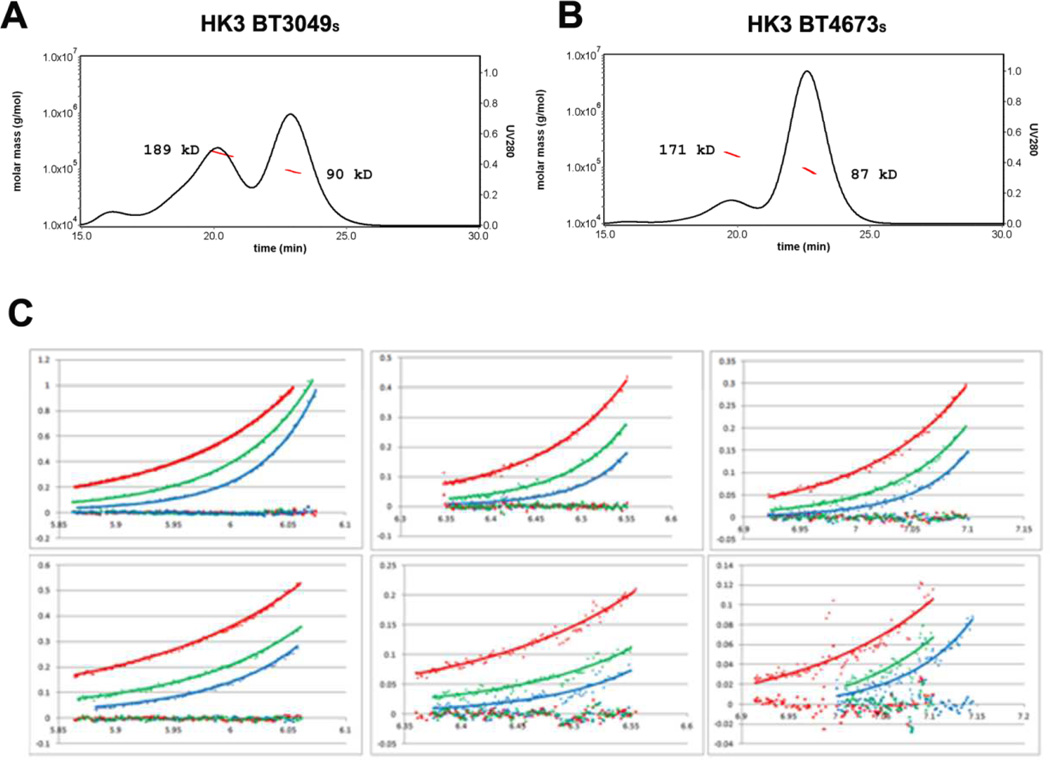
It is generally accepted that histidine kinase receptors are homodimers [9], with or without ligand binding. Size-exclusion chromatography coupled with static light scattering indicates that both sensor proteins appear to be in equilibrium among monomers, dimers and higher-order oligomers in aqueous solution of 20 mM Tris-HCl, pH8.5 (Figure 4A and 4B). The majority of both proteins are monomeric right after purification and the dimeric fraction increases after a few days sitting in the cold room at 4 °C. The dimeric form becomes dominant in two weeks and the proteins will crash out as precipitates after a month. The analytical ultracentrifuge profiles for both proteins can be best modeled to similar Kd at the level of 1 µM (Figure 4C). However, the number can only be considered as a rough estimation because of the slow transition of the oligomerization states.
Figure 4. Solution analyses of dimerization by HK3 sensor proteins.
(A, B) Dimerization analysis by SEC coupled with MALS for HK3 BT3049S and HK3BT4673S, respectively. Continuous black lines show the ultraviolet absorption at 280 nm and the red dotted lines show the molecular weights calculated from MALS. Both pictures were made by Astra V (Wyatt). Data were measured within 24 hours of purification.
(C) Analytical ultracentrifugation data and fittings for BT4673S (upper panels) and BT3049S (lower panels), showing measurements made at 2.00, 1.33 and 0.67 mg/mL in panels at left, middle and right, respectively. The colored diamonds represent measured absorbance (280 nm) at equilibrium after runs at 13000 (blue), 11000 (green), and 9000 (red) rpm. The colored squares represent the residues between measured absorbance (280 nm) and fitted data at 13000 (blue), 11000 (green), and 9000 (red) rpm, all of which are near baseline. Data were measured within three days of purification.
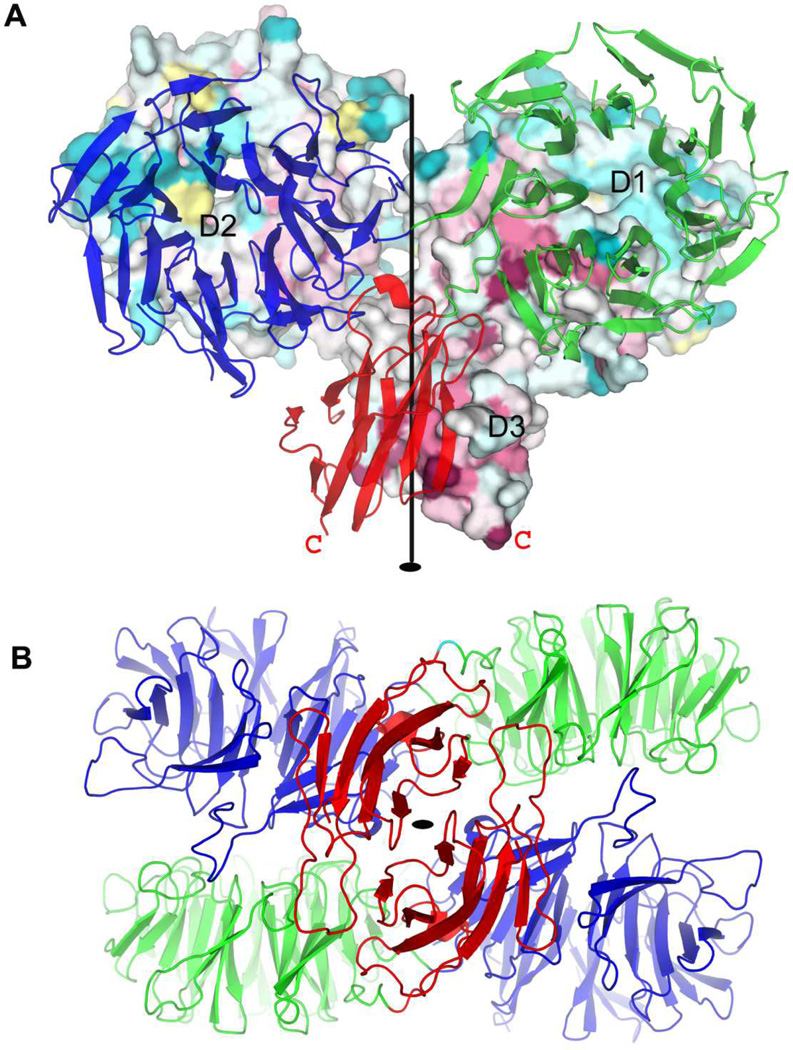
Two types of dimeric associations are observed in our crystal structures. That for BT3049S seems functionally relevant whereas that for BT4673S seems artifactual. In the structure of BT3049S (Figures 5 and 6), the protomers are related to one another by approximate two-fold symmetry (rotation χ = 177.7°, translation tχ = 0.62 Å for dimer AC; χ = 179.2°, tχ = 0.12 Å for dimer BD) with one D3:D3′ interface and two symmetric D1:D2′ and D1′:D2 interfaces, where prime stands for the partner protomer. The D3:D3′ interface is the biggest, burying 1886 Å2 of surface area, and each D1:D2′ interface buries 834 Å2 of surface area. The interfacial residues are at the most conserved patches on the protein surface, indicating their importance for the function of the sensor (Figure 5A). In addition, this dimerization has both C-termini poised to link to the transmembrane helices. Based on these observations, it seems likely that this dimeric structure is biologically relevant. Exceptionally however, as compared to other histidine kinase receptors, each HK3 protomer has only one transmembrane helix; the N-termini in this configuration are then remote from the membrane surface, and there is no predicted N-terminal helix for the preponderance of HK3 sensor domains in our bioinformatics analysis (Figures S1). In the BT4673S structure, the subunits are also related by quasi-diad symmetry; however, a dimeric membrane association does not seem feasible. Moreover, the dimeric interface here is smaller and the residues at the interface are not well conserved, implying that the conformer caught in this structure may be an artifact of ectodomain truncation.
Figure 5. Dimeric structure of HK3 sensor protein BT3049S.
(A) View of the HK3 BT3049S dimer from within the plane of the membrane. The orientation is such that both β-propeller domains are viewed down their propeller axes. Propeller top sides [37] are in apposition at the reciprocal D1:D2′ interfaces. One protomer is shown as surface representation with residues colored based on its conservation through all HK3 homologs in B. thetaiotaomicron [73] and with domains D1, D2, and D3 labeled. The other protomer is shown as a ribbon diagram colored by domain: D1 (blue), D2 (green) and D3 (red). The C-terminus of each protomer is labeled as a red letter C. The quasi two-fold rotation axis is shown as a black line.
(B) View of the HK3 BT3049S dimer out from the membrane surface, from bottom of (A). The structure is shown as a ribbon diagram with coloring as for the ribbon protomer in C: D1 (blue), D2 (green) and D3 (red). The quasi two-fold rotation axis is shown as a black oval. The picture was made using PyMol [69].
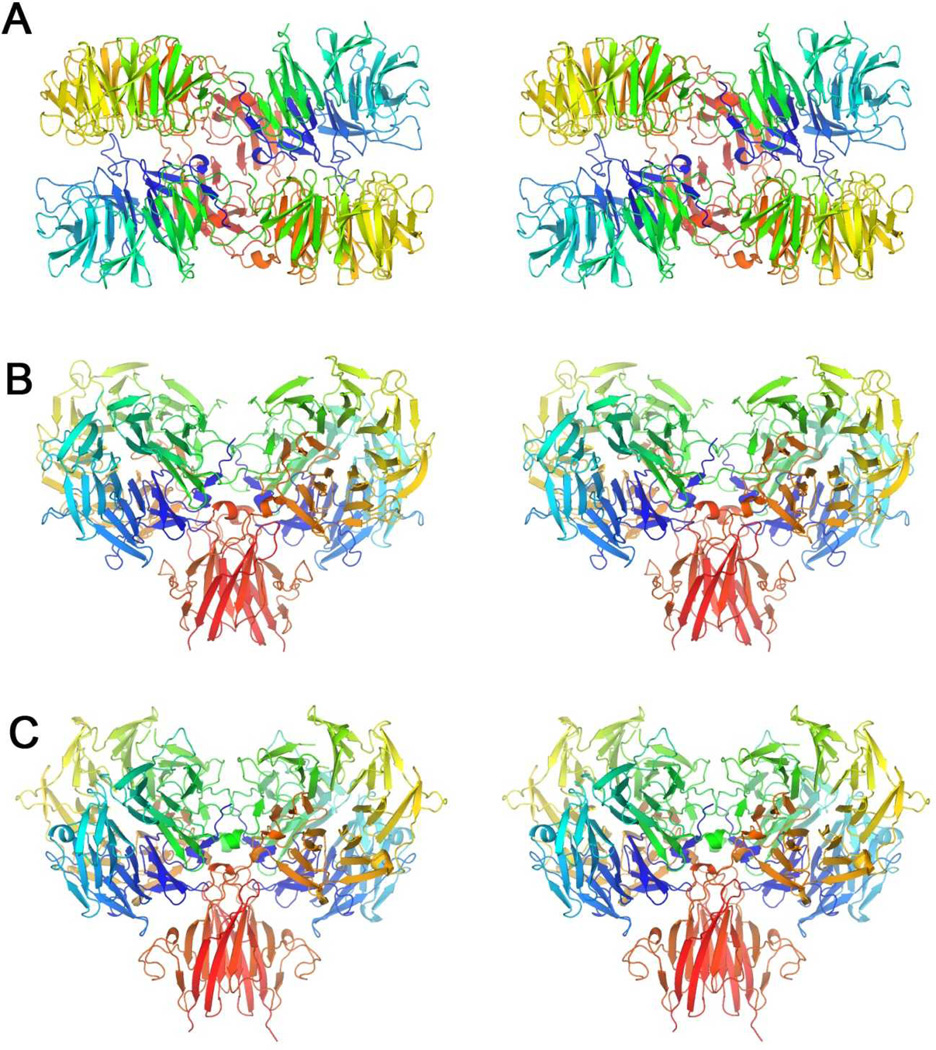
Figure 6. Stereoviews of productive B. thetaiotaomicron HK3 dimers.
(A) BT3049S dimer CD (PDBid 3V9F) viewed looking toward the membrane surface along the diad axis.
(B) BT3049S dimer CD viewed from the side, 90° from (A), with the putative membrane below.
(C) BT4663S dimer AB (PDBid 4A2L) viewed as in (B).
Each polypeptide chain is drawn in ribbon representation with spectral coloring, blue/green (N-D1) to yellow/orange (D2) to red (D3-C).
Potential ligand screening
The glycan-array screening did not generate any positive hit. We cannot conclude that the real ligand is not in the library because many other factors could have caused the binding assay to fail, including unfavorable buffer conditions, the imposition of immobilized substrates, or too stringent washing conditions.
We also tried to soak various sugar molecules into the crystals but all sugars under test, including D-(+)-glucose, sucrose, xylitol, D-sorbitol, myo-inositol, D-(+)-trehalose, D-(+)-galactose, heparin 3k and heparin 6k, significantly decreased or eliminated the diffraction of the crystals, making structural analysis a challenge. Nevertheless, the sensitivity of these protein crystals to sugar soaking suggests that an interaction has occurred, perhaps involving significant conformational change. The exact effect of these sugar molecules on the protein needs further evaluation.
Discussion
The very prevalence of HK3 sensors in association with diverse polysaccharide utilization genes in B. thetaiotaomicron is itself suggestive of importance in host-microbe symbiosis for this bacterium in the human gut. The direct wiring imparted by the marriage of each histidine kinase sensor with response regulator elements in the hybrid HTCS organization of these gene products further suggests a pre-defined control of responses to specific signals. B. thetaiotaomicron is known for its saccharide processing ability with a genome containing 246 glycolytic enzymes [40]. Interestingly, 61% of these glycosylhydrolases were predicted to locate in the periplasm, outer membrane or extracellular space, implying that processed polysaccharide products can be utilized by all members in the ecosystem including the host [31]. B. thetaiotaomicron is able to grow on plant dietary sources or host-derived glycans owing to a complement of PUL gene clusters. The B. thetaiotaomicron genome encodes 36 HK3 sensors, including 34 that have a complete HTCS domain architecture and 28 that are located within a PUL [5]. Three of these HTCS systems have been shown to regulate the expression of glycolytic enzymes in response to host diet [5, 41]. By implication, the signals for HK3 sensors appear to be identifying sugars from foodstuffs in need of processing.
Based on our previous bioinformatics analysis of transmembrane histidine kinase sensors, which numbered sensor domains in order of prevalence for groupings at PSI-BLAST level of E ≤ 10−3, these HK3 domains are the third most populated family. The canonical HK3-DHp-CA-REC-AraC architecture (Figure 1A) of the HK3 sensor proteins in B. thetaiotaomicron is also prevalent elsewhere. From our survey of more than 1000 HK3 domains, we found 77% of these proteins to be putative histidine-kinase receptors and, in turn, 82% of the HK-receptor genes also contain response-regulator REC domains and 75% have DNA-binding domains. A substantial minority of HK3 domains are associated with other kinds of transmitters, notably DGC and PP2C domains. Similar to four-helix bundle sensors [15] and PhoQ/DcuS/CitA (PDC)-type sensors [29, 42], HK3-like domains serve as evolutionary modules supporting a variety of receptor systems.
The HK3 sensors contain three structural domains: two β-propeller domains, D1 and D2, followed by a C-terminal β-sandwich domain D3. A Dali search indicated that there is no previous structure with the same domain organization (apart from that now reported from BT4663 [21]). Similar D1/D3 interfaces exist across all six copies in the asymmetric units of the two crystals. D2 is the least conserved domain and it appears to be quite flexible relative to the D1/D3 dual. Thus, the relative dispositions of D2 are very different in the BT3049S and BT4673S structures (Figures 1B and C). The differences are especially apparent in the dimeric organizations; whereas the BT4673S dimer is impossibly disposed for co-ordinated transmembrane passage from its two C-termini, the BT3049S dimer is appropriate for signal transduction. In the BT3049S structure, related by two-fold symmetry, each D1 β-propeller domain interacts face-on with the D2 β-propeller from its partner protomer and the two D3 β-sandwich domains pack against one another (Figures 5 and 6). This arrangement orients the two C-termini for connection to the transmembrane segment, even though they are still 25 Å apart. The N-termini are far apart and are pointing oppositely, making simultaneous membrane contact by N-termini impossible. This is consistent with the fact that the majority of HK3 sensors are predicted to have only one transmembrane helix per chain.
Hypothetically, by virtue of genetic associations, many HK3 histidine kinases function as environmental sugar sensors, and characteristics of the HK3 sensor domains are consistent with this hypothesis. Although many β-sandwich domains are able to bind saccharides [43], we are not aware of tenascin- or other FnIII-like domains that do so. There are, however, seemingly relevant examples of carbohydrate binding to β-propeller domains. Notably, sialidases bind substrates and the inhibitory sugar 2-deoxy-2,3-dehydro-N-acetyl-neuraminic acid (DANA) at the top side of this six-bladed β-propeller [44]. Since the D1/D2′ and quasi-symmetric D2/D1′ interfaces of BT3049S pack with top sides of these β-propellers facing one another (Figure 5), a prospective sialidase-like binding site for saccharides would be in this interface. Since the many HK3 sensors bind diverse sugars, the binding site would be expected to be at variable surfaces. In the BT3049S dimer, such variable surfaces are present at the top-side surfaces of D1 blades 4–6 and of D2 blades 1–4 (Figure 5A), contrasting with the more conserved surfaces at interdomain contacts nearer to the diad axis. Unfortunately, we have failed in our attempts to identify the possible ligands by glycan-array screening and by co-crystallization or soaking with various sugar molecules.
After our structures were deposited and released from the PDB, crystal structures were reported for another HK3 sensor domain, BT4663S. Lowe et al. [21] confirmed that unsaturated disaccharides derived from heparin and heparin sulfate are able to bind to BT4663, and they described structures with and without a bound disaccharide ligand. Similar to BT4673S and BT3049S reported in this work, BT4663S is also a quasi-two-fold symmetric dimer, both in its apo and ligated states. In keeping with a likely biological relevance, the apo-state dimers of BT3049 and BT4663 HK3 sensor proteins are very similar. First, the subunits themselves are similar, for example, the D1:D3 interfaces in BT4663 vs. BT3049 are related by rotations of 19.4° ± 1.3° (24 pairs); secondly, the dimeric associations are alike (Figure 6). With D1-D2 β-propeller units superimposed for one protomer each from two dimers, the corresponding D1′-D2′ units are brought into alignment by rotations of just 9.5° ± 1.5°. The D3:D3′ interfaces differ more appreciably, however; here rotations of 20.0° ± 1.1° are needed for superposition. Overall (D1-D2-D3), the BT3049S dimer appears to have a somewhat more intimate interface, burying 2720 A2 of surface area as compared to 1900A2 in apo BT4663S and 2140 A2 in ligated BT4663S. The sugar moiety in ligated BT4663S is located at the top-side interfaces of D1/D2′ β-propeller pairs, as suggested above from sugar-sialidase complexes and the location of HK3 hypervariability. The D1-D2/D1′-D2′ β-propeller pairs are similar for the apo and ligated states (15.2° ± 0.7° and 8.9° ± 0.7° comparing BT3049S and apo BT4663S, respectively, with ligated BT4663S), but the D3/D3′ interfaces are very different (33.7° ± 0.8° and 16.9° ± 1.0° for respectively the same comparisons).
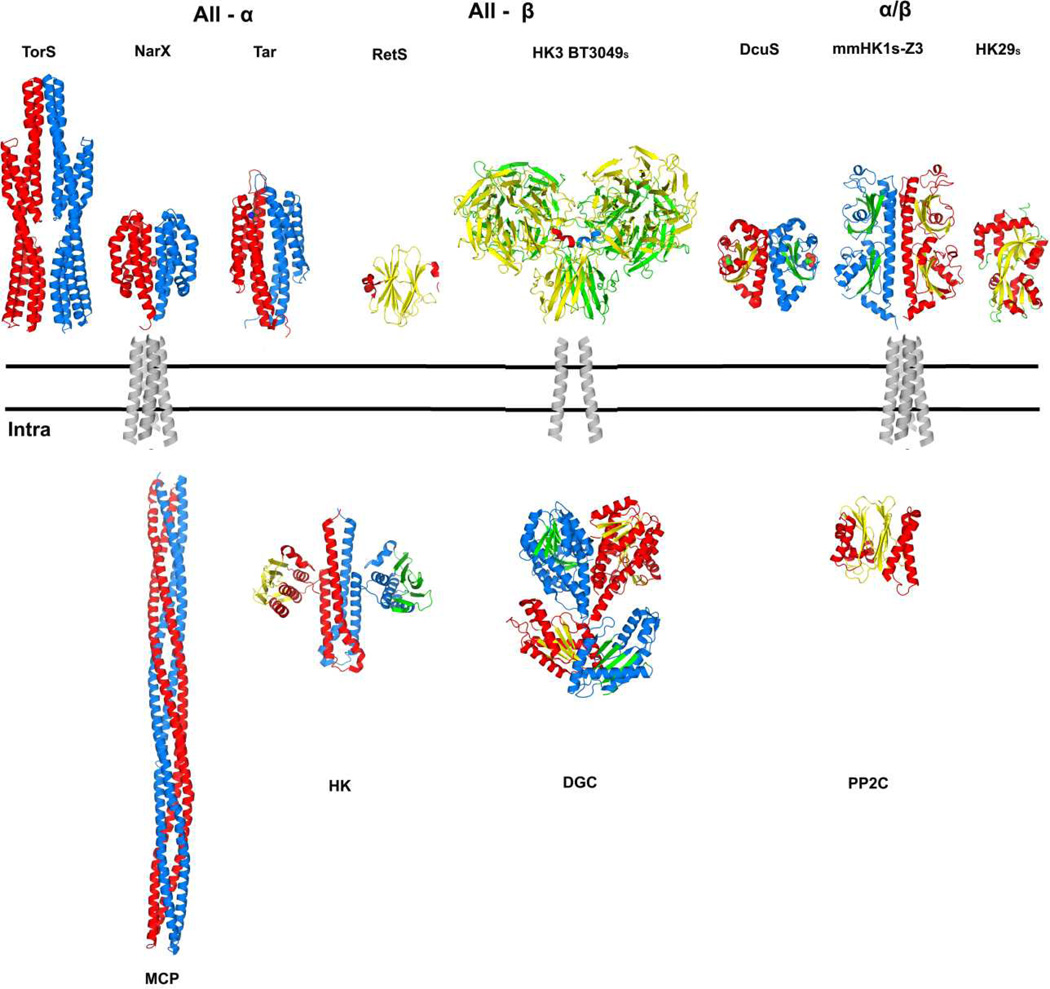
The nature of the signal that is produced upon ligand binding to HK and transduced across cell membrane has been controversial. On the one hand, the conservative DHp and kinase domains seem to support a universal signal transduction mechanism. On the other hand, various signal transduction mechanisms have been proposed, including a most recent suggestion for the HK3 system [21] and efforts to reconcile or unify them are fraught by the complexity of these receptor systems. The HK3 structures add further diversity to the nature of sensor domains from which the signals must emanate. There are three major groups of sensor structures, all-α, all-β and α/β (Figure 7). Among all-α fold sensors, NarX binds the ligand nitrate at the dimer interface [15] and the MCP receptor Tar binds one aspartate using each individual domain [14]. All reported all-α sensor proteins have four-helix bundle folds and they form quasi-four-helix bundles at the dimer interface, which connect to the transmembrane helices. Two types of all-β fold structures have been reported including the β-sandwich RetS [45] and the three-domain complexes reported here. TMHMM prediction indicates that these sensors may have only one TM helix. There are three types of α/β fold, which are single-PDC, double-PDC and periplasmic-binding-protein (PBP) like [29]. Like all-α fold sensors, these PDC fold sensors link to the TM domain using helices at both ends, while it is not known how the PBP-like sensor works.
Figure 7. Summary of diverse, variably associated transmembrane receptor systems.
The double lines represent the plasma membrane. Alternative extracellular / periplasmic sensor domains are shown above the membrane and alternative cytoplasmic signaling portions are shown below the membrane. A transmembrane domain connects the outside to the inside in diverse combinations. Structure figures were generated PyMol [69] from PDB files as indicated: TorSS (3O1H; [22]), NarXS (3EZH; [15]), TarS (2LIG; [74]), RetSS (3JYB; [45]), HK3 BT3049S (3V9F; this work), DcuSS (3BY8; [75]), HK1S-Z3 (3LIB; [29]), HK29s (3H7M; [76]), transmembrane domains (1H2S; [39]), MCP (2CH7; [77]), HK (2C2A; [23]), DGC (2WB4; [78]) and PP2C (2XZV; [79]). All structures are shown as dimers except for RetSS, PP2C and HK29S. The coloring for one protomer has helix (red) and sheet (yellow); that for the partner protomer has helix (blue) and sheet (green). The four-helix bundle transmembrane domain is shown as representative for conventional all-helix (e.g. NarX or Tar) and PDC-domain (e.g. DcuS or HK1) receptors; a two-helix transmembrane domain is shown for the unconventional HK3 receptors, which connect to various signaling components in the cytoplasm.
Differing sensors are not used exclusively by HK but are shared with other signaling molecules such as MCP, DGC or PP2C receptors. MCP proteins are believed to form trimers of dimers and they can transfer the extracellular signal to bound histidine kinases [46]. The active DGC proteins are dimers that are responsible for making the secondary messenger c-di-GMP from two GTP molecules in bacteria, thereby controlling diverse cellular functions [47]. PP2C domains, found ubiquitously through bacterium, plant and mammalian kingdoms, serve to dephosphorylate a diverse range of proteins in the regulation of many signal transduction pathways [48]. In contrast to other signaling modules, the PP2C domains are able to function as a monomer.
The striking diversity found in both the intracellular domains and extracellular domains (Figure 7) seems to contradict the hypothesis of a conservative signal transduction mechanism. Still, the successful construction of functional chimeric receptors strongly supports the idea [49, 50]. Further investigations are required to clarify the dilemma.
Experimental Procedures
Cloning
The periplasmic domain of BT4673 (HK3 BT4673S), which was predicted by PredictProtein at http://www.predictprotein.org/ [51], was amplified from B. thetaiotaomicron genomic DNA by PCR (primers are identified below). The amplified DNA fragment was inserted into modified plasmid pET22b+, where the NdeI site was mutated into an NcoI site. The insert is located between resulting NcoI and XhoI restriction sites in frame with the C-terminal His-tag. The predicted periplasmic domain of BT3049 (HK3 BT3049S) was amplified by PCR and cloned between BamH1 and Xho1 sites in plasmid pSMT3, which is designed to produce the target protein tagged by His-SUMO at its N-terminus.
| Primer 1 | Primer2 | |
| HK3BT4673s | AGGAGATATACCATGGAAGCTTATGG AAAAAAAGTAAATTATCAGCAATTCG |
GGTGGTGGTGCTCGAGCCACGGT GGAAG |
| HK3BT3049s | CAGATTGGTGGATCCGCCCACCCTTA CCTCATCC |
GTGGTGGTGCTCGAGTTATGTCA GCCAGAGAG |
Protein expression
Large-scale protein production was carried out from BL21(DE3) cells grown in 1 L Luria Broth (LB) medium that contained either ampicillin for HK3BT4673S-pET22b+ constructs or kanamycin for HK3BT3049S-pSMT3 constructs. Each culture medium was inoculated with 10 ml from an overnight starter culture and left shaking at 37 °C. The temperature was adjusted down to 20 °C when OD600 reached ~0.5. After 30 min, the induction was begun by adding IPTG to a final concentration of 0.5 mM for induction of protein expression. Cells were collected by centrifugation after overnight growth and stored in −80 °C until use. For the production of SeMet proteins, a minimal medium containing SeMet was used.
Purification
Proteins were purified by procedures similar to those described previously for HK1 family histidine kinase sensors [29]. Briefly, frozen cell pellets were melted and resuspended into lysis buffer, cells were broken by sonication, and cell debris was removed by centrifugation. Nickel-affinity purification was performed and eluted protein was followed by both OD280 and SDS-PAGE analysis. For HK3 BT4673S, pooled fractions were concentrated and applied to a size exclusion column. Eluted fractions corresponding to single peaks were then pooled, concentrated, and used directly for further characterizations in solution and for crystallization. For HK3 BT3049S, the SUMO-fusion protein was digested overnight by the ULP protease immediately after the metal-affinity chromatography step. The digested protein was then separated from the His-tagged SUMO and ULP proteins by reapplication to a nickel affinity column, and the flow-through was concentrated and further purified by size exclusion chromatography.
Crystallization and cryoprotection
Crystallization screening was carried out with Crystal Screen HT and Index kits from Hampton Research and with the Crystal Screen Wizard kit from Emerald Biosystems. Trials were made at two protein concentrations (OD280=10 and highest possible concentration) using a Mosquito crystallization robot. Crystals were optimized by hanging-drop screening at conditions around those for the initial hit. Finally, HK3 BT4673S was crystallized in 5% PEG MME 2K, 10% tacsimate pH7.0 plus 0.1 M cacodylate buffer at pH 5.5, and HK3 BT3049S was crystallized in 5% PEG MME 2K and 0.1 M Tris buffer at pH7.0. Microseeding was required for the growth of crystals to suitable sizes for both proteins. Cryoprotection was attained by sequential addition of increments of mother liquor supplemented with 25% (w/v) glycerol, followed by flash cooling in liquid nitrogen.
Structure determination and refinement
The structure of HK3 BT4673S was solved by tantalum SAD phasing [52]. Tantalum cluster powder was added to the mother liquor containing the crystals. After soaking for 1 hr, the crystals turned green and were transferred to cryoprotectant without tantalum cluster. An attempted three-wavelength Ta MAD experiment at the tantalum LIII-edge failed due to radiation damage; however, SAD measurements at the peak of the LIII-edge (λ=1.2544 Å) were successful from a single frozen crystal at the X4A beamline of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. Due to the existence of a long unit cell axis (c = 433 Å), the crystal was aligned with its c-axis roughly parallel to the rotational axis to reduce overlaps on the detector. The bending of a sagittal focusing crystal was also adjusted to make focused spots on the detector surface to further reduce overlaps. The XDS package [53–56] was used for data processing and the diffraction statistics are listed in Table 1. ShelxCD [57] was used to determine the tantalum substructure with data range between 5–2.3 Å. The substructure was then fed to SOLVE/RESOLVE [58] for substructure refinement, phasing, density modification and automated model building. The crude model from RESOLVE was completed by manual building in Coot [59] with the guidance of a Bijvoet-difference Fourier map calculated from a SeMet SAD dataset. The model was refined using REFMAC5 [60] from the CCP4 package [61] (Table 2). Water molecules and two Ta6Br12 clusters were added after models were well refined.
Table 1.
Diffraction Data Statistics
| Dataset | BT4673S Tantalum cluster |
BT3049S SeMet |
|---|---|---|
|
Beamline dmin (Å) |
NSLS X4A 2.3 |
APS 24-ID-C 3.3 |
| Space group | P41212 | P212121 |
| Multiplicity | 6.4 | 3.9 |
| Completenessa (%) | 99.9 (100) | 99.6 (97.5) |
| Rmergeb | 11.4 (82.9) | 10.1 (68.1) |
| CC½ (%) | 99.9 (87.5) | 99.8 (76.3) |
| <I/σ(I)> | 15.8 (2.7) | 12.8 (2.3) |
| γ (Å) | 1.2544 | 0.9793 |
Values in the outermost shell are given in parentheses.
Rmerge = (Σ |Ii − < Ii > |) / Σ |Ii|, where Ii is the integrated intensity of a given reflection.
Although BT3049S has the same domain components as BT4673S, the sequence alignment has an identity of only 19.7%; this, together with subsequently discovered conformational differences, made molecular replacement unsuccessful to solve the structure. The structure of BT3049s was determined by SeMet SAD phasing from a single-crystal dataset collected at the peak wavelength of the selenium K-edge at Advanced Photon Source (APS) beamline 24-ID-C (λ = 0.9793 Å). Due to the existence of an even larger unit cell axis (c = 487.5 Å) in this crystal, to reduce overlaps 720 diffraction images were collected with an oscillation angle of 0.25°. The XDS package was used for data processing and statistics are listed in Table 1. There are four molecules in the crystallographic asymmetric unit of P212121, corresponding to a solvent content of 50%. ShelxCD was used to find the SeMet substructure (20 sites) with data between 30 - 6 Å. SAD phasing and density modification were carried out by PHASER [62] and DM [63] with data between 30 - 3.3 Å. For density modification, 12-fold improper domain NCS calculated from the Se substructure was used. Model building and refinement were performed as for BT4673S structure except that B-factor sharpened, NCS averaged electron density maps were used for model building and tighter NCS restraints were enforced throughout the refinements. Statistics for the refinement are listed in Table 2.
B. thetaiotaomicron genome survey and sequence alignment
Psi-Blast [64] analyses were carried out at the NCBI website against the B. thetaiotaomicron genome with HK3 BT3049S as the query until convergence. This search found 36 sequences with at least 70% query sequence coverage. Searches with HK3 BT4673S led to the same result. The multiple sequence alignment from Psi-Blast provided an initial template, which was adjusted manually with the consideration of the 14 repeats from two seven-bladed β-propeller domains.
Database survey of HK3 family sensors
PsiBlast searches [64] were carried out against non-redundant database downloaded from NCBI with each sequence from our initial set of 38 HK3 sequences as the query, running until convergence with Evalue cutoff at 0.001. All hits having more than 700 amino acid residues were combined and redundant sequences were removed. The resulting sequences were analyzed for domain structures using the conserved domain search service from NCBI website and for transmembrane helices assignment with TMHMM prediction [65, 66]. The sequences without any other domains except the sensor domain or the β-propeller domain were removed. The final sequences were categorized based on the function of the other domains.
Structure superposition and RMSD calculation
Structures were superimposed by the program LSQMAN [67] from the Uppsala Software Factory. We used the following test for superposition: at least three contiguous Cα positions of one structure must be within 3.0 Å of counterparts in the other structure for residues to be considered superimposed. Superpositions were made domain by domain, rather than for the whole protein, so that the effects of domain movement would be removed. RMSD values for the whole were obtained from the sum of squared deviations for the three individually superimposed domains.
Assembly analysis
The oligomerization state of both proteins was analyzed by size-exclusion chromatography (SEC) coupled with Wyatt multi-angle light scattering (MALS). Both proteins (100 µl, 2 mg/ml) were loaded to GE S200 sizing column with 20 mM Tris, pH 8.5 as running buffer. OD280, scattering at 18 angles, and differential refractive index data were recorded and the molecular weight of each peak was calculated by Wyatt’s Astra 5.3.1.5 software (Wyatt).
Sedimentation equilibrium experiments were performed using a Beckman/Coulter XLI analytical ultracentrifuge with absorbance optics. Freshly purified HK3BT4673s and HK3BT3049s proteins were brought to concentrations of approximately 2 mg/ml, 1.33 mg/ml, and 0.67 mg/ml. Experiments were conducted and analyzed as reported previously for another histidine kinase sensor protein [17]. Each sample was sedimented to equilibrium at 9,000, 11,000, and 13,000 rpm at 20°C, and absorbance scans were taken at 1 hr intervals.
Glycan array screening
Both HK3BT4673s and HK3BT3049s were sent to the Protein-Glycan Interaction Core (H) at the Consortium for Functional Glycomics (CFG) for glycan array screening in order to identify possible ligands. There are 511 mammalian glycan targets in the array, which are printed onto N-hydroxysuccinimide (NHS)-activated glass microscope slides (SCHOTT Nexterion) through covalent amide linkages. Both proteins with His-tag were purified and concentrated to 2 mg/ml before sending to the facility, where both were diluted to 0.2 mg/ml in 20mM Tris-HCL (pH 7.4), 50 mM sodium chloride, 0.05% Tween 20, 1% BSA for final binding assay. Qiagen anti-penta His antibody was used to detect bound His-tagged protein. More information about the assay can be found at the website of the consortium (http://www.functionalglycomics.org).
Supplementary Material
Acknowledgements
We thank John Schwanof and Randy Abramowitz of NSLS X4 and Igor Kourinov and Frank Murphy from APS NE-CAT for help with synchrotron data collection; Erik Martinez-Hackert and Jonah Cheung for helpful discussions; Göran Ahlsen and Lawrence Shapiro for aid in the analytical ultracentrifuge experiment; and Jinfeng Liu of the Rost laboratory for the bioinformatics analysis that laid the foundation for this work. We also thank David F. Smith and Jamie Heimburg-Molinaro of Emory University School of Medicine for their effort in glycan-array screening for potential ligands of the sensors, even though the results turned out negative. This work was supported in part by NIH grants GM034102 and GM107462 (W.A.H.). Beamline X4A at the National Synchrotron Light Source (NSLS) of Brookhaven National Laboratory is supported by the New York Structural Biology Center.
Abbreviations
- CA
catalytic domain
- DGC
diguanylate cyclase
- DHp
dimerization and histidine-containing phosphotransfer domain
- HK
histidine kinase
- HK3
histidine kinase sensor family 3
- HTCS
hybrid two-component system
- MCP
methyl-accepting chemotaxis protein
- PDC
PhoQ/DcuS/CitA
- PP2C
type 2C protein phosphatase
- PDB
Protein Data Bank
- PUL
polysaccharide utilization locus
- REC
receiver domain
- RR
response regulator
- SeMet
selenomethionine
- SAD
single-wavelength anomalous diffraction
- TCS
two-component system
Footnotes
Accession codes
Structural data depositions for crystallographic results have PDB IDs: 3OTT, 3V9F with associations to individual structures as designated in Table 2.
Author contributions
ZZ and WAH planned the project; ZZ and QL performed experiments; ZZ, QL and WAH analyzed data; ZZ, QL and WAH wrote the paper.
References
- 1.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 3.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 7.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 9.Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-reviews3013. REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo FD, Silhavy TJ. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1993;1:306–310. doi: 10.1016/0966-842x(93)90007-e. [DOI] [PubMed] [Google Scholar]
- 13.Yeo WS, Zwir I, Huang HV, Shin D, Kato A, Groisman EA. Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol Cell. 2012;45:409–421. doi: 10.1016/j.molcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci U S A. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore JO, Hendrickson WA. Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure. 2009;17:1195–1204. doi: 10.1016/j.str.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Ottemann KM, Xiao W, Shin YK, Koshland DE., Jr A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 19.Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J Mol Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe EC, Basle A, Czjzek M, Firbank SJ, Bolam DN. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci U S A. 2012;109:7298–7303. doi: 10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore JO, Hendrickson WA. An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure. 2012;20:729–741. doi: 10.1016/j.str.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada S, Shiro Y. Structural basis of the signal transduction in the two-component system. Adv Exp Med Biol. 2008;631:22–39. doi: 10.1007/978-0-387-78885-2_3. [DOI] [PubMed] [Google Scholar]
- 25.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada S, Sugimoto H, Kobayashi M, Ohno A, Nakamura H, Shiro Y. Structure of PAS-linked histidine kinase and the response regulator complex. Structure. 2009;17:1333–1344. doi: 10.1016/j.str.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Cheung J, Hendrickson WA. Sensor domains of two-component regulatory systems. Curr Opin Microbiol. 2010;13:116–123. doi: 10.1016/j.mib.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry J, Koteva K, Wright G. Receptor domains of two-component signal transduction systems. Mol Biosyst. 2011;7:1388–1398. doi: 10.1039/c0mb00329h. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raghavan V, Groisman EA. Orphan and hybrid two-component system proteins in health and disease. Curr Opin Microbiol. 2010;13:226–231. doi: 10.1016/j.mib.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q, Zhang Z, Hendrickson WA. Multi-crystal anomalous diffraction for low-resolution macromolecular phasing, Acta crystallographica Section D . Biological crystallography. 2011;67:45–59. doi: 10.1107/S0907444910046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schleif R. AraC protein: a love-hate relationship. Bioessays. 2003;25:274–282. doi: 10.1002/bies.10237. [DOI] [PubMed] [Google Scholar]
- 34.Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 35.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leahy DJ, Hendrickson WA, Aukhil I, Erickson HP. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 38.Chen CK, Chan NL, Wang AH. The many blades of the beta-propeller proteins: conserved but versatile. Trends Biochem Sci. 2011;36:553–561. doi: 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 40.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci U S A. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang C, Tesar C, Gu M, Babnigg G, Joachimiak A, Pokkuluri PR, Szurmant H, Schiffer M. Extracytoplasmic PAS-like domains are common in signal transduction proteins. J Bacteriol. 2010;192:1156–1159. doi: 10.1128/JB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chavas LM, Tringali C, Fusi P, Venerando B, Tettamanti G, Kato R, Monti E, Wakatsuki S. Crystal structure of the human cytosolic sialidase Neu2. Evidence for the dynamic nature of substrate recognition. J Biol Chem. 2005;280:469–475. doi: 10.1074/jbc.M411506200. [DOI] [PubMed] [Google Scholar]
- 45.Jing X, Jaw J, Robinson HH, Schubot FD. Crystal structure and oligomeric state of the RetS signaling kinase sensory domain. Proteins. 2010;78:1631–1640. doi: 10.1002/prot.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 47.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 48.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 51.Rost B, Liu J. The PredictProtein server. Nucleic Acids Res. 2003;31:3300–3304. doi: 10.1093/nar/gkg508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knablein J, Neuefeind T, Schneider F, Bergner A, Messerschmidt A, Lowe J, Steipe B, Huber R. Ta6Br(2+)12, a tool for phase determination of large biological assemblies by X-ray crystallography. J Mol Biol. 1997;270:1–7. doi: 10.1006/jmbi.1997.1074. [DOI] [PubMed] [Google Scholar]
- 53.Kabsch W. Automatic indexing of rotation diffraction patterns. J Appl Cryst. 1988;21:67–72. [Google Scholar]
- 54.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 55.Kabsch W. Integration, scaling, space-group assignment and post-refinement, Acta crystallographica Section D . Biological crystallography. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabsch W. Xds, Acta crystallographica Section D . Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 58.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution, Acta crystallographica Section D . Biological crystallography. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics, Acta crystallographica Section D . Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method, Acta crystallographica Section D . Biological crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 61.The CCP4 suite: programs for protein crystallography. Acta crystallographica Section D, Biological crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 62.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cowtan K. ‘dm’: An automated procedure for phase improvement by density modification. CCP4/ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 64.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 66.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 67.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement, Acta crystallographica Section D . Biological crystallography. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 68.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography, Acta crystallographica Section D . Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeLano WL. The PyMOL Molecular Graphics System in, DeLano Scientific. CA, USA: Palo Alto; 2002. [Google Scholar]
- 70.Gouet P, Courcelle E. ENDscript: a workflow to display sequence and structure information. Bioinformatics. 2002;18:767–768. doi: 10.1093/bioinformatics/18.5.767. [DOI] [PubMed] [Google Scholar]
- 71.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 72.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 75.Cheung J, Hendrickson WA. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J Biol Chem. 2008;283:30256–30265. doi: 10.1074/jbc.M805253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheung J, Le-Khac M, Hendrickson WA. Crystal structure of a histidine kinase sensor domain with similarity to periplasmic binding proteins. Proteins. 2009;77:235–241. doi: 10.1002/prot.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 78.Wassmann P, Massa C, Zaehringer F, Schirmer T. Crystal Structure of Activated Pled, Identification of Dimerization and Catalysis Relevant Regulatory Mechanisms. 2009 http://dx.doi.org/10.2210/pdb2wb4/pdb.
- 79.Su J, Schlicker C, Forchhammer K. A Third Metal Is Required for Catalytic Activity of the Signal-transducing Protein Phosphatase M tPphA. J Biol Chem. 2011;286:13481–13488. doi: 10.1074/jbc.M109.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.