Summary
MicroRNAs (miRNAs) are essential for regulation of gene expression. Though numerous miRNAs have been identified by high throughput sequencing, few precursor miRNAs (pre-miRNAs) are experimentally validated. Here we report a strategy for constructing high-throughput sequencing libraries enriched for full-length pre-miRNAs. We find widespread and extensive uridylation of Argonaute bound pre-miRNAs, which is primarily catalyzed by two terminal uridylyltransferases: TUT7 and TUT4. Uridylation by TUT7/4 not only polishes pre-miRNA 3′ ends, but also facilitates their degradation by the exosome, preventing clogging of Ago with defective species. We show that the exosome exploits distinct substrate preferences of DIS3 and RRP6, its two catalytic subunits, to distinguish productive from defective pre-miRNAs. Furthermore, we identify a positive feedback loop formed by the exosome and TUT7/4 in triggering uridylation and degradation of Ago-bound pre-miRNAs. Our study reveals a pre-miRNA surveillance system that comprises TUT7, TUT4 and the exosome in quality control of miRNA synthesis.
Introduction
MicroRNA biogenesis begins with processing of primary miRNA transcripts (pri-miRNAs) by the Microprocessor, composed of the RNase III enzyme Drosha and DGCR8, into hairpin structured precursor miRNAs (pre-miRNAs) with 5′ phosphates and predominantly 2-nucleotide (nt) 3′ overhangs (Kim et al., 2009; Wilson and Doudna, 2013). Alternatively, the splicing machinery and the debranching enzyme, either directly or with help from unknown 5′ exonucleases or the exosome, liberate pre-miRNAs that reside close to one or both boundaries of introns (mirtrons) (Yang and Lai, 2011). Pre-miRNAs are further processed by a second RNase III enzyme Dicer, together with TRBP and Ago in the miRNA loading complex (miRLC), into miRNA duplexes (Kim et al., 2009; Wilson and Doudna, 2013). One strand of the duplex is loaded into one of the four Ago proteins to assemble the microRNP (miRNP) or the RNA Induced Silencing Complex (RISC) (Hammond et al., 2001; Mourelatos et al., 2002). Chaperones and co-chaperones assist miRNA loading to Agos (Iwasaki et al., 2010; Miyoshi et al., 2010). pre-miR-451, whose processing is Dicer-independent, is loaded into the miRNA precursor deposit complex (miPDC) (Liu et al., 2012), where it is cleaved by Ago2 and trimmed into mature miR-451 (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010).
As an important class of regulatory RNAs, miRNAs are also regulated (Obernosterer et al., 2006; Siomi and Siomi, 2010). One pathway involves untemplated addition of nucleotides to the 3′ end of miRNAs or pre-miRNAs by terminal uridylyltransferases (TUTs) (Kim et al., 2010). For instance, extensive base pairing between a miRNA and its target leads to destabilization of the miRNA via trimming and tailing (Ameres et al., 2010). In contrast, adenylation of miR-122 by TUT2 is crucial for maintaining its stability in the liver (Katoh et al., 2009). In embryonic stem (ES) cells, oligo-uridylation of pre-let-7 induced by Lin28a and TUT4 (ZCCHC11), sequesters pre-let-7 away from the pre-miRNA processing machinery, and attracts the exosome-independent 3′ exonuclease DIS3L2 for degradation (Chang et al., 2013; Hagan et al., 2009; Heo et al., 2008; Heo et al., 2009; Newman et al., 2008). On the contrary, in somatic cells, mono-uridylation of pre-let-7 family members with 1-nt 3′ overhangs by TUT7, TUT4, or TUT2 promotes their processing by Dicer (Heo et al., 2012).
The exosome is a major 3′–> 5′ ribonucleolytic complex that is found in both the nucleus and the cytoplasm and plays crucial roles in the processing, maturation, surveillance and degradation of many types of RNAs (Doma and Parker, 2007; Houseley and Tollervey, 2009). It is an evolutionarily conserved complex that is composed of an inert nine protein structural core and two catalytic subunits: DIS3 (RRP44) and RRP6 (EXOSC10) (Liu et al., 2006; Mitchell et al., 1997). DIS3 contains a PIN domain that functions as an endoribonuclease (Lebreton et al., 2008) and an RNase II/R homologous domain that functions as a processive, hydrolytic 3′–> 5′ exoribonuclease, which provides the predominant activity of DIS3 (Dziembowski et al., 2007). RRP6 is a distributive, hydrolytic 3′–> 5′ exoribonuclease (Briggs et al., 1998). The composition of mammalian exosomes is expanded by the presence of a DIS3 homolog DIS3L, which can substitute for DIS3 in the cytoplasm (Staals et al., 2010; Tomecki et al., 2010). RRP6 is localized in both the nucleus and the cytoplasm in mammals (Tomecki et al., 2010; van Dijk et al., 2007). Therefore, although exonucleolytic activities of nuclear exosomes are furnished by DIS3 and RRP6, those of cytoplasmic exosomes may be mediated by RRP6 with either DIS3 or DIS3L in mammals (Staals et al., 2010; Tomecki et al., 2010). The function of DIS3 depends largely on the integrity of the exosome structural core (Wasmuth and Lima, 2012), whereas RRP6 also has core exosome independent targets and functions (Ibrahim et al., 2010). In yeasts, the exonucleolytic activity of the nuclear exosome is stimulated by the TRAMP polyadenylation complex, which is composed of a noncanonical poly(A) polymerase Trf4p/Trf5p, a zinc-knuckle RNA binding protein Air1p/Air2p and the RNA helicase Mtr4p (LaCava et al., 2005; Vanacova et al., 2005; Wyers et al., 2005). In mammals, the TRAMP complex is composed of poly(A) polymerase TUT3, ZCCH7 and MTR4 (Lubas et al., 2011). The short tail added by the TRAMP complex facilitates the exosome in digesting RNA substrates with secondary structures. However, it is presently unknown whether there are similar factors that promote the exonucleolytic activity of mammalian exosomes in the cytoplasm.
MicroRNA biogenesis consists of multiple maturation events. Yet, it is largely unknown, particularly in mammals, how cells control the quality of this elaborate and thus error-prone process. One quality control pathway for miRNAs and siRNAs in plants or flies is via decay of small RNAs that lack 2′-O-methylation in their 3′ ends from Agos by noncanonical TUTs, such as HESO1 or MUT68 (Ameres et al., 2010; Ibrahim et al., 2010; Ren et al., 2012; Zhao et al., 2012). However, mammalian miRNAs are not methylated and the relevance of this surveillance pathway for mammals is not apparent.
Here, we report the first genome-wide profiling of full-length pre-miRNAs bound to Agos, key proteins in miRNA biogenesis and function. We find widespread and extensive uridylation of the 3′ end of Ago-bound pre-miRNAs that is primarily catalyzed by TUT7 and TUT4. Uridylation facilitates either processing or degradation of pre-miRNAs by the exosome to prevent clogging of Ago, which is found in limited quantities in cells (Grimm et al., 2010), by defective pre-miRNAs. The exosome exploits distinct substrate preferences of its two catalytic subunits DIS3 and RRP6 to distinguish productive from faulty pre-miRNAs, and forms a positive feedback loop with TUT7/4 in triggering degradation of Ago-bound pre-miRNAs. Our study reveals a pre-miRNA surveillance system that consists of TUT7, TUT4 and the exosome in quality control of miRNA synthesis.
Results
A strategy for constructing high-throughput sequencing libraries of full-length pre-miRNAs
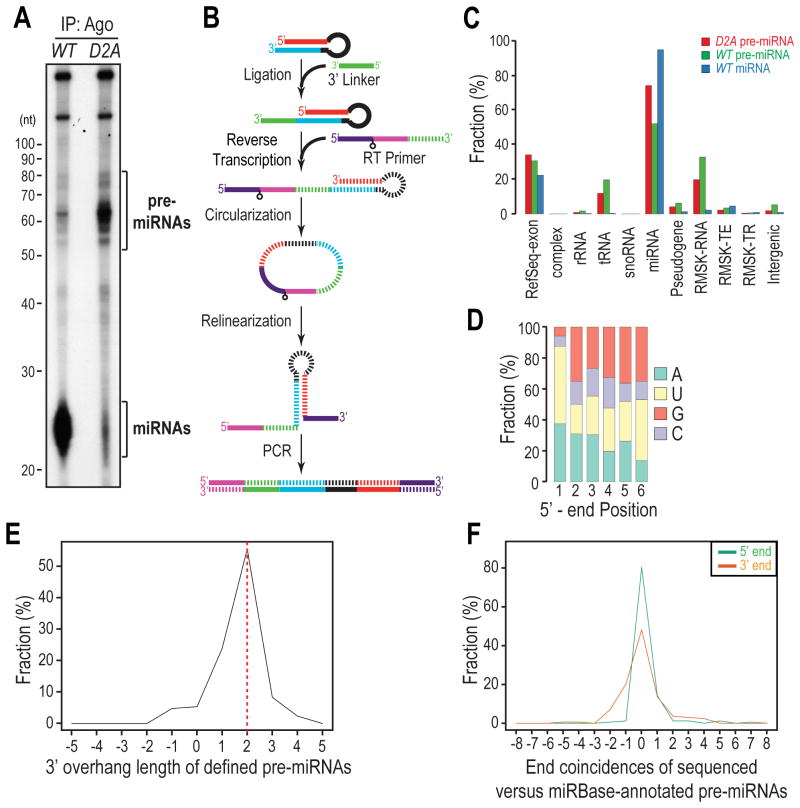
Although innumerable small RNAs have been identified by high-throughput sequencing (HITS) analyses, few full-length pre-miRNAs have been experimentally identified in vivo. pre-miRNAs are far fewer than miRNAs and their size overlaps that of other, far more abundant RNAs such as tRNAs and snRNAs. Indeed, pre-miRNAs constitute only a minute population (< 1%) from size-selected libraries prepared from total RNAs in HITS studies (Burroughs et al., 2012; Li et al., 2013). Furthermore, the extensive secondary structure of pre-miRNAs makes them poor substrates for RNA ligases. Agos are core protein components that bind to pre-miRNAs in both miPDC and miRLC (Liu et al., 2012). We exploited this property to purify pre-miRNAs and prepared HITS libraries from Ago immunoprecipitates (IPs) from Dicer Knock-Out mouse embryonic fibroblasts (MEFs), which have been transduced with either wild-type (WT) or catalytically inactive (D2A) Dicer (Liu et al., 2012; Tan et al., 2009). We surmised that pre-miRNAs should accumulate in D2A MEFs while miRNAs should be detected primarily in WT MEFs. Indeed, 50-80 nt RNAs, which match the size range of pre-miRNAs, accumulated in D2A MEFs, indicating that they were Dicer substrates (Figure 1A). Consistent with the essential role of Dicer in miRNA biogenesis, miRNAs were detected primarily in WT MEFs (Figure 1A).
Figure 1. Genome-wide profiling of Ago-bound, full-length precursor miRNAs; see also Figure S1.
A. Autoradiography of 5′-end radiolabeled RNAs isolated from Ago immunoprecipitates (IPs) from Dicer-null MEFs reconstituted with wild-type (WT) or catalytically inactive (D2A) Dicer transgenes.
B. Schematic of preparing pre-miRNA HITS libraries. Striped segments are complementary to solid segments with the same color.
C. Genomic annotation of mapped reads. Complex: mouse complex/cluster regions. RMSK-RNA: RepeatMasker annotated RNA-repeats, such as rRNAs, tRNAs, snRNAs, etc. RMSK-TE: transposable elements, including DNA transposons, LINEs, and SINEs. RMSK-TR: tandem repeats, including micro satellite, simple and low-complexity repeats.
D. Nucleotide composition for the first 6 positions of defined pre-miRNAs in the D2A library.
E. 3′ overhang length distribution of defined pre-miRNAs in the D2A library. Positive and negative values indicate 3′ and 5′ overhangs respectively. The same rule applies to all other figures.
F. Coincidence between annotated 5′ ends of 5p or 3′ ends of 3p miRNAs in miRBase and the defined 5′-or 3′-ends of pre-miRNAs sequenced from the D2A library. Positive and negative values indicate that the defined ends of pre-miRNAs are within and beyond the annotated ends of miRNAs in miRBase respectively.
Our initial attempts in constructing pre-miRNA HITS libraries using the classical small RNA cloning protocol were not successful as we failed in ligating the 5′ linker to pre-miRNAs due to their strong secondary structure and recessed 5′ ends, which made them inaccessible to the RNA ligase. To solve this problem, we adapted a method that attached the 5′ linker through CircLigase-mediated cDNA circularization (Ingolia et al., 2009) (Figure 1B), and prepared HITS libraries of miRNAs from WT MEFs and of pre-miRNAs (50-80 nt) from both WT and D2A MEFs (Figure 1A).
The majority of reads in pre-miRNA libraries represent bona fide pre-miRNAs
We obtained more than 10 million reads from each library that mapped to the genome. The length distribution of mapped reads resembled that in the Ago IPs for both pre-miRNA libraries (Figures 1A and S1A–B), suggesting that our library construction did not suffer from biases of capturing pre-miRNAs of different lengths. 50-80% of the mapped reads from the two pre-miRNA libraries were aligned to miRNA loci (Figure 1C and Table S1). To verify that these reads represented genuine pre-miRNAs, we first computationally defined their 5′ and 3′ ends by selecting one of the most frequently cloned and fully-mapped sequences for each pre-miRNA. Most defined ends of pre-miRNAs from the D2A library were aligned precisely to those from the WT library (Figure S1C), suggesting that inactivation of Dicer did not perturb the pri-miRNA processing machinery. The defined ends of pre-miRNA candidates display many characteristics of bona fide pre-miRNAs. For instance, > 85% of them begin with 5′-uridine/-adenosine (Figure 1D) and > 90% of them have 1-3 nt 3′ overhangs, with > 50% possessing the 2-nt 3′ overhang signature of Drosha cleavage (Figure 1E). Furthermore, > 85% of their 5′ ends and > 50% of their 3′ ends match the annotated 5′ end of 5p miRNAs and the 3′ end of 3p miRNAs in miRBase, respectively (Figure 1F). Interestingly, in contrast to defined 5′ ends, defined 3′ ends of pre-miRNAs are more likely to diverge from the annotated ends in miRBase, suggesting that 3′ ends of Ago-bound pre-miRNAs may be enzymatically polished after Drosha processing.
Widespread and extensive RNA modifications in Ago-bound pre-miRNAs
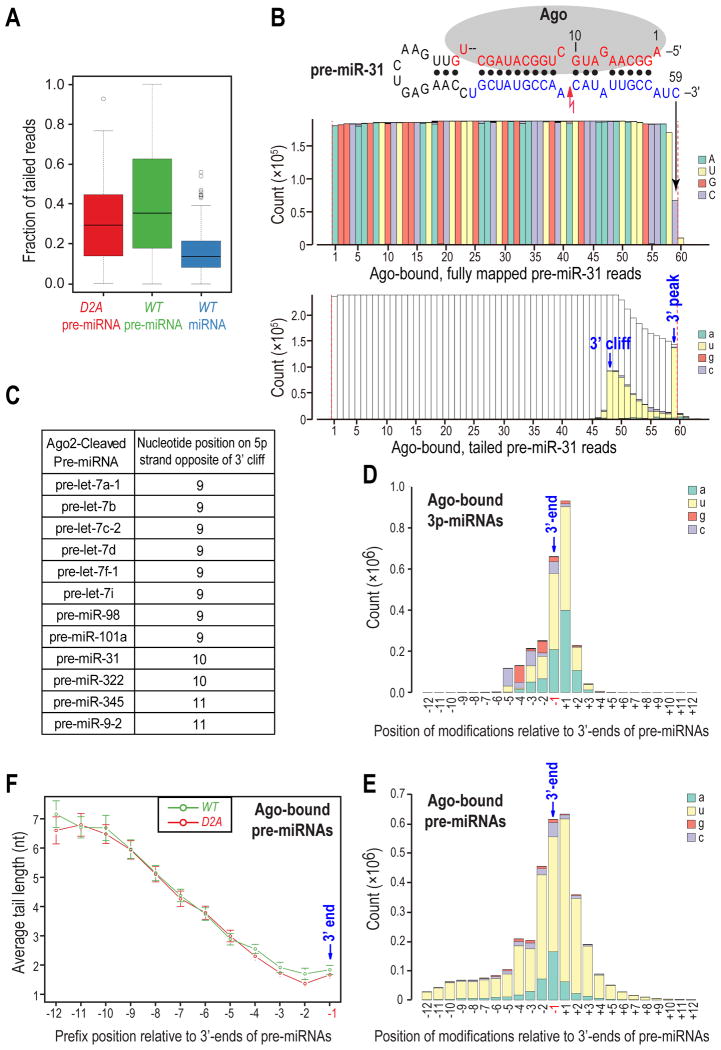
Interestingly, we found that more than half of the identified Ago-bound pre-miRNAs were represented by many sequencing reads (30-60% of total) with non-genome-matching nucleotides (RNA modifications) at their 3′ ends (tailed reads) (Figure 2A and Table S1). These extended reads contained predominantly uridines in their tails and some reads actually harbored oligo(U) tails as long as 6 nts (Figure S2A). RNA tails in pre-miRNA libraries form two patterns: a 3′ peak close to the 3′ end and a 3′ cliff that is > 10 nts away from the 3′ end (Figure 2B). Though the majority of pre-miRNAs have 3′-peaked tails, a small subset contains both types of tails (Figure 2C). Notably, reads with 3′-cliffed tails are found in both WT and D2A libraries, indicating that they are not dependent on the catalytic activity of Dicer. Among the pre-miRNAs with 3′ cliffed tails are all the pre-let-7 family members that are cleaved by Ago2 (Diederichs and Haber, 2007) (Figure 2C), indicating that 3′-cliffed tails are formed by addition of uridines to the 3′ end of Ago2-cleaved pre-miRNAs (ac-pre-miRNAs). In support of this notion, the majority of 3′ cliffs (12/14) are found opposite to nucleotides 9 to 11 of pre-miRNAs, which is consistent with Ago2 using the 5p strand of pre-miRNAs as a guide to cleave the 3p strand (Figures 2B, C). Furthermore, pre-miR-31, which was not known to be an Ago2 substrate, was cleaved by Ago2 both in vivo and in vitro (Figures S2B, C). Taken together, these results indicated that Ago2-cleaved pre-miRNAs are frequently tailed after the cleavage site.
Figure 2. Widespread and extensive post-transcriptional modifications in Ago-bound pre-miRNAs; see also Figure S2.
A. Box plot of uridylation ratio (fraction of tailed reads) for each pre-miRNA or miRNA in each library.
B. Nucleotide composition for every position of Ago-bound pre-miR-31 reads in WT pre-miRNA library. Red lightning bolt denotes Ago2-mediated cleavage site. Red dotted lines delineate the computationally defined 5′ and 3′ ends. Top panel: fully-mapped reads. Lower panel: tailed reads. Genome-matching nucleotides in tailed reads are not colored for clarity. Same depictions apply to all figures.
C. Ago2-cleaved pre-miRNAs identified by 3′ cliffs in both WT and D2A pre-miRNA libraries.
D, E. Nucleotide composition of RNA modifications surrounding the defined 3′ ends of 3p miRNAs (D) or pre-miRNAs (E) in WT libraries.
F. Average tail length of reads with the indicated prefix (last genome-matching) positions in pre-miRNA libraries. Error bars represent 95% confidence intervals (CIs).
Tails in the WT miRNA library contain similar amounts of adenosines and uridines (Figure S2D). Consistent with previous reports (Berezikov et al., 2011; Chiang et al., 2010), we found that tails of 3p miRNAs are enriched in both adenosines and uridines (Figure 2D), while 5p miRNAs predominantly have adenosines in their tails (Figure S2E). As 5p miRNAs can only acquire tails after Dicer processing whereas 3p miRNAs can also inherit tails from their precursors (Chiang et al., 2010), adenosines and uridines in 3p miRNAs' tails are primarily added after Dicer processing and inherited from their precursors respectively. When we plotted RNA modifications from WT (Figure 2E) or D2A pre-miRNA library (Figure S2F) and those from the 3p miRNA library (Figure 2D) by their relative positions to the defined 3′ end of pre-miRNAs, we found that the chance of transmission of RNA modifications from pre-miRNAs to their 3p miRNAs, as judged by the abundance of uridines, is position-dependent (Figures 2D, E, and S2F). The transmission is predominantly restricted within two positions; the 3′ end of a pre-miRNA (position -1) and the downstream position (+1), even though neighboring positions are also uridylated to comparable levels (Figures 2D, E, and S2F). Moreover, we found that the tail length of a pre-miRNA read depends on the position of its last genome-matching nucleotide (prefix position) relative to the defined 3′-end of the full-length pre-miRNA: the farther away the prefix position from the 3′ end, the longer the tail (Figure 2F). In general, tailed reads whose prefix positions reside close to the 3′ end of pre-miRNAs contain 1 to 2 additions. On the contrary, reads whose prefix positions lie further upstream from the 3′ end can harbor 6-7 additional nucleotides (Figure 2F). Collectively, these results suggested that Ago-bound pre-miRNAs that are mono-uridylated at or immediately downstream of their 3′-ends can give rise to miRNAs that carry these modifications and assemble functional miRNPs, which is consistent with previous findings for group II let-7 miRNAs (Heo et al., 2012). In contrast, Ago-bound pre-miRNAs with oligo(U) tails (≥ 2 nts), either extending out from the 3′ end, or added to severely trimmed 3′ ends cannot serve as Dicer substrates and fail to assemble miRNPs.
Uridylation of pre-miRNAs by TUT7 and TUT4 plays a dual role in miRNA biogenesis
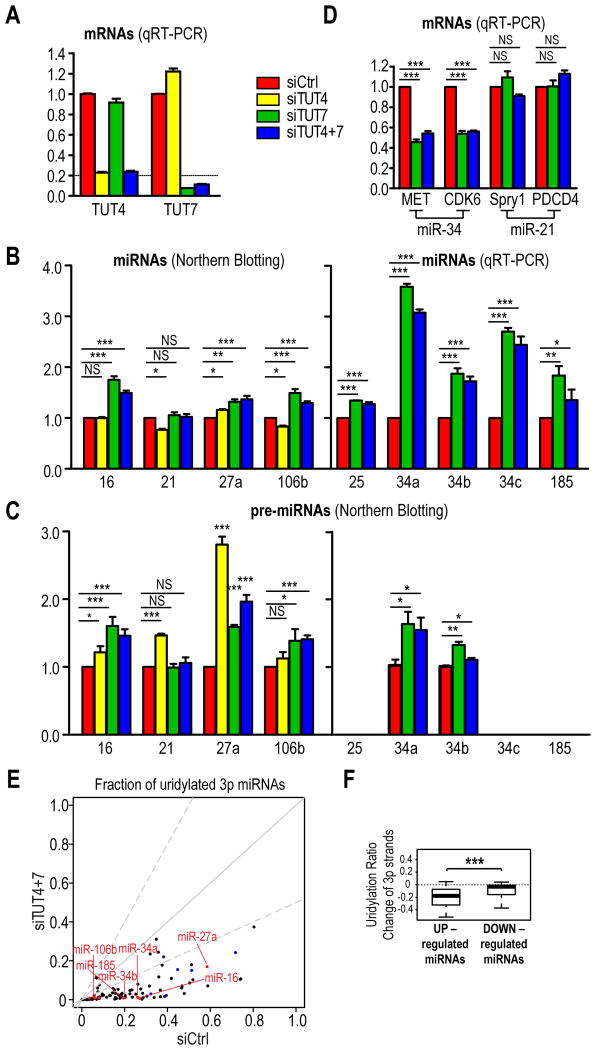
There are seven TUTs in mammals that have tailing activities towards many different types of RNAs. To test which TUTs can uridylate pre-miRNAs in vitro, we incubated immunopurified FLAG-TUTs 1-7 (Figure S3A) with 5′ radiolabeled pre-miRNA substrates. Consistent with a recent report (Heo et al., 2012), we found that TUT2, TUT4 and TUT7 displayed tailing activities towards pre-miRNAs (Figure S3B) and preferred UTP over other nucleotides in vitro (data not shown). Notably, such uridylation activity was not observed when catalytically inactive TUT4 or TUT7 were used in the assay (Figure S3C), suggesting that uridylation was not mediated by a contaminating enzyme. To find out which of the three candidate TUTs regulate miRNA biogenesis in vivo, we depleted TUT2, TUT4 or TUT7 from MEFs. Given the high similarity between TUT4 and TUT7, we also knocked down both TUTs simultaneously. Depletion of TUT2 or TUT4 alone had no appreciable effect on miRNA levels (Figures 3A, B, S3D and data not shown). In contrast, several mature miRNAs were significantly upregulated in TUT7- and TUT4/7-depleted cells (Figures 3A, B, S3D). Similar results were obtained with a second set of siRNAs against TUT4 or TUT7 (Figures S3F, G), demonstrating that our finding was not due to off-targeting effects. For most of the upregulated miRNAs except miR-34a and 34c, only their precursors but not their primary transcripts increased (Figures 3C and S3E), suggesting that the increase in most miRNAs resulted after Drosha processing but prior to Dicer cleavage. Interestingly, primary transcripts of miR-34a and 34c also increased in TUT7- and TUT4&7-depleted cells. Nevertheless, their fold changes were smaller than those of their mature miRNAs (Figures 3B and S3E), indicating that the increase of miR-34a/c in TUT7- and TUT4&7-depleted cells was contributed not only by stabilization of their precursors (Figure 3C), but also by increase in their primary transcripts. This is consistent with our observation that most miRNAs are modestly modulated by TUT7/4 in MEFs. Concomitant with an increase in miR-34 levels, known targets of miR-34 (MET and CDK6) (He et al., 2007; Migliore et al., 2008) were further repressed in TUT7- and TUT4&7-depleted cells (Figure 3D). In contrast, known targets of miR-21 (Spry1 and PDCD4) (Ma et al., 2011), which was not altered in TUT7 or TUT4&7 knockdown cells, remained unchanged (Figure 3D). Thus, TUT7/4 can modulate the activity of miRNAs by fine-tuning their biogenesis.
Figure 3. Uridylation of pre-miRNAs by TUT7 and TUT4 fine tunes select miRNA biogenesis; see also Figure S3.
A. mRNA levels of TUT4 and TUT7 in indicated knockdown cells, measured by quantitative real-time (qRT)-PCR. The standard error of the mean is from four experiments. siCtrl, non-targeting siRNA.
B, C. miRNA (B) and pre-miRNA (C) levels in indicated knockdown cells. The level of pre-miR-35 and pre-miR-185 are too low to be detected by NB. SEM is from at least three experiments. Statistical significance was calculated by one-tail t test. NS, non-significant; *, p<0.05; **, p<0.01; ***, p<0.001. Same depiction applies to all figures.
D. miR-34 or miR-21 target levels in indicated knockdown cells, measured by qRT-PCR. SEM is from three experiments. Statistical significance was calculated by t test.
E. Uridylation ratios of a subset of pre-miRNAs, as inferred from that of their 3p miRNAs, are dramatically diminished upon depletion of TUT4 and TUT7. Red dots indicate 3p strands of pre-miRNAs whose expression is verified by qRT-PCR or NB. Blue dots denote 3p strands of pre-let-7 family members.
F. Box Plots for uridylation ratio differences of 3p strands between control and TUT4&7-depleted cells for differentially expressed (⩾ 2 fold) miRNAs. Only guide strands (more abundant species within miRNA duplexes) were used for comparison of expression levels of miRNAs. Statistical significance was calculated by Welch's t-test.
HITS analysis of miRNAs from control or TUT4&7 knockdown cells showed that uridylation ratios (fractions of uridylated reads) of most 3p miRNAs, including miR-34-3p and miR-106b-3p, which share the same precursor with upregulated miRNAs, were significantly diminished in TUT4&7-depleted cells (Figure 3E and Table S2). As 3p miRNAs primarily inherit uridines from their precursors, this suggested that upregulated miRNAs are preferably uridylated by TUT7 and TUT4 in MEFs. A small subset of 3p miRNAs, including those from most pre-let-7 family members, had their uridylation ratios only modestly reduced in TUT4&7-depleted cells, presumably due to uridylation by TUT2 in the absence of TUT7 and TUT4 (Figure 3E). Taken together, these results suggested that TUT7 and TUT4 are the primary uridylyltransferases for a large number of pre-miRNAs in MEFs.
HITS analysis showed that a group of miRNAs were upregulated in TUT4&7-depleted MEFs (Figures S3H and Table S3). Strikingly, the uridylation ratio of their 3p miRNAs, which reflects the uridylation status of their parental pre-miRNAs, showed a statistically significant reduction in uridylation ratios of 3p miRNAs from pre-miRNAs whose miRNAs are upregulated, relative to those downregulated in TUT7- and TUT4&7-depleted cells (Figure 3F). Altogether, these results suggested that TUT7 either alone or in combination with TUT4 fine tunes the maturation of a subset of miRNAs through uridylation and destabilization of their precursors.
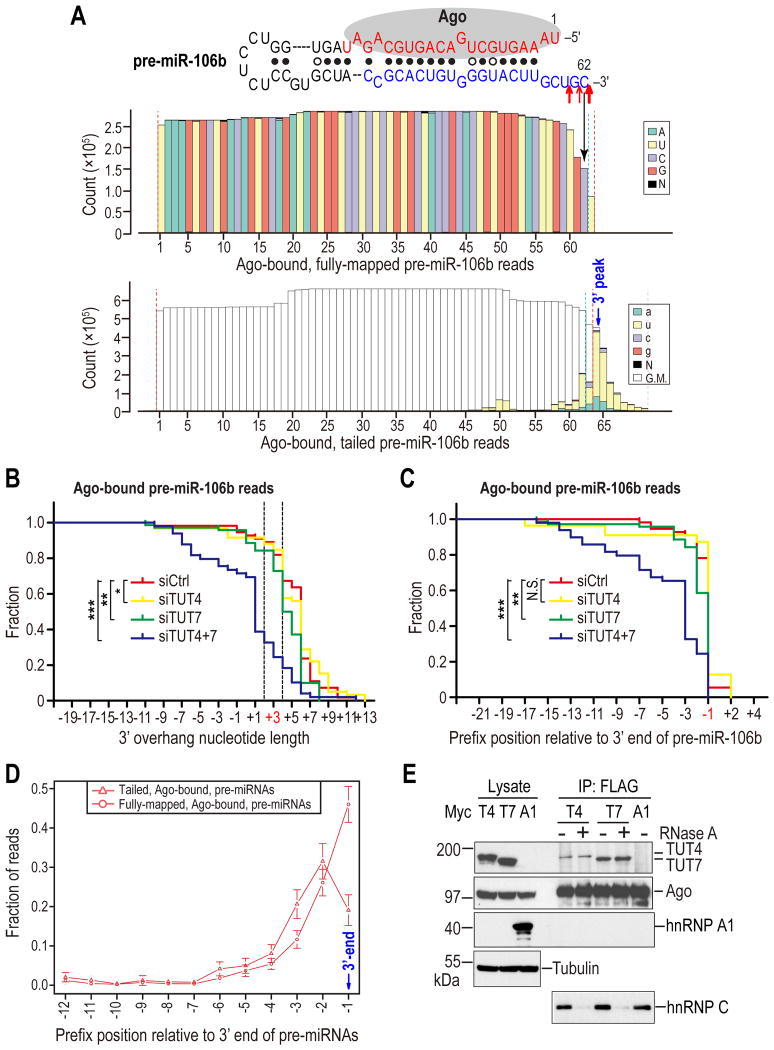
Uridylation of pre-miRNAs by TUT7 and TUT4 facilitates their decay from Ago-containing complexes
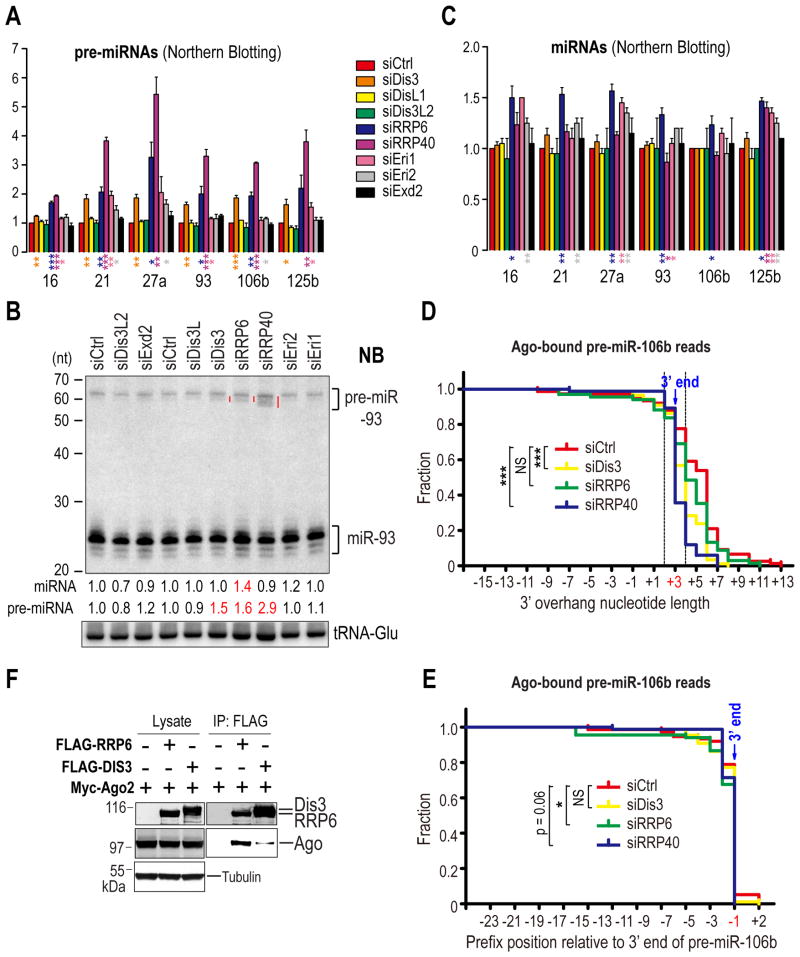
Pre-miR-106b is one of the most abundantly uridylated pre-miRNAs in our HITS libraries (Figure 4A). To further verify our analysis with miRNA HITS libraries, we cloned and sequenced pre-miR-106b, from Ago IPs from TUT4, TUT7, TUT4&7 or control knockdown cells. Although the majority of pre-miR106b reads contain a genome-encoded uridine right after the cytidine that forms the miRBase annotated 3′-end (Figure 4A), sequencing of in vitro Drosha processed pri-miR-106b products (Figure S4A) confirmed that the majority of Drosha cleavage sites is after the cytidine (Figure 4A). Notably, none of the Drosha processed products contained a uridine after the cytidine, suggesting that the last uridine is added after Drosha processing. Consistent with our finding from miRNA HITS libraries, the uridylation ratio and the tail length of Ago-bound pre-miR-106b reads are modestly and substantially reduced in TUT7- and TUT4&7-depleted cells respectively (Figure 4B and Table S4), verifying that TUT7 and TUT4 uridylate pre-miR-106b in vivo.
Figure 4. Uridylation of Ago-bound pre-miRNAs by TUT7 and TUT4 facilitates their degradation; see also Figure S4.
A. Nucleotide composition for every position of Ago-bound pre-miR-106b reads in WT pre-miRNA library. Red arrows indicate 3′ cleavage sites identified from in-vitro Drosha processed pri-miR-106b products. Size of arrows reflects the clone frequency such that large, medium, and small arrows represent 10/23, 7/23, and 5/23 respectively. Red dotted lines delineate the computationally defined 5′ and 3′ ends. Blue dotted lines define the manually corrected 3′ end.
B. Reverse cumulative plot of 3′ overhang lengths of pre-miR-106b reads (fully-mapped and tailed), sequenced from Ago IPs from indicated knockdown cells. The native 3′ overhang length of pre-miR-106b is 3 nts. Reads whose ends fall within the two dotted lines are optimal Dicer substrates as defined by in-vitro Dicer processing assay in Figure S4B. Statistical significance was calculated by Anderson-Darling (A-D) test.
C. Reverse cumulative plot of prefix positions of pre-miR-106b reads relative to its native 3′ end (position -1), sequenced from Ago IPs from indicated knockdown cells. Statistical significance was calculated by A-D test.
D. Average fraction of fully mapped or tailed reads for Ago-bound pre-miRNAs with prefix positions upstream of the defined 3′ end of pre-miRNAs. Error bars represent 95% CIs.
E. Association of TUT4 and TUT7 with Ago2. Western blots of lysates and FLAG IPs from 293T cells co-expressing FLAG-Ago2 with Myc-TUT4, Myc-TUT7 or Myc-hnRNPA1. HnRNP C, which was shown to associate with Ago2 in an RNA-dependent manner, served as a positive control for RNase A digestion. Tubulin: loading control.
Surprisingly, compared to Ago IPs from control knockdown cells, those from TUT4&7-depleted cells contained a substantially larger fraction of pre-miR-106b reads that appear to be degradation intermediates with blunt or 5′ overhangs (Figures 4B, C and Table S4), which were not processed by Dicer in vitro (Figure S4B). Similar results were also observed when we sequenced pre-miR-18a from Ago IPs (Figure S4C and Table S5). These findings suggest that uridylation of pre-miRNAs by TUT7 and TUT4 is required for complete decay of Ago-bound pre-miRNAs, as in their absence truncated fragments accumulate in Ago, which may lead to clogging of Ago with defective pre-miRNAs. Consistent with this hypothesis, there are is a higher percentage of tailed than fully-mapped reads for Ago-bound pre-miRNAs at positions upstream of the 3′ end of Ago-bound pre-miRNAs in our libraries (Figure 4D), indicating that tailing and trimming are intimately associated in decay of Ago-bound pre-miRNAs. Moreover, the peak of 3′-peaked tails, which represents an equilibrium site between tailing and trimming activities towards the 3′ end of pre-miRNAs, is preferentially situated at a fixed distance (3 nts) away from the hairpin region of the pre-miRNA (Figure S4D), suggesting that tailing by TUT7 and TUT4 may facilitate digestion of pre-miRNAs by providing a grip for an exonuclease(s). Taken together, these results suggest that uridylation of pre-miRNAs facilitates degradation of structured pre-miRNAs bound to Agos.
Where do TUT4 and TUT7 uridylate Ago-bound pre-miRNAs? To address this question, we first examined the subcellular localization of TUT4 and TUT7. Confocal microscopy showed that both TUT4 and TUT7 were localized to the cytoplasm (Figure S4E). As the amount of uridylated pre-miRNAs bound to Ago is drastically diminished in TUT4&7-depleted cells (Figures 4B, S4C, Tables S4 and S5), we speculated that at least a fraction of pre-miRNAs are uridylated by TUT7 and TUT4 when bound to Ago. In support of this hypothesis, TUT7 and TUT4 co-immunoprecipitated (co-IPed) with Ago2 in an RNA-independent manner (Figures 4E and S4F). On the contrary, no interaction was detected between an unrelated RNA-binding protein hnRNP A1 and Ago2 (Figure 4E), demonstrating the specificity of the association between TUT7/4 with Ago2.
Quality Control and turnover of pre-miRNAs by the exosome
To identify 3′ exonucleases that collaborate with TUT7 and TUT4 in degrading pre-miRNAs, we performed siRNA screening for candidates that included the catalytic subunits (DIS3, DIS3L, RRP6) and a core structural protein (RRP40) of the exosome (Houseley and Tollervey, 2009; Staals et al., 2010; Tomecki et al., 2010); an exosome-independent exonuclease that is homologous to DIS3 (DIS3L2) (Chang et al., 2013; Lubas et al., 2013; Malecki et al., 2013); and exonucleases that are involved in siRNA turnover or miRNA biogenesis (ERI1 and EXD2) (Han et al., 2011; Kennedy et al., 2004; Liu et al., 2011) (Figure S5A). Among the candidates tested, knock-down of DIS3, RRP6 or RRP40, exosome components, led to the most pronounced and consistent increase in precursors for all miRNAs tested, with the strongest accumulation of pre-miRNAs in RRP40-depleted cells (Figures 5A, B, and S5B), presumably owing to disassembly of the exosome and co-depletion of RRP6 upon depletion of RRP40 (Tomecki et al., 2010). The accumulation of pre-miRNAs could not be solely attributed to changes in their primary transcript levels, as the increase of pri-miRNAs (Figure S5C) was less than that of pre-miRNAs (Figure 5A). This suggested that pre-miRNAs may be direct targets of the exosome. Interestingly, except for RRP6, depletion of DIS3 or RRP40 did not result in appreciable increase in miRNA levels (Figure 5C). Instead, truncated species of some pre-miRNAs such as pre-miR-93 (Figure 5B), accumulated in DIS3-, RRP6- and in particular RRP40-depleted cells. These results suggested that the pre-miRNA population targeted by the exosome was nonfunctional and could not be processed by Dicer. Therefore, we speculated that the exosome might function in quality control of pre-miRNAs by removing defective species.
Figure 5. Quality control and turnover of pre-miRNAs by the exosome; see also Figure S5.
A - C. Pre-miRNA (A) or miRNA (C) levels in indicated knockdown cells. SEM is from three independent experiments. Statistical significance was calculated by unpaired two-tail t test. Representative NB of total RNAs from indicated knockdown cells (B). Red lines denote defective (truncated) pre-miR-93. Numbers indicate fold changes of pre-miRNAs and miRNAs relative to the control knockdown. tRNA-Glu: loading control.
D. Reverse cumulative plot of 3′ overhang lengths of pre-miR-106b reads (fully-mapped and tailed), sequenced from Ago IPs from indicated knockdown cells. Statistical significance was calculated by A-D test.
E. Reverse cumulative plot of prefix positions of pre-miR-106b reads relative to its native 3′ end (position -1), sequenced from Ago IPs from indicated knockdown cells. Statistical significance was calculated by A-D test.
F. Association of RRP6 and DIS3 with Ago2. Western blots of lysates and FLAG IPs from 293T cells co-expressing Myc-Ago2 with FLAG-RRP6 or DIS3-FLAG. Tubulin: loading control.
Surprisingly, Ago IPs from DIS3- or RRP40-depleted cells, had much fewer pre-miR-106b reads with uridine tails, than control knockdown cells (Figure 5D, E and Table S4). Notably, uridylation deficiency in Ago-bound pre-miRNAs is not a result of defects in the pre-miRNA tailing machinery as profiles of pre-miR-106b reads are indistinguishable between control cells and those with exosome component knockdown when sequenced from total RNA (Figures S5D, E and Table S6). As such, we speculated that the exosome might stimulate uridylation of Ago-bound pre-miRNAs by TUT7/4 (see below). Indeed, we found that DIS3 and RRP6 also co-IPed with Ago2 (Figures 5F and S5F, G, H). Taken together, these results suggest that the exosome may cooperate with TUT7 and TUT4 to function in turnover and quality control of pre-miRNAs.
Distinct preferences of DIS3 and RRP6 towards pre-miRNAs
To test whether pre-miRNAs can be directly decayed by the two catalytic subunits of the exosome, we immunoaffinity purified FLAG-tagged DIS3 or RRP6 from transfected 293T cells (Figure S6A), and incubated them with 5′ radiolabeled pre-miRNAs. Wild-type (WT), but not catalytically inactive mutant (Wasmuth and Lima), DIS3 or RRP6 protein, digested pre-miRNAs to end products of around 4-5 nts (Figures S6B, C), indicating that pre-miRNAs could serve as substrates for the two catalytic subunits of the exosome.
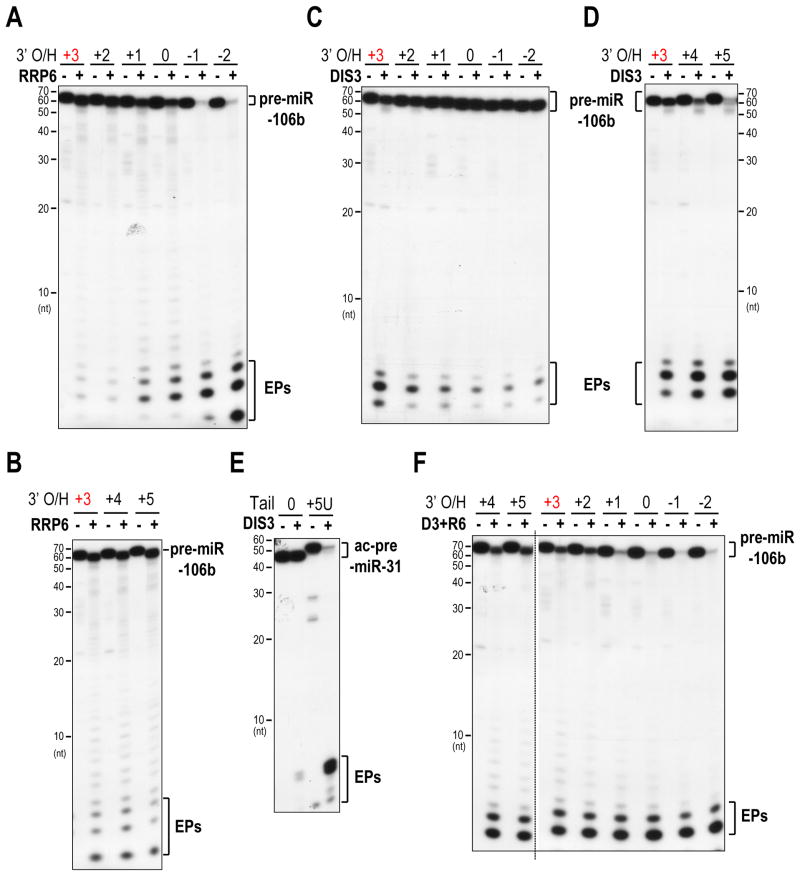
To test whether the exosome may participate in quality control of pre-miRNAs, we examined whether the two catalytic subunits of the exosome could distinguish pre-miRNA substrates with native 3′ overhangs from those with suboptimal or defective 3′ overhangs. Surprisingly, we found that RRP6 preferred degrading pre-miRNAs with < 2-nt 3′ overhangs, and especially those with 5′ overhangs (Figure 6A). Gradual addition of more uridines to the 3′ end of pre-miRNAs did not enhance the activity of RRP6 (Figure 6B). We obtained similar results when using pre-miR-16 as the substrate (Figure S6D), suggesting that the end preference of RRP6 is not pre-miRNA specific. However, pre-miR-106b is extensively uridylated (Figures 4A and S5E), so we speculated that another exonuclease within the exosome, whose activity is stimulated by uridylation, would be responsible for degradation of pre-miRNAs with uridine tails.
Figure 6. Distinct preferences of DIS3 and RRP6 towards pre-miRNAs; see also Figure S6.
A, B. Decay assay using RRP6 and 5′ radiolabeled pre-miR-106b with progressively shortened (A) or uridylated (B) 3′ overhangs. 3′ O/H, 3′ overhang. The native 3′ overhang is marked in red. Same depiction applies to all figures. Prolonged exposure was used for comparison of the end products in panel B.
C, D. Decay assay of pre-miR-106b with progressively shortened (C) or uridylated (D) 3′ overhangs using DIS3.
E. Decay assay of Ago2-cleaved pre-miR-31 (ac-pre-miR-31) with or without an oligo(U) tail using DIS3.
F. Decay assay of pre-miR-106b with 3′ overhangs of different lengths using a mixture of DIS3 and RRP6 (D3+R6).
To test this hypothesis, we incubated DIS3 with pre-miR-106b containing either the native, recessed or uridylated 3′ overhangs (Figures 6C, D). Remarkably, the exonuclease activity of DIS3 was greatly stimulated by uridine tails (Figure 6D). Further examination revealed that DIS3 preferred digesting pre-miRNAs with > 2-nt 3′ overhangs (Figures 6C, D). This end preference of DIS3 is not specific to pre-miR-106b, as DIS3 also showed similar activity towards ac-pre-miR-31 and pre-miR-21, when they were engineered to harbor the same tails as those found in our pre-miRNA libraries (Figures 6E and S6E). Moreover, a point mutation (U57A) that “unzips” the end structure of pre-miR-21 to create a longer dangling 3′-end, transformed pre-miR-21 into a better substrate for DIS3 (Figures S6F, G), indicating that DIS3 prefers pre-miRNAs with single-stranded 3′ ends. Overall, these findings indicate that RRP6 and DIS3 have distinct preferences towards pre-miRNAs: RRP6 prefers pre-miRNAs with < 2-nt 3′ overhangs while DIS3 favors pre-miRNAs with > 2-nt 3′ overhangs.
In accord with our findings, pre-miRNAs with 3′ overhangs of different lengths were digested more efficiently in the presence of both DIS3 and RRP6, than either alone, in vitro (Figure 6F). Taken together, these results suggested that 3′ ends of pre-miRNAs are monitored by the exosome, which exploits the distinct preferences of its two catalytic subunits to distinguish productive from faulty pre-miRNAs.
A positive feedback loop formed by the exosome and TUT7/4 in degradation of Ago-bound pre-miRNAs
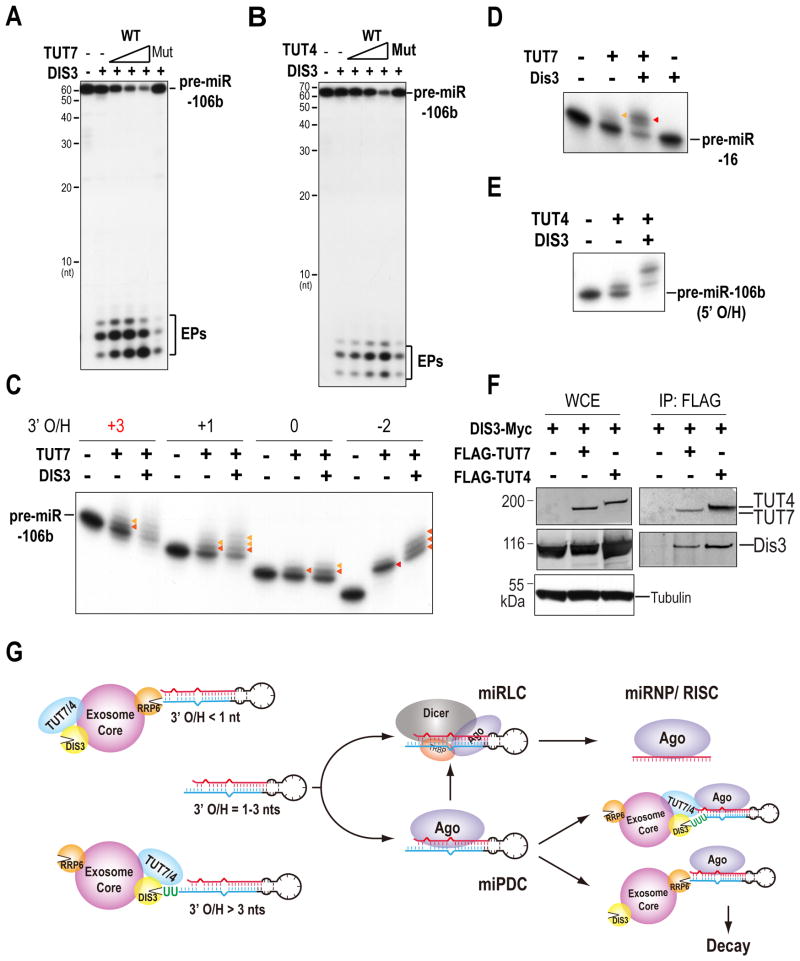
As DIS3 favors pre-miRNAs with > 2-nt 3′ overhangs, we postulated that pre-miRNAs, which predominantly harbor 2-nt 3′ overhangs, would first need tailing by TUT7/4 to become better substrates for DIS3. Indeed, we found that decay of pre-miR-106b by DIS3 was stimulated in vitro by addition of wild-type TUT7 or TUT4, but not catalytically-inactive mutants, indicating that tailing was required for stimulating decay of pre-miRNAs by DIS3 (Figures 7A, B).
Figure 7. A positive feedback loop constituted by TUT7, TUT4 and the exosome in uridylation and degradation of pre-miRNAs; see also Figure S7.
A, B. Decay assay of pre-miR-106b using mixtures of DIS3 with wild-type (WT) or catalytically inactive (Wasmuth and Lima) TUT7 (A) and TUT4 (B).
C. Uridylation assay using TUT7 and DIS3 with pre-miR-106b harboring 3′ overhangs of different lengths.
D. Uridylation assay using TUT7 and DIS3 with pre-miR-16.
E. Uridylation assay using TUT4 and DIS3 with pre-miR-106b with a 2-nt 5′ overhang (5′ O/H).
F. Interaction of TUT4 and TUT7 with DIS3. Western blots of lysates and FLAG IPs from 293T cells co-expressing DIS3-Myc with FLAG-TUT4 or FLAG-TUT7.
G. Model for a pre-miRNA surveillance network in quality control of mammalian miRNA synthesis
To test whether the exosome may stimulate uridylation of pre-miRNAs by TUT7/4, we conducted uridylation assay in the presence or absence of DIS3 or RRP6. Surprisingly, we found that the uridylyltransferase activity of TUT7 or TUT4 towards pre-miRNAs, regardless of their 3′ overhang length, was specifically stimulated in vitro by addition of immunoprecipitates from wild-type DIS3, but not RRP6 or catalytically inactive DIS3 (Figures 7C-E and S7A, B), suggesting that DIS3 cooperates with TUT7/4 to polish the 3′ end of suboptimal substrates for more efficient digestion. Uridylation of pre-miRNAs was not observed with addition of only DIS3 (Figure 7D), indicating that enhanced uridylation of pre-miRNAs by TUT7/4 in the presence of DIS3 was not due to a contaminating uridylyltransferase in our DIS3 protein preparation. In support of this hypothesis, the tail length of Ago-bound pre-miR-106b reads is shorter in DIS3- or RRP40-depleted cells than that in control knockdown cells (Figure 5D and Table S4), indicating that DIS3 is required for efficient uridylation of Ago-bound pre-miRNAs in vivo. This cross-stimulation of activities between the exosome and TUT7/4 prompted us to test whether TUT7/4 may associate with the exosome. Indeed, we found that both TUT7 and TUT4 co-IPed with DIS3 and RRP6 (Figures 7F and S7C, D). Taken together, these results suggest that a subset of Agos is in complex with TUT7/4 and the exosome, which form a positive feedback loop in degradation of Ago-bound pre-miRNAs.
Discussion
Multiple protein factors and mechanisms operate to ensure loading of mammalian Ago proteins with authentic miRNAs (Buhler et al., 2008; Wilson and Doudna, 2013). The structure, stem length, loop size, 5′ phosphate, and 3′ overhang length of pre-miRNAs are sensed by Dicer, while Ago quality controls incoming miRNAs or pre-miRNAs through preferentially accepting those with 5′-uridines/adenosines and 5′ phosphates (Liu et al., 2012; Wilson and Doudna, 2013). Our study has revealed a novel quality control mechanism in mammalian miRNA synthesis and identified a pre-miRNA surveillance network formed by TUT7, TUT4 and the exosome, which functions not only in polishing the 3′ end of pre-miRNAs, but also in pre-miRNA turnover and discriminating authentic from faulty pre-miRNAs (Figure 7G).
By adapting a novel strategy for isolating pre-miRNAs and constructing HITS libraries of full-length pre-miRNAs, we report the first genome-wide profile of pre-miRNAs bound to Ago. We find that in somatic cells a large fraction of Ago-bound pre-miRNAs contain extensively uridylated tails. We find that uridylation of pre-miRNAs by TUT7/4 has dual roles: Mono-uridylation that polishes the 3′ end of pre-miRNAs for a 2-nt 3′ overhang facilitates Dicer processing, as previously reported for group II let-7 miRNAs (Heo et al., 2012), and stabilizes pre-miRNAs against degradation by the exosome. On the other hand, mono-uridylation that deviates the 3′ overhang away from the optimal length (2 nt) or oligo-uridylation not only fine tunes select miRNA maturation by destabilizing their precursors, but also facilitates exonucleolytic digestion of pre-miRNAs bound to Ago, which prevents clogging of Ago with defective pre-miRNAs. This function of TUT7/4 is reminiscent of the role for the yeast noncononical poly(A) polymerase Cid14, which limits access of small RNAs derived from rRNAs or tRNAs to Ago (Buhler et al., 2008). Together, these suggest that uridylation may play distinct roles for different pre-miRNAs. We speculate that sequence and structural features of pre-miRNAs and/or protein cofactors that bind to specific pre-miRNAs likely influence the extent and effects of uridylation, and should prove an interesting subject for future studies.
We identify the exosome as a key player in quality control of pre-miRNAs in mammals. We find that DIS3 and RRP6, two of its catalytic subunits, display distinct preferences towards pre-miRNAs: RRP6 prefers pre-miRNAs with < 2-nt 3′ overhangs while DIS3 favors pre-miRNAs with > 2-nt 3′ overhangs. Pre-miRNAs typically possess 2-nt 3′ overhangs that are recognized by the PAZ domain of Dicer (Heo et al., 2012; Park et al., 2011). Our finding suggests one more explanation for the need of a 2-nt 3′ overhang for pre-miRNAs: pre-miRNAs with 2-nt 3′ overhangs are least vulnerable to degradation by RRP6 and DIS3. Therefore, it is possible that the stability of a pre-miRNA may be regulated by Drosha through its 3′ cleavage site selection. In that mode, the exosome may act as a sentinel for quality control of pre-miRNAs through sensing the 3′ end of pre-miRNAs and eliminating defective species, such as those with blunt or 5′ overhangs and other spurious RNAs with hairpin structures, while preserving productive Dicer substrates. Although depletion of DIS3L did not affect pre-miRNA levels in MEFs it is possible that DIS3L has a role in pre-miRNA surveillance or turnover under different conditions or in other cell types.
We find that in mammals, the exonuclease activity of DIS3 towards pre-miRNAs is stimulated by TUT7 or TUT4. Thus, in analogy to the nuclear TRAMP complex, TUT7/4 and DIS3 cooperate in the cytoplasm, where TUT7/4 resides, to stimulate degradation of RNA targets. However, as DIS3 and RRP6 are localized to both the nucleus and the cytoplasm in mammals, the exosome may also degrade pre-miRNAs in the nucleus, possibly assisted by other factors. We find that the uridyltransferase activity of TUT7 or TUT4 towards pre-miRNAs is promoted by their interaction with DIS3 or a DIS3-associated exosome component, indicating cross-stimulation of activities between TUT7/4 and the exosome. The positive feedback loop formed by the exosome and TUT7/4 may synergize the action of the processive exonuclease DIS3 with that of the distributive uridylyltransferase TUT7/4 for efficient digestion of structured pre-miRNAs, particularly when they are bound by Agos. Alternatively, this positive feedback loop may be designed to prevent the buildup of suboptimal pre-miRNAs with trimmed 3′ ends, which may compete with productive species for binding to Dicer or Ago.
In summary, our study has uncovered a pre-miRNA surveillance system composed of TUT7, TUT4 and the exosome. Our findings set the stage for future mechanistic studies to address how the activity of TUTs is stimulated by the exosome and vice versa; whether there are additional quality control and turnover mechanisms for specific pre-miRNAs or miRNAs.
Experimental Procedures
Cell lines
Dicer+/fl; Dicerfl/fl; HA-Dicer, Dicerfl/fl; HA-Dicer(D2A), Dicerfl/fl; Myc-Ago2, Dicerfl/fl; Myc-Ago2(D669A) cell lines were cultured as previously described (Liu et al., 2012).
RNA Immunoprecipitation
RNA immunoprecipitation was performed as in (Liu et al., 2012); Dynabeads Protein A (Life Technologies) and rabbit-anti-mouse IgG, Fc fragment-specific (Jackson Immunoresearch Labs) were used for immunoprecipitation of Agos with 2A8 antibody.
Construction of HITS Libraries Enriched for Full-Length Pre-miRNAs
The small RNA cloning protocol is adapted from (Ingolia et al., 2009) with a few modifications and is detailed in Supplemental Experimental Procedures.
Purification of Proteins, Uridylation and Decay assays
FLAG-tagged proteins were immunoaffinity purified from transfected 293T cells using anti-FLAG M2 beads (Sigma), followed by peptide elution, dialysis and concentration. They were used in uridylation and decay assays. Details are in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
A strategy for identifying full-length pre-miRNAs bound to Agos
Uridylation by TUT7/4 polishes pre-miRNAs and facilitates degradation by the exosome
Distinct features recognized by DIS3 and RRP6 in pre-miRNA degradation
A positive feedback loop formed by the exosome and TUT7/4 in pre-miRNA degradation
Acknowledgments
We thank F. Ibrahim for technical assistance with the uridylation assay and all members of Mourelatos lab for helpful discussions. This work was supported by NIH Grant GM072777 to Z.M. and by a Brody Family Fellowship to M.M.
Footnotes
Accession Number: Illumina sequencing data were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under study accession IDs: PRJEB6756, PRJEB6759.
Author Contributions: X.L. and Z.M. conceived and directed experiments. X.L. conceived the pre-miRNA sequencing method, prepared sequencing libraries and along with N.V. conducted all biochemical experiments. Q.Z., M.M. and P.A. performed all bioinformatical analyses. X.L., M.M, P.A., N.V, Q.Z., B.D.G and Z.M. analyzed data. X.L. and Z.M. wrote the manuscript with input and editing from M.M, P.A., N.V, Q.Z., B.D.G.
Conflict of Interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Kawano M, Ando Y, Daub CO, Hayashizaki Y. pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5′/3′ arm variation including concomitant cleavage and polyuridylation patterns. Nucleic Acids Res. 2012;40:1424–1437. doi: 10.1093/nar/gkr903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Grimm D, Wang L, Lee JS, Schurmann N, Gu S, Borner K, Storm TA, Kay MA. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- Li N, You X, Chen T, Mackowiak SD, Friedlander MR, Weigt M, Du H, Gogol-Doring A, Chang Z, Dieterich C, et al. Global profiling of miRNAs and the hairpin precursors: insights into miRNA processing and novel miRNA discovery. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Liu X, Jin DY, McManus MT, Mourelatos Z. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Mol Cell. 2012;46:507–517. doi: 10.1016/j.molcel.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Kumar M, Choudhury SN, Becker Buscaglia LE, Barker JR, Kanakamedala K, Liu MF, Li Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:10144–10149. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68:10128–10136. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17:1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Chen X, Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, Cuellar TL, McManus MT, Kiriakidou M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell. 2012;48:133–144. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.