Abstract
The transition from hunting and gathering to plant and animal domestication was one of the most important cultural and technological revolutions in human history. According to archeologists and paleoanthropologists, this transition triggered major demographic expansions. However, few genetic studies have found traces of Neolithic expansions in the current repartition of genetic polymorphism, pointing rather toward Paleolithic expansions. Here, we used microsatellite autosomal data to investigate the past demographic history of 87 African and Eurasian human populations with contrasted lifestyles (nomadic hunter-gatherers, semi-nomadic herders and sedentary farmers). Likely due to the combination of a higher mutation rate and the possibility to analyze several loci as independent replicates of the coalescent process, the analysis of microsatellite data allowed us to infer more recent expansions than previous genetic studies, potentially resulting from the Neolithic transition. Despite the variability in their location and environment, we found consistent expansions for all sedentary farmers, while we inferred constant population sizes for all hunter-gatherers and most herders that could result from constraints linked to a nomadic or semi-nomadic lifestyle and/or competition for land between herders and farmers. As an exception, we inferred expansions for Central Asian herders. This might be linked with the arid environment of this area that may have been more favorable to nomadic herders than to sedentary farmers. Alternatively, current Central Asian herders may descent from populations who have first experienced a transition from hunter-gathering to sedentary agropastoralism, and then a second transition to nomadic herding.
Keywords: lifestyle, demography, population genetics, Neolithic transition, BEAST, coalescent
Introduction
Reconstructing the demographic history of human populations is a major goal in many disciplines such as paleoanthropology, archeology and population genetics. The transition from hunting and gathering to plant and animal domestication, which occurred independently in several regions of the World during the Neolithic period (10 000–3000 years before present (YBP)), is of particular interest. This transition, which represents one of the most important cultural and technological revolutions in our history, affected many aspects of lifestyle (diet, technologies, social organization) and led to the sedentarization of many populations that is widely assumed by archeologists and paleoanthropologists to have driven recent human population expansions.1, 2, 3 Bocquet-Appel4 notably showed an increase in the number of enclosures and in the proportion of subadults in Eurasian burial sites during the Neolithic. This was interpreted as a proof of a natality increase, and thus of demographic expansion. However, this potential natality increase could have been counterbalanced to some extent by a mortality increase because of rapid changes in selective pressures, in particular a spread of infectious diseases catalyzed by sedentarization and the increase of population density.4, 5 At the same time, many other populations remained nomadic during this transition. Among these populations, some kept a hunter-gatherer lifestyle, but others developed new means of subsistence like nomadic herding.
Population genetic studies have provided some additional insights on the understanding of human demographic history. Numerical methods based on the coalescent theory6 allow inferring demographic parameters from current population genetic data.7, 8, 9 In particular, many studies based on non-recombining sex-specific markers, namely the Y chromosome and the hypervariable control region (HVS-I) of the mitochondrial DNA, have detected strong middle and upper Paleolithic expansions in both African and Eurasian post-Neolithic populations. With HVS-I data, these expansions were dated at 70 000 YBP in Africa,10 and between 63 000 YBP and 17 000 YBP in Eurasia.11 Estimates based on the analysis of the Y-chromosome polymorphism are generally more recent: between 15 000 and 12 000 YBP for Africa, and between 40 000 and 11 000 for Eurasia.11, 12 Nevertheless, these Y-chromosome-based estimates still predate the Neolithic transition.
Moreover, some authors have taken advantage of neutral autosomal DNA sequences by combining inferences from several independent loci. This has allowed refining the dating of Paleolithic expansions in Africa, with dates ranging between 50 000 and 25 000 YBP.13, 14 All these studies therefore pointed to expansions starting in the Paleolithic, without evidence for a subsequent Neolithic expansion. To our knowledge, only one study detected Neolithic expansions: by separately analyzing different mitochondrial haplogroups according to their time of origin, Gignoux et al15 showed demographic expansions starting at approximately 3000 YBP in Africa, 8000 YBP in Europe and 5000 YBP in Southeastern Asia.
Regarding the relationship between lifestyle and demographic history, a recent study based on complete mitochondrial sequences inferred constant population sizes for hunter-gatherer populations in Africa, but demographic expansions for farmers starting at 60 000 YBP.16 In a previous study, we also found contrasted demographic history between sedentary and nomadic populations predating the Neolithic transition in Africa and Eurasia, using resequencing data from both the mitochondrial HVS-I region and neutral autosomal sequences. This suggests that strong Paleolithic expansions may have ultimately favored the emergence of agriculture in some populations.17 However, as ancient expansions could have masked potential signals of more recent expansions, these findings do not preclude the possibility of a second expansion event resulting from the Neolithic transition. Indeed, strong ancient demographic events are more easily detected with coalescent-based methods, and, in some cases, they can mask the signals of more recent events, especially if the population has not yet reached genetic equilibrium.18
In this paper, we used autosomal microsatellites data that offer both the advantage of a high mutation rate19 and the possibility to analyze several loci as independent replicates to infer recent expansion events. Recent simulation and empirical studies on non-human species have shown that these markers can reveal very recent demographic events that more slowly evolving markers such as DNA sequences may fail to detect.20, 21, 22 Here we investigated the past demographic history of 60 African and Eurasian populations with contrasted lifestyles (ie, sedentary farmers, nomadic hunter-gatherers and nomadic herders), using a coalescent-based approach, in order to determine (1) whether autosomal microsatellite data allowed us to detect recent expansions and (2) whether such expansions occurred concurrently in populations characterized by different lifestyles.
First, we focused on Central Africa, where we studied a set of populations (historically called Pygmies) that are thought to have retained a nomadic lifestyle based on hunter-gathering, as well as neighboring farmer populations. These two groups of populations are genetically differentiated and their divergence was inferred to have occurred ∼60 000 years ago,23, 24 thus long before the Neolithic sedentarization of farmer populations (∼3500–5000 YPB in this area3, 25, 26). We also analyzed a set of semi-nomadic herder populations originating from different areas in Africa. Second, we analyzed a sample of populations from four distant geographical regions of Eurasia (Middle-East, Pamir, Russia and East Asia), where sedentary farmers coexist with nomadic herders, studying one herder and one farmer population from each region. In addition, we analyzed four hunter-gatherer populations (commonly called Negritos) from Philippines. Finally, we performed a more detailed study on farmer and herder populations from Central Asia. This area is of peculiar interest as it is characterized by a more arid climate than the rest of Eurasia that could have affected the development of farming and the demography of human groups.27 Moreover, although Central Asia is thought to have been a major corridor during the successive Eurasian migration waves,28, 29 many aspects of the demographic history of this area remain poorly understood. As detailed by Harris,30 plant growing (barley, wheat, etc) and animal domestication appeared at ∼10 000–11 000 BP in the Middle-East and then spread through Eurasia following three main directions: a first wave reached Pakistan at ∼9000 BP, the second one reached Central-Asia at ∼8000–9000 BP and the last one reached Europe at ∼7000–5000 BP. However, it is not known whether nomadic herders in Central Asia descended directly from hunter-gatherers or whether the ancestors of current nomadic herders have experienced two successive lifestyle shifts (ie, first from hunter-gathering to agropastoralism, and then from agropastoralism to nomadic herding).3
Materials and methods
Population sampling and marker sets
For Africa, we used the data set of Verdu et al24, 31 that consists of 337 individuals from 12 farmer populations and 281 individuals from 10 Pygmy hunter-gatherer populations from Central Africa (Gabon and Cameroon) (Supplementary Table S1). This data set contains 28 unlinked autosomal microsatellite loci located on 18 different chromosomes (Supplementary Table S2), from the ‘Screening Set 10' provided by the Marshfield Foundation Mammalian Genotyping Service (available at http://research.marshfieldclinic.org/genetics/GeneticResearch/screeningsets.asp).
In addition, we used 23 herder populations (503 individuals) from Cameroon, Kenya, Tanzania and Sudan, extracting the data for the same 28 microsatellite loci from the complete data set of Tishkoff et al32 (Supplementary Tables S1 and S2).
For Eurasia, we selected eight populations from the HGDP-CEPH Human Genome Diversity Cell Line Panel.33, 34 To avoid any bias of geographic structure in the sampling, we first identified four distant areas where farmer and herders coexist nowadays (Middle-East, Pakistan, Russia and East Asia), and then selected randomly one traditionally nomadic herder population and one neighboring sedentary farmer population from each of these regions (Han Chinese and Mongols from East Asia, Pathans and Balochi from Pamir, Palestinians and Bedouins from Middle-East and Russians and Yakut from Russia). This represented a total of 231 individuals (Supplementary Table S1). We used 21 autosomal microsatellites markers out of the 27 markers selected by Ségurel et al35 from the HGPD-CEPH panel (Supplementary Table S2). We removed indeed six loci (GATA22F11, GATA88H02, SE30, GATA28D01, GATA11B12 and GATA12A07) from the original data set as they presented compound repeat motives, and might thus not fit the assumption of a SSM (single stepwise model). We also analyzed four hunter-gatherer populations (118 individuals) from the Philippines (commonly called Negritos) genotyped by Heyer et al36 for the same 28 unlinked autosomal microsatellite loci as for the African populations (Supplementary Table S2).
For our detailed study of Central Asia, we used data from 30 populations from West Uzbekistan to East Kyrgyzstan: 17 herder populations, for a total of 584 individuals, and 13 farmer populations, for a total of 382 individuals (Supplementary Table S1). Among these populations, 26 had been genotyped in previous studies35, 37 for the same 21 markers that we used for the HGDP Eurasian populations (Supplementary Table S2). The four other populations (KIB, TAB, KIM and TAM) were genotyped for this study, using the same PCR and genotyping procedures (see below). All sampled individuals were healthy donors from whom informed consent was obtained, and the study was approved by appropriate Ethic Committees and scientific organizations in all countries where samples have been collected.
Genotyping
Regarding both the data from Verdu et al24, 31 for Africa and the data from Heyer et al36 for Philippines, PCR amplifications were performed in a final volume of 20 μl containing 1 × buffer, 0.25 mM of each desoxyribonucleotide triphosphate (dNTP), 0.06 U Taq DNA polymerase, 125 nM of each primer and 20 ng DNA under the following conditions for all markers: initial denaturation at 94 °C for 10 min, followed by 35 cycles of 94 °C for 30 s, annealing at 55 °C for 75 s, 72 °C for 20 s and 72 °C for 10 min as final extension.
Regarding both the data from Ségurel et al35 and our newly genotyped data for Central Asia, PCR amplifications were performed in a 20 ml final volume composed of 16 Eppendorf buffer, 125 mM of each dNTP, 0.5 U Taq DNA polymerase, 125 nM of each primer and 10 ng DNA. The reactions were performed with an initial denaturation step at 94 °C for 5 min, followed by 36 cycles at 94 °C for 30 s, annealing at 55 °C for 30 s, 72 °C for 20 s and 72 °C for 10 min as final extension.
All reactions were further analyzed by capillary electrophoresis (ABI 310, Applied Biosystems, Foster City, CA, USA). Alleles were scored with the software GeneMarker version 1.6 (SoftGenetics LLC, State College, PA, USA), and each allele call was manually verified. All ambiguous allelic states, as well as all alleles called only once in a single individual (singletons), were reprocessed and verified independently several times.
Regarding both the data set from the HGDP-CEPH Human Genome Diversity Cell Line Panel33, 34 for Eurasia and the data set from Tishkoff et al32 for Africa, the genotyping was performed by the Marshfield Foundation Mammalian Genotyping38 Service (http://research.marshfieldclinic.org/).
Data analysis
We computed the likelihood distribution of demographic parameters39 using the MCMC algorithm implemented in BEAST v1.7.4.40 We tested four demographic models implemented in this software: constant effective population size (N0) (‘Constant model'), exponential variation of N0 (‘Exponential model'), logistic variation of N0 (‘Logistic model') and the ‘expansion model', in which N0 is the present day population size, N1 the population size that the model asymptotes to going into the distant past, and g the exponential growth rate that determines how fast the transition is from near the N1 population size to N0 population size. This allowed us to infer the current effective population sizes (N0) and growth rates (g) for all analyzed populations. For the ‘Expansion model', we also inferred the ancestral population size (N1). We then inferred the dates of expansion onsets (t) using the formula: t=(1/g) × ln(N1/N0) that makes the approximate assumption that the population started to grow from its initial size N1 to reach its final size N0 after t generations. This formula was applied to each step of the MCMC algorithm. We used a single step mutational model (SSM) that takes homoplasy into account. The implementation of microsatellite models in BEAST v1.7.4 is described in Wu and Drummond.41
We performed three runs of 2.8 × 108 steps per population and per demographic model for the African and Philippine populations, and three runs of 2.1 × 108 steps for the others Eurasian and Central Asian populations (that corresponded for all populations to three runs of 107 steps per locus). We recorded one tree every 1000 steps, resulting in a total of 105 trees per locus and per run. We then removed the first 10% of each run (burn-in period) and combined the runs to obtain reasonably high effective sample sizes (ESS of 100 or above, see, eg, Gignoux et al15). The convergence of these runs was assessed by visual inspection of traces using Tracer v1.542 to check for concordance between runs, and also by the computation of the Gelman and Rubin's convergence diagnostic43 using R v2.14.144 with the function ‘gelman.diag' available in the add-on package coda.45
In order to facilitate a large exploration of the parameter space, for the autosomal sequences, we chose uniform priors between 1 and 106 for 2Ne and between −0.1 and 0.1 per generation for g. In agreement with previous studies,19, 24 we assumed a uniform prior between 10−4 and 10−3 per generation for the mean mutation rate (μ), and the mutation rate of each locus was drawn independently from a Gamma distribution (mean=μ and shape=2). This high value for mutation rate is consistent with previous estimates based on pedigree data.46 As in previous studies,11, 14, 16, 23 we assumed a generation time of 25 years. For each population and demographic model, we obtained the mode and the 95% highest probability density (HPD) interval of each parameter, inferred from the posterior distributions obtained using the add-on package Locfit47 in R v2.14.1. Then, we computed the Deviance Information Criteria (DIC) of each model, as the model with lower DIC is considered as the best-fitting model for the observed data.48 DIC is computed as DIC=pD +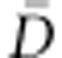 , where pD represents the effective number of parameters of the model, defined as
, where pD represents the effective number of parameters of the model, defined as  − D(
− D(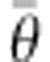 ). D (
). D (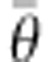 ) is the deviance parameter calculated as −2 log (p(y| θ)), where p(y| θ) represents the likelihood function, substituted here with the probability of the data (y) given the posterior mean of the parameter values (θ).
) is the deviance parameter calculated as −2 log (p(y| θ)), where p(y| θ) represents the likelihood function, substituted here with the probability of the data (y) given the posterior mean of the parameter values (θ). 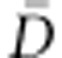 is the expectation of the likelihood associated with the means of posterior parameters distributions (
is the expectation of the likelihood associated with the means of posterior parameters distributions (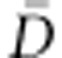 =E
θ [D (
=E
θ [D (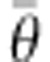 )]). To obtain
)]). To obtain 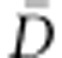 , we repeated all analyses using the posterior mean of each parameter as punctual priors. A difference of five points or more in DIC was considered as significant.48, 49
, we repeated all analyses using the posterior mean of each parameter as punctual priors. A difference of five points or more in DIC was considered as significant.48, 49
Results
Contrasting demographic histories for sedentary and nomadic populations
For Africa, the expansion model had the lowest DIC value for all sedentary farmer populations, whereas the constant model had the lowest DIC value for all nomadic hunter-gatherer populations and semi-nomadic herder populations (Table 1 and Supplementary Figure S1). As differences in DIC values between the best-fitting model and the others were higher than five points for all populations except for one farmer population (Ewondo), we could, therefore, conclude that these models significantly best fitted the data as compared with the others for these populations.48, 49 Conversely, for the Ewondo population, the Expansion and the Constant model could not be distinguished (Supplementary Table S3).
Table 1. Mean values of the modes and 95% HPD limits for the inferred current effective population size (N 0), ancestral effective population.
| Area | Lifestyle | BF model | N1 lower | N1 mode | N1 upper | N0 lower | N0 mode | N0 upper | g lower | g mode | g upper |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Central Africa | Sedentary farmers | Expansiona | 17 | 18 429 | 93 394 | 39 | 40 370 | 186 263 | 1.39 × 10−03 | 3.47 × 10−03 | 1.50 × 10−02 |
| Central Africa | Nomadic HG | Constant | − | _ | _ | 9873 | 15 859 | 19 026 | _ | _ | _ |
| Africa | Semi-nomadic herders | Constant | _ | _ | _ | 14 395 | 20 437 | 35 005 | _ | _ | _ |
| Eurasia | Sedentary farmers | Expansion | 12 181 | 15 853 | 22 404 | 19 099 | 25 600 | 37 769 | 2.47 × 10−04 | 1.96 × 10−03 | 3.49 × 10−03 |
| Eurasia | Nomadic herders | Constant | _ | _ | _ | 21 969 | 26 651 | 32 552 | _ | _ | _ |
| Eurasia (Philippines) | Nomadic HG | Constant | _ | _ | _ | 4999 | 6208 | 7926 | _ | _ | _ |
| Central-Asia | Sedentary farmers | Expansion | 11 592 | 24 988 | 47 875 | 23 713 | 32 662 | 49 933 | 2.27 × 10−04 | 1.85 × 10−03 | 3.28 × 10−03 |
| Central-Asia | Nomadic herders | Expansion | 11 042 | 21 706 | 41 412 | 21 752 | 28 804 | 42 180 | 2.41 × 10−04 | 1.90 × 10−03 | 3.48 × 10−03 |
For each area, we report only the mean values over populations. Detailed results for each population separately are presented in Supplementary Table S4.
Note that for one African population (Ewondo), the Expansion model and a Constant model could not be distinguished from their DIC values (Supplementary Table S3).
For Eurasia, among the eight populations from the HGDP-CEPH panel, we found that the expansion model best fitted the data for sedentary farmers, whereas this was the constant model for nomadic herders (Table 1 and Supplementary Table S3 and Supplementary Figure S1). The constant model also best fitted the data for the four Philippine nomadic hunter-gatherer populations (Table 1 and Supplementary Table S3 and Supplementary Figure S1). Conversely, when focusing on Central-Asian populations, we found that the expansion model best fitted the data for both sedentary farmer and nomadic herder populations (Table 1 and Supplementary Table S3 and Supplementary Figure S1).The modes and 95% HPD of all inferred parameters for each population separately are provided in Supplementary Table S4.
Expansion timings
For sedentary farmer populations from Africa, we inferred expansion signals starting between 3036 and 6939 (1217–23 054) YBP, concomitantly with the Neolithic transition in this area, at ∼3500–5000 YPB3, 25, 26 (Table 2 and Supplementary Table S4 and Supplementary Figure S1). For all Eurasians farmers, as well as for Central Asian nomadic herder populations, we estimated expansion onset times from 5111 (2669–30 851) YBP for Palestinians to 2864 (1609–12 842) YBP for Han Chinese (Table 2 and Supplementary Table S4 and Supplementary Figure S1). In all cases, the modal estimates of the expansion onsets fell after the Neolithic transition but the 95% HPD included both the Neolithic transition and the upper Paleolithic period (Table 2).
Table 2. Modes and 95% HPD of expansion onset times (t) estimated under the expansion model for expanding populations, compared with datations of the emergence of farming reported by archeologists and paleoanthropologists.
| Area | Analyzed populations | Lifestyle | Emergence of farming/ herding | t, 95% lower | t, mode | t, 95% upper |
|---|---|---|---|---|---|---|
| Africa | Central-African farmersa | Sedentary farmers | 3500–5000 YBP3, 25, 26 | 2905 | 5339 | 17 801 |
| East-Asia | Han Chinese | Sedentary farmers | 7000–9000 YBP3,30 | 1608 | 2864 | 12 842 |
| Middle-East | Palestinians | Sedentary farmers | 10 000–11 000 YBP3,30 | 2668 | 5111 | 30 851 |
| Pamir | Pathans | Sedentary farmers | 8000–9000 YBP3,30 | 2153 | 4588 | 22 691 |
| Russia | Russians | Sedentary farmers | 7000 YBP3,30 | 2415 | 4489 | 21896 |
| Central-Asia | Indo-Iranian farmersa | Sedentary farmers | 8000–9000 YBP3,30 | 2399 | 4760 | 23 406 |
| Central-Asia | Turko-Mongol herdersa | Nomadic herders | 8000–9000 YBP3,30 | 2160 | 4280 | 21 163 |
For these areas, we report the mean values over populations. Detailed results for each population separately are presented in Supplementary Table S4.
Discussion
In this study, we have found genetic signals of demographic expansions for almost all sedentary farmer populations analyzed, and we inferred constant population sizes for all nomadic hunter-gatherer populations and most nomadic herder populations. As an exception, the nomadic herders from Central Asia displayed an expansion signal similar to that of the farmer populations in the same area. For both African and Eurasian expanding populations, the modal estimate of expansion times were consistent with an onset of these expansions during or after the Neolithic expansion, even though the 95% HPD could not exclude late Paleolithic expansions (up to ∼30 000 years ago).
It is important to note that the method used in this study makes the assumption that populations are isolated and panmictic, and this is questionable for human populations. Nevertheless, we analyzed a large set of populations sampled in very distant geographical regions (ie, Central Africa, Middle-East, Central Asia, Pamir, Russia, Southeast Asia and East Asia), and the main conclusions of this study are consistent across these different regions. It therefore seems unlikely that local processes, such as admixture, could have similarly biased estimates in each of these regions, as these admixture processes are likely to vary in nature and intensity across regions. As we used many independent loci, we can also be confident to have avoided confounding effects of selection.
Similarly, note that we compared genetically based inferences of past effective population sizes with archeological and paleoanthropological records that should reflect census population sizes. However, the aim of this study was to compare trends in evolution of population size through time rather than inferring the population size itself. In this context, even if effective and census population sizes usually differ in their absolute values, they are likely to follow the same trend over time. Local demographic processes such as differences in reproductive success among individuals may lead to variations in the ratio between effective and census population sizes. However, as we analyzed a large set of populations sampled in very distant geographical regions and found consistent results, it appears thus that our result are robust to such variations.
Contrasted demographic histories associated with different lifestyles
Despite cultural and environmental variability, we inferred similar patterns of expansion for all studied sedentary farmer populations in both Africa and Eurasia, except for one African farmer population (Ewondo) for which we found a constant population size. Moreover, except for Central Asia, we found constant population sizes for both nomadic hunter-gatherers and nomadic herders.
These findings are consistent with paleoanthropological and archeological records that suggest that the growth of farmer populations after the Neolithic resulted from the processes of sedentarization.1, 3 Conversely, both herders and hunter-gatherer populations may have remained at constant size because of constraints associated with their nomadic lifestyle. For instance, birth intervals are generally longer (at least by 4 years) in nomadic populations than in sedentary populations.50 These results are also consistent with Cerny et al,51 who found much lower expansion signals for nomadic pastoralists than for sedentary farmers in the West African Sahel area, using a set of neutrality tests.
The fact that the Ewondo population stands out as an exception has been already observed in our previous study on autosomal and mitochondrial sequence data17 that also revealed Paleolithic expansions for all African farmer populations except the Ewondo population. The reasons why this population seems to have remained at constant size during both the Paleolithic and the Neolithic period will need to be further investigated.
Finally, note that some previous studies have revealed a recent bottleneck signal in African hunter-gatherer populations. For instance, Patin et al23 said that ‘a bottleneck beginning 2500–25 000 years ago with an 80% decrease in population size, followed by a recovery starting 125 years later with a size increase of between 100% and 400%' fits the data significantly better than a constant size model for West-African hunter-gatherer populations. Using a set of complete mitochondrial sequences, Batini et al16 also showed a recent bottleneck (4000–650 BP) for these populations. Accordingly, Excoffier and Schneider52 used simulation-based analyses to show that a post-Neolithic bottleneck event could explain the fact that they did not detect Pleistocene expansions for the ancestors of contemporary hunter-gatherer populations in Africa. They suggest that this bottleneck event may result from competition against farmers for land that may have led to a reduction in, and fragmentation of, available habitat and resources.52 The fact that we could not detect this bottleneck signal here could result from the parametric nature of our method that allows adequately detecting general demographic tendencies but cannot offer enough flexibility to detect short consecutive events.
It is interesting to note that we detected expansion signals for both sedentary farmers and nomadic herders in Central Asia. This could be because of the arid continental climate in this area that has strongly affected the cultural development and limited the spread of farmer civilizations up to 3000 YBP.27 The limitation of opportunities for farmers to expand in this area may have allowed herder population size to increase more than elsewhere. Alternatively, as suggested by Renfrew53 from linguistic data, current Central Asian herders may descend from sedentary agropastoralist populations (Jeitun culture). In this case, these populations may have experienced two successive transitions, the first occurring from hunter-gathering to agropastoralism and having potentially led to demographic expansions, and the second occurring from agropastoralism to nomadic herding.
Autosomal microsatellites allow inferring more recent expansions as compared with other markers
Previous population genetic studies also found demographic expansion events in farmers but not in hunter-gatherers in Africa using autosomal DNA sequences14, 23 or complete mitochondrial DNA sequences.16 Similarly, our previous work on HVS-1 data demonstrated strong signals of expansion in both African and Eurasian sedentary farmer populations, weaker expansion signals in Eurasian herders and no signal in African hunter-gatherers.17 All these studies reported expansions that started during the Paleolithic. Here, using autosomal microsatellite data, although confidence intervals were quite large, we inferred modal estimates for the expansion onsets during or after the Neolithic transition for all farmer populations in Africa and Eurasia, as well as for herder populations in Central Asia. This finding appears to be robust to the uncertainty of generation time in humans. Indeed, using a generation time of 29 years54 instead of 25 years11, 14, 23 yielded estimates that were only 1.2 times more ancient, thus still after the Neolithic transition (results not shown).
Our finding of recent expansions is consistent with Gignoux et al15 who detected Neolithic demographic expansions using mtDNA data. Indeed, by separately analyzing different haplogroups according to their time of origin, they reported expansions starting at approximately 3000 YBP in Africa, 8000 YBP in Europe and 5000 YBP in Southeastern Asia for the most recent haplogroups. They inferred demographic patterns at the continental scale by simultaneously analyzing individuals from multiple populations with various lifestyles. By studying a large set of populations, we were able to show here that (1) different expansion patterns are associated with different lifestyles, (2) demographic patterns (and their timing) are consistent between populations of the same group (ie, nomadic or sedentary) within each area and (3) peculiar regions such as Central Asia display atypical patterns. Moreover, as we pointed out above, mtDNA data can be affected by selection. The fact that we obtained similar expansion patterns from autosomal microsatellite data strongly suggests that these inferred expansions truly reflect demographic processes and not selective sweeps. Note that, for Eurasia, the inferred modal estimates for the expansion onsets are more recent than the Neolithic transition for all the studied populations, with no correlation between these inferred estimates and the datations of plant and animal domestications reported by Bocquet-Appel and Bar-Yosef3 (Table 2). This result could be explained by an initial increase in mortality because of the rapid changes in environmental pressures that occurred along with the Neolithic transition (eg, new diet, spread of infectious diseases, etc).5 Thus, population expansions may have not started right at the beginning of the Neolithic period, but rather at a later stage when, among other things, technological innovations and a better immunological resistance to the new pathogens allowed substantial demographic expansions.
Otherwise, most previous population genetics studies focusing on the past demographic history of African and Eurasian populations have inferred upper or middle Paleolithic expansions, but no Neolithic expansions. For Africa, using mitochondrial or autosomal sequences data, many authors inferred expansion times ranging from 80 000 to 25 000 YBP.10, 13, 14, 16 Using Y-chromosome microsatellites, Pritchard et al12 estimated more recent but still Paleolithic expansion times, between 15 000 and 12 000 YBP. Analyzing separately different mitochondrial haplogroups, Atkinson et al55 found several successive Paleolithic expansion waves occurring at different times depending upon the population, but earlier than 8000 YBP in all cases. For Eurasia, Chaix et al11 showed estimations of expansion times ranging between 63 000 and 17 000 YBP using HVS-I data, and between 40 000 and 11 000 YBP using Y-chromosome microsatellites (see also Pritchard et al12). Finally, one study showed expansion events in early Europeans farmers, starting at least 12 000 YBP according to an unbiased sample of complete mitochondrial DNA sequences.56 Altogether, these results suggest that different type of markers can be informative at different timescales. Furthermore, we found more recent expansion events in East-Asia than in the rest of Eurasia, unlike a previous study showing a Eurasian East-to-West gradient of Paleolithic expansions onsets.11 It suggests that we may have detected another set of expansion events, potentially resulting from the Neolithic transition. These events therefore do not appear to have followed the same East-to-West process as the Paleolithic expansions.
Finally, note that the fact that microsatellites may allow detecting more recent events than other type of markers with coalescent-based methods has previously been demonstrated with simulation-based analyses20 and empirical data analyses on non-human organisms.20, 21 However, note that Y-chromosome microsatellite-based studies,11, 13 including our own studies on Y-chromosome microsatellite data collected in the same populations as the ones investigated here (C Aimé et al, unpublished results), also pointed toward more ancient expansions. Thus, the higher sensitivity to recent events of autosomal microsatellites cannot only be explained by their relatively high mutation rates, as then similar results would be expected for these two types of markers. We suggest the increased sensitivity of autosomal microsatellites to recent demographic events may reflect the combination of a high mutation rate and of the fact that, unlike Y-chromosome microsatellites, autosomal microsatellites are independent loci and therefore provide independent replicates of the coalescent process. Indeed, mutations can accumulate on the different Y-chromosome haplotypes, and even if homoplasy may punctually reduce the differences between haplotypes, these differences are expected to increase with time. Conversely the autosomal microsatellites evolve independently and therefore this process of accumulation does not occur.
Conclusion and further work
Using microsatellite autosomal data for a large set of populations from several distant geographic areas, we have shown contrasted demographic histories that correlate with lifestyle differences. Benefiting from the high mutation rate of microsatellites and the simultaneous analysis of multiple independent loci, we detected more recent expansion events than some previous studies that may result from the sedentarization of farmer populations during the Neolithic transition. Finally, comparing our results with previous studies, we have shown that different types of markers can be informative at different timescales. It will be therefore interesting to extend those analyses to other types of genetic markers, such as genome-wide data. Simulation studies may also help to better understand the specific responses of each type of genetic marker to past demographical events.
Data accessibility
The data from Tishkoff et al32 (Human diversity panel) and the data from Cann et al33 (HGDP-CEPH Human Genome diversity cell line panel) come from the Marshfield clinic database. These data are available at http://research.marshfieldclinic.org/genetics/genotypingData_Statistics/humanDiversityPanel.asp. The data from Tishkoff et al32 are also available at www.med.upenn.edu/tishkoff/Supplemental/files.html and at http://chgr.mc.vanderbilt.edu/page/supplementary-data. Other data used in this study has been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted by the EBI, under accession number EGAS00001000652.
Acknowledgments
We warmly thank all volunteer participants. We are also grateful to Michael Fontaine for his help on some data analyses, and Friso Palstra for his assistance with English language use. We also thank the editor and two anonymous reviewers for helpful comments and suggestions. All computationally intensive analyses were run on the Linux cluster of the ‘Museum National d'Histoire Naturelle' (administrated by Julio Pedraza). This work was supported by ATM-MNHN grant ‘Les relations Sociétés-Natures dans le long terme', by the ANR grant ‘Altérité culturelle' (ANR-10-ESVS-0010) and by the ANR grant DemoChips (12-BSV7-0012). CA was financed by a PhD grant from the ‘Centre National de la Recherche Scientifique'. CA and FA received a travel grant from the NEFREX project funded by the European Union (People Marie Curie Actions, International Research Staff Exchange Scheme, call FP7-PEOPLE-2012-IRSES) to visit Alexei Drummond's group (developer of Beast) in New Zealand.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Bar-Yosef O, Belfer-Cohen A.From sedentary hunter-gatherers to territorial farmers in the LevantIn: Gregg SA (ed):: Between Bands and States Carbondale, IL, USA: Center for Archaeological Investigations, University at Carbondale; 1991181–202. [Google Scholar]
- Mazoyer M, Roudart L. A History of World Agriculture: From the Neolithic Age to the Current Crisis. London; 2006. [Google Scholar]
- Bocquet-Appel JP, Bar-Yosef O. The Neolithic Demographic Transition and Its Consequences. Springer; 2008. [Google Scholar]
- Bocquet-Appel JP. When the world's population took off: the springboard of the Neolithic demographic transition. Science. 2011;333:560–561. doi: 10.1126/science.1208880. [DOI] [PubMed] [Google Scholar]
- Caldwell JC, Caldwell BK. Was there a Neolithic mortality crisis. J Popul Res. 2003;20:153–168. [Google Scholar]
- Kingman JFC. The coalescent. Stochast Proc App. 1982;13:235–248. [Google Scholar]
- Emerson BC, Paradis E, Thebaud C. Revealing the demographic histories of species using DNA sequences. Trends Ecol Evol. 2001;16:707–716. [Google Scholar]
- Excoffier L, Heckel G. Computer programs for population genetics data analysis: a survival guide. Nat Rev Genet. 2006;7:745–758. doi: 10.1038/nrg1904. [DOI] [PubMed] [Google Scholar]
- Kuhner MK. Coalescent genealogy samplers: windows into population history. Trends Ecol Evol. 2008;24:86–93. doi: 10.1016/j.tree.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Alshamali F, Pereira JB, et al. The Expansion of mtDNA Haplogroup L3 within and out of Africa. Mol Biol Evol. 2012;29:915–927. doi: 10.1093/molbev/msr245. [DOI] [PubMed] [Google Scholar]
- Chaix R, Austerlitz F, Hegay T, Quintana-Murci L, Heyer E. Genetic traces of east-to-west human expansion waves in Eurasia. Am J Phys Anthropol. 2008;136:309–317. doi: 10.1002/ajpa.20813. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol Biol Evol. 1999;16:1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- Cox MP, Morales DA, Woerner AE, et al. Autosomal resequence data reveal late stone age signals of population expansion in Sub-Saharan African foraging and farming populations. PLoS One. 2009;4:e6366. doi: 10.1371/journal.pone.0006366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval G, Patin E, Barreiro LB, Quintana-Murci L. Formulating a historical and demographic model of recent human evolution based on resequencing data from noncoding regions. Plos One. 2010;5:e10284. doi: 10.1371/journal.pone.0010284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gignoux CR, Henn BM, Mountain JL. Rapid, global demographic expansions after the origins of agriculture. Proc Natl Acad Sci USA. 2011;108:6044–6049. doi: 10.1073/pnas.0914274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batini C, Lopes J, Behar DM, et al. Insights into the demographic history of African pygmies from complete mitochondrial genomes. Mol Biol Evol. 2011;28:1099–1110. doi: 10.1093/molbev/msq294. [DOI] [PubMed] [Google Scholar]
- Aime C, Laval G, Patin E, et al. Human genetic data reveal contrasting demographic patterns between sedentary and nomadic populations that predate the emergence of farming. Mol Biol Evol. 2013;30:2629–2644. doi: 10.1093/molbev/mst156. [DOI] [PubMed] [Google Scholar]
- Lavery S, Moritz C, Fielder DR. Genetic patterns suggest exponential population growth in a declining species. Mol Biol Evol. 1996;13:1106–1113. [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Ravigne V, Estoup A. Inference on population history and model checking using DNA sequence and microsatellite data with the software diyabc (v1.0) BMC Bioinformatics. 2010;11:401. doi: 10.1186/1471-2105-11-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Guo Y, Qi D, et al. Genetic structuring and recent demographic history of red pandas (Ailurus fulgens) inferred from microsatellite and mitochondrial DNA. Mol Ecol. 2011;20:2662–2675. doi: 10.1111/j.1365-294X.2011.05126.x. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Snirc A, Frantzis A, et al. History of expansion and anthropogenic collapse in a top marine predator of the black sea estimated from genetic data. Proc Natl Acad Sci USA. 2012;109:E2569–E2576. doi: 10.1073/pnas.1201258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin E, Laval G, Barreiro LB, et al. Inferring the demographic history of African farmers and Pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P, Austerlitz F, Estoup A, et al. Origins and genetic diversity of Pygmy hunter-gatherers from Western Central Africa. Curr Biol. 2009;19:312–318. doi: 10.1016/j.cub.2008.12.049. [DOI] [PubMed] [Google Scholar]
- Phillipson D. African Archaeology. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- Oslisly R.The history of human settlement in the Middle Ogooue Valleyin Weber W, White LJT, Vedder A, Naughton-Treves L (eds): African Rain Forest Ecology and Conservation: An Interdisciplinary Perspective New Haven (Connecticut) and London: Yale University Press; 2001. Chapter 7, pp101–118. [Google Scholar]
- Dirksen VG, Van Geel B.Mid to late Holocene climate change and its influence on cultural development in South Central Siberiain Scott EM, Alekseev AY, Zaitseva GI (eds): Impact of the Environment on Human Migration in Eurasia. NATO Science Series: IV: Earth and Environmental Sciences Vol 42Kluver Academic Publishers; 2004291–307. [Google Scholar]
- Nei M, Roychoudhury AK. Evolutionary relationships of human populations on a global scale. Mol Biol Evol. 1993;10:927–943. doi: 10.1093/oxfordjournals.molbev.a040059. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton University Press: Princeton; [Google Scholar]
- Harris DR.The origins and spread of agriculture and pastoralism in Eurasia: an overviewIn: Harris DR (ed): The Origins and Spread of Agriculture and Pastoralism in Eurasia London, UK: UCL Press University College; 1996 [Google Scholar]
- Verdu P, Becker NS, Froment A, et al. Sociocultural behavior, sex-biased admixture, and effective population sizes in Central African Pygmies and non-Pygmies. Mol Biol Evol. 2013;30:918–937. doi: 10.1093/molbev/mss328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, De Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Ségurel L, Martinez-Cruz B, Quintana-Murci L, et al. Sex-specific genetic structure and social organization in central Asia: insights from a multi-locus study. PLoS Genet. 2008;4:e1000200. doi: 10.1371/journal.pgen.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Georges M, Pachner M, Endicott P. Genetic diversity of four Filipino Negrito populations from Luzon: comparison of male and female effective population sizes and differential integration of immigrants into Aeta and Agta communities. Hum Biol. 2013;85:189–208. doi: 10.3378/027.085.0310. [DOI] [PubMed] [Google Scholar]
- Heyer E, Balaresque P, Jobling MA, et al. Genetic diversity and the emergence of ethnic groups in Central Asia. Bmc Genet. 2009;10:49. doi: 10.1186/1471-2156-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, Broman KW. Genotyping for human whole-genome scans: past, present, and future. Adv Genet. 2001;42:77–96. doi: 10.1016/s0065-2660(01)42016-5. [DOI] [PubMed] [Google Scholar]
- Kuhner MK, Yamato J, Felsenstein J. Estimating effective population size and mutation rate from sequence data using Metropolis-Hastings sampling. Genetics. 1995;140:1421–1430. doi: 10.1093/genetics/140.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Drummond AJ. Joint inference of microsatellite mutation models, population history and genealogies using transdimensional Markov Chain Monte Carlo. Genetics. 2011;188:151–164. doi: 10.1534/genetics.110.125260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ.Tracer, ver. 1.5. Available at http://beast.bio.ed.ac.uk/Tracer , 2007
- Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences (with discussion) Stat Sci. 1992;7:457–511. [Google Scholar]
- R Development Core Team . Vienna: R Foundation for Statistical Computing; 2011. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. Coda: convergence diagnosis and output analysis for MCMC. R News. 2006;6:7–11. [Google Scholar]
- Dib C, Faure S, Fizames C, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Loader C. Local Regression and Likelihood. New-York: Springer; 1999. [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BR, Van Der Linde A. Bayesian measures of model complexity and fit. J R Stat Soc B Stat Meth. 2002;64:583–616. [Google Scholar]
- Bolker B. Ecological Models and Data in R. Princeton: Princeton University Press; 2008. [Google Scholar]
- Short R. The biological basis for the contraceptive effects of breast feeding. Int J Gynaecol Obstet. 1982;25:207–217. doi: 10.1016/0020-7292(87)90406-1. [DOI] [PubMed] [Google Scholar]
- Cerny V, Pereira L, Musilova E, et al. Genetic structure of pastoral and farmer populations in the African Sahel. Mol Biol Evol. 2011;28:2491–2500. doi: 10.1093/molbev/msr067. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Schneider S. Why hunter-gatherer populations do not show signs of Pleistocene demographic expansions. Proc Natl Acad Sci USA. 1999;96:10597–10602. doi: 10.1073/pnas.96.19.10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew C.Language families and the spread of farmingin Harris DR (ed): The Origins and Spread of Agriculture and Pastoralism in Eurasia London: UCL Press; 199670–92. [Google Scholar]
- Tremblay M, Vezina H. New estimates of intergenerational time intervals for the calculation of age and origins of mutations. Am J Hum Genet. 2000;66:651–658. doi: 10.1086/302770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson QD, Gray RD, Drummond AJ. Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa. Proc Biol Sci. 2009;276:367–373. doi: 10.1098/rspb.2008.0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Rudan P, Paabo S, Krause J. Complete mitochondrial genomes reveal Neolithic expansion into Europe. PLoS One. 2012;7:e32473. doi: 10.1371/journal.pone.0032473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from Tishkoff et al32 (Human diversity panel) and the data from Cann et al33 (HGDP-CEPH Human Genome diversity cell line panel) come from the Marshfield clinic database. These data are available at http://research.marshfieldclinic.org/genetics/genotypingData_Statistics/humanDiversityPanel.asp. The data from Tishkoff et al32 are also available at www.med.upenn.edu/tishkoff/Supplemental/files.html and at http://chgr.mc.vanderbilt.edu/page/supplementary-data. Other data used in this study has been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted by the EBI, under accession number EGAS00001000652.


