Abstract
Increased immunoglobulin G (IgG) response to dietary antigens can be associated with gastrointestinal dysfunction and autoimmunity. The underlying processes contributing to these adverse reactions remain largely unknown, and it is likely that genetic factors play a role. Here we estimate heritability and attempt to localize genetic factors influencing IgG antibody levels against food-derived antigens using an integrative genomics approach. IgG antibody levels were determined by ELISA in >1300 Mexican Americans for the following food antigens: wheat gliadin; bovine casein; and two forms of bovine serum albumin (BSA-a and BSA-b). Pedigree-based variance components methods were used to estimate additive genetic heritability (h2), perform genome-wide association analyses, and identify transcriptional signatures (based on 19,858 transcripts from peripheral blood lymphocytes). Heritability estimates were significant for all traits (0.15-0.53), and shared environment (based on shared residency among study participants) was significant for casein (0.09) and BSA-a (0.33). Genome-wide significant evidence of association was obtained only for antibody to gliadin (p=8.57×10-8), mapping to the human leukocyte antigen II region, with HLA-DRA and BTNL2 as the best candidate genes. Lack of association of known celiac disease risk alleles HLA-DQ2.5 and -DQ8 with anti-gliadin antibodies in the studied population suggests a separate genetic etiology. Significant transcriptional signatures were found for all IgG levels except BSA-b. These results demonstrate that individual genetic differences contribute to food antigen antibody measures in this population. Further investigations may elucidate the underlying immunological processes involved.
Keywords: IgG antibody, gliadin, integrative genomics, association, transcriptional profiling, pedigree study
Introduction
Adverse reactions to dietary substances are common in the U.S. general population, with most of the affected individuals experiencing gastrointestinal, skin, and/or respiratory symptoms. Reactions include IgE-mediated allergies that affect an estimated 4-6% of children and 1-2% of adults [Patel, et al. 2011] and, more commonly, food intolerances, which can be either immune or non-immune mediated. Food intolerances are often dose-dependent and generally take longer to become symptomatic than allergic reactions [Skypala 2011]. Food allergies and intolerances have been associated with atopic diseases (e.g., asthma, rhinitis and eczema) and autoimmune disorders (such as celiac disease, CD) [Briani, et al. 2008; Tan and Corren 2011]. CD is an autoimmune enteropathy triggered by gluten proteins of wheat and related cereal grains in genetically susceptible individuals that affects an estimated 2-3 million people in the U.S [Fasano, et al. 2003].
Individuals are thought to be universally exposed to a wide variety of food proteins (although exposure to some food items varies by culture). Differences in the level of antibodies produced in response to food antigens are therefore likely due in part to genetic differences. Previous research indicates that a hyperactive immune response to certain food proteins tends to run in families, and genetic factors have been implicated in some instances [Hong, et al. 2009;Liu, et al. 2009; Tsai, et al. 2009]. For example, certain human leukocyte antigen (HLA) alleles are present in higher frequencies in allergic individuals than in controls (e.g. peanut allergy (HLA-DRB1, HLA-DQB1, and HLA-DPB1 gene polymorphisms) and apple allergy (HLA-DRB1*07 allele) [Howell, et al. 1998; Senechal, et al. 1999]) and HLA-DQ variants have long been known to predispose to CD [Louka and Sollid 2003]. There is also evidence suggesting that CD14, IL10, IL13, and SPINK5 gene polymorphisms may predispose to food allergy and/or sensitization in general [Campos Alberto, et al. 2008; Kusunoki, et al. 2005; Woo, et al. 2003]. However, the underlying disease processes contributing to adverse reactions to many food proteins, especially those of non-allergic etiology, remain largely unknown.
Materials and Methods
Participants
Individuals participating in this study consisted of 1367 members of randomly ascertained, extended Mexican American families from San Antonio, TX, who were recruited for participation in the San Antonio Family Heart Study (SAFHS), which seeks to identify cardiovascular disease risk factors [Mitchell, et al. 1996]. Up to 6 generations and 63 families are represented in the sample, as previously described [Rubicz, et al. 2013]. Initial recruitment took place during the years 1991-1995. Participants range in age from 16 to 94 years (with a mean of 39 years) and they consist of 816 women and 551 men. The Institutional Review Board of the University of Texas Health Science Center at San Antonio approved the study protocols, and all participants signed statements of informed consent.
Serology
Following an overnight fast, blood samples were collected from participants using EDTA vacutainers at the time of recruitment (1991-1995). Frozen plasma aliquots were obtained as previously described [Cheng, et al. 1986] and carefully stored at -80°C, until their recent use for antibody determinations. ELISA kits were used to determine IgG antibody titers to: gliadin [Samaroo, et al. 2010]; bovine casein [Niebuhr, et al. 2011; Severance, et al. 2011]; and two forms of bovine serum albumin consisting of complete BSA (BSA-a) and a more-purified form derived from Cohn Fraction V (BSA-b) [Sheridan and Simmons 1983]. Antigens for these assays were obtained from the Sigma-Aldrich Chemical Company, St. Louis, MO (catalogue numbers are: casein C7078; gliadin G3375; BSA-a A9647; and BSA-b 85040C).
SNP Genotyping
DNA samples extracted from lymphocytes were typed for SNPs using several versions of Illumina's SNP genotyping BeadChip microarrays (HumanHap550v3, HumanExon510Sv1, Human1Mv1, Human1M-Duov3) according to the Illumina Infinium Protocol (Illumina, San Diego, CA) and underwent stringent quality control measures prior to analysis, as previously described [Rubicz, et al. 2013]. SNPs were excluded if they had a low call rate, were monomorphic, had a minor allele in <10 individuals, and if Hardy-Weinberg test statistics (calculated in SOLAR [Almasy and Blangero 1998]) were p ≤ 10-4. SNPs overlapping between the different microarray versions were kept for further processing. SNP genotypes were checked for Mendelian consistency with SimWalk2 [Sobel, et al. 2002], and the most likely incorrect genotype was blanked. Subsequently, the blanked genotypes were re-imputed using MERLIN [Abecasis, et al. 2002] (based on local relatives' genotypes, or if uncertain then using a weighted average of possible genotypes). Allele frequencies were calculated for the resulting 944,565 SNPs using maximum likelihood estimates in SOLAR [Almasy and Blangero 1998].
Statistical methodology
Variance components (VC) methods were used to estimate heritability (h2), defined as the aggregate additive autosomal genetic effects, on the antibody levels to each food antigen using SOLAR [Almasy and Blangero 1998]. Prior to analysis, the quantitative antibody level measurements were transformed using a rank-based inverse normalization, as VC analysis can be sensitive to non-normality, particularly high kurtosis. The covariates sex, age, and their interactions were included in all statistical analyses. In addition, a household variance component (based on co-habitation of family members at the time of the study, including both genetically related and unrelated individuals) was used to model the influence of shared environmental factors [Spence, et al. 1977].
To determine if there is overlap in the additive genetic factors influencing the food antigen IgG measurements, bivariate heritability analyses were conducted in SOLAR for each pair of antibody traits [Almasy and Blangero 1998; Boehnke, et al. 1986; Lange and Boehnke 1983]. Genome-wide association analyses were run in SOLAR [Almasy and Blangero 1998] using 944,565 SNPs. Association analyses were conducted using an additive measured genotype model, allowing for non-independence of family members due to shared genes and shared environment. We empirically estimated the genome-wide significance threshold (at α=0.05) for our sample to be p≤1.3×10-7 using a large number of genome-wide analyses on simulated traits unlinked to the genotyped SNPS, using the true pedigree structures and genome-wide SNP genotypes for analysis of each replicate. Principal components analysis (PCA) was conducted on SNP genotypes to identify principal components (PCs) that can be used to correct for differences in ancestral ethic contributions to study participants, as population substructure may produce spurious results in association analyses [Price, et al. 2006]. PCA (using R princomp) [R Development Core Team 2011] was run on all individuals contributing independent haplotypes to the sample, including all founders; offspring were assigned the average of both their parents' values for each PC. This was done so that the PCA would not unknowingly capture differences between pedigrees per se, as real differences between pedigrees (in particular among rare variation) may exist and aid in the localization of genetic effects. We included the first five PCs as additional covariates in all statistical analyses.
Transcriptional Profiles
Gene expression data that were generated from peripheral blood lymphocyte samples collected from participants at the same time as the plasma samples used for antibody determinations (as previously described [Goring, et al. 2007]) were also analyzed. Microarray-based expression data were available for 1,243 study participants, and the raw and normalized expression values are available under accession number E-TABM-305 at http://www.ebi.ac.uk/arrayexpress. Sample quality was assessed by comparing the number of expressed probes (at p ≤ 0.05), the mean expression across expressed probes, and the mean correlation across expressed probes with other samples. 20,643 significantly expressed probes were identified at a false discovery rate (FDR) of ≤ 0.05, using a one-sided binomial test based on counts of successful compared to unsuccessful detection at p ≤ 0.05. Significantly expressed transcripts next underwent background noise correction, log2 transformation, and quantile normalization. Of these, 19,858 transcripts mapped to known genes and were included in the analyses here. We examined whether any SNPs that were significantly associated with the food antigen antibody traits were also associated with expression levels of neighboring transcripts, which would suggest these SNPs may be cis-regulatory variants. In addition, we tested more generally whether the expression levels of any transcripts were significantly correlated with food antigen antibody levels. A regression model was implemented in SOLAR [Almasy and Blangero 1998] with transcript level as the focal covariate for IgG antibody level, while simultaneously taking into account the non-independence of related individuals. Significance was tested using a likelihood ratio statistic. Prior to correlating expression levels with the food antigen antibody level traits, the relationship between each of the top 50 expression PCs and the antibody level traits were examined (by regression analysis) and were regressed out except for those that were significantly correlated, so that any true connections between transcripts and food antigen antibody traits were not removed. Transcripts significantly correlated with the antibody traits (using an FDR of 0.05 to account for multiple testing) were analyzed using Ingenuity Pathway Analysis (IPA) version 6.3 [Ingenuity Systems] to identify possible connections (pathways) between identified transcripts.
Results
Antibody levels
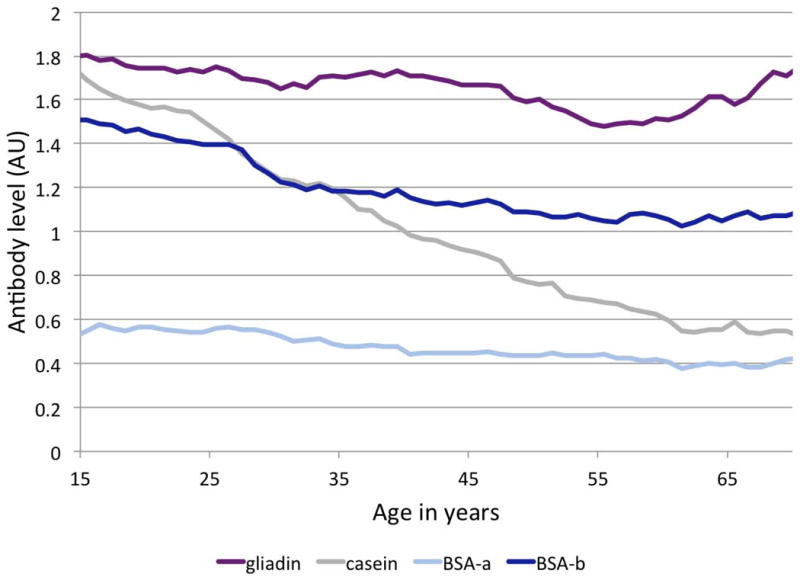
IgG antibody levels to four food antigens (gliadin, casein, BSA-a, BSA-b) were measured in a sample of 1367 Mexican Americans participating in the San Antonio Family Heart Study. Figure 1 shows antibody levels across individuals of different ages in this cross-sectional study, with most antibody levels decreasing over the examined age range of 16 to 94 years, and a slight increase for some antibody levels in the older age categories. IgG levels to casein demonstrated the largest difference between young and old age categories. All four antibodies were detected in a substantial number of study participants, suggesting that food intolerances to these four food antigens are not rare in the studied Mexican American population.
Figure 1.
Heritability estimates
We investigated whether additive genetic factors in the aggregate influenced the levels of antibodies. Heritability estimates were significant for antibodies against all of the food antigens (Table I). Estimates ranged from h2 = 0.15 for BSA-a to h2 = 0.53 for gliadin. Shared household, a proxy for shared environmental exposures based on co-habitation at the same residence at the time of sample collection, was significant for only casein (0.09) and BSA-a (0.33). With the exception of BSA-a, the shared household effects were smaller in magnitude than the estimated heritabilities. We performed bivariate heritability analysis to determine whether and to what extend genetic factors influencing the different IgG antibody levels might overlap. Genetic factors are significantly positively correlated for most pairs of food antigen antibody measures (supplementary Table S1), ranging from 0.22 to 0.52, indicating that a substantial proportion of the underlying genetic variation is shared among these four food IgG antibodies.
Table I.
Heritability estimates of food antigen IgG measurements with household effects
| Trait | h2 (heritability) | p-value (h2) | c2 (household effects) | p-value (c2) |
|---|---|---|---|---|
| gliadin | 0.53 | 2.3×10 -25 | 0.06 | 0.04 |
| casein | 0.44 | 2.4×10-13 | 0.09 | 0.01 |
| BSA-a | 0.15 | 1.5×10-3 | 0.33 | 1.0×10-7 |
| BSA-b | 0.21 | 1.1×10-4 | 0.09 | 0.02 |
Bold=significant after correction for multiple testing (p=0.05/4 traits = 0.0125)
Genome-wide association analysis
To localize specific genetic factors influencing these traits, we conducted genome-wide association analysis. We estimated empirically that a pointwise p ≤ 1.3×10-7 corresponds to a genome-wide p-value of ≤0.05 for our Mexican American cohort, which consists of large multigenerational families. Genome-wide significant results were obtained for antibodies against gliadin in the HLA II region of chromosome 6, with HLA-DRA and BTNL2 being the closest candidate genes (top SNP rs3135350, p = 8.6×10-8) (Table II, Figure 2). As seen in the regional plot of the extended HLA region, several other SNPs are in LD with rs3135350, including rs3129860 (r2 = 0.88, p = 8.8×10-8, Figure 3). After conditional association analysis on rs3135350, no other SNPs are significantly associated with antibody to gliadin, suggesting a single locus rather than multiple nearby loci. The quantile-quantile plot of observed p-values shows that the HLA region accounts for the deviation from the diagonal line of observed versus expected p-values under the null hypothesis, as there is no evidence of inflation of significant levels after excluding the HLA region from the plot (supplementary Figure S1). We examined whether the four HLA SNPs that were significantly associated with anti-gliadin IgG levels were also significantly associated with the other food antigens. Non-significant p-values, after correcting for multiple testing, were obtained for casein and BSA-a for two gliadin SNPs (rs3135350 and rs3129860) (Table III). However, the p-values for casein and BSA-a were quite small (<0.1) and the effect was in the same direction, suggesting that the locus may not be unique to gliadin.
Table II.
Significant and suggestive results of genome-wide association analysis for food antigen IgG measurements
| Trait | SNP | SNP location: chromosome | SNP location: base pair | Nearest gene | Minor allele | Minor allele frequency* | p-value (ßSNP)** |
|---|---|---|---|---|---|---|---|
| gliadin | rs4848780 | 2 | 122940870 | TSN | T | 0.335 | 8.25×10-7 (-0.21) |
| rs3130320 | 6 | 32331236 | NOTCH4 | T | 0.197 | 4.48×10-7 (0.27) | |
| rs3117116 | 6 | 32474995 | BTNL2 | C | 0.069 | 1.00×10-6 (0.42) | |
| rs3135352 | 6 | 32500884 | HLA-DRA/BTNL2 | G | 0.049 | 1.12×10-7(0.50) | |
| rs3135350 | 6 | 32500959 | HLA-DRA/BTNL2 | G | 0.046 | 8.57×10-8(0.51) | |
| rs3129971 | 6 | 32501213 | HLA-DRA/BTNL2 | G | 0.049 | 3.40×10-7 (0.48) | |
| rs3129860 | 6 | 32509057 | HLA-DRA | A | 0.060 | 8.76×10-8(0.47) | |
| rs3135391 | 6 | 32518965 | HLA-DRA | T | 0.051 | 1.39×10-7 (0.49) | |
| rs8084 | 6 | 32519013 | HLA-DRA | A | 0.403 | 6.34×10-7 (0.21) | |
| rs7192 | 6 | 32519624 | HLA-DRA | T | 0.389 | 6.50×10-7 (0.21) | |
| rs7194 | 6 | 32520458 | HLA-DRA | G | 0.391 | 4.31×10-7 (0.21) | |
| rs7195 | 6 | 32520517 | HLA-DRA | A | 0.391 | 4.31×10-7 (0.21) | |
| rs3135388 | 6 | 32521029 | HLA-DRA | T | 0.049 | 1.12×10-7(0.50) | |
| rs2213586 | 6 | 32521072 | HLA-DRA | T | 0.391 | 4.31×10-7 (0.21) | |
| rs2213585 | 6 | 32521128 | HLA-DRA | C | 0.391 | 4.31×10-7 (0.21) | |
| rs2227139 | 6 | 32521437 | HLA-DRA | C | 0.391 | 4.31×10-7 (0.21) | |
| rs3129889 | 6 | 32521523 | HLA-DRA | G | 0.049 | 4.31×10-7 (0.50) | |
| casein | rs11635085 | 15 | 90847640 | C15orf32 | G | 0.107 | 6.47×10-7 (-0.30) |
| BSA-a | rs7562116 | 2 | 231816490 | ARMC9 | G | 0.223 | 1.30×10-6 (0.22) |
| rs9677476 | 2 | 231834534 | ARMC9 | G | 0.274 | 1.11×10-6 (0.21) | |
| rs13415839 | 2 | 231834778 | ARMC9 | T | 0.270 | 1.29×10-6 (0.21) | |
| BSA-b | rs2360969 | 2 | 208081241 | CREB1 | T | 0.413 | 1.24×10-6 (-0.21) |
| rs1372356 | 14 | 87585702 | GPR65 | C | 0.489 | 5.81×10-7 (-0.20) | |
| rs2236267 | 14 | 87702074 | KCNK10 | C | 0.462 | 4.13×10-7 (0.20) |
Bold=genome-wide significant at p≤1.3×10-7; significance threshold was estimated based on our sample of Mexican American participants. The threshold for suggestive evidence for association used here is p≤1.3×10-6.
Minor allele frequency was estimated for our study sample using maximum likelihood methods and taking family relationships into account.
The direction of effect of the regression coefficient is relative to the minor SNP allele (e.g., for SNP rs3135350, the minor allele is associated with an increase in IgG anti-gliadin antibody level).
Figure 2.
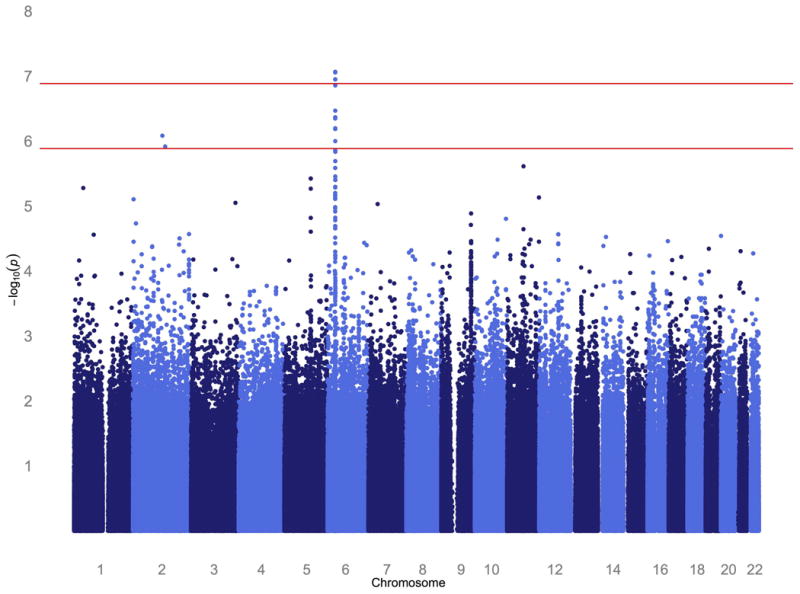
Figure 3.
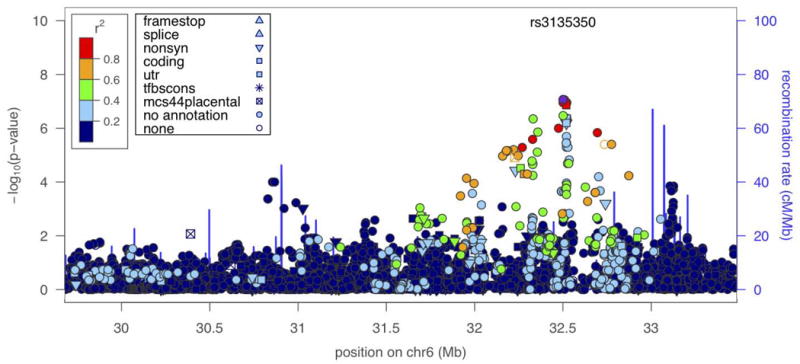
Table III.
Association results for food antigen IgG traits for significant gliadin SNPs
| Trait | rs3135352 p-value (ßSNP)* | rs3135350 p-value (ßSNP)* | rs3129860 p-value (ßSNP)* | rs3135388 p-value(ßSNP)* |
|---|---|---|---|---|
| gliadin | 1.12×10-7(0.50) | 8.57×10-8(0.51) | 8.76×10-8(0.47) | 1.12×10-7(0.50) |
| casein | 0.07 (0.16) | 0.04 (0.19) | 0.05 (0.16) | 0.07 (0.16) |
| BSA-a | 0.07 (0.16) | 0.05 (0.18) | 0.05 (0.17) | 0.07 (0.16) |
| BSA-b | 0.24 (0.11) | 0.30 (0.10) | 0.13 (0.14) | 0.24 (0.11) |
Note that results for casein and BSA-a antigen traits are not significant after applying a stringent Bonferroni correction for multiple testing (p = 0.05/4 food antigen traits = 0.0125). However, the fact that the p-values are fairly small, with effect sizes in the same direction, suggests that it may be possible that the locus is not unique to gliadin.
Transcript analysis
To pinpoint genes that may influence the measures of antibody to food antigens, we took an integrative genomics approach in which we analyzed gene expression profiles from peripheral blood lymphocytes collected at the same time as the plasma samples used for antibody determinations. We investigated whether the four significant SNPs associated with the anti-gliadin antibody trait (rs3135352, rs3135350, rs3129860, and rs3135388) were also significantly associated with the expression levels of any neighboring transcripts, which would indicate that these SNPs are potential cis-acting regulatory variants. There were 150 expressed transcripts in the HLA region, yielding 600 SNP-transcript pairs. After correcting for multiple testing (p = 0.05/600 = 8.33×10-5) there were no significantly associated SNP-transcript pairs. The smallest p-value was p = 2.20×10 -3 for rs3129860 and the expression of HLA-DQB1.
We also used the transcriptional profiles as a separate way to identify genes related (though not necessarily causally) to the measured antibodies, by investigating whether transcript levels were correlated with IgG level, while simultaneously modeling shared genetic and environmental factors among relatives and household members. We identified significant transcriptional correlates for three antibody traits (supplementary Table S2). After applying Bonferroni correction for multiple testing (p = 0.05/19858 transcripts = 2.52×10-6), 21 transcripts were significantly associated with anti-gliadin antibody levels, two with anti-casein antibodies, 175 with anti-BSA-a antibody level, and none for BSA-b. There was little overlap of significant transcripts among the different IgG antibody measures. At a less stringent 0.05 FDR, there were 387 significantly correlated transcripts for gliadin, 413 for casein, 10,402 for BSA-a, and 0 for BSA-b. The discrepancy between the two different measures of IgG antibodies against BSA may be due to BSA-a containing more impurities.
The FDR 0.05 significant transcripts were then subjected to network analysis using IPA. An FDR 0.01 level was used for BSA-a to deal with a more manageable number of transcripts (1,231) for pathway analysis. Supplementary Table S3 gives highly significant functional categories of transcripts and numbers of genes involved. Highly significant functional categories across the food antigen traits were inflammatory response, cell death, cellular proliferation and differentiation, and autoimmunity (including rheumatoid arthritis for BSA-a). Results of IPA canonical pathways assignments (according to generalized pathways from the IPA library) for the transcript analyses are provided in supplementary Table S4, along with the number of genes in each pathway. Several canonical pathway categories are involved in inflammatory response, including IL-10 and CD40 signaling.
Relationship of gliadin trait to celiac disease
Given that gliadin is the prime environmental trigger for celiac disease and the condition is closely associated with antibody response to gliadin, we investigated whether published genome-wide significant susceptibility loci for CD show evidence for association with anti-gliadin antibodies in this study (supplementary Table S5). After applying a Bonferroni correction for examining 55 CD-related SNPs, a single SNP (rs10188217, p = 3.5×10-4) was significantly associated with the gliadin trait. This SNP, rs10188217, is located on chromosome 2, in the PUS10 (pseudouridylate synthase 10) gene. A nearby SNP, rs13003464, also showed a fairly small p-value (p = 5.2×10-3), and is in LD with rs10188217 (r2 = 0.87).
Discussion
This study demonstrates that individual genetic differences contribute to anti-food antigen IgG levels in this sample of Mexican Americans. Shared household is also a significant contributing factor to some antibody measures, probably due to similar exposure to dietary antigens given that individuals residing in the same household are likely to share meals and dietary habits. Our results are in agreement with other studies that demonstrate adverse reaction to food antigens is due to both genetic and environmental factors [Nistico, et al. 2006]. However, to our knowledge, heritability estimates have not been published for these IgG measures. IgG antibody response to dietary antigens has been suggested as a measure of food sensitivity in some individuals. However, it contrasts greatly with IgE antibody response, which is involved in classic allergic reaction to food antigens (i.e., immediate immune reaction that occurs within minutes to hours of exposure) [Vojdani 2009]. Previously published significant heritability estimates for IgE antibodies against milk (h2=0.15) and wheat (h2=0.35) antigens [Tsai, et al. 2009] are somewhat lower than our estimates for IgG antibodies to casein (h2=0.44) and gliadin (h2=0.53). Our bivariate analysis indicates that some of the underlying genetic factors appear to be shared between these IgG antibody traits.
Individuals with food sensitivities are reported to be at greater risk for atopic diseases, including asthma, allergic rhinitis and eczema, which tend to cluster in families [Kiyohara, et al. 2008; Kusunoki, et al. 2005; Liu, et al. 2009; Vojdani 2009]. In addition, there is overlap between genetic risk factors for adverse food reactions and autoimmune diseases such as inflammatory bowel disease [Parmar, et al. 2012]. The inflammatory response and symptoms associated with CD, another autoimmune condition, are triggered by the ingestion of gluten proteins, and it is estimated that 15-20% of individuals with CD will develop other autoimmune diseases [Cosnes, et al. 2008]. Our analysis of gene expression data in this study provides further support for common mechanisms of these conditions, as we identified significant functional pathways related to autoimmune disease (rheumatoid arthritis) for the BSA-a IgG antibody trait, and dermatological conditions for anti-gliadin IgGs.
Our genome-wide investigation for genetic factors influencing food antigen IgG antibody levels identified a significant association of HLA class II genes HLA-DR and BTNL2 at 6p21.3 with anti-gliadin antibodies. Although gliadin is the main environmental trigger for CD and elevated levels of antibodies against gluten are associated with the disease, the genes that we identify here are not known CD risk factors: HLA-DRA is involved in binding peptides derived from antigens for recognition by CD4 T-cells; and BTNL2 has been identified as a risk factor for sarcoidosis and ulcerative colitis (both are diseases with an autoimmune background) that likely functions to co-stimulate T-cells [Rybicki, et al. 2005]. CD requires priming of CD4 T-cells by gliadin peptides, which then accumulate in the lamina propria where they produce the cytokine IFN-γ that contributes to intestinal inflammation, and it is possible that the genes identified here are involved in this process. On the other hand, these genetic factors may be associated with the IgG antibody response to gluten in non-celiac gluten sensitivity [Lundin and Alaedini 2012]. We did not successfully map major loci using our modest sample size (by GWAS standards for commonly measured traits and diseases) for the remaining food antigen IgG measures, which suggests a complex genetic architecture underlying these traits.
Among individuals with CD, an estimated 95% are known to carry at least one copy of the HLA-DQ2.5 and -DQ8 risk haplotypes, coded for by alleles DQA1*05/DQB1*02 and DQA1*0505/DQB1*0301, respectively [Monsuur, et al. 2008; Sollid, et al. 1989; Sollid, et al. 2012]. In our study we did not find a significant association of level of antibodies against gliadin antigen with these CD risk haplotypes using HLA tagging SNPs rs2187668 (for DQ2.5) and rs7454108 (for DQ8) as described by Monsuur et al. 2008. As most individuals with these risk alleles will not develop CD upon exposure to gluten, other factors must also be involved, and additional loci, particularly in the HLA region, are implicated in CD development and severity [Cosnes, et al. 2008; Henderson, et al. 2007; Kim, et al. 2004]. Meta-analyses and dense genotyping using the Immunochip have increased the number of CD risk loci to >40 [Dubois, et al. 2010; Trynka, et al. 2011]. We found evidence for association of the anti-gliadin antibody trait with CD susceptibility locus rs10188217 on chromosome 2 in PUS10 (also a risk locus for ulcerative colitis and Crohn's disease [Festen, et al. 2011; McGovern, et al. 2010]) and our analysis of the gene expression data identified an overlap with CD susceptibility gene BACH2 [McGovern, et al. 2010]. These results point to some genetic overlap between anti-gliadin antibody levels and CD. However, the non-significant findings for the major CD risk haplotypes indicates that although there may be similar underlying mechanisms leading to these disease states, they in fact likely have largely separate etiologies. Shared factors such as increased intestinal permeability could contribute to both gluten sensitivity and CD, but the immune response in these two disease processes might be directed at different antigenic determinants (i.e., different gluten proteins and/or epitopes) [Samaroo, et al. 2010].
To summarize, here we demonstrated that genetic and environmental (e.g., shared residence) factors significantly contributed to levels of IgG antibodies against several food antigens, and many pairs of antibody traits appear to be influenced by shared genetic factors. We identified a significant association of anti-gliadin IgG with the HLA II region, which is involved in immune function, and we report some genetic overlap between anti-gliadin antibodies and CD risk alleles. Our analysis of gene expression data (from peripheral blood lymphocytes) identified significant correlations between transcripts with several IgG traits, as well as a number of functional categories that appeared to be related to levels of these antibodies, though the directionality of effect is uncertain at the moment. Our study has a number of limitations. It would be useful to have dietary information to examine the relationship between antibody presence and level and exposure to the studied food antigens, but detailed exposure information was not available. Also, the size of our study is moderate, in particular when compared to current meta-analytic GWAS on common diseases and commonly measured quantitative traits. Hopefully this investigation will be a stepping-stone towards future investigations and combining different data sources to increase the sample size, permitting identification of the specific causal variants and the mechanisms by which they contribute to the food antigen sensitivity and possibly atopic and/or autoimmune diseases.
Supplementary Material
Acknowledgments
We thank the participants of the San Antonio Family Heart Study. Grants from the US National Heart, Lung and Blood Institute (HL045522 and HL080149) and the National Institute for Mental Health (MH0708143, MH078111 and MH083824) supported the data collection and analysis. Serological assays were supported by the Stanley Medical Research Institute. Transcriptional profiling was supported by the Azar and Shepperd families. The statistical computer package SOLAR is supported by a grant from the US National Institute of Mental Health (MH059490). Analyses were conducted in facilities constructed with support from the National Center for Research Resources (RR017515) and a gift from the SBC Foundation. The NIH LRP health disparities division provided additional support. The authors declare no conflict of interest.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke M, Moll PP, Lange K, Weidman WH, Kottke BA. Univariate and bivariate analyses of cholesterol and triglyceride levels in pedigrees. Am J Med Genet. 1986;23(3):775–792. doi: 10.1002/ajmg.1320230306. [DOI] [PubMed] [Google Scholar]
- Briani C, Samaroo D, Alaedini A. Celiac disease: from gluten to autoimmunity. Autoimmun Rev. 2008;7(8):644–650. doi: 10.1016/j.autrev.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Campos Alberto EJ, Shimojo N, Suzuki Y, Mashimo Y, Arima T, Matsuura T, Inoue Y, Yamaide A, Tomiita M, Fujii K, et al. IL-10 gene polymorphism, but not TGF-beta1 gene polymorphisms, is associated with food allergy in a Japanese population. Pediatr Allergy Immunol. 2008;19(8):716–721. doi: 10.1111/j.1399-3038.2007.00709.x. [DOI] [PubMed] [Google Scholar]
- Cheng ML, Woodford SC, Hilburn JL, VandeBerg JL. A novel system for storage of sera frozen in small aliquots. J Biochem and Biophys Methods. 1986;13(1):47–51. doi: 10.1016/0165-022x(86)90007-2. [DOI] [PubMed] [Google Scholar]
- Cosnes J, Cellier C, Viola S, Colombel JF, Michaud L, Sarles J, Hugot JP, Ginies JL, Dabadie A, Mouterde O, et al. Incidence of autoimmune diseases in celiac disease: protective effect of the gluten-free diet. Clin Gastroenterol Hepatol. 2008;6(7):753–758. doi: 10.1016/j.cgh.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, Zhernakova A, Heap GA, Adany R, Aromaa A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano A, Berti I, Gerarduzzi T, Not T, Colletti RB, Drago S, Elitsur Y, Green PH, Guandalini S, Hill ID, et al. Prevalence of celiac disease in at-risk and not-at-risk groups in the United States: a large multicenter study. Arch Intern Med. 2003;163(3):286–292. doi: 10.1001/archinte.163.3.286. [DOI] [PubMed] [Google Scholar]
- Festen EA, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G, Dubois PC, Lagace C, Stokkers PC, Hommes DW, et al. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn's disease and celiac disease. PLoS Genet. 2011;7(1):e1001283. doi: 10.1371/journal.pgen.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39(10):1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Henderson KN, Tye-Din JA, Reid HH, Chen Z, Borg NA, Beissbarth T, Tatham A, Mannering SI, Purcell AW, Dudek NL, et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity. 2007;27(1):23–34. doi: 10.1016/j.immuni.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Hong X, Tsai HJ, Wang X. Genetics of food allergy. Curr Opin Pediatr. 2009;21(6):770–776. doi: 10.1097/MOP.0b013e32833252dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell WM, Turner SJ, Hourihane JO, Dean TP, Warner JO. HLA class II DRB1, DQB1 and DPB1 genotypic associations with peanut allergy: evidence from a family-based and case-control study. Clin Exp Allergy. 1998;28(2):156–162. doi: 10.1046/j.1365-2222.1998.00224.x. Ingenuity Systems, http://www.ingenuity.com. [DOI] [PubMed] [Google Scholar]
- Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci USA. 2004;101(12):4175–4179. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara C, Tanaka K, Miyake Y. Genetic susceptibility to atopic dermatitis. Allergol Int. 2008;57(1):39–56. doi: 10.2332/allergolint.R-07-150. [DOI] [PubMed] [Google Scholar]
- Kusunoki T, Okafuji I, Yoshioka T, Saito M, Nishikomori R, Heike T, Sugai M, Shimizu A, Nakahata T. SPINK5 polymorphism is associated with disease severity and food allergy in children with atopic dermatitis. J Allergy Clin Immunol. 2005;115(3):636–638. doi: 10.1016/j.jaci.2004.12.1114. [DOI] [PubMed] [Google Scholar]
- Lange K, Boehnke M. Extensions to pedigree analysis. IV. Covariance components models for multivariate traits. Am J Med Genet. 1983;14(3):513–524. doi: 10.1002/ajmg.1320140315. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang S, Tsai HJ, Hong X, Wang B, Fang Y, Pongracic JA, Wang X. Genetic and environmental contributions to allergen sensitization in a Chinese twin study. Clin Exp Allergy. 2009;39(7):991–998. doi: 10.1111/j.1365-2222.2009.03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louka AS, Sollid LM. HLA in coeliac disease: unravelling the complex genetics of a complex disorder. Tissue Antigens. 2003;61(2):105–117. doi: 10.1034/j.1399-0039.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- Lundin KE, Alaedini A. Non-celiac gluten sensitivity. Gastrointest Endosc Clin N Am. 2012;22(4):723–734. doi: 10.1016/j.giec.2012.07.006. [DOI] [PubMed] [Google Scholar]
- McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42(4):332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94(9):2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- Monsuur AJ, de Bakker PI, Zhernakova A, Pinto D, Verduijn W, Romanos J, Auricchio R, Lopez A, van Heel DA, Crusius JB, et al. Effective detection of human leukocyte antigen risk alleles in celiac disease using tag single nucleotide polymorphisms. PloS One. 2008;3(5):e2270. doi: 10.1371/journal.pone.0002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr DW, Li Y, Cowan DN, Weber NS, Fisher JA, Ford GM, Yolken R. Association between bovine casein antibody and new onset schizophrenia among US military personnel. Schizophr Res. 2011;128(1-3):51–55. doi: 10.1016/j.schres.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Nistico L, Fagnani C, Coto I, Percopo S, Cotichini R, Limongelli MG, Paparo F, D'Alfonso S, Giordano M, Sferlazzas C, et al. Concordance, disease progression, and heritability of coeliac disease in Italian twins. Gut. 2006;55(6):803–808. doi: 10.1136/gut.2005.083964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar AS, Lappalainen M, Paavola-Sakki P, Halme L, Farkkila M, Turunen U, Kontula K, Aromaa A, Salomaa V, Peltonen L, et al. Association of celiac disease genes with inflammatory bowel disease in Finnish and Swedish patients. Genes Immun. 2012;13(6):474–480. doi: 10.1038/gene.2012.21. [DOI] [PubMed] [Google Scholar]
- Patel DA, Holdford DA, Edwards E, Carroll NV. Estimating the economic burden of food-induced allergic reactions and anaphylaxis in the United States. J Allergy Clin Immunol. 2011;128(1):110–115. doi: 10.1016/j.jaci.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. Vienna, Austria: 2011. R: A language and environment for statistical computing R Foundation for Statistical Computing. URL http://www.R-project.org/ [Google Scholar]
- Rubicz R, Yolken R, Drigalenko E, Carless MA, Dyer TD, Bauman L, Melton PE, Kent JW, Jr, Harley JB, Curran JE, et al. A genome-wide integrative genomic study localizes genetic factors influencing antibodies against Epstein-Barr virus nuclear antigen 1 (EBNA-1) PLoS Genetics. 2013;9(1):e1003147. doi: 10.1371/journal.pgen.1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicki BA, Walewski JL, Maliarik MJ, Kian H, Iannuzzi MC. The BTNL2 gene and sarcoidosis susceptibility in African Americans and Whites. Am J Hum Genet. 2005;77(3):491–499. doi: 10.1086/444435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaroo D, Dickerson F, Kasarda DD, Green PH, Briani C, Yolken RH, Alaedini A. Novel immune response to gluten in individuals with schizophrenia. Schizophr Res. 2010;118(1-3):248–255. doi: 10.1016/j.schres.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal H, Geny S, Desvaux FX, Busson M, Mayer C, Aron Y, Oster JP, Bessot JC, Peltre G, Pauli G, et al. Genetics and specific immune response in allergy to birch pollen and food: evidence of a strong, positive association between atopy and the HLA class II allele HLA-DR7. J Allergy Clin Immunol. 1999;104:395–401. doi: 10.1016/s0091-6749(99)70384-2. [DOI] [PubMed] [Google Scholar]
- Severance EG, Lin J, Sampson HA, Gimenez G, Dickerson FB, Halling M, Gressitt K, Haile L, Stallings CR, Origoni AE, et al. Dietary antigens, epitope recognition, and immune complex formation in recent onset psychosis and long-term schizophrenia. Schizophr Res. 2011;126(1-3):43–50. doi: 10.1016/j.schres.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Sheridan JW, Simmons RJ. Pancreatin-EDTA treatment affects buoyancy of cells in Cohn fraction V protein density gradients without residual effect on cell size. Aust J Exp Biol Med Sci. 1983;61:727–737. doi: 10.1038/icb.1983.68. [DOI] [PubMed] [Google Scholar]
- Skypala I. Adverse food reactions--an emerging issue for adults. J Am Diet Assoc. 2011;111(12):1877–1891. doi: 10.1016/j.jada.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70(2):496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J Exp Med. 1989;169(1):345–350. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid LM, Qiao SW, Anderson RP, Gianfrani C, Koning F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics. 2012;64(6):455–460. doi: 10.1007/s00251-012-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence MA, Westlake J, Lange K. Estimation of the variance components for dermal ridge count. Ann Hum Genet. 1977;41(1):111–115. doi: 10.1111/j.1469-1809.1977.tb01968.x. [DOI] [PubMed] [Google Scholar]
- Tan RA, Corren J. The relationship of rhinitis and asthma, sinusitis, food allergy, and eczema. Immunol Allergy Clin North Am. 2011;31(3):481–491. doi: 10.1016/j.iac.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, Bakker SF, Bardella MT, Bhaw-Rosun L, Castillejo G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43(12):1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HJ, Kumar R, Pongracic J, Liu X, Story R, Yu Y, Caruso D, Costello J, Schroeder A, Fang Y, et al. Familial aggregation of food allergy and sensitization to food allergens: a family-based study. Clin Exp Allergy. 2009;39(1):101–109. doi: 10.1111/j.1365-2222.2008.03111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery BP, Lin J, Kulis M, Fu Z, Steele PH, Jones SM, Scurlock AM, Gimenez G, Bardina L, Sampson HA, et al. Peanut oral immunotherapy modifies IgE and IgG4 responses to major peanut allergens. J Allergy Clin Immunol. 2013;131(1):128–134. doi: 10.1016/j.jaci.2012.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojdani A. Detection of IgE, IgG, IgA and IgM antibodies against raw and processed food antigens. Nutr Metab. 2009;12:6–22. doi: 10.1186/1743-7075-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JG, Assa'ad A, Heizer AB, Bernstein JA, Hershey GK. The -159 C-->T polymorphism of CD14 is associated with nonatopic asthma and food allergy. The J Allergy Clin Immunol. 2003;112(2):438–444. doi: 10.1067/mai.2003.1634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


