Summary
CAG/CTG trinucleotide repeats are unstable, fragile sequences that strongly position nucleosomes, but little is known about chromatin modifications required to prevent genomic instability at these or other structure-forming sequences. We discovered that regulated histone H4 acetylation is required to maintain CAG repeat stability and promote gap-induced sister chromatid recombination. CAG expansions in the absence of H4 HATs NuA4 and Hat1 and HDACs Sir2, Hos2, Hst1 depended on Rad52, Rad57, and Rad5, and were therefore arising through homology-mediated post-replication repair (PRR) events. H4K12 and H4K16 acetylation were required to prevent Rad5-dependent CAG repeat expansions, and H4K16 acetylation was enriched at CAG repeats during S-phase. Genetic experiments placed the RSC chromatin remodeler in the same PRR pathway, and Rsc2 recruitment was coincident with H4K16 acetylation. Here we have utilized a repetitive DNA sequence that induces endogenous DNA damage to identify histone modifications that regulate recombination efficiency and fidelity during post-replication gap-repair.
Keywords: CAG trinucleotide repeat, histone H4K16 acetylation, post-replication repair, template switch, sister chromatid recombination, RSC chromatin remodeling
Introduction
CAG/CTG trinucleotide repeats can expand beyond a stable threshold of approximately 35 repeats and cause several heritable neurodegenerative diseases, including Huntington’s disease, myotonic dystrophy type 1, and many spinocerebellar ataxias (Mirkin, 2007). Once expanded, the repeat becomes increasingly unstable and prone to further expansion, exacerbating the disease phenotypes in successive generations (McMurray, 2010). Repeat expansions can occur during DNA processing events in which DNA is rendered transiently single stranded, allowing the repeats opportunity to form stable secondary structures that can interfere with nick repair, Okazaki fragment ligation, and replication fork progression (Mirkin, 2007). In addition, expanded repeats are fragile sites and breakage of the CAG repeat is highly length-dependent (Freudenreich, 2007). Repeat instability can occur via misalignment during DNA repair, 3′ strand slippage events, or incorporation of 5′ hairpin structures (McMurray, 2010; Mirkin, 2007; Panigrahi et al., 2005).
Histone modifications occur during different types of repair, but those that occur after double-strand break (DSB) induction are best understood. At the occurrence of a DSB, the MRX complex (MRN) binds the broken DNA ends and activates the Mec1 (ATR) and Tel1 (ATM) kinases, which will in turn phosphorylate histone H2A (H2AX in mammals), creating a γH2AX domain (Bao, 2011). The γH2AX domain acts as a platform for further recruitment of repair-associated proteins, including the NuA4 histone acetyltransferase (HAT) complex that acetylates histones flanking the break site (Downs et al., 2004; Murr et al., 2006). Histone H3 and H4 N-terminal tail acetylation peaks at 0.6 kb from a nuclease induced DSB in budding yeast two hours after HO-induction (Tamburini and Tyler, 2005), and can be detected to approximately 1.5 kb from a DSB break site in mammalian cells (Xu et al., 2010). In yeast, the catalytic component of the NuA4 complex is the HAT Esa1. Along with the HATs Hat1 and Gcn5, Esa1 is recruited to an HO DSB during homologous recombination (HR) (Tamburini and Tyler, 2005; Parthun et al., 2007), and also contributes to non-homologous end-joining (NHEJ) (Bird et al., 2002; Choy and Kron, 2002; Lin et al., 2008). In mammalian cells, the NuA4 complex homolog includes the HAT Tip60, the scaffold protein Trrap, the chromatin remodeler p400, and the helicases Ruvbl1 and Ruvbl2 (Seeber et al., 2013). As repair of a DSB progresses, histone deacetylases (HDACs) are recruited to the lesion, presumably to reestablish the original chromatin structure. In yeast, the HDACs Rpd3, Hst1, and Sir2 are recruited to an HO break, appearing as the repair process is completed (Tamburini and Tyler, 2005; Lin et al., 2008). In mammalian cells, the Sir2 ortholog SIRT1 is recruited to an I-SceI break, and HDAC1 and HDAC2 of the NuRD complex are required for efficient HR and NHEJ (Oberdoerffer et al., 2008; Bao, 2011).
Though it is well-established that histone acetylation occurs at a DSB, the consequence of histone acetylation to repair outcome or fidelity of the repair event remains unclear. Additionally, much less is known about the contribution of histone acetylation to other types of repair, such as HR at a single-stranded DNA gap or during post-replication repair (PRR), and the subset of HATs and HDACs that contribute to these repair pathways remain to be elucidated. CAG repeats create site-specific replication and repair difficulties due to the formation of hairpin structures. The inherent instability of the repeat tract allows repair fidelity to be assessed by monitoring expansions and contractions of the repeat (Fig. 1A). Therefore, CAG repeats present an interesting sequence at which to study the contribution of histone acetylation and chromatin factors to repair outcome.
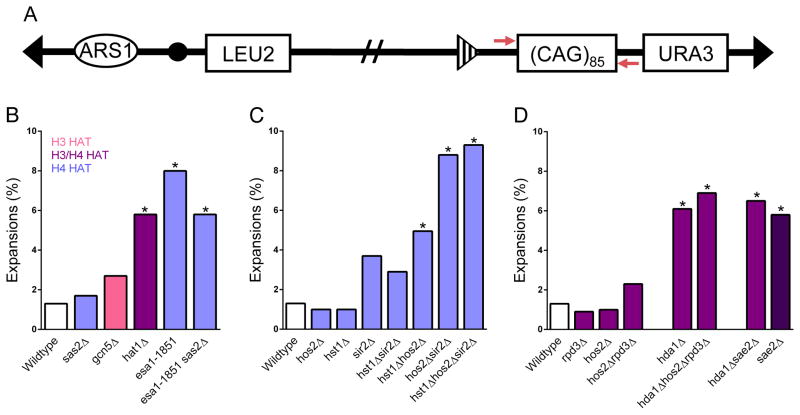
Figure 1. H4 HDACs and HATs protect against (CAG) 85 repeat expansions.
(A) A (CAG)85 tract is on the right arm of YAC CF1. Repeat length was measured by PCR amplification using primers (red) up- and downstream from the repeat. The frequency of (CAG)85 expansions was measured in strains lacking (B) histone H3 and H4 HATs, (C) histone H4 HDACs, (D) histone H3 HDACs and the endonuclease Sae2. Expansion frequencies were tested for significant deviation from wildtype frequency using Fisher’s exact test, *p<0.05. See also Table S1.
Here, an analysis of HATs and HDACs in Saccharomyces cerevisiae revealed that CAG repeat expansion frequency is significantly increased in mutants defective in either histone H4 N-tail acetylation or deacetylation, and this increase is dependent on homology-mediated recombination events as well as the error-free PRR protein Rad5, suggesting that endogenous damage at the CAG repeat requires dynamic H4 acetylation and deacetylation to promote PRR with fidelity. The RSC2 chromatin remodeler is recruited to the CAG repeat concurrent with H4K16 acetylation. Further, H4 acetylation by the NuA4 complex and RSC2 are both required for gap-induced sister chromatid recombination (SCR). We propose that acetylation of the H4 tail promotes gap-induced SCR through recruitment of RSC2 chromatin remodeling activity, and this is a general mechanism that ensures fidelity of post-replication recombination events.
Results
Histone H4 HATs Esa1 and Hat1 prevent CAG/CTG repeat expansions
Considering that CAG repeats strongly bind nucleosomes (Wang, 2007) and create a site-specific challenge to replication and repair, we sought to determine whether CAG stability was dependent on regulated histone acetylation. We utilized an expanded (CAG)85 repeat on a yeast artificial chromosome (YAC) and detected the frequency of repeat expansions and contractions by PCR (Fig. 1A; Table S1). While CAG expansions were not increased in the absence of Gcn5, which targets H3 and H2B N-tail lysines and is recruited to DSBs, or in the absence of Sas3, a HAT that together with Gcn5 is responsible for most H3 acetylation (Tamburini and Tyler, 2005; Lin et al., 2008; Kimura et al., 2002; Millar and Grunstein, 2006), CAG repeats were destabilized in the absence of H4 HATs Esa1 and Hat1 (Fig. 1B; Table S1).
Esa1 is the catalytic subunit of the NuA4 complex that acetylates all four lysines in the histone H4 tail as well as H2A N-terminal tail lysine residues (Millar and Grunstein, 2006; Smith et al., 1998; Allard et al., 1999), and is a primary HAT required for H4 acetylation in vivo (Millar and Grunstein, 2006; Smith et al., 1998; Allard et al., 1999; Clarke et al., 1999; Bird et al., 2002). ESA1 is essential in S. cerevisiae, therefore we used two mutant alleles to study Esa1 function: esa1-1851, which is hypersensitive to DNA-damaging agents and shows a loss of H4 acetylation at 30°C; and esa1-L357H, which is not DNA damage sensitive and displays wildtype levels of H4 acetylation at 30°C (Bird et al., 2002). CAG repeat expansions were significantly increased in the esa1-1851 mutant (Fig. 1B) but not in cells with the esa1-L357H allele at 30°C (Table S1), implicating H4 acetylation in preventing repeat expansions. Expansion frequency was not increased in an H2A-NΔ mutant (Table S1). Thus, H4 appears to be the relevant acetylation target of Esa1 for CAG repeat maintenance.
Hat1 acetylates newly synthesized H4 on lysines 5 and 12 (Ma et al., 1998). The hat1Δ mutant displayed a 4.5-fold increase in CAG repeat expansions over the wildtype (Fig. 1B). Deletion of Sas2, a HAT responsible for acetylation of H4K16 proximal to telomeres (Kimura et al., 2002), did not affect repeat stability (Fig. 1B). Thus, Esa1 and Hat1 are the primary H4 HATs necessary to prevent CAG expansions.
H4 HDACs protect against CAG repeat instability
Since efficient acetylation of histone H4 lysine residues is important to maintain CAG repeats, we next characterized the effect of histone deacetylation by examining CAG stability in HDAC mutants. Expansion frequency was slightly elevated in the sir2Δ and hst1Δsir2Δ mutants while the hst1Δhos2Δ, hos2Δsir2Δ, and hst1Δhos2Δsir2Δ mutants all displayed significant increases in CAG repeat expansion frequency (Fig. 1C). Since all three HDACs have specificity for H4K16 (Millar and Grunstein, 2006), this residue would not be efficiently deacetylated in the triple hst1Δhos2Δsir2Δ mutant, implicating H4K16 modification as an important contributor to CAG repeat stability. Interestingly, Hos2 and Hst1 appear to be working independently of the Set3 complex to prevent CAG repeat instability, as deleting other Set3 complex members or Sum1 (Pijnappel et al., 2001) did not increase CAG repeat instability (Table S1).
Rpd3 is a global HDAC that, redundantly with Hos2, catalyzes deacetylation of H3 and H4 N-terminal lysine residues, with the exception of H4K16 (Millar and Grunstein, 2006). Notably, neither the rpd3Δ nor the rpd3Δhos2Δ mutant showed an increase in CAG repeat expansions compared to wildtype cells (Fig. 1D); therefore this HDAC is not involved in protection against CAG expansions. Interestingly, loss of Rpd3 did lead to a significant decrease in contraction frequency (Table S1), similar to the suppression of (CAG)20 expansions observed after Rpd3L knockdown, which was attributed to stabilization of the Sae2 nuclease (Debacker et al., 2012). Hda1 also targets both histone H3 and the Sae2 nuclease (Robert et al., 2011), and we showed previously that Sae2 is required to prevent (CAG)70 expansions (Sundararajan et al., 2010). Since the expansion frequency in the hda1Δ mutant was equivalent to both the hda1Δhos2Δrpd3Δ and hda1Δsae2Δ mutants (Fig. 1D), we conclude that the relevant Hda1 target is most likely Sae2.
Taken together with the HAT mutant instability, our data show that H4 N-tail lysines must be both acetylated and deacetylated to prevent CAG expansions. This result suggests that a dynamic process is required, rather than a particular histone modification state. Of note, the HAT mutants esa1-1851, esa1-1851 sas2Δ and hat1Δ, as well as the HDAC mutants hst1ΔhosΔ2 and hst1Δhos2Δsir2Δ exhibited a significant or borderline significant increase in contractions as well, whereas the other tested HATs and HDACs did not (Table S1). The profile of HATs and HDACs involved in preventing CAG instability is overlapping but not identical to those found to play a role at an HO-induced break (Tamburini and Tyler, 2005; Lin et al., 2008). Thus, the primary type of damage caused by the expanded CAG repeat that requires H4 acetylation to be repaired with fidelity may not be a DSB.
Modifiable H4 N-terminal lysines prevent CAG repeat expansions
The HATs and HDACs that are most important in preventing CAG repeat expansions have overlapping specificity at H4 N-terminal lysines, but also have targets on H3 and potentially on proteins other than histones. To directly test if H3 or H4 N- tail lysines are important for CAG repeat stability we tested expansion frequency in a series of H3 and H4 mutants. Notably, the H3-NΔ mutant did not show a significant increase in expansions (Fig. 2A), demonstrating that H3 N-terminal lysine acetylation is not significantly affecting CAG repeat stability. To test the contribution of the H4 N-terminal lysine residues, repeat stability was measured in strains with unacetylatable arginine substitutions. Expansions were significantly increased in both the H4K12R and H4K16R mutants, but only modestly increased in the H4K5,8R mutant (Fig. 2A), establishing that H4K12 and H4K16 acetylation are of primary importance in maintaining CAG repeat stability. H4K16Q and H4K5,8,12Q strains, with glutamine mutations that mimic acetylated lysine, also displayed elevated levels of repeat expansions, confirming that deacetylation of the H4 tail is also important (Table S1). The presence of a single acetylatable residue in the H4 N-terminal tail rescued hypersensitivity to the DSB-inducing agent CPT and this rescue was independent of the position of the lysine on the N-terminus (Bird et al., 2002). Our results, however, reveal that acetylation of H4K12 and H4K16 are both vital to maintaining fidelity of repair within the CAG repeat, since loss of either acetylatable residue was almost as severe as the loss of three or all four acetylation sites (Fig. 2A). Nucleosome deposition in yeast is normal unless H4K5,8, and 12 residues are all mutated, along with the H3 N-terminus (Ma et al., 1998; Parthun, 2007), therefore the defects observed in the K12R and K16R mutants cannot be attributed to a deposition defect. The small additional increase in expansions in the H4K5,8,12R and H4K5,8,12,16R mutants could be due to an added effect due to a chromatin assembly defect present in these mutants but not in the single mutants, or alternatively indicate a minor level of redundancy in the repeat stability function of the H4 N-terminal lysines.
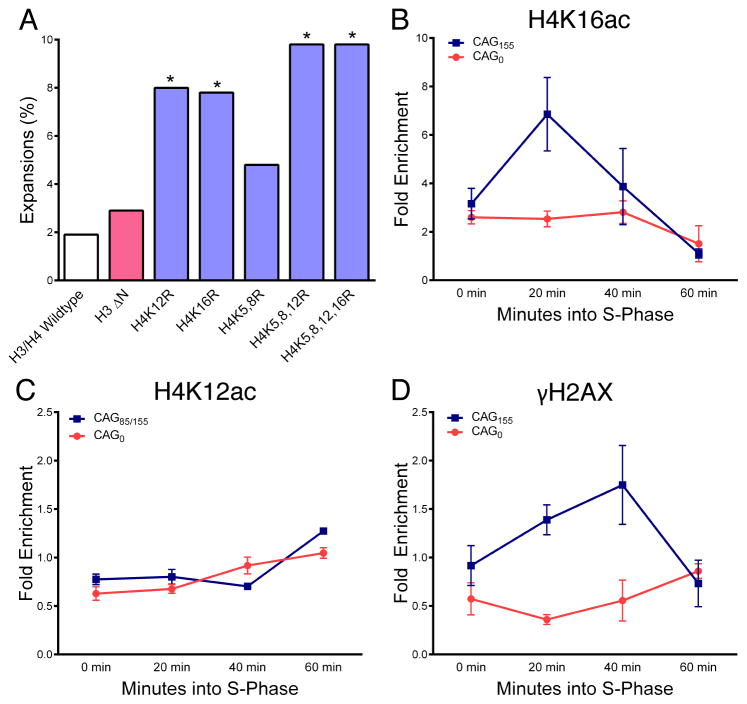
Figure 2. Acetylatable H4K12 and H4K16 maintain (CAG)85 repeat stability; H4K16 is acetylated at a CAG repeat tract.
(A) CAG tract lengths were measured by the PCR-based stability assay (Fig. 1A) in H3 or H4 mutant strains. In these cells, the chromosomal copies of H3 and H4 have been deleted and mutant H3 and H4 are expressed from a plasmid. Histone mutants are compared to a strain containing wildtype H3 and H4 expressed from the plasmid for statistical analysis by Fisher’s exact test, *p<0.05. (B-D) Histone H4K16ac, H4K12ac, and H2AS129 phosphorylation were detected in α-factor synchronized cultures by ChIP at time points after release into fresh media; qPCR used primers 0.4 and 0.6 kb upstream of the CAG repeat. Fold enrichment was calculated relative to an ACT1 internal control. The averages and standard errors (SEM) for at least three independent experiments are plotted. See also Figure S1 and S2.
H4K16 acetylation is enriched at an expanded CAG repeat during S-phase
ChIP analysis was used to determine whether acetylation of H4K12 and H4K16 residues was directly detectable at an expanded CAG repeat tract. The level of DNA damage at CAG repeats is relatively low: 10–20% of cells with an expanded CAG tract show some sign of stress (Sundararajan and Freudenreich, 2011), whereas an HO-induced DSB occurs in ~90% of cells (Tamburini and Tyler, 2005). Thus, we used cells with a longer (CAG)155 repeat tract to maximize the chance of detecting histone modification enrichment compared to a no repeat (CAG)0 control. Histone H4K16 acetylation was significantly increased at the CAG tract 20 minutes after cells were released from G1 phase (early S phase) whereas this enrichment was not observable at the no repeat control (Fig. 2B, Fig. S1A, B). The H4K16ac enrichment was not due to a change in histone levels, as histone H3 levels remained relatively constant over the time course (Fig. S1C). In contrast, H4K12 and H4K5 acetylation were not enriched at the CAG repeat compared to the no repeat control (Fig. 2C, S1D). Thus, though the genetic data shows that H4K12 acetylation is important for stability of the repeat, it does not appear to be specifically induced by repair. Interestingly, γH2AX levels were elevated at the CAG repeat coincident with H4K16 acetylation at 20 min into S phase but with a longer persistence, peaking at 40 min, mid-S phase (Fig. 2D; S1B). The level of γH2AX enrichment is consistent with the level observed at other impediments to replication (e.g. tRNA genes or LTRs, ~1.4-fold) (Szilard et al., 2010).
The transient nature of the H4K16 modification at the repeat during S-phase along with the relatively modest level of H2A-S129 phosphorylation and the known ability of CAG/CTG repeats to impede fork progression (Kerrest et al., 2009; Pelletier, 2003), suggested that dynamic histone acetylation is associated with a replication-coupled event. We were unable to detect enrichment of H4K16 acetylation at a fork stalled by hydroxyurea (HU) treatment by ChIP (Fig. S2); therefore, the H4K16 acetylation enrichment observed at the CAG repeat is likely not due to fork stalling alone. Instead, H4K16 acetylation could mark other fork-associated events that commonly occur at CAG repeats, including fork reversal, nicks, gaps, or sister chromatid recombination (Fouché et al., 2006; Panigrahi et al., 2005; Kerrest et al., 2009).
Histone H4 acetylation state maintains CAG repeat stability by promoting sister chromatid recombination during post-replication repair events
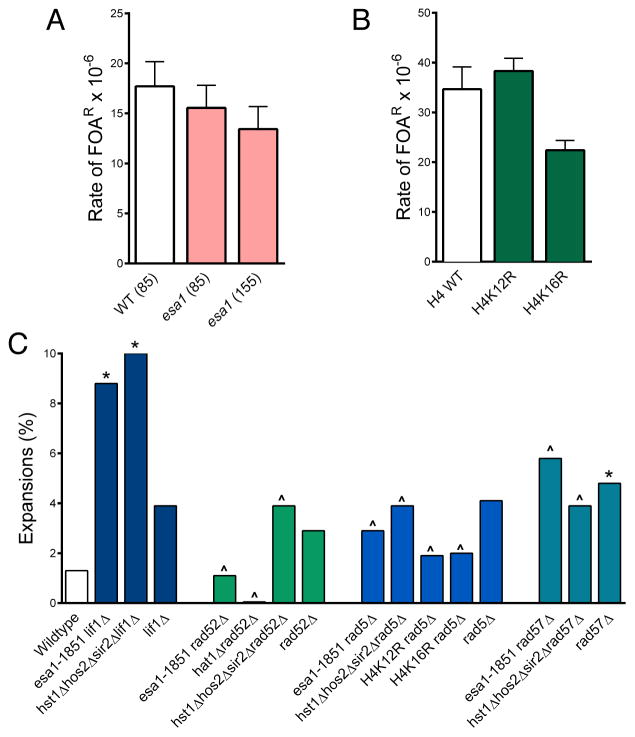
To determine if the absence of Esa1 or Hat1 activity results in increased DSBs or a defect in repairing DSBs occurring at the repeat, we tested chromosomal fragility of the CAG repeat in the esa1-1851, hat1Δ, and H4 K-to-R single mutants. CAG repeats did not show increased fragility in any of these mutants, including an esa1-1851 strain with a longer (CAG)155 repeat (Fig. 3A; Fig. 3B; Table S2). Therefore, any double-strand breaks that occur at the CAG repeat in the absence of H4 acetylation can be repaired with normal efficiency.
Figure 3. Expansions in the absence of H4 HATs and HDACs occur during homology-mediated repair of (CAG)85 repeats.
(A)Fragility of YAC CF1 (Fig. 1A) with 85 or 155 CAG repeats, as indicated, was measured by fluctuation assays (Sundararajan, 2010). The average of a minimum of three replicates per strain background is shown with standard error (SEM); esa1 refers to the esa1-1851 allele. Statistical deviation from wildtype was tested by the Student’s t-test. (B) As in (A) except using strains with the indicated mutated histone copies on a plasmid, compared to a strain with the wildtype H3/H4 genes on a plasmid and 85 CAG repeats on the YAC. (C) CAG tract lengths were measured by the PCR-based stability assay (Fig. 1); *p<0.05 to wildtype, ^p<0.05 suppression from HAT or HDAC mutant, Fisher’s exact test. See also Tables S2 and S1.
We next examined if expansions in the absence of H4 N-tail lysine acetylation and deacetylation are arising during NHEJ or HR. If expansions arise during a repair event, they should be suppressed in the absence of that pathway. Expansions in the esa1-1851 HAT or hst1Δhos2Δsir2Δ HDAC mutants were still elevated when Lif1, a DNA ligase IV complex member, was deleted, indicating that they are not arising during NHEJ-dependent events (Fig. 3C). In contrast, the absence of Rad52 suppressed repeat expansion and contraction frequency in both HAT and HDAC mutants to levels at or below the wildtype or rad52Δ single deletion levels (Fig. 3C; Table S1).
Considering that expansions were Rad52-dependent and the S-phase timing of H4K16 acetylation at the repeat, we examined the contribution of error-free post-replication repair and sister-chromatid recombination to CAG repeat maintenance. The error-free branch of PRR repairs replication-associated lesions using homology-mediated template switch, whereby the newly synthesized sister chromatid is used as a template for repair. The mechanism remains poorly understood, but error-free PRR and the generation of a template-switch intermediate requires the action of Rad5, a DNA-dependent ATPase and PCNA ubiquitin ligase, as well as the Rad51 paralog Rad57 (Vanoli et al., 2010; Minca and Kowalski, 2010). The increased repeat expansion frequency in the H4 HAT and HDAC mutants was suppressed in the absence of Rad5 or Rad57, and expansions occurring in the H4K12R and H4K16R mutants were also dependent on Rad5 (Fig. 3C). We conclude that CAG repeat expansions that occur in the absence of regulated H4 acetylation are arising during a Rad52-, Rad5- and Rad57-dependent event, such as the template switch that is required for homology-mediated post-replication repair.
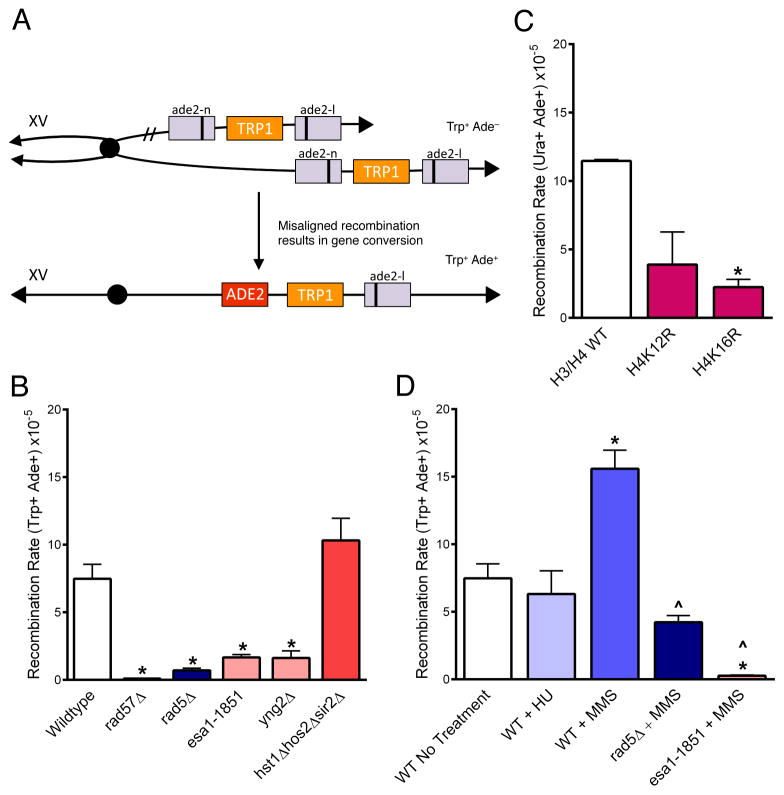
To evaluate if H4 acetylation is necessary to promote SCR in non-repetitive DNA, we took advantage of an assay that measures the rate of unequal SCR as an indicator of overall SCR rates (Mozlin et al., 2008; Fig. 4A; Table S3). We found that both Rad57 and Rad5 are required for SCR (Fig. 4B), in agreement with previous reports (Mozlin et al., 2008; Zhang and Lawrence, 2005). Interestingly, SCR rates were not decreased in the hst1Δhos2Δsir2Δ mutant, but rather were slightly increased (Fig. 4B). In contrast, SCR rates were significantly decreased from the wildtype in both the esa1-1851 mutant and a strain deleted for Yng2, a NuA4 complex member critical for histone H4 acetylation (Loewith et al., 2000; Choy and Kron, 2002) (Fig. 4B). Therefore, the HAT activity of NuA4 is required to promote wild-type levels of spontaneous SCR. To determine if the critical NuA4 substrate promoting SCR was the acetylated H4 tail as was the case for CAG stability, SCR rates were measured in the H4K12R and H4K16R mutants. SCR was suppressed in both mutants; the suppression in the H4K16R strain was significant and equivalent to the esa1-1851 and yng2Δ mutants (Fig. 4C). Thus, acetylation of the H4 tail, in particular K16 acetylation, is critical for spontaneous SCR.
Figure 4. Sister chromatid recombination depends on Esa1, Rad5, and H4K16 acetylation.
(A) Unequal sister chromatid gene conversion from Trp+ Ade− to Trp+ Ade+ can be used as a measure of sister chromatid recombination rates; Ade+ recombinants can also arise via intra-chromatid gene conversion but selection for Trp+ (or Ura+ in (C)) eliminates intrachromatid pop-out recombinants (Mozlin, 2008). Only gene conversion of the lower chromatid and the resulting product is shown. (B) Rates of spontaneous SCR. (C) SCR was measured in H4 point mutants using a Ura+ Ade- construct (URA3 in place of TRP1). Point mutants were expressed from a plasmid in cells in which the endogenous H3 and H4 genes were deleted. Point mutants were compared to a strain that expresses the H3/H4 wildtype from the plasmid for statistical analysis. (D) SCR rates were measured in cells treated for one hour with MMS (0.033%), or HU (0.2M). Statistical deviation using the Student’s t-test is indicated, *p<0.05 from wildtype, ^p<0.05 from wildtype MMS treated. Data are the average of at least 3 experiments +/− SEM. See also Table S3.
NuA4 is required for MMS-induced sister chromatid recombination
Based on our data, it seemed most likely that CAG expansions occurring when H4-tail acetylation is defective were arising during an SCR event initiated by damage at the repeat. We sought to determine if stalled forks or single-stranded DNA gaps could initiate SCR, and with what protein requirements. Cells were treated for 1 hr with HU to stall replication forks or methyl methanesulfonate (MMS) to induce single-stranded gaps; the short treatment favored the formation of single-stranded lesions as opposed to DSBs. While stalling replication forks with HU was not sufficient to induce SCR, single-stranded gaps created by MMS induced a significant increase in SCR (Fig. 4D), in agreement with previous results (Fasullo et al., 2001; Choy and Kron, 2002). The MMS-induced increase was suppressed in a rad5Δ strain, indicating that this treatment is inducing template switch events (Fig. 4D). NuA4 activity is also required for the MMS-induced increase in SCR, as it was significantly reduced in the esa1-1851 mutant (Fig. 4D). This suggests that H4 acetylation marks gaps that form when the replication fork bypasses a lesion (or CAG hairpin), and that this modification is required for efficient template switch recombination.
RSC1 and RSC2 promote CAG repeat stability and RSC2 is needed for gap-induced sister chromatid recombination
Acetylation dynamics could be influencing the efficiency and fidelity of post-replication repair by promoting one or more aspects of daughter strand gap repair. Acetylated histones can disrupt higher-order chromatin structure to facilitate repair protein binding (Shogren-Knaak et al., 2006; Murr et al., 2006). However, if the role of H4K12 and H4K16 acetylation was primarily to relieve chromatin condensation to facilitate repair, the HAT and HDAC mutants would be predicted to have opposite effects on CAG repeat stability. As this was not the case (Fig. 1), an alternative hypothesis is that acetylated residues could directly recruit one or more acetyl-lysine binding proteins that are important for gap induced HR repair. To test the direct recruitment model, we examined CAG repeat stability in the absence of proteins with bromodomains, which bind acetyl-lysine.
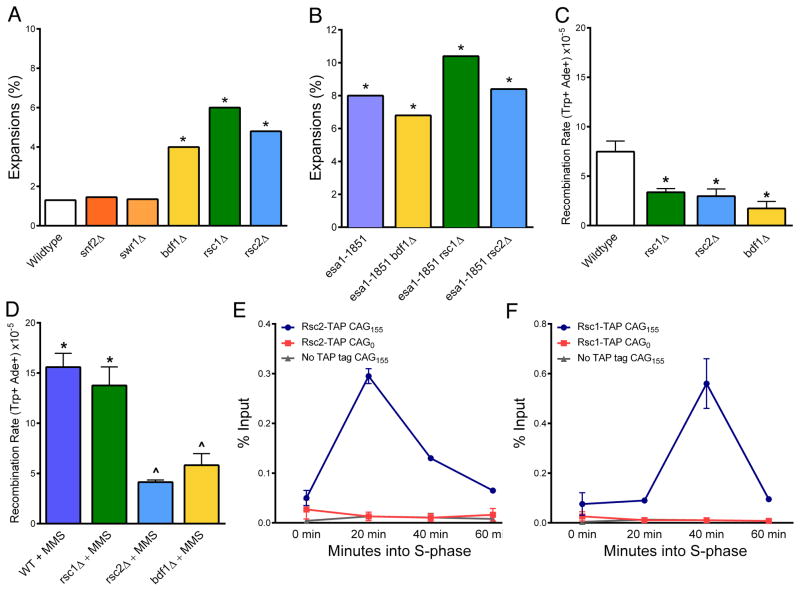
RSC is a multisubunit chromatin remodeler that exists in at least two different isoforms which contribute differentially to DNA repair: while RSC1 is required for nucleosome sliding at a DSB (Chambers et al., 2012), RSC2 is localized by ChIP to stalled replication forks and is required for PCNA ubiquitination after MMS and UV damage, as is the RSC homolog BAF180 in human cells (Niimi et al., 2012). We tested CAG stability in the absence of two RSC bromodomain-containing subunits, Rsc1 and Rsc2. CAG expansion frequency is significantly increased in both the rsc1Δ and rsc2Δ mutants (Fig. 5A), demonstrating the requirement of both RSC complex members in CAG repeat maintenance. In contrast, Snf2, the bromodomain-containing catalytic component of the SWI/SNF chromatin remodeler that is recruited to DSBs (Seeber et al., 2013), is not required for CAG repeat stability (Fig. 5A).
Figure 5. The RSC chromatin remodelers are required to prevent CAG repeat expansions, facilitate SCR, and are recruited to the CAG repeat.
(A, B) Instability was measured as in Fig. 1A; *p<0.05 to wildtype, Fisher’s exact test. (C) Spontaneous SCR rates were measured and analyzed as in Fig. 4A, B. (D) MMS-induced SCR as in Fig. 4D. (E–F) Recruitment of TAP-tagged Rsc proteins to 0.6 kb from the CAG repeat was measured by ChIP in α-factor synchronized cells at time points after release into fresh media; qPCR was run as in Fig 2B. Background levels were determined by IgG pull-down in a CAG-155 strain containing no TAP tag; percent of input sample precipitated is shown with no normalization to another locus. Averages and standard errors (SEM) for 2–4 independent replicates for each time point are shown.
Finally we tested Bdf1, a double bromodomain-containing protein that can directly interact with acetylated histone H3 and H4 N-terminal tails and can target the SWR1 histone exchange complex to chromatin (Kurdistani et al., 2004; Altaf et al., 2010). The absence of Bdf1 led to a significant increase in CAG expansions; however, a deletion of Swr1 or Htz1 had no effect (Fig. 5A; Table S1). Thus the action of Bdf1 in preventing CAG instability is not due to its role in SWR1 recruitment and histone exchange, and instead may be due to its ability to bind acetylated H4 tails.
If H4 acetylation is recruiting a bromodomain-containing protein to the CAG repeat, we expect to see epistasis between esa1-1851 and a mutant of that protein. Expansions in the esa1-1851 rsc1Δ, esa1-1851 rsc2Δ, and esa1-1851 bdf1Δ mutants are equivalent to the esa1 single mutant, consistent with Rsc1, Rsc2, and Bdf1 working within the same pathway as Esa1 to maintain CAG repeats (Fig. 5B). Also, similar to H4 acetylation mutants, expansions in both the rsc1Δ and rsc2Δ mutants were suppressed in the absence of Rad5 (Table S1), supporting that these remodelers play a role in the PRR template switch pathway. Strains mutant for Esa1, Rsc1, Rsc2, and Bdf1 have all been shown to be MMS-sensitive, with esa1 and rsc2 mutants showing the greatest sensitivity (Bird et al., 2002; Oum et al., 2011; Garabedian et al., 2012).
To assess if these proteins are contributing to SCR, we measured rates of unequal sister chromatid recombination in the absence of Rsc1, Rsc2, and Bdf1. Rsc1 and Rsc2 are required for wild-type levels of spontaneous SCR, consistent with a previous report using a different assay (Oum et al., 2011), and Bdf1 is also required (Fig. 5C). However, while MMS treatment leads to a significant increase in SCR in the rsc1Δ mutant, SCR is suppressed in the MMS-treated rsc2Δ and bdf1Δ mutants (Fig. 5D), similar to the esa1-1851 and rad5Δ mutants (Fig. 4D). This suggests that the RSC2 isoform, specifically, and Bdf1 are required for gap-induced SCR.
To further explore the difference between the function of the RSC2 and RSC1 remodelers during gap repair, we used ChIP to determine recruitment of the Rsc2 and Rsc1 subunits to the CAG repeat. Significantly, the Rsc2 protein was recruited to the CAG tract with the same timing as H4K16 acetylation, peaking 20 minutes into S phase, which supports the idea that H4K16ac could be recruiting RSC2. The Rsc1 protein was also strongly recruited to the repeat but with later timing, consistent with the data that it plays a role in preventing expansions and promoting SCR, yet with a different profile than esa1-1851, rsc2Δ and bdf1Δ strains with respect to repair of MMS-induced damage.
Discussion
Histone H4K16 acetylation is a novel marker of post-replication HR repair
Here we have identified a novel role for histone H4 acetylation in maintaining genomic stability during post-replication gap repair. Thus, like DSB repair, the error-free branch of PRR is regulated by histone modification, but with a different profile of HATs and HDACs involved. In particular, our data show that H4K16 acetylation by NuA4 is a repair-induced modification that is important for both the efficiency of sister chromatid recombination and the fidelity of repair. Deacetylation of H4K16 by the Hos2, Sir2, and Hst1 HDACs is also important as defects in this step impact repair fidelity and increase the likelihood of a CAG expansion; deacetylation is not required for normal levels of sister chromatin recombination, suggesting a role during the resolution step of repair.
A CAG or CTG hairpin structure can impair polymerase progression leading to a fork stall or a gap behind the fork that requires SCR to be resolved (Kerrest et al., 2009; Pelletier et al., 2003). Indeed, previous 2D analysis of replication through a (CTG)55 repeat on a yeast chromosome revealed the formation of joint molecules that could represent reversed forks or other template switch events between sister chromatids (Kerrest et al., 2009). Though DSBs can also occur at expanded CAG repeats, our results support that the pathway described here is primarily related to template switching and gap-induced recombination.
Role of chromatin remodeling at H4 acetylated regions
Our results indicate that both acetylation by HATs and deacetylation by HDACs are important for maintaining CAG stability. Therefore, though chromatin decompaction may be one important outcome of H4K16ac, high-fidelity repair requires dynamic acetylation. By deleting proteins containing a bromodomain that could act as an acetyl-lysine binding pocket, we identified Rsc2 as an attractive candidate for direct recruitment to H4 acetyl-lysines at a gap. Indeed, maximal Rsc2 recruitment coincides with maximal H4K16 acetylation at the CAG repeat.
The RSC remodeling complex has previously been implicated in NHEJ, HR, and spontaneous sister chromatid exchange (Seeber et al., 2013; Oum et al., 2011). Here we have shown that while both Rsc1 and Rsc2 are required for spontaneous SCR, Rsc2 is specifically required for MMS-induced SCR. In this respect, Rsc2 is similar to Esa1 and Rad5, suggesting cooperation with the Nu4A complex during repair of gaps. Rsc1 and Rsc2 are also recruited to the repeat at different times during the repair process: Rsc2 is recruited early, whereas Rsc1 is recruited later, when H2A phosphorylation peaks. The RSC remodeling complex was proposed to promote SCR by cohesin loading (Oum et al., 2011). However, in a previous study, CAG repeat instability was not increased in the scc1-73 cohesin mutant or the scc2-4 cohesin loading mutant (Gellon et al., 2011). Though we cannot rule out a role for RSC in cohesin loading as the temperature sensitive Scc1 and Scc2 mutants may retain some cohesion function, we favor a role for RSC2 in remodeling chromatin at the CAG repeat to promote repair.
The bdf1Δ mutant also exhibited increased CAG repeat expansions and was required for the MMS-induced increase in SCR, similar to Esa1 and Rad5. Free histone H4 is acetylated at lysines 5, 8, and 12 by Hat1, and this pattern is required for proper histone deposition during replication and repair (Ma et al., 1998; Parthun, 2007), and would therefore already exist on newly replicated DNA. Indeed, we did not detect a repeat-specific increase in acetylated H4K5 or K12. Bdf1 preferentially binds H4 acetylated on lysines 8 and 12 but not 16, thus as proposed (Millar et al., 2004), there could be a switch between an initial chromatin state where H4K12 is acetylated, H4K16 is not, and Bdf1 is bound, and a damage-induced state where H4K16 acetylation would disfavor Bdf1 binding and allow access by other factors (such as Rsc2) to the H4 tail. One possibility is that the H4K12R mutation, the absence of Hat1, or the absence of Bdf1 all impair this switch and thus the downstream events needed for efficient template switch recombination, explaining their phenotypes.
A model for H4 acetylation during post-replication gap repair
We suggest a model in which post-replicative lesions, such as a gap left behind after the fork encounters a lesion or hairpin that impedes replication, require acetylation of H4K16 for efficient Rad52-, Rad5-, and Rad57-dependent repair off the sister chromatid; for example, by stabilizing the template switch intermediate or facilitating D-loop progression (Fig. 6, left). At an HO-break, histone acetylation and NuA4 recruitment was abolished in the absence of Rad52 or Rad51, respectively, indicating that acetylation is triggered by an event during HR (Tamburini and Tyler, 2005; Bennett et al., 2013). Our data are also consistent with H4 acetylation occurring during recombination since expansions were suppressed to wildtype levels in the esa1-1851 rad52Δ and hat1Δrad52Δmutants (Fig. 3C). On the other hand, deacetylation likely occurs later in the process as repair is completed, since the H4K16ac mark had disappeared by late S/G2. We propose that dynamic acetylation promotes recombinational repair by acting as a locus-specific marker for efficient targeting of the chromatin remodeler RSC2 to the gap to promote efficient D-loop progression (Fig. 6; left). NuA4 binding may be facilitated by interaction with γH2AX (Downs et al., 2004), consistent with ChIP data showing that Mre11 and γH2AX are both present at an expanded CAG tract by the time of H4K16ac (Sundararajan et al., 2010; this study), or may be recruited by the Rad51-ssDNA filament as suggested by (Bennett et al., 2013). Our results do not rule out that H4-tail acetylation could affect the recruitment of additional repair factors, or that structural changes in chromatin induced by H4 acetylation could also contribute to the overall repair process.
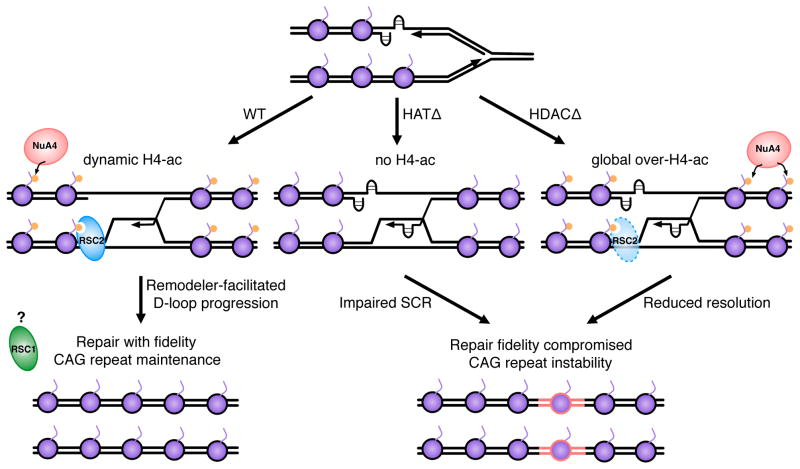
Figure 6. A model for dynamic histone H4K16 modification at CAG repeats in response to endogenous DNA damage.
At the occurrence of a replication-associated, single-stranded DNA gap, histone H4 is acetylated at lysine 16 (orange dots) to mark the lesion. Alternatively, a stalled or reversed fork could be the initiating event (not shown). These modifications promote high-fidelity sister chromatid recombination and repair, through recruitment of Rsc2 (left panel). Rsc1 may contribute to a later step, such as resolution. In the absence of H4 acetylation by NuA4, remodeler recruitment is impaired, leading to inefficient and low fidelity SCR (middle panel). In the absence of H4 deacetylation by the HDACs, SCR still occurs, but reduced resolution or multiple invasions results in repeat additions (red DNA).
In the context of repetitive DNA, the absence of proper acetylation and remodeling could lead to instability by impeding D-loop extension during gap repair. Therefore, in the absence of functional NuA4, a lack of H4K16 acetylation would lead to impaired recruitment of RSC2, inefficient SCR, and low fidelity repair (Fig. 6; middle). The dissociation of the 3′ end of the invading strand would allow opportunity for secondary structure formation in the repetitive DNA. In the orientation studied, a gap on the lagging strand template would be associated with a CTG repeat on the invading nascent lagging strand, which could form a hairpin leading to an expansion. Hairpin formation on one of the template strands, followed by failure to be copied, could lead to contractions. Alternatively, premature dissociation of the invading strand would allow opportunity for misalignment in a subsequent reinvasion, leading to a repeat length change (not shown).
H4 deacetylation, on the other hand, may either be important to promote repair factor dynamics at the site of the lesion or to designate when repair is complete, restoring chromatin compaction and inhibiting reinvasion by the Rad51 filament. The absence of the H4 HDACs would result in a global increase in H4 acetylation, leading to loss of the locus-specific signal of damage, impairing timely recruitment of repair factors to the lesion, and resulting in low fidelity repair (Fig. 6, right). Alternately, since SCR levels were normal or even slightly elevated in the hst1Δhos2Δsir2Δst rain, recruitment may be normal but resolution of the repair process may be impaired, leading to excessive D-loop extension or Rad51 filament reinvasions, and thereby more opportunities for CAG repeat expansions.
Balance between productive repair and repeat instability during post-replication HR
To maintain genomic stability, lesions must be repaired in a fashion appropriate to lesion type and stage of the cell cycle. Homologous recombination and post-replication template switching are mechanisms by which a lesion can be repaired using a homologous template to ensure no loss of genetic material; therefore these repair pathways are protective to genome integrity. In fact, both pathways have been demonstrated to prevent CAG repeat fragility and stability (Sundararajan et al., 2010; Daee et al., 2007; this study, Fig. 3C), presumably because when not available, other inappropriate repair pathways are engaged that lead to poor repair or repeat length changes. However, although protective for genome stability, if repair proceeds inappropriately or in a compromised manner, expansions can arise via HR. Recent studies in srs2Δ,mre11Δ, and ctf18Δ/dcc1Δ mutants showed that repeat expansions in these backgrounds were generated in a Rad51- or Rad52-dependent process (Kerrest et al., 2009; Sundararajan et al., 2010; Gellon et al., 2011), and those proteins all have important roles in controlling SCR. In addition, Rad5 was found to promote expansion of GAA and ATTCT repeats which was proposed to occur because of misalignment during template switching, resulting in addition of repeat units during synthesis (Shishkin et al., 2009; Cherng et al., 2011).
Altogether, these results indicate that Rad5-dependent template switching coupled to Rad52-dependent SCR is a common pathway for repairing lesions that occur at expanded repeats or other structure-forming sequences, but in a manner prone to generating repeat length changes if not properly regulated. The present study introduces the new finding that H4K16 acetylation is required to ensure accurate repair during Rad5-dependent template switches.
Conclusions
H4K16 acetylation has a demonstrated role in DNA repair in mice and human cells (Krishnan et al., 2011; Li et al., 2010; Sharma et al., 2010) and the loss of H4K16 acetylation has been associated with human cancers (Fraga et al., 2005; Pfister et al., 2008). Studies in human cells support that H4 acetylation is important in promoting HR events; for example, TIP60-dependent H4 acetylation has been shown to diminish 53BP1 binding and promote BRCA1-dependent HR (Tang et al., 2013), and depletion of MOF leads to deficient recruitment of repair factors to IR foci and a decrease in sister chromatid exchanges (Sharma et al., 2010; Li et al., 2010). Therefore, a role for H4 acetylation in promoting SCR may be conserved from yeast to humans. Given our results, this modification is predicted to be particularly important during post-replicative repair events in human cells, at both non-repetitive sequences and structure-forming repeats.
Our results add to the understanding of how the cell can use histone modification to direct repair of non-DSB lesions and to facilitate the fidelity of the repair process. Promoting repair fidelity will conserve genomic integrity and prevent loss of heterozygosity, protecting from cancer and disease. In addition, our data suggest that it is not only the marks themselves but the dynamic regulation of the histone modifications that is important to prevent mutagenic repair. A change in the balance of this regulation via expression differences of the HATs and HDACs involved or by other means could also influence the probability of trinucleotide repeat expansion, which varies widely between tissues and developmental windows.
Experimental Procedures
Yeast strains and plasmids
Yeast strains used in this study are listed in Table S4. Mutants were created using one-step gene replacement and gene disruptions were confirmed by PCR. Introduction of the esa1 alleles and H4 point mutations was confirmed by sequencing. Additional information is available in the Supplemental Experimental Procedures. All experiments were performed at 30°C.
CAG fragility and stability assays
Stability and fragility assays were performed on the YAC CF1 depicted in Fig. 1A as previously described (Sundararajan et al., 2010). Tract length changes were assessed using high-resolution gel images of PCR products in Photoshop. All expansion and contraction data are in Table S1; fragility assay data are in Table S2.
Sister chromatid recombination assays
Assays were performed as previously described (Mozlin et al., 2008) with the following modifications: strains were grown in 5ml YEPD at 30°C until saturation; total viable cell count was calculated by plating 10−5 dilutions on YC; recombinants were selected by plating 10−2 dilutions on YC-Ade-Trp or YC-Ade-Ura. For drug treatment, colonies freshly grown on solid YEPD were resuspended in 1ml YEPD with MMS (0.033%) or HU (0.2M), exposed for 1 hour at 30°C, and washed twice with distilled water before beginning the assay. Each assay was performed in at least triplicate; outliers were calculated using Grubb’s test and removed before statistical analysis using the Student’s t-test. All SCR rate data are in Table S3.
Chromatin immunoprecipitation
ChIP was performed 2–3 times per strain as previously described (Aparicio et al., 2005) using anti-phospho H2A-S129 (Abcam) and residue-specific H4 acetyl lysine antibodies that do not react to peptides with the corresponding lysine to arginine substitution (Fig. 2, Fig. S1, Upstate; Fig. S2, Millipore); Dynabeads (Novagen) or IgG agarose beads (Sigma) were used for immunoprecipitation. TAP-tagged Rsc proteins were isolated using IgG agarose (Sigma) or IgG sepharose 6 Fast Flow beads (GE Healthcare), and samples were washed in FA lysis buffer (Aparicio et al., 2005) with increased NaCl concentration (1M) to reduce background signal. DNA levels were quantified by qPCR using SYBR green PCR mastermix (Roche); qPCR reactions were run in duplicate or triplicate. PCR conditions and primer sequences are available upon request.
Supplementary Material
Highlights.
H4 HATs and HDACs promote post-replication HR repair to prevent CAG repeat expansions
Histone H4K16 acetylation is enriched during S-phase at an expanded CAG repeat tract
Gap-induced sister chromatid recombination requires H4K16ac and Esa1 of NuA4
The RSC2 remodeler contributes to gap repair and fidelity of template switch events
Acknowledgments
We thank L.F. Pemberton, A.L. Kirchmaier, O.J. Rando, and J.S. Thompson for providing the H4 plasmids, M. Grunstein for the H3-NΔ plasmid, F. Winston for the H2A-NΔ plasmid, M.F. Christman for the esa1 mutants, C.L. Peterson for a myc-tagged RFA1 strain, C. Wu for TAP-tagged RSC strains, and L. Symington for the SCR assay system. This work was supported by the National Institutes of Health (awards R01GM063066 and P01GM105473), Tufts University (awards to CF; NH; JY; SW; JM), and Sigma Xi (NH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18(18):5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Cote J. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. JBC. 2010;285(21):15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O, Geisberg JV, Sekinger E, Yang A, Moqtaderi Z, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Current Protocols in Molecular Biology. 2005;69:21.3.1–21.3.33. doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed] [Google Scholar]
- Bao Y. Chromatin response to DNA double-strand break damage. Epigenomics. 2011;3(3):307–321. doi: 10.2217/epi.11.14. [DOI] [PubMed] [Google Scholar]
- Bennett G, Papamichos-Chronakis M, Peterson CL. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nature Commun. 2013;(4):2084. doi: 10.1038/ncomms3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419(6905):411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Chambers AL, Brownlee PM, Durley SC, Beacham T, Kent NA, Downs JA. The two different isoforms of the RSC chromatin remodeling complex play distinct roles in DNA damage responses. PloS One. 2012;7(2):e32016. doi: 10.1371/journal.pone.0032016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19(4):2525–26. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherng N, Shishkin AA, Schlager LI, Tuck RH, Sloan L, Matera R, Mirkin SM. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. PNAS. 2011;108(7):2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Kron SJ. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol Cell Biol. 2002;22(23):8215–8225. doi: 10.1128/MCB.22.23.8215-8225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daee DL, Mertz T, Lahue RS. Postreplication repair inhibits CAG.CTG repeat expansions in saccharomyces cerevisiae. Mol Cell Biol. 2007;27(1):102–110. doi: 10.1128/MCB.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacker K, Frizzell A, Gleeson O, Kirkham-McCarthy L, Mertz T, Lahue RS. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biology. 2012;10(2) doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16(6):979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Fasullo M, Giallanza P, Dong Z, Cera C, Bennett T. Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics. 2001;158(3):959–972. doi: 10.1093/genetics/158.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature Genetics. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Fouche N, Ozgur S, Roy D, Griffith JD. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34(20):6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenreich CH. Chromosome fragility: Molecular mechanisms and cellular consequences. Frontiers in Bioscience. 2007;12:4911–4924. doi: 10.2741/2437. [DOI] [PubMed] [Google Scholar]
- Garabedian MV, Noguchi C, Ziegler MA, Mukund MD, Singh T, Nakamura T, Noguchi E. The double-bromodomain proteins Bdf1 and Bdf2 modulate chromatin structure to regulate S-phase stress response in S. pombe. Genetics. 2012;190:487–500. doi: 10.1534/genetics.111.135459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L, Razidlo DF, Gleeson O, Verra L, Schulz D, Lahue RS, Freudenreich CH. New functions of Ctf18-RFC in preserving genome stability outside its role in sister chromatid cohesion. PLoS Genetics. 2011;7(2) doi: 10.1371/journal.pgen.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrest A, Anand RP, Sundararajan R, Bermejo R, Liberi G, Dujon B, Richard GF. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nature Struct Mol Biol. 2009;16(2):159–167. doi: 10.1038/nsmb.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nature Genetics. 2002;32(3):370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Chow MZ, Wang Z, Zhang L, Liu B, Liu X, Zhou Z. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. PNAS. 2011;108(30):12325–12330. doi: 10.1073/pnas.1102789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Travasoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117(6):721–33. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Dou Y. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30(22):5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes and Development. 2008;22(15):2062–2074. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Bio. 2000;20(11):3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. PNAS. 1998;95(12):6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nature Rev Genet. 2010;11(11):786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nature Rev Mol Cell Biol. 2006;7(9):657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Millar CB, Kurdistani SK, Grunstein M. Acetylation of yeast histone H4 lysine 16: A switch for protein interactions in heterochromatin and euchromatin. Cold Spring Harb Symp Quant Biol. 2004;69:193–200. doi: 10.1101/sqb.2004.69.193. [DOI] [PubMed] [Google Scholar]
- Minca EC, Kowalski D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell. 2010;38(5):649–661. doi: 10.1016/j.molcel.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447(7147):932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Mozlin AM, Fung CW, Symington LS. Role of the saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics. 2008;178(1):113–126. doi: 10.1534/genetics.107.082677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature Cell Biol. 2006;8(1):91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Niimi A, Chambers AL, Downs JA, Lehmann AR. A role for chromatin remodellers in replication of damaged DNA. Nucleic Acids Res. 2012;40(15):7393–7403. doi: 10.1093/nar/gks453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oum JH, Seong C, Kwon Y, Ji JH, Sid A, Ramakrishnan S, Shim EY. RSC facilitates Rad59-dependent homologous recombination between sister chromatids by promoting cohesin loading at DNA double-strand breaks. Mold Cell Biol. 2011;31(19):3924–3937. doi: 10.1128/MCB.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12(8):654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- Parthun MR. Hat1: The emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26(37):5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Tam M, Wang YH, Montgomery SE, Dar AC, Cleary JD, Nichol K. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: Slipped-out repeats and slip-out junctions. Nucleic Acids Research. 2002;30(20):4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM. Replication and expansion of trinucleotide repeats in yeast. Mol Cell Biol. 2003;23(4):1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S, Rea S, Taipale M, Mendrzyk F, Straub B, Ittrich C, Lichter P. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer. 2008;122(6):1207–1213. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15(22):2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Foiani M. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471(7336):74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Hauer M, Gasser SM. Nucleosome remodelers in double-strand break repair. Curr Opin Genetics Dev. 2013;23(2):174–184. doi: 10.1016/j.gde.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Pandita TK. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30(14):3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishkin AA, Voineagu I, Matera R, Cherng N, Chernet BT, Krasilnikova MM, Mirkin SM. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol Cell. 2009;35(1):82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. PNAS. 1998;95(7):3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan R, Freudenreich CH. Expanded CAG/CTG repeat DNA induces a checkpoint response that impacts cell proliferation in saccharomyces cerevisiae. PLoS Genetics. 2011;7(3):e1001339. doi: 10.1371/journal.pgen.1001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan R, Gellon L, Zunder RM, Freudenreich CH. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in saccharomyces cerevisiae. Genetics. 2010;184(1):65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, Jacques PE, Laramee L, Cheng B, Galicia S, Bataille AR, Durocher D. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nature Structural and Molecular Biology Struct Mol Biol. 2010;17(3):299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25(12):4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature Structural & Molecular Biology Struct Mol Biol. 2013;20(3):317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genetics. 2010;6(11):e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH. Chromatin structure of repeating CTG/CAG and CGG/CCG sequences in human disease. Frontiers in Bioscience. 2007;12:4731–4741. doi: 10.2741/2422. [DOI] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191(1):31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. PNAS. 2005;102(44):15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BO, Wang SS, Zhang Y, Fu XH, Dang W, Lenzmeier BA, Zhou JQ. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genetics. 2011;7(1):e1001272. doi: 10.1371/journal.pgen.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.