Abstract
Runx1 is required for definitive hematopoiesis and is well-known for its frequent chromosomal translocations and point mutations in leukemia. Runx1 regulates a variety of genes via Ets1 activation on an Ets1•Runx1 composite DNA sequence. The structural basis of such regulation remains unresolved. To address this problem, we determined the crystal structure of the ternary complex containing Runx11-242 and Ets1296-441 bound to T cell receptor alpha (TCRα) enhancer DNA. In the crystal, an Ets1-interacting domain of Runx1 is bound to the Ets1 DNA-binding domain and displaced an entire autoinhibitory module of Ets1, revealing a novel mechanism of Ets1 activation. The DNA binding and transcriptional studies with a variety of structure-guided Runx1 mutants confirmed a critical role of direct Ets1•Runx1 interaction in Ets1 activation. More importantly, the discovered mechanism provides a plausible explanation for how the Ets1•Runx1 interaction effectively activates not only a wild-type Ets1, but also a highly inhibited phosphorylated form of Ets1.
Keywords: AML, Runx1, Ets1, TCRα, crystal structure, transcription
INTRODUCTION
Regulation of gene expression in eukaryotes is an extremely complex process1. Cis-regulatory sequences on promoters and enhancers of target genes facilitate the cooperative assembly of transcriptional regulatory complexes1. In addition, multiple signaling cascades contribute to more elaborate regulation of gene expression by post-translational modifications of transcriptional factors (TFs)2. In spite of recent breakthroughs in high-resolution methods for studies of TFs genomics, the details of TFs partnerships, especially with contributions of post-translational modifications, are poorly understood due to insufficient structural data1. Ets1•Runx1 partnership provides a perfect basis for filling the gap. The two TFs regulate T cell receptor alpha (TCRα) and a variety of other genes via binding to a composite Ets1•Runx1 sequence3-7.
Ets1 transcription factor participates in embryonic development, lymphoid differentiation, proliferation, apoptosis and angiogenesis8. It is amplified and rearranged in a variety of cancers.9 DNA binding of Ets1 is regulated by an inhibitory serine rich region (SRR) and inhibition regulatory module (IRM) flanking its DNA-binding Ets domain (ED) (Fig. 1a)10. IRM comprises of inhibitory helices HI1 and HI2 N-terminal to the ED and H4 and H5 C-terminal to ED11, 12. The IRM and the SRR interact with the ED, resulting in a 40-fold reduction in DNA binding affinity.13 DNA binding of Ets1 is also regulated by an Ca2+ signaling-mediated phosphorylation of the serines in SRR13-15. Partner proteins activate Ets1 and among them is a Runx1 transcription factor. Runx1 acts by targeting the IRM of Ets1 via direct physical interaction between the two proteins4, 10, 16, 17.
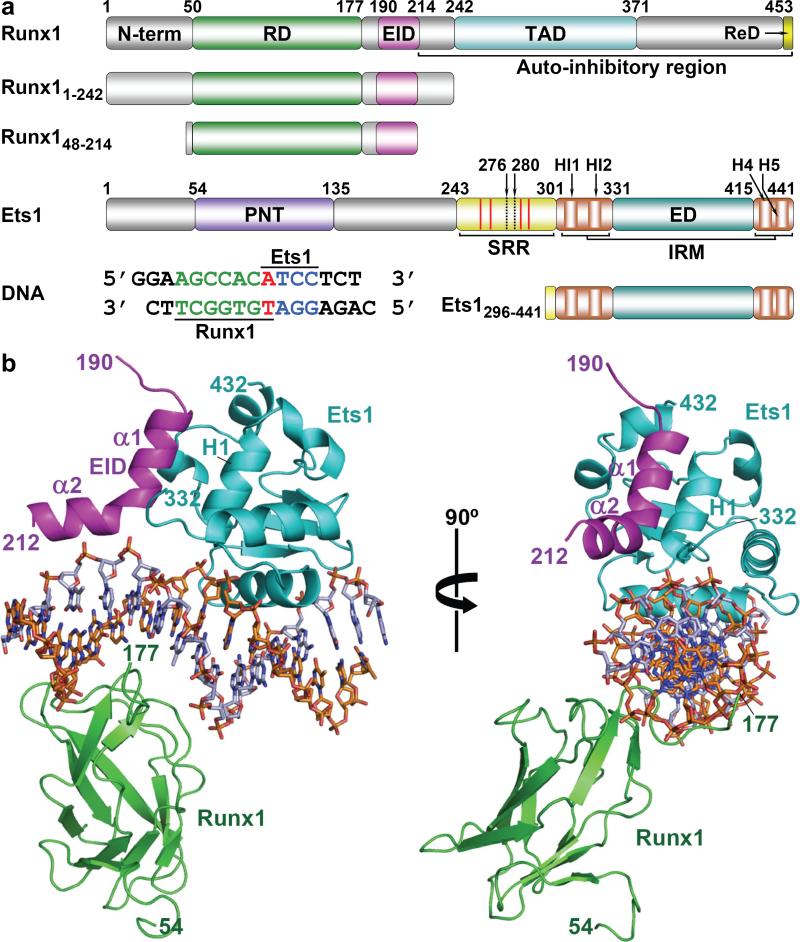
Figure 1. Overall structure of ternary complex.
(a) Domain organization of full-length Runx1 and Ets1 proteins and their fragments (Runx11-242, Runx148-214 and Ets1296-441) used for crystallization. The highlighted domains are: N-terminal, Runt domain (RD), Ets1 interacting domain (EID), transactivation domain (TAD) and repression domain (ReD) for Runx1; and pointed domain (PNT), serine rich region (SRR), inhibition regulatory module (IRM) and ETS domain (ED) for Ets1. The inhibitory helices HI1, HI2 and H4 of IRM are also marked. The red lines within SRR indicate the location of phosphorylation sites. Two dotted lines represent the N terminal of the constructs used for binding studies (Ets1280-441 and Ets1276-441). TCRα enhancer DNA used for crystallization contains Runx1 (green) and Ets1 (blue) binding sites with one overlapping base pair (red). (b) Overall structure of Runx11-242•Ets1296-441•TCRα in two orientations. Runx1 (green), Ets1 (cyan) and EID of Runx1 (magenta) are drawn as ribbons and DNA is drawn as sticks.
Runx1 is required for definitive hematopoiesis and is well-known for its frequent chromosomal translocations and point mutations in leukemia18. DNA binding of Runx1 is auto-inhibited by sequences flanking its DNA-binding Runt domain (RD) (Fig. 1a)16, 17. Core binding factor β (CBFβ) binds RD and enhances Runx1 DNA binding without interacting with DNA19. In contrast, the Runx1•Ets1 cooperation requires the binding of both factors to a composite Runx1•Ets1 DNA motif. Genome wide analysis for the co-occupancy of Ets1 and Runx1 revealed such composite motifs in a number of genes6, 7. Among the well-characterized motifs is a GGATGTGG motif of T cell receptor alpha (TCRα) and beta (TCRβ) gene enhancers3-5. In our previous studies, we revealed an allosteric mechanism of Runx1 activation by CBFβ20. Here we expand our studies to the mechanism of Ets1 activation by Runx1.
MATERIALS AND METHODS
Construction of plasmids, expression and purification
The coding region for different Runx1 constructs (Runx11-242, Runx148-214, Runx11-214, Runx11-190 and Runx148-242) were PCR amplified from pVL1392-AML1.FL plasmid and cloned into pET3a vector (Novagen). The resulting constructs were expressed in E. coli strain Rosetta-2(DE3) at 18 °C for 16 h following induction with 0.3 mM isopropyl β-D-thiogalactopyranoside (IPTG) at A600 = 1. Cells were harvested and resuspended in buffer A (20 mM Tris-HCl pH 7.2, 1 M NaCl, 1 mM ETDA, 10 mM β-mercaptoethanol and 10% glycerol) and lysed with an EmulsiFlex-C5 homogenizer (Avestin). Polyethyleneimine was added to the supernatant at a final concentration of 0.07% and pellet was removed by centrifugation. Proteins were enriched by ammonium sulphate precipitation at 30% saturation. Each pellet was resuspended in buffer A100 containing 100 mM NaCl and dialyzed against the same buffer for 3 h. The proteins were purified using HiTrap SP HP (GE Healthcare) followed by HiTrap Heparin HP column (GE Healthcare). Pooled fractions were diluted to half with 1.5 M ammonium sulfate in buffer A100 and purified using HiTrap Phenyl HP column (GE Healthcare). Purity of fractions was checked by SDS PAGE and stored at -80 °C.
Single and multiple site mutagenesis protocol21 was used to create point mutants of Runx1. Mutations were first introduced in full-length Runx1 plasmid (pCDN3-Myc3-Runx1) and the resultant mutants were further subcloned in vector pET3a. All mutants were verified by DNA sequencing and were expressed and purified as described above.
Ets1280-441 was purified according to reported protocols22. Cloning, expression and purification conditions for Ets1296-441 were similar to those of the Ets1280-441 protein, except supernatant obtained from 50% ammonium sulfate saturation was used for purification. Coding region for Ets1276-441 was cloned in a ligation independent vector in fusion with affinity His6-tagged small ubiquitin related modifier (SUMO) protein.23 His6-SUMO-Ets1276-441 was expressed in E. coli strain BL21(DE3) at 37 °C for 5 h following induction with 0.5 mM IPTG at A600 = 0.5. Harvested expressed cells were resuspended in 20 mM Tris-HCl pH 7.2, 1 M NaCl, 1 mM β-mercaptoethanol, 5 mM Imidazole and 12% glycerol. Initial treatment for purification of His6-SUMO-Ets1276-441 was similar to other Ets1 constructs except the pellet obtained by 40-65% of ammonium sulfate fractionation was used for purification. The pellet was resuspended in 20 mM Tris-HCl pH 7.2, 750 mM NaCl, 1 mM β-mercaptoethanol, 5 mM Imidazole and 5% glycerol and purified by HisTrap HP column (GE Healthcare). His6-SUMO was cleaved with His6-tagged dtUD1 (doubly tagged UD1) protease that was added to protein at 1/5,000 mass ratio and incubated for 3 h at 4 °C before loading onto Ni-IDA column (Bio-Rad). Flow through from the column containing Ets1276-441 was collected and stored at -80 °C.
Ets1276-441 was phosphorylated using the published protocol13. Peak fractions were pooled and analyzed by SDS-PAGE and MALDI TOF/TOF (Supplementary Fig. 1). CaMKIIα used in the kinase reaction was purified according to reported protocol24.
Crystallization and diffraction data collection
Double-stranded TCRα enhancer DNA (TCRα) was prepared by annealing synthetic oligonucleotides 5′-GGAAGCCACATCCTCT-3′ and 5′-CAGAGGATGTGGCTTC-3′. Oligonucleotides were purified using Mono Q™ 5/50 GL column (GE Healthcare) and annealed by heating to 95 °C for 5 min and cooling gradually to room temperature. Annealed DNA was purified using Mono Q™ 5/50 GL. The DNA solution was desalted dried and dissolved in 10 mM Tris-HCl pH 7.5.
For Runx11-242•Ets1296-441•TCRα complex preparation, the frozen Runx11-242 and Ets1296-441 samples were thawed on ice, dialyzed against 10 mM Tris-HCl pH 7.5 and 20 mM DTT buffer and concentrated using Millipore Ultrafree centrifugal devices to 7 and 10 mg•ml-1, respectively. At first a Runx11-242•TCRα was prepared and then Ets1296-441 was added to reach an equal molar ratio of components. The ternary complex was concentrated up to 7 mg•ml-1. The state of sample aggregation at every step was monitored by dynamic light scattering using DynoPro (Wyatt Technology). The Runx148-214•Ets1296-441•TCRα complex was prepared in a similar way.
All crystallization screenings were performed by sitting-drop vapor-diffusion method using Natrix screen kit from Hampton Research. Runx11-242•Ets1296-441•TCRα produced square-bipyramid and plate shaped crystals in the same drop within 48 hours. Initial crystallization condition was further optimized as 100 mM KCl, 15 mM magnesium chloride hexahydrate, 25 mM MES pH 5.6, 14% v/v PEG MME 550 and 6% v/v glycerol (Supplementary Fig. 2a). The size of either crystal was improved by macroseeding after 12 h of equilibration. Growth of the seeded crystals was completed in 10 days (Supplementary Figs 2b and 2c). The crystals were cryoprotected in reservoir solutions containing additions of 3% v/v PEG MME 550 and 18% v/v of ethylene glycol for square-bipyramid crystals and 15% v/v of glycerol for plate shaped crystals.
The Runx148-214•Ets1296-441•TCRα also produced two types of crystals from the same drop of Natrix crystal screen, one deformed cube type and another rod type. The optimum growth of crystals was achieved from a 2.5% w/v PEG 4000, 5 mM magnesium chloride hexahydrate, 25 mM MES pH 5.6 and 5% v/v glycerol (Supplementary Figs 2d and 2e). The cryoprotectant for deformed cube crystals contained additions of 2.5% w/v PEG 4000, 12.5% v/v PEG 200 and 11.5% v/v ethylene glycol. The rod shaped crystals diffracted poorly to 7 Å resolution and were excluded from further studies.
The diffraction data sets were collected using synchrotron radiation at the Advanced Photon Source on the Northeastern Collaborative Access Team beamline BL24ID-E. To minimize the radiation damage, each complete dataset was obtained from one crystal exposed at several different positions. All intensity data were indexed, integrated, and scaled using the HKL2000 program package25 (Table 1).
Table 1.
Data collection and refinement statistics.
| Complex | Runx11-242•Ets1296-441•TCRα | Runx148-214•Ets1296-441•TCRα | |
|---|---|---|---|
| Crystal shape | plate | square-bipyramid | deformed cube |
| Data collection | |||
| Space group | C2221 | C2 | P21 |
| Cell dimensions: | |||
| a (Å) | 102.622 | 87.343 | 66.153 |
| b (Å) | 138.945 | 100.140 | 100.032 |
| c (Å) | 98.439 | 67.225 | 72.064 |
| β (°) | 90 | 120.49 | 110.23 |
| Resolution (Å)* | 50-2.5 (2.54-2.5) | 30-2.7 (2.75-2.7) | 30-2.3 (2.34-2.3) |
| Unique reflections | 24213 (1203) | 13577 (674) | 35376 (1128) |
| Rmerge (%)* | 5.6 (43.3) | 4.3 (46.2) | 5.6 (39.2) |
| I/σ(I) | 29.7 (2.3) | 42.9 (2.1) | 26.7 (2.1) |
| Completeness (%) | 98.0 (98.0) | 98.4 (97.1) | 92.4 (60.3) |
| Redundancy | 2.9 (2.7) | 3.2 (2.5) | 3.2 (2.6) |
| Temperature (K) | 100 | 100 | 100 |
| Mosaicity (°) | 0.28-0.76 | 0.79-1.11 | 0.59-0.95 |
| Refinement | |||
| Resolution (Å) | 29.67-2.5 | 29.63-2.7 | 29.64-2.3 |
| No. reflections | 23493 | 13278 | 33721 |
| Rwork / Rfree | 22.5/24.5 | 22.0/27.2 | 23.1/28.0 |
| No. atoms | |||
| Runx1/Ets1/DNA | 988/880/650 | 1142/851/650 | 2198/1788/1300 |
| Water/Glycerol | 66/0 | 27/0 | 143/6 |
| Residues in model | |||
| Runx1 | 50-177 | 54-177, 190-212 | 53-177, 189-205/52-177,190-204 |
| Ets1 | 334-437 | 332-432 | 333-439/332-437 |
| B-factors (Å2) | |||
| From Wilson plot | 35.1 | 44.0 | 49.0 |
| Mean value | 43.1 | 61.7 | 52.9 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.007 | 0.008 |
| Bond angles (°) | 1.4 | 1.2 | 1.4 |
| Ramachandran plot | |||
| Favored (%) | 85.9 | 81.3 | 88.3 |
| Allowed (%) | 13.6 | 18.2 | 11.0 |
| Generous (%) | 0.5 | 0.5 | 0.7 |
Values in parentheses are for the last resolution shell.
Structure determination and refinement
The structure of deformed cube crystal was determined by the molecular replacement method starting with the coordinates of ED from Ets1 structure (PDB code 1gvj)26 and RD from Runx1•DNA structure (PDB code 1hjc)20. The asymmetric unit contained two molecules of Runx148-214•Ets1296-441•TCRα. Addition of a DNA molecule and major manual rebuilding of the initial model was performed with TURBO-FRODO software. Application of zonal scaling27 and bulk solvent correction improved the quality of electron density maps, enabling EID of Runx1 to be clearly traced and most of the protein side chains to be well fitted. A model was refined using standard protocols. Non-crystallographic two-fold symmetry (NCS) restrains for backbone atoms were applied during the refinement. The structure determination of square-bipyramid and plate shaped crystals was similar to that of cube crystal, but without NCS restraints. The final refinement statistics for all three structures are provided in Table 1. CNS version 1.1 was used for all crystallographic computing28. The figures with electron density maps were prepared with TURBO-FRODO software and all remaining figures displaying the protein structures were prepared with PyMol software from Delano Scientific.
SPR experiments
Surface Plasmon Resonance (SPR) studies were performed using Biacore 3000 biosensor system (GE Healthcare) at 25 °C. A 26 bp double stranded DNA corresponding to TCRα enhancer sequence 5′-AAGCAGAAGCCACATCCTCTGGAAAG-3′ with a covalently linked biotin at 5′ end was captured on SA sensor chip (GE Healthcare) at ~100 response units. Just before initiating the experiment, each protein or its mutants were purified through gel filtration column (Superose 12 10/300 GL, GE healthcare) to remove protein aggregates if present. Kinetic runs were conducted at 30 μl•min-1 to eliminate mass transport and rebinding artifacts. As analytes, each protein or its mutants were diluted serially in the running buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 2 mM MgCl2, 2 mM DTT and 0.005% Tween 20) to concentrations shown in Supplementary Tables 1-3 and injected over the immobilized ligand surface for 120 sec. For experiments where the binding of one protein was measured in the presence of another, 100 nM of second protein was included in running buffer. Dissociation of analytes was then measured by injecting running buffer or running buffer containing protein. The surface was ‘regenerated’ with a pulse of 1 M NaCl at the end of each cycle. Duplicate injections of the same concentration in each experiment were superimposable, demonstrating no loss of activity after regenerating the surface. Buffer injection subtracted graphs were analyzed with BIA evaluation version 4.1 using a nonlinear least squares method to obtain the association and dissociation rate constants (ka and kd, respectively). For these calculations, the global fitting of the association and dissociation phases of the response curves using a 1:1 binding model was applied. The fittings were considered satisfactory if χ2 < 2. The equilibrium dissociation constants (KD) were calculated from the equation KD = kd/ka. Standard deviations were calculated from three experiments.
Transient transfection assays
A 98 bp nucleotide sequence from minimal human TCRα enhancer (12-109)4 was cloned upstream to the SV40 promoter in pGL3-Basic vector (where the SV40 promoter is inserted upstream to Luciferase gene). pCDN3-Myc3-Runx1/mutants and pCMV-Tag2a-FlagEts1 were used as an expression vector for Runx1 and Ets1 respectively. Thr286 of pEGFP-C1-CamKIIα was mutated to Asp for constitutive enzymatic activity24. Expression of constitutively active CamKIIα was confirmed by observing the GFP fluorescence in the cells.
Stock cultures of 293T cells were maintained as described earlier22. For transcriptional assays, 293T cells were seeded at 50,000 cells per well of 6-well plates and transfected in duplicate the following day using X-treme gene HP DNA transfection reagent following manufacturer's protocol. In addition to 2 μg of the promoter/reporter construct, the cells were cotransfected with 25 ng of pTK-RL (Promega) to normalize for any differences in transfection efficiency. Expression levels of wild type Ets1 and Runx1 as well as Runx1 mutants were determined by transiently transfecting 293T cells separately with expression vector harboring construct for the corresponding protein. To test cooperativity in binding of these proteins, cotransfection of Ets1 and Runx1/mutants was performed. Equal amounts of total DNA in each transfection was maintained by adding empty vector CMV5 as needed. Luciferase activity was measured 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega).
RESULTS
Crystal structures
We crystallized and solved the structures of a ternary complex of Runx1 (residues 1-242), Ets1 (296-441) and 16 base pair DNA fragment of TCRα enhancer (Runx11-242•Ets1296-441•TCRα) in square-bipyramid and plate crystal forms and also Runx148-214•Ets1296-441•TCRα in a deformed cube form (Table 1). In all three structures, the ED and RD are bound to respective binding sites positioned opposite relative to each other (Fig. 1b and Supplementary Figs. 3a, 3b and 3c). Distributions of temperature factors in these complexes are shown Supplementary Figs. 3d, 3e and 3f. The excellent electron density maps of Runx148-214•Ets1296-441•TCRα (Supplementary Figs. 4a and 4b) unambiguously revealed the structure of residues 189-205 from an Ets1-interaction domain (EID) of Runx1, which includes the helix α1 (194-203). In addition to helix α1, the main-chain tracing of EID helix α2 (204-211) was clearly defined in square-bipyramid Runx11-242•Ets1296-441•TCRα crystal (Supplementary Fig. 4c). Explanation of the crystal packing effects resulting in disorder of Runx1 Phe194 side-chain in square-bipyramid Runx11-242•Ets1296-441•TCRα crystal, in disorder of EID α2 in Runx148-214•Ets1296-441•TCRα crystal, and in disorder of an entire EID in Runx11-242•Ets1296-441•TCRα plate crystal are provided in Supplementary Fig. 5.
Runx1 EID interactions with Ets1 and DNA
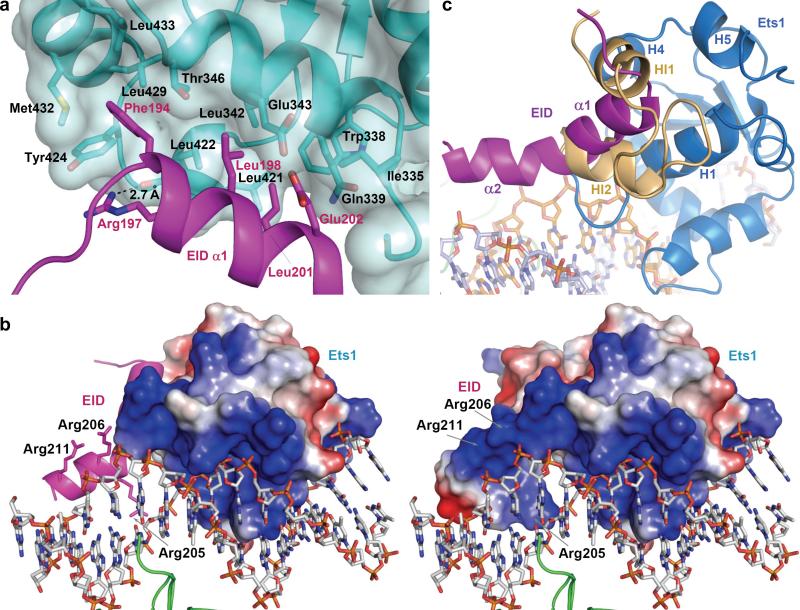
The Runx1 helix α1 is packed on Ets domain parallel to H1 of Ets1, whereas Runx1 α2 is packed parallel to DNA (Fig. 1b). EID interaction buries a total surface area of 1146 Å2. The EID•Ets1 interactions are dominated by hydrophobic amino acid residues (Supplementary Table 4). The EID Phe194, Leu198 and Leu201 side chains are packed into the wide hydrophobic depression at Ets1 surface (Fig. 2a). In addition, EID Arg197 forms a direct hydrogen bond with backbone oxygen of Ets1 Leu422 (Fig. 2a). At the EID•DNA interface, a positively charged area of EID formed by Arg205, Arg206 and Arg208, is positioned against the negatively charged phosphates of DNA (Fig. 2b) providing the long-range protein•DNA electrostatic interactions that further stabilize the structure of EID•Ets1•DNA. In addition, an Arg205 side chain electron density in Runx148-214•Ets1296-441•TCRα crystal (Supplementary Fig. 4a) indicates a direct interaction of Arg205 with DNA backbone (Supplementary Fig. 6).
Figure 2. EID interactions with Ets1 and DNA.
(a) Packing of hydrophobic residues at the EID•Ets1 interface. Ets1 and EID are shown as blue and magenta ribbons. The side chains of participating residues are shown in sticks. An Ets1 surface is semitransparent. A hydrogen bond between EID Arg197 side chain and backbone oxygen of Ets1 Leu422 is also shown here by a dashed line. (b) Packing of positively charged area of EID•Ets1 against DNA. On the left the surface calculation includes Ets1 and on the right the surface calculation includes both EID and Ets1. The positively and negatively charged surface areas are in blue and red, respectively. (c) Comparison of EID position in Runx11-242•Ets1296-441•TCRα (magenta) with the positions of IRM helices HI1 and HI2 in autoinhibited free Ets1 (orange) (PDB access code 1R36) after the superimposition of Ets1 molecules. Ets1 of ternary complex is omitted for clarity.
Runx1 EID displaces the IRM of Ets1
The structures of ED and RD and their modes of DNA recognition in Runx11-242•Ets1296-441•TCRα complex are consistent with the previously reported crystal structures (Supplementary Fig. 7).22,26, 29-33 However, a prominent difference exists in folding of IRM within Ets1 that is bound to DNA. The HI1 is unfolded in Ets1•Pax5•DNA and is folded in (Ets1 dimer)•DNA, while HI2 is folded in both complexes (Supplementary Fig. 8). In contrast, the entire inhibitory module of Ets1, including IRM, is disordered in Runx11-242•Ets1296-441•TCRα. Comparison of Ets1•EID structure from our ternary complex with the solution structure of autoinhibited Ets112 shows an overlapping of the space occupied by EID of Runx1 and IRM of Ets1 (Fig. 2c). This demonstrates that EID of Runx1 displaces IRM of Ets1. Thus, the crystal structure of Runx11-242•Ets1296-441•TCRα provides a novel mechanism of Ets1 cooperative binding to DNA in which the EID of DNA-bound Runx1 displaces the IRM of Ets1 and consequently disrupts the inhibitory function of the residues N-terminal to IRM (Fig. 3). This is consistent with previously reported constitutively active mutants of Ets1 that disrupt the IRM docking to ED and also lead to Ets1 activation.15
Figure 3. Mechanism of Ets1 activation by Runx1.
(a) Ets1 binding to TCRα in the absence of Runx1. Normally, Ets1 is partially inhibited in the absence of Runx1. IRM plays a major role in stabilization of SRR inhibitory confirmation. DNA binding results in disorder of IRM helix HI1 and disruption of SRR inhibitory conformation. Phosphorylation of SRR serines by CamKIIα dramatically enhances an inhibitory conformation of SRR, resulting in a highly inhibited form of Ets1. (b) Ets1 binding to TCRα in the presence of DNA-bound Runx1. EID of DNA-bound Runx1 is exposed and disordered. Upon approaching Ets1, EID of Runx1 binds to ED and DNA by forming helices α1 and α2, and displacing both IRM helices HI1 and HI2. This results in disorder of IRM and destabilization of SRR inhibitory confirmation, producing a fully active Ets1. Similar to activation of wild-type Ets1, Runx1 also fully activates the phosphorylated form of Ets1. In both panels, the red stars indicate the presence of phosphorylated serines in SRR.
Runx1 EID contributes to cooperative binding of Ets1 to DNA
To validate the functional relevance and specificity of the observed EID•ED and EID•DNA interactions, we decided to prepare a variety of Runx1 deletions and mutations and analyze their DNA-binding by SPR.
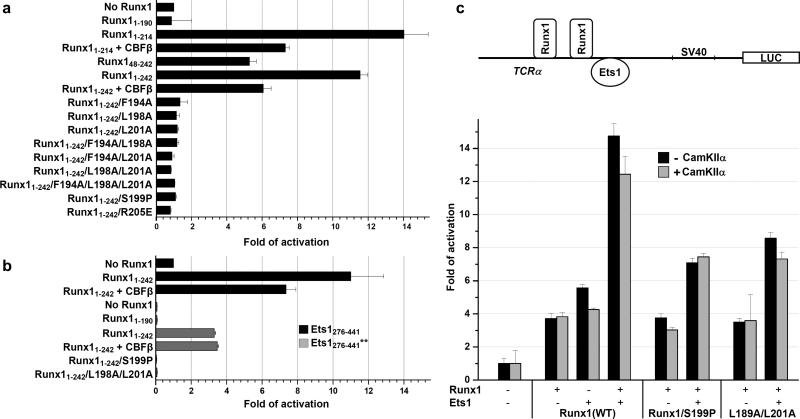
First, we tested DNA binding of Runx1 proteins (Supplementary Table 1). Runx11-214 exhibited 7 fold lower KD of 0.47 nM than Runx11-242 KD of 3.3 nM. These data confirm the presence of autoinhibitory sequences C-terminal to 214.17 However, deletion of 47 residues from N-terminus of Runx1 had little impact of DNA binding (KD of Runx148-242 was 2.9 nM). All EID mutants bound DNA with KDs (2.05 nM to 4.91 nM) that are comparable to KD of Runx11-214 (3.3 nM). In summary, all mutated and truncated Runx1 bound to DNA with KDs less than 5 nM and are useful for studies of Ets1 cooperative binding to the TCRα enhancer.
Next, we tested the effect of Runx1 deletions on Ets1 DNA-binding activation (Fig. 4a and Supplementary Table 2). KD of Ets1 is 30 nM, however in the presence of DNA-bound Runx11-242 the DNA binding activity of Ets1280-441 is enhanced 11.5 fold (KD=2.6 nM). C-terminal truncation of Runx1 up to residue 214 (Runx11-214) had minimal effect on Ets1 activation, which is consistent with the disordered state of residues 213-242 in crystal. In contrast, further C-terminal truncation up to residue 190 (Runx11-190) resulted in a complete loss of Ets1 activation (KD=34.4 nM). This series of mutants maps the region between 190 and 214 as essential for cooperativity with Ets1, as previously shown.17 Absence of Ets1 activation by Runx11-190 confirms that, in spite of partial overlapping of Ets1 and Runx1 binding sites, the cooperation through DNA effect does not occur and points to a direct participation of EID in the cooperative binding of Ets1 to the TCRα enhancer. The absence of cooperation through DNA conformational changes is consistent with the studies by Goetz et al10 using a different approach. In order to eliminate through DNA effect, in their electrophoretic mobility shift assays (EMSA) they used DNA duplexes containing a nick on one strand between the Runx1 and Ets1 binding sites. They found that Runx1 retained Ets1 activation even with a nicked DNA and concluded the absence of Runx1•Ets1 cooperation through changes in DNA conformation.10 In combination, our biochemical and structural data demonstrate that a direct protein interaction explains the cooperative DNA binding between these two factors.
Figure 4. Ets1 activation by Runx1.
(a) DNA binding of Ets1280-441 to TCRα enhancer DNA in presence of various Runx1 proteins or Runx1•CBFβ1-141 complex. (b) DNA binding of Ets1276-441 and phosphorylated Ets1276-441** in presence of various Runx1 proteins or Runx1•CBFβ1-141 complex. In panels (a) and (b) the fold of Ets1 DNA binding activation is calculated from the average of dissociation constants obtained from three individual SPR measurements (Supplementary Tables 2 and 3). (c) Transient transfection assay showing transactivation of TCRα enhancer. Luciferase gene expression is driven by TCRα gene enhancer in 293T cell in presence and absence of constitutively active CaMKIIα. Fold of activation is calculated relative to the reporter only control. The standard deviations are calculated from three independent experiments with P < 0.05. The reporter construct used is current assay is shown above the graph.
Interaction of Runx1 EID with both Ets1 and DNA is highly specific
We analyzed the effect of EID point mutations in its Ets1- and DNA-interacting interfaces (Fig. 4a and Supplementary Table 2). We mutated each of three hydrophobic Ets1-interacting amino acid residues Phe194, Leu198 and Leu201 to Ala in Ets1-interacting helix α1 and also prepared double and triple Ala mutants. Furthermore, we introduced a kink in helix α1 by a Ser199Pro mutation. Any of these mutations destabilize EID•Ets1 interaction and result in loss of Ets1 DNA binding activation. To evaluate the contribution of electrostatic interactions between the EID helix α2 and DNA, we prepared the Arg205Glu mutant. As a result, the positive charge reduction in helix α2 prevented the cooperative DNA binding by Ets1. In summary, SPR experiments confirm that interaction of EID with both Ets1 and DNA is highly specific and necessary for activation of Ets1 DNA binding.
Concerted binding of Runx1 and Ets1 to TCRα enhancer
Order of addition DNA binding experiments indicate that Runx1 and Ets1 form a ternary complex with TCRα enhancer in a highly concerted manner, consistent with previous observations.17 First, Runx1 binds DNA, and only then does EID recruit Ets1 by binding to ED and displacing the IRM of Ets1. As a consequence, Ets1 becomes active by over 11-fold. In case of reverse order of addition experiments, the TCRα enhancer-bound Ets1 was not capable of Runx1 stimulation (Supplementary Table 1). SPR experiments also show that the addition of CBFβ, a heterodimeric partner of Runx proteins, does not enhance Ets1 recruitment (Fig. 4a and Supplementary Table 2). This is in line with the model of Ets1•Runx1•CBFβ•TCRα quaternary complex revealing absence of physical interaction between CBFβ and Ets1 (Supplementary Fig. 9). A similar role of CBFβ and “order of addition” effect was observed with Runx1 and Ets1 binding to a so-called SC1/core DNA having different spacing between Runx1 and Ets1 binding sites10, 17. The Runx1•Ets1 cooperative action in two different composite sites points to a flexibility of the linker between the RD and EID, which is consistent with the disorder of this linker in our crystals.
Runx1 overcomes an inhibitory effect of Ets1 phosphorylation
Ca2+-dependent phosphorylation of serines in the flexible inhibitory region of Ets1 was found to further stabilize the inhibitory conformation and consequently reinforce Ets1 auto-inhibition up to 50 fold.13, 15 To test whether Runx1 is also capable of overcoming Ets1 autoinhibition after the phosphorylation, we phosphorylated Ets1276-441 at two critical sites, Ser282 and Ser285 (Ets1276-441**, Fig. 1a and Supplementary Fig. 1). SPR experiments revealed a 14-fold reduction in Ets1276-441 DNA-binding affinity after the phosphorylation (Fig. 4b and Supplementary Table 3). Remarkably, in presence of prebound Runx11-242 or Runx11-242•CBFβ complex, the DNA-binding of Ets1276-441** was dramatically enhanced and reached the same order (only about 3-fold less) as a wild type Ets1276-441. To the contrary, Ets1276-441** DNA-binding was not enhanced in presence of prebound Runx11-190 lacking EID. To confirm that EID is responsible for the enhancement of Ets1276-441** DNA binding, we evaluated the effect of single Ser199Pro and double Leu198Ala/Leu201Ala mutations in α2 helix of EID. Both mutations disrupt EID•Ets1 interaction and result in the loss of Ets1276-441** DNA binding enhancement. These experiments confirm that Runx1 EID overcomes an inhibitory effect of Ets1 phosphorylation by displacing the IRM of Ets1.
Synergistic trans-activation of TCRα gene by Runx1•Ets1 cooperation
Furthermore, we examined the effect of Runx1 and Ets1 cooperative binding and trans-activation of TCRα gene enhancer fragment by transient transfection assay. To determine the impact of Ets1 phosphorylation, the experiments were also performed in presence of constitutively active CaMKIIα. Synergistic trans-activation of TCRα gene enhancer was observed both with wild-type and phosphorylated Ets1. However, disrupting EID•Ets1 interaction with Runx1 mutations eliminated the synergy. The results of the SPR (Fig. 4b) and transient transfection (Fig. 4c) experiments provide additional support for a mechanism of Ets1 activation that is based on displacement of an entire N-terminal inhibitory module of either wild type or phosphorylated Ets1 by EID of DNA-bound Runx1 (Fig. 3).
DISCUSSION
Comparison of Runx1•Ets1 cooperation with other modes of Ets proteins cooperation
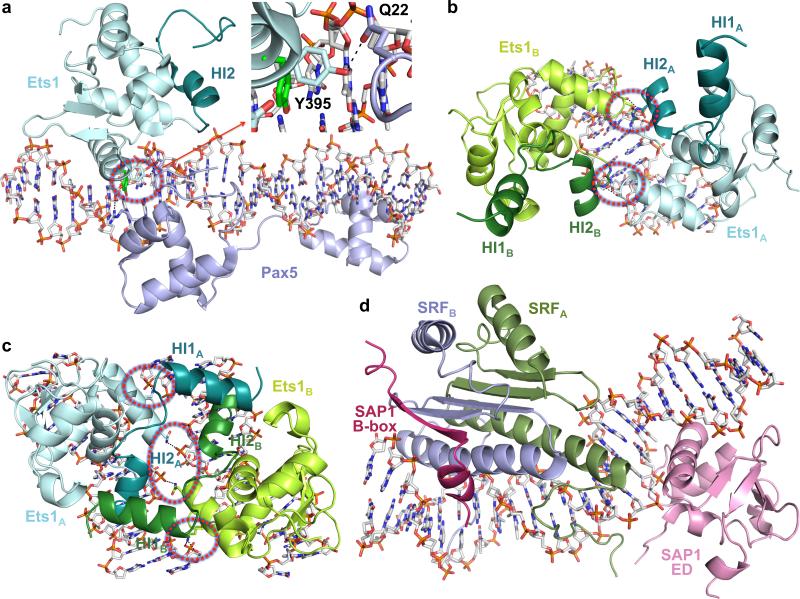
Analysis of Runx1•Ets1•TCRα structure and other Ets1 ternary complex crystal structures shows that Ets1 can adopt a variety of different mechanisms for formation of high-order complexes on DNA. For example, Ets1 binds poorly to suboptimal 5′-GGAG-3′ sequence on mb-1 promoter, however, in presence of Pax5 it binds mb-1 promoter with high affinity34. Crystal structure of Pax5•Ets1•mb-1 revealed a direct interaction between DNA-binding domains of Pax5 and Ets1 that altered the DNA-binding surface of Ets1, mainly by conformational switch of its Tyr395 side chain (Fig. 5a)29, 30. Another example is Ets1 cooperative binding to stromelysin-1 promoter containing palindromic head-to-head Ets-binding sites separated by four base pairs35. Crystal structure shows that cooperative binding to stromelysin-1 promoter is facilitated by DNA-mediated homodimerization of Ets1 (Fig. 5b)22, 36. Recently we reported also the crystal structure of Ets1 in complex with TCRα enhancer DNA26. In this structure Ets1 binds as a homodimer to parallel pieces of dsDNA having Ets-binding sites with opposite orientation (Fig. 5c)26. Observation of additional intermolecular Ets1•DNA interactions within this complex (Fig. 5c) indicates that Ets1 binding is cooperative26. Among these Ets1 complexes the most dramatic effect on Ets1 inhibitory IRM-SRR sequences we can see in Runx1•Ets1•TCRα where an IRM is fully disordered. In the Pax5•Ets1•mb-1 only HI2 of IRM remains bound to Ets domain, however, HI1 is unfolded and partially structured. HI2 remains well folded also in the DNA-bound Ets1 homodimers. Moreover, an Ets domain in these structures also favors the binding of HI1 from a symmetry related molecule.
Figure 5. High-order complexes of Ets1 and Sap1.
Crystal structures of (a) Pax5•Ets1•mb-1 (PDB code 1mdm), (b) (Ets1)2•stromelysin-1 (3mfk), (c) (Ets1)2•2TCRα (3ri4), and (d) SAP1•SRF•SRE (1hbx). The areas with intermolecular interactions leading to Ets1 cooperative DNA-binding are circled by dashed red lines. Zoomed area in panel (a) demonstrates a conformational switch of Ets1 Tyr395 side chain induced by interaction with Gln22 of Pax5. The Tyr395 side chain from a superimposed Runx1•Ets1•TCRα is shown in green color.
It is also interesting to compare mechanism of Ets1 activation by Runx1 with the mechanism of activation of another Ets family member, SAP1. Like Ets1, DNA binding of SAP1 is also autoinhibited. SAP1 B-box sequences that are attached by a flexible linker to the C-terminal of Ets domain participate in autoinhibition by interacting with Ets domain. However, SAP1 became active and binds to c-fos serum response element (SRE) in presence of serum response factor (SRF)37, 38. The crystal structure of SAP1•SRF•SRE shows how an autoinhibition of SAP1 was relieved by switching the B-box from Ets domain to the surface of SRF (Fig. 5d)39. Furthermore, the cooperative binding is enforced by direct interaction between DNA binding domains of SAP1 and SRF39, 40. The crystal structures highlight the differences between Ets1 activation by Runx1 and SAP1 activation by SRF. In case of Ets1, binding of Runx1 EID to Ets domain disrupts the autoinhibitory IRM-SRR region packing. However, in case of SAP1 activation its autoinhibitory B-box is moved away from Ets domain by interacting with SRF.
Conclusion
Protein-protein interactions and post-translational modifications play an important role in combinatorial regulation of transcription. During different stages of regulation the same transcription factor may interact with different sets of partners. For example, studies of Runx1 partners during different stages of megakaryocytic differentiation revealed its interactions with either GATA, or AP-1 or Ets proteins41. Such a dynamic nature of Runx1 interactions and transcriptional factors in general, often precludes in-depth structural characterization of the underlying mechanisms of action. Here we succeeded in discovery of a novel mechanism of Ets1 activation by DNA-bound Runx1 that involves the displacement of entire inhibitory module of Ets1 by EID of Runx1. Moreover, our data revealed that such displacement efficiently counteracts the strong inhibitory effect of Ets1 phosphorylation. This novel mechanism of phosphorylated Ets1 activation by Runx1 partnership appears to be unique. First, unlike the widely used reversible phosphorylation-dependent regulation of transcription factors42, it does not require Ets1 dephosphorylation. Second, not every Ets1 cooperative DNA binding can overcome the inhibitory effect of Ets1 phosphorylation. For example, contrary to the Runx1•Ets1 partnership, Ets1•Ets1 cooperative binding to palindromic Ets-binding sites on stromelysin-1 promoter DNA results in additional stabilization of IRM by intermolecular interactions22. However, these Ets1•Ets1 interactions are not capable of counteracting the inhibitory effect of phosphorylation, and upon phosphorylation, Ets1 loses its ability to bind the stromelysin-1 promoter35. It remains to be seen whether Runx1 is a unique activator of phosphorylated Ets1 or if a variety of other protein partners (e.g. Pax5, AP-1, USF1, etc.)8 are also capable of activating phosphorylated Ets1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jeff Lovelace and Gloria E. Borgstahl for maintenance and management of the Eppley Institute's X-ray Crystallography Facility; Dmitry G. Vassylyev for the zonal scaling instruction files; Altair M. Ioffe for participation in Runx1 mutants preparations and SPR experiments; Nancy A. Speck, Ulli Bayer and Walter Chazin for providing cDNAs for Runx1 and CBFβ, CaMKIIα and Calmodulin, respectively.
The Sources of Support: This work is supported by NIH grant GM082923 and Nebraska Department of Health and Human Services grant LB506 to THT. The Eppley Institute's core facilities are supported by the Cancer Center Support Grant P30CA036727. This work is also based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team Beamlines, which are supported by grants from the National Center for Research Resources (5P41RR015301-10) and the National Institute of General Medical Sciences (8 P41 GM103403-10) from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Cloning, expression and purification of Runx1, Ets1 and CBFβ were performed by KM, TS and OIB. Crystallizations were performed by THT, KM, NDB and TS. SPR, transient transfection experiments and writing the corresponding paragraphs of methods were performed by TS. Cryoprotection of crystals and diffraction data collection was performed by NDB and THT. AR provided advice for transient transfection experiments. THT initiated, planned and managed the project, solved and analyzed the crystal structures, and wrote the manuscript.
PROTEIN DATA BANK ACCESSION NUMBERS
Atomic coordinates and structure factors for Runx11-242•Ets1296-441•TCRα plate and square-bipyramid crystal structures and Runx148-214•Ets1296-441•TCRα deformed cube crystal structure have been deposited in the Protein Data Bank with accession numbers 4L0Y, 4L0Z and 4L18, respectively.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Lelli KM, Slattery M, Mann RS. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012 Dec 15;46:43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tootle TL, Rebay I. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays. 2005 Mar;27(3):285–298. doi: 10.1002/bies.20198. [DOI] [PubMed] [Google Scholar]
- 3.Wotton D, Ghysdael J, Wang S, Speck NA, Owen MJ. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994 Jan;14(1):840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995 Apr 15;9(8):995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 5.Sun W, Graves BJ, Speck NA. Transactivation of the Moloney murine leukemia virus and T-cell receptor beta-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995 Aug;69(8):4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007 Aug 1;21(15):1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009 Dec;5(12):e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003 Aug 20;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaikhibrahim Z, Wernert N. ETS transcription factors and prostate cancer: the role of the family prototype ETS-1 (review). Int J Oncol. 2012 Jun;40(6):1748–1754. doi: 10.3892/ijo.2012.1380. [DOI] [PubMed] [Google Scholar]
- 10.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol Cell Biol. 2000 Jan;20(1):81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalicky JJ, Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996 Feb;5(2):296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, et al. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem. 2005 Feb 25;280(8):7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 13.Lee GM, Pufall MA, Meeker CA, Kang HS, Graves BJ, McIntosh LP. The affinity of Ets-1 for DNA is modulated by phosphorylation through transient interactions of an unstructured region. J Mol Biol. 2008 Oct 17;382(4):1014–1030. doi: 10.1016/j.jmb.2008.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, et al. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005 Jul 1;309(5731):142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 15.Cowley DO, Graves BJ. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev. 2000 Feb 1;14(3):366–376. [PMC free article] [PubMed] [Google Scholar]
- 16.Kim WY, Sieweke M, Ogawa E, Wee HJ, Englmeier U, Graf T, et al. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. Embo J. 1999 Mar 15;18(6):1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu TL, Goetz TL, Graves BJ, Speck NA. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1). Mol Cell Biol. 2000 Jan;20(1):91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004 May 24;23(24):4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, et al. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993 May;194(1):314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 20.Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001 Mar 9;104(5):755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babayeva ND, Wilder PJ, Shiina M, Mino K, Desler M, Ogata K, et al. Structural basis of Ets1 cooperative binding to palindromic sequences on stromelysin-1 promoter DNA. Cell Cycle. 2010 Aug 1;9(15):3054–3062. doi: 10.4161/cc.9.15.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif. 2007 May;53(1):40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singla SI, Hudmon A, Goldberg JM, Smith JL, Schulman H. Molecular characterization of calmodulin trapping by calcium/calmodulin-dependent protein kinase II. J Biol Chem. 2001 Aug 3;276(31):29353–29360. doi: 10.1074/jbc.M101744200. [DOI] [PubMed] [Google Scholar]
- 25.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006 Aug;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 26.Babayeva ND, Baranovskaya OI, Tahirov TH. Structural basis of Ets1 cooperative binding to widely separated sites on promoter DNA. PLoS One. 2012;7(3):e33698. doi: 10.1371/journal.pone.0033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007 Jul 12;448(7150):157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998 Sep 1;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J Biol Chem. 2002 Nov 22;277(47):45529–45536. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 30.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell. 2001 Dec;8(6):1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 31.Bartfeld D, Shimon L, Couture GC, Rabinovich D, Frolow F, Levanon D, et al. DNA recognition by the RUNX1 transcription factor is mediated by an allosteric transition in the RUNT domain and by DNA bending. Structure. 2002 Oct;10(10):1395–1407. doi: 10.1016/s0969-2126(02)00853-5. [DOI] [PubMed] [Google Scholar]
- 32.Backstrom S, Wolf-Watz M, Grundstrom C, Hard T, Grundstrom T, Sauer UH. The RUNX1 Runt domain at 1.25A resolution: a structural switch and specifically bound chloride ions modulate DNA binding. J Mol Biol. 2002 Sep 13;322(2):259–272. doi: 10.1016/s0022-2836(02)00702-7. [DOI] [PubMed] [Google Scholar]
- 33.Bravo J, Li Z, Speck NA, Warren AJ. The leukemia-associated AML1 (Runx1)--CBF beta complex functions as a DNA-induced molecular clamp. Nat Struct Biol. 2001 Apr;8(4):371–378. doi: 10.1038/86264. [DOI] [PubMed] [Google Scholar]
- 34.Fitzsimmons D, Hodsdon W, Wheat W, Maira SM, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996 Sep 1;10(17):2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 35.Baillat D, Begue A, Stehelin D, Aumercier M. ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J Biol Chem. 2002 Aug 16;277(33):29386–29398. doi: 10.1074/jbc.M200088200. [DOI] [PubMed] [Google Scholar]
- 36.Lamber EP, Vanhille L, Textor LC, Kachalova GS, Sieweke MH, Wilmanns M. Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. Embo J. 2008 Jul 23;27(14):2006–2017. doi: 10.1038/emboj.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang SH, Shore P, Willingham N, Lakey JH, Sharrocks AD. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. Embo J. 1999 Oct 15;18(20):5666–5674. doi: 10.1093/emboj/18.20.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. Embo J. 1992 Dec;11(12):4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassler M, Richmond TJ. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. Embo J. 2001 Jun 15;20(12):3018–3028. doi: 10.1093/emboj/20.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mo Y, Ho W, Johnston K, Marmorstein R. Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J Mol Biol. 2001 Nov 30;314(3):495–506. doi: 10.1006/jmbi.2001.5138. [DOI] [PubMed] [Google Scholar]
- 41.Pencovich N, Jaschek R, Tanay A, Groner Y. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood. 2011 Jan 6;117(1):e1–14. doi: 10.1182/blood-2010-07-295113. [DOI] [PubMed] [Google Scholar]
- 42.Holmberg CI, Tran SE, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002 Dec;27(12):619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.