Abstract
Context
The CX3CR1 gene is implicated as a candidate gene for age-related macular degeneration (AMD) through several lines of evidence. There is uncertainty, however, as to whether common genetic variants in CX3CR1 alter risk of AMD, since prior studies have been inconsistent and mostly limited to evaluation of two non-synonymous variants, T280M (rs3732378) and V249I (rs3732379).
Objective
We aimed to determine if common variants in CX3CR1 predict future risk of AMD.
Design
Prospective nested case-control study within five large study populations with long-term follow-up.
Participants
We measured genotypes for T280M, V249I and 13 other common single-nucleotide polymorphisms (SNPs) of the CX3CR1 gene among people who developed AMD (N=1110, including N=369 with neovascular AMD) and 2532 age- and sex-matched controls.
Main outcome measures
We determined the incidence rate ratios (RR) and 95% confidence intervals (CI) for incidence of AMD for each variant, and examined interactions with other AMD-associated variants and modifiable risk factors.
Results
In additive genetic models, we identified non-significant associations with AMD for T280M (RR=0.87, P=0.074) and three other SNPs, rs2853707 (RR=0.88, P=0.069), rs12636547 (RR=0.85, P=0.098), and rs1877563 (RR=0.84, P=0.056), one of which, rs2853707, is positioned in the CX3CR1 promoter region and was associated with neovascular AMD (RR=0.75, P=0.028). We observed that a recessive model was a better fit to the data for some SNPs, with associations between rs11715522 and AMD (RR=1.27, P=0.034), and between rs2669845 (RR=3.10, P=0.035), rs2853707 (RR=0.48, P=0.050) and rs9868689 (RR=0.31, P=0.017) and neovascular AMD. Moreover, in exploratory analyses we identified a number of possible interactions including between V249I and rs2669845 and dietary intake of omega-3 fatty acids (P=0.004, and P=0.009, respectively) for AMD; between rs2669845 and obesity (P=0.031) for neovascular AMD; between T280M and complement component 3 (C3) R102G for AMD (P=0.027); between rs2669845 and Y402H in complement factor H (CFH) for AMD (P=0.037); and between rs2669845, rs2853707, and V249I and C3 R102G for neovascular AMD (P=0.008, 0.039 and 0.002, respectively).
Conclusion
This study failed to identify significant associations between common CX3CR1 variants and AMD after considering the number of SNPs analyzed and multiple comparisons. However, we observed evidence consistent with recessive modes of association, and that an effect of CX3CR1 variants may depend on other factors including dietary intake of omega-3 fatty acids, obesity, and genotypes at CFH Y402H and C3 R102G. If replicated in other populations, these findings would support a role for CX3CR1 in AMD, but also suggest that its role may involve mechanisms that are independent of the T280M/V249I variations.
Introduction
The leading cause blindness among whites in the US and other industrialized countries,1 AMD is a complex disease of aging caused by the interplay between predisposing genetic factors and exposure to environmental and lifestyle risk factors such as diet, cigarette smoking, and obesity.2, 3 Vast strides in AMD research primarily over the past decade have resulted in the identification of a group of genes for which common variants exhibit very strong associations with AMD, providing what is perhaps one of the best examples in support of the “common disease-common variants” hypothesis.4, 5
At the same time, continued research has underlined the complexity behind the genetic epidemiology of AMD. Part of the complexity of the AMD pathogenesis relates to evidence that even strongly associated single common genetic variants are, on their own, probably not sufficient to cause AMD.6 Instead, evidence suggests that a person’s risk of AMD is determined by the interplay of multiple genetic variants with each other and with environmental and lifestyle factors (i.e. it is determined by a combination of gene-gene and gene-environment interactions).7 Developing a more complete understanding of the genetic epidemiology of AMD is needed to lay the groundwork for the identification of more effective and targeted treatment or preventive strategies and to improve the accuracy of risk prediction models.7, 8
There is clear evidence for a pivotal role of immune and inflammatory pathway alterations in age-related macular degeneration (AMD). Common variants within a handful of complement pathway genes alter risk of AMD, including variants in complement factor H (CFH) (1q32), complement component 2 (C2), complement factor B (CFB), and complement component 3 (C3).2, 6 Some studies have also described significant associations with AMD for the functionally relevant T280M polymorphism in the CX3C chemokine receptor 1 (CX3CR1) gene,9-11 and there are several other lines of evidence to support a role in AMD for CX3CR1 and its ligand CX3CL1 (a.k.a. fractalkine), including human, animal, and laboratory studies.12-18 However, the largest genetic epidemiology study of CX3CR1 and AMD recently found no evidence for association of this single nucleotide polymorphism (SNP) with the particularly visually devastating neovascular form of AMD.19
In the present study, we investigated the two previously identified non-synonymous coding variants in CX3CR1 (T280M and V249I) as well as a set of 13 common variants across CX3CR1 in a prospective nested case-control study within five large study populations. We aimed to test for associations with AMD, and to investigate potential inter-relationships of this gene and other genetic and non-genetic risk factors.
Methods
The study population consisted of nested case-control samples of participants in 5 prospective studies: the Women’s Health Study (WHS), the Physicians’ Health Study (PHS), the Women’s Antioxidant and Folic Acid Cardiovascular Study (WAFACS), the Nurse’s Health Study (NHS), and the Health Professional’s Follow-up Study (HPFS). Study methods and characteristics of the study populations have been widely published.2, 20-28 The WHS, PHS, and WAFACS were all randomized, placebo-controlled trials designed to investigate the effect of aspirin and anti-oxidants on cardiovascular or cancer outcomes. The PHS is a population of initially healthy male physicians, whereas the WHS comprises initially healthy female health professionals. WAFACS participants are female health professionals at high risk of cardiovascular disease (CVD), with a prior history of myocardial infarction or at least three major risk factors for CVD. The HPFS and NHS are observational cohorts that were not restricted based on initial health status. The HPFS consists of male dentists, pharmacists and other health professionals, and the NHS of female nurses. Together, these study populations include a total of >100,000 men and women with stored baseline blood samples. The research protocol was approved by the institutional review boards at Brigham & Women’s Hospital and the Harvard School of Public Health. A description of the contributing study populations is provided in Table 1.
Table 1. Summary of study study populations contributing cases and controls for the study.
| Female Only Study populations | Male Only Study populations | ||||
|---|---|---|---|---|---|
| Study | NHS | WHS | WACS | PHS | HPFS |
| Baseline | 1989-90 | 1993-96 | 1993-96 | 1982-84 | 1993-94 |
| Design | Cohort | RCT | RCT | RCT | Cohort |
| Participants | 121,700 | 39,876 HCPs | 8,171 HCP | 22,071 MDs | 51,529 HCPs |
| RNs ≥43yo | ≥40yo | ≥40yo w/ CVD or 3 CVD RFs |
≥40yo | ≥40yo | |
| N with Blood | 32,826 | 28,345 | 5,922 | 15,124 | 18,018 |
| N Cases | 309 | 185 | 92 | 363 | 161 |
| N Controls | 710 | 396 | 209 | 827 | 390 |
NHS=Nurses’ Health Study
WHS=Women’s Health Study
WACS=Women’s Antioxidant and Cardiovascular Disease Study
PHS=Physicians’ Health Study
HPFS=Health Professionals Follow-up Study
RN=registered nurse
RCT=randomized clinical trial
HCP=health care professional
CVD=cardiovascular disease
RF=risk factor
MD=medical doctor
We defined baseline as the time when participants provided a baseline blood sample and completed a mailed questionnaire on which they reported demographic information as well as a medical history and personal information on a number of lifestyle factors, including height, weight, and cigarette-smoking history. We excluded any participants with prevalent AMD at baseline, as well as subjects who did not provide a baseline blood specimen, and >99% of all study participants reported their ethnicity as white. We followed participants with yearly followup questionnaires (every 2 years in the NHS and HPFS) to obtain information on newly developed diseases including AMD.
Confirmation of AMD
We have previously described and validated procedures for our 2-stage documentation of incident AMD, which are nearly identical in each study population.22, 25, 29-31 On each study questionnaire, we asked participants to report any new diagnosis of AMD, including the month and year of diagnosis as well as the name and address of the diagnosing eye doctor, and for signed permission to review medical records. For each report of AMD, we sent a letter to the participant’s ophthalmologist or optometrist to obtain information from the medical record on the date of diagnosis, best-corrected visual acuity, and the chorioretinal lesions present (drusen; retinal pigment epithelial [RPE] changes including atrophy, hypertrophy, and RPE detachment; geographic atrophy; subretinal neovascular membrane; or disciform scar), as well as treatment history. For the present study, we included confirmed cases of AMD associated with a visual acuity loss of 20/30 or worse. In those cases in which other ocular anomalies were also present, we asked the eye doctor to judge whether the visual acuity would be expected to be 20/30 or worse as a result of AMD alone. We defined neovascular AMD as the documented presence of an RPE detachment, sub-retinal neovascular membrane, or disciform scar that was not due to other causes (e.g., histoplasmosis or choroidal rupture). Dry AMD included cases with the documented presence of drusen and/or retinal pigment epithelial changes but with no signs of neovascular AMD. We classified participants based on the most severely affected eye.
We selected two controls for each case of dry AMD and three controls for each case of neovascular AMD at random from study participants in the same study population as the case who were still at risk of AMD at the time the case was diagnosed, who were of the same age within 1 year, and who reported having an eye exam in the past two years.
Genotyping
We examined 15 single nucleotide polymorphisms (SNPs) in or flanking CX3CR1 including two previously identified coding SNPs (rs3732378 [T280M], and rs3732379 [V249I]) and 13 tagSNPs selected using the phase II HapMap data from the CEU populations (www.hapmap.org; release 22). Tagging was done with the tagger module in Haploview, with criteria that each SNP must have had 1) a minor allele frequency of 5% or greater, and 2) an r2 value of at least 0.8 for pairwise tagging.32 The position of these SNPs within the CX3CR1 gene is delineated in Figure 1. We also genotyped the following highly replicated AMD-associated SNPs: rs1061170 (CFH Y402H), rs2230199 (C3 R102G), and rs10490924 (ARMS2 A69S), all of which demonstrated highly significant associations with AMD in this study population, Table 2. DNA was extracted from the buffy coat fraction of centrifuged blood specimens using the QIAmp Blood Kit (Qiagen). The genotyping was carried out in a custom 384 SNP multiplex using an Illumina Golden Gate assay. Automated genotype calling was carried out using Illumina GenomeStudio version 2010.2, which uses GenCall software to automatically cluster, call genotypes and assign confidence scores. All laboratory personnel were blinded to case/control status.
Figure 1. Relative position of the set of 15 single nucleotide polymorphisms in CX3CR1.
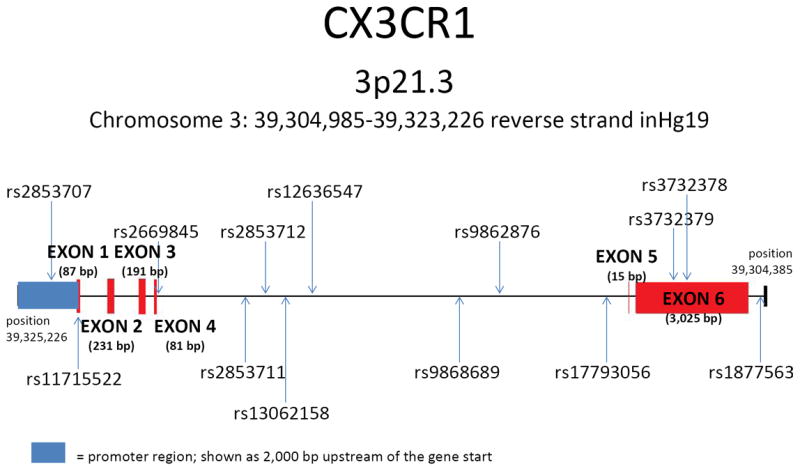
The CX3CR1 gene is located on Chromosome 3 at position 3p21.3. The figure was prepared based on the reverse strand inHg19 covering Chromosome 3: 39,304,985-39,323,226 bp. The position and relative size of exons are depicted in red and the promoter region in blue.
*SNP rs1014638 lies 3,057 bp 5′ of the CX3CR1 gene and SNP rs17038640 lies 4,828 bp 3′ of the CX3CR1 gene
Table 2. Associations of complement factor H (CFH) Y402H, ARMS2 A69S, and complement component 3 (C3) R102G with AMD in the study population.
| SNP | Variant | Genotype | OR | 95% CI | P-value | |
|---|---|---|---|---|---|---|
| CFH Y402H | YH | TC | 1.99 | 1.77 | 2.22 | |
| HH | CC | 3.94 | 3.14 | 4.95 | 4.40 × 10-30 | |
| ARMS2 A69S | AS | GT | 2.04 | 1.82 | 2.29 | |
| SS | TT | 4.08 | 3.30 | 5.26 | 2.10 × 10-29 | |
| C3 R102G | RG | CG | 1.39 | 1.21 | 1.57 | |
| GG | GG | 1.93 | 1.46 | 2.46 | 4.30 × 10-06 |
Statistical Analysis
We examined allele distributions for each SNP and used chisquare tests for Hardy-Weinberg equilibrium (HWE), and estimated linkage disequilibrium (LD) (both r2 and D’) between each pair of SNPs. We initially fit logistic regression models under a additive genetic model to estimate the incidence rate ratios (RR) and 95% confidence intervals (CI) for each genotype adjusted for other risk factors. We first obtained separate estimates of the IRR in each study population and tested for heterogeneity using Cochrane’s Q test. As there was no statistical evidence for heterogeneity between study populations (P for each SNP≥0.61), we present only pooled data from the study populations. Controlling for age, sex, cigarette smoking and obesity, we modeled the allelic effects using a multiplicative (i.e. additive) coding scheme using a single variable for each SNP coded 0 for subjects homozygous for the major allele (or, alternatively for the candidate SNPs, the allele previously found to be associated with the lowest risk of AMD), 1 for heterozygotes, and 2 for subjects homozygous for the minor allele (or, alternatively for the candidate SNPs, the allele previously found to be associated with increased risk of AMD). We next fit models using separate indicator variables for subjects who were heterozygous, and subjects who were homozygous for the risk allele; as well as recessive models using an indicator variable for subjects who were homozygous for the minor allele at each locus in reference to subjects who were homozygous for the major allele or heterozygotes. To arrive at the best-fitting model, we compared these alternative models using Akaike’s Information Criteria (AIC).33 As a rule of thumb, two models are statistically indistinguishable if the AIC difference is less than 2. We then proceeded to fit both additive and recessive models for each SNP also adjusting for complement factor H (CFH) Y402H, ARMS2 A69S, and complement component 3 (C3) R102G, three SNPs with very well-validated associations with AMD.
For T280M and V249I, as well as any SNPs that were found to be associated with AMD with a p-value of <0.10 we next tested one by one for interactions with CFH Y402H, ARMS2 A69S, and C3 R102G, as well as for possible gene-environment interactions with cigarette smoking, obesity, and omega-3 fatty acid intake (known AMD risk factors). We used additive coding for CFHY402H, ARMS2 A69S, and C3 R102G and included product terms to test for interactions with other CX3CR1 SNPs using either additive or recessive coding. We used permutation tests to confirm the analytic significance tests for interactions, randomizing case versus control status within matched sets and performing association testing over 10,000 iterations.
Results
The study population included the 1110 cases of incident AMD matched with 2532 controls, including 369 cases with neovascular AMD. Cases and controls were matched on age in each study population (mean age at baseline was 62 y), but not on other risk factors such as cigarette smoking and obesity (Table 3). Of the 15 CX3CR1 SNPS, genotype data were successfully obtained for each SNP in ≥97% of cases and controls. Genotype and allele frequencies are provided in Table 4. We found no significant departures from Hardy-Weinberg equilibrium for any of the 15 SNPs among the control group (each P>0.1).
Table 3. Baseline characteristics of the combined study population.
| Cases | Controls | P-value | |
|---|---|---|---|
| Age, y (mean +/- SD) | 62.4 +/- 6.9 | 62.4 +/- 6.9 | 0.94 |
| Smoking (N, %) | |||
| Current | 171 (15.5%) | 239 ( 9.5%) | <0.0001 |
| Past | 478 (43.4%) | 1042 (41.5%) | 0.27 |
| Obesity (N, %) | 146 (13.2%) | 255 (10.1%) | 0.007 |
| Omega-3 FA Intake, g/d (mean +/- SD) | 0.74 +/- 0.77 | 0.75 +/- 0.73 | 0.83 |
Table 4. Genotype and allele frequencies for 15 SNPs in CX3CR1, by case/control status.
| SNP | Dry AMD Cases | Dry AMD Controls | Neovascular Cases | Neovascular Controls | All Cases | All Controls | ||
|---|---|---|---|---|---|---|---|---|
| rs3732378 | Minor Allele | T | 0.160 | 0.177 | 0.172 | 0.175 | 0.164 | 0.176 |
| Genotype | CC | 0.704 | 0.676 | 0.693 | 0.682 | 0.700 | 0.678 | |
| CT | 0.274 | 0.295 | 0.270 | 0.287 | 0.273 | 0.292 | ||
| TT | 0.023 | 0.030 | 0.037 | 0.031 | 0.027 | 0.030 | ||
| rs3732379 | Minor Allele | A | 0.288 | 0.282 | 0.275 | 0.282 | 0.284 | 0.282 |
| Genotype | GG | 0.507 | 0.507 | 0.516 | 0.507 | 0.510 | 0.507 | |
| GA | 0.411 | 0.423 | 0.419 | 0.421 | 0.413 | 0.422 | ||
| AA | 0.083 | 0.070 | 0.065 | 0.071 | 0.077 | 0.071 | ||
| rs2669845 | Minor Allele | T | 0.143 | 0.126 | 0.136 | 0.122 | 0.141 | 0.124 |
| Genotype | CC | 0.727 | 0.765 | 0.754 | 0.763 | 0.735 | 0.764 | |
| CT | 0.260 | 0.220 | 0.221 | 0.229 | 0.247 | 0.224 | ||
| TT | 0.014 | 0.016 | 0.026 | 0.008 | 0.017 | 0.013 | ||
| rs2853712 | Minor Allele | C | 0.443 | 0.436 | 0.451 | 0.443 | 0.446 | 0.439 |
| Genotype | TT | 0.311 | 0.317 | 0.307 | 0.315 | 0.310 | 0.316 | |
| TC | 0.491 | 0.494 | 0.485 | 0.483 | 0.489 | 0.489 | ||
| CC | 0.198 | 0.189 | 0.208 | 0.202 | 0.201 | 0.194 | ||
| rs11715522 | Minor Allele | G | 0.397 | 0.379 | 0.382 | 0.373 | 0.392 | 0.377 |
| Genotype | TT | 0.361 | 0.378 | 0.394 | 0.372 | 0.372 | 0.376 | |
| TG | 0.485 | 0.487 | 0.448 | 0.509 | 0.473 | 0.496 | ||
| GG | 0.154 | 0.136 | 0.158 | 0.119 | 0.155 | 0.129 | ||
| rs2853711 | Minor Allele | T | 0.277 | 0.283 | 0.277 | 0.279 | 0.277 | 0.281 |
| Genotype | GG | 0.528 | 0.512 | 0.530 | 0.509 | 0.529 | 0.511 | |
| GT | 0.390 | 0.410 | 0.386 | 0.424 | 0.389 | 0.416 | ||
| TT | 0.082 | 0.078 | 0.085 | 0.067 | 0.083 | 0.074 | ||
| rs1014638 | Minor Allele | C | 0.288 | 0.283 | 0.274 | 0.276 | 0.284 | 0.280 |
| Genotype | AA | 0.518 | 0.513 | 0.520 | 0.516 | 0.519 | 0.514 | |
| AC | 0.388 | 0.409 | 0.412 | 0.415 | 0.396 | 0.411 | ||
| CC | 0.094 | 0.079 | 0.068 | 0.069 | 0.086 | 0.075 | ||
| rs13062158 | Minor Allele | C | 0.291 | 0.293 | 0.281 | 0.284 | 0.288 | 0.289 |
| Genotype | TT | 0.508 | 0.492 | 0.531 | 0.506 | 0.516 | 0.498 | |
| TC | 0.402 | 0.430 | 0.376 | 0.420 | 0.393 | 0.426 | ||
| CC | 0.090 | 0.078 | 0.093 | 0.074 | 0.091 | 0.076 | ||
| rs2853707 | Minor Allele | C | 0.223 | 0.230 | 0.222 | 0.242 | 0.223 | 0.235 |
| Genotype | TT | 0.609 | 0.592 | 0.594 | 0.570 | 0.604 | 0.583 | |
| TC | 0.336 | 0.356 | 0.369 | 0.377 | 0.346 | 0.365 | ||
| CC | 0.055 | 0.052 | 0.037 | 0.053 | 0.049 | 0.053 | ||
| rs9862876 | Minor Allele | G | 0.251 | 0.240 | 0.238 | 0.257 | 0.247 | 0.247 |
| Genotype | CC | 0.571 | 0.573 | 0.561 | 0.547 | 0.567 | 0.562 | |
| CG | 0.357 | 0.374 | 0.403 | 0.393 | 0.372 | 0.382 | ||
| GG | 0.073 | 0.053 | 0.037 | 0.060 | 0.061 | 0.056 | ||
| rs12636547 | Minor Allele | G | 0.089 | 0.105 | 0.092 | 0.088 | 0.090 | 0.098 |
| Genotype | CC | 0.832 | 0.802 | 0.820 | 0.830 | 0.828 | 0.813 | |
| CG | 0.159 | 0.187 | 0.177 | 0.163 | 0.165 | 0.177 | ||
| GG | 0.009 | 0.012 | 0.003 | 0.007 | 0.007 | 0.010 | ||
| rs17793056 | Minor Allele | G | 0.477 | 0.502 | 0.483 | 0.482 | 0.479 | 0.494 |
| Genotype | AA | 0.273 | 0.239 | 0.279 | 0.266 | 0.275 | 0.250 | |
| AG | 0.499 | 0.518 | 0.476 | 0.505 | 0.492 | 0.513 | ||
| GG | 0.227 | 0.243 | 0.245 | 0.229 | 0.233 | 0.237 | ||
| rs17038640 | Minor Allele | C | 0.337 | 0.322 | 0.328 | 0.331 | 0.334 | 0.326 |
| Genotype | TT | 0.441 | 0.462 | 0.472 | 0.446 | 0.451 | 0.455 | |
| TC | 0.445 | 0.433 | 0.401 | 0.447 | 0.431 | 0.439 | ||
| CC | 0.115 | 0.105 | 0.127 | 0.107 | 0.119 | 0.106 | ||
| rs9868689 | Minor Allele | A | 0.229 | 0.232 | 0.233 | 0.235 | 0.230 | 0.233 |
| Genotype | GG | 0.595 | 0.576 | 0.557 | 0.581 | 0.583 | 0.578 | |
| GA | 0.351 | 0.385 | 0.420 | 0.369 | 0.374 | 0.379 | ||
| AA | 0.054 | 0.039 | 0.023 | 0.050 | 0.044 | 0.044 | ||
| rs1877563 | Minor Allele | T | 0.104 | 0.121 | 0.104 | 0.112 | 0.104 | 0.117 |
| Genotype | CC | 0.804 | 0.773 | 0.800 | 0.787 | 0.803 | 0.778 | |
| CT | 0.184 | 0.213 | 0.192 | 0.203 | 0.186 | 0.209 | ||
| TT | 0.012 | 0.014 | 0.008 | 0.011 | 0.011 | 0.013 |
Main effects of CX3CR1 Variants
All AMD
In logistic regression models controlling for age, sex, cigarette smoking status, and obesity, we observed no significant association between T280M or V249I with AMD in additive (RR=0.92, CI=0.80-1.06, P=0.24 for T280M; RR=1.01, CI=0.90-1.14, P=0.82 for V249I), dominant, or recessive models, Table 5. Models testing for associations of the other 13 SNPs showed a non-significant association of modest magnitude in additive models for rs2669845 (RR=1.15, CI=0.99-1.35, P=0.069) and rs1877563 (RR=0.85, CI=0.73-1.00, P=0.056), with similar results in the co-dominant models for these SNPs, Table 5. Further adjustment for CFH Y402H, ARMS2 A69S, and C3 R102G generally strengthened associations. In these models, the association with T280M was most consistent with a modest protective effect of the rare allele (RR=0.87, CI=0.75-1.01, P=0.074). Comparison of AIC statistics identified two SNPs for which the recessive model was favored, revealing an increased risk for those with two copies of the rare ‘G’ allele at rs11715522 (RR=1.27, CI=1.02-1.58, P=0.034) and a non-significant increased risk for participants with two copies of the rare ‘C’ allele at rs13062158 (RR=1.18, CI=0.88-1.58, P=0.27).
Table 5. Results of logistic regression models for log-additive versus recessive associations between 15 CX3CR1 SNPs with age-related macular degeneration using pooled data from 5 study populations.
| Additive Models | Recessive Models | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Adjusted for age, sex, smoking, and obesity | Also adjusted for CFH, ARMS2, C3 | Adjusted for age, sex, smoking, and obesity | Also adjusted for CFH, ARMS2, C3 | |||||||
|
| ||||||||||
| SNP | OR | P-value | OR | P-value | Permutation P-value | OR | P-value | OR | P-value | Permutation P-value |
| rs3732378 | 0.92 | 0.24 | 0.87 | 0.074 | 0.062 | 0.92 | 0.7 | 0.92 | 0.72 | 0.75 |
| rs3732379 | 1.02 | 0.80 | 1.01 | 0.85 | 0.86 | 1.09 | 0.54 | 1.09 | 0.58 | 0.59 |
| rs2669845 | 1.15 | 0.069 | 1.14 | 0.12 | 0.14 | 1.38 | 0.28 | 1.28 | 0.43 | 0.61 |
| rs2853712 | 1.00 | 0.97 | 1.00 | 0.96 | 0.99 | 1.01 | 0.91 | 0.99 | 0.93 | 0.91 |
| rs11715522 | 1.08 | 0.18 | 1.11 | 0.073 | 0.084 | 1.26 | 0.027 | 1.27 | 0.034 | 0.038 |
| rs2853711 | 0.99 | 0.89 | 0.96 | 0.52 | 0.42 | 1.14 | 0.33 | 1.12 | 0.43 | 0.52 |
| rs1014638 | 1.02 | 0.67 | 1.02 | 0.77 | 0.79 | 1.15 | 0.30 | 1.09 | 0.58 | 0.54 |
| rs13062158 | 1.01 | 0.80 | 1.05 | 0.46 | 0.52 | 1.23 | 0.13 | 1.18 | 0.27 | 0.25 |
| rs2853707 | 0.91 | 0.15 | 0.88 | 0.069 | 0.066 | 0.89 | 0.50 | 0.83 | 0.31 | 0.27 |
| rs9862876 | 1.00 | 0.94 | 0.99 | 0.82 | 0.84 | 1.04 | 0.80 | 1.08 | 0.64 | 0.55 |
| rs12636547 | 0.87 | 0.12 | 0.85 | 0.098 | 0.10 | 0.62 | 0.28 | 0.60 | 0.32 | 0.20 |
| rs17793056 | 0.94 | 0.25 | 0.93 | 0.21 | 0.19 | 0.98 | 0.83 | 0.93 | 0.43 | 0.41 |
| rs17038640 | 1.04 | 0.45 | 1.07 | 0.25 | 0.21 | 1.13 | 0.29 | 1.14 | 0.31 | 0.28 |
| rs9868689 | 0.97 | 0.62 | 0.97 | 0.68 | 0.71 | 0.95 | 0.77 | 1.00 | 0.98 | 0.95 |
| rs1877563 | 0.85 | 0.056 | 0.84 | 0.056 | 0.060 | 0.80 | 0.56 | 0.89 | 0.76 | 0.79 |
Neovascular AMD
For the subset of neovascular AMD cases, we observed a non-significant association with the CX3CR1 promoter SNP rs2853707 (RR=0.82, CI=0.66-1.02, P=0.074) in an additive model adjusting for age, sex, cigarette smoking, and obesity; and this relationship appeared somewhat stronger after additional control for CFH Y402H, ARMS2 A69S, and C3 R102G (RR=0.75, CI=0.59-0.97, P=0.028), Table 6. Comparison of AIC statistics identified four SNPs for which the recessive model was favored. In the recessively coded models adjusted for age, sex, cigarette smoking and obesity, three of these CX3CR1 SNPs showed nominally significant associations with neovascular AMD, and one SNP showed borderline significance (Table 6). After further adjustment for CFH Y402H, ARMS2 A69S, and C3 R102G, participants with two copies of the minor ‘T’ allele at rs2669845 had a 3-fold higher risk of development of neovascular AMD compared with participants with one or no copies of this allele (RR=3.10, CI=1.08-8.87, P=0.035). Increased risk of neovascular AMD was also observed for participants with two copies of the ’G’ allele at rs11715522 (RR=1.47, CI=1.03-2.10, P=0.033), but this was attenuated after adjustment for CFH Y402H, ARMS2 A69S, and C3 R102G (RR=1.42, CI=0.96-2.10, P=0.083). Protective effects were seen for participants with 2 copies of the minor ‘A’ allele of rs9868689 and for participants with 2 copies of the minor ‘C’ allele at the rs2853707 promoter SNP. After full adjustment, the RR for association with neovascular AMD were 0.31, CI=0.12-0.82 (P=0.017) for rs9868689, and 0.48, CI=0.23-1.00 (P=0.050) for rs2853707, Table 6.
Table 6. Results of logistic regression models for log-additive versus recessive association between 15 CX3CR1 SNPs with neovascular age-related macular degeneration using pooled data from 5 study populations.
| Additive Models | Recessive Models | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Adjusted for age, sex, smoking, and obesity | Also adjusted for CFH, ARMS2, C3 | Adjusted for age, sex, smoking, and obesity | Also adjusted for CFH, ARMS2, C3 | |||||||
|
| ||||||||||
| SNP | OR | P-value | OR | P-value | Permutation P-value | OR | P-value | OR | P-value | Permutation P-value |
| rs3732378 | 0.97 | 0.82 | 0.98 | 0.86 | 0.86 | 1.24 | 0.53 | 1.42 | 0.35 | 0.33 |
| rs3732379 | 0.96 | 0.71 | 1.03 | 0.80 | 0.85 | 0.95 | 0.85 | 1.14 | 0.65 | 0.63 |
| rs2669845 | 1.16 | 0.29 | 1.19 | 0.25 | 0.28 | 3.20 | 0.018 | 3.10 | 0.035 | 0.020 |
| rs2853712 | 0.99 | 0.89 | 0.96 | 0.69 | 0.64 | 0.98 | 0.88 | 0.94 | 0.75 | 0.66 |
| rs11715522 | 1.07 | 0.45 | 1.11 | 0.34 | 0.36 | 1.47 | 0.033 | 1.42 | 0.083 | 0.087 |
| rs2853711 | 1.01 | 0.90 | 1.02 | 0.86 | 0.81 | 1.31 | 0.24 | 1.44 | 0.16 | 0.15 |
| rs1014638 | 1.04 | 0.73 | 1.01 | 0.94 | 0.94 | 1.04 | 0.88 | 1.06 | 0.84 | 0.82 |
| rs13062158 | 1.03 | 0.77 | 1.07 | 0.57 | 0.56 | 1.36 | 0.18 | 1.34 | 0.25 | 0.21 |
| rs2853707 | 0.82 | 0.074 | 0.75 | 0.028 | 0.026 | 0.61 | 0.13 | 0.48 | 0.05 | 0.039 |
| rs9862876 | 0.88 | 0.22 | 0.88 | 0.30 | 0.26 | 0.54 | 0.053 | 0.64 | 0.2 | 0.18 |
| rs12636547 | 1.01 | 0.95 | 0.94 | 0.75 | 0.78 | 0.38 | 0.36 | 0.47 | 0.52 | 0.55 |
| rs17793056 | 1.01 | 0.92 | 1.03 | 0.80 | 0.81 | 1.14 | 0.39 | 1.10 | 0.57 | 0.55 |
| rs17038640 | 1.00 | 0.98 | 0.99 | 0.95 | 0.97 | 1.25 | 0.25 | 1.19 | 0.42 | 0.40 |
| rs9868689 | 0.98 | 0.87 | 0.96 | 0.76 | 0.72 | 0.41 | 0.025 | 0.31 | 0.017 | 0.011 |
| rs1877563 | 0.89 | 0.40 | 0.84 | 0.28 | 0.32 | 0.79 | 0.72 | 1.01 | 0.99 | 0.99 |
Linkage disequilibrium (LD) measures (Figure 2) showed D’=1.0 and r2=0.54 between T280M and V249I. Measures of LD were lower between these two SNPs and the other CX3CR1 SNPs showing significant or borderline associations with AMD. For example, LD was estimated at D’=0.48 and r2=0.03 between T280M and rs11715522, and D’=0.026 and r2=0.00 between T280M and rs2853707 suggesting that the observed associations of the other CX3CR1 SNPs with AMD are not due to LD with T280M.
Figure 2. Linkage disequilibrium (LD) between SNPs in the CX3CR1 gene.
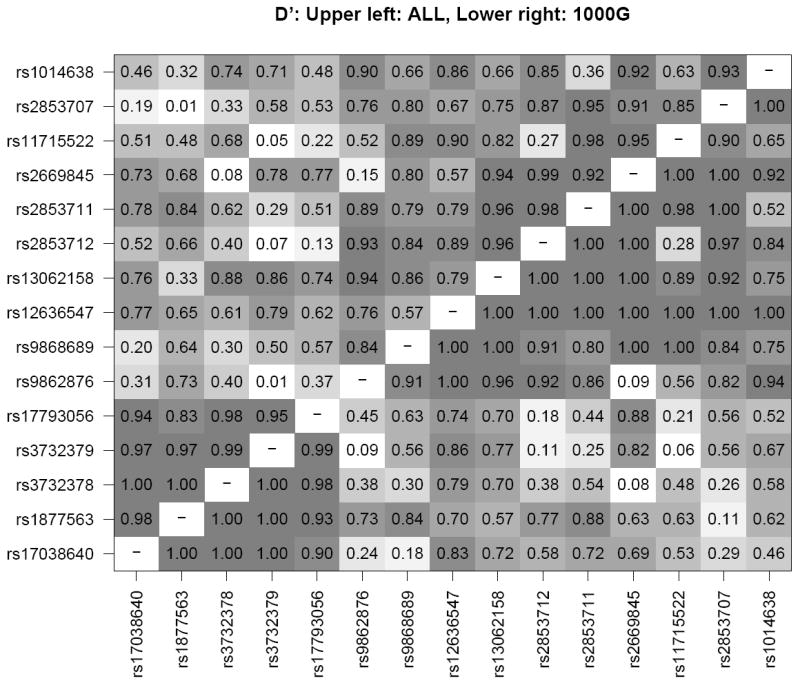
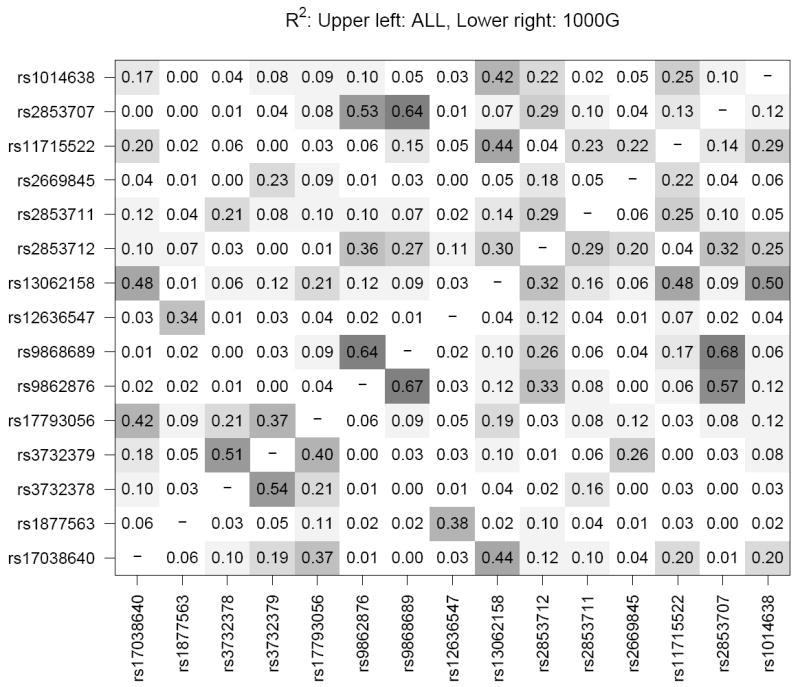
R2 and D’ LD measures were calculated using Haploview32 for the combined case/control samples from 5 study populations (WHS, PHS, WAFACS, NHS, HPFS). In each panel, the upper left triangle shows LD for the study sample while the lower right triangle shows LD for the same SNPs in the 1000 genomes European panel (381 individuals, release 11/23/2010) for comparison.55 Shading corresponds to the superimposed numeric estimates of LD for each pair of SNPs.
Interaction effects of CX3CR1
Gene-environment interactions
Investigation of potential gene-environment interactions identified suggestive interactions between some CX3CR1 SNPs and dietary intake of omega-3 fatty acids and obesity, Table 7, but not with cigarette smoking (data not shown). Specifically, the increased risk of AMD associated with obesity appeared to be due to stronger associations in the subgroups of participants with at least one risk allele at either rs2669845 (RR=1.83, CI=1.12 - 2.98), or rs11715522 (RR=2.23, CI=1.00 - 4.99). Interactions with omega-3 fatty acids tended toward a protective effect of omega-3 fatty acids in the strata with the rare alleles (T and A, respectively) at rs3732379 and rs2669845; however for rs11715522 we observed an increased risk of neovascular AMD associated with higher intake of omega-3 fatty acid intake among the subgroup carrying at least one copy of the rarer ‘G’ allele (RR=1.62, CI=1.01 - 2.61).
Table 7. Gene-environment interactions for selected SNPs within CX3CR1 and risk of macular degeneration.
| Model | Outcome | Risk Factor | CX3CR1 SNP | P-value |
|---|---|---|---|---|
| Additive | All AMD | Omega-3 | rs2669845 | 0.069 |
| All AMD | Omega-3 | rs11715522 | 0.057 | |
| Wet AMD | Obesity | rs2669845 | 0.031 | |
| Wet AMD | Obesity | rs11715522 | 0.085 | |
| Recessive | All AMD | Omega-3 | rs3732379 | 0.004 |
| All AMD | Omega-3 | rs2669845 | 0.009 |
Gene-gene interactions
Examination of gene-gene interactions, Table 8, suggested a possible interaction between rs2853707 and ARMS2 A69S (P for interaction = 0.077) for all AMD cases versus controls assuming additive effects at rs2853707. Separate models for those with versus without at least one ARMS2 A69S risk allele revealed that rs2853707 was associated with a reduced risk of AMD only among people with no ARMS2 A69S risk alleles (RR=0.73, CI=0.58 - 0.92 for TC; RR=0.53, CI=0.34 - 0.85 for CC), whereas there was no effect among those with at least one ARMS2 A69S risk allele (RR=0.97, CI=0.74 -1.28 for TC; RR=0.94, CI=0.55 - 1.64 for CC). For all AMD cases versus controls we also identified one statistically significant interaction assuming recessive effects, between rs2669845 and CFH Y402H (P for interaction=0.037), as well as two interactions of borderline significance (Table 8).
Table 8. Gene-gene interactions for CFH Y402H, ARMS2 A69S, C3 R102G and selected SNPs within CX3CR1 and risk of macular degeneration.
| Model | Outcome | SNP | CX3CR1 SNP | P-value | Permutation P-value |
|---|---|---|---|---|---|
| Additive | All AMD | ARMS2 | rs2853707 | 0.077 | 0.063 |
| Wet AMD | CFH | rs11715522 | 0.078 | 0.077 | |
| Wet AMD | C3 | rs2669845 | 0.008 | 0.008 | |
| Wet AMD | C3 | rs2853707 | 0.039 | 0.039 | |
| Wet AMD | C3 | rs3732378 | 0.051 | 0.047 | |
| Wet AMD | C3 | rs3732379 | 0.002 | 0.002 | |
| Recessive | All AMD | CFH | rs2669845 | 0.037 | 0.006 |
| All AMD | C3 | rs9868689 | 0.064 | 0.067 | |
| All AMD | C3 | rs9862876 | 0.058 | 0.090 | |
| Wet AMD | ARMS2 | rs3732378 | 0.087 | 0.063 | |
| Wet AMD | C3 | rs3732378 | 0.027 | 0.012 |
We also observed possible interactions for neovascular AMD assuming additive genetic effects of the CX3CR1 SNPs; specifically, between rs11715522 and CFH Y402H (P for interaction=0.078), and between T280M and C3 R102G (P for interaction=0.051). In stratified models, the increased risk of neovascular AMD associated with rs11715522 appeared strongest among those who carried at least one CFH Y402H risk allele (RR=1.28, CI=0.99 - 1.66 for TG; RR=1.64, CI=0.98 - 2.76); whereas the 280M allele conferred decreased risk of neovascular AMD among the subset of participants with no C3 R102G risk alleles (RR=0.53, CI=0.29 - 0.98 for 280T/M; RR=0.28, CI=0.08 - 0.96). There were also three possible interactions with C3 R102G, specifically for rs2669845 (P for interaction=0.008), rs2853707 (P for interaction=0.039), and V249I (P for interaction = 0.002), Table 8. Increased risk of neovascular AMD associated with rs2669845 was limited to participants who also carried at least one C3 R102G risk allele; whereas the minor allele of rs2853707 was associated with a decreased risk of AMD in the same subgroup. The V249I SNP tended toward a protective effect among those with 249I and no C3 R102G risk alleles, but an increased risk when 249I was present along with at least one C3R102G risk allele (Table 9). In models assuming a recessive effect for the CX3CR1 SNP, we observed possible interactions between T280M and C3 R102G (P for interaction=0.027), and between T280M and ARMS2 A69S (P for interaction=0.087). Permutation tests confirmed the magnitude of the P-values for all gene-gene interactions.
Table 9. Effects of selected CX3CR1 SNPs on risk of neovascular AMD within strata defined by risk at C3 R102G.
| Genotype | No C3 R102G Risk | C3 R102G Risk | |||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | ||||
| rs2669845 | CC | 1.00 | 1.00 | ||||
| CT | 0.95 | 0.50 | 1.78 | 1.74 | 1.08 | 2.80 | |
| TT | 0.90 | 0.25 | 3.17 | 3.03 | 1.17 | 7.84 | |
| rs2853707 | TT | 1.00 | 1.00 | ||||
| TC | 0.78 | 0.45 | 1.35 | 0.48 | 0.32 | 0.73 | |
| CC | 0.61 | 0.20 | 1.82 | 0.23 | 0.10 | 0.53 | |
| rs3732378 (T280M) | CC | 1.00 | 1.00 | ||||
| CT | 0.53 | 0.29 | 0.98 | 1.18 | 0.79 | 1.77 | |
| TT | 0.28 | 0.08 | 0.96 | 1.39 | 0.62 | 3.13 | |
| rs3732379 (V249I) | GG | 1.00 | 1.00 | ||||
| GA | 0.65 | 0.40 | 1.06 | 1.39 | 0.96 | 2.01 | |
| AA | 0.42 | 0.16 | 1.13 | 1.92 | 0.92 | 4.03 | |
Discussion
Similar to findings from a recent clinic-based cross-sectional study including 1093 cases with neovascular AMD,19 we failed to observed a statistically significant association of the T280M loss of function CX3CR1 SNP with incident AMD; and the data from this prospective evaluation tended in the direction of a modest protective association in contrast to the increased risks observed in some small clinic-based cross-sectional studies. We also observed no overall association between the V249I SNP and AMD. However, unlike previous studies, we examined not only the T280M and V249I SNPs, but included 13 other common SNPs in CX3CR1 including rs2853707, which lies within the CX3CR1 promoter region. Among these, we identified one SNP (rs11715522) that was nominally associated with incident AMD overall, three SNPs (rs2669845, rs2853707, and rs9868689) that showed associations with incident neovascular AMD, though these would not reach significance if adjusted for multiple testing. Further exploratory investigation suggested several possible interactions between CX3CR1 variants and CFH Y402H, C3 R102G, obesity and dietary intake of omega-3 fatty acids.
Although this study is limited by our inability to perform standardized clinical assessments of retinal status among participants of these large geographically dispersed study populations of male and female health professionals, we have demonstrated that our case ascertainment method has high specificity,29, 34 which minimizes bias in a prospective study.35 Furthermore, the consistency of our findings in these populations linking CFH Y402H and ARMS2 A69S with AMD, as well as our prior work on modifiable risk factors for AMD,29, 34, 36, 37 and the strong associations demonstrated in the present data for CFH Y402H, ARMS2 A69S, and C3 R102G with AMD provide further reassurance of the validity of the findings for variants within CX3CR1. Findings of the study must also be interpreted in light of the large number of tests performed and the modest p-values observed in the models for the primary tests of association of each SNP with AMD, none of which would be significant after adjustment for multiple testing, as well as the lack of replication in additional independent study populations.
CX3CR1 is the receptor for the CX3 chemokine ligand (CX3CL1, fractalkine). Fractalkine is an unusually large chemokine that induces directed chemotaxis in nearby responsive immune cells. Fractalkine is expressed on numerous cells, including activated endothelial cells and macrophages, and its expression is enhanced in the presence of inflammatory stimuli and in atherosclerosis. It is also the only one of the 200 or so known chemokines to exhibit not only a soluble but also a transmembrane form, the latter giving it the ability to mediate leukocyte adhesion to cells such as microglia (the resident macrophages in the retina) that express CX3CR1. The CX3CL1/CX3CR1 axis also appears to have novel functions in regard to cell adhesion, antiapotosis, and cell proliferation.38
CX3CR1-expressing microglia appear to have a complex role in relation to the evolution of neurodegenerative diseases. Evidence suggests microglia may both contribute to and limit tissue injury during the chronic low-level inflammation and neuro-degeneration, and these effects may vary within different organs and diseases, as well as with age.38-41 There may be a precarious balance between beneficial and detrimental effects, which may depend at least in part on presumed alterations in CX3CR1 that result from common genetic variation as well as on interactions with other key AMD-associated factors (both genetic and environmental). A recent investigation based on a rat model of blue light-induced photoreceptor degeneration combined with cell culture studies demonstrated that apoptosis of retinal photoreceptors was followed by increased levels of fractalkine and CX3CR1 in conjunction with microglial activation and migration and upregulation of pro-inflammatory factors such as IL1-beta and TNF-alpha. Blockage of soluble fractalkine decreased the inflammatory response, whereas over-expression of the membrane-bound form was associated with neuroprotection.15 In human AMD donor eyes, others have observed CX3CR1 deposits in drusen, and CX3CR1-positive microglial cells interposed between choroidal vessels and in close contact with neovascular lesions.14 There are two known nonsynonymous SNPs in CX3CR1, and laboratory studies have shown defective migration of retinal microglial cells isolated from individuals carrying the 280M allele, and this effect was shown to be related to interactions with the membrane bound form of fractalkine. 14 Aged CX3CR1 deficient mice show accentuated subretinal accumulation of microglial cells, drusen-like lesions (shown to be lipid-bloated microglial cells), and outer retinal degeneration, which could be prevented by raising the mice in a light restricted environment. Laser injury to CX3CR1 deficient mice resulted in significantly greater choroidal neovascularization.14 When interpreting this literature, it is important to consider that there may be important differences between mouse and human CX3CL1/CX3CR1.42
Previous epidemiological studies have largely been limited to studies of the possible association between one or both of the nonsynonymous SNPs (T280M and 249I) and AMD, yielding inconsistent results. The first few reports on the association of the T280M and V249I CX3CR1 variants in AMD suggested a 2-3-fold increased risk of AMD among people carrying the minor alleles at each locus.9,10,14 More recently, however, Zerbib and colleagues recruited 1093 patients with neovascular AMD from 4 retinal centers in France along with a group of 396 controls with normal retinal examination findings, and found no association of 280M with AMD (OR=0.9, CI=0.6 - 1.3 for 280T/M; OR=0.6, CI=0.3 -1.4 for 280M/M).19 Similarly, Brion and colleagues reported no association with T280M or 4 tag SNPs in CX3CR1 in a study of 385 cases of AMD (225 with atrophic AMD, 57 with neovascular AMD, and 71 with mixed AMD) and 282 age-matched controls.43 Finally, a small study of copy number variation in CX3CR1 in relation to AMD demonstrated a mildly protective effect that was diminished after adjustment for age.44
To our knowledge, there have been no prior studies of rs2853707, which lies within the CX3CR1 promoter region. Because of it’s position, we used MatInspector (www.genomatrix.de) to explore the possibility that this SNP may have functional relevance, and found that the minor allele is predicted to create a binding site for Myb-like transcriptional regulators. The Myb gene family encodes a number of nuclear transcriptional proteins including c-Myb, a DNA-binding transcription factor that functions in apoptosis, proliferation, and differentiation. Alterations in c-Myb activity can alter the balance of inflammatory activity in the context of tissue injury or vasculoproliferative disease; and increased c-Myb activity increases survival rates of certain cell types.45 Given the fact that fractalkine/CX3CR1 has been implicated in these same biological pathways,38 it seems plausible that the present findings of significant associations between rs2853707 and AMD may be related to functionally relevant variation within the CX3CR1 promoter region.
Because the genes involved in AMD have pleiotropic effects throughout the body, variants in AMD-associated genes could also alter the clinical course or survival of individuals who carry certain alleles. Particularly when the alleles are common, such effects could introduce selection bias when prevalent rather than incident cases are studied.35 This is potentially quite important for studies of CX3CR1, since people with the 280M and 249I CX3CR1 variants have been shown to have a reduced risk of cardiovascular disease. At the same time, although evidence is more limited, 280M and/or 249I have also been associated with reduced risk of asthma;46 as well as with modestly increased risks of metabolic syndrome, diabetes, higher waist circumference and obesity.47 Other evidence links the CX3CL1/CX3CR1 axis with Alzheimer’s disease (reduced risk in CX3CR1 deficient mice),48 polymyositis and dermatomyositis (patients have increased serum fractalkine levels),49 rheumatoid arthritis, and other conditions.50 The present study, using a validated prospective nested case-control methodology should therefore help clarify associations of common variants of CX3CR1 gene with risk of AMD, since the population that gives rise to the cases is clearly established, rates of loss to follow-up are extremely low in all the study populations, and the control selection and analysis methodology inherently account for censoring due to death or loss to follow-up.
The findings of this investigation also point to an additional level of complexity in understanding the possible role of CX3CR1 in AMD. Prior epidemiological studies have revealed that risk of AMD is affected by a combination of environmental effects plus strong effects of variants within several genes, especially CFH, ARMS2, and C3; and there is a growing body of evidence suggesting that these genetic and non-genetic risk factors interact with each other in a variety of ways to determine individual AMD risk.2, 3, 51, 52 Our data suggesting possible interactions between common variants in CX3CR1 and omega-3 fatty acids, obesity, and three strongly AMD-associated variants (CFH Y402H, ARMS2 A69S, and C3 R102G) provide further details regarding the interplay of common AMD-associated variants, and suggest that more work is needed to fully understand the intricacies of the genetic and non-genetic contributors to this blinding disease.
We identified possible interactions of CX3CR1 SNPs and obesity for neovascular AMD, and there is evidence showing associations between variation at CX3CR1 and obesity.47, 53 Together such observations are consistent with the idea that the impact of obesity on risk of AMD may be different depending on an individual’s genetic underpinnings. We also observed possible interactions between three genetic variants in CX3CR1 and dietary intake of omega-3 fatty acids, which has been associated with risk of AMD in a number of epidemiological studies, and with slowed progression of retinal lesions in a Ccl2(-/-)/Cx3Cr1(-/-) murine double knock-out model.54 The present study also points to possible interactions between variants in CX3CR1 and other common strongly AMD-associated variants. These include suggested interactions between the rs2853707 promoter region SNP and ARMS2 A69S as well as rs11715522 and CFH Y402H; between rs2669845 and CFH Y402; and a number of interactions with C3 R102G, including with rs2669845, rs2853707, T280M, and V249I.
In summary, this prospective study provides new information on the potential association between common variation within CX3CR1 and risk of AMD that fails to confirm prior findings of associations between the 280M or 249I alleles with AMD. The data suggest the possibility of a more minor role for CX3CR1 (as compared, e.g. to CFH, ARMS2, and C3) that may include an intricate relationship between common variants in this gene, common variants in CFH, ARMS2, and C3, obesity, and dietary intake of omega-3 fatty acids with risk of AMD. The findings are consistent with a protective association of the 280M allele within subgroups of the population defined by ARMS2 and C3 genotypes, and protection from AMD associated with the promoter SNP rs2853707, though these observations require further confirmation. If replicated in other large populations, such observations suggest that mechanisms other than those previously demonstrated in studies of the CX3CR1 T280M variant could be involved in some cases of AMD.
Acknowledgments
This work was supported by NIH grants EY017362, EY013834, EY009611, EY014458, CA87969, CA49449, HL35464, CA34944, CA40360, HL26490, HL34595, CA47988, HL43851, HL46959.
References
- 1.Congdon N, O’Colmain B, Klaver CC, et al. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004 Apr;122(4):477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 2.Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007 Jan;125(1):55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 3.Schaumberg DA, Chasman D, Morrison MA, et al. Prospective study of common variants in the retinoic acid receptor-related orphan receptor alpha gene and risk of neovascular age-related macular degeneration. Arch Ophthalmol. 2010 Nov;128(11):1462–1471. doi: 10.1001/archophthalmol.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES. The new genomics: global views of biology. Science. 1996 Oct 25;274(5287):536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 5.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001 Sep;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 6.Hageman GS, Gehrs K, Lejnine S, et al. Clinical validation of a genetic model to estimate the risk of developing choroidal neovascular age-related macular degeneration. Hum Genomics. 2011 Jul 1;5(5):420–440. doi: 10.1186/1479-7364-5-5-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feigl B, Morris CP. The challenge of predicting macular degeneration. Curr Med Res Opin. 2011 Sep;27(9):1745–1748. doi: 10.1185/03007995.2011.603301. [DOI] [PubMed] [Google Scholar]
- 8.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009 Feb;5(2):e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuo J, Smith BC, Bojanowski CM, et al. The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. Faseb J. 2004 Aug;18(11):1297–1299. doi: 10.1096/fj.04-1862fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Hu J, Zhang J, Guan H. Polymorphisms in CFH, HTRA1 and CX3CR1 confer risk to exudative age-related macular degeneration in Han Chinese. Br J Ophthalmol. 2010 Sep;94(9):1211–1214. doi: 10.1136/bjo.2009.165811. [DOI] [PubMed] [Google Scholar]
- 11.Chan CC, Tuo J, Bojanowski CM, Csaky KG, Green WR. Detection of CX3CR1 single nucleotide polymorphism and expression on archived eyes with age-related macular degeneration. Histol Histopathol. 2005 Jul;20(3):857–863. doi: 10.14670/hh-20.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Sheets KG, Knott EJ, et al. Cellular and 3D optical coherence tomography assessment during the initiation and progression of retinal degeneration in the Ccl2/Cx3cr1-deficient mouse. Exp Eye Res. 2011 Nov;93(5):636–648. doi: 10.1016/j.exer.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Ohno-Matsui K, Nakahama K, et al. Amyloid beta enhances migration of endothelial progenitor cells by upregulating CX3CR1 in response to fractalkine, which may be associated with development of choroidal neovascularization. Arterioscler Thromb Vasc Biol. 2011 Jul;31(7):e11–18. doi: 10.1161/ATVBAHA.110.215517. [DOI] [PubMed] [Google Scholar]
- 14.Combadiere C, Feumi C, Raoul W, et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007 Oct;117(10):2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Xu G, Liu W, Ni Y, Zhou W. Role of Fractalkine/CX3CR1 Interaction in Light-Induced Photoreceptor Degeneration through Regulating Retinal Microglial Activation and Migration. PLoS One. 2012;7(4):e35446. doi: 10.1371/journal.pone.0035446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raoul W, Auvynet C, Camelo S, et al. CCL2/CCR2 and CX3CL1/CX3CR1 chemokine axes and their possible involvement in age-related macular degeneration. J Neuroinflammation. 2010;7:87. doi: 10.1186/1742-2094-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raoul W, Feumi C, Keller N, et al. Lipid-bloated subretinal microglial cells are at the origin of drusen appearance in CX3CR1-deficient mice. Ophthalmic Res. 2008;40(3-4):115–119. doi: 10.1159/000119860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raoul W, Keller N, Rodero M, Behar-Cohen F, Sennlaub F, Combadiere C. Role of the chemokine receptor CX3CR1 in the mobilization of phagocytic retinal microglial cells. J Neuroimmunol. 2008 Jul 31;198(1-2):56–61. doi: 10.1016/j.jneuroim.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Zerbib J, Puche N, Richard F, et al. No association between the T280M polymorphism of the CX3CR1 gene and exudative AMD. Exp Eye Res. 2011 Oct;93(4):382–386. doi: 10.1016/j.exer.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Steering Committee of the Physicians’ Health Study Research Group. Final report on the aspirin component of the ongoing Physicians’ Health Study. N Engl J Med. 1989;321(3):129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 21.A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119(10):1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christen WG, Glynn RJ, Manson JE, Ajani UA, Buring JE. A prospective study of cigarette smoking and risk of age-related macular degeneration in men. JAMA. 1996;276(14):1147–1151. [PubMed] [Google Scholar]
- 23.Cook NR, Lee IM, Gaziano JM, et al. Low-dose aspirin in the primary prevention of cancer: the Women’s Health Study: a randomized controlled trial. Jama. 2005 Jul 6;294(1):47–55. doi: 10.1001/jama.294.1.47. [DOI] [PubMed] [Google Scholar]
- 24.Rexrode KM, Lee IM, Cook NR, Hennekens CH, Buring JE. Baseline characteristics of participants in the Women’s Health Study. J Womens Health Gend Based Med. 2000 Jan-Feb;9(1):19–27. doi: 10.1089/152460900318911. [DOI] [PubMed] [Google Scholar]
- 25.Schaumberg DA, Christen WG, Buring JE, Glynn RJ, Rifai N, Ridker PM. High-sensitivity C-reactive protein, other markers of inflammation, and the incidence of macular degeneration in women. Arch Ophthalmol. 2007 Mar;125(3):300–305. doi: 10.1001/archopht.125.3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassuk SS, Albert CM, Cook NR, et al. The Women’s Antioxidant Cardiovascular Study: design and baseline characteristics of participants. J Womens Health (Larchmt) 2004 Jan-Feb;13(1):99–117. doi: 10.1089/154099904322836519. [DOI] [PubMed] [Google Scholar]
- 27.Belanger CF, Hennekens CH, Rosner B, Speizer FE. The Nurses’ Health Study. Am J Nurs. 1978 Jun;78(6):1039–1040. [PubMed] [Google Scholar]
- 28.Colditz GA, Rimm EB, Giovannucci E, Stampfer MJ, Rosner B, Willett WC. A prospective study of parental history of myocardial infarction and coronary artery disease in men. Am J Cardiol. 1991 May 1;67(11):933–938. doi: 10.1016/0002-9149(91)90163-f. [DOI] [PubMed] [Google Scholar]
- 29.Cho E, Hung S, Willett WC, et al. Prospective study of dietary fat and the risk of age-related macular degeneration. Am J Clin Nutr. 2001 Feb;73(2):209–218. doi: 10.1093/ajcn/73.2.209. [DOI] [PubMed] [Google Scholar]
- 30.Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA. 1996;276(14):1141–1146. [PubMed] [Google Scholar]
- 31.Christen WG, Glynn RJ, Chew EY, Albert CM, Manson JE. Folic acid, pyridoxine, and cyanocobalamin combination treatment and age-related macular degeneration in women: the Women’s Antioxidant and Folic Acid Cardiovascular Study. Arch Intern Med. 2009 Feb 23;169(4):335–341. doi: 10.1001/archinternmed.2008.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Akaike H. Information theory and an extension of the maximum likelihood principle. Paper presented at: Second International Symposium on Information Theory; 1973; Budapest. [Google Scholar]
- 34.Seddon JM, Rosner B, Sperduto RD, et al. Dietary fat and risk for advanced age-related macular degeneration. Arch Ophthalmol. 2001 Aug;119(8):1191–1199. doi: 10.1001/archopht.119.8.1191. [DOI] [PubMed] [Google Scholar]
- 35.Rothman KJ, Greenland S. Modern epidemiology. Second ed. Philadelphia: Lipincott-Raven; 1998. pp. 133–134. [Google Scholar]
- 36.Cho E, Seddon JM, Rosner B, Willett WC, Hankinson SE. Prospective study of intake of fruits, vegetables, vitamins, and carotenoids and risk of age-related maculopathy. Arch Ophthalmol. 2004 Jun;122(6):883–892. doi: 10.1001/archopht.122.6.883. [DOI] [PubMed] [Google Scholar]
- 37.Cho E, Hankinson SE, Willett WC, et al. Prospective study of alcohol consumption and the risk of age-related macular degeneration. Arch Ophthalmol. 2000 May;118(5):681–688. doi: 10.1001/archopht.118.5.681. [DOI] [PubMed] [Google Scholar]
- 38.White GE, Greaves DR. Fractalkine: a survivor’s guide: chemokines as antiapoptotic mediators. Arterioscler Thromb Vasc Biol. 2012 Mar;32(3):589–594. doi: 10.1161/ATVBAHA.111.237412. [DOI] [PubMed] [Google Scholar]
- 39.Savarin-Vuaillat C, Ransohoff RM. Chemokines and chemokine receptors in neurological disease: raise, retain, or reduce? Neurotherapeutics. 2007 Oct;4(4):590–601. doi: 10.1016/j.nurt.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prinz M, Priller J. Tickets to the brain: role of CCR2 and CX3CR1 in myeloid cell entry in the CNS. J Neuroimmunol. 2010 Jul 27;224(1-2):80–84. doi: 10.1016/j.jneuroim.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Damani MR, Zhao L, Fontainhas AM, Amaral J, Fariss RN, Wong WT. Age-related alterations in the dynamic behavior of microglia. Aging Cell. 2011 Apr;10(2):263–276. doi: 10.1111/j.1474-9726.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis CN, Harrison JK. Proline 326 in the C terminus of murine CX3CR1 prevents G-protein and phosphatidylinositol 3-kinase-dependent stimulation of Akt and extracellular signal-regulated kinase in Chinese hamster ovary cells. J Pharmacol Exp Ther. 2006 Jan;316(1):356–363. doi: 10.1124/jpet.105.093039. [DOI] [PubMed] [Google Scholar]
- 43.Brion M, Sanchez-Salorio M, Corton M, et al. Genetic association study of age-related macular degeneration in the Spanish population. Acta Ophthalmol. 2011 Feb;89(1):e12–22. doi: 10.1111/j.1755-3768.2010.02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu MM, Agron E, Chew E, et al. Copy number variations in candidate genes in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011 May;52(6):3129–3135. doi: 10.1167/iovs.10-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farrell KA, Withers SB, Holt CM. C-Myb function in the vessel wall. Front Biosci (Elite Ed) 2011;3:968–977. doi: 10.2741/e302. [DOI] [PubMed] [Google Scholar]
- 46.Tremblay K, Lemire M, Provost V, et al. Association study between the CX3CR1 gene and asthma. Genes Immun. 2006 Dec;7(8):632–639. doi: 10.1038/sj.gene.6364340. [DOI] [PubMed] [Google Scholar]
- 47.Sirois-Gagnon D, Chamberland A, Perron S, Brisson D, Gaudet D, Laprise C. Association of common polymorphisms in the fractalkine receptor (CX3CR1) with obesity. Obesity (Silver Spring) 2011 Jan;19(1):222–227. doi: 10.1038/oby.2010.125. [DOI] [PubMed] [Google Scholar]
- 48.Lee S, Varvel NH, Konerth ME, et al. CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer’s disease mouse models. Am J Pathol. 2010 Nov;177(5):2549–2562. doi: 10.2353/ajpath.2010.100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki F, Kubota T, Miyazaki Y, et al. Serum level of soluble CX3CL1/fractalkine is elevated in patients with polymyositis and dermatomyositis, which is correlated with disease activity. Arthritis Res Ther. 2012 Mar 6;14(2):R48. doi: 10.1186/ar3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark AK, Staniland AA, Malcangio M. Fractalkine/CX3CR1 signalling in chronic pain and inflammation. Curr Pharm Biotechnol. 2011 Oct;12(10):1707–1714. doi: 10.2174/138920111798357465. [DOI] [PubMed] [Google Scholar]
- 51.Ho L, van Leeuwen R, Witteman JC, et al. Reducing the genetic risk of age-related macular degeneration with dietary antioxidants, zinc, and omega-3 fatty acids: the Rotterdam study. Arch Ophthalmol. 2011 Jun;129(6):758–766. doi: 10.1001/archophthalmol.2011.141. [DOI] [PubMed] [Google Scholar]
- 52.Jun G, Nicolaou M, Morrison MA, et al. Influence of ROBO1 and RORA on risk of age-related macular degeneration reveals genetically distinct phenotypes in disease pathophysiology. PLoS One. 2011;6(10):e25775. doi: 10.1371/journal.pone.0025775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah R, Hinkle CC, Ferguson JF, et al. Fractalkine is a novel human adipochemokine associated with type 2 diabetes. Diabetes. 2011 May;60(5):1512–1518. doi: 10.2337/db10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuo J, Ross RJ, Herzlich AA, et al. A high omega-3 fatty acid diet reduces retinal lesions in a murine model of macular degeneration. Am J Pathol. 2009 Aug;175(2):799–807. doi: 10.2353/ajpath.2009.090089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]


