Significance
Cyanobacteria, as well as chloroplasts of plants and algae, are the sites of photosynthesis that produces oxygen. Photosynthetic membranes, also known as thylakoid membranes, in these organisms contain galactolipids, without exception, as the major components. Galactolipids are thus believed to be important for photosynthesis or at least for the formation of the flattened shape of thylakoid membranes. The biosynthetic pathway of galactolipids is definitely different in plants and cyanobacteria. Here we identified the final piece of the long-sought gene for the galactolipid synthesis in cyanobacteria, namely monoglucosyldiacylglycerol epimerase, which converts glucolipid to galactolipid. Cyanobacterial mutants in which this gene is disrupted still formed authentic thylakoid membranes without galactolipids and kept the ability to grow photosynthetically, indicating that galactolipids are not essential for either oxygenic photosynthesis or the development of the thylakoid membrane structure.
Keywords: chromatophore, cluster analysis, endosymbiosis, evolution of photosynthesis, NAD(P)H-dependent oxidoreductase
Abstract
The thylakoid membranes of oxygenic photosynthetic organisms are dominated by the galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG). In cyanobacteria, MGDG is synthesized via monoglucosyldiacylglycerol (GlcDG). However, the putative epimerase involved in the conversion of GlcDG to MGDG has not been identified. Here we report the identification of the gene for the glucolipid epimerase (mgdE) by comparative genomic analysis. Knockout mutants of mgdE in Synechocystis sp. PCC 6803 lacked both MGDG and DGDG and accumulated GlcDG. The mutants did possess thylakoid membranes and showed normal maximal photosynthetic activity, albeit with reduced utilization of light energy. These results cast doubt on the long-standing belief that oxygenic photosynthesis is absolutely dependent on galactolipids.
The galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) are prevalent membrane lipids in organisms that carry out oxygenic photosynthesis, such as land plants, algae, and cyanobacteria (1). Therefore these lipids are believed to be essential for oxygenic photosynthesis. In fact, crystallographic analysis of photosystem complexes revealed that stoichiometric amounts of both types of galactolipids are bound in the interior of photosystem complexes (2, 3), and a galactolipid-deficient mutant in the higher plant, Arabidopsis thaliana, has no photosynthetic activity (4). MGDG and DGDG constitute about 50% and 30%, respectively, of the total lipids in the majority of photosynthetic membranes. Because of the substantial biomass of photosynthetic organisms, these lipids are thought to be the most abundant on Earth (5).
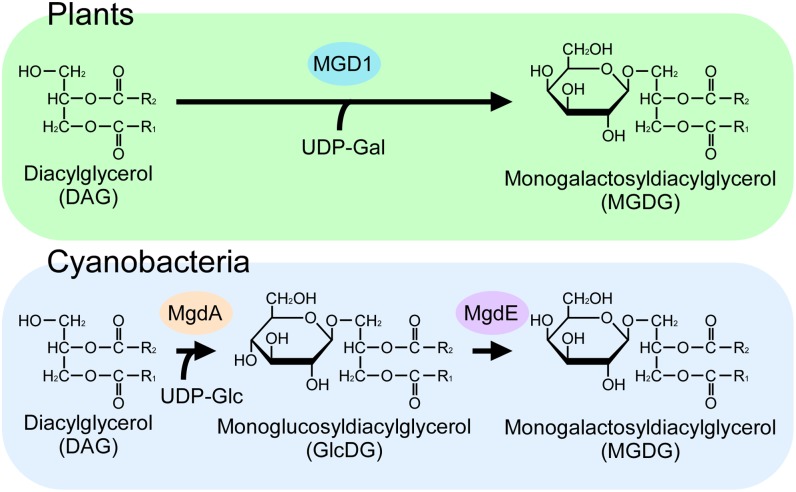
Despite the conservation of galactolipids in oxygenic organisms, their biosynthetic pathways differ in plants and cyanobacteria (Fig. 1). Plants use UDP-galactose and diacylglycerol as substrates for MGDG synthase, which transfers a galactose unit to diacylglycerol by a single reaction (6). In cyanobacteria, on the other hand, metabolic experiments using an isotope tracer suggest that monoglucosyldiacylglycerol (GlcDG) is synthesized initially by the transfer of a glucose unit from UDP-glucose to diacylglycerol and that GlcDG then undergoes isomerization to yield MGDG via the action of GlcDG epimerase (7, 8). The second transfer of a galactose unit to MGDG for the synthesis of DGDG also involves distinct enzymes in plants and cyanobacteria. We previously identified the genes for GlcDG synthase and DGDG synthase in cyanobacteria using comparative genomic analysis (9–12). However, further efforts to identify the epimerase gene were unsuccessful.
Fig. 1.
Difference in galactolipid synthesis in plants and cyanobacteria. (Upper) The MGDG synthesis pathway conserved in red algae, green algae, and higher plants. (Lower) The pathway conserved in cyanobacteria and chromatophores of Paulinella chromatophora.
To identify the epimerase gene, we adopted cluster analysis for extraction of the candidate genes. By analysis of knockout mutants and coexpression in Escherichia coli, we found that one of these genes, sll1376, encodes GlcDG epimerase. The mutants accumulated GlcDG instead of MGDG and DGDG, indicating that a glucolipid can compensate for the functions of the galactolipids. These results cast doubt on the long-standing belief that galactolipids are absolutely required for oxygenic photosynthesis.
Results and Discussion
Identification of the GlcDG Epimerase Gene in Synechocystis sp. PCC 6803.
The genomic sequence of the chromatophore (a chloroplast-like organelle that probably originated from a Prochlorococcus-like cyanobacterium) in the photosynthetic cercozoan Paulinella chromatophora (13) provided a starting point for identifying the epimerase. The genes for all identified enzymes involved in the membrane lipid biosynthesis in cyanobacteria are conserved in the 867 protein-coding genes of the chromatophore genome. To identify the putative epimerase gene, we used computational cluster analysis (14) to look for genes in cyanobacteria that (i) are conserved in the chromatophore genome of P. chromatophora; (ii) also are conserved among most cyanobacterial species; and (iii) encode oxidoreductase or a related motif that could accomplish the epimerase reaction [e.g., NAD(P)-bd or Aldo/ket_red of InterPro, European Molecular Biology Laboratory-European Bioinformatics Institute]. Three candidate genes fit these criteria.
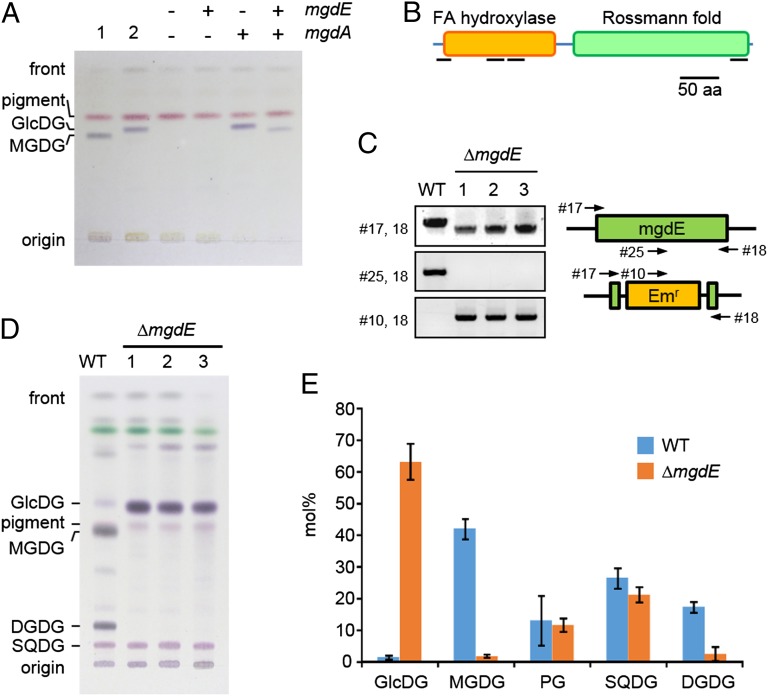
To investigate whether the candidate genes encoded the epimerase, we first knocked them out individually in the cyanobacterium Synechocystis sp. PCC 6803. To circumvent the possibility that the epimerase gene may be essential and the mutation lethal, we also introduced a plant-type MGDG synthase gene from cucumber (CsMGD1) to bypass the endogenous galactolipid synthetic pathway (Fig. S1). One of the knockout mutants showed an accumulation of GlcDG, suggesting that the disrupted gene, sll1376, encodes the epimerase (Fig. S1). This gene then was expressed in E. coli cells with GlcDG synthase (mgdA). As expected, E. coli expressing only mgdA accumulated GlcDG in their membranes, whereas the cells coexpressing the candidate gene and mgdA accumulated a lipid that comigrated with authentic MGDG by TLC (Fig. 2A). Based on these results, we concluded that this candidate gene encodes the enzyme responsible for the epimerization, and we designated it “GlcDG epimerase” (mgdE).
Fig. 2.
Analysis of glucolipids and galactolipids in engineered E. coli and Synechocystis mutants. (A) Coexpression of mgdA and mgdE in E. coli. Lipid extracts from E. coli overexpressing plant MGDG synthase (lane 1) and cyanobacterial GlcDG synthase (lane 2) were used as standards. The pink band in all lanes is an endogenous E. coli pigment. (B) Predicted structure of MgdE. Black bars under the domain boxes indicate predicted membrane-spanning domains. FA, fatty acid. (C) The genomic mgdE region of wild-type and ΔmgdE strains in which mgdE was replaced by the erythromycin-resistance gene, Emr, as shown schematically on the right, was amplified by PCR with the indicated primer pairs (numbers and arrows) and visualized by agarose gel electrophoresis. (D) TLC analysis of lipids in the wild-type and ΔmgdE mutant cells of Synechocystis sp. PCC 6803. (E) Detailed lipid composition of the wild-type and ΔmgdE mutant cells. Values are the average (wild-type, n = 6; ΔmgdE, n = 5) ± the SD.
The predicted MgdE protein contains a fatty acid hydroxylase domain and a Rossmann-fold domain and belongs to the short-chain dehydrogenase/reductase family of proteins (Fig. 2B) (15). The Rossmann fold functions as an oxidoreductase domain (16). Interestingly, in some cyanobacteria, such as Prochlorococcus, MgdE lacks the hydroxylase domain (Fig. S2).
Galactolipids Are Not Essential for Functions of the Thylakoid Membranes.
To address the intriguing question of whether oxygenic photosynthesis can proceed without galactolipids, we constructed a knockout mutant of mgdE (ΔmgdE) that was expected to accumulate GlcDG instead of MGDG if the deletion is not lethal. We isolated several ΔmgdE strains (Fig. 2C) that grew more slowly than the wild type, having an approximately twofold longer doubling time under optimal growth conditions. The deletion strains were essentially devoid of MGDG and DGDG, which were totally replaced by GlcDG (Fig. 2 D and E). To ensure that the deletion mutant did not contain minute amounts of MGDG, we analyzed the composition of the sugar polar head of the TLC fraction corresponding to monohexosyldiacylglycerols. No galactose moiety was detected in the deletion mutant by GC-MS analysis, confirming that MGDG comprised <0.4% of total membrane lipids.
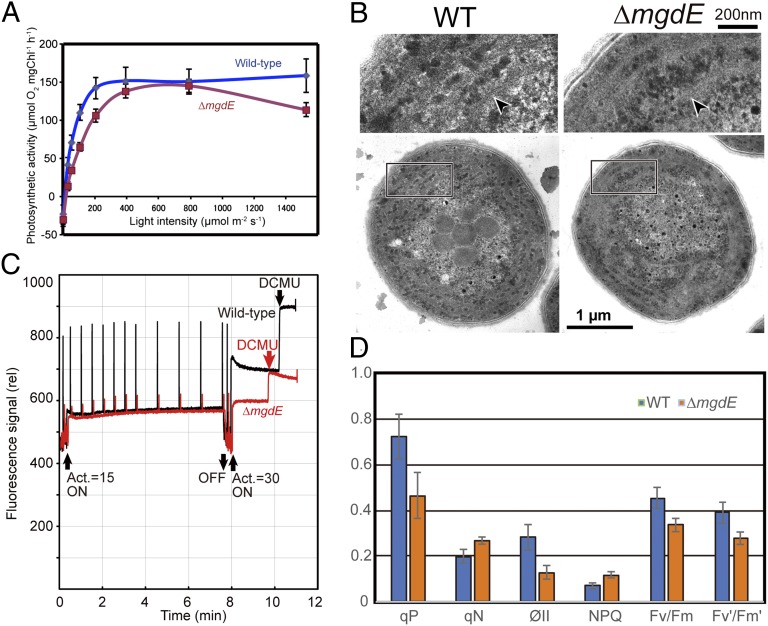
The ΔmgdE mutants retained ∼70% of the chlorophyll content of the wild type (wild type: 4.10 ± 0.5 μg chlorophyll in 1 ml of culture at OD730 = 1.0; ΔmgdE: 2.81 ± 0.66 μg chlorophyll in 1 ml of culture at OD730 = 1.0) and retained photosynthetic activity (Fig. 3A). In addition, electron microscopy clearly indicated that the mutants contain normal-looking thylakoid membranes (i.e., lamellar photosynthetic membranes; Fig. 3B). The maximum oxygen evolution activity at saturating light intensity was almost identical for the ΔmgdE and wild-type strains, but the light intensity for half saturation was significantly higher for ΔmgdE than for wild type (130 vs. 60 µmol⋅m−2⋅s−1, respectively). This inefficiency of the ΔmgdE mutant at low light can be attributed to higher energy dissipation, as evidenced by pulse amplitude modulation fluorescence [parameters coefficient of nonphotochemical quenching (qN) and nonphotochemical quenching parameter (NPQ) in Fig. 3D]. The rate of electron transport also was reduced in ΔmgdE, as indicated by lower Fv/Fm ratios, which is a measure of maximum quantum efficiency of photosystem II, coefficient of photochemical quenching (qP), and quantum efficiency of photosystem II (ΦII) (Fig. 3D). These results suggest that galactolipids (MGDG and DGDG) are not required for the formation of photosynthetically active membranes if glucolipids are available to the cells, although optimal photosynthesis requires membranes containing galactolipids.
Fig. 3.
Photosynthetic activity of mgdE mutant cells. (A) Oxygen evolution rates in wild-type (blue trace) and ΔmgdE (red trace) cells as a function of light intensity. Chl, chlorophyll. (B) Presence of the thylakoid membranes in ΔmgdE mutant cells. Ultrathin sections of wild-type (Left) and ΔmgdE mutant (Right) cells were examined by electron microscopy. Enlarged images of the boxed regions highlight the thylakoid membranes (arrowheads). (C) Pulse amplitude modulation fluorescence analysis of wild-type (black trace) and ΔmgdE (red trace) cells. Saturating pulses were applied during the actinic light irradiation (Act.). At the end of each measurement, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; 10 µM) was added to obtain the maximal fluorescence. (D) Comparison of parameters obtained in the pulse amplitude modulation fluorescence analysis for wild-type (blue bars) and ΔmgdE (orange bars) cells. Values presented are the average (wild-type, n = 15; ΔmgdE, n = 10) ± SE.
Distribution of the mgdE Gene in Cyanobacteria.
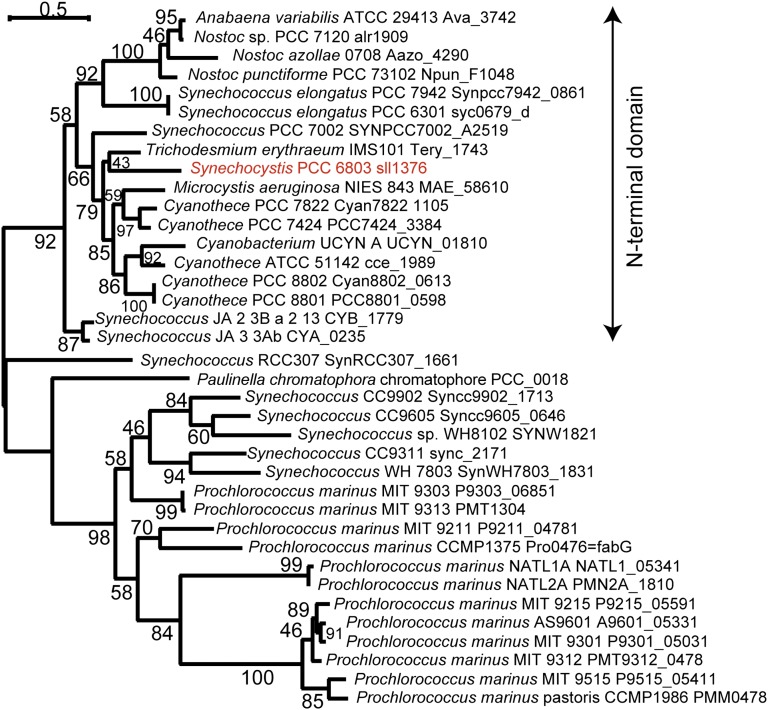
MgdE is a previously unrecognized type of epimerase, having a distinct oxidoreductase domain, that is conserved only within cyanobacteria and the chromatophore of Paulinella (Fig. 4). No homolog of mgdE was identified in any other prokaryotic or eukaryotic phylum by BLAST search. In many species, mgdE and mgdA are not located in the same genomic region (Table S1). Apparent synteny of these genes was found only in Microcystis aeruginosa NIES-843 and Cyanothece sp. PCC 8801, PCC 8802, and PCC 7822; Synechocystis sp. PCC 6803 appears to have an unrelated gene, duf95, in an inverse orientation between mgdE and mgdA.
Fig. 4.
Phylogenetic tree of MgdE proteins. Homologs of Sll1376 proteins were selected from the clustered data in a Gclust database specially prepared for this analysis by adding Paulinella chromatophore sequences. The phylogenetic tree was estimated with the maximum likelihood method using TreeFinder (version March 2008, http://www.treefinder.de/). The number at each branch indicates the confidence level.
Notably, no mgdE was detected in the entire genomes of Gloeobacter violaceus and Thermosynechococcus elongatus, but mgdA was conserved (Table S1). There are several possible explanations for this observation. First, another oxidoreductase could serve as the glucolipid epimerase in these cyanobacteria. In fact, we found that E. coli expressing mgdA and mgdE were sensitive to high temperature and did not accumulate MGDG when grown at 37 °C. On the other hand, MgdA is activated by high temperature (17). Thus, at least for T. elongatus, which grows in hot springs at around 55 °C, MgdE may have been evolutionarily optimized for high temperature, or a different, more thermostable epimerase may be used that would be present in other cyanobacteria only as a minor enzyme. Another possibility is that GlcDG and MGDG may be synthesized independently in G. violaceus and T. elongatus. The presence of a homolog of the plant-type MGDG synthase gene in the genome of G. violaceus (18) supports this possibility. Further study is necessary to understand better the mechanisms of galactolipid synthesis in these organisms.
We next examined the partial mgdE homologs identified in the Prochlorococcus genome (Fig. S2). The Prochlorococcus marinus MED4 mgdA and mgdE homologs were expressed in E. coli as described above, but neither GlcDG nor MGDG accumulated. However, further optimization of the expression system is necessary before we can firmly conclude that other genes or factors are necessary for these partial homologs in P. marinus.
Requirement of Glycolipids in the Photosynthetic Membranes.
The results described above show that galactolipids are not essential for photosynthesis. However, we cannot rule out the possibility that very small amounts of MGDG and/or DGDG still exist in the membranes of ΔmgdE and that these galactolipids are necessary to maintain the function of photosynthesis. This consideration does not apply to DGDG, because the strain of Synechocystis sp. PCC 6803 with disruption of the DGDG synthase gene is devoid of DGDG but grows as rapidly as the wild-type strain in optimal growth conditions (10, 11). The growth was better in the mgdE mutants expressing plant MGDG synthase than in the ΔmgdE strain. The mutants expressing plant MGDG synthase accumulated GlcDG but also had detectable amounts of MGDG (Fig. S1). Thus, it is likely that this small amount of MGDG is a key for the better growth of cyanobacteria. This MGDG might be important for the functions of photosynthetic protein rather than for development of the membrane structure because the amount of such MGDG is stoichiometrically too low to construct the membrane structure. MGDG is known to bind in the interior of photosystem proteins (2), and the replacement of MGDG by GlcDG might affect their functions in ΔmgdE. To test this hypothesis, crystallographic analysis of photosystem proteins, such as photosystem II, from ΔmgdE will be required.
Conclusions
Two major conclusions can be drawn from our results. First, galactolipids do not appear to be absolutely required for oxygenic photosynthesis in Synechocystis. Kobayashi et al. (19) showed that the mgd1 mutant of Arabidopsis does not develop photosynthetically active chloroplasts. In this case, galactolipids appear to be both structural elements of thylakoid membranes and specific functional components of the photosystems. In contrast, our findings suggest that glucolipids can replace galactolipids as membrane components in cyanobacteria without notable effects on membrane structure. Currently we are attempting to express mgdA in the mgd1 mutant to see whether GlcDG also can replace the galactolipids in plants. The difference in the fluorescence properties of wild-type and ΔmgdE cells might be explained by a specific role of photosystem-bound galactolipids that cannot be replaced by glucolipids.
The second major conclusion is that all the genes encoding the galactolipid biosynthesis pathway in cyanobacteria, as proposed based on tracer experiments (8), now have been identified. Epimerization is the only possibility that can account for the single-step conversion of GlcDG to MGDG. Additionally, 13C tracer experiments are being performed to confirm that lipid structure is not substantially remodeled during this conversion. Considering all these results, we conclude that extant cyanobacteria and chloroplasts of plants and algae use different pathways to synthesize galactolipids, although they likely share a single common ancestor.
Methods
Comparative Genomic Analysis.
The genomic data of the chromatophore of Paulinella was added to the CyanoClust4 dataset (20) and reclustered with the Gclust software (14). Multiple alignment and phylogenetic analyses were performed as described (21) using the amino acid sequences of MgdE from 36 species of cyanobacteria.
Coexpression of mgdA and mgdE in E. coli.
The mgdE sequence of Synechocystis sp. PCC 6803 was amplified by PCR and cloned into the pCox1 vector, which is a derivative of pACYC184 (see vector construction, Fig. S3A). The resultant construct and pET24a carrying mgdA (9) were introduced into E. coli strain BL21 (DE3), and expression was induced at 30 °C for 9 h with 1 mM isopropyl β-d-1-thiogalactopyranoside. Lipids of the cells were extracted as described (22) and separated by TLC using acetone:toluene:H2O in a ratio of 90:30:6. Lipid extracts from E. coli expressing mgdA (9) or CsMGD1 (6) were used as standards. (Note that gene names are written in lowercase in prokaryotes and in uppercase in plants.)
Disruption of mgdE in Synechocystis.
Flanking regions of mgdE were amplified and cloned using the In fusion system (Clontech) into pMobEm1, a plasmid vector constructed for this research (see vector construction, Fig. S3C). The resultant plasmid was introduced into wild-type Synechocystis sp. PCC 6803 or the strain overexpressing CsMGD1 under the control of the light-inducible psbA promoter described below. The transformants were selected on BG11 agar plates with erythromycin (Wako Pure Chemical Industries) at a final concentration of 10 μg/mL. Candidate knockout mutants were segregated at least three times and subjected to analysis of genotype. Lipids were extracted as described (22), separated by TLC using chloroform:methanol:28% ammonium (65:35:5) or hexane:tetrahydrofuran:2-propanol:H2O (50:0.5:35:3) (23), and visualized with α-naphthol by spraying and heating at 120 °C for 10 min. GC was carried out using a Shimadzu GC-2014 chromatograph equipped with a flame ionization detector on a capillary column (BPX90, 60 m × 0.25 mm; SGE Analytical Science). The column temperature was increased gradually from 140 °C to 215 °C at a rate of 5 °C/min. Both the injector and detector temperatures were 250 °C. The linear velocity of the carrier gas (He) was 25 cm/min.
Vector Construction.
The bacterial expression vector pCox1 was constructed by the In fusion cloning system with a DNA fragment containing the lacI gene and the lac operator, T7 promoter, a multicloning site from pET24a (primers 1 and 2 in Table S2), and another fragment containing the p15A replication origin and chloramphenicol-resistant gene from pACYC184 (primers 3 and 4). A vector for gene disruption in Synechocystis sp. PCC 6803, pMobEm1, was constructed with DNA fragments including oriVT (primers 5 and 6) from pRL271 (obtained from C. P. Wolk, Michigan State University, East Lansing, MI) and SacB (primers 7 and 8) from pK18mobSacB (obtained from the National BioResource Project of Japan). The erythromycin-resistance gene was first amplified (primers 9 and 10) from pRL271 and subcloned into the HindIII site of pBluescript II SK+; then it was reamplified with multiple cloning sites (primers 11 and 12) and used to construct pMobEm1. pSEM1 is a derivative of pSynExp2 (provided by Edgar Cahoon, University of Nebraska, Lincoln, NE) (24). Oligonucleotides 13 and 14 were annealed, blunt-ended with KOD-Plus-Neo DNA polymerase (Toyobo), and cloned into the NdeI site of pSynExp2.
The N-terminally GST-fused plant-type MGDG synthase gene from cucumber, CsMGD1, was amplified with primers no. 15 and 16 from pGEX-MGD1 (6) and cloned into the NruI site of pSEM1 vector using SmaI restriction sites conferred by the primers. The sll1376 ORF (mgdE) was amplified with primers 17 and 18 and inserted between the NdeI and HindIII sites of pCox1 with the In Fusion system. The 5′ region of mgdE was amplified with primers 19 and 20 and cloned using the In Fusion system into the SmaI site of pMobEm1. The PCR-amplified 3′ region of mgdE (primers 21 and 22) was inserted into the ApaI site of the resultant plasmid to make a knockout vector of mgdE. All PCR fragments, which were amplified with KOD-Plus-Neo DNA polymerase, were confirmed by sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) and the Life Technologies 3130 Genetic Analyzer.
Genotype of Transformants.
Genomic DNA from wild-type strains and transformants were used as templates for PCR with the primers described in Table S2 and Hybripol DNA polymerase (Bioline). To confirm the integration of CsMGD1 into the genome, PCR-based confirmation of gene disruption was performed using primers 23 and 24. Primers 17 and 18 were used to amplify full-length mgdE, primers 18 and 25 were used to detect the deletion of the central part of mgdE, and primers 10 and 18 were used to confirm the insertion of the erythromycin-resistance gene into mgdE.
Chlorophyll Content, Oxygen Evolution Rates and Pulse Amplitude Modulation Fluorescence.
Chlorophyll content was measured as described (25). The oxygen evolution rate of intact cells was measured with a Clark-type oxygen electrode (Hansatech Instruments Ltd.). Cells were suspended in BG-11 medium containing 10 mM NaHCO3 and were illuminated with a halogen lamp with infrared cutoff filters. Pulse amplitude modulation fluorescence analysis was performed as described (26).
Transmission Electron Microscopy.
Cyanobacterial cells were fixed with 2.5% (wt/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 2 h and were postfixed overnight with 1% osmium tetroxide (Wako Pure Chemical Industries) in 0.1 M cacodylate buffer. The cells then were dehydrated with an increasing ethanol series and propylene oxide and finally were embedded in Epon resin (Nisshin EM). Ultrathin sections were stained with uranyl acetate and lead acetate. Images were obtained with an electron microscope model 1200EX (JOEL) operated at 80 kV.
Supplementary Material
Acknowledgments
We thank Dr. Haruki Hashimoto at the Graduate School of Arts and Sciences, University of Tokyo for assistance with electron microscopy. This work was supported in part by Grants-in-Aid for Young Scientists (B) 19770025 and 21770033 and Grant-in-Aid for Scientific Research (C) 24570043 from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by the Precursory Research for Embryonic Science and Technology and Core Research for Evolutional Science and Technology programs of the Japanese Science and Technology Agency.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403708111/-/DCSupplemental.
References
- 1.Dörmann P. Galactolipids in plant membranes. Encyclopedia of Life Sciences. 2013 doi: 10.1002/9780470015902.a0020100.pub2. [DOI] [Google Scholar]
- 2.Jones MR. Lipids in photosynthetic reaction centres: Structural roles and functional holes. Prog Lipid Res. 2007;46(1):56–87. doi: 10.1016/j.plipres.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473(7345):55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA. 2007;104(43):17216–17221. doi: 10.1073/pnas.0704680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gounaris K, Barber J. Monogalactosyldiacylglycerol: The most abundant polar lipid in nature. Trends Biochem Sci. 1983;8(10):378–381. [Google Scholar]
- 6.Shimojima M, et al. Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc Natl Acad Sci USA. 1997;94(1):333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feige G, Heinz E, Wrage K, Cochems N, Ponzelar E. Biogenesis and Function of Plant Lipids. Amsterdam: Elsevier/North Holland Biomedical; 1980. [Google Scholar]
- 8.Sato N, Murata N. Lipid biosynthesis in the blue-green alga, Anabaena variabilis: I. Lipid classes. Biochim Biophys Acta. 1982;710(3):271–278. [Google Scholar]
- 9.Awai K, et al. Comparative genomic analysis revealed a gene for monoglucosyldiacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol. 2006;141(3):1120–1127. doi: 10.1104/pp.106.082859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Awai K, Watanabe H, Benning C, Nishida I. Digalactosyldiacylglycerol is required for better photosynthetic growth of Synechocystis sp. PCC6803 under phosphate limitation. Plant Cell Physiol. 2007;48(11):1517–1523. doi: 10.1093/pcp/pcm134. [DOI] [PubMed] [Google Scholar]
- 11.Sakurai I, Mizusawa N, Wada H, Sato N. Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol. 2007;145(4):1361–1370. doi: 10.1104/pp.107.106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuzawa Y, et al. Cyanobacterial monogalactosyldiacylglycerol-synthesis pathway is involved in normal unsaturation of galactolipids and low-temperature adaptation of Synechocystis sp. PCC 6803. Biochim Biophys Acta. 2014;1841(4):475–483. doi: 10.1016/j.bbalip.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Nowack EC, Melkonian M, Glöckner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18(6):410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- 14.Sato N. Gclust: Trans-kingdom classification of proteins using automatic individual threshold setting. Bioinformatics. 2009;25(5):599–605. doi: 10.1093/bioinformatics/btp047. [DOI] [PubMed] [Google Scholar]
- 15.Kramm A, Kisiela M, Schulz R, Maser E. Short-chain dehydrogenases/reductases in cyanobacteria. FEBS J. 2012;279(6):1030–1043. doi: 10.1111/j.1742-4658.2012.08494.x. [DOI] [PubMed] [Google Scholar]
- 16.Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 17.Shimojima M, Tsuchiya M, Ohta H. Temperature-dependent hyper-activation of monoglucosyldiacylglycerol synthase is post-translationally regulated in Synechocystis sp. PCC 6803. FEBS Lett. 2009;583(14):2372–2376. doi: 10.1016/j.febslet.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 18.Yuzawa Y, et al. Phylogeny of galactolipid synthase homologs together with their enzymatic analyses revealed a possible origin and divergence time for photosynthetic membrane biogenesis. DNA Res. 2012;19(1):91–102. doi: 10.1093/dnares/dsr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi K, et al. Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J. 2013;73(2):250–261. doi: 10.1111/tpj.12028. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki NV, Sato N. 2010. CyanoClust: Comparative genome resources of cyanobacteria and plastids. Database 2010:bap025Available at: http://database.oxfordjournals.org/
- 21.Sato N. Phylogenomic and structural modeling analyses of the PsbP superfamily reveal multiple small segment additions in the evolution of photosystem II-associated PsbP protein in green plants. Mol Phylogenet Evol. 2010;56(1):176–186. doi: 10.1016/j.ympev.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 23.Hölzl G, Zähringer U, Warnecke D, Heinz E. Glycoengineering of cyanobacterial thylakoid membranes for future studies on the role of glycolipids in photosynthesis. Plant Cell Physiol. 2005;46(11):1766–1778. doi: 10.1093/pcp/pci189. [DOI] [PubMed] [Google Scholar]
- 24.Sattler SE, Cahoon EB, Coughlan SJ, DellaPenna D. Characterization of tocopherol cyclases from higher plants and cyanobacteria. Evolutionary implications for tocopherol synthesis and function. Plant Physiol. 2003;132(4):2184–2195. doi: 10.1104/pp.103.024257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnon DI, McSwain BD, Tsujimoto HY, Wada K. Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim Biophys Acta. 1974;357(2):231–245. doi: 10.1016/0005-2728(74)90063-2. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa M, Fujiwara M, Sonoike K, Sato N. Orthogenomics of photosynthetic organisms: Bioinformatic and experimental analysis of chloroplast proteins of endosymbiont origin in Arabidopsis and their counterparts in Synechocystis. Plant Cell Physiol. 2009;50(4):773–788. doi: 10.1093/pcp/pcp027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.