Abstract
Refractive error (RE) is a complex, multifactorial disorder characterized by a mismatch between the optical power of the eye and its axial length that causes object images to be focused off the retina. The two major subtypes of RE are myopia (nearsightedness) and hyperopia (farsightedness), which represent opposite ends of the distribution of the quantitative measure of spherical refraction. We performed a fixed effects meta-analysis of genome-wide association results of myopia and hyperopia from 9 studies of European-derived populations: AREDS, KORA, FES, OGP-Talana, MESA, RSI, RSII, RSIII and ERF. One genome-wide significant region was observed for myopia, corresponding to a previously identified myopia locus on 8q12 (p = 1.25×10−8), which has been reported by Kiefer et al. as significantly associated with myopia age at onset and Verhoeven et al. as significantly associated to mean spherical-equivalent (MSE) refractive error. We observed two genome-wide significant associations with hyperopia. These regions overlapped with loci on 15q14 (minimum p value = 9.11×10−11) and 8q12 (minimum p value 1.82×10−11) previously reported for MSE and myopia age at onset. We also used an intermarker linkage- disequilibrium-based method for calculating the effective number of tests in targeted regional replication analyses. We analyzed myopia (which represents the closest phenotype in our data to the one used by Kiefer et al.) and showed replication of 10 additional loci associated with myopia previously reported by Kiefer et al. This is the first replication of these loci using myopia as the trait under analysis. “Replication-level” association was also seen between hyperopia and 12 of Kiefer et al.'s published loci. For the loci that show evidence of association to both myopia and hyperopia, the estimated effect of the risk alleles were in opposite directions for the two traits. This suggests that these loci are important contributors to variation of refractive error across the distribution.
Introduction
Refractive errors (RE) are etiologically complex, multifactorial disorders characterized by a mismatch between the optical focal length of the eye and its axial length. This optical mismatch causes images to be focused away from the retina. The two major subtypes of spherical RE are myopia (nearsightedness) and hyperopia (farsightedness). Clinically significant myopia affects at least 25% of individuals over age 40 in the United States and western Europe, while hyperopia affects about 10% of individuals in this same age group [1]. Recent reports show that the prevalence of myopia has increased significantly in the United States over the last 3 decades; myopia of 2 (D) diopters or more was estimated to afflict 41.6% of Americans aged 12 to 54 years in 1999–2004, compared to only 25% in 1971–1972 [2]. The myopia epidemic is most acute in East Asia, where prevalence estimates of myopia (of at least 0.5 D) routinely surpass 70% among late teenagers and young adults [3], [4], [5]. A recent study of 19 year-old male military conscripts from Seoul, Korea, found that a staggering 96.5% were myopic [6].
The causes of RE are complex and are a combination of environmental and genetic factors [7]. Twin studies have reported a heritability greater than 0.50 for RE [8]. Several studies have calculated the heritability to be as high as 0.98 for myopia and 0.75 for hyperopia [9], [10], [11], [12]. The search for environmental factors influencing RE have mostly focused on myopia. These include near work and time spent outdoors during childhood and teenage years [13], [14], [15], [16].
Genome-wide association studies have become an essential tool in the study of traits such as RE, and to date there have been 67 published loci for refraction phenotypes [17]. In particular, Kiefer et al. [18] performed a genome-wide association study of myopia using self-reported age at onset in 45,771 participants and found 22 significant genome-wide associations. Verhoeven et al. [19] performed a genome wide association of the quantitative trait mean spherical equivalent (MSE) and found 24 significant genome-wide associations (2 of which were replications of previously published loci). [19]. Thirteen loci were genome-wide significant in both the Kiefer et al. and Verhoeven et al. studies [20].
Here we present the results of a genome-wide association meta-analysis of 2 dichotomous RE traits, myopia and hyperopia (adjusted for age, sex and years of education), in 9 populations: the Age-Related Eye Disease Study (AREDS), the Cooperative Health Research in the Region of Augsburg (KORA) the Framingham Eye Study (FES), Ogliastra Genetic Park-Talana (OGP-Talana) Study, the Multi-ethnic Study of Atherosclerosis (MESA), the Rotterdam Eye Studies I, II and III (RSI, RSII, RSIII) and the Erasmus Rucphen Family Study (ERF). These are termed the discovery meta-analyses of myopia and hyperopia hereafter. Eight of the discovery samples were previously included in the meta-analysis of refractive error by Verhoeven et al. [19]. One sample, the MESA study, was not included in either Kiefer et al. [18]or Verhoeven et al.'s [19], [21]studies. We attempted replication of significant and suggestive associations from the discovery meta-analyses through meta-analysis of association studies using these same trait definitions to these selected regions in 8 additional studies: the 1958 British Birth Cohort, the Blue Mountains Eye Study (BMES), the CROATIA-Vis Island Study, the CROATIA-Korcula Study, the Diabetes Control and Complications Trial (DCCT), the Orkney Complex Disease Study (ORCADES), the TwinsUK Study, and the Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR). All of these studies were previously included in the meta-analysis of refractive error by Verhoeven et al. [19]. Finally, we examined the results of our discovery meta-analyses of myopia and hyperopia in the regions found to be associated with myopia age at onset by Kiefer et al. [18]. In genetic association studies, the term replication is generally used to mean detection of statistical association of the same trait to the same associated genetic locus in an independent set of data. Here, we also use the term replication when discussing the results of our myopia trait (adjusted for age at examination, sex and years of education) since it is expected to be quite similar to the age at onset of myopia trait used by Kiefer et al. [18] in their study. We show independent replication of 11 of Kiefer et al.'s loci for myopia age at onset [18], and while our myopia trait is not exactly the same as that of Kiefer et al. [18], it is the closest phenotype available in our data. We also examined these same regions for association to hyperopia. The association to hyperopia would not constitute a “replication” of Kiefer et al.'s myopia findings, but association with this related trait may help to clarify the complex genetic underpinnings of refractive error.
Materials and Methods
Populations
The nine GWASs meta-analyzed in the discovery GWAS portion of this study included subjects aged 35–84 years from the Cooperative Health Research in the Region of Augsburg Study (KORA F3, Southern Germany), subjects aged 55–80 from the Age-related Eye Study (AREDS), unrelated subjects aged 28–84 from the Framingham Eye Study (FES), subjects aged 46-86 from the Multi-Ethnic Study of Atherosclerosis (MESA) study, and subjects aged 18–88 from the Ogliastra Genetic Park-Talana (OGP-Talana) study in Sardinia, subjects aged 55 and older from the Rotterdam Eye Study I, subjects aged 55 and older from the Rotterdam Eye Study II, subjects aged 45 and older from the Rotterdam Eye Study III, and subjects aged 18–86 from the ERF study, resulting in a total sample size of 16,830 individuals for the myopia analyses and 14,981 for the hyperopia analyses. All individuals were of European ancestry. This study involved meta-analysis of aggregate statistics from multiple studies. Approval was obtained by the local ethics committees for all studies, all studies were conducted according to the principles expressed in the Declaration of Helsinki and informed consent was obtained from the study participants at all study sites.
Study design
GWAS analyses of genotype data imputed to HapMap-II were performed for the traits myopia and hyperopia (adjusted for age at examination, sex and years of education) in 9 studies: the Age-Related Eye Disease Study (AREDS), the “Kooperative Gesundheitsforschung in der Region Augsburg” (KORA, “Cooperative Health Research in the Region of Augsburg”), the Framingham Eye Study (FES), the Ogliastra Genetic Park – Talana (OGP-Talana) study, the Multiethnic Study of Atherosclerosis (MESA) and the Rotterdam Eye Studies RSI, RSII, RSIII and the Erasmus Rucphen Family Study (ERF). The results from these analyses were then combined into a discovery meta-analysis GWAS of each trait. Fixed effects meta-analyses were performed with METAL [22] using p values and the effective sample size for each population. METAL calculates a genomic control value [23] for each population and then adjusts each population's results using the corresponding λ value. The discovery meta-analysis genome-wide significance threshold was taken to be 5×10−8.
In an attempt to replicate our discovery meta-analysis results and to increase the power of the analyses using our discovery dataset, we obtained association results from 8 other studies, the Blue Mountains Eye Study (BMES), CROATIA-Split, CROATIA-Vis Island, CROATIA-Korcula studies, the Diabetes Control and Complications Trial (DCCT), and the Orkney Complex Disease Study (ORCADES) (Supplemental Methods), just for 30 genomic regions that contained SNPs with association p-values less than 1×10−5 to either myopia (11 regions) or hyperopia (14 regions) or both (5 regions) in our discovery meta-analysis (the previously well-replicated association region on chromosome 15q14 was excluded). These studies all performed association of SNPs in these regions with myopia and hyperopia (adjusted for age at examination, sex, years of education when available and up to three principal components when there was significant evidence of population stratification in the data). A replication meta-analysis was performed using the same methods as above on association results in the novel genome-wide significant region for the hyperopia trait in these 8 additional datasets. An additional meta-analysis was then performed in these 30 regions combining results from the discovery datasets and these 8 additional studies. All 8 of these additional datasets were part of the Verhoeven et al. study of mean spherical equivalent. This additional analysis and these datasets are described in Materials S1–S3.
Quality control of discovery datasets
AREDS and KORA
Quality control measures are described elsewhere [24] but in brief: Individuals with chromosome abnormalities and sex discrepancies were removed. Cryptic relatedness was estimated by calculating pairwise identical by descent (IBD) coefficients. For each pair with a kinship coefficient of 0.125 or greater, one member of the pair was dropped based on genotyping rate and trait phenotype, preferring to retain the person with higher genotyping rates and more extreme phenotypes. Population stratification was assessed using principal components. Batch effects and patterns of missingness were eliminated by testing each batch against the others using Fisher's Exact test. As AREDS was a multi-center study, we also tested for differences between collection sites. Samples were dropped for poor performance on the array or a genotyping rate of <98%. SNPs were also removed from a population if its call rate was below 99%, its minor allele frequency was below 0.01, or if its distribution departed significantly from Hardy-Weinberg expectations (p<1×10−4) in a single population. We additionally dropped SNPs in both populations where HWE p <1×10−4 in 1 population and HWE p <1×10−3 in the other. SNPs were also excluded if they showed more than one genotype inconsistency between HapMap control samples and the consensus genotype in the HapMap database or investigator-provided duplicate samples.
Framingham Eye Study
Quality control measures are described elsewhere [24] but in brief: Samples were chosen based on pedigree information and genotyping quality. Samples with a genotypic call rate below 95% were not chosen for analysis. The mean call rate for analyzed samples was 99.2% (SD = 0.4%). The final marker list contained 436,494 high-quality SNPs with a minor-allele frequency> = 0.01, a Mendelian error rate below 2% across all pedigrees, a genotype call rate above 95%, and whose distribution was consistent with Hardy-Weinberg expectations (P>1×10−4).
MESA
For the MESA dataset, SNPs with MAF less than 0.02 or HWE p value less than 0.001 were removed from the analysis. Genotyping was performed using the Affymetrix Genome-Wide Human SNP Array 6.0. IMPUTE version 2.1.0 was used to perform imputation for the MESA Caucasian participants (chromosomes 1–22) using HapMap Phase I and II - CEU as the reference panel (release #24 - NCBI Build 36 (dbSNP b126)). SNPs with genotype call rate less than 0.95, MAF less than 0.02, HWE p value less than 0.001, or oevar less than 0.3 were removed from the analysis. Association tests were performed by SNPTEST v2 (Marchini et al., 2007).
OGP-Talana
Quality control of the SNP data was performed using the GenABEL software package in R. Samples with overall SNP call rate <93%, with minor allele frequency <0.01, with Hardy-Weinberg P value>10−6, showing excess heterozygosity, or being classified as outliers by allelic identity-by-state (IBS) clustering analysis, were excluded.
Rotterdam eye studies I,II and III
Subjects with cataracts and history of cataract or refractive surgery were excluded from the study. DNA was extracted from blood leucocytes according to standard procedures. Genotyping of SNPs was performed using the Illumina Infinium II HumanHap550 chip v3.0 array (RS-I); the HumanHap550 Duo Arrays and the Illumina Human610-Quad Arrays (RS-II), and the Illumina Human 610 Quad Arrays (RS-III). Samples with low call rate (<97.5%), with excess autosomal heterozygosity (>0.336), or with sex-mismatch were excluded, as were outliers identified by the identity-by-state clustering analysis (outliers were defined as being>3 s.d. from population mean or having identity-by-state probabilities>97%). GWAS analyses were performed using GRIMP.
Erasmus rucphen family study
Subjects with cataracts and history of cataract or refractive surgery were excluded from the study. DNA was genotyped on one of four different platforms (Illumina 6k, Illumina 318K, Illumina 370K and Affymetrix 250K). Samples with low call rate (<97.5%), with excess autosomal heterozygosity (>0.336), or with sex-mismatch were excluded, as were outliers identified by the identity-by-state clustering analysis (outliers were defined as being>3 s.d. from population mean or having identity-by-state probabilities>97%). GWAS analyses were performed using the ProbABEL package from the ABEL set. A lambda correction was performed to adjust for cryptic relationship.
Genotype imputation of data
To produce a consensus set of genotypes for imputing to the HapMap-II, AREDS and KORA high quality SNPs were filtered to those present on HapMap-II. Imputation to the HapMap-II reference panel (CEU population release 22, NCBI build 36) was performed in MACH [22], [25] in 2 stages. Stage one was the model parameter estimation stage which used a random sample of 300 individuals from each population, using the greedy option which only uses the reference haplotypes (supplied here from the HapMap) and 100 Markov Chain iterations. Stage two is the actual imputation stage and uses the model parameters estimated in stage one to speed up the imputation of the genotypes. After imputation, the remaining high quality genotyped SNPs were merged back in with the SNPs from the imputation procedure for the AREDS and KORA data. For the FES data, genotype imputation to the HapMap-II reference panel (CEU population release 22, NCBI build 36) was carried out in a two-step process using the Markov Chain Haplotyping (MACH version 1.0.16.a) software. First, crossover and error-rate maps were built using 400 unrelated individuals (200 male and 200 female) sampled from FHS subjects. Second, genotype imputations of approximately 2.5 million autosomal HapMap-II SNPs were carried out on the entire FHS dataset using parameters estimated from step 1. For MESA, IMPUTE version 2.1.0 was used to perform imputation for the Caucasian participants (chromosomes 1-22) using HapMap Phase I and II - CEU as the reference panel (release #24 - NCBI Build 36 (dbSNP b126)). For OGP-Talana, using the phase II CEU HapMap individuals (release 22, NCBI build 36) as reference panel for imputation, genotypes were imputed for nearly 2.5 million SNPs using MACH. SNPs imputed with Rsq <0.3 were excluded. For RSI,II and III and ERF, a set of genotyped input SNPs with call rate>98%, with minor allele frequency>0.01, and with Hardy-Weinberg P value>10−6 was used for imputation. We used the Markov Chain Haplotyping (MACH) package version 1.0.15 software (Rotterdam, The Netherlands; imputed to plus strand of NCBI build 36, HapMap release #22) for the analyses. For each imputed SNP, a reliability of imputation was estimated as the ratio of the empirically observed dosage variance to the expected binomial dosage variance (O/E ratio).
Data analysis
Genetic association was estimated by fitting a logistic regression model separately to the traits myopia and hyperopia. To create the dichotomous traits, we calculated mean spherical equivalent (MSE) as the average of spherical equivalent (SE) of refraction between the two eyes, or the single SE value for persons with only a single SE measurement. For myopia, cases were defined as MSE <−1D, controls>0D and individuals between 0D and −1D coded as unknown. For hyperopia, cases were defined as MSE>+1D, controls <0D and individuals between 0D and +1D coded as unknown. A general additive genetic model was used to code the SNP effect (i.e. SNPs were coded according to the number of minor alleles [0,1,2] for each person); covariates included age; sex; and years of education. For AREDS, KORA and FES, this was accomplished using the PLINK (version 1.07) statistical software (http://pngu.mgh.harvard.edu/~purcell/plink) [26]. For AREDS analyses, the first three principal components (eigenvectors) of the EIGENSTRAT analysis were also included along with the covariates listed above. For MESA, these association tests were performed by SNPTEST v2.52. For OGP-Talana, all regression models were run using the ProbABEL package from the ABEL set of programs which adjusts jointly for cryptic relationship and population stratification. For RSI, II and III and ERF, we used genomic control [23] to obtain optimal and unbiased results and applied the inverse variance method of each effect size estimated for both autosomal SNPs that were genotyped and imputed in both cohorts.
Association analyses were performed for both traits and a genome-wide meta-analysis was performed on the 9 populations and 8 replication data sets (Blue Mountains Eye Study, Croatia Vis Island Study, Croatia Korcula Study, Diabetes Control and Complications Trial, Orkney Complex Disease Study, UK Twins Study, 1958 British Birth Cohort, Wisconsin Epidemiologic Study of Diabetic Retinopathy). Details of the genome-wide analyses of the individual discovery datasets and the replication analyses are shown in the supplemental methods and results including QQ-plots and Manhattan plots for each of the discovery cohorts in Figures S1-S9. Figure S10 is a flowchart showing the workflow of the entire study.
SNP selection for replication
Thirty genomic regions that contained SNPs with association p-values less than 1×10−5 to either myopia (11) or hyperopia (14) or both (5) in our discovery meta-analysis (excluding the 15q14 region) were chosen for replication or further study in the 8 additional datasets. We analyzed all SNPs within a 500 kb window centered on the most significant SNP in each region from the discovery meta-analysis.
For the comparison of our discovery meta-analysis results with the myopia age at onset loci from the Kiefer et al. [18] study, a list of strongly associated variants that were genome-wide significant (p≤5×10−8) or suggestive (p<1×10−6) in Kiefer et al. [18] was selected. We analyzed all SNPs within a 500 kb window centered on these replication SNPs in our data.
Calculation of effective number of tests and replication significance thresholds
It has become increasingly clear that only attempting to replicate the exact SNPs found to be genome-wide significant in a discovery GWAS can produce a failure to replicate due to underlying differences in linkage disequilibrium (LD) and allele frequencies [27], [28], even in populations self-identified as having the same ethnicity. Ioannidis et al. [29] have shown that restricting replication efforts to only a few of the most significant SNPs from an associated region leads to less robust information for those loci. The resulting failure to replicate may be because those selected SNP(s) are not necessarily more informative or closer to the causal variant than other SNPs in the region. Several approaches to this problem have been proposed, including incorporating linkage information [30], pathway-based association [31] and other methods which use multiple SNPs in the analysis [32], [33], [34], [35], [36], [37], [38], [39]. A linkage disequilibrium (LD) based binning strategy, proposed by Christoferou [39] may prove to be the most useful. However, the issues of handling SNPs which map to more than one gene due to overlapping reading frames and the correlations between genes and derivative gene scores still need to be resolved. Until that problem has a solution, it may be more powerful to study a dense panel of SNPs from each associated region, and utilize imputation to the latest version of 1000 Genomes data to provide additional genotypes to harmonize available SNPs across studies even when genotyped on different platforms. Here we selected all SNPs that were within a specified window of the original SNP and used the method of Ramos et al. [40] to model the LD structure in one of the replication populations to calculate the effective number of independent tests being performed across all of our replication regions. Traditional methods of correcting for multiple comparisons, such as the widely used Bonferroni correction considering all SNPs tested, are notoriously conservative because they do not take intermarker correlation fully into account but treat all the tests as independent. By using the effective number of independent tests in a Bonferroni correction, Type I error is still controlled and power is improved. Various approaches to calculating the effective number of independent tests when using such a regional replication strategy have been proposed since many of the SNPs in such a region are in LD with each other and do not represent independent tests [41], [42], [43], [44], [45], although many of these approaches are still overly conservative. The Ramos et al. [40] approach properly accounts for SNP interdependence, allows computation of the effective number of independent tests for very large numbers of highly correlated SNPs and is less computationally intensive than permutation-based methods. We used the method of Ramos et al. [40] to calculate the number of effective tests (Neff) in all the replication regions and divided α by this effective number of tests to calculate the significance threshold separately in the AREDS, KORA and Framingham datasets. The Ramos method calculates Neff by first estimating the KxK covariance matrix for the K SNPs in the replication regions using the genotype data. Then the covariance matrix is spectrally decomposed to calculate the eigenvalues. The effective number of tests is then estimated using the relationship
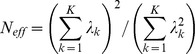 |
in which λk is the kth eigenvalue of the K×K covariance matrix for the K SNPs [46]. The Bonferroni-corrected significance threshold is then calculated as α/Neff.
The markers in each region are very densely spaced, with high levels of LD between markers in each block. The calculations from the AREDS data gave the largest effective number of tests and thus the most conservative Bonferroni-corrected significance threshold; thus this was chosen as our significance threshold for our replication studies. However, the Bonferroni-corrected thresholds derived by applying this method to the KORA and Framingham data were only slightly less conservative than the threshold derived from the AREDS data.
Results
After all quality control measures and appropriate association analyses, genome-wide association results from Caucasian participants in the AREDS, KORA, FES, OGP-Talana, MESA, RSI, RSII, RSIII and ERF studies were combined in a genome-wide discovery meta-analysis totaling 16,830 individuals for myopia and 14,981 individuals for hyperopia. Table 1 describes the characteristics of the populations after classifying participants into myopia, hyperopia, control or unknown categories.
Table 1. Baseline Characteristics of the nine populations.
| AREDS | KORA | FES | MESA | OGP-TALANA | RS1 | RS2 | RS3 | ERF | Total | |
| N | 1877 | 1869 | 1389 | 1462 | 683 | 5238 | 2009 | 1970 | 2028 | 18525 |
| Mean Age (SD) | 68.0 (4.7) | 55.6 (11.8) | 55.6 (8.9) | 61.9 (9.4) | 42.2 (19.1) | 68.5 (8.6) | 64.2 (7.4) | 60.8 (5.5) | 48.5 (14.3) | |
| N Myopia1 Cases | 346 | 550 | 348 | 486 | 71 | 763 | 395 | 594 | 370 | 3923 |
| Myopia Cases MSE2 | −2.81 | −2.72 | −3.08 | −3.20 | −4.41 | −3.21 | −3.08 | −3.22 | −3.03 | |
| N Myopia Controls | 1333 | 840 | 773 | 731 | 428 | 3964 | 1374 | 1056 | 1197 | 11696 |
| Myopia Controls MSE2 | 1.59 | 1.38 | 1.60 | 1.70 | 1.38 | 1.88 | 1.72 | 1.39 | 1.24 | |
| N Myopia Unknown | 198 | 479 | 268 | 245 | 184 | 601 | 240 | 320 | 461 | |
| N Hyperopia3 Cases | 854 | 424 | 426 | 506 | 64 | 2779 | 919 | 556 | 540 | 7068 |
| Hyperopia Cases MSE2 | 2.56 | 2.30 | 2.31 | 2.21 | 2.42 | 2.48 | 2.33 | 2.23 | 2.29 | |
| N Hyperopia Controls | 600 | 1010 | 654 | 714 | 153 | 1350 | 627 | 907 | 829 | 6844 |
| Hyperopia Controls MSE2 | −1.76 | −1.79 | −1.92 | −2.32 | −2.56 | −2.00 | −2.09 | −2.21 | −1.52 | |
| N Hyperopia Unknown | 423 | 435 | 309 | 242 | 466 | 1109 | 463 | 507 | 659 | |
| Sex (% Male) (Myopia/Hyperopia) | 41/40 | 50/49 | 42/41 | 54/43 | 41/40 | 48/39 | 49/44 | 46/43 | 53/62 | |
| Myopia λ | 1.001 | 0.997 | 1.004 | 1.024 | 1.085 | 1.020 | 1.018 | 1.015 | 1.323 | 1.038 |
| Hyperopia λ | 1.020 | 1.023 | 0.997 | 1.017 | 1.156 | 1.041 | 1.024 | 1.010 | 1.254 | 1.046 |
1. For myopia, cases were defined as MSE <−1D, controls>0D and individuals between 0D and −1D coded as unknown.
2. Average MSE of all cases or controls used in the analyses.
3. For hyperopia, cases were defined as MSE>+1D, controls <0D and individuals between 0D and +1D coded as unknown.
Testing for population stratification using EIGENSOFT and principal components analysis found no evidence of population stratification in KORA, but some evidence of substructure was detected in the AREDS, FES and MESA studies. These were adjusted for in the genome-wide association analyses by including the first three principal components from the PCA as covariates in our regression models. The OGP-Talana data were also adjusted for cryptic relatedness using the ProbABEL R package. For ERF and RS1–3, the population was assumed to be homogeneous and outliers excluded. Genomic control [23] values (λ) calculated by METAL [47] for each population prior to meta-analysis for each trait are given in Table 1. These values were used by METAL to adjust each population's results before including in the fixed effects meta-analysis. The QQ plots of the meta-analysis p values (Figure 1a and Figure 2a) showed some deviation from the null. However, the genomic control method [23] was used to further control for population stratification and inter-population differences in the final meta-analysis. The variance inflation factors calculated by METAL [47] for the final meta-analysis across the nine cohorts for myopia and hyperopia were 1.038 and 1.046 respectively. Lambda values ranging from approximately 0.95 to 1.1 are considered desirable.
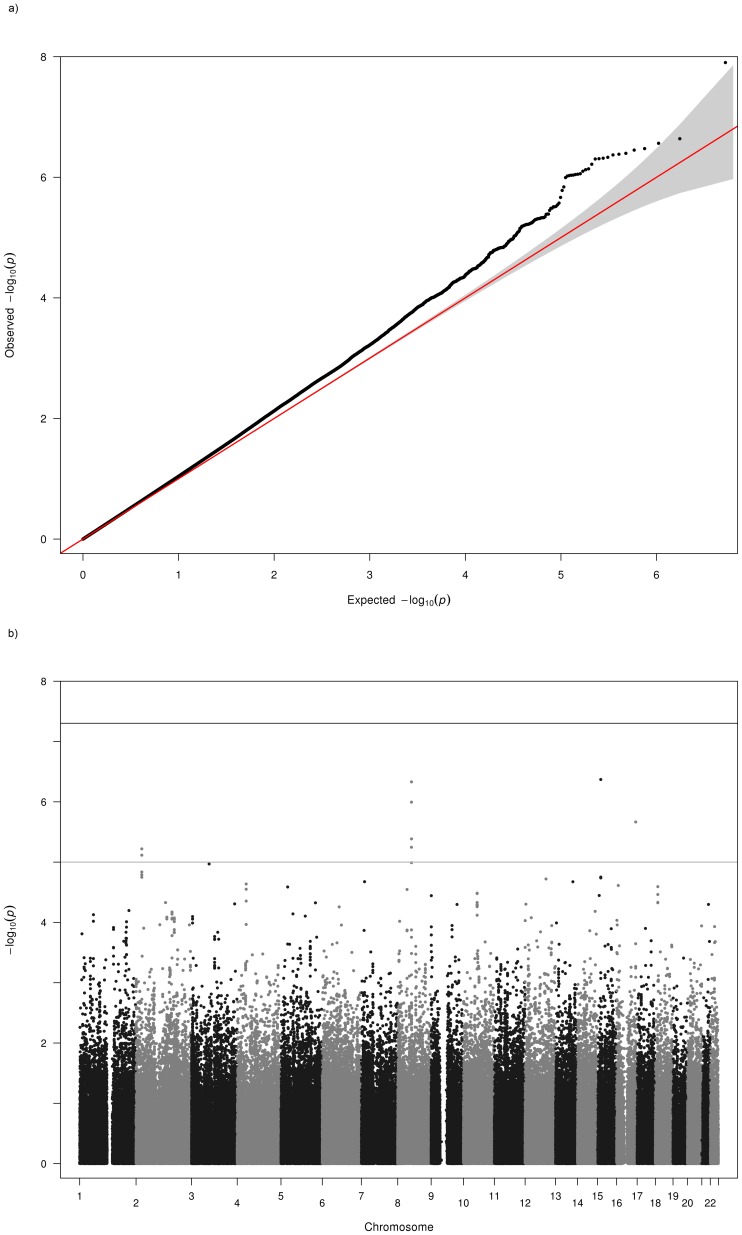
Figure 1. Q-Q and Manhattan Plots for the myopia analysis of all cohorts.
a) Q-Q plot for association between all SNPs analyzed and myopia in the meta-analysis. Each dot represents an observed statistic (defined as -log10 P) versus the corresponding expected statistic. The red line corresponds to the null distribution. b) Manhattan plot for association between all SNPs analyzed and myopia in the meta-analysis. Each dot represents an observed statistic (defined as -log10 P). The darker gray line corresponds to the genome-wide significance threshold and the lighter gray line represents the suggestive threshold.
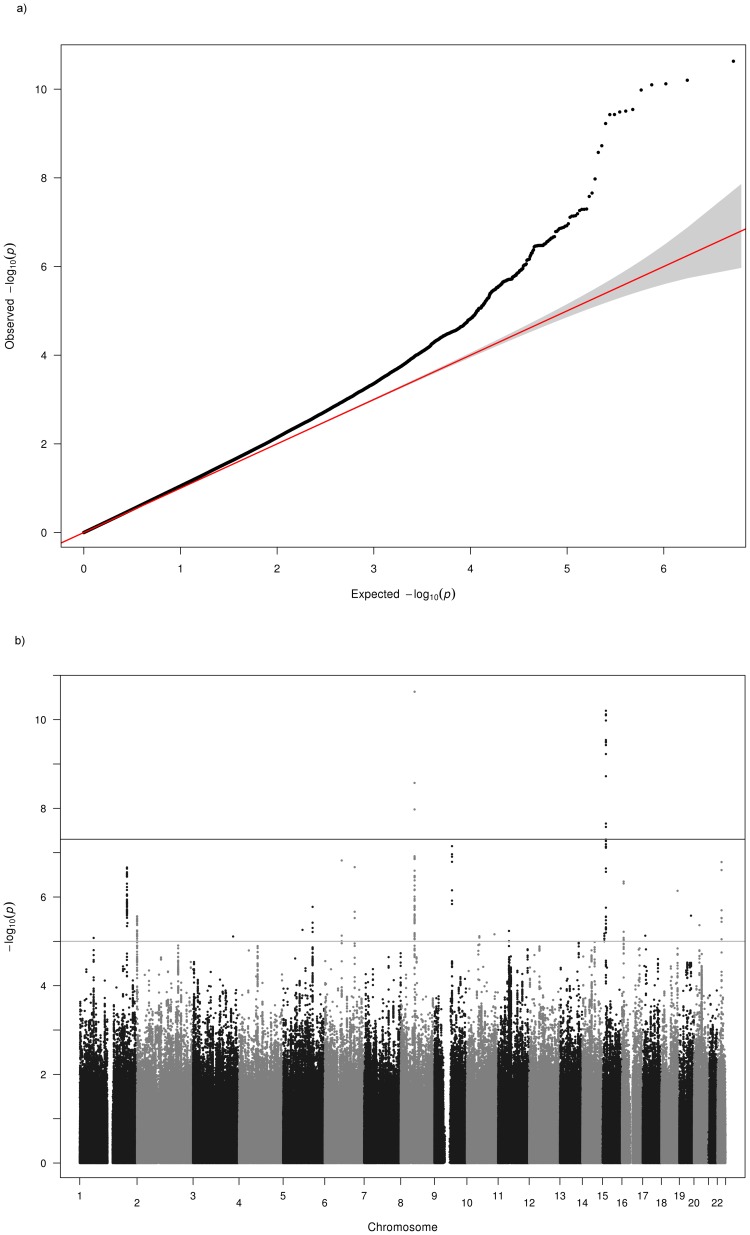
Figure 2. Q-Q and Manhattan Plots for the hyperopia analysis of all cohorts.
a) Q-Q plot for association between all SNPs analyzed and hyperopia in the meta-analysis. Each dot represents an observed statistic (defined as -log10 P) versus the corresponding expected statistic. The red line corresponds to the null distribution. b) Manhattan plot for association between all SNPs analyzed and hyperopia in the meta-analysis. Each dot represents an observed statistic (defined as -log10 P). The darker gray line corresponds to the genome-wide significance threshold and the lighter gray line represents the suggestive threshold.
Results of the genome-wide meta-analyses are shown in Figure 1b and Figure 2b and results for each sample separately are given in Figure S1 (AREDS), Figure S2 (KORA), Figure S3 (FES), Figure S4 (MESA), Figure S5 (OGPT), Figure S6 (RS-I), Figure S7 (RS-II), Figure S8 (RS-III), Figure S9 (ERF). Eight additional studies (1958 British Birth Cohort, BMES, CROATIA-Vis, CROATIA-Korcula, DCCT, ORCADES, TwinsUK and WESDR) were used for replication and baseline characteristics of these studies can be found in Table S3. Results of further meta-analyses of genomic regions that exhibited suggestive evidence of association with myopia or hyperopia using regional results from the 8 additional studies listed above are given in Tables S6 and S7. Meta-analyses combining the replication region association results from the 9 discovery datasets and the 8 replication datasets did not result in genome-wide significant results, except for the 8q12 locus (results not shown) that was already genome-wide significant in the discovery dataset.
To determine if our discovery meta-analyses showed evidence of association in any of 35 loci (Table S1) reported to exhibit genome-wide significant or suggestive (p<1×10−6) association with myopia age at onset by Kiefer et al.
[18], a total of 33,591 SNPs overlapping all associated loci were selected (Table S2). These included the most significant discovery SNP plus all available genotyped and imputed SNPs within 500kb of the most significant discovery SNP (Table S2). Accounting for all the LD in each region reduced the effective number of tests, Neff, to 475.71. The replication significance threshold, calculated while taking into account this LD structure in replication regions [40], was 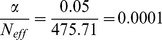 .
.
Myopia
Results of the discovery meta-analysis (Figure 1, Table S4) shows one genome-wide significant marker corresponding to a previously identified myopia age at onset [18] and refractive error [19] locus on 8q12 (rs10113215, p = 1.25×10−8). We also observed association to the well-replicated locus on 15q14 (near GJD2) that was close to genome-wide significant (rs1370156, p = 2.29×10−7). No attempt was made to replicate the chromosome 15q14 region since it has been well replicated. SNPs in the 8q12 replication region did not reach the replication threshold (for rs10113215, replication p = 0.02; top replication p-value in the region was p = 0.0022 for rs6995115). For the discovery meta-analysis suggestive regions, one of the selected SNPs achieved the replication threshold for myopia (rs4326350 on 8p23, p = 6.1×10−5). However, it should be remembered that this region did not exhibit genome-wide significant association in the discovery meta-analysis (replication p-values in Table S6).
In addition to the 8q12 locus, 10 other myopia age at onset regions from the Kiefer et al. study [18] showed significant evidence of replication in our discovery meta-analysis (Table 2). Eight of these loci have also been reported as associated with MSE by Verhoeven et al. [19]. However, two of the regions we replicated were not reported significantly associated with MSE by Verhoeven et al. [19]. On chromosome 3p26, rs2587916 reached the replication threshold in our discovery meta-analysis (p = 2.79×10−5). This SNP is 256 bp away from the SNP reported in this region by Kiefer et al. [18], rs1843303 (which had p = 6.32×10−4 in our data, Table 2). These two SNPs exhibit strong linkage disequilibrium with an R2 of 0.963 and a D′ of 1 in our data. The most significant SNP at the second locus on chromosome 6 is the same SNP as reported by Kiefer et al. [18], rs7744813 (p = 6.07×10−6, Table 2).
Table 2. Results of the replication of regions significantly associated with myopia age at onset by Kiefer et al. [18] showing meta-analysis association results for each chosen SNP with myopia in our data.
| Replication SNP1 | Chromosome | Position | Replication P value2 | Best SNP3,6 | Offset4,6 | P value5,6 | Nearest Gene(s)7 | Reported by Verhoeven et al. |
| rs6702767 | 1 | 200844547 | 1.12E-01 | rs4471299 | 391129 | 1.92E-04 | No | |
| rs11681122 | 2 | 146786063 | N/A | rs10928276 | 661 | 4.61E-04 | No | |
| rs17428076 | 2 | 172851936 | 7.13E-02 | rs3821093 | 157350 | 7.50E-03 | No | |
| rs1898585 | 2 | 178660450 | N/A | rs1405645 | 192929 | 1.47E-03 | No | |
| rs1550094 | 2 | 233385396 | N/A | rs1656404 | 5456 | 3.72E-05 | PRSS56 | Yes |
| rs1843303 | 3 | 4185124 | 6.32E-04 | rs2587916 | 256 | 2.79E-05 | SUMF1/SETMAR | No |
| rs7624084 | 3 | 141093285 | 2.93E-02 | rs1007118 | 247701 | 3.53E-03 | No | |
| rs1031004 | 4 | 80516849 | N/A | rs1440853 | 10203 | 4.09E-04 | No | |
| rs5022942 | 4 | 81959966 | N/A | rs1353387 | 12783 | 6.16E-05 | BMP3 | Yes |
| rs7744813 | 6 | 73643289 | 6.07E-06 | KCNQ5 | No | |||
| rs12193446 | 6 | 129820038 | 8.74E-06 | LAMA2 | Yes | |||
| rs9365619 | 6 | 164251746 | 5.26E-01 | rs6900149 | 211224 | 2.34E-02 | No | |
| rs2137277 | 8 | 40734662 | 2.84E-05 | rs4736884 | 5031 | 1.78E-05 | ZMAT4 | Yes |
| chr8:60178580 | 8 | 60178580 | N/A | rs10113215 | 46386 | 1.25E-08 | TOX | Yes |
| rs10963578 | 9 | 18338649 | N/A | rs10115405 | 17893 | 8.99E-04 | No | |
| rs11145746 | 9 | 71834380 | 1.12E-02 | rs3002374 | 35408 | 2.88E-04 | No | |
| rs4245599 | 10 | 60365755 | 5.75E-05 | rs12264028 | 87616 | 2.57E-05 | BICC1 | Yes |
| rs6480859 | 10 | 79081948 | 5.36E-02 | rs16933964 | 457642 | 1.00E-03 | No | |
| rs745480 | 10 | 85986554 | 6.88E-03 | rs4244950 | 34147 | 2.12E-04 | No | |
| rs4367880 | 10 | 114795256 | N/A | rs7071843 | 316234 | 1.11E-03 | No | |
| rs11602008 | 11 | 40149305 | N/A | rs7924805 | 61948 | 1.02E-03 | No | |
| chr11:65348347 | 11 | 65348347 | N/A | rs610037 | 198510 | 5.94E-03 | No | |
| rs10736767 | 11 | 84637065 | 6.61E-02 | rs1940124 | 18791 | 6.49E-04 | No | |
| rs6487748 | 12 | 9435768 | N/A | rs12822596 | 125774 | 1.83E-03 | No | |
| rs3138142 | 12 | 56115585 | 6.68E-02 | rs2291615 | 219566 | 3.18E-03 | No | |
| rs4291789 | 13 | 100672921 | N/A | rs8000506 | 3929 | 2.98E-05 | ZIC2/ZIC5 | Yes |
| rs61988414 | 14 | 42313443 | N/A | rs12878452 | 2013 | 1.61E-03 | No | |
| chr14:54413001 | 14 | 54413001 | N/A | rs12147340 | 493078 | 1.43E-03 | No | |
| rs524952 | 15 | 35005886 | 8.74E-05 | rs1370156 | 21004 | 2.29E-07 | GJD2 | Yes |
| rs4778882 | 15 | 79382019 | N/A | rs925114 | 323501 | 6.84E-04 | No | |
| rs17648524 | 16 | 7459683 | 3.03E-06 | rs4581716 | 1549 | 1.65E-06 | RBFOX1 | Yes |
| rs2908972 | 17 | 11407259 | 4.10E-03 | rs4792105 | 295899 | 1.79E-03 | No | |
| rs10512441 | 17 | 31239645 | 2.47E-03 | rs17780981 | 120609 | 5.52E-04 | No | |
| rs9902755 | 17 | 47220726 | 1.51E-01 | rs7222737 | 31323 | 2.16E-03 | No | |
| chr17:79585492 | 17 | 79585492 | N/A | rs11651296 | 232337 | 8.53E-03 | No |
1. SNPs which are either genome-wide significant or meet our replication threshold are highlighted in bold text. Allele frequencies for these SNPs in each of our discovery populations can be found in Table S8.
2. For each SNP reported by Kiefer et al., Replication P value is the P value of that SNP in our analysis. If that SNP was not genotyped or imputed in our data, it is indicated with N/A.
3. For regions where the most significant SNP in our analysis is not the original reported SNP, that SNP is reported as Best SNP.
4. Offset is the absolute distance in base pairs to the original SNP and the P value associated with Best SNP.
5. Z scores and direction of effect for all SNPs are in Table S2.
6. This column left blank where the original SNP is the most significant SNP in the region.
7. Nearest Gene(s) indicates the closest gene by physical position for these SNPs.
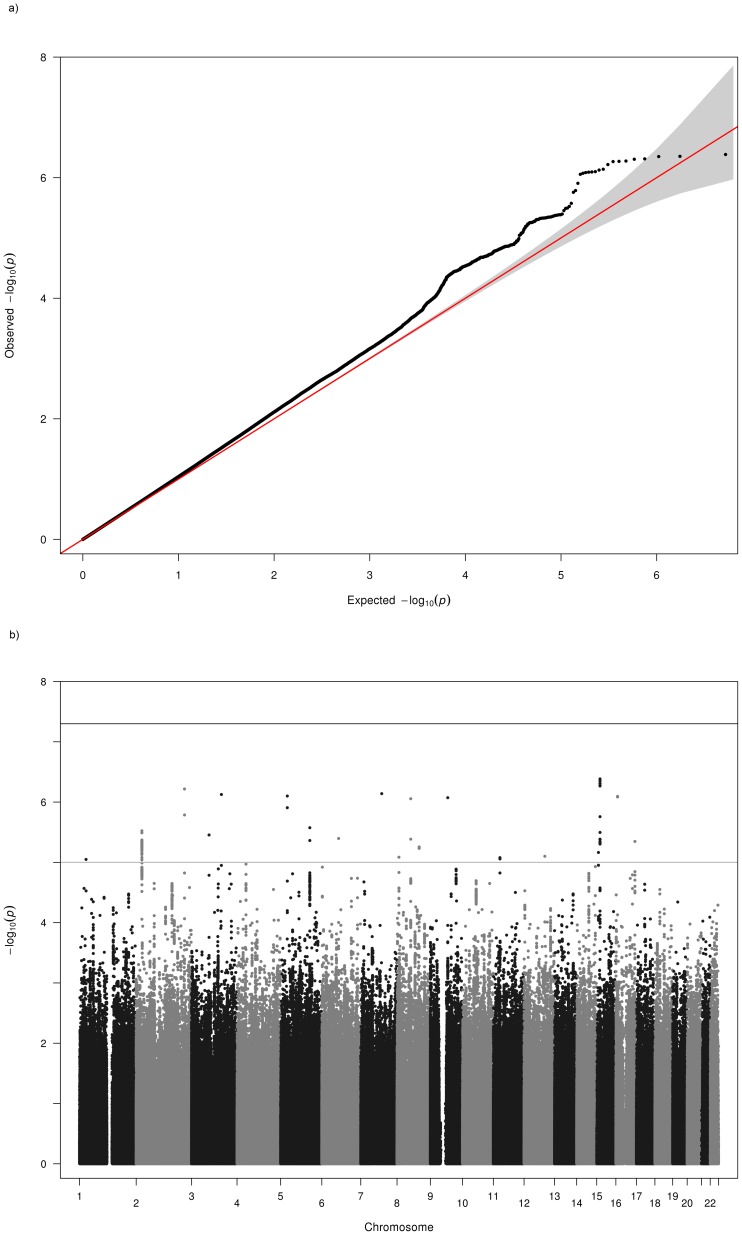
Due to the high genomic control values for OGP-Talana and ERF (Table 1), we examined QQ plots of only the common SNPs (MAF>0.2) to see if this made an improvement, since all the associated SNPs reported here have high MAFs. In OGP-Talana this improved the QQ plots (Figure S9) but it made no difference for ERF. Therefore, we dropped ERF from the analysis and re-examined the results (Figure 3). For most loci this made minimal difference to the p values. However, for 3 loci there was a considerable difference. The genome-wide significant result for myopia on chromosome 8 was no longer genome-wide significant (p = 8.8×10−7), although it still remained well below our replication significance threshold. The loci on 2q37 and 3p26 were no longer below our replication threshold.
Figure 3. Q-Q and Manhattan Plots for the myopia analysis excluding the ERF cohort a) Q–Q plot for association between all SNPs analyzed and myopia in the meta-analysis excluding the ERF cohort.
Each dot represents an observed statistic (defined as -log10 P) versus the corresponding expected statistic. The red line corresponds to the null distribution. b) Manhattan plot for association between all SNPs analyzed and myopia in the meta-analysis excluding the ERF cohort. Each dot represents an observed statistic (defined as -log10 P). The darker gray line corresponds to the genome-wide significance threshold and the lighter gray line represents the suggestive threshold.
Hyperopia
Meta-analysis results showed two genome-wide significant associations with hyperopia (Figure 2, Table S5). These regions overlapped with loci on 15q14 (rs11073060, p = 9.11×10−11) and 8q12 (rs10089517,p = 1.82×10−11) previously reported for MSE in Verhoeven et al. [19] and for myopia age at onset in Kiefer et al. [18]. No attempt was made to replicate the 15q14 locus since it has been well replicated for MSE. None of the SNPs selected to attempt replication of the discovery meta-analysis genome-wide significant association with hyperopia on chromosome 8q12 achieved the replication threshold (rs10089517, p = 0.08; top replication p-value in the region was 0.014 at rs11778476) (Table S7). In addition, for the discovery meta-analysis suggestive regions, one SNP achieved the replication threshold for hyperopia (rs12660628 on 6q21, p = 7.7×10−5). However, it should be remembered that this region did not exhibit genome-wide significant association in the discovery meta-analysis (replication p-values in Table S7).
In addition to the 15q14 and 8q12 loci, 10 other regions (Table 3) that were genome-wide significant in the Kiefer et al. [18] analysis of myopia age at onset exhibited p values for association with hyperopia that met our “replication” threshold for these regions. Given this is a different but related trait, this finding is interesting. Five of these regions have been replicated using myopia as the trait in our data here (three of which were also found to be significantly associated with MSE by Verhoeven et al. [19]). Verhoeven et al. [19] also found that 1 more of these 10 regions (Table 3) showed significant association with MSE. Of the remaining 4 regions from Table 3 the most significant of these 4 SNPs was rs1371993 (p = 1.13×10−5), a SNP on chromosome 4, 35Kb from the SNP reported by Kiefer et al. [18] for myopia age at onset (rs1031004, not available in our data).
Table 3. Results of the hyperopia analyses in the regions that were significantly associated with myopia age at onset by Kiefer et al. [18] showing meta-analysis association results for each chosen SNP.
| Replication SNP1 | Chromosome | Position | Replication P value2 | Best SNP3,6 | Offset4,6 | P value5,6 | Nearest Gene(s)7 | Reported by Verhoeven et al. |
| rs6702767 | 1 | 200844547 | 1.60E-01 | rs6703834 | 264384 | 4.58E-03 | No | |
| rs11681122 | 2 | 146786063 | N/A | rs17412774 | 12116 | 1.50E-04 | No | |
| rs17428076 | 2 | 172851936 | 6.43E-03 | rs3821093 | 157350 | 2.44E-04 | No | |
| rs1898585 | 2 | 178660450 | N/A | rs6718702 | 84399 | 1.47E-05 | PDE11A | No |
| rs1550094 | 2 | 233385396 | N/A | rs1881494 | 12631 | 4.63E-05 | PRSS56 | Yes |
| rs1843303 | 3 | 4185124 | 1.98E-05 | rs795294 | 826 | 1.18E-05 | SUMF1/SETMAR | No |
| rs7624084 | 3 | 141093285 | N/A | rs9821337 | 2901 | 1.88E-04 | No | |
| rs1031004 | 4 | 80516849 | N/A | rs1371993 | 35034 | 1.13E-05 | GK2 (MIM:137028) | No |
| rs5022942 | 4 | 81959966 | N/A | rs2201544 | 30290 | 4.94E-03 | Yes | |
| rs7744813 | 6 | 73643289 | 7.00E-08 | KCNQ5 | No | |||
| rs12193446 | 6 | 129820038 | 1.84E-07 | LAMA2 | Yes | |||
| rs9365619 | 6 | 164251746 | 2.67E-01 | rs2759387 | 412079 | 9.50E-03 | No | |
| rs2137277 | 8 | 40734662 | 2.72E-02 | rs6474290 | 94596 | 2.42E-03 | Yes | |
| chr8:60178580 | 8 | 60178580 | N/A | rs10089517 | 141 | 1.82E-11 | TOX | Yes |
| rs10963578 | 9 | 18338649 | N/A | rs10115405 | 17893 | 2.54E-04 | No | |
| rs11145746 | 9 | 71834380 | 8.33E-03 | rs10481782 | 22378 | 2.71E-04 | No | |
| rs4245599 | 10 | 60365755 | 1.16E-03 | rs1866168 | 4194 | 8.11E-04 | Yes | |
| rs6480859 | 10 | 79081948 | 1.45E-02 | rs16933964 | 457642 | 4.35E-04 | No | |
| rs745480 | 10 | 85986554 | 3.26E-01 | rs17103281 | 25190 | 1.06E-04 | No | |
| rs4367880 | 10 | 114795256 | N/A | rs7914029 | 215000 | 3.40E-04 | No | |
| rs11602008 | 11 | 40149305 | N/A | rs10837366 | 75045 | 7.61E-05 | LRRC4C (MIM:608817) | No |
| chr11:65348347 | 11 | 65348347 | N/A | rs11820062 | 81589 | 7.56E-03 | No | |
| rs10736767 | 11 | 84637065 | 1.99E-01 | rs10898278 | 303825 | 3.05E-03 | No | |
| rs6487748 | 12 | 9435768 | N/A | rs7305636 | 157088 | 9.29E-04 | No | |
| rs3138142 | 12 | 56115585 | 4.32E-02 | rs12828230 | 230568 | 5.87E-04 | No | |
| rs4291789 | 13 | 100672921 | N/A | rs1347190 | 24823 | 6.65E-06 | ZIC2/ZIC5 | Yes |
| rs61988414 | 14 | 42313443 | N/A | rs10149831 | 125528 | 1.35E-03 | No | |
| chr14:54413001 | 14 | 54413001 | N/A | rs17127526 | 444960 | 1.26E-03 | No | |
| rs524952 | 15 | 35005886 | 3.07E-08 | rs11073060 | 16036 | 9.11E-11 | GJD2 | Yes |
| rs4778882 | 15 | 79382019 | N/A | rs1443658 | 4348 | 2.88E-03 | No | |
| rs17648524 | 16 | 7459683 | 4.86E-07 | RBFOX1 | Yes | |||
| rs2908972 | 17 | 11407259 | 1.39E-04 | rs12602611 | 166838 | 1.26E-05 | SHISA6 | No |
| rs10512441 | 17 | 31239645 | 4.78E-03 | rs17183113 | 210521 | 2.40E-03 | No | |
| rs9902755 | 17 | 47220726 | 2.81E-01 | rs8064938 | 439898 | 1.73E-03 | No | |
| chr17:79585492 | 17 | 79585492 | N/A | rs6565596 | 60374 | 1.13E-02 | No |
1. SNPs which are either genome-wide significant or meet our replication threshold are highlighted in bold text. Allele frequencies for these SNPs in each of our discovery populations can be found in Table S8.
2. For each SNP reported by Kiefer et al. , Replication P value is the P value of that SNP in our analysis. If that SNP was not genotyped or imputed in our data, it is indicated with N/A.
3. For regions where the most significant SNP in our analysis is not the original reported SNP, that SNP is reported as Best SNP.
4. Offset is the absolute distance in base pairs to the original SNP and the P value associated with Best SNP.
5. Z scores and direction of effect for all SNPs are in Table S2.
6. This column left blank where the original SNP is the most significant SNP in the region.
7. Nearest Gene(s) indicates the closest gene by physical position for these SNPs.
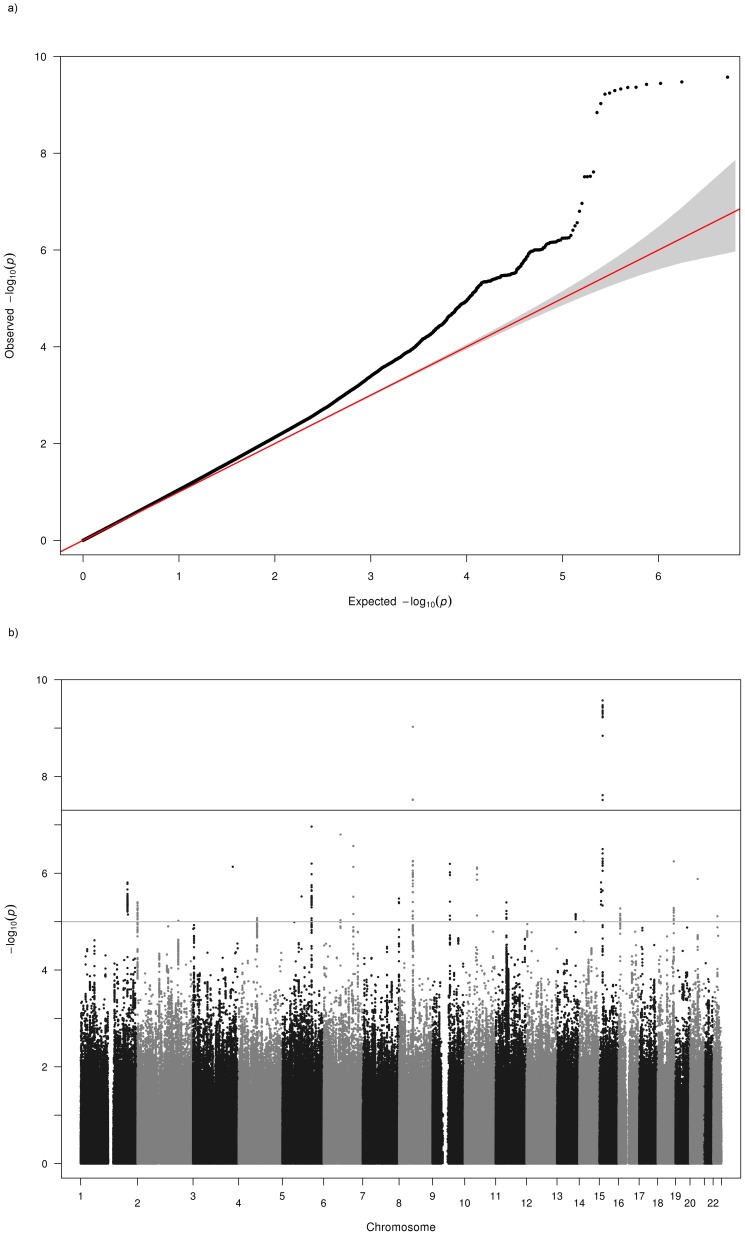
Due to the high genomic control values for OGP-Talana and ERF (Table 1), we examined QQ plots of only the common SNPs (MAF>0.2) to see if this made an improvement, since all the SNPs reported here have high MAFs. In OGP-Talana this improved the QQ plots (Figure S9) but it made no difference for ERF. Therefore, we dropped ERF from the analysis and re-examined the results (Figure 4). For all loci this made minimal difference to the p values and did not change the conclusions.
Figure 4. Q-Q and Manhattan Plots for the hyperopia analysis excluding the ERF cohort a) Q-Q plot for association between all SNPs analyzed and hyperopia in the meta-analysis excluding the ERF cohort.
Each dot represents an observed statistic (defined as -log10 P) versus the corresponding expected statistic. The red line corresponds to the null distribution. b) Manhattan plot for association between all SNPs analyzed and hyperopia in the meta-analysis excluding the ERF cohort. Each dot represents an observed statistic (defined as -log10 P). The darker gray line corresponds to the genome-wide significance threshold and the lighter gray line represents the suggestive threshold.
Discussion
We conducted a meta-analysis of 9 myopia and hyperopia genome-wide association studies. We detected the known loci on chromosomes 8q12 and 15q14. The locus on chromosome 8q12 has been reported associated with mean spherical equivalent in an analysis which included many of the cohorts in this study [19], and myopia age at onset in an independent study [18]. The locus on chromosome 15q14 was discovered in some of the cohorts included in this analysis [48] and has been well replicated in studies of both MSE [21] and myopia age at onset [18]. These findings were therefore expected. However, the signal for 15q14 is only genome-wide significant in the hyperopia analysis here. In addition, although the 8q12 locus was genome-wide significant in the myopia analysis, it was more significant in the hyperopia analysis. Nonetheless, the direction of effect of these SNPs is exactly opposite in the myopia and hyperopia analyses – suggesting that the causal mechanisms being tagged by these SNPs are operating across the spectrum of refractive error.
We also examined the results of our discovery meta-analyses of myopia (which were adjusted for age at examination and years of education) to attempt targeted “replication” of 35 GWAS-identified loci that have previously been reported by Kiefer et al. to be associated with age at onset of myopia [18]. Since age at onset was not available in all our study samples, it was not possible to perform an exact replication of the Kiefer et al. [18] trait on which they performed survival analysis of myopia age at onset. Our analyses, where we included age at exam and years of education, is the closest phenotype we had available. We also examined evidence for association with hyperopia in these same regions of the genome, since myopia and hyperopia represent opposite ends of the distribution of refractive error. It is reasonable that loci that affect the variability of MSE as a whole may therefore affect risk of both myopia and hyperopia.
Our analysis provides evidence for replication of a number of loci identified by Kiefer et al. [18]. Those which were replicated using the myopia trait (Table 2) represent the closest phenotype available from all of our samples to the one used in their analysis. In particular, this study presents the first report of replication of 11 regions associated with myopia. Of note, nine of these regions also showed genome-wide significant evidence of association to MSE by Verhoeven et al. [49]: chromosome 2 near PRSS56 (MIM: 609995), chromosome 4 near BMP3 (MIM:112263), chromosome 6 near LAMA2 (MIM:156225), chromosome 8 near ZMAT4 (40734662 bp), chromosome 8 near TOX (MIM:606863, 60178580 bp), chromosome 10 near BICC1 (MIM: 612717), chromosome 13 near ZIC2(MIM:603073)/ZIC5, chromosome 15 near GJD2 (MIM:607058) and chromosome 16 near RBFOX1(MIM:605104). The candidate genes in these 9 regions have been discussed by both Kiefer et al. [18] and Verhoeven et al. [19]. The two remaining Kiefer et al. loci that were not reported as significantly associated with MSE in Verhoeven et al. [19] were on 3p26.1 and 6q13. The SNP reported by Kiefer et al. [18] in the 3p26.1 region did not meet our replication threshold but another SNP, only 256bp away and in strong linkage disequilibrium with this SNP, did meet our threshold. Kiefer et al. [18] proposed the nearby gene SETMAR (MIM:609834), a histone methylation and DNA repair gene as a candidate to explain their observed association with myopia. However, both the SNP detected in our study and the SNP reported by Kiefer et al. [18] are intronic to one transcript of SUMF1 (MIM:607939), which codes for an enzyme that catalyzes the hydrolysis of sulfate esters. Mutations in this gene are known to cause the lysosomal storage disorder multiple sulfatase deficiency. This multisystem syndrome has been reported to have ocular phenotypes, in the form of retinal degeneration and nystagmus [50]. However, this signal on 3p26.1 was no longer a significant replication when the ERF study results were removed from the analysis. While the Q-Q plot of the ERF study results shows some deviation from expected, it does not appear to exhibit overall inflation of the false positive rate for this sample. Thus the replication of this 3p26 locus using all 9 studies may be valid but additional evidence from a larger study will be useful in determining the importance of this locus to risk of myopia. In the 6q13 region, our study replicated the exact same SNP that was reported to have the strongest association with myopia age at onset in the Kiefer et al. [18] study and this result did not change with the removal of the ERF study results from our meta-analysis. This associated SNP is in an intron of the KCNQ5 gene (potassium voltage-gated channel, KQT-like subfamily, member 5, MIM:607357), which is a member of the KCNQ potassium channel gene family. KCNQ5 has been shown to be differentially expressed in subregions of the brain and in skeletal muscle [51]. Voltage-dependent potassium channels are important regulators of the resting membrane potential and affect the excitability of electrically active cells (MIM: 607357). KCNQ5 is also expressed in the retinal pigment epithelium (RPE) and neural retina. These potassium channels are believed to affect ion flow across the RPE [52] and the function of cone and rod photoreceptors [52], [53].
Other regions that were found to be significantly associated with myopia by Kiefer et al. [18] showed some evidence of association with hyperopia but not with myopia in our data. The significance levels of these associations reached our “replication” threshold. This intriguing result suggests that these loci may not be myopia specific. However, much larger sample sizes will be required to further investigate this issue.
One of the Kiefer et al. [18] loci that did not replicate in the analysis of myopia and was not previously reported as significantly associated with MSE was a locus on 2q31.2. This locus showed evidence of association with hyperopia in our data that reached our “replication” threshold. Kiefer et al. suggested that this association might be due to variants in the phosphodiesterase 11A gene (PDE11A, MIM:604961), which as a known cell signaling molecule is a good candidate gene for development of refractive errors, given the importance of neural signaling in the control of eye growth. However, the signal in our hyperopia analysis stretches across 3 genes: PDE11A; tetratricopeptide repeat domain 30A (TTC30A) protein; and alkylglycerone phosphate synthase (AGPS, MIM:603051). Mutations in AGPS are associated with rhizomelic chondrodysplasia punctata, type 3, a multisystem developmental disorder in which patients frequently develop cataracts [54].
For the locus on chromosome 4 that showed some evidence of association with hyperopia in our data, Kiefer et al. [18] suggested that ANTXR2 (MIM:106490), a gene involved in extracellular matrix adhesion was the best candidate, but other good candidates exist in this region such as BMP2 inducible kinase (BMP2K) and annexin A3 (ANXA3, MIM:106490) a gene involved in regulation of cell growth and signal transduction pathways. Two other bone morphogenic proteins whose genes are located elsewhere in the genome have been identified as candidate genes by Kiefer et al. [18] and Verhoeven et al. [19] and have also been observed in animal models of myopia [55], [56]. The role of this group of genes in growth regulation is well known [57].
Given that hyperopia and myopia are the extreme ends of the refractive error distribution, it is tempting to assume that the same risk factors must affect the risk of developing both traits equally. However, it is not yet clear whether those environmental and genetic factors which increase the risk of developing myopia necessarily affect the risk of hyperopia. The results presented here provide some tantalizing evidence that some genetic factors may be important in both traits whereas others may be more important in driving myopization than hyperopization or vice versa. It has now been shown that 9 regions (2q37, 4q21, 6q22, 8p11, 8q12, 10q21, 13q32, 15q14, 16p13) show association to age at onset of myopia [18], myopia adjusted for age at exam, sex and years of education (results presented here) and mean spherical equivalent [19]. However, we observed replication-level association with myopia for an additional 2 loci (6q13 and 8p11) which were not genome-wide significant for mean spherical equivalent [19] but were genome-wide significant for myopia age at onset [18]. An additional four regions that were genome-wide significant in the Kiefer et al. analysis of age at onset of myopia [18] have only been “replicated” in our hyperopia analyses. These results indicate that the genetic underpinnings of refractive errors are quite complex and that analyses of both the qualitative and quantitative phenotypes may add to our understanding of refractive error causation. The study participants whose data were analyzed here were not selected for extreme or “high” myopia (typically defined as SE <-6D) and there were very few individuals with high myopia in any of these datasets. Future studies to examine whether any of the loci that show association to myopia, hyperopia and mean spherical equivalent in the population-based studies also show evidence of association to high myopia would be interesting and should be pursued.
Some of the other loci that showed significant association with myopia in the Kiefer et al. [18] study did not replicate in our current study. Dichotomizing the trait from spherical equivalent to myopia or hyperopia in each population did reduce sample size for each population compared to the number of individuals with measurements of spherical equivalent. This consequent reduction in power was the reason we added additional populations to our discovery meta-analysis compared to our refractive error meta-analysis [24], to offset the lower sample size. This current study is still, however, smaller than the Kiefer et al. [18] study we were attempting to replicate and so some of the other loci may yet replicate in a larger study.
In summary, we have provided evidence in favor of replication of 11 loci involved in causation of myopia. Twelve loci that have been shown to be associated with myopia age at onset [18] showed “replication-level” association with hyperopia here (7 of these loci also showed replication-level association with the myopia trait; 5 loci only showed this level of association with hyperopia). Further research is required to determine whether any of the candidate genes identified near these associated SNPs are truly causing the development of refractive errors, or whether the actual causal variant is located in another nearby gene or other functional locus in high LD with the SNPs associated with the trait. Evidence for expression of many of these genes have indicated that they are active in the eye [19] and investigation of the ENCODE data suggests many loci have regulatory functions, which is consistent with the current hypothesis of regulation of eye growth through a visually-evoked signaling cascade. However, more research using in vitro and in vivo models is necessary to elucidate the underlying mechanisms of normal emmetropization and how it can be disrupted to produce refractive errors.
Supporting Information
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in AREDS.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) for KORA.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) for Framingham Eye Study.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs associated with myopia (A,C) and hyperopia (B,D) in MESA.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in OGP-Talana.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in ERF.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in RS-I.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in RS-II.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in RS-III.
(TIF)
Flowchart showing the analysis workflows of the entire study.
(TIF)
Chromosomal regions selected to represent loci reported to exhibit genome-wide significant or suggestive association with myopia age at onset by Kiefer et al. (2013).
(XLSX)
Association results in our discovery meta-analysis for the complete set of SNPs selected to represent loci reported to exhibit genome-wide significant or suggestive association with myopia age at onset by Kiefer et al. (2013).
(XLSX)
Baseline Characteristics of Samples Used in the Replication of Our Discovery Meta-analysis Results.
(XLSX)
Most significant associations with myopia in the discovery GWAS meta-analysis.
(XLSX)
Most significant associations with hyperopia in the discovery GWAS meta-analysis.
(XLSX)
Original Myopia Discovery Meta-analysis p-value and Replication p-value from Meta-analysis of Regional Results in 8 Replication Samples.
(XLSX)
Original Hyperopia Discovery Meta-analysis p-value and Replication p-value from Meta-analysis of Regional Results in 8 Replication Samples.
(XLSX)
Comparison of minor allele frequencies (MAF) for each of the Kiefer et al. SNPs for the discovery populations.
(XLSX)
PRISMA Checklist.
(DOCX)
Replication Study Participants, Genotyping, Quality Control and Imputation.
(DOCX)
Association Analysis of Discovery Samples.
(DOCX)
Supplementary References.
(DOCX)
Acknowledgments
Contributors
The Framingham Heart Study data were obtained from the NIH repository dbGaP (accession numbers phs000007/HMB-IRB-MDS and phs000007/HMB-IRB-NPU-MDS). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. The following persons and institutions participated in the DCCT/EDIC Study Research Group: Study Chairmen - S. Genuth, D.M. Nathan, B. Zinman (vice-chair), O. Crofford (past); Albert Einstein College of Medicine - J. Crandall, M. Reid, J. Brown-Friday, S. Engel, J. Sheindlin, H. Martinez (past), H. Shamoon (past), H. Engel (past), M. Phillips Case Western Reserve University - R. Gubitosi-Klug, L. Mayer, S. Pendegast, H. Zegarra, D. Miller, L. Singerman, S. Smith-Brewer, M. Novak, J. Quin (past), W. Dahms (deceased), Saul Genuth (past), M. Palmert (past); Cornell University Medical Center - D. Brillon, M.E. Lackaye, S. Kiss, R. Chan, V. Reppucci (past), T. Lee (past), M. Heinemann (past) Henry Ford Health System - F. Whitehouse, D. Kruger, J. K. Jones, M. McLellan (past), J.D. Carey, E. Angus, A. Thomas, A. Galprin (past); International Diabetes Center - R. Bergenstal, M. Johnson, M. Spencer (past), K. Morgan, D. Etzwiler (deceased), D. Kendall (past) Joslin Diabetes Center - Lloyd Paul Aiello, E. Golden, A. Jacobson (past), R. Beaser, O. Ganda, O. Hamdy, H. Wolpert, G. Sharuk, P. Arrigg, D. Schlossman, J. Rosenzwieg (past), L. Rand (past); Massachusetts General Hospital - D.M. Nathan, M. Larkin, M. Ong, J. Godine, E. Cagliero, P. Lou, K. Folino, S. Fritz (past), S. Crowell (past), K. Hansen (past), C. Gauthier-Kelly (past); Mayo Foundation - J. Service, G. Ziegler Medical University of South Carolina - L. Luttrell, S. Caulder, M. Lopes-Virella (past), J. Colwell (past), J. Soule (past), J. Fernandes, K. Hermayer, S. Kwon, M. Brabham (past), A. Blevins, J. Parker, D. Lee (past), N. Patel, C. Pittman, P. Lindsey (past), M. Bracey (past), K. Lee, M. Nutaitis, A. Farr (past), S. Elsing (past), T. Thompson (past), J. Selby (past), T. Lyons (past), S. Yacoub-Wasef (past), M. Szpiech (past), D. Wood (past), R. Mayfield (past); Northwestern University - M. Molitch, B. Schaefer, L. Jampol, A. Lyon, M. Gill, Z. Strugula, L. Kaminski, R. Mirza, E. Simjanoski, D. Ryan; University of California, San Diego - O. Kolterman, G. Lorenzi, M. Goldbaum University of Iowa - W. Sivitz, M. Bayless; University of Maryland School of Medicine - D. Counts, S. Johnsonbaugh, M. Hebdon (past), P. Salemi, R. Liss, T. Donner (past), J. Gordon (past), R. Hemady (past), A. Kowarski (past), D. Ostrowski (past) S. Steidl (past), B. Jones (past); University of Michigan - W.H. Herman, C.L. Martin, R. Pop-Busui, A. Sarma, J. Albers, E. Feldman, K. Kim, S. Elner, G. Comer, T. Gardner, R. Hackel, R. Prusak, L. Goings, A. Smith, J. Gothrup, P. Titus, J. Lee, M. Brandle, L. Prosser, D.A. Greene (past), M.J. Stevens (past), A. K. Vine (past); University of Minnesota - J. Bantle, N. Wimmergren, A. Cochrane, T. Olsen (past), E. Steuer (past), P Rath (past), B. Rogness (past); University of Missouri - D. Hainsworth, D. Goldstein, S. Hitt, J. Giangiacomo; University of New Mexico - D.S. Schade, J.L. Canady, J.E. Chapin, L.H. Ketai C; University of Pennsylvania – S. Braunstein, P.A. Bourne, S. Schwartz (past), A. Brucker, B.J. Maschak-Carey (past), L. Baker (deceased); University of Pittsburgh - T. Orchard, N. Silvers, C. Ryan, T. Songer, B. Doft, S. Olson, R.L. Bergren, L. Lobes, P. Paczan Rath, D. Becker, D. Rubinstein, P.W. Conrad, S. Yalamanchi, A. Drash (past); University of South Florida - A. Morrison, M.L. Bernal, J. Vaccaro-Kish (past), J. Malone, P.R. Pavan, N. Grove, M.N. Iyer, A.F. Burrows, E.A. Tanaka (past), R. Gstalder (past); University of Tennessee - S. Dagogo-Jack, C. Wigley, H. Ricks, A. Kitabchi, M. B. Murphy (past), S. Moser (past), D. Meyer (past), A. Iannacone (past), E. Chaum, S. Yoser (past), M. Bryer-Ash (past), S. Schussler (past), H. Lambeth (past); The University of Texas Southwestern Medical Center at Dallas - P. Raskin, S. Strowig; University of Toronto - B. Zinman, A. Barnie, R. Devenyi, M. Mandelcorn, M. Brent, S. Rogers, A. Gordon; University of Washington - J. Palmer, S. Catton, J. Brunzell, H. Wessells, I. H. de Boer, J. Hokanson, J. Purnell, J. Ginsberg, J. Kinyoun, S. Deeb, M. Weiss, G. Meekins, J. Distad, L. Van Ottingham (past); University of Western Ontario - J. Dupre, J. Harth, D. Nicolle, M. Driscoll, J. Mahon, C. Canny; Vanderbilt University - M. May, J. Lipps, A. Agarwal, T. Adkins, L. Survant, R. L. Pate, G. E. Munn, R. Lorenz (past), S. Feman (past); Washington University, St. Louis - N. White, L. Levandoski, I. Boniuk, G. Grand, M. Thomas, D. D. Joseph, K. Blinder, G. Shah, Boniuk (past), Burgess (past), J. Santiago (deceased); Yale University School of Medicine - W. Tamborlane, P. Gatcomb, K. Stoessel, K. Taylor (past)J. Goldstein (past), S. Novella (past), H. Mojibian (past), D. Cornfeld (past); Clinical Coordinating Center (Case Western Reserve University) - R. Gubitosi-Klug, J. Quin, P. Gaston, M. Palmert (past), R. Trail (past), W. Dahms (deceased); Data Coordinating Center (The George Washington University, The Biostatistics Center) - J. Lachin, P. Cleary, J. Backlund, W. Sun, B. Braffett, K. Klumpp, K. Chan (past), L. Diminick, D. Rosenberg (past), B. Petty (past), A. Determan (past), D. Kenny (past), B. Rutledge (past), Naji Younes (past), Williams (past), L. Dews, M. Hawkins; National Institute of Diabetes and Digestive and Kidney Disease Program Office - C. Cowie, J. Fradkin, C. Siebert (past), R. Eastman (past); Central Fundus Photograph Reading Center (University of Wisconsin) - R. Danis, S. Gangaputra, S. Neill, M. Davis (past), L. Hubbard (past), H. Wabers, M. Burger, J. Dingledine, V. Gama, R. Sussman; Central Biochemistry Laboratory (University of Minnesota) - M. Steffes, J. Bucksa, M. Nowicki, B. Chavers; Central Carotid Ultrasound Unit (New England Medical Center) - D. O′Leary, J. Polak, A. Harrington, L. Funk (past); Central ECG Reading Unit (University of Minnesota) – R. Crow (past), B. Gloeb (past), S. Thomas (past), C. O′Donnell (past); Central ECG Reading Unit (Wake Forest University) – E. Soliman, Z.M. Zhang, R. Prineas (past), C. Campbell; Central Neuropsychological Coding Unit – C. Ryan, D. Sandstrom, T. Williams, M. Geckle, E. Cupelli, F. Thoma, B. Burzuk, T. Woodfill; Central ANS Reading Unit (Mayo Clinic) – P. Low, C. Sommer, K. Nickander; Computed Tomography Reading Center (Harbor UCLA Research and Education Institute) – M. Budoff, R. Detrano (past), N. Wong, M. Fox, L. Kim (past), R. Oudiz; Johns Hopkins Medical Institutions – J. Lima, D. Bluemke, E. Turkbey, R. J. van der Geest, C. Liu, A. Malayeri, A. Jain, C. Miao (past), H. Chahal (past), R. Jarboe (past); External Evaluation Committee– G. Weir (Chairman), M. Espeland, B. Klein, T Manolio, L. Rand, D. Singer, M. Stern, A.E. Boulton, C. Clark (past), R. D′Agostino (past); Molecular Risk Factors Program Project (Medical University of South Carolina) - M. Lopes-Virella, W.T. Garvey (past), T.J. Lyons, A. Jenkins, R. Klein, G. Virella, A. Jaffa, Rickey Carter, D. Lackland (past), M. Brabham (past), D. McGee (past), D. Zheng (past), R. K. Mayfield (past); Genetic Studies Group (Hospital for Sick Children) - A. Paterson, A. Boright, S. Bull, L. Sun, S. Scherer (past), B. Zinman (past); SCOUT (Veralight) – J. Maynard; Epigenetics (Beckman Research Institute of City of Hope Medical Center) – R. Natarajan, F. Miao, L. Zhang, Z. Chen; Editor, EDIC Publications - D.M. Nathan. For the current paper, the Andrew D. Paterson serves as the DCCT/EDIC lead author and contact person and is a named co-author of this paper.
We thank numerous clinicians, clinical staff, patients, and their families for their participation in and dedication to the project; the Ogliastra population and all the individuals who participated in this study. We are very grateful to the municipal administrators for their collaboration to the project and for economic and logistic support. We are extremely appreciative of the support and wisdom provided by Dr. Hemin Chin of the National Eye Institute. The Center for Inherited Disease Research, fully funded through a federal contract (HHSN268200782096C) from National Institutes of Health to The Johns Hopkins University, performed genotyping of the AREDS and KORA cohorts. Clinical data and DNA from the DCCT/EDIC study will be made available through the National Institute of Diabetes and Digestive and Kidney Diseases repository at https://www.niddkrepository.org/niddk/home.do. This manuscript was not prepared under the auspices of and does not represent analyses or conclusions of the NIDDK Central Repositories, or the NIH. Rotterdam Study and ERF thank Ada Hooghart, Corina Brussee, Riet Bernaerts-Biskop, Patricia van Hilten, Pascal Arp, Jeanette Vergeer, Marijn Verkerk and Sander Bervoets.
Funding Statement
This work was funded in part by the Intramural Research Program of the National Human Genome Research Institute (JEBW, RW, CLS) and the National Eye Institute (MFC, EC), National Institutes of Health, and NIH R01EY020483 (DS, TM, JEBW). The KORA Study is supported by funds from Helmholtz Center Munich and the German Federal Ministry of Education and Research (BMBF). The Multi-Ethnic Study of Atherosclerosis (MESA) and MESA SNP Health Association Resource (SHARe) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support is provided by grants and contracts N01 HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and RR-024156. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-6-4278. Funding for the collection of refractive error data was supported by the Intramural Research Program of the National Eye Institute (ZIAEY000403). Support was also provided by the National Center for Research Resources, Grant UL1RR033176, and is now at the National Center for Advancing Translational Sciences, Grant UL1TR000124. The Blue Mountains Eye Study was supported by the Australian National Health & Medical Research Council (NHMRC) project grants (IDs 974159, 991407, 211069 and 457349) and Centre for Clinical Research Excellence (CCRE) in Translational Clinical Research in Eye Diseases, CCRE in TCR-Eye (ID 529923). The Blue Mountains Eye Study GWAS and genotyping costs were supported by Australian NHMRC project grants (IDs 512423, 475604, 529912 and 590204), and the Wellcome Trust, United Kingdom, as part of Wellcome Trust Case Control Consortium 2 (grant IDs 085475/B/08/Z and 085475/08/Z). EGH (631096), PNB (1028444) and JJW (358702 and 632909) are supported by the NHMRC fellowship scheme. The Centre for Eye Research Australia receives Operational Infrastructure Support from the Victorian government. OGP-Talana was supported by grants from the Italian Ministry of Education, University and Research (MIUR) no. 5571/DSPAR/2002 and (FIRB) D.M no. 718/Ric/2005. The DCCT Research Group is sponsored through research contracts from the National Institute of Diabetes, Endocrinology and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, N01-DK-6-2204, R01-DK-077510) and the National Institutes of Health. ADP holds a Canada Research Chair in the Genetics of Complex Diseases. The Rotterdam Study and ERF were supported by the Netherlands Organisation of Scientific Research (NWO) (Vidi 91796357); Erasmus Medical Center and Erasmus University, Rotterdam, The Netherlands; Netherlands Organization for Health Research and Development (ZonMw); UitZicht; the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); the Municipality of Rotterdam; the Netherlands Genomics Initiative/NWO; Center for Medical Systems Biology of NGI; Lijf en Leven; M. D. Fonds; Henkes Stichting; Stichting Nederlands Oogheelkundig Onderzoek; Swart van Essen; Bevordering van Volkskracht; Blindenhulp; Landelijke Stichting voor Blinden en Slechtzienden; Rotterdamse Vereniging voor Blindenbelangen; OOG; Algemene Nederlandse Vereniging ter Voorkoming van Blindheid; the Rotterdam Eye Hospital Research Foundation; and Topcon Europe. The Croatian studies were funded by grants from the Medical Research Council (United Kingdom), from the Republic of Croatia Ministry of Science, Education and Sports (108-1080315-0302). The authors acknowledge the Wellcome Trust Clinical facility (Edinburgh) for the genotyping of the CROATIA-Vis study, an EU framework 6 project EUROSPAN (contract no LSHG-CT-2006-018947) for the genotyping of the CROATIA-Korcula study that was performed at the Helmholtz Zentrum Munchen (Munich, Germany). ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society, the Medical Research Council Human Genetics Unit and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). The authors acknowledge the Wellcome Trust Clinical facility (Edinburgh) for DNA extraction for the ORCADES study and Peter Lichner and the Helmholtz Zentrum Munchen genotyping staff (Munich, Germany) for genotyping. The GWAS of the 1958 British birth cohort was funded by the Wellcome Trust. This work was carried out at the UCL Institutes of Child Health and Institutes of Ophthalmology which also receive funding from the NIHR Biomedical Research Centres in Child Health and Ophthalmology respectively. The Wisconsin Epidemiologic Study of Diabetic Retinopathy was funded by NIH grant R01EY016379. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kempen JH, Mitchell P, Lee KE, Tielsch JM, Broman AT, et al. (2004) The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol 122: 495–505. [DOI] [PubMed] [Google Scholar]
- 2. Vitale S, Sperduto RD, Ferris FL 3rd (2009) Increased prevalence of myopia in the United States between 1971–1972 and 1999–2004. Arch Ophthalmol 127: 1632–1639. [DOI] [PubMed] [Google Scholar]
- 3. He M, Zeng J, Liu Y, Xu J, Pokharel GP, et al. (2004) Refractive error and visual impairment in urban children in southern china. Invest Ophthalmol Vis Sci 45: 793–799. [DOI] [PubMed] [Google Scholar]
- 4. Lin LL, Shih YF, Hsiao CK, Chen CJ (2004) Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singapore 33: 27–33. [PubMed] [Google Scholar]
- 5. Edwards MH, Lam CS (2004) The epidemiology of myopia in Hong Kong. Ann Acad Med Singapore 33: 34–38. [PubMed] [Google Scholar]
- 6. Lee JH, Jee D, Kwon JW, Lee WK (2013) Prevalence and risk factors for myopia in a rural korean population. Invest Ophthalmol Vis Sci 54: 5466–5471. [DOI] [PubMed] [Google Scholar]
- 7. Wojciechowski R (2011) Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet 79: 301–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanfilippo PG, Hewitt AW, Hammond CJ, Mackey DA (2010) The heritability of ocular traits. Surv Ophthalmol 55: 561–583. [DOI] [PubMed] [Google Scholar]
- 9. Teikari J, Koskenvuo M, Kaprio J, O′Donnell J (1990) Study of gene-environment effects on development of hyperopia: a study of 191 adult twin pairs from the Finnish Twin Cohort Study. Acta Genet Med Gemellol (Roma) 39: 133–136. [DOI] [PubMed] [Google Scholar]
- 10. Teikari JM, Kaprio J, Koskenvuo MK, Vannas A (1988) Heritability estimate for refractive errors–a population-based sample of adult twins. Genet Epidemiol 5: 171–181. [DOI] [PubMed] [Google Scholar]
- 11. Wojciechowski R, Congdon N, Bowie H, Munoz B, Gilbert D, et al. (2005) Heritability of refractive error and familial aggregation of myopia in an elderly American population. Invest Ophthalmol Vis Sci 46: 1588–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peet JA, Cotch MF, Wojciechowski R, Bailey-Wilson JE, Stambolian D (2007) Heritability and familial aggregation of refractive error in the Old Order Amish. Invest Ophthalmol Vis Sci 48: 4002–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ip JM, Rose KA, Morgan IG, Burlutsky G, Mitchell P (2008) Myopia and the urban environment: findings in a sample of 12-year-old Australian school children. Invest Ophthalmol Vis Sci 49: 3858–3863. [DOI] [PubMed] [Google Scholar]
- 14. Ip JM, Saw SM, Rose KA, Morgan IG, Kifley A, et al. (2008) Role of near work in myopia: findings in a sample of Australian school children. Invest Ophthalmol Vis Sci 49: 2903–2910. [DOI] [PubMed] [Google Scholar]
- 15. Dirani M, Tong L, Gazzard G, Zhang X, Chia A, et al. (2009) Outdoor activity and myopia in Singapore teenage children. British Journal of Ophthalmology 93: 997–1000. [DOI] [PubMed] [Google Scholar]
- 16. Rose KA, Morgan IG, Ip J, Kifley A, Huynh S, et al. (2008) Outdoor activity reduces the prevalence of myopia in children. Ophthalmology 115: 1279–1285. [DOI] [PubMed] [Google Scholar]
- 17. Stambolian D (2013) Genetic susceptibility and mechanisms for refractive error. Clin Genet 84: 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kiefer AK, Tung JY, Do CB, Hinds DA, Mountain JL, et al. (2013) Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet 9: e1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verhoeven VJ, Hysi PG, Wojciechowski R, Fan Q, Guggenheim JA, et al. (2013) Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet 45: 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wojciechowski R, Hysi PG (2013) Focusing in on the complex genetics of myopia. PLoS Genet 9: e1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhoeven VJ, Hysi PG, Saw SM, Vitart V, Mirshahi A, et al. (2012) Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Human Genetics. [DOI] [PMC free article] [PubMed]
- 22. Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, et al. (2008) Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet 40: 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- 24. Stambolian D, Wojciechowski R, Oexle K, Pirastu M, Li X, et al. (2013) Meta-analysis of genome-wide association studies in five cohorts reveals common variants in RBFOX1, a regulator of tissue-specific splicing, associated with refractive error. Hum Mol Genet 22: 2754–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, et al. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ioannidis JP (2007) Non-replication and inconsistency in the genome-wide association setting. Hum Hered 64: 203–213. [DOI] [PubMed] [Google Scholar]
- 28. Kraft P, Zeggini E, Ioannidis JP (2009) Replication in genome-wide association studies. Stat Sci 24: 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ioannidis JP, Thomas G, Daly MJ (2009) Validating, augmenting and refining genome-wide association signals. Nat Rev Genet 10: 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roeder K, Bacanu SA, Wasserman L, Devlin B (2006) Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet 78: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang K, Li M, Bucan M (2007) Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 81: 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M, Wang K, Grant SF, Hakonarson H, Li C (2009) ATOM: a powerful gene-based association test by combining optimally weighted markers. Bioinformatics 25: 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gauderman WJ, Murcray C, Gilliland F, Conti DV (2007) Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol 31: 383–395. [DOI] [PubMed] [Google Scholar]
- 34. Wang T, Elston RC (2007) Improved power by use of a weighted score test for linkage disequilibrium mapping. Am J Hum Genet 80: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, et al. (2010) Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet 86: 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP (2008) A powerful and flexible multilocus association test for quantitative traits. Am J Hum Genet 82: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang K, Abbott D (2008) A principal components regression approach to multilocus genetic association studies. Genet Epidemiol 32: 108–118. [DOI] [PubMed] [Google Scholar]
- 38. Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, et al. (2010) A versatile gene-based test for genome-wide association studies. Am J Hum Genet 87: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Christoforou A, Dondrup M, Mattingsdal M, Mattheisen M, Giddaluru S, et al. (2012) Linkage-disequilibrium-based binning affects the interpretation of GWASs. Am J Hum Genet 90: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramos E, Chen G, Shriner D, Doumatey A, Gerry NP, et al. (2011) Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia 54: 783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dudbridge F, Gusnanto A (2008) Estimation of significance thresholds for genomewide association scans. Genetic Epidemiology 32: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE (2008) Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 9: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. American Journal of Human Genetics 74: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheverud JM (2001) A simple correction for multiple comparisons in interval mapping genome scans. Heredity 87: 52–58. [DOI] [PubMed] [Google Scholar]
- 45. Li J, Ji L (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95: 221–227. [DOI] [PubMed] [Google Scholar]
- 46. Bretherton CS, Widmann M, Dymnikov VP, Wallace JM, Blade I (1999) The effective number of spatial degrees of freedom of a time-varying field. Journal of Climate 12: 1990–2009. [Google Scholar]
- 47. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, et al. (2010) A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nature Genetics 42: 897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Verhoeven VJ, Hysi PG, Wojciechowski R, Fan Q, Guggenheim JA, et al. (2013) Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet 45: 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blanco-Aguirre ME, Kofman-Alfaro SH, Rivera-Vega MR, Medina C, Valdes-Flores M, et al. (2001) Unusual clinical presentation in two cases of multiple sulfatase deficiency. Pediatr Dermatol 18: 388–392. [DOI] [PubMed] [Google Scholar]
- 51. Lerche C, Scherer CR, Seebohm G, Derst C, Wei AD, et al. (2000) Molecular cloning and functional expression of KCNQ5, a potassium channel subunit that may contribute to neuronal M-current diversity. The Journal of biological chemistry 275: 22395–22400. [DOI] [PubMed] [Google Scholar]
- 52. Pattnaik BR, Hughes BA (2012) Effects of KCNQ channel modulators on the M-type potassium current in primate retinal pigment epithelium. American journal of physiology Cell physiology 302: C821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang X, Yang D, Hughes BA (2011) KCNQ5/K(v)7.5 potassium channel expression and subcellular localization in primate retinal pigment epithelium and neural retina. American journal of physiology Cell physiology 301: C1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Itzkovitz B, Jiralerspong S, Nimmo G, Loscalzo M, Horovitz DD, et al. (2012) Functional characterization of novel mutations in GNPAT and AGPS, causing rhizomelic chondrodysplasia punctata (RCDP) types 2 and 3. Hum Mutat 33: 189–197. [DOI] [PubMed] [Google Scholar]
- 55. Wang Q, Zhao G, Xing S, Zhang L, Yang X (2011) Role of bone morphogenetic proteins in form-deprivation myopia sclera. Mol Vis 17: 647–657. [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Y, Liu Y, Wildsoet CF (2012) Bidirectional, optical sign-dependent regulation of BMP2 gene expression in chick retinal pigment epithelium. Invest Ophthalmol Vis Sci 53: 6072–6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reddi AH, Reddi A (2009) Bone morphogenetic proteins (BMPs): from morphogens to metabologens. Cytokine Growth Factor Rev 20: 341–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in AREDS.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) for KORA.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) for Framingham Eye Study.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs associated with myopia (A,C) and hyperopia (B,D) in MESA.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in OGP-Talana.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in ERF.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in RS-I.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in RS-II.
(TIF)
QQ and Genome-wide Manhattan plot of genotyped and imputed SNPs for association with myopia (A,C) and hyperopia (B,D) in RS-III.
(TIF)
Flowchart showing the analysis workflows of the entire study.
(TIF)
Chromosomal regions selected to represent loci reported to exhibit genome-wide significant or suggestive association with myopia age at onset by Kiefer et al. (2013).
(XLSX)
Association results in our discovery meta-analysis for the complete set of SNPs selected to represent loci reported to exhibit genome-wide significant or suggestive association with myopia age at onset by Kiefer et al. (2013).
(XLSX)
Baseline Characteristics of Samples Used in the Replication of Our Discovery Meta-analysis Results.
(XLSX)
Most significant associations with myopia in the discovery GWAS meta-analysis.
(XLSX)
Most significant associations with hyperopia in the discovery GWAS meta-analysis.
(XLSX)
Original Myopia Discovery Meta-analysis p-value and Replication p-value from Meta-analysis of Regional Results in 8 Replication Samples.
(XLSX)
Original Hyperopia Discovery Meta-analysis p-value and Replication p-value from Meta-analysis of Regional Results in 8 Replication Samples.
(XLSX)
Comparison of minor allele frequencies (MAF) for each of the Kiefer et al. SNPs for the discovery populations.
(XLSX)
PRISMA Checklist.
(DOCX)
Replication Study Participants, Genotyping, Quality Control and Imputation.
(DOCX)
Association Analysis of Discovery Samples.
(DOCX)
Supplementary References.
(DOCX)