Abstract
Gastric cancer is a leading cause of cancer deaths, but analysis of its molecular and clinical characteristics has been complicated by histological and aetiological heterogeneity. Here we describe a comprehensive molecular evaluation of 295 primary gastric adenocarcinomas as part of The Cancer Genome Atlas (TCGA) project. We propose a molecular classification dividing gastric cancer into four subtypes: tumours positive for Epstein–Barr virus, which display recurrent PIK3CA mutations, extreme DNA hypermethylation, and amplification of JAK2, CD274 (also known as PD-L1) and PDCD1LG2 (also known as PD-L2); microsatellite unstable tumours, which show elevated mutation rates, including mutations of genes encoding targetable oncogenic signalling proteins; genomically stable tumours, which are enriched for the diffuse histological variant and mutations of RHOA or fusions involving RHO-family GTPase-activating proteins; and tumours with chromosomal instability, which show marked aneuploidy and focal amplification of receptor tyrosine kinases. Identification of these subtypes provides a roadmap for patient stratification and trials of targeted therapies.
Supplementary information
The online version of this article (doi:10.1038/nature13480) contains supplementary material, which is available to authorized users.
Subject terms: Gastric cancer, Cancer genomics
The Cancer Genome Atlas reports on molecular evaluation of 295 primary gastric adenocarcinomas and proposes a new classification of gastric cancers into 4 subtypes, which should help with clinical assessment and trials of targeted therapies.
Supplementary information
The online version of this article (doi:10.1038/nature13480) contains supplementary material, which is available to authorized users.
Four classes of gastric adenocarcinoma
This contribution from The Cancer Genome Atlas (TCGA) project describes the molecular evaluation of 295 primary gastric adenocarcinomas. Based on the results, the authors propose a novel classification separating gastric cancers into four subtypes according to: Epstein–Barr virus positive status, microsatellite instability, chromosomal instability or genomic stability. Given the histologic and etiologic heterogeneity of gastric cancer identification of these subtypes, using a schema that can readily be applied to patient samples should help with patient stratification and trials of targeted therapies.
Supplementary information
The online version of this article (doi:10.1038/nature13480) contains supplementary material, which is available to authorized users.
Main
Gastric cancer was the world’s third leading cause of cancer mortality in 2012, responsible for 723,000 deaths1. The vast majority of gastric cancers are adenocarcinomas, which can be further subdivided into intestinal and diffuse types according to the Lauren classification2. An alternative system, proposed by the World Health Organization, divides gastric cancer into papillary, tubular, mucinous (colloid) and poorly cohesive carcinomas3. These classification systems have little clinical utility, making the development of robust classifiers that can guide patient therapy an urgent priority.
The majority of gastric cancers are associated with infectious agents, including the bacterium Helicobacter pylori4 and Epstein–Barr virus (EBV). The distribution of histological subtypes of gastric cancer and the frequencies of H. pylori and EBV associated gastric cancer vary across the globe5. A small minority of gastric cancer cases are associated with germline mutation in E-cadherin (CDH1)6 or mismatch repair genes7 (Lynch syndrome), whereas sporadic mismatch repair-deficient gastric cancers have epigenetic silencing of MLH1 in the context of a CpG island methylator phenotype (CIMP)8. Molecular profiling of gastric cancer has been performed using gene expression or DNA sequencing9,10,11,12, but has not led to a clear biologic classification scheme. The goals of this study by The Cancer Genome Atlas (TCGA) were to develop a robust molecular classification of gastric cancer and to identify dysregulated pathways and candidate drivers of distinct classes of gastric cancer.
Sample set and molecular classification
We obtained gastric adenocarcinoma primary tumour tissue (fresh frozen) from 295 patients not treated with prior chemotherapy or radiotherapy (Supplementary Methods S1). All patients provided informed consent, and local Institutional Review Boards approved tissue collection. We used germline DNA from blood or non-malignant gastric mucosa as a reference for detecting somatic alterations. Non-malignant gastric samples were also collected for DNA methylation (n = 27) and expression (n = 29) analyses. We characterized samples using six molecular platforms (Supplementary Methods S2–S7): array-based somatic copy number analysis, whole-exome sequencing, array-based DNA methylation profiling, messenger RNA sequencing, microRNA (miRNA) sequencing and reverse-phase protein array (RPPA), with 77% of the tumours tested by all six platforms. Microsatellite instability (MSI) testing was performed on all tumour DNA, and low-pass (∼6× coverage) whole genome sequencing on 107 tumour/germline pairs.
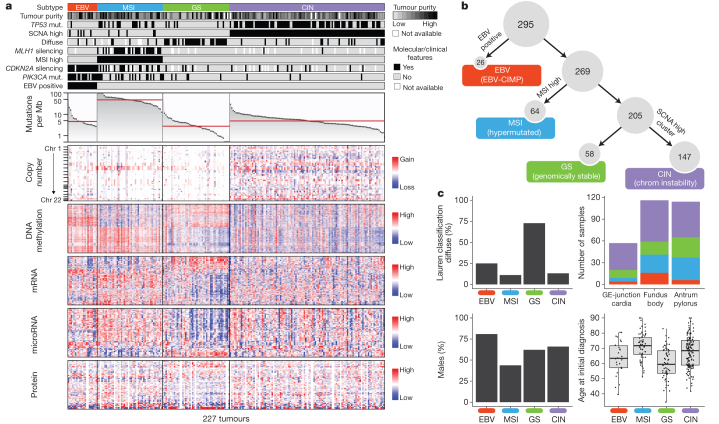
To define molecular subgroups of gastric cancer we first performed unsupervised clustering on data from each molecular platform (Supplementary Methods S2–S7) and integrated these results, yielding four groups (Supplementary Methods S10.2). The first group of tumours was significantly enriched for high EBV burden (P = 1.5 × 10−18) and showed extensive DNA promoter hypermethylation. A second group was enriched for MSI (P = 2.1 × 10−32) and showed elevated mutation rates and hypermethylation (including hypermethylation at the MLH1 promoter). The remaining two groups were distinguished by the presence or absence of extensive somatic copy-number aberrations (SCNAs). As an alternative means to define distinct gastric cancer subgroups, we performed integrative clustering of multiple data types using iCluster13 (Supplementary Methods S10.3). This analysis again indicated that EBV, MSI and the level of SCNAs characterize distinct subgroups (Supplementary Fig. 10.3). Based upon these results from analysis of all molecular platforms, we created a decision tree to categorize the 295 gastric cancer samples into four subtypes (Fig. 1a, b) using an approach that could more readily be applied to gastric cancer tumours in clinical care. Tumours were first categorized by EBV-positivity (9%), then by MSI-high status, hereafter called MSI (22%), and the remaining tumours were distinguished by degree of aneuploidy into those termed genomically stable (20%) or those exhibiting chromosomal instability (CIN; 50%).
Figure 1. Molecular subtypes of gastric cancer.
a, Gastric cancer cases are divided into subtypes: Epstein–Barr virus (EBV)-positive (red), microsatellite instability (MSI, blue), genomically stable (GS, green) and chromosomal instability (CIN, light purple) and ordered by mutation rate. Clinical (top) and molecular data (top and bottom) from 227 tumours profiled with all six platforms are depicted. b, A flowchart outlines how tumours were classified into molecular subtypes. c, Differences in clinical and histological characteristics among subtypes with subtypes coloured as in a, b. The plot of patient age at initial diagnosis shows the median, 25th and 75th percentile values (horizontal bar, bottom and top bounds of the box), and the highest and lowest values within 1.5 times the interquartile range (top and bottom whiskers, respectively). GE, gastroesophageal.
Evaluation of the clinical and histological characteristics of these molecular subtypes revealed enrichment of the diffuse histological subtype in the genomically stable group (40/55 = 73%, P = 7.5 × 10−17) (Fig. 1c), an association not attributable to reduced SCNA detection in low purity tumours (Supplementary Fig. 2.8). Each subtype was found throughout the stomach, but CIN tumours showed elevated frequency in the gastroesophageal junction/cardia (65%, P = 0.012), whereas most EBV-positive tumours were present in the gastric fundus or body (62%, P = 0.03). Genomically stable tumours were diagnosed at an earlier age (median age 59 years, P = 4 × 10−7), whereas MSI tumours were diagnosed at relatively older ages (median 72 years, P = 5 × 10−5). MSI patients tended to be female (56%, P = 0.001), but most EBV-positive cases were male (81%, P = 0.037), as previously reported14. We did not observe any systematic differences in distribution of subtypes between patients of East Asian and Western origin (Supplementary Methods S1.8). Initial outcome data from this cohort did not reveal survival differences between the four subgroups (Supplementary Information S1.7)
EBV-associated DNA hypermethylation
EBV is found within malignant epithelial cells in 9% of gastric cancers14. EBV status was determined using mRNA, miRNA, exome and whole-genome sequencing, yielding highly concordant results (Supplementary Fig. 9.7). By contrast, we detected only sporadic evidence of H. pylori, which may reflect the decline of bacterial counts accompanying the progression from chronic gastritis to subsequent carcinoma, as well as technical loss of luminal bacteria during specimen processing. Unsupervised clustering of CpG methylation performed on unpaired tumour samples revealed that all EBV-positive tumours clustered together and exhibited extreme CIMP, distinct from that in the MSI subtype8, consistent with prior reports15 (Fig. 2a). Differences between the EBV-CIMP and MSI-associated gastric-CIMP methylation profiles of tumours mirrored differences between these groups in their spectra of mutations (Fig. 1a) and gene expression (Supplementary Fig. 10.6a). EBV-positive tumours had a higher prevalence of DNA hypermethylation than any cancers reported by TCGA (Supplementary Fig. 4.6). All EBV-positive tumours assayed displayed CDKN2A (p16INK4A) promoter hypermethylation, but lacked the MLH1 hypermethylation characteristic of MSI-associated CIMP16. Genes with promoter hypermethylation most differentially silenced in EBV-positive gastric cancer are shown in Supplementary Table 4.3.
Figure 2. Molecular characteristics of EBV-positive gastric cancers.
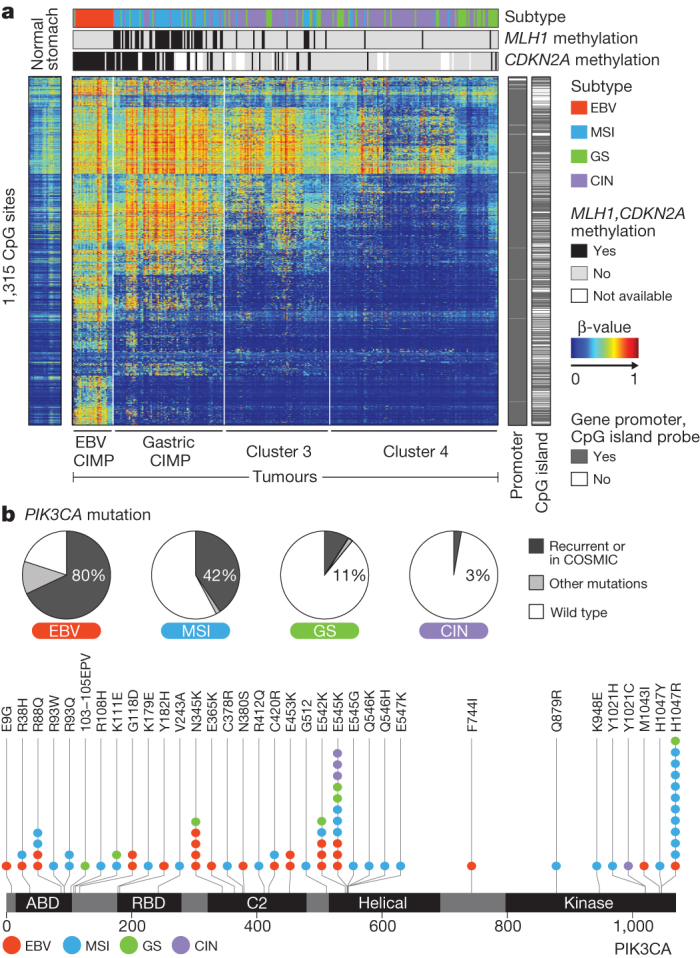
a, The heatmap represents unsupervised clustering of DNA methylation at CpG sites for 295 tumours into four clusters: EBV-CIMP (n = 28), Gastric-CIMP (n = 77), cluster 3 (n = 73) and cluster 4 (n = 117). Profiles for non-malignant gastric mucosa are to the left of the tumours. b, The proportion of tumours harbouring PIK3CA mutation in the molecular subtypes with mutations at sites noted recurrently in this data set or in the COSMIC database marked separately (top). Locations of PIK3CA mutations with the subtype of the sample with each mutation colour-coded (bottom).
We observed strong predilection for PIK3CA mutation in EBV-positive gastric cancer as suggested by prior reports17,18, with non-silent PIK3CA mutations found in 80% of this subgroup (P = 9 × 10−12), including 68% of cases with mutations at sites recurrent in this data set or in the COSMIC repository. In contrast, 3 to 42% of tumours in the other subtypes displayed PIK3CA mutations. PI(3)-kinase inhibition therefore warrants evaluation in EBV-positive gastric cancer. PIK3CA mutations were more dispersed in EBV-positive cancers, but localized in the kinase domain (exon 20) in EBV-negative cancers (Fig. 2b). The most highly transcribed EBV viral mRNAs and miRNAs fell within the BamH1A region of the viral genome (Supplementary Fig. 9.8) and showed similar expression patterns across tumours, as reported separately19.
Somatic genomic alterations
To identify recurrently mutated genes, we analysed the 215 tumours with mutation rates below 11.4 mutations per megabase (Mb) (none of which were MSI-positive) separately from the 74 ‘hypermutated’ tumours. Within the hypermutated tumours, we excluded from analysis 11 cases with a distinctly higher mutational burden above 67.7 mutations per Mb (including one tumour with an inactivating POLE mutation20,21) (Supplementary Information S3.2–3.3), because their large numbers of mutations unduly influence analysis. We used the MutSigCV22 tool to define recurrent mutations in the 63 remaining hypermutated tumours by first evaluating only base substitution mutations, identifying 10 significantly mutated genes, including TP53, KRAS, ARID1A, PIK3CA, ERBB3, PTEN and HLA-B (Supplementary Table 3.5). We found ERBB3 mutations in 16 of 63 tumours, with 13 of these tumours having mutations at recurrent sites or sites reported in COSMIC. MutSigCV analysis including insertions/deletions expanded the list of statistically significant mutated genes to 37, including RNF43, B2M and NF1 (Supplementary Fig. 3.9). Similarly, HotNet analysis of genes mutated within MSI tumours revealed common alterations in major histocompatibility complex class I genes, including B2M and HLA-B (Supplementary Fig. 11.5–11.7). B2M mutations in colorectal cancers and melanoma result in loss of expression of HLA class 1 complexes23, suggesting these events benefit hypermutated tumours by reducing antigen presentation to the immune system.
Through MutSigCV analysis of the 215 non-hypermutated tumours, we identified 25 significantly mutated genes (Fig. 3). This gene list again included TP53, ARID1A, KRAS, PIK3CA and RNF43, but also genes in the β-catenin pathway (APC and CTNNB1), the TGF-β pathway (SMAD4 and SMAD2), and RASA1, a negative regulator of RAS. ERBB2, a therapeutic target, was significantly mutated, with 10 of 15 mutations occurring at known hotspots; four cases had the S310F ERBB2 mutation that is activating and drug-sensitive24.
Figure 3. Significantly mutated genes in non-hypermutated gastric cancer.
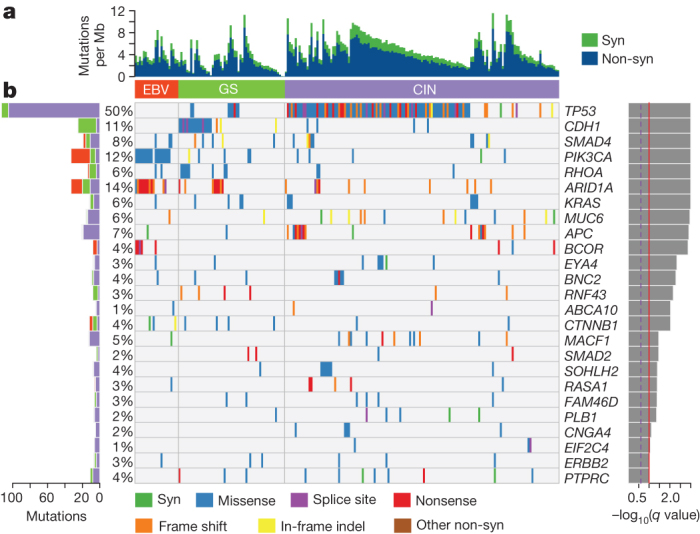
a, Bars represent somatic mutation rate for the 215 samples with synonymous and non-synonymous mutation rates distinguished by colour. b, Significantly mutated genes, identified by MutSigCV, are ranked by the q value (right) with samples grouped by subtype. Mutation colour indicates the class of mutation.
In addition to PIK3CA mutations, EBV-positive tumours had frequent ARID1A (55%) and BCOR (23%) mutations and only rare TP53 mutations. BCOR, encoding an anti-apoptotic protein, is also mutated in leukaemia25 and medulloblastoma26. Among the CIN tumours, we observed TP53 mutations in 71% of tumours. CDH1 somatic mutations were enriched in the genomically stable subtype (37% of cases). CDH1 germline mutations underlie hereditary diffuse gastric cancer (HDGC). However, germline analysis revealed only two CDH1 polymorphisms, neither of which is known to be pathogenic. As in the EBV-subtype, inactivating ARID1A mutations were prevalent in the genomically stable subtype. We identified mutations of RHOA almost exclusively in genomically stable tumours, as discussed below.
We analysed the patterns of base changes within gastric cancer tumours and noted elevated rates of C to T transitions at CpG dinucleotides. We observed an elevated rate of A to C transversions at the 3′ adenine of AA dinucleotides, especially at AAG trinucleotides, as reported in oesophageal adenocarcinoma27. The A to C transversions were prominent in CIN, EBV and genomically stable, but as previously observed27, not in MSI tumours (Supplementary Fig. 3.10).
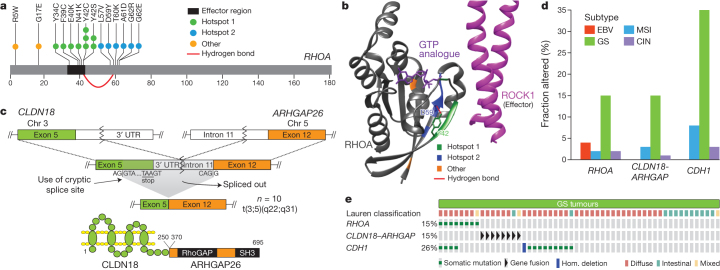
We identified RHOA mutation in 16 cases, and these were enriched in the genomically stable subtype (15% of genomically stable cases, P = 0.0039). RHOA, when in the active GTP-bound form, acts through a variety of effectors, including ROCK1, mDIA and Protein Kinase N, to control actin-myosin-dependent cell contractility and cellular motility28,29 and to activate STAT3 to promote tumorigenesis30,31. RHOA mutations were clustered in two adjacent amino-terminal regions that are predicted to be at the interface of RHOA with ROCK1 and other effectors (Fig. 4a, b). RHOA mutations were not at sites analogous to oncogenic mutations in RAS-family GTPases. Although one case harboured a codon 17 mutation, we did not identify the dominant-negative G17V mutations noted in T-cell neoplasms32,33. Rather, the mutations found in this study may act to modulate signalling downstream of RHOA. Biochemical studies found that the RHOA Y42C mutation attenuated activation of Protein Kinase N, without abrogated activation of mDia or ROCK134. RHOA Y42, mutated in five tumours, corresponds to Y40 on HRAS, a residue which when mutated selectively reduces HRAS activation of RAF, but not other RAS effectors35. Given the role of RHOA in cell motility, modulation of RHOA may contribute to the disparate growth patterns and lack of cellular cohesion that are hallmarks of diffuse tumours.
Figure 4. RHOA and ARHGAP6/26 somatic genomic alterations are recurrent in genomically stable gastric cancer.
a, Missense mutations in the GTPase RHOA, including residues Y42 and D59, linked via hydrogen bond (red arc). b, Mutated regions (coloured as in panel a) mapped on the structures of RHOA and ROCK1. c, A schematic of CLDN18–ARHGAP26 translocation is shown for the fusion transcript and predicted fusion protein. SH3 denotes SRC homology 3 domain. d, The frequency of RHOA and CDH1 mutations, CLDN18–ARHGAP6 or ARHGAP26 fusions are shown across gastric cancer subtypes. e, RHOA mutations and CLDN18–ARHGAP6 or ARHGAP26 fusions are mutually exclusive in genomically stable tumours.
Dysregulated RHO signalling was further implicated by the discovery of recurrent structural genomic alterations. Whole genome sequencing of 107 tumours revealed 5,696 structural rearrangements, including 74 predicted to produce in-frame gene fusions (Supplementary Information S3.7–3.8). De novo assembly of mRNA sequencing data confirmed 170 structural rearrangements (Supplementary Information S5.4a), including two cases with an interchromosomal translocation between CLDN18 and ARHGAP26 (GRAF). ARHGAP26 is a GTPase-activating protein (GAP) that facilitates conversion of RHO GTPases to the GDP state and has been implicated in enhancing cellular motility34. CLDN18 is a component of the tight junction adhesion structures36. RNA sequencing data from tumours without whole genome sequencing identified CLDN18–ARHGAP26 fusions in 9 additional tumours, with two more cases showing CLDN18 fusion to the homologous GAP encoded by ARHGAP6 totalling 13 cases with these rearrangements (Supplementary Table 5.6).
The fusions linked exon 5 of CLDN18 to exon 2 (n = 2) of ARHGAP6, to exon 10 (n = 1), or to exon 12 (n = 10) of ARHGAP26 (Fig. 4c). As these fusions occur downstream of the CLDN18 exon 5 stop codon, they appeared unlikely to enable translation of fusion proteins. However, mRNA sequencing revealed a mature fusion transcript in which the ARHGAP26 or ARHGAP6 splice acceptor activates a cryptic splice site within exon 5 of CLDN18, before the stop codon, yielding an in-frame fusion predicted to maintain the transmembrane domains of CLDN18 while fusing a large segment of ARHGAP26 or ARHGAP6 to the cytoplasmic carboxy terminus of CLDN18. These chimaeric proteins retain the carboxy-terminal GAP domain of ARHGAP26/6, potentially affecting ARHGAP’s regulation of RHOA and/or cell motility. Furthermore, these fusions may also disrupt wild-type CLDN18, impacting cellular adhesion. The CLDN18–ARHGAP fusions were mutually exclusive with RHOA mutations and were enriched in genomically stable tumours (62%, P = 10−3) (Fig. 4d). Within the genomically stable subtype, 30% of cases had either RHOA or CLDN18–ARHGAP alterations. Evaluation of gene expression status in pathways putatively regulated by RHOA using the Paradigm-Shift algorithm predicted activation of RHOA-driven pathways (Supplementary Fig. 11.4a–c), suggesting that these genomic aberrations contribute to the invasive phenotype of diffuse gastric cancer.
SCNA analysis using GISTIC identified 30 focal amplifications, 45 focal deletions, and chromosome arms subject to frequent alteration (Supplementary Figs 2.3–2.9). Focal amplifications targeted oncogenes such as ERBB2, CCNE1, KRAS, MYC, EGFR, CDK6, GATA4, GATA6 and ZNF217. Additionally, we saw amplification of the gene that encodes the gastric stem cell marker CD44 and a novel recurrent amplification at 9p24.1 at the locus containing JAK2, CD274 and PDCD1LG2. JAK2 encodes a receptor tyrosine kinase and potential therapeutic target. CD274 and PDCD1LG2 encode PD-L1 and PD-L2, immunosuppressant proteins currently being evaluated as targets to augment anti-tumour immune response. Notably, these 9p amplifications were enriched in the EBV subgroup (15% of tumours), consistent with studies showing elevated PD-L1 expression in EBV-positive lymphoid cancers37,38. Evaluation of mRNA revealed elevated expression of JAK2, PD-L1 and PD-L2 in amplified cases (Supplementary Fig. 2.10). More broadly, PD-L1/2 expression was elevated in EBV-positive tumours, suggesting that PD-L1/2 antagonists and JAK2 inhibitors be tested in this subgroup. Focal deletions were identified at the loci of tumour suppressors such as PTEN, SMAD4, CDKN2A and ARID1A. Additional GISTIC analysis on the four molecular subtypes is detailed in Supplementary Figs 2.5–2.6.
Gene expression and proteomic analysis
Our analysis of each of the expression platforms revealed four mRNA, five miRNA and three RPPA clusters (Supplementary Methods S5–S7). Some expression clusters are similar across platforms (Supplementary Methods S10) and/or have correspondence with specific molecular subtypes. For example, mRNA cluster 1, miRNA cluster 4 and RPPA cluster 1 have substantial overlap and are strongly associated with genomically stable tumours, both individually and as a group; the 34 cases with all three assignments were predominantly genomically stable (20/34, P = 2 × 10−8). Similarly, mRNA cluster 3, miRNA cluster 2 and RPPA cluster 3 are similar and are associated with the MSI subtype as a group (12/22, P = 5 × 10−4). However, absolute correspondence between expression clusters and molecular subtypes was not always seen. For example, RPPA cluster 3 showed moderate association with both MSI and EBV (P = 0.018 and P = 0.038, respectively), and miRNA clusters each had similar proportions of CIN (no associations with P < 0.05). Overall, the expression data recapitulate features of the molecular classification, pointing to robustness of this taxonomy.
We analysed mRNA sequence data for alternative splicing events, finding MET exon 2 skipping in 82 of 272 (30%) cases, associated with increased MET expression (P = 10−4). We also found novel variants of MET in which exons 18 and/or 19 were skipped (47/272; 17%; Supplementary Fig. 5.5). Intriguingly, the exons removed by these alterations encode regions of the kinase domain.
Through supervised analysis of RPPA data, we observed 45 proteins whose expression or phosphorylation was associated with the four molecular subtypes (Supplementary Fig. 7.2). Phosphorylation of EGFR (pY1068) was significantly elevated in the CIN subtype, consistent with amplification of EGFR within that subtype. We also found elevated expression of p53, consistent with frequent TP53 mutation and aneuploidy in the CIN subtype.
Integrated pathway analysis
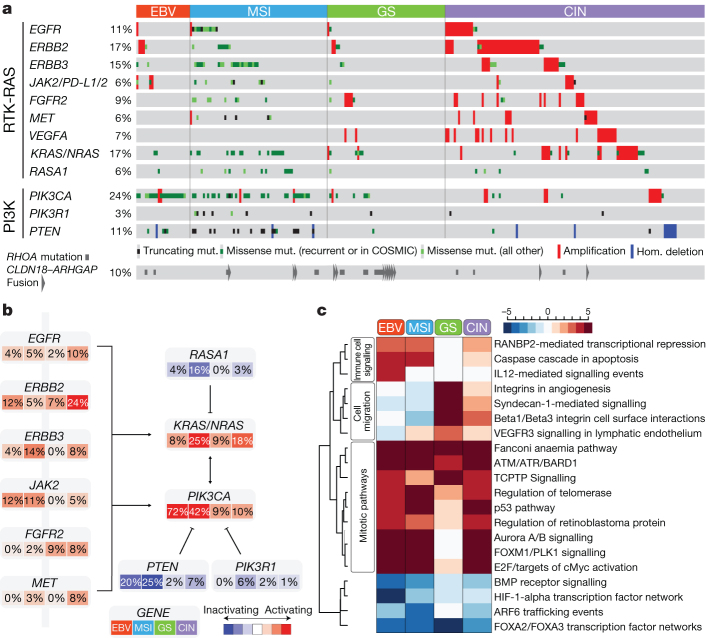
We integrated SCNA and mutation data to characterize genomic alterations in known signalling pathways, including candidate therapeutic targets (Fig. 5a, b). We focused on alterations in receptor tyrosine kinases (RTKs) and RAS and PI(3)-kinase signalling. EBV-positive tumours contained PIK3CA mutations and recurrent JAK2 and ERBB2 amplifications. Although MSI cases generally lacked targetable amplifications, mutations in PIK3CA, ERBB3, ERBB2 and EGFR were noted, with many mutations at ‘hotspot’ sites seen in other cancers (Supplementary Fig. 11.14). Absent from MSI gastric cancers were BRAF V600E mutations, commonly seen in MSI colorectal cancer39. Although the genomically stable subtype exhibited recurrent RHOA and CLDN18 events, few other clear treatment targets were observed. In CIN tumours, we identified genomic amplifications of RTKs, many of which are amenable to blockade by therapeutics in current use or in development. Recurrent amplification of the gene encoding ligand VEGFA was notable given the gastric cancer activity of the VEGFR2 targeting antibody ramucirumab40. Additionally, frequent amplifications of cell cycle mediators (CCNE1, CCND1 and CDK6) suggest the potential for therapeutic inhibition of cyclin-dependent kinases (Supplementary Fig. 11.15).
Figure 5. Integrated molecular description of gastric cancer.
a, Mutations, copy-number changes and translocations for select genes are shown across samples organized by molecular subtypes. Mutations that are recurrent in this data set or in the COSMIC repository are distinguished by colour. Alteration frequencies are expressed as a percentage of all cases. b, Alterations in RTK/RAS and RTK/PI(3)K signalling pathways across molecular subtypes. Red denotes predicted activation; blue denotes predicted inactivation. c, The heatmap shows NCI-PID pathways that are significantly elevated (red) or decreased (blue) in each of the four subtypes as compared with non-malignant gastric mucosa.
We compared expression within each subtype to that of the other subtypes, and to non-malignant gastric tissue (n = 29) (Supplementary Fig. 11.2). We computed an aggregate score for each pathway of the NCI pathway interaction database41 and determined statistical significance by comparison with randomly generated pathways (Supplementary Methods S11). Hierarchical clustering of samples and pathways (Fig. 5c) revealed several notable patterns, including elevated expression of mitotic network components such as AURKA/B and E2F, targets of MYC activation, FOXM1 and PLK1 signalling and DNA damage response pathways across all subtypes, but to a lesser degree in genomically stable tumours. In contrast, the genomically stable subtype exhibited elevated expression of cell adhesion pathways, including the B1/B3 integrins, syndecan-1 mediated signalling, and angiogenesis-related pathways. These results suggest additional candidate therapeutic targets, including the aurora kinases (AURKA/B) and Polo-like (PLK) family members. The strength of IL-12 mediated signalling signatures in EBV-positive tumours suggests a robust immune cell presence. When coupled with evidence of PD-L1/2 overexpression, this finding adds rationale for testing immune checkpoint inhibitors in EBV-positive gastric cancer.
Discussion
Through this study of the molecular and genomic basis of gastric cancer, we describe a molecular classification (Fig. 6) that defines four major genomic subtypes of gastric cancer: EBV-infected tumours; MSI tumours; genomically stable tumours; and chromosomally unstable tumours. This classification may serve as a valuable adjunct to histopathology. Importantly, these molecular subtypes showed distinct salient genomic features, providing a guide to targeted agents that should be evaluated in clinical trials for distinct populations of gastric cancer patients. Through existing testing for MSI and EBV and the use of emerging genomic assays that query focused gene sets for mutations and amplifications, the classification system developed through this study can be applied to new gastric cancer cases. We hope these results will facilitate the development of clinical trials to explore therapies in defined sets of patients, ultimately improving survival from this deadly disease.
Figure 6. Key features of gastric cancer subtypes.
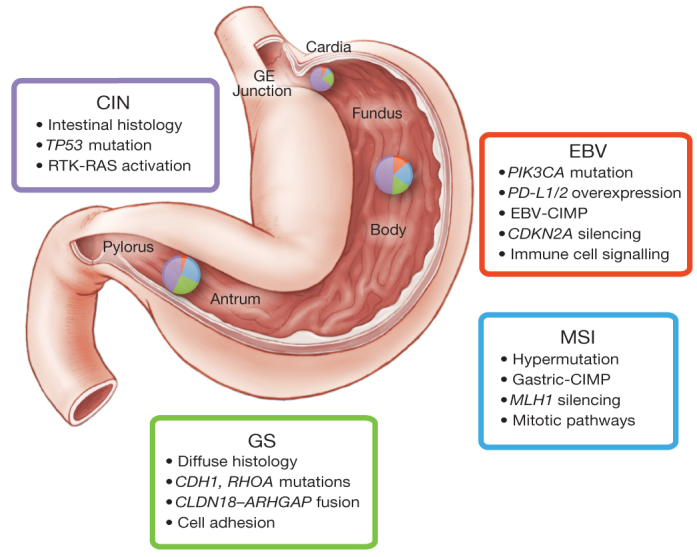
This schematic lists some of the salient features associated with each of the four molecular subtypes of gastric cancer. Distribution of molecular subtypes in tumours obtained from distinct regions of the stomach is represented by inset charts.
Methods Summary
Fresh frozen gastric adenocarcinoma and matched germline DNA samples were obtained from 295 patients under IRB approved protocols. Genomic material and (when available) protein were subjected to single nucleotide polymorphism array somatic copy-number analysis, whole-exome sequencing, mRNA sequencing, miRNA sequencing, array-based DNA methylation profiling and reverse-phase protein arrays. A subset of samples was subjected to whole-genome sequencing. Initial analysis centred on the development of a classification scheme for gastric cancer. Subsequent analysis identified key features from each of the genomic/molecular platforms, looking both for features found across gastric cancer and those characteristic of individual gastric cancer subtypes. Primary and processed data are deposited at the Data Coordinating Center (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp); primary sequence files are deposited in CGHub (https://cghub.ucsc.edu/). Sample lists, and supporting data can be found at (https://tcga-data.nci.nih.gov/docs/publications/stad_2014/).
Supplementary information
This file contains Supplementary Information – see contents pages 1-4 for full details. (PDF 9697 kb)
Acknowledgements
We are grateful to all the patients and families who contributed to this study and to C. Gunter and J. Weinstein for scientific editing, to M. Sheth for administrative support and to L. Omberg for support with the Sage Bionetworks Synapse platform. This work was supported by the Intramural Research Program and the following grants from the United States National Institutes of Health: 5U24CA143799, 5U24CA143835, 5U24CA143840, 5U24CA143843, 5U24CA143845, 5U24CA143848, 5U24CA143858, 5U24CA143866, 5U24CA143867, 5U24CA143882, 5U24CA143883, 5U24CA144025, U54HG003067, U54HG003079, U54HG003273 and P30CA16672.
PowerPoint slides
Author Contributions
The Cancer Genome Atlas Research Network contributed collectively to this study. Biospecimens were provided by the tissue source sites and processed by the Biospecimen Core Resource. Data generation and analyses were performed by the genome-sequencing centres, cancer genome-characterization centres and genome data analysis centres. All data were released through the Data Coordinating Center. The NCI and NHGRI project teams coordinated project activities. The following TCGA investigators of the Stomach Analysis Working Group contributed substantially to the analysis and writing of this manuscript. Project leaders, A. J. Bass, P. W. Laird, I. Shmulevich; data coordinator, V. Thorsson; analysis coordinators, V. Thorsson, N. Schultz; manuscript coordinator, M. Sheth; graphics coordinator, T. Hinoue; DNA sequence analysis, A. Taylor-Weiner, A. Pantazi, C. Sougnez, K. Kasaian; mRNA analysis, R. Bowlby, A. J. Mungall; miRNA analysis, A. Chu, A. Gordon Robertson, D. Yang; DNA methylation analysis, T. Hinoue, H. Shen, P. W. Laird; copy number analysis, A. Cherniack; protein analysis, J.-S. Lee, R. Akbani; pathway/integrated analysis, N. Weinhold, S. Reynolds, C. Curtis, R. Shen, S. Ng, B. Raphael, H.-T. Wu, Y. Liu, V. Thorsson, N. Schultz; pathology expertise and clinical data, A. Boussioutas, B. G. Schneider, J. Kim, J. E. Willis, M. L. Gulley, K. Garman, M. Blanca Piazuelo, V. Thorsson, K. M. Leraas, T. Lichtenberg, J. A. Demchok, A. J. Bass; microbiome analysis, C. S. Rabkin, M. L. Gulley, R. Bowlby, A. J. Mungall, A. Chu and C. Pedamallu.
Competing interests
The author declare no competing financial interests.
Footnotes
The primary and processed data used to generate the analyses presented here can be downloaded from The Cancer Genome Atlas at (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). All of the primary sequence files are deposited in CGHub and all other data are deposited at the Data Coordinating Center (DCC) for public access (http://cancergenome.nih.gov/) and (https://cghub.ucsc.edu/). Additional sample data and supporting data are available from (https://tcga-data.nci.nih.gov/docs/publications/stad_2014/).
Change history
9/10/2014
A minor correction was made to the author list
Contributor Information
The Cancer Genome Atlas Research Network:
Adam J. Bass, Vesteinn Thorsson, Ilya Shmulevich, Sheila M. Reynolds, Michael Miller, Brady Bernard, Toshinori Hinoue, Peter W. Laird, Christina Curtis, Hui Shen, Daniel J. Weisenberger, Nikolaus Schultz, Ronglai Shen, Nils Weinhold, David P. Kelsen, Reanne Bowlby, Andy Chu, Katayoon Kasaian, Andrew J. Mungall, A. Gordon Robertson, Payal Sipahimalani, Andrew Cherniack, Gad Getz, Yingchun Liu, Michael S. Noble, Chandra Pedamallu, Carrie Sougnez, Amaro Taylor-Weiner, Rehan Akbani, Ju-Seog Lee, Wenbin Liu, Gordon B. Mills, Da Yang, Wei Zhang, Angeliki Pantazi, Michael Parfenov, Margaret Gulley, M. Blanca Piazuelo, Barbara G. Schneider, Jihun Kim, Alex Boussioutas, Margi Sheth, John A. Demchok, Charles S. Rabkin, Joseph E. Willis, Sam Ng, Katherine Garman, David G. Beer, Arjun Pennathur, Benjamin J. Raphael, Hsin-Ta Wu, Robert Odze, Hark K. Kim, Jay Bowen, Kristen M. Leraas, Tara M. Lichtenberg, Stephanie Weaver, Michael McLellan, Maciej Wiznerowicz, Ryo Sakai, Gad Getz, Carrie Sougnez, Michael S. Lawrence, Kristian Cibulskis, Lee Lichtenstein, Sheila Fisher, Stacey B. Gabriel, Eric S. Lander, Li Ding, Beifang Niu, Adrian Ally, Miruna Balasundaram, Inanc Birol, Reanne Bowlby, Denise Brooks, Yaron S. N. Butterfield, Rebecca Carlsen, Andy Chu, Justin Chu, Eric Chuah, Hye-Jung E. Chun, Amanda Clarke, Noreen Dhalla, Ranabir Guin, Robert A. Holt, Steven J. M. Jones, Katayoon Kasaian, Darlene Lee, Haiyan A. Li, Emilia Lim, Yussanne Ma, Marco A. Marra, Michael Mayo, Richard A. Moore, Andrew J. Mungall, Karen L. Mungall, Ka Ming Nip, A. Gordon Robertson, Jacqueline E. Schein, Payal Sipahimalani, Angela Tam, Nina Thiessen, Rameen Beroukhim, Scott L. Carter, Andrew D. Cherniack, Juok Cho, Kristian Cibulskis, Daniel DiCara, Scott Frazer, Sheila Fisher, Stacey B. Gabriel, Nils Gehlenborg, David I. Heiman, Joonil Jung, Jaegil Kim, Eric S. Lander, Michael S. Lawrence, Lee Lichtenstein, Pei Lin, Matthew Meyerson, Akinyemi I. Ojesina, Chandra Sekhar Pedamallu, Gordon Saksena, Steven E. Schumacher, Carrie Sougnez, Petar Stojanov, Barbara Tabak, Amaro Taylor-Weiner, Doug Voet, Mara Rosenberg, Travis I. Zack, Hailei Zhang, Lihua Zou, Alexei Protopopov, Netty Santoso, Michael Parfenov, Semin Lee, Jianhua Zhang, Harshad S. Mahadeshwar, Jiabin Tang, Xiaojia Ren, Sahil Seth, Lixing Yang, Andrew W. Xu, Xingzhi Song, Angeliki Pantazi, Ruibin Xi, Christopher A. Bristow, Angela Hadjipanayis, Jonathan Seidman, Lynda Chin, Peter J. Park, Raju Kucherlapati, Rehan Akbani, Shiyun Ling, Wenbin Liu, Arvind Rao, John N. Weinstein, Sang-Bae Kim, Ju-Seog Lee, Yiling Lu, Gordon Mills, Peter W. Laird, Toshinori Hinoue, Daniel J. Weisenberger, Moiz S. Bootwalla, Phillip H. Lai, Hui Shen, Timothy Triche Jr, David J. Van Den Berg, Stephen B. Baylin, James G. Herman, Gad Getz, Lynda Chin, Yingchun Liu, Bradley A. Murray, Michael S. Noble, r B. Arman Askoy, Giovanni Ciriello, Gideon Dresdner, Jianjiong Gao, Benjamin Gross, Anders Jacobsen, William Lee, Ricardo Ramirez, Chris Sander, Nikolaus Schultz, Yasin Senbabaoglu, Rileen Sinha, S. Onur Sumer, Yichao Sun, Nils Weinhold, Vésteinn Thorsson, Brady Bernard, Lisa Iype, Roger W. Kramer, Richard Kreisberg, Michael Miller, Sheila M. Reynolds, Hector Rovira, Natalie Tasman, Ilya Shmulevich, Sam Ng, David Haussler, Josh M. Stuart, Rehan Akbani, Shiyun Ling, Wenbin Liu, Arvind Rao, John N. Weinstein, Roeland G. W. Verhaak, Gordon B. Mills, Mark D. M. Leiserson, Benjamin J. Raphael, Hsin-Ta Wu, Barry S. Taylor, Aaron D. Black, Jay Bowen, Julie Ann Carney, Julie M. Gastier-Foster, Carmen Helsel, Kristen M. Leraas, Tara M. Lichtenberg, Cynthia McAllister, Nilsa C. Ramirez, Teresa R. Tabler, Lisa Wise, Erik Zmuda, Robert Penny, Daniel Crain, Johanna Gardner, Kevin Lau, Erin Curely, David Mallery, Scott Morris, Joseph Paulauskis, Troy Shelton, Candace Shelton, Mark Sherman, Christopher Benz, Jae-Hyuk Lee, Konstantin Fedosenko, Georgy Manikhas, Olga Potapova, Olga Voronina, Dmitry Belyaev, Oleg Dolzhansky, W. Kimryn Rathmell, Jakub Brzezinski, Matthew Ibbs, Konstanty Korski, Witold Kycler, Radoslaw Łaźniak, Ewa Leporowska, Andrzej Mackiewicz, Dawid Murawa, Pawel Murawa, Arkadiusz Spychała, Wiktoria M. Suchorska, Honorata Tatka, Marek Teresiak, Maciej Wiznerowicz, Raafat Abdel-Misih, Joseph Bennett, Jennifer Brown, Mary Iacocca, Brenda Rabeno, Sun-Young Kwon, Robert Penny, Johanna Gardner, Ariane Kemkes, David Mallery, Scott Morris, Troy Shelton, Candace Shelton, Erin Curley, Iakovina Alexopoulou, Jay Engel, John Bartlett, Monique Albert, Do-Youn Park, Rajiv Dhir, James Luketich, Rodney Landreneau, Yelena Y. Janjigian, David P. Kelsen, Eunjung Cho, Marc Ladanyi, Laura Tang, Shannon J. McCall, Young S. Park, Jae-Ho Cheong, Jaffer Ajani, M. Constanza Camargo, Shelley Alonso, Brenda Ayala, Mark A. Jensen, Todd Pihl, Rohini Raman, Jessica Walton, Yunhu Wan, John A. Demchok, Greg Eley, Kenna R. Mills Shaw, Margi Sheth, Roy Tarnuzzer, Zhining Wang, Liming Yang, Jean Claude Zenklusen, Tanja Davidsen, Carolyn M. Hutter, Heidi J. Sofia, Robert Burton, Sudha Chudamani, and Jia Liu
References
- 1.Ferlay, J. et al. GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide. IARC CancerBase 11, http://globocan.iarc.fr, accessed on January 15, 2014 (2013)
- 2.Laurén P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Acta Pathol., Microbiol. Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 3.WHO Classification of Tumours of the Digestive System 4th edn. (International Agency for Research on Cancer, 2010)
- 4.Uemura N, et al. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 5.Bertuccio P, et al. Recent patterns in gastric cancer: a global overview. International J. Cancer. 2009;125:666–673. doi: 10.1002/ijc.24290. [DOI] [PubMed] [Google Scholar]
- 6.Richards FM, et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum. Mol. Genet. 1999;8:607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 7.Keller G, et al. Analysis for microsatellite instability and mutations of the DNA mismatch repair gene hMLH1 in familial gastric cancer. International J Cancer. 1996;68:571–576. doi: 10.1002/(SICI)1097-0215(19961127)68:5<571::AID-IJC3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 8.Toyota M, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–5442. [PubMed] [Google Scholar]
- 9.Tan IB, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476–485. doi: 10.1053/j.gastro.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei Z, et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–565. doi: 10.1053/j.gastro.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Boussioutas A, et al. Distinctive patterns of gene expression in premalignant gastric mucosa and gastric cancer. Cancer Res. 2003;63:2569–2577. [PubMed] [Google Scholar]
- 12.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nature Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 13.Shen R, Olshen AB, Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–2912. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy G, Pfeiffer R, Camargo MC, Rabkin CS. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology. 2009;137:824–833. doi: 10.1053/j.gastro.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsusaka K, et al. Classification of Epstein–Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–7197. doi: 10.1158/0008-5472.CAN-11-1349. [DOI] [PubMed] [Google Scholar]
- 16.Geddert H, Zur Hausen A, Gabbert HE, Sarbia M. EBV-infection in cardiac and non-cardiac gastric adenocarcinomas is associated with promoter methylation of p16, p14 and APC, but not hMLH1. Anal. Cell. Pathol. 2010;33:143–149. doi: 10.1155/2010/453764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, et al. High-throughput mutation profiling identifies frequent somatic mutations in advanced gastric adenocarcinoma. PLoS ONE. 2012;7:e38892. doi: 10.1371/journal.pone.0038892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sukawa Y, et al. Alterations in the human epidermal growth factor receptor 2-phosphatidylinositol 3-kinase-v-Akt pathway in gastric cancer. World J. Gastroenterology. 2012;18:6577–6586. doi: 10.3748/wjg.v18.i45.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strong MJ, et al. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog. 2013;9:e1003341. doi: 10.1371/journal.ppat.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature487, 330–337 (2012) [DOI] [PMC free article] [PubMed]
- 21.Levine Douglas A. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernal M, Ruiz-Cabello F, Concha A, Paschen A, Garrido F. Implication of the β2-microglobulin gene in the generation of tumor escape phenotypes. Cancer Immunol. Immunother. 2012;61:1359–1371. doi: 10.1007/s00262-012-1321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greulich H, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc. Natl Acad. Sci. USA. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grossmann V, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118:6153–6163. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- 26.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nature Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 29.Thumkeo D, Watanabe S, Narumiya S. Physiological roles of Rho and Rho effectors in mammals. Eur. J. Cell Biol. 2013;92:303–315. doi: 10.1016/j.ejcb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Aznar S, et al. Simultaneous tyrosine and serine phosphorylation of STAT3 transcription factor is involved in Rho A GTPase oncogenic transformation. Mol. Biol. Cell. 2001;12:3282–3294. doi: 10.1091/mbc.12.10.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nature Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 32.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nature Genet. 2014;46:166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 34.Doherty GJ, et al. The endocytic protein GRAF1 is directed to cell-matrix adhesion sites and regulates cell spreading. Mol. Biol. Cell. 2011;22:4380–4389. doi: 10.1091/mbc.e10-12-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joneson T, White MA, Wigler MH, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of RAS. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 36.Türeci O, et al. Claudin-18 gene structure, regulation, and expression is evolutionary conserved in mammals. Gene. 2011;481:83–92. doi: 10.1016/j.gene.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Chen BJ, et al. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clinical Cancer Res. 2013;19:3462–3473. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green MR, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clinical Cancer Res. 2012;18:1611–1618. doi: 10.1158/1078-0432.CCR-11-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature487, 330–337 (2012) [DOI] [PMC free article] [PubMed]
- 40.Fuchs CS, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383:31–39. doi: 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 41.Schaefer CF, et al. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Information – see contents pages 1-4 for full details. (PDF 9697 kb)