Abstract
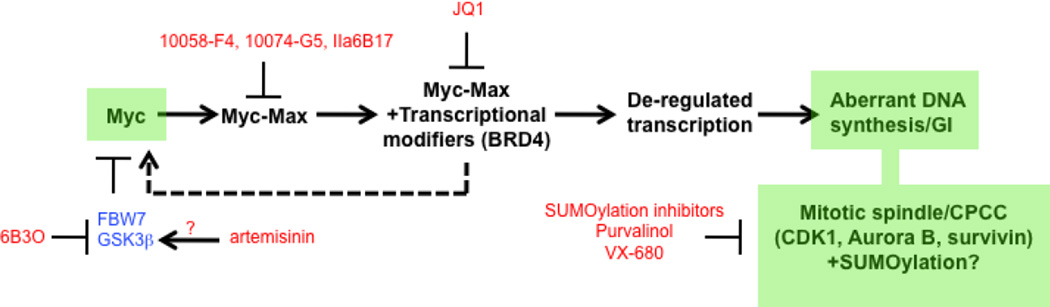
The c-Myc (Myc) oncoprotein is among the most attractive of cancer targets given that is deregulated in the majority of tumors and that its inhibition profoundly affects their growth and/or survival. However, its role as a seldom-mutated transcription factor, its lack of enzymatic activity for which suitable pharmaceutical inhibitors could be crafted and its expression by normal cells have largely been responsible for its being viewed as “undruggable”. Work over the past several years, however, has begun to reverse this idea by allowing us to view Myc within the larger context of global gene regulatory control. Thus, Myc and its obligate heterodimeric partner, Max, are integral to the coordinated recruitment and post-translational modification of components of the core transcriptional machinery. Moreover, Myc over-expression re-programs numerous critical cellular functions and alters the cell’s susceptibility to their inhibition. This new knowledge has therefore served as a framework upon which to develop new pharmaceutical approaches. These include the continuing development of small molecules which act directly to inhibit the critical Myc-Max interaction, those which act indirectly to prevent Myc-directed post-translational modifications necessary to initiate productive transcription and those which inhibit vital pathways upon which the Myc-transformed cell is particularly reliant.
Introduction
A myriad collection of correlative human studies and transgenic animal models has established beyond any reasonable doubt that deregulation of c-Myc (Myc) underlies the pathogenesis of numerous cancers and in many cases contributes to their aggressiveness (1–6). Moreover, the frequency with which this aberrant expression occurs is virtually unmatched, thus placing MYC into contention for the most frequently deregulated oncogene in human tumors. Myc amplification is the most frequent somatic copy number increase seen in tumor cells (7) and the range of neoplasms in which Myc is otherwise deregulated is wide. It includes, but is hardly confined to, many hematopoietic tumors and cancers of the central nervous system, GI track, breast, prostate and lung. Even what appears to be normally regulated Myc has been found to be linked to and critical for executing the transforming programs of upstream oncogenes (8–11). This suggests that human tumorigenesis is much more dependent upon the proper functioning of Myc than would be gleaned simply by noting its level of expression in various tumors. Thus inhibiting Myc, even when it appears to be properly behaved, may significantly impair tumor development and strongly supports the idea that Myc is an important factor upon which many oncogenic signaling pathways converge and upon which tumor growth depends (11–15). The notion that Myc is a linchpin for tumor survival and/or proliferation (14, 6,17) is one major reason why such intense interest in its therapeutic targeting has developed as it suggests that potent pharmacologic agents should have widespread utility irrespective of cancer type (18,19). This contrasts sharply with more conventional forms of targeted therapies, which are typically effective only in tumors driven by oncoproteins with specific mutations. Typical examples include tyrosine kinase inhibitors directed against Bcr-Abl and mutant forms of Jak2 in CML and myelodysplastic syndromes, respectively and serine/threonine (Ser/Thr) kinase inhibitors directed against mutant forms of B-Raf or other members of the BRAF/MEK/ERK pathway in melanoma (20,21).
A second reason that pharmacologic inhibition of Myc is a particularly compelling concept is that, in addition to its role in tumor cells, Myc is now appreciated as being necessary to sustain a healthy tumor matrix. In model systems of Myc-driven neoplasms, expression of the oncoprotein by the tumor has been shown to be required for tumor neo-vascularization and presumably operates by up-regulating the expression of genes encoding proteins such as VEGF and FGF to encourage and sustain this process (22,23). Proliferating cancer cells, presumably Myc-dependent if not necessarily Myc-driven, can also secrete factors such as CSF1 and IL4, which are necessary for the recruitment for macrophages and endothelial precursors from bone marrow sources (23,24–27).
The requirement for Myc to support the extracellular matrix also extends to its expression by these non-neoplastic cellular constituents. For example, the alternative activation pathway through which tumor-associated macrophages produce tumor-promoting and pro-invasive factors such as VEGF, TGF-β and MMP9 is highly dependent on their expression of endogenous Myc (23,25,26). Similarly, the proliferation and expansion of tumor-supporting cellular components including smooth muscle cells, pericytes and fibroblasts are all undoubtedly dependent on their properly controlled regulation of Myc to ensure that they keep apace with the neoplasm’s growth (23,24,28,29). Interestingly, the expression of Myc by normal endothelial cells does not appear to be required for their proliferation and participation in vasculogenesis but is required for the genesis of endothelial precursors from bone marrow-derived progenitors (30–32).
One example of the reliance on tumor cell Myc expression for maintaining the tumor matrix comes from work showing that the genetic silencing of endogenous tumor cell Myc in SV40-large T and small t-antigen-driven pancreatic β-cell tumors is associated with a widespread collapse of the tumor vasculature and loss of infiltrating inflammatory cells, both of which precede any overt tumor apoptosis and regression (13). This suggests that the failure to maintain a proper extratumoral environment is a cause rather than an effect of tumor regression, a claim that was substantiated by microarray analyses demonstrating that inhibiting Myc genetically in the β–cell tumors was associated with a rapid dysregulation of numerous tumor associated cytokines, chemokines and other inflammatory mediators that likely fortify the extratumoral cellular environment. Taken together, these studies suggest that pharmacologic targeting of Myc would directly affect separate yet absolutely essential compartments comprised on the one hand of the tumor itself and on the other by its matrix and associated normal cell populations.
Interaction between Myc, Max and DNA
The monomeric form of Myc’s bHLH-ZIP dimerization domain is largely unstructured in solution and possesses only about 27% α–helical content (33–35). Such regions of intrinsic disorder (ID) are common and occur in approximately one-third of all eukaryotic proteins and as many as 60–80% of those involved in normal signal transduction and neurodegenerative disorders (36–39). ID regions are characterized by extensive backbone flexibility and an absence of anything more than transient tertiary structure (40). The amino acid sequences comprising ID regions are both different from and less complex than those from more ordered regions (40–42). It has been suggested that the extended nature of ID regions, and the consequent solvent-exposed surface area, allows them to participate in coupled folding and binding reactions and to efficiently form large interfaces with diverse targets (36,37,43,44). This results in relatively weak but highly specific interactions such that only the correct partner supplies the required complementary surface to generate sufficient enthalpic gain to offset the entropic loss associated with folding (45). These interactions can occur through two distinct but non-mutually exclusive mechanisms in which a disordered protein can acquire order either prior to or after binding to its target (the so-called “conformational selection” and “induced fit” models, respectively (46). This versatile and highly adaptive behavior is also efficient and economical and is well illustrated in the case of Max whose bHLH domain is also highly disordered and whose interaction with Myc as well as other Myc family members (N-Myc and L-Myc), Mnt, Mga and each of the four members of the Mad/Mxd bHLH-ZIP family could well be determined and facilitated by this ID-dependent structural flexibility to promote distinct tertiary structures (39–41). A more striking ID-mediated interaction between Myc’s bHLH-ZIP domain and even less related proteins such as AP2, Brca1, Miz1, and Nmi (50–52) might also explain the former protein’s highly variable but specific binding preferences. Such structural adaptability toward its binding partners also suggests a means to leverage the anticipated susceptibility of ID regions to small molecule binding. In such a model, it can be envisioned that this interaction would “lock in” one or more structures, thereby exploiting the entropic gain of a disordered system to inhibit the interaction of the protein with one member of its partner repertoire. Protein conformations prone to interacting with a desired target would, upon binding to a small molecule, be removed from the ensemble of potentially reactive structures without affecting those ID conformations capable of interacting with other protein targets and thus providing considerable specificity.
Upon interacting with Max, Myc’s bHLH-ZIP domain undergoes coupled folding and binding that leads to the formation of a parallel, left-handed, four-helix bundle where each monomer forms two -helices separated by a ca. 10 residue loop. The overall Myc α-helical content of this stable and relatively rigid structure is approximately 70% and its thermal stability is considerably greater than that of the Max homodimer (ca. 55°C vs. 38°C) (33). Further increases in heterodimer α–helical content and stability are achieved upon binding to DNA containing the consensus CACGTG E-box motif (70% to 84% and 55°C to 73°C, respectively) in keeping with the behavior of other dimeric transcription factors (33,55–58). Although it seems likely that Myc-Max heterotetramers (54) and the various proteins that associate with Myc in its chromatin-bound, transcriptionally active form provide even further stability, the degree to which this occurs has not been investigated although ample precedent for this type of influence, both positive and negative, has been documented in the case of other proteins (48).
Heterodimers comprised exclusively of the Zip domains of Myc and Max form readily but are much less stable than homodimers and heterodimers formed by the “pure” pZIP proteins Jun, Fos and GCN4 (59–61) and are more in keeping with the weaker homodimers formed by the ZIP domain of TFEB, an E-box-binding transcription factor that, like Myc, also contains an adjacent bHLH domain (62). This suggests that the individual HLH and ZIP domains act cooperatively to facilitate one another’s relatively poor dimerization capabilities into a stable structure whose free energy of association is greater than that of its individual interacting domains. It seems likely that the instability of each of these segments facilitated the discovery of small molecules that could prevent and/or disrupt the dimerization process (62). Moreover, because a minor part of the dimer interface may contribute disproportionately to the affinity between the proteins (63,64), targeting these “hot spots” and restricting their conformational freedom may well be sufficient for inhibiting protein–protein interactions.
Control of transcription complex assembly by Myc
The binding of Myc-Max heterodimers to DNA initiates the organized recruitment of well over 20 associated factors into a large, chromatin-associated complex who precise composition and stoichiometry likely depend upon a number of non-mutually exclusive factors including the cell type, its proliferative and metabolic status, the identities and activities of other neighboring chromatin-associated proteins, whether the bound E-boxes are “high affinity” or “low affinity”, the spacing between adjacent sites, and the interaction between Myc-Max dimers and their associated factors bound distally and brought into contact by chromatin looping (65–71). The net result of this multiprotein complex recruitment is a global change in chromatin architecture and function such that at least one-third of actively transcribed genes are bound by Myc within 1 kb of their transcriptional initiation site (71,72). With few exceptions, the precise protein-protein contacts of each of these factors and their temporal order of their associations have not been clearly elucidated. The most abundant and well-characterized factors include TRRAP, GCN5, TIP48, TIP49, TIP60, BAF53, CBP and p300 (66,73–76). Among the functions associated with these proteins are ATPase activities of TIP48 and TIP 49 and histone acetyl transferase activities of GCN5, TIP60, CBP and p300. The latter four enzymes tend to be quite specific for the acetylation of histones H3 and H4 and their respective lysine 4 residues. However, they also acetylate Myc itself and enhance its stability by at least three-fold although the precise acetylation patterns inmparted by each enzyme are distinct (75,76). pTEFb (transcription elongation factor b), an RNA Pol II pause release factor comprised of cyclins T1 and T2 and CDK9 and TFII-H an RNA Pol II promoter clearance factor are also components of this complex with pTEFb binding directly to Myc (56,77,78). Both pTEFb and TFII-H possess intrinsic kinase activities and phosphorylate the C-terminal domain of RNA Pol II (79–82). pTEFb also appears to be responsible for guiding other histone modifications such as histone 2B ubiquitination and histone H3 methylation (83). The pause factors DSIF and NELF also associate with and are phosphorylated and inhibited by pTEFb (68,71). These are then recruited to the complex by BRD4, an acetylated lysine (KAc)-recognizing member of the bromodomain and extra-terminal (BET) family (84–80).
At least part of BRD4’s ability to recruit pTEFb is likely explained by the fact that the cyclin T1 component of pTEFb is acetylated on at least 3 lysine residues, thus promoting its interaction with the BRD4 bromodomain (90). pTEFb suppresses abortive Pol II-mediated negative strand transcriptional initiation while relieving positive strand pausing and facilitating uni-directional read-through of authentic transcripts (71, 91–95). Direct Myc inhibition leads to a loss of Pol II-mediated elongation without affecting the amount of promoter proximal-bound Pol II. It also reduces Ser2-phosphorylated Pol II, which is the form associated with elongation, but has minimal effects on Ser5-phosphorylated Pol II, the form associated with initiation (71). It has been suggested that Myc overexpression or deregulation leads to a more efficient recruitment of pTEFb at bound promoters via their direct interaction (65, 68,71) thus relieving transcriptional block by permitting positive strand Pol II read-through and repressing antisense transcription (71).
Many of the above components, notably DSIF and pTEFb are members of other Myc-independent multiprotein transcriptional complexes, thus attesting to their dynamic nature and versatility (95). Additional histone modification mediated by Myc and its associated factors has been reported to occur at the level of histone H3K9 di- and tri- de-methylation and involves the recruitment of one or more demethylases by members of the JARID1 family of proteins which bind directly to the bHLH-ZIP domain of Myc (96,97). The JARID1 family consists of a single ortholog in Drosophila, where it is known as Lid and four members in mammalian cells, which are expressed in a tissue-specific manner. Myc inhibits the activity of the JARID1-demethylase complex, thus forming a positive feedback loop to ensure high levels of H3K4 methylation during periods of robust Myc-mediated transcription (96,97).
In summary, Myc appears to be a universal but unequal amplifier of gene expression that functions less to regulate the actual amount of RNA Pol II bound to actively transcribed genes than to reduce anti-sense transcription and relieve sense strand pausing. This process is orchestrated by the complex and dynamic interplay amongst a large complex of Myc-associated proteins whose precise constituents may be determined by a variety of both cell- and growth-specific conditions. Post-translational acetylation of histones, primarily H3 and H4, is instrumental in this process as is the phosphorylation of the C-terminal domain of RNA PolII. Loss of Myc is associated with the rapid loss of these modifications and transcriptional attenuation. The transforming function of Myc is likely due to the over-expression of genes containing high affinity binding sites that respond to both physiologic and pathologically elevated levels of Myc as well as to genes with low affinity sites that respond only to the highest levels of the oncoprotein generated by its deregulation.
KAc as a Determinant of Myc Binding and Transcription
In addition to the intrinsic affinity of a binding site for Myc-Max heterodimers, it is known that binding is also facilitated by the level of pre-existing chromatin acetylation, which tends to cluster in so-called euchromatic islands located at the 5’-ends of actively transcribed genes that are also associated with methylated histones (67,98–100). Acetylation but not methylation is further enhanced upon DNA binding by Myc via the mechanisms discussed above and Myc itself appears to be acetylated and stabilized as a result (67,75,76,102). An important question is whether the pre-existing KAc tags are functionally different from those that appear subsequent to Myc-Max binding. It is tempting to speculate that the former tend to be represented by histone H3 acetylation and that they serve as epigenetic landmarks or molecular signposts that guide Myc-Max complexes to their intended binding sites so as to minimize both the need for chromatin scanning and the possibility that transcription will initiate at unintended sites. In contrast, KAc tags appearing after the Myc-Max dimer and its accessory proteins are bound by DNA, would provide the opportunity for BRD4 to engage acetylated pTEFb (and possibly Myc itself) and perhaps allow a more robust acetylation of the surrounding chromatin and the transcriptional complex by virtue of a highly functional assembly of multiple acetylases.
The recognition by BRD4 of acetylated histones and pTEFb is a critical step in the transcriptional control of Myc-regulated genes as siRNA-mediated suppression of BRD4 in acute myeloid leukemia cells leads to terminal myeloid differentiation and the elimination of myeloid stem cells (85) in a manner that was precisely phenocopied by a small molecule inhibitor of KAc recognition by BRD4 (see below).
Inhibition of Myc: Challenges and Opportunities
The foregoing summary reveals the complex and dynamic molecular anatomy underlying Myc-driven transcriptional regulation. In doing so, it provides a framework within which to consider how the function of this machinery can potentially be impaired in highly specific and perhaps complementary ways. It also highlights some of the theoretical and practical barriers to the design and implementation of strategies designed to target Myc. Recent insights suggest that, while some of these barriers will be difficult to overcome, they are not insurmountable. We discuss some of these impediments below and how they have been, or are being, addressed
First, unlike the case for many other oncoproteins which drive transformation as a result of protein mutations, Myc-driven cancers are virtually always caused by the oncoprotein’s overexpression and/or de-regulation. Myc coding region mutations, when they occur at all, are largely confined to a relatively small subgroup of Burkitt’s and AIDS-related lymphomas and generally serve only to stabilize an otherwise normal Myc protein (104–108). In virtually all other cases, no opportunities exist to target a cancer-specific mutant form of the oncoprotein as there are, for example, in melanomas with B-Raf mutations, non-small cell lung cancers with EGFR mutations or CML with Bcr-Abl translocations. Second, unlike these typical examples, all of which are protein kinases and all of which are more susceptible to inhibition by small molecules than their wild-type counterparts, Myc possesses no intrinsic enzymatic activity. Therefore, the design of Myc-specific decoy substrates, based upon principles that have guided targeted therapeutic strategies against other oncoproteins for the better part of two decades (109) is not a viable option for the development of direct Myc inhibitors. Moreover, approaches that target the kinases associated with the Myc transcriptional complex such with pTEFb or TFII-H would likely be associated with unacceptable toxicities given the ubiquity with which these factors regulate normal gene expression. Together, these observations underlie the third concern, namely that because normal and cancer cells express identical forms of Myc, is it possible that Myc inhibitors might simply be too non-specific and toxic to be used effectively? We will address the first and last of these concerns together given that without a satisfactory way to allay these potential problems, Myc inhibitors, no matter how ingeniously designed, would likely have limited appeal.
Many and perhaps all Myc-driven tumors seem to be particularly reliant on or “addicted” to high levels of the oncoprotein (110,111). Evidence for this is provided by studies showing that controlled shutdown of the high-level transgenic Myc expression that drives tumorigenesis in certain mouse models is associated with rapid tumor regression and/or apoptosis (1). Indeed, in some cases, only transient Myc inhibition is necessary for sustained tumor regression whereas in others, re-expression, rather than leading to tumor re-growth, is associated with massive apoptosis (112). Other models in which the expression levels of Myc can be fine-tuned have shown that even slight increases in its level can promote proliferation in otherwise normal tissues (113). Given recent insights into the way that Myc controls metabolism, it seems likely that such sensitivity has a foundation in the high reliance of all rapidly proliferating cells on glycolysis and glutamine as sources of anabolic substrates and oxidatively-generated energy, respectively (110,114,115). Collectively, these findings suggest that Myc inhibition in tumor cells might be more consequential than an equivalent degree of inhibition in normal cells. With regard to this latter point, it is well know that Myc is generally expressed at low to undetectable levels in most organs consistent with the quiescent nature of most of their component cells. This would be expected to provide a relatively large therapeutic window within which to launch an attack on the aberrantly expressed Myc that is reactivated in tumor cells and that, if expressed at all in normal cells, would otherwise be confined to a slowly proliferating sub-population. Of course, the exception to this simplistic model is that highly proliferative cellular compartments do in fact exist in hematopoietic and gastro-intestinal precursor cell populations. Thus, Myc inhibitors might at the very least be expected to have side effects that would manifest themselves as bone marrow aplasia and loss of intestinal villus integrity. These toxicities would be akin to those encountered following treatment with conventional cytotoxic agents that non-specifically target all proliferating cells; in fact, they are actually seen following the organism-wide inhibition of Myc by genetic means in mice (9). Surprising, and as yet unexplained, is that these side effects are not only relatively mild but are transient, even in the face of chronic Myc suppression (9). These experiments support the idea that many tumors are much more sensitive to Myc inhibition than their normal cell counterparts, that their eradication might actually be enhanced by intermittent therapies, and that the anticipated side effects would not only be entirely predictable but well-tolerated as well.
What about the lack of any demonstrable Myc enzymatic activity for which inhibitors could be designed? This arguably presents the greatest challenge as it closes the door on an approach that has led to the greatest advancements in targeted therapy. Unfortunately, there are only a limited number of ways around this obstacle, which are described below in their individual sections. They include disrupting protein-protein interactions (PPIs), either those between Myc and Max (direct inhibitors) or between Myc-Max and other proteins (indirect inhibitors). Alternatively, one might capitalize on unique susceptibilities of tumor cells that arise specifically as a result of Myc deregulation (synthetic lethality).
Disruption of PPIs: practical considerations
The selective disruption of PPIs by small molecules has long been considered a therapeutic Holy Grail, made even more tantalizing by the importance of PPIs in normal cellular signaling pathways and abnormal PPIs in oncologic, cardiovascular and neurodegenerative disorders (38–46). Yet the attainment of this goal has been tempered by a number of caveats, some of which are quite compelling. First, unlike enzyme inhibitors, which typically compete with natural substrates for binding to naturally engineered pockets that are shielded from their aqueous environment, small molecule inhibitors of PPIs must prevent or disrupt interactions that occur over relatively large surface areas (generally in the range of 1000–3000 Å2) that are associated with high free energies of association and whose interfaces are relatively flat, featureless and devoid of the more geographically prominent regions that tend to accommodate small molecules readily (116–119). The problem is further amplified in the case of Myc specifically because of the inherent disorder of its dimerization domain (120). Additionally, the key interacting residues that initiate the PPIs at such sites are generally not immediately apparent even if crystallographic structures are used as guides. For these reasons, PPIs have long been considered to be “undruggable” (119,121,122). Adding relevancy to these concerns for the purpose of this specific review is the fact that the crystal structure of the Myc-Max heterodimer also fails to reveal any obvious targetable pockets along the contact interface (54). These concerns have been partly tempered by the finding that some if not all PPIs may be initiated by a small (~500 Å2) area within the larger interacting region whose constituent amino acids contribute disproportionately to the free energy of binding of the entire interacting surface. These domains tend to occur near the geometric center of the protein-protein interface and to be comprised of aromatic amino acids. This suggests that small molecules that bind to and distort such regions might exert a disproportionate effect on PPIs (63,123). In fact, small compounds of this nature have now been shown to be relatively potent PPI inhibitors. These include the nutlins, which interfere with the interaction between TP53 and its suppressor HDM2, and ABT-737 (and its derivatives ABT-263 and ABT-199), which serves as a BH3 mimetic to prevent the interaction between the pro-survival members of the Bcl-2 family such as Bcl-2 itself, Bcl-xL and Bcl-w, and the pro-apoptotic Bcl-2 members such as Bax and Bak (124,125). Additional small molecules that inhibit Stat3 PPIs have recently shown considerable promise as well (126,127). Finally, small molecules designed to mimic aspects of protein secondary structure such as α-helices, β–sheets, and β–strands have also been shown promise as PPI inhibitors (127,128).
Direct Inhibitors
Peptidomimetic inhibitors
The first small molecule direct Myc inhibitors were described by Berg et al (129) who monitored the dissociation of purified, recombinant MycCFP and MaxYFP fusion proteins using an elegant FRET-based assay. They screened a small (7000 member) peptidomimetic library and identified five active compounds, four of which were evaluated further by ELISA- and electrophoretic mobility shift (EMSA)-based assays of Myc-Max interaction. Using a single concentration of each compound (25 µM) they were able to consistently obtain low (12–%) but reproducible inhibition of heterodimerization in the FRET assay, and IC50s of 75–250 µM and 50–162 µM in the ELISA and EMSA assays, respectively. Disruption of DNA binding by one of the compounds could not be demonstrated.
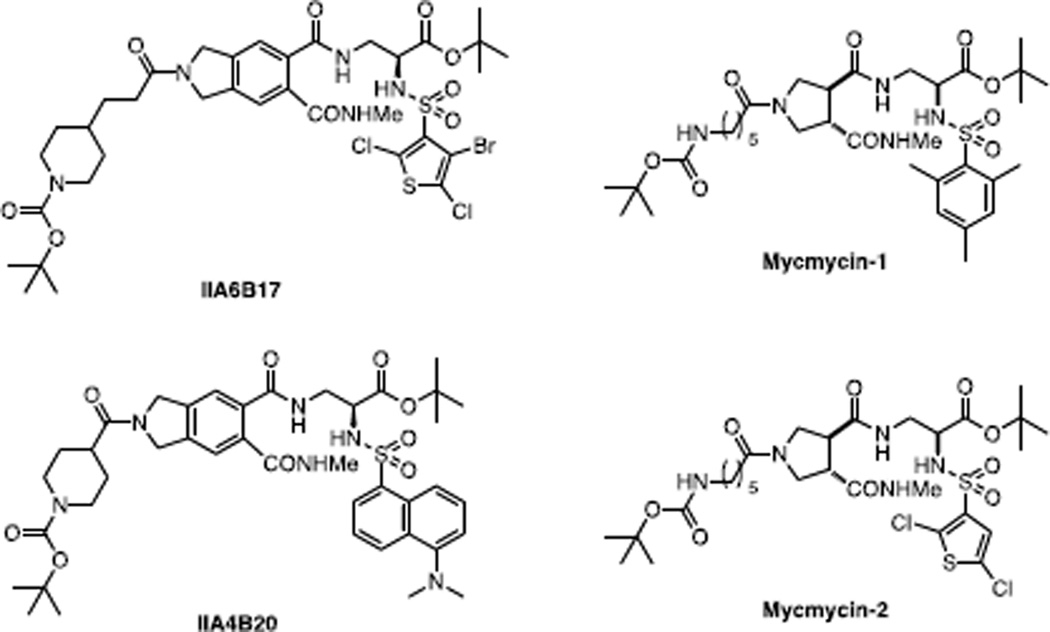
Having identified three compounds that gave consistent results in three independent assays of Myc-Max dimerization, Berg et al (129) then asked if they could selectively abrogate the appearance of transformed foci in chick embryo fibroblasts (CEFS) in response to Myc overexpression versus Src and Jun overexpression. The best of these compounds (IIA6B17 and IIA4B20) showed Myc/Src/Jun IC50s of 18 µM/>35 µM/18 µM and 20 µM/>35 µM/35 µM, respectively) (Fig. 1). At the highest concentrations tested (30–35 µM) both compounds, particularly IIA6B17 also significantly reduced the growth of normal CEFs. Collectively, these findings raise a number of points. First, given the relatively poor performance in FRET, ELISA and EMSA assays, their relative potency in cell-based assay was surprising. However, as described below for other Myc inhibitors (11,130), imperfect correlations among these assays tends to be the norm and likely relate to issues of cellular uptake, distribution and stability that are not accounted for in the former assays.
Fig. 1.
Structures of IIA6B17 and IIA4B20 (129) and their analogs mycmycin-1 and mycmycin-2 (132).
Second, the finding of some selectivity for Myc-mediated transformation can likely be explained by “oncogene” addiction of transformed cells which are more reliant on high levels of Myc to maintain this state than they are to maintain normal functions. In other words, lowering Myc levels in these cells even modestly exerts a disproportionate effect on transformation relative to “normal” cellular functions under its purview. On the other hand, the inhibition of Jun transformation and normal cell proliferation could be explained by a common mechanism, namely the reliance of all cells, transformed or otherwise, on endogenous levels of Myc for rapid growth under any circumstance. Somewhat more surprising was the relative resistance or Src-transformed cells given that transformation by this oncogene is well-known to rely on functional Myc (131). However, it is quite possible that, under the conditions of these experiments, Src-transformed cells utilize alternate and Myc-independent pathways to overcome the effect of these inhibitors.
In a follow up to the study of Berg et al (129), Shi et al (132) derived 120 analogs each of IIA6B17 and IIA4B20 and screened them at a single concentration (20 µM) for their ability to inhibit colony formation by Myc-transformed CEFs. Two of the compounds thus screened by this biological assay, “mycmycin-1” and “mycmycin-2” (Fig. 1) were found to be highly specific (IC95=20 µM) and had no effect on colony formation by either Src- or Jun-transformed CEFs. As had been the case for IIA6B17 and IIA4B20, both mycmycin-1 and mycmycin-2 were able to prevent the association of recombinant Myc and Max proteins.
“Credit card” inhibitors
In a novel, structure-based, rational approach, Xu et al (133) capitalized on the fact that small regions within ID protein-protein interfaces, particularly those enriched for amino acids such as Trp, Tyr, His and other hydrophobic residues, contribute a disproportionate fraction of the free energy of binding (63,64). They viewed these amino acids as slots or “card readers” within an otherwise flat surface and hypothesized that the insertion of a planar or “credit card”-like molecule would be a potentially effective way to block the PPI. Xu et al (133) then designed a library of planar structures with hydrophobic cores that was predicted to make favorable enthalpic contributions from van der Walls interactions, π–stacking and desolvation as well as favorable entropy gains arising from hydrophobic effects. Despite the small size of the library (285 structures), 40 hits were obtained using the same FRET-based assay used by Berg et al (129) and four were evaluated in greater depth. After showing that these functioned to disrupt Myc-Max heterodimer DNA binding in a standard EMSA assay (IC50s =17–36 µM), Xu et al (133) then employed an elegant far UV CD-based assay using synthetic Myc and Max bHLH-ZIP peptides tethered by thioester ligation. Surprisingly, they found the overall α–helical content of the highly structured dimer to be conserved upon addition of the compounds. They speculated that the compounds might disrupt the inter-helix contacts between Myc and Max thereby giving a similar per-residue normalized helicity signal even in the absence of a PPI. Another explanation is that the credit card molecules do not actually cause Myc-Max dissociation but rather perturb the structure of the dimer in a way that preserves it overall α-helical integrity but abrogates DNA binding.
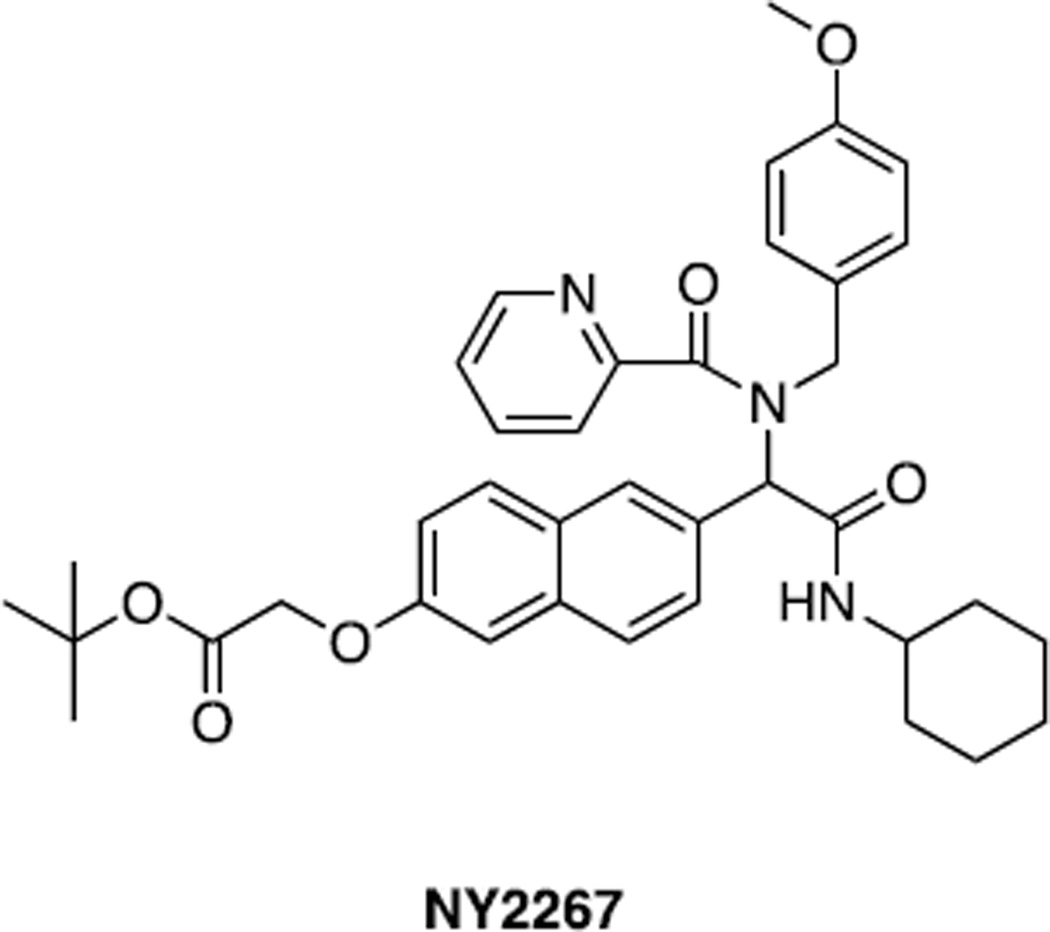
Xu et al (133) also evaluated the above compounds for their ability to inhibit oncogenic transformation. One in particular, NY2267 (Fig. 2) shared significant selectivity for Myc transformed CEFs versus cells transformed by Src, Jun or PI3 kinase. Interestingly, and despite this seemingly selective effect, tests of the compounds with Myc, Jun and NF-κB reporter vectors indicated that NY2267 was unable to discriminate between Myc and Jun luciferase reporters.
Fig. 2.
Structure of NZY2267 (see ref. 133)
Mycro1 and Mycro2
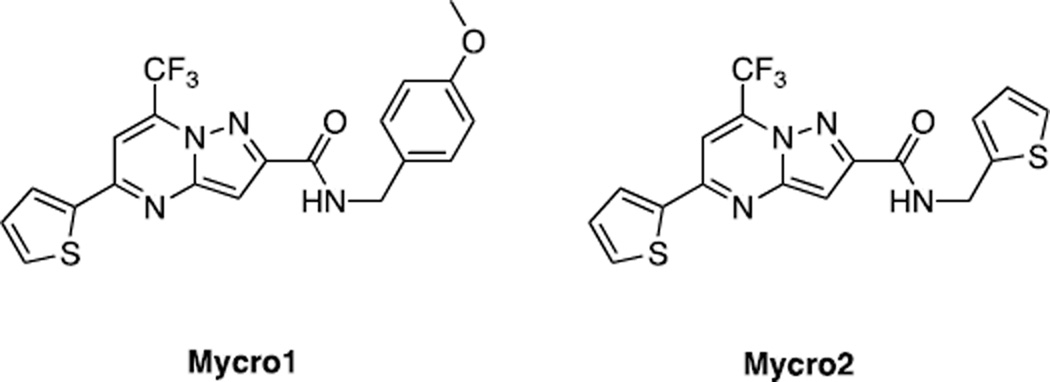
Kiessling et (134) al. developed a fluorescence polarization (FP)-based assay that allowed quantification of Myc-Max bHLH-ZIP heterodimer binding to a fluorophore-tagged, E-box-containing oligonucleotide and the identification of small molecule inhibitors of this interaction. A screen of a 17,298 member library identified a pyrazolo[1,5-α]pyrimidine dubbed “Mycro1” and a subsequent analysis of structure-activity relationships with commercially available derivatives identified an analog, Mycro2, that were subsequently evaluated in more depth. Mycro1 and Mycro2 (Fig. 3) inhibited DNA binding with IC50s of 30 µM and 23 µM, respectively. The IC50s for Max homodimeric binding to the same oligonucleotide were 2–3-fold higher thus indicating reasonably good selectivity for the heterodimer. Further evaluation against CEBPα and jun bZIP homodimers confirmed the compound’s specificity (IC50s >100 µM for each), particularly in the case of Mycro2 and neither compound affected the interactions between two SH2 domains. EMSA assays confirmed these results. Using CFP-Myc and GST-Max fusion proteins in standard pulldown experiments Kiessling et al (134) also provided evidence that the Mycro1- and Mycro2-mediated loss of DNA binding was actually due to the loss of heterodimerization.
Fig. 3.
Structures of Mycro1 and Mycro2.
Testing in a series of cell lines showed that Mycro1 and Mycro2 inhibited proliferation in a dose-dependent manner in Burkitt lymphoma, breast cancer and osteogenic sarcoma and non-transformed NIH3T3 cells. For both cell types, IC50s after 5–7 days of treatment were nearly identical for the two compounds (10–20 µM). Interestingly, they had no effect on the proliferation of PC12 pheochromocytoma cells which contain no Max protein and are therefore thought to be Myc-Max independent (135). As discussed above, the relative inability to discriminate between transformed and non-transformed cells is to be expected for direct inhibitors. Somewhat surprising was that the authors did not report any evidence of apoptosis in their treated cell lines. They attributed this to a lack of non-specific toxicity, although more likely it was due to incomplete intracellular Myc-Max association so as to inhibit proliferation while still allowing sufficient heterodimer association to maintain viability.
In additional studies, Kiessling et al (134) showed that, at the same concentrations used to achieve proliferative arrest, both Mycro1 and Mycro2 selectively inhibited the expression of an E-box-containing reporter vector relative to similar reporters bearing AP1 or serum response elements.
Finally, Mycro1 and Mycro2 inhibited anchorage-independent growth of Myc-transformed fibroblasts by as much as 60% while exerting little effect on Src-transformed cells. Not determined from these studies was whether the inhibition of colony formation was due to a Myc-specific property of transformation or simply to an overall inhibition of proliferation as described earlier for NIH3T3 fibroblasts. Similarly, the resistance of Src-transformed cell growth in soft agar suggested that this oncogene rendered cells relatively Myc-independent.
In a follow-up study, the same group reported the results of a screen of a ~1700 member library of pyrazolo[1,5-α]pyrimidines based upon the structures of Mycro1 and Mycro2 (136). Five compounds-designated 1–5-were identified whose IC50s in the above-described FRET assay ranged from 29–64 µM but whose average selectively for Myc-Max heterodimers versus Max homodomers was only 2-fold for 4 of the compounds (range 1.7–2.6). The fifth compound was more selective with only 24% inhibition of heterodimer binding being observed at the highest tested concentration (100 µM). The assessment of these compounds in a single osteosarcoma cell line indicated an IC50 for compound 1 of approximately 10 µM without noticeably affecting the growth of PC12 cells. All remaining compounds either inhibited PC12 cells or demonstrated evidence of non-specific toxicity. Further assessment of 1’s activity profile showed it to be comparable to Mycro1 and Mycro2 in EMSA, soft-agar colony and luciferase reporter assays without demonstrating any “off target” effects against fos-jun. Based on the apparent high specificity, the authors suggested that 1 could prove to be a promising scaffold for future drug development.
10058-F4 and 10074-G5
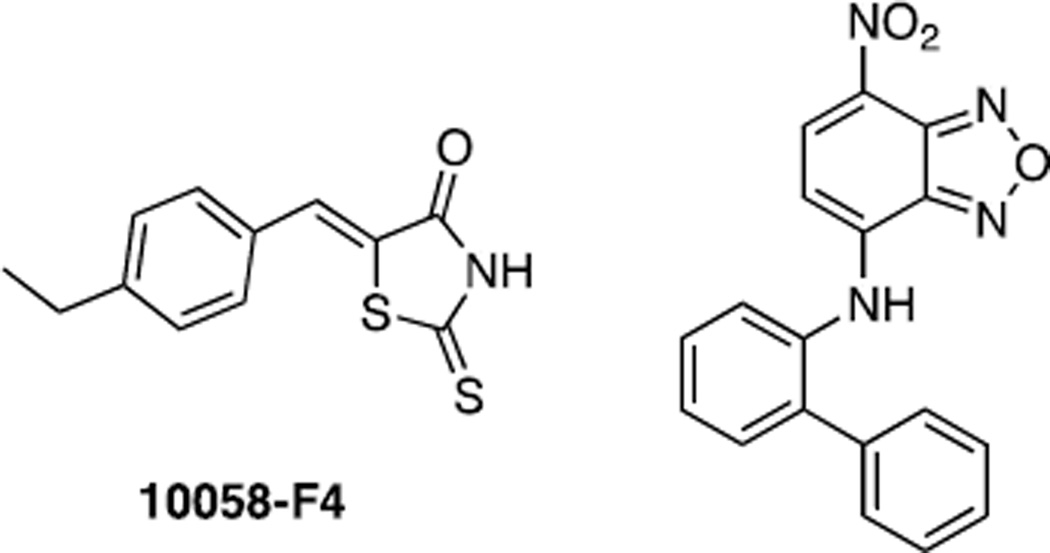
Closely following the report by Berg et al (129), Yin et al (130) reported the identification of a distinct set of Myc inhibitors. The approach taken by this group was somewhat different from that used by Berg et al (129) and others and was employed as a way of circumventing the inevitable identification of molecules that, while effective and preventing heterodimerization by purified Myc and Max proteins, would be unlikely chemotherapeutic candidates due to poor cell penetration and/or non-specific toxicity. Yin et (130) al utilized a yeast two-hybrid-based assay in which the bHLH-ZIP domains of Max and Myc were fused to the transcriptional activation domain and the DNA binding domain, respectively, of the yeast Gal4 transcription factor. Functional reconstitution of Myc-Max heterodimers in s. cerevesiae containing a Gal4-responsive β-galactosidase reporter gene strongly induced the expression of the enzyme. This assay also afforded an additional advantage in that any identified hits were likely to be molecules that actually inhibited Myc-Max association and not DNA binding. The yeast strain was then employed in a survey of ~10,000 small molecules that were selected based on 3D pharmacophore analysis to represent a broad spectrum of biologically relevant pharmacophore diversity space. An initial control yeast strain employing similarly constructed expression plasmids for the bHLH-containing partner proteins Id2 and E47 was screened in parallel to identify compounds that were selective for Myc-Max heterodimers. Eventually, seven such compounds were identified, all of which conformed well with Lipinski’s rules (116) and all of which inhibited Myc-Max association by ≥75% and Id2-E47 association by <25% without any significant cellular toxicity. Further specificity was established by testing each of the compounds against 32 additional yeast strains harboring various known bHLH, bZIP and bHLH-ZIP combinatorial protein pairs. Of the 231 possible non-Myc-Max protein pair + drug combinations that were compared in this manner, only 3% showed any significant evidence of “off target” effects. Indeed, three compounds, including two designated 10058-F4 and 10074-G5 (Fig. 4), showed complete specificity for Myc-Max. Interestingly, of the off target effects that were seen with the remaining Myc inhibitors, 42% occurred with combinations of Max and Mad family members whose HLH-ZIP sequences and interactions most closely approximate those between Myc and Max. None of the candidate compounds inhibited Max homodimerization in this assay. Because the initial yeast-based screen also identified compounds that specifically inhibited the control Id2-E47 interaction, but not the Myc-Max interaction, 10 of these compounds were further tested against the entire yeast two-hybrid battery. As was the case for Myc-Max, off target effects were observed in 2.7% of the 330 possible non Id2-E47 interactions with five of the compounds demonstrating absolute specificity against their intended target. These results indicated that yeast two hybrid approaches could be used as simple and robust cell-based assays to identify cell-penetrable and non-toxic compounds capable of disrupting and/or preventing various PPIs with high specificity.
Fig. 4.
Structures of 10058-F4 and 10074-G5
Further analyses of the seven identified Myc compounds provided additional confirmation of their specificity and efficacy. These studies included demonstrating the in vitro disruption of recombinant Myc-Max heterodimers in standard GST-pulldown-type experiments, the specific inhibition of two Myc responsive genes and the lack of inhibition of a gene that is typically responsive to combinations of myogenic HLH proteins. Testing of the Myc-specific compounds in rat fibroblasts showed that cells expressing either endogenous levels of Myc or deregulated levels were highly susceptible to each of the compounds whereas a myc−/− cell line showed a relative lack of response (138). The initial cellular responses consisted of Go/G1 arrest followed by eventual apoptotic death. Transient exposure of the Myc-over-expressing cells to several of the compounds followed by injection into recipient immuno-deficient animals significantly slowed tumor growth.
Subsequent work by this group used computer-assisted searches of >500,00 compounds for molecules with chemical similarity as well as directed synthetic approaches to identify more potent 10058-F4 analogs (139). The approach taken was to search for related compounds whose structures differed from that of 10058-F4 in either the six-member or five-member ring (Fig. 4) and which did not alter the sub-structure by >15%. A total of 63 such compounds were identified which were then prioritized by comparing their IC50s to that of 10058-F4 in a cell-based proliferation assay with HL60 human promyelocytic leukemia cells which over-express Myc due to gene amplification. Of the 48 six-member-substituted analogs, four were found to have IC50s comparable to that of 10058-F4, ranging from 23–51 µM (versus 49 µM for 10058-F4). Of the 15 five-member ring analogs, four were found to be significantly more potent (range 4.6–18 µM). All of the analogs were also able to disrupt Myc-Max heterodimers in HL60 cells as measured by co-IP assays and to promote protein-DNA dissociation in EMSA-based assays. Combining the best attributes of each group so as to generate 17 analogs with both 5- and 6-member “optimized” ring substitutions did not result in any further improvements in cell proliferation assays. In retrospect, these non-additive properties were consistent with the predicted behavior of ID protein regions, which would be capable of binding multiple structures. Thus the conformations best suited to binding a dual five- and six-member ring-substituted analog might be quite different from those which bind either of the singly substituted analogs.
Wang et al (139) next showed that 10058-F4 and all of its most potent analogs bound directly to c-Myc’s bHLH-ZIP domain by employing a fluorescence polarization assay that capitalized on the inherent fluorescent nature of most of these small molecules. They calculated observed affinities for nine of the compounds that ranged from 0.6 to 8.6 times that of 10058-F4 (kobs=2.3 +/− 0.7 µmol/L). The modest correlation between the compounds’ anti-proliferative effects and their binding to Myc and disruption of Myc-Max heterodimers were proposed to be due to factors that affect in cellulo behavior such as uptake, subcellular distribution and intracellular stability.
In a subsequent study by the same group, Follis et al (140) employed a series of deletion and point mutations within the Myc bHLH-ZIP domain to map the precise binding sites for both 10058-F4 and 10074-G5. In the case of 10058-F4, this corresponded to a region spanning the Helix 2-ZIP junction (residues 402–412) whereas for 10074-G5 the binding site mapped to region centered around the N-terminal region of Helix 1 (residues ~363–381). Binding of 10058-F4 was abrogated by several single and double point mutations that included L404P, Q407K and V406A/E409V. Similarly, binding by 10074-G5 was eliminated by point mutations R367G and E369K/L370P. Interestingly, each of these mutations would be predicted to lead to significant charge and/or structure perturbations within their respective binding sites that could easily prevent compound binding.
The above findings were confirmed using synthetic peptides, which encompassed only a single binding site, and by monitoring structural changes using circular dichroism (CD) and 1H and 13C NMR spectroscopy. A control, bacterially-expressed peptide corresponding to the entire bHLH-ZIP domain (residues 353–437) was also employed to confirm that any observed changes in the smaller peptides were being recapitulated in the context of the full-length dimerization domain. The CD spectra of this full-length peptide remained essentially unchanged in the presence of 10058-F4 or 10074-G5, demonstrating under both conditions the expected large degree of ID (33) and indicating that any change in structure mediated by small molecule binding must be confined to relatively small regions. In contrast, the CD spectra of both synthetic peptides were markedly altered to more rigid-appearing conformations upon addition of their cognate ligands. In NMR studies, the similar behaviors of the peptides and the full-length bHLH-ZIP domain confirmed that the structural alterations being imposed by the small molecules were geographically localized. A docking simulation performed between the inhibitors and their bound structures showed that in neither case was the peptide conformation representative of that found in the Myc-Max crystal structure (54). Together, these studies indicated that the product of a coupled folding and binding reaction is not necessarily helpful in predicting either potential small molecule binding sites or the conformations these sites will assume. Moreover, the conformations deemed most likely to be occurring in the presence of 10058-F4 and 10074-G5 and their respective binding sites did not appear to be compatible with the formation of the interface needed to associate with Max. Additional analysis of the entire Myc bHLH-ZIP domain with a disorder-predicting program (141) revealed that the two small molecule binding sites each mapped to separate regions associated with abrupt transitions in predicted ID from high to low. It has been proposed that such regions, particularly those that are relatively hydrophobic and non-conserved may be particularly capable of binding small molecules. These properties have also been observed for ID regions involved in partner protein recognition (38).
In follow-up to the above work, Hammoudeh et al (142) examined the remaining compounds originally identified by Yin et al (130) Using the same techniques employed to map the binding sites for 10058-F4 and 10074-G5, they identified a third independent binding site for the compound 10074-A4 immediately C-terminal to and possibly overlapping somewhat with that of the 10074-G5 binding site. The affinity of 10074-A4 for its cognate site in the full-length bHLH-ZIP domain was significantly lower than that for 10058-F4 and 10074-G5 to their sites (21+/−2 µM, vs. 5.3+/−0.7 µM for 10058-F4 and 2.8+/−0.7µM for 10074-G5). All three compounds competing with 10058-F4 bound to the site with at least three-fold less affinity. In contrast, 10050-C10, the only other compound binding to the 10074-G5 site, did so with three-fold higher affinity. CD analysis showed that each compound induced distinctive conformational changes in their respective synthetic peptide binding sites, consistent with the idea that different ligands selected and stabilized distinct structures from the entire ensemble of available ID structures. Although the above-calculated values indicate that the affinities of the small molecules for Myc are rather low, they are nonetheless in the range of the 1 µM Kd reported for Myc-Max heterodimers (121) even before considering the possibility that the binding sites may represent entropically favored points of dimerization initiation.
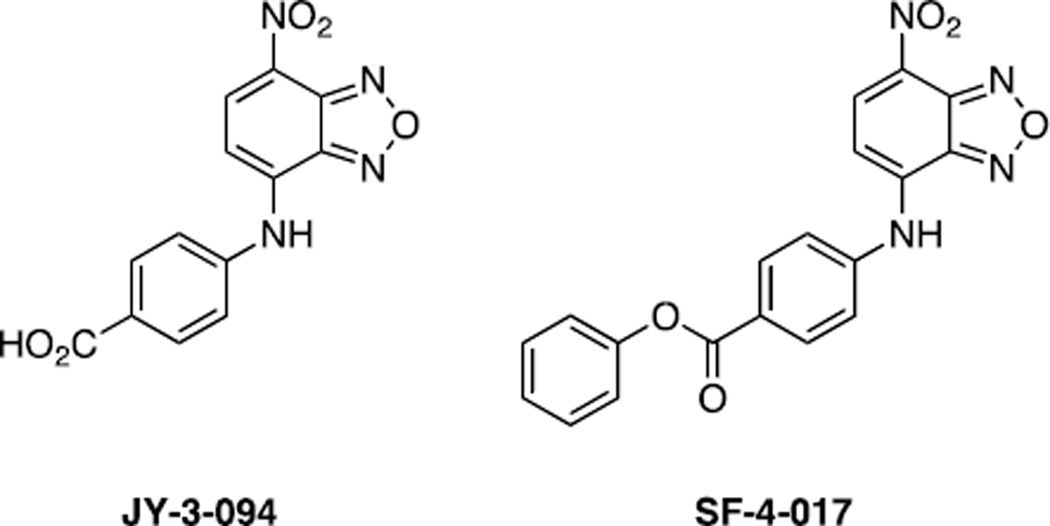
Yap et al (144) conducted a structure–activity relationship analysis of 10074-G5 in which the NMR model of its Myc binding site was used as a guide. The electron-rich nitro group and furazan ring were found to be critical to Myc inhibitory activity, which is consistent with these groups binding the basic residues R366, R367 and R372 as proposed above. In addition, the ortho-biphenyl moiety was also found to be essential and likely due to its occupancy of a hydrophobic domain created by F375 and I381. It was also discovered that the ortho-phenyl ring of the biphenyl could be replaced with a para-carboxylic acid that actually led to enhanced activity relative to the parent compound 10074-G5. This may be due to the resulting compound, JY-3-094 (Fig. 5), engaging in electrostatic interactions with R378. An EMSA assay revealed that JY-3-094 disrupted Myc–Max dimerization with an IC50 value of 33 µM, which is almost five times as potent as 10074-G5 (IC50 = 146 µM). Although JY-3-094 exhibited no cytotoxicity to HL60 and Daudi cells, this was presumably due to its ionizable carboxylic acid. The observed lack of activity of JY-3-094 was remedied by esterification of its carboxylic acid to afford a panel of ester prodrugs that exhibited IC50s in the low micromolar range in both HL60 and Daudi cells (145). LC/MS quantification revealed that the ester pro-drugs were metabolized to the Myc inhibitor JY-3-094 in cells thus confirming that the cytotoxicities, at least in part, stem from JY-3-094. Interestingly, while blockage of the carboxylic acid of JY-3-094 generally led to a loss in Myc inhibitory activity as anticipated through loss of the electrostatic interaction with R378, a phenol ester, SF-4-017 (Fig. 5), proved as potent as JY-3-094 in vitro and a co-IP demonstrated SF-4-017 to be an active inhibitor of Myc–Max dimerization in cells. SF-4-017 represents a new lead Myc inhibitor that should be resistant to both serum and intracellular esterases.
Fig. 5.
Structures of JY-3-094 and SF-4-017.
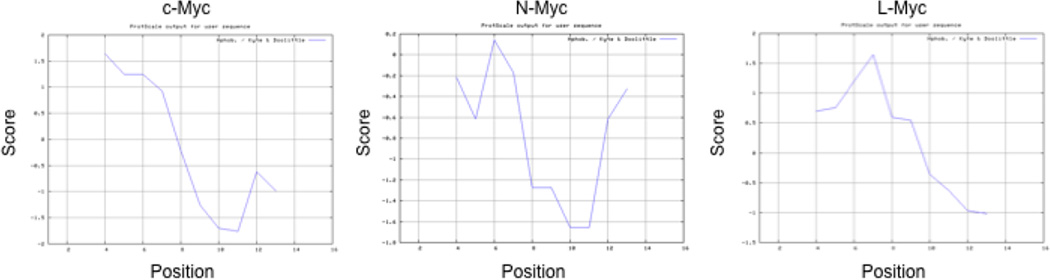
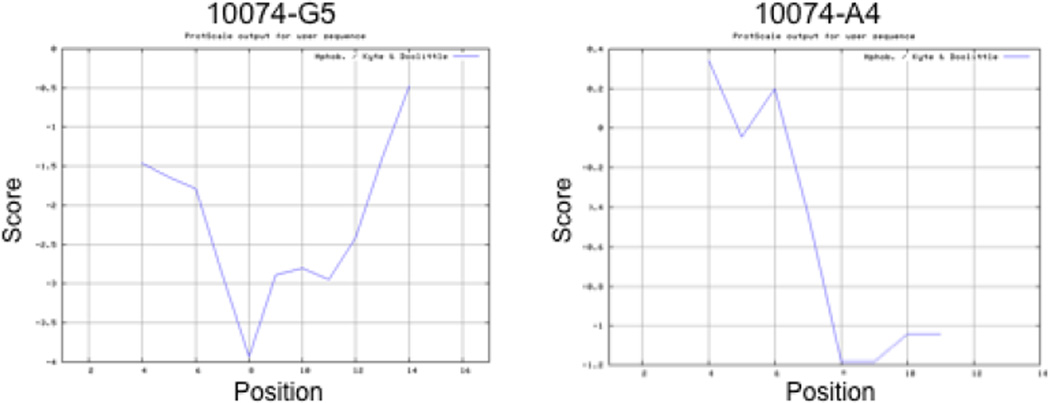
In order to gain more insight into how the Myc402–412 domain interacts 10058-F4, Cuchillo and Michel (146) computed the structures of the binding subset from within the entire conformational ensemble using classical force fields and explicit solvent metadynamics molecular simulations (125,126). Their study revealed a large amount of conformational heterogeneity in both the apo and holo strucures of the peptide. Most importantly, it suggested that rather than there being any single peptide conformation that best represents the interaction with 10058-F4, the small molecule interacts relatively weakly with and stabilizes a large number of conformations. Cuchillo and Michel (146) also found that 10058-F4 made preferential contact with Tyr402, Ile402 and Leu404, which lie within the most hydrophobic segment of the entire bHLH-ZIP domain and define a point of abrupt transition to greater hydrophilicity. In contrast, no such segment was identified in Max. Thus, a simple model would suggest that 10058-F4’s specificity lies in its ability to recognize these points of sharp hydrophobic-hydrophilic transition. This model is consistent with the overall hydrophobicity profiles of the presumptive sites of 10058-F4 binding on N-Myc and L-Myc (Fig. 6) and with recent findings that N-Myc-Max and L-Myc-Max heterodimerization is also susceptible to disruption by 10058-F4 (149 and H. Wang & EVP, unpublished). In fact, this model would also seem to apply in the cases of 10074-G5 and 10074-A4 whose minimal binding sites share the same sharp hydrophobic-hydrophilic transition (Fig. 7). On the other hand, the specificity of 10058-F4 for Myc-Max bHLH-ZIP dimers (130), coupled with its highly Myc-specific gene knockdown profiles (71,149), suggests that this model is over-simplified. This is supported by the finding that the affinity of 10058-F4 for its cognate binding site more than doubles when the minimal peptide binding site (residues 402–412) is extended to include the entire bHLH-ZIP domain (Kd=5.3 µM vs 13 µM) (140). A similar increase in affinity also occurs in the case of 10074-G5 as well (Kd=2.8 µM vs. 4.4 µM) (140). This suggests that the “fine tuning” of site recognition by small molecules, as well as a reinforcement of binding affinity, arises from residues that extend somewhat beyond the minimal binding sites previously defined by synthetic peptides.
Fig. 6.
Kyte-Doolittle hydrophobicity plots for residues 400–415 of Myc and the corresponding residues of N-Myc and L-Myc that are the presumptive sites for 10058-F4 binding. A window setting of 7 was used for all three plots (http://web.expasy.org/protscale/). Note that proteins have regions of comparable hydropbobicity that lie at the N-terminus of the core 10058-F4 binding site and that are associated with abrupt hydrophilic transitions.
Fig. 7.
Kyte-Doolittle plots for Myc binding sites for 10074-G5 (residues 359–375) and 10074-A4 (residues 373–386) (140,142). All settings were identical to those described in Fig. 6. Note the abrupt transitions from relatively hydrophobic sites to hydrophilic sites in both cases.
Two studies to date have confirmed the specificity of 10058-F4 on Myc or N-Myc target genes or their products. In the first, exposure of non-transformed embryonal stem cells to 10058-F4 for a short time (6 hr) was associated with the loss of expression of Myc target genes but not of non-Myc target genes (71). On a more global level, this was also associated with a loss of Pol II Ser2 phosphorylation, which is necessary for transcriptional elongation. In contrast minimal or no effect was seen on Ser5 phosphorylation, which is associated with initiation. Accompanying this finding was the additional observation that 10058-F4 treatment led to a loss of occupancy of Pol II in the actively transcribed bodies of Myc-regulated genes but not at initiation sites. In contrast, non-Myc-transcribed genes show no evidence for Pol II loss at either group of sites. Lending conviction to these results was the finding of similar effects when Myc inhibition was mediated by shRNA knockdown (71). These findings were consistent with the model described above that Myc functions to control transcriptional pausing and release and the read-through of its target genes whose relative sensitivities to Myc are nonetheless varied and complex.
In a related study, Zirath et al (149), compared the proteomic profiles of a human N-Myc-amplified neuroblastoma cell line exposed to 10058-F4 or shN-Myc RNA. Gene ontology analysis showed a common core group of biological processes to be affected by both manipulations most notably those involving mitochondrial structure and function (i.e TCA cycle enzymes and electron transport chain proteins), glycolysis and fatty acid β–oxidation. Roughly half the total genes knocked down as a result of exposure to 10058-F4 an snN-Myc shRNA had been previously reported as being Myc target genes. These findings were particularly notable given the well-documented relationship between both Myc expression, glycolysois and mitochondrial structure and function (150,151). Finally, Zirath et al (149) examined microarry data from 251 primary human neuroblastomas. Expression profiles for 20 transcripts corresponding to the metabolic proteins knocked down by 10058-F4 were available for this data set and 70% correlated positively with N-Myc expression by the tumors and negatively with event-free survival and overall survival.
Holien et al (152) have studied the effect of the compound in multiple myeloma, which typically shows high-level deregulation of Myc for a variety of reasons (152–155). The authors tested 10058-F4 against 6 human myeloma cell lines and 12 patient-derived primary samples, and found that, irrespective of their origin, their responses could be classified as showing either relative sensitivity to the inhibitor (IC50 < 50 µM) or relative resistance (IC50s 50->100 µM). Although the correlation was imperfect, there was a tendency for those lines and primary cells expressing the highest Myc levels to fall within the latter group and, indeed the cells that were the least sensitive to 10058-F4 expressed the highest Myc levels. Holien et al (152) also demonstrated that culturing the cells on a monolayer of bone marrow stromal cells did not alter their sensitivity to 10058-F4 in contrast to IL-6 where such co-cultivation increased IL-6 resistance by 8-fold.
Similarities among direct Myc Inhibitors
Is it possible to say anything about the relationships amongst the direct Myc inhibitors? Other than those reported by Follis et al (140) and Hammoudeh et al (142), no studies have been performed to identify sites on Myc to which these compounds bind or, for that matter, whether they even bind to Myc (although this seems highly likely). However, the study of Hammoudeh et al (142) does suggest some similarities among Myc inhibitors. In addition to identifying the above-described three sites for Myc inhibitors, the investigator also showed that a dual-specific compound, 10019-D3, that disrupted both Myc-Max and Id2-E47 heterodimers (130) also bound to the 10058-F4 site (Myc402–412) with a Kd=11+/=4 µM. Interestingly 10019-D3 contains the same core pyrazolo[1,5-a]pyrimidine- 2-carboxamide core structure as Mycro1 and Mycro2, (134). Thus it is likely that Mycro1 and Mycro2 share 10058-F4’s binding site and, quite possibly, may also disrupt the Id2-E47 heterodimer. Taken together, these results suggest that there are only a limited number of binding sites on the Myc bHLH-ZIP domain capable of binding small molecules and preventing is association with Max.
In vivo studies of direct Myc inhibitors
Studies by Guo et al (156) and Claussen et al (157) have described the pharmacokinetics of 10058-F4 and 10074-G5 in mice while also determining their efficacies against human DU145 and PC3 prostate cancer and Burkitt’s lymphoma xenografts, respectively. In the first case, 10058-F4 was found to achieve peak plasma concentrations of ~300 µM following a single i.v. dose of 20 mg/Kg with a terminal half-life of approximately 60 min. Peak tumor concentrations were approximately 10-fold lower and at least eight distinct metabolites were identified. No significant effects on tumor growth were observed. Similarly, 20 mg/Kg of 10074-G5 administered as a single i.v. dose to mice bearing Daudi Burkitt’s lymphoma xenografts showed the compound to reach a peak plasma concentration of 58 µM and to have a plasma half-life of approximately 37 min. As in the case with 10058-F4, peak tumor concentrations were about 10-fold lower than obtained in the serum. As many as 20 metabolites were observed with a number of these appearing to be glucuronide derivatives of hydroxylated or nitro-reduced 10074-G5, thus suggesting that this parental compound is rapidly metabolized by hepatic phase I and phase II enzymes. Taken together, these studies indicated that, while both 10058-F4 and 10074-G5 are well-tolerated, their in vivo efficacy, at least against the tumors tested, were limited due to their rapid metabolism. Continued testing of selected analogs of 10058-F4 and 10074-G5 described above are currently underway
Despite these initial disappointing results, there is some cause for renewed optimism, particularly in the case of 10058-F4. This is based on the recent report of Zirath et al (149) who have shown that this compound, when administered at 20 mg/Kg as a daily ip dose, can double the survival of mice genetically engineered to develop neuroblastoma due to neuroectodermal targeting of N-Myc (158). These results were compatible with the group’s finding that 10058-F4 promotes a dose-dependent dissociation of N-Myc-Max heterodimers that mimics that obtained with Myc-Max. It is also entirely consistent with our own studies indicating that 10058-F4 can disrupt the association between L-Myc and Max and is therefore capable of targeting all Myc family members (H. Wang and EVP, unpublished). Preliminary in vitro studies performed in human neuroblastoma cell lines have indicated that several 10058-F4 analogs are 3–4-fold more potent than 10058-F4 as would have been predicted from the previous findings of Wang et al (139) (M. A. Henriksson and EVP, unpublished). Although we are currently unable to account for these different responses of neuroblastoma and prostate cancer to 10058-F4 (149,156), possible non-mutually exclusive explanations include differential sensitivities of Myc and N-Myc over-expressing cells, strain-specific differential metabolism of the compound or differences in the model systems (i.e. tumor xenografts versus transgenically-driven tumors). Arguing in favor of this latter possibility was the finding that 10058-F4 administration to mice bearing human neuroblastoma xenografts did not significantly affect tumor growth (149). Regardless of the basis, these experiments have revealed for the first time the in vivo efficacy of direct Myc inhibitors. Moreover, the effects appear comparable or even superior to those seen in other model systems with the indirect inhibitor JQ1 (see below).
Indirect Myc Inhibitors
Inhibitors of KAc recognition
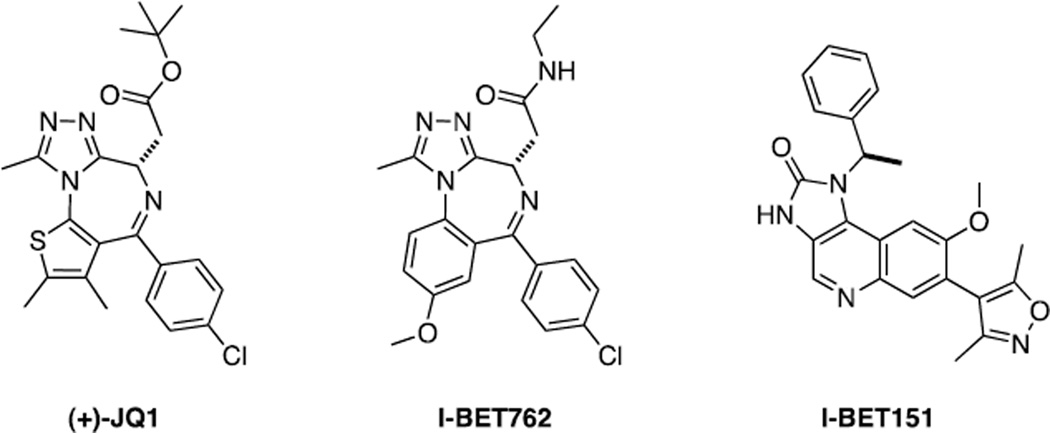
The finding that DNA-bound Myc-Max heterodimers associate with pTEFb and relieve promoter proximal pausing, that this is associated with changes in the histone acetylation profile of bound genes and that KAc side chains are recognized by BET subfamily members (BRD2, BRD3, BRD4 and BRDT) provided reason to suspect that targeting the association between BET proteins and KAc might be a rational way of inhibiting Myc-driven transcription (65,68,71,102,159,160). Indeed a direct role for BRD4 in oncogenic signaling had been previously established by demonstrating its involvement in a highly aggressive and incurable form of squamous cancer known as NUT midline carcinoma in which a t(15;19) chromosomal translocation results in an in-frame fusion between both N-terminal BRD4 bromodomains and the NUT protein (nuclear protein in testis) (102,161,162). Modeling ligand binding within the first bromodomain of BRD4, Filippakopoulos et al (163) and Miyoshi et al (164) developed (+)-JQ1, a thieno-triazolobenzo-1,4-diazepine that binds to the KAc-binding domain of all bromodomains of the BET family but not to the bromodomains of non-BET family members. A key feature of (+)-JQ1 is the 9-methyltriazole ring that functions as a bioisostere of KAc. Importantly, the tert-butyl ester group at C6 was deliberately incorporated to limit the interaction of (+)-JQ1 with other known benzodiazepine-binding proteins. Although (+)-JQ1 is arguably the most studied bromodomain inhibitor, there are now a large number of such compounds in the literature that can be classified based on their KAc mimetics (165). In particular, there are two main KAc mimetic chemotypes: the triazolobenzodiazepines, such as (+)-JQ1 and I-BET762, and the 3,5-dimethylisoxazoles, such as IBET-151 (Fig. 8). It was recently demonstrated that JQ1 displaced BRD4 from acetylated chromatin in cells and that it induced squamous differentiation and cell cycle arrest in a NUT cell line. Moreover, it also dramatically reduced the growth of three NUT xenografts in vivo as well as the growth of a xenograft obtained from newly derived patient material (163). Calculated in vitro IC50s were as low as 4 nM. Moreover, the compound was found to be 49% orally bioavailable.
Fig. 8.
Structures of BET domain inhibitors
Zuber et al (103) extended the above findings in a study that initially set out to identify epigenetic susceptibilities in AML. They used a relatively small (1094 member) doxycline-regulatable shRNA library directed against 243 known chromatin regulators and transduced this as a single pool into a AML cell line driven by MLL-AF9 and NrasG12D. Deep sequencing of the surviving cell population indicated shRNAs directed against BRD4 to be among the most consistently depleted. Zuber et al (103) then asked whether JQ1 might have the same effect and, indeed, found that 13 of 14 AML cell lines were highly susceptible to JQ1 (IC50s ≤500 nM) as were 15 of 18 primary AML samples, including three infant leukemias with MLL gene rearrangements. In all cases, JQ1 treatment led to apoptosis and/or monocytic maturation. Consistent with these findings, gene set enrichment analysis showed that JQ1-treated AML cells markedly up-regulated macrophage-specific genes and down-regulated those for leukemic stem cell marker genes. They also observed a down-regulation of Myc transcripts and protein that in some cases occurred within 60 min of JQ1’s addition, thus suggesting a direct effect. Both BRD4 shRNA expression and JQ1 treatment further down-regulated sets of genes that contained a broadly overlapping subset of known Myc targets. JQ1’s effect on Myc correlated with the loss of a BDR4-enriched chromosomal domain located approximately 2 kb upstream of the Myc transcriptional start site. Interestingly, the expression of ectopic Myc in JQ1-treated AML cells prevented many of the above-described phenotypic changes except for apoptosis. This suggests that even though the primary effect of JQ1 is to block KAc recognition by BRD4, the phenotypes that ensue are largely due to Myc down-regulation. Thus, the various phenotypic consequences of JQ1 treatment appear to require both BRD4 inhibition and Myc down-regulation. As long as Myc expression can be maintained in the face of JQ1 exposure, cells may utilize alternative means of recognizing KAc-tagged chromatin. A prediction of this model is that JQ1 and related agents should be ineffective against tumors driven by N-Myc (i.e neuroblastomas) unless the MYCN locus, like that of MYCC, contains BRD4 binding sites that positively regulates its expression. JQ1 should also be a much less efficient Myc inhibitor when Myc is subject to the control of exogenous promoters that do not harbor BRD4 binding sites such as those associated with some Ig enhancers or papilloma virus sequences. Parenteral JQ1 administration also prolonged the life span of mice bearing MLL-AF9 and NrasG12D AML xenografts although the increase in survival was <50% (103).
A subsequent study by Mertz et al (166) confirmed the sensitivity of most AML cell lines to JQ1 and extended the efficacy profile to multiple myeloma, which tends to be quite dependent on deregulated Myc over-expression (4,153,155). 14 of 15 myeloma cell lines were growth inhibited at JQ1 concentrations ranging from 100–500 nM. Gene expression arrays were performed in a Burkitt lymphoma and myeloma line after only several hours of exposure to JQ1 in order to demonstrate the earliest and most direct targets. Importantly, in control studies the transcriptional response to the inactive enantiomer of JQ1 was minimal, thus emphasizing the importance of KAc-binding by the active molecule. In contrast, treatment with the latter molecule was associated with the down regulation of a large number of genes that had been previously demonstrated to be E-box-containing Myc targets. The authors confirmed the findings of Zuber et al (103) that Myc down-regulation is a primary mediator of JQ1’s effects. Administration of JQ1 to animals bearing Burkitt lyphoma xenografts significantly slowed tumor growth rates and survival although, as had been previously seen by Zuber et al (103), the latter was extended by only about 50%. Somewhat more encouraging were the results of a tumor xenograft study using an AML cell line that had proven to be the most sensitive to JQ1 in in vitro experiments. Here, and at the optimal dose and timing employed, tumor xenografts essentially remained quiescent during the JQ1 treatment period. Although survival studies were not provided, it appears that the inhibition of tumor growth was of sufficient magnitude so as to profoundly affect animal longevity. These studies suggest that a minority of AML cells lines (and perhaps primary tumors as well) may be disproportionately susceptible to BET inhibitors. Finally, these authors also examined the response of N-Myc to JQ1 in a single MYCN-amplified neuroblastoma cell line. Consistent with the idea posited above that the N-Myc promoter may not contain BRD4-enriched sites, N-Myc transcript levels were reduced by only 50–60% versus the more typical >90% down-regulation observed in Myc-dependent tumors.
In parallel with the studies reported by Mertz et al. (166), Delmore et al (167) also examined the effects of JQ1 on multiple myeloma and provided additional evidence to potentially explain the disproportionate sensitivity to BRD4 inhibition in this disease. First, they found that BRD4 expression positively correlated with disease progression and that the BRD4 locus is frequently amplified. In at least one cell line, up-regulation of BRD4 was seen upon co-culturing the cells with bone marrow stromal cells suggesting that this higher level expression might be involved in imparting the characteristic osteotrophic behavior of multiple myeloma in the context of the bone marrow microenvironment. They then used global transcriptional profiling and unbiased gene set enrichment analysis to define the consequences of JQ1-mediated BET inhibition in three cell lines in which Myc was dysregulated by different mechanisms. In all cases, JQ1 mediated a selective effect on Myc-regulated gene expression as well a down-regulation of Myc itself. Known Myc-dependent biological modules such as ribosomal biogenesis and glycolysis, were also significantly down-regulated by JQ1. Moreover, and in a variation on the theme described Zuber et al (103), JQ1 treatment led to a loss of BRD4 concentrated on Ig enhancers in close proximity to translocated MYCC target genes as well as at transcriptional start sites. Additionally, 23 of 25 myeloma cell lines demonstrated high sensitivities to JQ1, (IC50s 68–500 nM) that was little changed when the cells were propagated in bone marrow stromal cells, which is known to confer resistance to diverse chemotherapeutic agent. (154,168). Three of five primary patient-acquired MM samples also underwent apoptosis with IC50s ranging from 400–800 nM. Finally, JQ1 was tested in several myeloma xenografts where, in addition to slowing tumor growth (based on bioluminescent imaging), it also reduced the abundance of serum M protein. However, the mean prolongation of survival between untreated and treated groups, while statistically significant, was only about 60% (ca. 22 days vs. 36 days).
Ott et al (169) extended the above findings to examples of pre-B ALL, the most common hematologic malignancy of childhood. An examination of 9 pre-B ALL cell lines with diverse chromosomal aberration showed all to be quite sensitive to JQ1 with some exquisitely so (ranges of IC50s=31–263 nM. In contrast to AML where JQ1 treatment led to a cell cycle arrest followed by eventual apoptosis, treated ALL cells experienced a more rapid onset of apoptosis and a more delayed and attenuated cell cycle arrest. As previously seen in AML cases, JQ1 treatment produced a rapid displacement of BRD4 from the MYCC promoter and the down-regulation of Myc RNA and protein. An examination of 32 validated Myc target genes showed most to be significantly down-regulated after as little as 4 hr of exposure to JQ1.
Two of most sensitive cell lines in the study by Ott et al (169) harbored translocations between the CRLF4 gene and the IgH region. CRLF4-IL7R heterodimers normally respond to thymic stromal lymphopoietin via activation of the JAK-Stat pathway and mutations along this entire pathway are not only relatively frequent in pediatric ALL but in some cases identify a particularly high-risk group (170). By analogy to multiple myeloma, where Myc down-regulation in response to JQ1 treatment can be traced to a loss of BRD4 binding to the IgH enhancer (167), Ott et al (169) failed to find similar sites in the translocated CRLF4 gene. However, they did find the IL7R gene promoter to be bound by BRD4, which was displaced after a relatively short period of exposure to JQ1 leading to a marked decline in IL7R cell surface protein. As expected, this effect was entirely unrelated to the status of the CRLF4 gene as it was seen in several cell lines with CRL4 rearrangements. These findings have important implications for future JQ1-directed therapies as they suggest that the responses of tumors (or lack thereof) may be highly contextual in nature and significantly determined by BRD4-dependent genes other than MYCC.
The full molecular consequences of IL7R role as a JQ1 target were more fully appreciated upon genome-wide expression profiling. This revealed that a series of genes containing Myc binding sites as well as a second set containing Stat5 binding sites were down-regulated in response to JQ1. In contrast, genes harboring NF-κB sites remained largely unaffected, thus attesting to JQ1’s specificity. Finally, in an elegant xenograft model that involved primary pre-B ALL cells with a P2RY5-IL7R translocation and no other mutations along the CRLF4-IL7R pathway, treatment with JQ1 for five days resulted in a marked reduction in phospho-Stat5 and decreases in both MYC and IL7R transcripts, with no significant changes in CRLF2 expression. Unfortunately, while JQ1 treatment normalized peripheral blood counts, it did not lead to a remission and treated mice experienced only a ca. 30% prolongation in survival. A likely explanation for this sub-optimal response was provided by an examination of splenic tissues from the JQ1-treated animals, which showed high level expression of both Myc and phospho-Stat5. Because only this single in vivo example was reported, it is not clear whether it represents the ultimate fate of other treated cases of ALL. Very clearly however, it provided the first evidence for the development of JQ1 resistance occurring via a “predictable” mechanism.
Very recently, Segura et (171) al have studied the effect of BRD4 inhibition in melanoma, a disease in which approximately 50% of advanced stage tumors are associated with B-Raf point mutations (typically V300E) that portend a highly unfavorable prognosis (172). A microarray analysis of 22 melanoma cell lines and 44 primary tumors showed a significantly elevated expression of Brd4 in tumors relative to that detected in either immortalized melanocytes or benign nevi. Moreover, although there was considerable overlap between transformed and benign samples, none of the latter showed levels of BRD4 expression higher than the mean of the former. A similar pattern was observed for BRD2. Approximately half the cell lines examined also showed evidence for BRD4 or BRD2 gene amplification (typically 2.5–4-fold). Two JQ1-related BRD4 inhibitors, MS417 and MS436, were next tested against 2 melanoma cells lines in which they promoted G0/G1 arrest and differentiation without apparent regard for B-Raf status. These results were confirmed by studies demonstrating that siRNA or shRNA-mediated suppression of BRD4, but not BRD2 or BRD3, were associated with significant cytostatic effect and loss of colony-forming ability in vitro and that the xenografts from the latter cells were smaller and had reduced Ki67 staining. Consistent with these findings, MS417 treatment of animals bearing melanoma xenografts led to a ca. 4-fold reduction in total tumor burden and reduced metastatic dissemination, although survival results were not presented. Transcriptional profiling performed on 3 melanoma cell lines prior to and after treatment with MS436 showed the expected downregulation of Myc RNA and protein and up-regulation of the negative cell cycle regulators and negative Myc targets p21CIP1 and p27KIP. Although gene ontology analysis indicated the most frequently affected genes were those involving cell cycle regulation, vascular development and chromatin assembly, it was not clear whether these were actual Myc targets.
Interestingly, the results in melanoma cells reported by Segura et al (171) showed wide variations in their sensitivities to MS417 and MS436. Moreover, MS417, which was the more potent, demonstrated the submicromolar IC50s typical for JQ1 (103,166,167) in only 3 of 7 tested cell lines and the IC50s for MS436 ranged from approximately 7–62 µM. Whether these disparities reflect a relative resistance of melanomas to BRD4 inhibitors in general or simply to these particular compounds remains to be determined.
Lack of response to JQ1
Despite their impressive effects against certain tumor types, there appear to be numerous cancer cell lines that are unaffected by or only partially responsive to BRD4 inhibitors. These include numerous ALLs, breast cancer, melanomas, osteosarcomas, and cervical adenocarcinomas (103,166). Although the reasons for such resistance have not been thoroughly explored, several explanations are possible. First, given the important role played by Myc down-regulation in response to JQ1, it is possible that that BRD4 binding locus in the MYCC gene promoter may simply not be as important in regulating gene transcription as it is more susceptible cells. It is also possible that some tumors are less reliant on BRD4 for the “reading” of epigenetic tags or on Myc itself for providing the scaffolding that supports transcriptional regulation.
Despite the initial excitement surrounding the initial identification of BET domain inhibitors as a potential treatment for Myc-driven cancers, considerable work remains. First, the tumor responses to these agents, while significant, have tended to be relatively minor (generally a <50% prolongation in survival and/or disease progression in experimental xenografts). To a large extent, this may reflect the current lack of in vivo optimization of these compounds. For example, the serum half-life of JQ1 is only about one hr, i.e comparable to that of 10058-F4 (103,156) and it remains unknown how well the compound penetrates tumors. It seems likely that medicinal chemistry-based approaches will be successful in increasing both the in vivo potencies and half-lives of new analogs.
Synthetic lethal Myc inhibitors
The basis of synthetic lethal interactions
Unlike the foregoing discussion which has centered around the direct disruption of Myc-Max complexes or the indirect inhibition of Myc by targeting acetylation, there are several recent reports in the literature of intriguing “synthetic lethal”-based approaches to targeting Myc. The topic of synthetic lethality has been recently reviewed (173,174) and will therefore not be discussed in detail here other than to remind the reader that, classically, it is defined as a state in which two mutant genes (typically within the same or closely cooperating pathways), either of which is compatible with survival, become incompatible when co-expressed. The definition has been adapted to cancer chemotherapy to explain how oncogenic mutations may sensitize cancer cells to certain chemotherapeutic agents while sparing normal cells. A classic example of synthetic lethality occurs in Brca1-mutated forms of breast cancer which, due to defective DNA repair, tend to be exquisitely sensitive to chemotherapeutic agents that inflict additional genotoxicity and thus further stress an already compromised pathway (175). In fact, such synthetic lethality need not necessarily be confined to cancer. For example, patients with Fanconi’s anemia can harbor heterogeneous mutations in the large group of proteins comprising the so-called FA complex, which cooperates with BRCA1 at stalled replication forks to excise and repair intra-strand links. Such defective DNA repair makes these patients highly susceptible to other DNA damaging agents, an observation that is used as part of the diagnostic evaluation for the disease (176).
To date, synthetic lethal screens have been performed primarily by siRNA or shRNA screening-based approaches. The anticipated outcome of such screens is that they would identify highly susceptible targets (e.g. proteins with enzymatic activity) for which small molecule inhibitors could then be readily devised or perhaps re-purposed from the current pharmacopoeia. Discussed below are several such examples indicating that the approach is not only feasible but can be used to identify novel targets and exciting therapeutic opportunities.
Inhibitors of GSK3β
As applied to Myc, the synthetic lethal approach was first used by Rottman et al (177) who sought to identify proteins that cooperate with the cell death receptor DR5, whose activation by the TNF-α–related protein TRAIL promotes Myc-induced apoptosis. The authors performed a rather limited siRNA screen of 624 siRNAs largely directed against the human kinome and sought to identify siRNAs that sensitized non-transformed Myc-over-expressing HA1E human kidney epithelial cells to an agonist monoclonal antibody against DR5. Among the 13 siRNAs that potentiated cell death by >2-fold were 2 that were directed against GSK3β. The differential effect on cell survival was confirmed with additional siRNAs as well as with 6-bromoindirubin-3-oxime (6B3O) and LiCl, both of which are known small molecule inhibitors of GSK3β. GSK3β, a Ser/Thr kinase, plays an important role in regulating Myc protein stability by directly phosphorylating Thr58. In turn, this forms a key recognition signal for FBW7, an F-box-containing component of the SCF (Skp–Cullin–F-box) ubiquitin ligase complex and a known tumor suppressor (178). Lack of Thr58 phosphorylation due to GSK3β inhibition would therefore stabilize Myc protein, as occurs in some Burkitt lymphoma-associated Myc mutations involving Thr58 (104). This higher level of Myc expression might be expected to sensitize cells to apoptosis mediated by an exogenous agent. Indeed, the authors found that siRNAs directed against GSK3β both reduced Myc Thr58 phosphorylation and increased Myc protein levels, a finding that was precisely recapitulated by siRNA-mediated knockdown of FBW7. The investigators also showed that a human colon cancer cell line with Myc deregulation (179) was also sensitized (both in vitro and in vivo) to DR5 agonists by GSK3β inhibition, but only if it was able to express FBW7. The authors speculated that the GSK3β–FBW7 axis acts as a type of buffer to limit the phenotypic consequences of deregulated Myc and further proposed that novel agents targeted against proteins in this pathway might serve as effective therapeutic agents for those tumors that over-express Myc. On the other hand it is well known that the ability of Myc to promote apoptosis is highly dependent on the normal function of the p53 pathway (180,181) thus suggesting that this strategy might face severe limitations given the frequency with which this pathway is impaired in human cancer (182).
MYRA-A and MYRA-B
Shortly following the report by Rottman et al (177) Mo and Henriksson (183) identified two structurally unrelated compounds termed MYRA-A and MYRA-B (“Myc pathway response agents”) that caused selective growth inhibition and/or apoptosis in Myc overexpressing cells (Fig. 9). The compounds were identified using mouse fibroblasts with a tetracycline-regulatable Myc transgene that allowed the compounds to be assessed in an otherwise isogenic background. Further evaluation showed the relative IC50s for both MYRA-A and MYRA-B to be ~5-fold lower in Burkitt lymphoma cells versus normal lymphoblastoid cell lines. Similar results were seen in rat fibroblasts where, as expected, myc−/− cells were highly resistant to the compounds, particularly MYRA-B. The authors also demonstrated that, in combination with Myc over-expression, MYRA-A and MYRA-B appeared to exert their selective effects by promoting apoptosis rather than proliferative arrest.
Fig. 9.
Structures of MYRA-A and MYRA-B
A closer examination of the molecular mechanisms underlying the above effects indicated distinct differences between MYRA-A and MYRA-B. For example, EMSA experiments revealed that MYRA-A did not, in fact, behave as a synthetic lethal but rather directly inhibited DNA binding by Myc-Max as well as by Mnt-Max and Max-Max, and inhibited Myc reporter vectors. In contrast, MYRA-B showed no activity against Myc-Max in either EMSA or reporter assays. Interestingly, despite MYRA-A’s inhibition of Myc-Max DNA binding, it did not interfere with the proteins’ dimerization suggesting that its effects on DNA binding were mediated by a structural alteration in the dimer formed between Max and its partners that prevents its proper recognition of an E-Box. Although structurally similar to the anthracyclines, MYRA-A apparently does not function through intercalation of DNA (and, therefore, as a topoisomerase II poison) since it did not affect the DNA binding of E-box-binding upstream stimulating factor (USF). Perhaps MYRA-A operates via the mechanism proposed for “credit card” inhibitors, in which small molecules binding to the Myc-Max heterodimer distorts its rigid structure just enough to cause a loss of DNA binding without altering PPI enough to promote dissociation.
Further interesting differences emerged when MYRA-A and MYRA-B were tested in two different classical Myc soft agar transformation assays using single-hit transformation of Rat1a fibroblasts by Myc alone or transformation of primary rat fibroblasts by co-expression of Myc+Ras. MYRA-A was highly potent at inhibiting colony formation and did so by >90% even at concentrations as low as 3 µM. Similar high potency effects were seen with MYRA-B against the former cell type whereas the latter cells were quite resistant, requiring concentrations as high as 50–100 µM to achieve noticeable suppression of colony formation. This suggested that the deregulated expression of Ras in the face of concurrent Myc over-expression could overcome any synthetic lethal advantage of MYRA-B, thus further implying that MYRA-B might target the MAPK/ERK pathway.
CDK1 inhibitors
Major cell cycle transitions are controlled by the cyclin-dependent Ser/Thr kinases (CDKs), particularly CDK1 and CDK2, in association with various cyclins such as D, E, A and B (184). Because cyclin B/CDK1 is particularly crucial for spindle assembly, chromatin condensation, nuclear envelope breakdown and mitosis Th’ng et al (185), Goga et al (186) examined the effect of small molecule CDK1 inhibitors on a panel of oncogene-transformed Rat1a fibroblasts. In response to the inhibitor purvalanol, they showed that Myc-transformed cells arrested in G2/M and underwent massive apoptosis whereas other cells tended to arrest in G2/M only. Similar studies performed in human retinal pigmented epithelial cells showed that, individually, both CDK1 inhibition and Myc over-expression were well-tolerated for as long as 5 days (<20% apoptosis) but that the combination lead to >70% apoptosis. Goga et al (186) then found that, while neither Ras over-expression nor TP53 status affected purvalanol+Myc-mediated apoptosis, it was completely rescued by Bcl-2 over-expression. A similar relationship between CDK1 inhibition and Myc deregulation was seen in several Myc-over-expressing human tumor cell lines in which the degree of apoptosis appeared to be directly related to the level of Myc expression. Additional evidence that the effects of small molecule CDK1 inhibitors were truly acting as assumed was provided by experiments showing that the inactivation of a temperature-sensitive CDK1 in a mutant cell line also led to massive apoptosis when Myc was concurrently over-expressed.
Goga et al (186) also examined primary lymphoma cells arising as a result of transgenic Eµ-Myc deregulation or of hepatomas arising in Tet-o-MYC/LAP-tTA mice. They demonstrated that exposure of the cells in vitro to purvalanol resulted in apoptotic cell death and that purvalanol treatment of mice bearing transplanted tumor cells significantly prolonged survival. Similar treatment of Tet-o-MYC/LAP-tTA mice led to a >3-fold reduction in macroscopically visible hepatomas and large increases in TUNEL-positive hepatocytes.
Realizing that the synthetic lethal combination of purvalanol and Myc was lowering the apoptotic threshold even further than occurs with Myc over-expression alone, Goga et al (186) next sought mechanistic insight by examining several members of the “inhibitors of apoptosis” (AIP) family of proteins which interfere with some of the same apoptosis pathways that are utilized by Myc (163). The found that exposure to purvalanol led to a rapid disappearance of one IAP, survivin, irrespective of Myc status. This finding agreed with previous work that CDK1-mediated phosphorylation stabilizes survivin (188) and was extended by showing that shRNA-mediated surviving depletion greatly sensitized RPE-Myc cells to apoptosis. Taken together, these studies suggested that CDK inhibitors, particularly those targeting CDK1, might prove to be useful and novel anti-neoplastic agents, particularly for tumors that express high levels of Myc, irrespective of their TP53 status. To date, however, the identification of specific and non-toxic CDK inhibitors that exert more than marginal effect as monotherapies have been slow to be developed (189).
Aurora B kinase Inhibitors
Given the above-discussed link between Cdk1 and survivin, Yang et al (190) have identified synthetic lethal relationship between Myc and Aurora B, a Ser/Thr kinase that, along with INCEP, Borealin and survivin, comprises the chromosomal passenger protein complex (CPPC) that is involved in the regulation of cytokinesis, spindle checkpoint function and chromosome segregation (191). They found that a small molecule inhibitor of Aurora B, VX-680, or siRNA knockdown of Aurora B or other components of the CPPC caused a profound apoptotic crisis in several Myc over-expressing cells, as well as cells that over-expressed adenovirus E1a, papillomavirus E6 and Notch, all of which deregulate Myc. A similar phenotype was observed in N-Myc-expressing cells. Cell death was TP53-independent and due to a combination of initial apoptosis followed by a latter phase of lethal autophagy. Over the course of their demise, the cells became multi-nucleated and polyploid and also showed evidence of centrosome amplification. They attributed this phenomenon to Myc’s well-known ability to promote illicit DNA synthesis and S-phase progression in the absence of an intervening mitosis (192,193) and demonstrated that blocking DNA synthesis with aphidicolin or density arrest strongly attenuated apoptosis. Finally, Yang et (190) al demonstrated that the administration of VX-680 to mice bearing Myc-driven T- or B-cell lymphomas increased survival and that this was associated with large increases in tumor cell apoptosis, polyploidy and autophagy. The authors emphasized that a potential advantage of the large role played by autophagy in promoting cellular demise in response to CPPC inhibition and Myc over-expression is that, unlike apoptosis, it is TP53-independent and might therefore affect a broader spectrum of tumors. Taken together, these studies highlight the potential of Aurora B kinase inhibitors for the treatment of cancers with clear evidence of Myc deregulation. Indeed, a number of such inhibitors are currently in various stages of clinical trials where they appear to be further advanced and to hold more promise than similar trials with CDK inhibitors (194).
Inhibitors of SUMOylation
Kessler et al (195) have recently conducted a genome-wide screen to identify targets whose inhibition promoted selective death in human mammary epithelial cells expressing a 4-hydroxytamoxifen (4HT)-inducibe MycER fusion protein (Myc-ER-HMECs). Approximately 75,000 bar-coded shRNAs in retroviral backbones were transduced in the above cells in the absence of 4HT followed by selection and expansion of the stably transduced cells for several generations in its presence. Comparison of the +/− 4HT populations by array hybridization of PCR-amplified barcodes indicated depletion of >400 shRNAs in response to Myc deregulation. Gene ontology analysis indicated the depleted population to be biased for shRNAs directed against ion channels and enzymes functioning in SUMOylation and mitotic spindle function. Interestingly, included among the latter group were shRNAs directed against GSK3β and FBW7 (see discussion above and refs. 177 & 190).
Kessler et al (195) next examined the SUMOylation pathway in more detail and, in particular SUMO-activating enzyme 2 (SAE2), and its heterodimeric partner protein SAE1 whose corresponding shRNAs were amongst the most highly depleted in Myc-ER-HMECs. Numerous shRNAs against SAE2 and SAE1 were found to profoundly affect the proliferation and survival of Myc-ER HMEC in the presence of 4HT. shRNAs targeting the SUMO E2-conjugating enzyme Ubc9, were also noted to be synthetically lethal with activated Myc. That the synthetic lethal phenotype of SAE2 could be rescued by constitutive over-expression of a wild-type, shRNA-resistant SAE2 cDNA, but not by its catalytically inactive counterpart, indicated that the inhibition of SUMOylation itself is the basis of the synthetic lethal effect.
More thorough exploration of the mechanisms underling the synthetic lethal relationship between SUMOylation and Myc over-expression came from studies showing that, in Myc-ER-HMECs treated with 4HT, cells arrested in G2/M, developed polyploidy and also accumulated typical features of mitotic stress including the acquisition of multipolar and abnormal mitotic spindles and lagging chromosomes that were strikingly reminiscent of the mitotic defects described above by Yang et al (190) in Myc over-expressing cells exposed to Aurora B kinase inhibitors.
Transcriptional profiling provided some insight into the underlying basis for SAE2’s relationship to Myc over-expression. Kessler at al (195) found that MycER activation in HMECs lead to changes in over 600 transcripts but that, in the face of SAE2 depletion, 86 (~22%) of the positively regulated transcripts in this collection were either not up-regulated or were actually repressed. Perhaps not surprisingly, 17 of the transcripts within this group encoded proteins with known function in mitotic spindle structure or function and further evaluation showed that shRNA-mediated depletion of 3 of the top 4 transcripts in this group were synthetically lethal with Myc over-expression.
Kessler et al (195) then evaluated the effects of SAE2 depletion in two breast cancer cell lines whose clonogenicity was adversely affected by Myc depletion and two where this was not the case. They found the conditional expression of SAE2 shRNA in the former cell lines to significantly compromise clonogenicity whereas this manipulation had no effect on the Myc-independent cell lines. Similarly, xenograft growth in immuno-compromised mice from the former cell lines, but not the latter, was also suppressed by SAE2 shRNA expression.
Finally, the workers compiled microarray data on nearly 1300 breast cancer patients, stratified it according to tumor expression of Myc, SAE1 and SAE2 expression and then correlated it with metastasis-free survival. Consistent with their previous results, they found that patients with the highest levels of Myc and the lowest levels of SAE1 and SAE2 had the longest survival. Indeed, compared to the cohort with high levels of all three transcripts, the prolongation in survival was remarkable: 70% of the patients in the former group survived 12 yr whereas comparable survival in the latter group was only five years. In contrast, levels of SAE1 and SAE2 transcripts were not correlated with survival in tumors with low levels of Myc expression.
Although the results of Kessler et al (195) point to the need for SUMOylation pathways to be intact to allow cancer cells to overcome high levels of Myc over-expression, the precise mechanism through which this occurs remains unclear. The simplest explanation would be that SUMOylation of some particular component of the mitotic spindle machinery is necessary to allow it to adapt to and withstand the incessant aberrant DNA replication and sustained mitotic demands imposed by Myc over-expression. In the absence of this post-translational modification, the mitotic apparatus is simply not up to the job, leading to the replicative mayhem that drives cellular demise. It will be of interest to learn whether these results and those of Yang et al (190) are eventually shown to share a common mechanism.
The use of mitotic spindle poisons in cancer chemotherapy has a long and distinguished history and includes older drugs such as the vinca alkaloids as well new agents such the taxanes (196). Unfortunately, the inability of these agents to distinguish between normal and neoplastic cells provides them with quite narrow therapeutic windows. The results provided by Yang et al (190) and Kessler et al (195) suggest that more refined and nuanced targeting of the mitotic spindle apparatus, particularly at upstream points, may be able to overcome this limitation in a manner that takes advantage of one of the molecular abnormalities that is so central to the cancer phenotype.
Anti-malarial compounds
Artemisinin and its derivatives are potent anti-malarial agents that have enjoyed widespread use because of their high safety and efficacy. They also display a remarkable ability to induce apoptosis in a large spectrum of cancer cell lines (197,198). Lu et al (199) screened the NCI-55 cancer cell panel with 4 artemisinin analogs and found artesunate to be particularly potent. They compared gene expression profiles in this cell line collection and found a strong correlation between Myc levels and sensitivity to artesunate as well as to 3 related but less potent artemisinin analogs, artemisinin, artemether and arteether and the common metabolite of all artemisinin compounds dihydroartemisinin (DHA). Further evaluation confirmed that HL60 promyelocytic leukemia cells and HCT-116 colo-rectal cancer cells, both of which express high levels of Myc, were exquisitely sensitive to DHA (IC50s 0.1–1 µM). In keeping with the findings of other small molecule Myc inhibitors, treatment of both cell lines was associated with drastic reductions in Myc protein levels and a Go/G1 arrest. The susceptibility HCT-116 cells to DHA was dramatically abrogated when endogenous Myc levels were reduced by shRNA-mediated knockdown. Conversely, enforced expression of Myc in the non-transformed NIH3T3 mouse fibroblasts sensitized them to the pro-apoptotic effects of DHA. An intriguing finding was that the removal of DHA after a 24 hr exposure did not lead to the re-expression of Myc or re-entry into the cell cycle; however the cells were followed for only 24 hr. Certainly, it would have been of interest to determine how long this effect persisted.
Further investigation of the mechanism underlying DHAs suppression of Myc protein came from the observation that the compound did not affect Myc transcript levels but rather destabilized Myc protein in a proteasome-dependent manner. Consistent with this mechanism, phosphorylation of Thr58 was increased upon treatment of HCT-116 cells with DHA even though total Myc protein declined. Inhibition of GSK3β with LiCl reversed Thr58 phosphorylation and stabilized Myc protein as did pharmacologic blockade of proteasome function. Collectively, these findings are consistent with the idea that artemisinin-like compounds increase the efficiency of GSK3β-mediate Myc phosporylation and promote its degradation. Because tumors with the highest levels of Myc are more likely to be “addicted” to the oncoprotein, even relatively small changes in Myc levels would be expected to be selectively detrimental to survival. These studies also emphasize the fact that the distinction between what we have classified as indirect Myc inhibitors and those working in a synthetic lethal manner may become blurred on occasion.
Max stabilizers
Jiang et al (200) have identified a unique group of indirect Myc inhibitors by capitalizing on the unique interaction profiles of Myc and Max. This is based on the fact that Myc homodimers do not form under physiologic conditions whereas Myc-Max heterodimers and Max homodimers form quite readily with the latter being the less stable (33,54). Jiang et al (200) used the AutoDock program (201) to screen the 1,700 chemical members of the NCI ChemDiv set for potential ligands for the Max homodimer. Several such compounds with low docking energies were identified and subsequent FRET analysis (129,132,200) indicated that they stabilized the Max homdomer, thereby potentially restricting its ability to provide a ready source of Myc partners. One compound in particular, termed NSC13728 (Fig. 11), strongly interfered with Myc-mediated transformation of CEFs and demonstrated not only considerable potency (IC50= 3 µM) but, at concentrations near this dose, as much as 100-fold greater specificity toward Myc-mediated transformation versus that mediated by Scr, Jun or PI-3-kinase. Co-immunoprecipitation experiments showed that NSC13728 reduced the abundance of Myc-Max heterodimers in cells and down regulated transcripts for several Myc-regulated genes such as those encoding cyclin D1, CDK4 and ornithine decarboxylase
Fig. 11.
Structure of the Max–Max homodimer stabilizer NSC13728.
Points of action of Myc inhibitors and the potential for synergy
The foregoing discussion should make clear that there are different modes of action for many small molecule Myc inhibitors. This leads to the obvious hypothesis that different classes of these agents may well work cooperatively (Fig. 10). For example, an agent such as JQ1 that inhibits MYCC gene transcription could be combined with one that interferes with Myc protein half life or with its dimerization in order to ensure a more complete and durable reduction of function protein levels, particularly in those cases where the tumor is known to be “Myc addicted”. This could be attained by virtue of the latter compounds serving to “mop up” any residual Myc protein that might escape MYCC’s transcriptional inhibition by the former agent. Combination therapy might also be potentially beneficial by allowing clinicians to minimize potential side effects by employing lower doses.
Fig. 10.
Summary of the distinct points of action and mechanisms of action of small molecule Myc inhibitors. Bold lettering depicts the time- and context-dependent changes in Myc and its downstream targets and function. Thus, the initial synthesis of Myc is followed by its heterodimerization with Max, its binding to chromatin and its association with and regulation of the transcriptional machinery to relive RNA PolII pausing. Deregulated transcription leads to the phenotypic changes that are characteristic of Myc-specific transformation, including aberrant DNA synthesis and the promotion of genomic instability. Green boxes indicate points where synthetic lethal interactions have been identified. Representative pharmacologic agents from each of the different categories of Myc inhibitors are depicted in red. The dotted arrow indicates that transcription of the MYCC gene itself is BRD4-dependent. Also note that synthetic lethal interactions can occur as a result of either increasing or decreasing Myc protein levels. In the first case, this is achieved by inhibiting GSK3β with compounds such as 6-bromoindirubin-3-oxime (6B3O). Increased Myc levels then render cells more sensitive to certain agents such as DR5 agonists. Myc degradation is promoted by agents such as artemisinin and its analogs. Although these increase Myc Thr58 phosphorylation, it is not clear if this is accomplished by a direct stiulation of GSK3β. See text for details as to the mechanism of action of specific inhibitors.
Indirect Consequences of Myc Inhibitor Treatment
In addition to incapacitating Myc-directed transcriptional regulation, small molecule Myc inhibitors have another consequence that reinforces their primary effect. Both direct inhibitors that target Myc and indirect inhibitors that target BRD4 ultimately lead to significant declines in total Myc protein and mRNA levels. While some of this might be attributable to a negative feedback on Myc expression that usually accompanies cells as they enter a non-proliferating quiescent state, other causes have been described. For example, we have already discussed the positive feedback control that BRD4 exerts on the MYCC gene promoter that explains the profound down-regulation of Myc mRNA and protein that accompanies treatment with JQ1. In Burkitt’s lymphoma both 10058-F4 and siRNA directed against Myc have been shown to induce a nearly 5-fold increase in the microRNA let-7a, which in turn negatively regulates Myc, presumably by targeting its stability and/or translation (202). This let-7a-mediated effect on Myc mRNA appears to be relatively common as it has also as occurring been described in both hepatoblastomas and AML (203,204). Interestingly, in the latter instance, let-7a was suppressed by activating the CXCR4 chemokine receptor by its ligand stromal-derived factor 1 leading to an up-regulation of Myc and an acquired resistance to the chemotherapeutic agent cytarabine (204). Consistent with all these findings are several reports implicating let-7a as a tumor suppressor whose expression is inversely correlated with that of Myc (205–207).
A final mechanism to account for the loss of Myc in response to both direct and indirect inhibitors may occur through the loss of its acetylation. In fully functional Myc transcriptional complexes that are associated with acetylated histone H3 and H4, Myc itself in acetylated by p300, GCN5 and TIP60 resulting in 3–4-fold increases in its stability (75,76,181). This suggests that there may be two separate pools of Myc protein within cells that are identifiable by virtue of their acetylation status: one pool is acetylated and stable and is bound to target genes whereas the second pool is unacetylated, unstable and unbound. It can be imagined that Myc inhibitors, irrespective of their mechanism of action, should reduce the level of the former, thus dramatically decreasing the half-life of the protein. It remains to be seen whether this plays a significant role is determining Myc stability and whether these three different and presumably non-mutually exclusive, mechanisms, function cooperatively.
Prospects for the future
Work on Myc inhibitors has progressed rapidly over the past several years. The earliest efforts, which were directed at identifying small molecules that prevent or disrupt the Myc-Max interaction, have progressed to the point of defining how these inhibitors interact with Myc at near atomicscale resolution. Significantly improved analogs of some of these compounds have been identified and efforts to improve their in vivo stability continue to progress. An outstanding question is whether synergistic efficacy can be attained by combinatorial approaches with inhibitors that target geographically distinct regions of Myc. An improved understanding of Myc’s role in coordinating the assembly of the transcriptional machinery has revealed a key susceptibility in the form of AcK and its recognition by BRD4. Improved analogs of current inhibitors will likely be a key focus in efforts to advance them to clinical trials. In addition, the realization that proper Myc-directed, unidirectional transcriptional read-through of Pol II-regulated target genes is reliant on the function of and cross-talk between numerous acetylases and kinases, has uncovered new potential indirect Myc targets. While the development of new inhibitors is never totally straight forward, the mere fact that these factors are enzymes suggests that the development of useful inhibitors may an easier task than has been the development of direct inhibitors. Finally, work showing that Myc-transformed cells are susceptible to synthetic lethality holds promise for the development of inhibitors against factors that regulate pathways which, in some cases, had already been known to be involved in Myc function. In most cases, however, the precise mechanisms by which these synthetic lethal compounds function to selectively target and eliminate Myc transformed cells remain murky but will likely be clarified in coming years. Additional combinatorial studies using collections of direct, indirect and synthetic lethal inhibitors could potentially be an intriguing way to wring every last drop of therapeutic benefit from this emerging field.
Highlights.
Myc is a nearly universal cancer target given its high expression in tumors
Because Myc is a transcription factor, its inhibition poses unique challenges
Two broad classes of Myc inhibitors have been identified: direct and indirect
Direct inhibitors interfere with the interaction between Myc and Max
Indirect inhibitors interfere with downstream Myc-dependent pathways
Acknowledgements
We thank members of both the Fletcher and Prochownik laboratories for their comments on this review. Our work in this area was supported by funding from the American Cancer Society and the American Chemical Society to SF, and by NIH grant R01 140624, the State of Pennsylvania Tobacco Fund and by NExT-CBC Project ID 562 all to EVP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin. Cancer Biol. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Delgado MD, León J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–616. doi: 10.1177/1947601910377495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and prostate cancer. Genes Cancer. 2010;1:617–628. doi: 10.1177/1947601910379132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 5.Pelengaris S, Khan M. The many faces of c-MYC. Arch. Biochem. Biophys. 2003;416:129–136. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Chen Y, Olopade OI. MYC and breast cancer. Genes Cancer. 2010;1:629–640. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukazawa T, Maeda Y, Matsuoka J, Yamatsuji T, Shigemitsu K, Morita I, Faiola F, Durbin ML, Soucek L, Naomoto Y. Inhibition of Myc effectively targets KRAS mutation-positive lung cancer expressing high levels of Myc. Anticancer Res. 2010;30:4193–4200. [PubMed] [Google Scholar]
- 9.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soucek L, Whitfield JR, Sodir NM, Massó-Vallés D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, Gudkov AV, Prochownik EV, Nikiforov MA. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene. 2008;27:1905–1915. doi: 10.1038/sj.onc.1210823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lefevre G, Calipel A, Mouriaux F, Hecquet C, Malecaze F, Mascarelli F. Opposite longterm regulation of c-Myc and p27Kip1 through overactivation of Raf-1 and the MEK/ERK module in proliferating human choroidal melanoma cells. Oncogene. 2003;22:8813–8822. doi: 10.1038/sj.onc.1207099. [DOI] [PubMed] [Google Scholar]
- 13.Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang D, Mannava S, Grachtchouk V, Tang WH, Patil S, Wawrzyniak JA, Berman AE, Giordano TJ, Prochownik EV, Soengas MS, Nikiforov MA. c-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27:6623–6634. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 17.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl. Acad. Sci. USA. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg T. Inhibition of transcription factors with small organic molecules. Curr. Opin. Chem. Biol. 2008;12:464–471. doi: 10.1016/j.cbpa.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 19.Prochownik EV, Vogt PK. Therapeutic Targeting of Myc. Genes Cancer. 2010;1:650–659. doi: 10.1177/1947601910377494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisen A, Claxton DF. Tyrosine kinase targeted treatment of chronic myelogenous leukemia and other myeloproliferative neoplasms. Adv. Exp. Med. Biol. 2013;779:179–196. doi: 10.1007/978-1-4614-6176-0_8. [DOI] [PubMed] [Google Scholar]
- 21.Kudchadkar RR, Smalley KS, Glass LF, Trimble JS, Sondak VK. Targeted therapy in melanoma. Clin. Dermatol. 2013;31:200–208. doi: 10.1016/j.clindermatol.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Nyberg P, Salo T, Kalluri R. Tumor microenvironment and angiogenesis. Front Biosci. 2008;13:6537–6553. doi: 10.2741/3173. [DOI] [PubMed] [Google Scholar]
- 25.Pello OM, De Pizzol M, Mirolo M, Soucek L, Zammataro L, Amabile A, Doni A, Nebuloni M, Swigart LB, Evan GI, Mantovani A, Locati M. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012;119:411–421. doi: 10.1182/blood-2011-02-339911. [DOI] [PubMed] [Google Scholar]
- 26.Pello OM, Chèvre R, Laoui D, De Juan A, Lolo F, Andrés-Manzano MJ, Serrano M, Van Ginderachter JA, Andrés V. In vivo inhibition of c-MYC in myeloid cells impairs tumor-associated macrophage maturation and pro-tumoral activities. PLoS One. 2012;7:e45399. doi: 10.1371/journal.pone.0045399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shchors K, Shchors E, Rostker F, Lawlor ER, Brown-Swigart L, Evan GI. The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1beta. Genes Dev. 2006;20:2527–2538. doi: 10.1101/gad.1455706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol. Cancer Res. 2012;10:1403–1418. doi: 10.1158/1541-7786.MCR-12-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 30.Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, Davis AC, Ihle JN, Cleveland JL. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He C, Hu H, Braren R, Fong SY, Trumpp A, Carlson TR, Wang RA. c-Myc in the hematopoietic lineage is crucial for its angiogenic function in the mouse embryo. Development. 2008;135:2467–2477. doi: 10.1242/dev.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitfield JR, Soucek L. Tumor microenvironment: becoming sick of Myc. Cell. Mol. Life Sci. 2012;69:931–934. doi: 10.1007/s00018-011-0860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fieber W, Schneider ML, Matt T, Kräutler B, Konrat R, Bister K. Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J. Mol. Biol. 2001;(307):1395–1410. doi: 10.1006/jmbi.2001.4537. [DOI] [PubMed] [Google Scholar]
- 34.Harvey SR, Porrini M, Stachl C, MacMillan D, Zinzalla G, Barran PE. Small-molecule inhibition of c-MYC:MAX leucine zipper formation is revealed by ion mobility mass spectrometry. J Am. Chem. Soc. 2012;134:19384–19392. doi: 10.1021/ja306519h. [DOI] [PubMed] [Google Scholar]
- 35.Michel J, Cuchillo R. The impact of small molecule binding on the energy landscape of the intrinsically disordered protein c-Myc. PLoS One. 2012;7:e41070. doi: 10.1371/journal.pone.0041070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 38.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 39.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: Introducing the D(2) concept. Annu. Rev. Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 40.Romero PR, Obradovic Z, Dunker AK. The complexity of disorder. Biophys. J. 1999;76(Part 2 of 2):A1–A525. [Google Scholar]
- 41.Cumberworth A, Lamour G, Babu MM, Gsponer J. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. Biochem. J. 2013;(454):361–369. doi: 10.1042/BJ20130545. [DOI] [PubMed] [Google Scholar]
- 42.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys. J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem. Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 44.Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr. Opin. Chem. Biol. 2010;14:481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 46.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr. Opin. Struct. Biol. 2011;21:441–446. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baudino TA, Cleveland JL. The Max network gone mad. Mol. Cell. Biol. 2001;21:691–702. doi: 10.1128/MCB.21.3.691-702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurlin PJ, Steingrìmsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–7028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahlström T, Henriksson M. Mnt takes control as key regulator of the myc/max/mxd network. Adv Cancer Res. 2007;97:61–80. doi: 10.1016/S0065-230X(06)97003-1. [DOI] [PubMed] [Google Scholar]
- 50.Bao J, Zervos AS. Isolation and characterization of Nmi, a novel partner of Myc proteins. Oncogene. 1996;12:2171–2176. [PubMed] [Google Scholar]
- 51.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 1995;4:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herkert B, Eilers M. Transcriptional repression: the dark side of myc. Genes Cancer. 2010;1:580–586. doi: 10.1177/1947601910379012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J, Banerjee A, Gross DJ. Assembly of b/HLH/z proteins cMyc, Max and Mad1 with Cognate DNA: Importance of Protein-Protein and Protein-DNA Interactions. Biochemistry. 2005;44:11855–11863. doi: 10.1021/bi050206i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 55.Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.John M, Leppik R, Busch SJ, Granger-Schnarr M, Schnarr M. DNA binding of Jun and Fos bZip domains: homodimers and heterodimers induce a DNA conformational change in solution. Nucleic Acids Res. 1996;24:4487–4494. doi: 10.1093/nar/24.22.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sánchez M, Jennings PA, Murre C. Conformational changes induced in Hoxb-8/Pbx-1 heterodimers in solution and upon interaction with specific DNA. Mol. Cell. Biol. 1997;17:5369–5376. doi: 10.1128/mcb.17.9.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muhle-Goll C, Nilges M, Pastore A. The leucine zippers of the HLH-LZ proteins Max and c- Myc preferentially form heterodimers. Biochemistry. 1995;34:13554–113564. doi: 10.1021/bi00041a035. [DOI] [PubMed] [Google Scholar]
- 60.O'Shea EK, Rutkowski R, Kim PS. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- 61.Thompson KS, Vinson CR, Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32:5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 62.Muhle-Goll C, Gibson T, Schuck P, Schubert D, Nalis D, Nilges M, Pastore A. The dimerization stability of the HLH-LZ transcription protein family is modulated by the leucine zippers: a CD and NMR study of TFEB and c-Myc. Biochemistry. 1994;33:11296–11306. doi: 10.1021/bi00203a027. [DOI] [PubMed] [Google Scholar]
- 63.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 64.Wells JA. Binding in the growth hormone receptor complex. Proc. Natl. Acad. Sci. USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 66.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 67.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell. Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 68.Gargano B, Amente S, Majello B, Lania L. P-TEFb is a crucial co-factor for Myc transactivation. Cell Cycle. 2007;6:2031–2037. doi: 10.4161/cc.6.16.4554. [DOI] [PubMed] [Google Scholar]
- 69.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faiola F, Wu YT, Pan S, Zhang K, Farina A, Martinez E. Max is acetylated by p300 at several nuclear localization residues. Biochem. J. 2007;403:397–407. doi: 10.1042/BJ20061593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 75.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, Blobel GA, McMahon SB. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang K, Faiola F, Martinez E. Six lysine residues on c-Myc are direct substrates for acetylation by p300. Biochem. Biophys. Res. Commun. 336:274–280. doi: 10.1016/j.bbrc.2005.08.075. (200) [DOI] [PubMed] [Google Scholar]
- 77.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 78.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 80.Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell. Biol. 2004;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 81.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell. Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 82.Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr. Opin. Cell. Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pirngruber J, Shchebet A, Johnsen SA. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle. 2009;8:3636–3642. doi: 10.4161/cc.8.22.9890. [DOI] [PubMed] [Google Scholar]
- 84.Zeng I, Zhou M-M. Bromodomain an acetyl lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 85.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 86.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 87.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 88.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000. Biol. Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schröder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnölzer M, Verdin E, Zhou MM, Ott M. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J. Biol. Chem. 2012;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cho S, Schroeder S, Ott M. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle. 2010;9:1697–1705. doi: 10.4161/cc.9.9.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lenasi T, Barboric M. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 2010;7:145–150. doi: 10.4161/rna.7.2.11057. [DOI] [PubMed] [Google Scholar]
- 94.Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol. Cell. Biol. 2013;33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 96.Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for Myc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Secombe J, Eisenman RN. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle. 2007;6:1324–1328. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- 98.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 99.Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol. Cell. Biol. 2005;25:1804–1820. doi: 10.1128/MCB.25.5.1804-1820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat. Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 105.Yano T, Sander CA, Clark HM, Dolezal MV, Jaffe ES, Raffeld M. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt’s lymphoma. Oncogene. 1993;8:2741–2748. [PubMed] [Google Scholar]
- 106.Clark HM, Yano T, Otsuki T, Jaffe ES, Shibata D, Raffeld M. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 1994;54:3383–3386. [PubMed] [Google Scholar]
- 107.Bahram F, von der Lehr N, Cetinkaya C, Larsson JG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 108.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;399:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 110.Dang CV. MYC, microRNAs and glutamine addiction in cancers. Cell Cycle. 2009;8:3243–3245. doi: 10.4161/cc.8.20.9522. [DOI] [PubMed] [Google Scholar]
- 111.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 112.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 113.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stites WE. Protein-protein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 118.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 119.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 120.Cheng Y, LeGall T, Oldfield CJ, Mueller JP, Van YY, Romero P, Cortese MS, Uversky VN, Dunker AK. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 121.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Curr. Opin. Chem. Biol. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meireles LM, Mustata G. Discovery of modulators of protein-protein interactions: current approaches and limitations. Curr. Top. Med. Chem. 2011;11:248–257. doi: 10.2174/156802611794072632. [DOI] [PubMed] [Google Scholar]
- 123.Gårdsvoll H, Danø K, Ploug M. Mapping part of the functional epitope for ligand binding on the receptor for urokinase-type plasminogen activator by site-directed mutagenesis. J. Biol. Chem. 1999;274:37995–38003. doi: 10.1074/jbc.274.53.37995. [DOI] [PubMed] [Google Scholar]
- 124.Billard C. BH3 Mimetics: Status of the field and new developments. Mol. Cancer Ther. 2013;12:1691–1700. doi: 10.1158/1535-7163.MCT-13-0058. [DOI] [PubMed] [Google Scholar]
- 125.Patel S, Player MR. Small-molecule inhibitors of the p53–HDM2 interaction for the treatment of cancer. Expert. Opin. Investig. Drugs. 2008;17:1865–1882. doi: 10.1517/13543780802493366. [DOI] [PubMed] [Google Scholar]
- 126.Yap JL, Chauhan J, Jung KY, Chen L, Prochownik EV, Fletcher S. Small-molecule inhibitors of dimeric transcription factors: Antagonism of protein–protein and protein–DNA interactions. Med. Chem. Commun. 2012;3:541–551. [Google Scholar]
- 127.Lanning M, Fletcher S. Recapitulating the α-helix: nonpeptidic, low-molecular-weight ligands for the modulation of helix-mediated protein-protein interactions. Future Med. Chem. 2013;5:2157–2174. doi: 10.4155/fmc.13.176. [DOI] [PubMed] [Google Scholar]
- 128.Adler MJ, Jamieson AG, Hamilton AD. Hydrogen-bonded synthetic mimics of protein secondary structure as disruptors of protein-protein interactions. Curr. Top. Microbiol. Immunol. 2011;348:1–23. doi: 10.1007/82_2010_91. [DOI] [PubMed] [Google Scholar]
- 129.Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. USA. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 131.Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, Irby R, Yeatman T, Courtneidge SA, Jove R. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg. Med. Chem. Lett. 2009;19:6038–6041. doi: 10.1016/j.bmcl.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, Janda KD. A credit-card library approach for disrupting protein-protein interactions. Bioorg. Med. Chem. 2006;14:2660–2673. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 134.Kiessling A, Sperl B, Hollis A, Eick D, Berg T. Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem. Biol. 2006;13:745–751. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 135.Hopewell R, Ziff EB. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner Max. Mol. Cell. Biol. 1995;15:3470–3478. doi: 10.1128/mcb.15.7.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kiessling A, Wiesinger R, Sperl B, Berg T. Selective inhibition of c-Myc/Max dimerization by a pyrazolo[1,5-a]pyrimidine. Chem Med Chem. 2007;2:627–630. doi: 10.1002/cmdc.200600294. [DOI] [PubMed] [Google Scholar]
- 137.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 138.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–48. [PubMed] [Google Scholar]
- 139.Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, Prochownik EV. Improved low molecular weight Myc-Max inhibitors. Mol. Cancer Ther. 2007;6:2399–2408. doi: 10.1158/1535-7163.MCT-07-0005. [DOI] [PubMed] [Google Scholar]
- 140.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 2008;15:1149–1155. doi: 10.1016/j.chembiol.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 141.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2000;61(Suppl 7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 142.Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 2009;131:7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- 143.Follis AV, Hammoudeh DI, Daab AT, Metallo SJ. Small-molecule perturbation of competing interactions between c-Myc and Max. Bioorg. Med. Chem. Lett. 2009;19:807–810. doi: 10.1016/j.bmcl.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 144.Yap JL, Wang H, Hu A, Chauhan J, Jung K–Y, Gharavi RB, Prochownik EV, Fletcher S. Pharmacophore identification of c-Myc inhibitor 10074-G5. Bioorg Med Chem Lett. 2013;23:370–374. doi: 10.1016/j.bmcl.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang H, Chauhan J, Hu A, Pendleton K, Yap JL, Sabato PE, Jones JW, Perri M, Yu J, Cione E, Kane MA, Fletcher S, Prochownik EV. Disruption of Myc-Max heterodimerization with improved cell-penetrating analogs of the small molecule 10074-G5. Oncotarget. 2013;4:936–947. doi: 10.18632/oncotarget.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cuchillo R, Michel J. Mechanisms of small-molecule binding to intrinsically disordered proteins. Biochem. Soc. Trans. 2012;40:1004–1008. doi: 10.1042/BST20120086. [DOI] [PubMed] [Google Scholar]
- 147.Barducci A, Bonomi M, Parrinello M. Metadynamics. Wiley Interdisciplinary. Reviews-Computational Molecular Science. 2011;1:826–843. [Google Scholar]
- 148.Piana S, Laio A. A bias-exchange approach to protein folding. J. Phys. Chem. B. 2007;111:4553–4559. doi: 10.1021/jp067873l. [DOI] [PubMed] [Google Scholar]
- 149.Zirath H, Frenzel A, Oliynyk G, Segerström L, Westermark UK, Larsson K, Munksgaard Persson M, Hultenby K, Lehtiö J, Einvik C, Påhlman S, Kogner P, Jakobsson PJ, Henriksson MA. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. USA. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, Prochownik EV. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS One. 2012;7:e37699. doi: 10.1371/journal.pone.0037699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Holien T, Våtsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120:2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 153.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, Mulligan G, Chesi M, Bergsagel PL, Fonseca R. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat. Rev. Cancer. 2007;7:585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 155.Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc. Natl. Acad. Sci. USA. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Guo J, Parise RA, Joseph E, Egorin MJ, Lazo JS, Prochownik EV, Eiseman JL. Efficacy, pharmacokinetics, tisssue distribution, and metabolism of the Myc-Max disruptor 10058-F4 [Z,E]-5-[4-ethylbenzylidine]-2-thioxothiazolidin-4-one, in mice. Cancer Chemother. Pharmacol. 2009;63:615–625. doi: 10.1007/s00280-008-0774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Clausen DM, Guo J, Parise RA, Beumer JH, Egorin MJ, Lazo JS, Prochownik EV, Eiseman JL. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics, and metabolism of 10074-G5, a novel small-molecule inhibitor of c-Myc/Max dimerization. J. Pharmacol. Exp. Ther. 2010;335:715–727. doi: 10.1124/jpet.110.170555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Lüscher B. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.French CA, Miyoshi I, Aster JC, Kubonishi I, Kroll TG, Dal Cin P, Vargas SO, Perez-Atayde AR, Fletcher JA. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19) Am. J. Pathol. 2001;159:1987–1992. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer research. 2003;63:304–307. [PubMed] [Google Scholar]
- 163.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miyoshi S, Ooike S, Iwata K, Hikawa H, Sugaraha K. International Patent No. PCT/JP2008/073864 (WO/2009/084693) 2009
- 165.Hewings DS, Rooney TP, Jennings LE, Hay DA, Schofield CJ, Brennan PE, Knapp S, Conway SJ. Progress in the development and application of small molecule inhibitors of bromodomain-acetyl-lysine interactions. J. Med. Chem. 2012;55:9393–9413. doi: 10.1021/jm300915b. [DOI] [PubMed] [Google Scholar]
- 166.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, Mitsiades N, Schlossman RL, Munshi NC, Kung AL, Griffin JD, Richardson PG, Anderson KC, Mitsiades CS. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat. Med. 2010;16:483–489. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, Kang H, Borowitz MJ, Camitta BM, Carroll AJ, Devidas M, Pullen DJ, Payne-Turner D, Tasian SK, Reshmi S, Cottrell CE, Reaman GH, Bowman WP, Carroll WL, Loh ML, Winick NJ, Hunger SP, Willman CL. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2012;119:3512–3522. doi: 10.1182/blood-2011-11-394221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, Gonzalez-Gomez P, Morante M, Jubierre L, Zhang W, Darvishian F, Ohlmeyer M, Osman I, Zhou M, Hernando E. BRD4 sustains proliferation and represents a new target for epigenetic therapy in melanoma. Cancer Res. 2013;73:6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ugurel S, Thirumaran RK, Bloethner S, Gast A, Sucker A, Mueller-Berghaus J, Rittgen W, Hemminki K, Becker JC, Kumar R, Schadendorf D. B-RAF and N-RAS mutations are preserved during short time in vitro propagation and differentially impact prognosis. PLoS One. 2007;2:e236. doi: 10.1371/journal.pone.0000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 174.Nijman SM, Friend SH. Cancer. Potential of the synthetic lethality principle. Science. 2013;342:809–811. doi: 10.1126/science.1244669. [DOI] [PubMed] [Google Scholar]
- 175.Comen EA, Robson M. Poly(ADP-ribose) polymerase inhibitors in triple-negative breast cancer. Cancer J. 2010;16:48–52. doi: 10.1097/PPO.0b013e3181cf01eb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kee Y, D'Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J. Clin. Invest. 2012;122:3799–3806. doi: 10.1172/JCI58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rottmann S, Wang Y, Nasoff M, Deveraux QL, Quon KC. A TRAIL receptor-dependent synthetic lethal relationship between MYC activation and GSK3beta/FBW7 loss of function. Proc. Natl. Acad. Sci. USA. 2005;102:15195–15200. doi: 10.1073/pnas.0505114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 179.Take Y, Kumano M, Teraoka H, Nishimura S, Okuyama A. DNA-dependent protein kinase inhibitor (OK-1035) suppresses p21 expression in HCT116 cells containing wild-type p53 induced by adriamycin. Biochem. Biophys. Res. Commun. 1996;221:207–212. doi: 10.1006/bbrc.1996.0575. [DOI] [PubMed] [Google Scholar]
- 180.Wang Y, Szekely L, Okan I, Klein G, Wiman KG. Wild-type p53-triggered apoptosis is inhibited by bcl-2 in a v-myc-induced T-cell lymphoma line. Oncogene. 1993;8:3427–3431. [PubMed] [Google Scholar]
- 181.Henriksson M, Selivanova G, Lindström M, Wiman KG. Inactivation of Myc-induced p53-dependent apoptosis in human tumors. Apoptosis. 2001;6:133–137. doi: 10.1023/a:1009644716727. [DOI] [PubMed] [Google Scholar]
- 182.Feng Z, Hu W, Rajagopal G, Levine AJ. The tumor suppressor p53: cancer and aging. Cell Cycle. 2008;7:842–847. doi: 10.4161/cc.7.7.5657. [DOI] [PubMed] [Google Scholar]
- 183.Mo H, Henriksson M. Identification of small molecules that induce apoptosis in a Myc-dependent manner and inhibit Myc-driven transformation. Proc. Natl. Acad. Sci. USA. 2006;103:6344–6349. doi: 10.1073/pnas.0601418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Malumbres M, Barbacid M. Cell cycle kinases in cancer. Curr. Opin. Genet. Dev. 2007;17:60–65. doi: 10.1016/j.gde.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 185.Th'ng JP, Wright PS, Hamaguchi J, Lee MG, Norbury CJ, Nurse P, Bradbury EM. The FT210 cell line is a mouse G2 phase mutant with a temperature-sensitive CDC2 gene product. Cell. 1990;63:313–324. doi: 10.1016/0092-8674(90)90164-a. [DOI] [PubMed] [Google Scholar]
- 186.Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat. Med. 2007;13:820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- 187.Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.O'Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc. Natl. Acad. Sci. USA. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Stone A, Sutherland RL, Musgrove EA. Inhibitors of cell cycle kinases: recent advances and future prospects as cancer therapeutics. Crit. Rev. Oncog. 2012;17:175–198. doi: 10.1615/critrevoncog.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 190.Yang D, Liu H, Goga A, Kim S, Yuneva M, Bishop JM. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc. Natl. Acad. Sci. USA. 2010;107:13836–13841. doi: 10.1073/pnas.1008366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.van der Waal MS, Hengeveld RC, van der Horst A, Lens SM. Cell division control by the chromosomal passenger complex. Exp. Cell. Res. 2012;318:1407–1420. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 192.Yin XY, Grove L, Datta NS, Long MW, Prochownik EV. c-Myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene. 1999;18:1177–1184. doi: 10.1038/sj.onc.1202410. [DOI] [PubMed] [Google Scholar]
- 193.Yin XY, Grove L, Datta NS, Katula K, Long MW, Prochownik EV. Inverse regulation of cyclin B1 by c-Myc and p53 and induction of tetraploidy by cyclin B1 overexpression. Cancer Res. 2001;61:6487–6493. [PubMed] [Google Scholar]
- 194.Farag SS. The potential role of Aurora kinase inhibitors in haematological malignancies. Br. J. Haematol. 2011;155:561–579. doi: 10.1111/j.1365-2141.2011.08898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, Rao M, Yu P, Dominguez-Vidana R, Liang AC, Solimini NL, Bernardi RJ, Yu B, Hsu T, Golding I, Luo J, Osborne CK, Creighton CJ, Hilsenbeck SG, Schiff R, Shaw CA, Elledge SJ, Westbrook TF. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dumontet C, Sikic BI. Mechanisms of action of and resistance to antitubulin agents: microtubule dynamics, drug transport, and cell death. J. Clin. Oncol. 1999;17:1061–1070. doi: 10.1200/JCO.1999.17.3.1061. [DOI] [PubMed] [Google Scholar]
- 197.Efferth T, Dunstan H, Sauerbrey A, Miyachi H, Chitambar CR. The anti-malarial artesunate is also active against cancer. Int. J. Oncol. 2001;18:767–773. doi: 10.3892/ijo.18.4.767. [DOI] [PubMed] [Google Scholar]
- 198.Efferth T, Olbrich A, Bauer R. mRNA expression profiles for the response of human tumor cell lines to the antimalarial drugs artesunate, arteether, and artemether. Biochem. Pharmacol. 2002;64:617–623. doi: 10.1016/s0006-2952(02)01221-2. [DOI] [PubMed] [Google Scholar]
- 199.Lu JJ, Meng LH, Shankavaram UT, Zhu CH, Tong LJ, Chen G, Lin LP, Weinstein JN, Ding J. Dihydroartemisinin accelerates c-MYC oncoprotein degradation and induces apoptosis in c-MYC-overexpressing tumor cells. Biochem. Pharmacol. 2010;80:22–30. doi: 10.1016/j.bcp.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 200.Jiang H, Bower KE, Beuscher AE, 4th, Zhou B, Bobkov AA, Olson AJ AJ, Vogt PK. Stabilizers of the Max homodimer identified in virtual ligand screening inhibit Myc function. Mol Pharmacol. 2009;76:491–502. doi: 10.1124/mol.109.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Biol. 1998;19:1639–1662. [Google Scholar]
- 202.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 203.Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM, Buendia MA. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc. Natl. Acad. Sci. USA. 2010;107:20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J. Clin. Invest. 2013;123:2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Akao Y, Nakagawa Y, Naoe T. Let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 206.Long XB, Sun GB, Hu S, Liang GT, Wang N, Zhang XH, Cao PP, Zhen HT, Cui YH, Liu Z. Let-7a microRNA functions as a potential tumor suppressor in human laryngeal cancer. Oncol. Rep. 2009;22:1189–1195. doi: 10.3892/or_00000554. [DOI] [PubMed] [Google Scholar]
- 207.Liu Y, Yin B, Zhang C, Zhou L, Fan J. Hsa-let-7a functions as a tumor suppressor in renal cell carcinoma cell lines by targeting c-myc. Biochem. Biophys. Res. Commun. 2012;417:371–375. doi: 10.1016/j.bbrc.2011.11.119. [DOI] [PubMed] [Google Scholar]