Summary
Microbial-derived natural products provide the foundation for most of the chemotherapeutic arsenal available to contemporary medicine. In the face of a dwindling pipeline of new lead structures identified by traditional culturing techniques and an increasing need for new therapeutics, surveys of microbial biosynthetic diversity across environmental metabiomes have revealed enormous reservoirs of as yet untapped natural products chemistry. In this review we touch on the historical context of microbial natural product discovery and discuss innovations and technological advances that are facilitating culture-dependent and culture-independent access to new chemistry from environmental microbiomes with the goal of re-invigorating the small molecule therapeutics discovery pipeline. We highlight the successful strategies that have emerged and some of the challenges that must be overcome to enable the development of high-throughput methods for natural product discovery from complex microbial communities.
Introduction
Genetically encoded small molecules comprise the single greatest source of available chemotherapeutics. Since 1981, 80% of anti-cancer agents and roughly 50% of all FDA-approved drugs have been or were derived from natural products (Newman and Cragg, 2012). In addition to their clinical utility, natural products are invaluable tools for advancing basic biological research. With diverse modes of action ranging from antibiotic to anticancer to immunosuppressive and beyond, novel natural products hold promise for the future of medicine, especially for diseases with limited treatment options.
Although the systematic chemical characterization of natural products commenced in the 19th century, the use of environmentally derived substances for medicinal purposes likely predates recorded human history (Solecki, 1975). Early medical texts from Mesopotamia dating to 2600 BC describe plant-derived substances, and Ancient Greek and Chinese cultures documented the application of botanical extracts for medicinal purposes in the 4th century BC (Cragg and Newman, 2005; Ji et al., 2009; Newman et al., 2000). Thus, humans have long recognized that natural products provide an important arsenal with which to combat disease and augment healing (reviewed in (Ji et al., 2009)).
Antibiotic properties of Penicillium fungi have been observed as early as 1875 by John Tyndall (Tyndall, 1876), but it was Alexander Fleming’s serendipitous re-discovery and partial purification of penicillin (1) in 1928 that first demonstrated that microbes, in this case the Penicillium rubens fungus, produce antibacterial substances (Fleming, 1929). This discovery needs little introduction; penicillin ultimately changed the course of human civilization curing previously debilitating and fatal illnesses. However, it is Selman Waksman who is credited with being the first to systematically explore microbial sources for novel natural products (Sakula, 1988). Waksman’s work culminated in 1943 with the isolation of streptomycin from the gram-positive, soil-dwelling actinomycete Streptomyces griseus, while working with a graduate assistant Albert Schatz (Schatz et al., 1944; Schatz and Waksman, 1944). This antibiotic was the first curative therapy for tuberculosis.
The recognition that natural products of microbial origin held clinical potential spurred the discovery of myriad antibacterial natural products from readily cultured environmental bacteria, including tetracycline (Streptomyces aureofaciens, 1945); chloramphenicol (Streptomyces venezulae, 1947); erythromycin (Streptomyces erythraea, 1949, ELilly); vancomycin (2) (Amycolatopsis orientalis, 1953, Eli Lilly); and rifamycin (Streptomyces mediterranei, 1957). The advances in natural product discovery in the 1940s and 1950s fostered programs focusing on cultivable environmental microorganisms in the pharmaceutical industry. These programs were successful not only for antibiotics, but also for the development of immunomodulatory compounds such as rapamycin (Vezina et al., 1975) and cyclosporin (Stahelin, 1996), as well as statin development (Endo, 1992), among others. However, as much of the low hanging fruit had been picked, discovery rates inevitably slowed and re-discovery became an inevitable impediment to bringing new natural products to market, leading to a waning interest from the pharmaceutical industry by the end of the 20th century (Baker et al., 2007).
Regrettably, the de-emphasis of natural products in the pharmaceutical industry coincided with major innovations in the biological sciences that could have prevented the lapse in the discovery pipelines had the effort been sustained. In the 1980s, advances in genetics and phylogenetic analysis, specifically, the use of ribosomal RNA sequencing as a microbial species profiling tool (Olsen et al., 1986), began to uncover a vast diversity of novel microbes. A combination of 16S rRNA profiling, visual analysis using fluorescent in situ hybridization (FISH) (Langer-Safer et al., 1982), and DNA reassociation kinetics (Torsvik et al., 1990a; Torsvik et al., 1990b) estimated up to 105 largely uncultured bacterial species per gram of soil and unearthed an array of unique, uncultured bacterial species in marine and thermophilic environments (Britschgi and Giovannoni, 1991; Giovannoni et al., 1990; Liesack and Stackebrandt, 1992; Stahl et al., 1984; Torsvik et al., 1990a; Torsvik et al., 1990b; Ward et al., 1990; Weller et al., 1991). Such biodiversity profiling analyses demonstrated that in environmental metabiomes, uncultured species outnumber the cultured ones by two to three orders of magnitude (Rappe and Giovannoni, 2003; Torsvik et al., 1998; Torsvik et al., 1996). Thus, largely comprised of the uncultured majority, these metabiomes represent a vast reservoir of uncharacterized biodiversity and, by extension, untapped natural product chemistry. These observations led to the development of the field of metagenomics (Handelsman et al., 1998), where DNA extracted directly from environmental samples (environmental DNA, eDNA) is cloned into model cultured hosts for detailed analysis. This approach to studying environmental bacteria has turned out to be a particularly appealing method to use in the search for novel natural products because the genes that encode the biosynthesis of individual secondary metabolites are generally clustered on bacterial chromosomes.
Concurrent with the identification of large numbers of novel environmental bacterial species, molecular biology was also undergoing rapid expansion, largely attributable to polymerase chain reaction (PCR) and DNA sequencing technologies. Whole genome sequencing in the mid-1990s paved the way for the development of the genomics discipline whereby DNA sequences could be analyzed and subsequently probed for phenotypes. While forward genetic approaches had already shown the tendency for bacterial natural product pathways to be encoded by genes clustered together within genomes (Hopwood and Merrick, 1977; Martin and Liras, 1989), genomics enabled reverse genetic and bioinformatic inquiries into natural product biosynthesis. A subsequent, steep drop in DNA sequencing cost and rapid advances in sequencing technologies, driven by the growth of the genomics field, transformed the natural product drug discovery process. The ever-growing number of available bacterial genome sequences (>500/month, most partial, deposited at NCBI in 2013 (NCBI, 2013)), made clear that significant fractions of prokaryotic genomes are devoted to secondary metabolism. Strikingly, genomic analyses have also revealed a large number of cryptic (i.e. silent) gene clusters, even within the genomes of bacterial species that have been extensively studied with respect to natural product chemistry (Bentley et al., 2002; Ikeda et al., 2003).
The realization that the majority of bacterial species remain uncultured and that even previously cultured species contain many cryptic clusters has led to a resurgence in attempts to access environmental biosynthetic diversity. In this review we discuss new culturing approaches, genomic and metagenomic techniques, and synthetic biology techniques that are being applied to the revitalization of natural product discovery pipelines and outline the future challenges and the promise of mining the metabiomes for biomedically relevant lead structures.
Rethinking Culture-Based Approaches In The Genomics Age
Historically, culture-based approaches followed the same general discovery protocol - bacteria were isolated from the environment, grown in monoculture fermentation broths, subjected to organic extraction, and screened for bioactivity. To maximize the variety of bacteria that could be grown in the laboratory, culturing conditions were varied with respect to parameters like broth composition, pH, and temperature. As the pool of easily accessible cultured bacteria has dwindled, the field refocused on developing more involved approaches to facilitate cultivation of previously uncultured bacteria and to induce activation of silent gene clusters in previously cultured microbes.
Improving Culturing Techniques
Recognizing that uncultured organisms are likely not intrinsically “unculturable”, several groups have attempted to simulate the native habitat of isolated bacterial species to enable their cultivation in the laboratory. In situ cultivation by enclosing the bacteria within a semipermeable membrane, which allows the passage of nutrients and growth factors but not cells, and introducing the membrane-enclosed chambers into their native environment, simulated in a laboratory setting, up to 40% of species from marine biofilms were grown as pure culture micro-colonies in one such experiment (Kaeberlein et al., 2002). The same group extended this simulation to culture rarely accessed aerobic and anaerobic species from subsurface sediment (Bollmann et al., 2010), and a commercial venture founded on these approaches has reported a number of novel metabolites in the patent literature, including two novel glycosylated macrolactam antibiotics NOVO3 and NOVO4 (3) with potent activity against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE) with minimal in vitro toxicity to mammalian cells (Lewis et al., 2009; Peoples et al., 2011). Furthermore, by focusing the screening efforts on identifying small molecules with a narrow range of activity against a specific pathogen, the collection of extracts from in situ and prolonged, up to 18 months (Buerger et al., 2012), cultivation resulted in the identification of lassomycin, a ribosomal cyclic peptide with potent activity against Mycobacterium tuberculosis, from the extract of Lentzea kentuckyensis (Gavrish et al., 2014). Other, related techniques that have been used to culture isolated bacteria in simulated native environments in the laboratory include culturing on supporting membranes suspended on a soil slurry (Ferrari et al., 2005) and embedding single cells in agarose microdroplets and culturing them in flowing seawater (Zengler et al., 2002) or directly on living coral (Ben-Dov et al., 2009).
Activating Cryptic Metabolism By Fermentation
While the cryptic biosynthetic gene clusters present in the genomes of readily cultured bacteria represent a sizable pool of low hanging fruit for drug discovery, forcing expression of these gene clusters has not been trivial. Mirroring the difficulties associated with culturing environmental microbes, conditions required for the activation of cryptic biosynthetic pathways have been difficult to identify and so far there does not appear to be a “silver bullet” approach for inducing the expression of previously silent gene clusters. One attractive, inexpensive approach to induce expression of cryptic biosynthetic clusters involves media supplementation. While investigating effects of rare earth metals on bacterial physiology, lanthanides scandium and lanthanum were discovered to increase or induce expression of silent gene clusters in several Streptomyces spp. through an as yet undetermined, guanosine pentaphosphate (pppGpp) dependent pathway (Kawai et al., 2007; Tanaka et al., 2010). Addition of N-acetylglucosamine (GlcNAc), a major component of fungal cell walls, was also shown to stimulate production of several cryptic antibiotic-producing clusters by acting on a pleiotropic transcriptional repressor DasR in Streptomyces in minimal media (Rigali et al., 2008). The short chain fatty acids sodium butyrate and valproic acid, known histone deacetylase (HDAC) inhibitors, have been used to induce expression of biosynthetic genes from Streptomyces coelicolor under normally non-permissive conditions (Moore et al., 2012). Although HDAC homologues are widespread in bacterial species, the mechanism of secondary metabolism upregulation by HDAC inhibitors is not yet understood. Interestingly, a screen of the Canadian Compound Collection for small molecules that upregulate pigment production in S. coelicolor identified at least one set of molecules that function by inhibiting fatty acid biosynthesis (Craney et al., 2012). Other enzyme inhibitors have also been observed to influence activation of biosynthetic pathways. In the fungus Sphaeropsidales sp. F-24’707, supplementation with tricyclozole, a 1,8-dihydroxynaphthalene biosynthesis inhibitor, resulted in the re-discovery of mutolide (4), a rare fungal macrolide originally discovered by UV mutagenesis of the wild type strain (Bode et al., 2002; Bode et al., 2000).
Co-culturing has been yet another effective approach for the induction of cryptic secondary metabolite pathways. As with media supplementation, it aims to elicit production of secondary metabolites in a target organism by providing a missing regulatory cue. Often, this technique has employed two specific bacterial strains that are known to interact. This was the case for identification of pestalone (5), which was produced by a marine fungus of Pestalotia sp. only in the presence of a marine deuteromycete CNL-365 (Cueto et al., 2001) and for athepsin K inhibitors F-9775A and F-9775B, whose production was induced in Aspergillus nidulans in the presence of soil actinomycetes (Cueto et al., 2001; Schroeckh et al., 2009). A more general method was recently developed by Onaka et al. who found that mycolic acid-producing bacteria Tsukamurella pulmonis TP-B0596, Rhodococcus erythropolis, and Corynebacterium glutamicum alter metabolite biosynthesis in over 85% of Streptomyces strains tested (Onaka et al., 2011). Although these percentages reflect the combined increase and decrease in secondary metabolite production, these three species were sufficient to induce biosynthesis of over 35, 34 and 23 different metabolites, respectively.
Exploiting Genomic Sequence Data In Cultured Bacteria
While fermentation based activation approaches represent largely trial and error efforts to activate cryptic metabolism, a number of groups have worked to take advantage of full genome sequence data to carryout more directed gene cluster induction and molecule detection studies. The ready availability of sequence data has led to the development of user-friendly bioinformatics tools, like antiSMASH, np.searcher and ClustScan, which identify biosynthetic gene clusters in assembled genomes and predict structural elements of their molecular products based upon substrate specificities of their conserved biosynthetic domains (Li et al., 2009; Medema et al., 2011; Starcevic et al., 2008). This type of analysis is now routinely applied to newly assembled genomes, allowing researchers to rapidly prioritize silent gene clusters for molecule discovery efforts. For example, bioinformatic analysis of the draft genome of Salinaspora tropica CNB-440 predicted the presence of biosynthetic clusters for a broad range of unknown polyketide synthase systems, as well as nonribosomal peptide synthetases and hybrid clusters (Udwary et al., 2007). Structure predictions afforded by bioinformatics are likely to become an ever more important part of future efforts to identify structurally and functionally novel natural products.
In addition to revealing the presence of silent biosynthetic gene clusters in cultured bacteria, bioinformatic analysis of sequence data is an effective tool for identification and characterization of their undetected products. For example, genomic sequence analysis of Salinispora tropica CNB-440 predicted that a cryptic 10-module PKS gene cluster would incorporate polyene units into the final product, producing characteristic UV absorption. Subsequent screening of culture broth extracts by UV absorption led to the isolation of a series of new polyene macrolactams, exemplified by salinilactam A (6) (Udwary et al., 2007). In a different study, bioinformatic analysis of silent NRPS and PKS-NRPS hybrid gene clusters encoded in the Pseudomonas fluorescens Pf-5 gene enabled the prediction of the amino acid precursors used as substrates by these cryptic clusters. By feeding stable isotope labeled precursors to P. fluorescens Pf-5 cultures and then tracking their incorporation by 2D NMR, this well studied bacterium was found to produce the orfamides (7), a novel family of macrolytic lipopeptides (Gross et al., 2007). Similarly, bioinformatic prediction of a cryptic tetrapeptide siderophore sequence in the genome of the industrially important bacterium Saccharopolyspora erythraea facilitated the use of radiolabeled precursors to identify the hydroxamate-type siderophore antibiotic erythrochelin A (Robbel et al., 2010).
In the case of NRPS peptides, a systematic genome mining technique, termed natural product peptidogenomics (NPP) has been developed that combines the predictive power of bioinformatics with the detection sensitivity of tandem mass spectrometry (MSn) (Kersten et al., 2011). This methods extends the repertoire of imaging mass spectrometry (IMS) based techniques that have been long used to study bacterial chemistry and, in particular, antibacterial peptides (extensively reviewed in (Watrous and Dorrestein, 2011)). It relies on comparing the peptide structures, determined from the MSn fragmentation patterns, with the sequence-based predictions to match the secondary metabolites present in the sample with the biosynthetic clusters responsible for their production. In proof of principle NPP experiments, detection of novel ribosomal and nonribosomal peptide natural products by MALDI-TOF imaging combined with de novo MSn structure elucidation allowed researchers to link known molecules to the gene clusters responsible for their production in previously studied bacteria, as well as identify 14 new peptide natural products from various sequenced streptomycetes (Kersten et al., 2011). Moreover, this technique was shown to work for parallel identification of three new peptide natural products from daptomycin-producing Streptomycies roseosporus NRRL 15998, demonstrating its potential for high-throughput small molecule discovery. More recently, a similar approach has been applied to glycosylated metabolites with the development of natural product glycogenomics (NPG). This technique further extends the application of IMS to the identification of novel glycosylated natural products and their corresponding biosynthetic gene clusters via MSn characterization of O- and N-glycosyl groups (Kersten et al., 2013).
Developing culture independent approaches (metagenomics)
Metagenomics provides the means of accessing bioactive small molecules synthesized by environmental bacteria through the direct cloning of DNA from essentially any environmental source, thereby bypassing the requirement of cultivation. Metagenomic approaches for natural product discovery fall into two general categories that are generically referred to as sequence-based and function-based. As the names imply, sequence-based methods rely on the sequencing and subsequent bioinformatics analysis of a metagenomic samples to identify biosynthetic gene clusters of interest, while functional methods rely on the direct functional screening of metagenomic clones for discrete activities to identify clones of interest.
Sequence-Driven Metagenome Mining
Many of the bioinformatics tools employed for genome mining have also been successfully applied to partially assembled DNA sequences from metagenomic sources to predict natural product compounds encoded within the genomes of uncultured bacteria. However, high degree of sequence similarity and the repetitious nature of biosynthetic domains render the assembly of sequences corresponding to natural product gene clusters from metagenomic sequences difficult and this has limited these straightforward sequence-based genome mining applications to the lower complexity metagenomes of uncultured symbionts, albeit with considerable success. Using a combination of targeted PCR screening of crude DNA isolates from zooids inhabiting the marine tunicate Lissoclinum patella and shotgun sequencing of the fraction containing genes associated with patellazole biosynthesis, a symbiotic α-proteobacterium Candidatus Endolissoclinum faulkneri was identified as a previously unknown source for a family of these potent cytotoxins (Kwan et al., 2012). Whole genome sequencing (WGS) of the lichen Peltigera membranacea led to the identification of nosperin, a novel polyketide of the pederin family of anti-cancer compounds, produced by its photobiont Nostoc sp. (Kampa et al., 2013). This finding expanded both the structural diversity and the known distribution of this family of secondary metabolites. In another example, a biosynthetic pathway encoding for the clinically promising cytotoxin apratoxin (8) was identified from a complex marine microbiome associated with the cyanobacterium Lyngbya bouillonii by physical micromanipulation of the symbiont to dislodge individual cells and whole genome amplification (WGA) to obtain partial genomic sequence from single cells (Grindberg et al., 2011). In addition to identifying new metabolites, metagenomic sequence data assembly recently led to the identification of a ‘metabolically talented’ marine sponge endosymbiont genus Entotheonella, which is believed to be responsible for nearly all bioactive molecules that have been isolated from its host (Wilson et al.,2014).
Unfortunately, even partial genome assembly has been difficult for metagenomes of higher complexity, which may contain as many as 104–105 unique species, as is the case for a typical soil metagenome (Pop, 2009; Rappe and Giovannoni, 2003; Torsvik et al., 1990a). However, biosynthetic capacity of complex environments can be mined using a PCR-based, “sequence tag” approach (Seow et al., 1997). Sequence tag approaches take advantage of the fact that much of the biosynthetic diversity arises from a relatively small number of biosynthetic classes (e.g. nonribosomal peptide synthase [NRPS], polyketide synthase [PKS], isoprene, sugar, shikimic acid, alkaloid, ribosomal peptide) related through common, highly conserved biosynthetic domains (Dewick, 2009) and focuses next-generation sequencing efforts on PCR amplicons of the most commonly used biosynthetic domains (i.e. sequence tags). In general, gene clusters encoding structurally related metabolites are predicted to share common ancestry and, therefore, high sequence identity among conserved domains. Correspondingly, a high degree of identity among sequence tags predicts that the biosynthetic domains they correspond to are involved in biosynthesis of structurally related small molecules. By extension, sequence tags with no close relatives might be predicted to arise from gene clusters encoding structurally novel classes of metabolites (Reddy et al., 2014). These correlations form the basis of bioinformatics tools, eSNaPD and NaPDoS, developed to facilitate functional classification of novel secondary metabolite biosynthetic gene clusters from metagenomic data (Owen et al., 2013; Reddy et al., 2014; Ziemert et al., 2012). Remarkably, short sequence tags (~400 base pairs) accurately predict the structural family of the molecule encoded by a gene cluster, as validated by the recovery and subsequent analysis of metagenomic DNA clones predicted to produce novel glycopeptide, lipopeptide, and bis-intercalator natural products (Owen et al., 2013).
The sequence tag approach is a powerful tool for rapid, low cost profiling of biosynthetic capacity of (meta)genomes, without the requirement for complete genome sequencing or chemical isolation. It has been used to genotype uncultured marine Streptomycetaceae, as well as to profile a collection of cultured Actinobacteria from marine sediment (Gontang et al., 2010; Prieto-Davo et al., 2013) and it is scalable to profiling metagenomes of any complexity. The true utility of the sequence tag approach, though, is in its ability to identify gene clusters encoding congeners of valuable bioactive compounds and potentially novel chemistries that have remained hidden in the environment (Charlop-Powers et al., 2014; Owen et al., 2013; Reddy et al., 2014). In a direct comparison of NRPS and PKS biosynthetic domain detection from metagenomic samples, this PCR-based method was shown to be 10 to 100 times more sensitive than shotgun sequencing in identifying unique sequences (Woodhouse et al., 2013). Sequence tag-based profiling of soil metagenomes revealed thousands of unique adenylation and ketosynthase domains, with only minor overlap among distinct microbiomes (Charlop-Powers et al., 2014; Owen et al., 2013). Profiling of environmental samples can be performed on purified metagenomic DNA, facilitating identification of the most biosynthetically rich samples for library construction.
The sequence tag strategy has been used to identify and heterologously express gene clusters from both soil and marine environments that encode known natural products, congeners of high value natural products and completely novel natural products. This includes the tryptophan dimer-based cytotoxins borregomycin and erdasporine (Chang and Brady, 2013; Chang et al., 2013); congeners of a potent antitumor compound pederin (Zimmermann et al., 2009); the pharmacologically important antitumor polyketide psymberin (Fisch et al., 2009); arimetamycin A (9), a natural doxorubicin analog, that is active against multidrug resistant cancer cell lines (Kang and Brady, 2013); a number of variants of the highly selective protease inhibitor microviridin (Gatte-Picchi et al., 2014; Ziemert et al., 2010) and structurally diverse aromatic polyketides including the MRSA active, tetarimycin A (10) as well as the MRSA and VRE active fasamycins A and B (Feng et al., 2012; Kallifidas et al., 2012); and a penatacyclic compound erdacin, with a novel ring system (King et al., 2009).
Functional Metagenome Mining
Sequence-based (meta)genome mining is rapidly becoming a relatively mature field with a plethora of tools at its disposal for identifying, capturing, and expressing cryptic clusters from sequenced genomes and novel biosynthetic clusters from metagenomes. In contrast, functional metagenomics methods have developed more slowly. Functional approaches aim to accelerate the rate of bioactive small molecule discovery from metagenomes by directly screening environmental DNA clones for bioactivities of interest. These approaches are akin to high-throughput screening efforts of broth extracts from culture collections, with the added dimension of roughly two to three orders of magnitude deeper biosynthetic diversity potential (Rappe and Giovannoni, 2003; Torsvik et al., 1998).
Early functional metagenomic studies and their derivatives
To date, functional metagenomics has largely consisted of very simple screens preformed on cosmid libraries hosted in a small number of model bacterial hosts. In one of the earliest efforts, direct HPLC-ESIMS screening of fermentation broths derived from a small (1020-member) soil DNA library hosted in Streptomyces lividans yielded terragine A and a family of related molecules - the first novel family of small molecules from a metagenomic source (Wang et al., 2000). Quickly, it became apparent that this kind of screen was impractical for the larger metagenomic libraries that would be required in order to identify diverse molecules from metagenomic sources. Consequently, the majority of functional assays for secondary metabolites have since relied on easily identifiable phenotypic traits that are commonly associated with secondary metabolite production, such as pigmentation and antibiosis. Screening of approximately 700,000 clones from soil metagenomic mega-library in E. coli for antibiosis against B. subtilis in overlay assays produced a large family of long-chain N-acyl amino acids, which were the first novel antibiotics discovered using functional metagenomics (Brady and Clardy, 2000). The same approach was used to identify long-chain N-acyl palmitoylputrescine from a bromeliad tank water metagenome (Brady and Clardy, 2004) and an isocyanide antibiotic (11) from a soil metagenome (Brady and Clardy, 2005). In a screen of soil metagenomic libraries, hosted in E. coli, a combination of pigmentation and activity against B. subtilis helped identify metagenomic clones encoding production of indirubin, indigo blue and other related molecules (Lim et al., 2005; MacNeil et al., 2001). In a chrome azurol S (CAS) colorimetric assay (Schwyn and Neilands, 1987), designed to report on siderophores production, new gene clusters encoding known siderophores vibrioferrin (12) and bisucaberin were found in libraries constructed from marine sources (Fujita et al., 2011; Fujita et al., 2012).
Improving Functional Access To Natural Products
Functional metagenomics is a potentially powerful approach for identification of bioactive natural products encoded by environmental microbiomes in a high throughput manner. However, to reach the full potential of this approach a number of methodological advances are required. Improvements in the size of cloned environmental DNA are needed to facilitate the capture of complete gene clusters on individual clones. Diversification of the heterologous hosts is required to enable native expression of a more diverse collection of genes clusters. Finally, enrichment of metagenomic libraries for biosynthetic clones is necessary to increase the efficiency of the screening process.
A number of groups have recently reported advances geared at solving these problems. Several groups have reported BAC libraries from soil samples, with average insert sizes of up to 90 kb (Liles et al., 2008; Liu et al., 2010; Yu et al., 2008); however, the reported libraries remain small in size and are not yet capable of saturating coverage of the environments from which they are derived. A variety of broad host-range vectors have been developed to facilitate heterologous expression in diverse model hosts. For example, vectors based on ΦC31 and ΦBT1 phage, site-specific integrase systems are able to facilitate integrative transfer of cloned DNA into a variety of Streptomyces spp. (Gregory et al., 2003; Kuhstoss and Rao, 1991). For transfer of libraries into diverse alpha-, beta-, and gammaproteobacterial species, several RK2-derived vectors have been recently developed (Aakvik et al., 2009; Kakirde et al., 2011). The utility of these vectors was demonstrated by parallel functional metagenomic library screens carried out in Agrobacterium tumefaciens, Burkholderia graminis, Caulobacter vibrioides, Escherichia coli, Pseudomonas putida, and Ralstonia metallidurans, using a single broad-host-range vector. This study identified orthogonal sets of clone-specific small molecules expressed in each strain with minimal overlap (Craig et al., 2010). On average, less than 2% of the bacterial metagenome is devoted to secondary metabolism (Garcia et al., 2011), which means that metagenomic libraries are only sparsely populated with gene clusters encoding bioactive small molecules. Increasing the biosynthetic density of metagenomic libraries will therefore translate directly into more efficient screening. The majority of existing enrichment techniques rely on some variation of probe hybridization (Kalyuzhnaya et al., 2006; Meyer et al., 2007; Morimoto and Fujii, 2009; Uchiyama et al., 2005; Zhang et al., 2009). Recently, enrichment strategies based on genetic complementation have shown promise as tools for library scale enrichments. In particular, the promiscuity of 4’-phosphopanthetheinyl transferases (PPTases) and their universal requirement to generate holo-NRPS and PKS enzymes has been used to develop successful gene cluster enrichment strategies (Charlop-Powers et al., 2013; Owen et al., 2012; Yin et al., 2007). While all of these methods remain works in progress, recent advances in the field suggest that functional metagenomics may one day reach the promise envisioned for it at the outset (Handelsman et al., 1998).
Activating silent pathways through host and cluster engineering
From minimally invasive techniques that employ minor modifications to the native host to induce gene cluster activation, to more invasive heterologous expression and gene cluster refactoring approaches – molecular biology, bacterial genetics and more recently synthetic biology have proved essential to many aspects of the modern natural product discovery process. When applied to the natural product discovery, these methods represent a diverse and growing toolbox for activating biosynthetic pathways that are identified by either genomic approaches in cultured bacteria (i.e. cryptic) or metagenomic approaches.
Cryptic Cluster Activation in Native Hosts
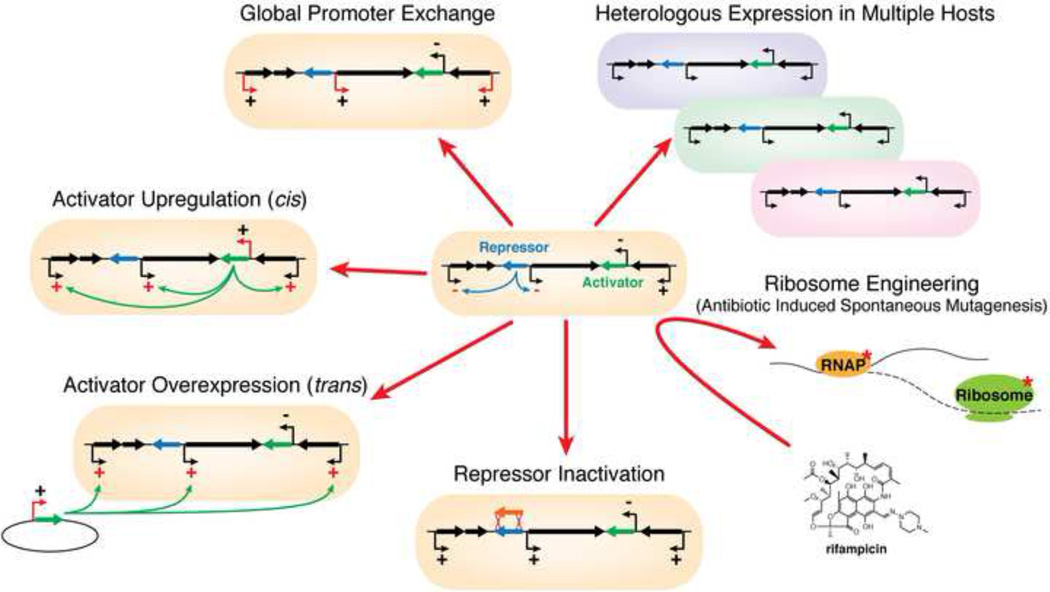
The simplest and the most effective approaches to activating cryptic secondary metabolite pathways rely on minimal modifications made directly in the native host organism. When sequence data is available it is possible to perform a detailed bioinformatic analysis of the cryptic cluster of interest and design rational strategies for their potential activation. Strategic insertion of strong constitutive promoters in front of either biosynthetic or regulatory genes in cryptic clusters has been one such productive strategy for activating biosynthesis. For example, insertion of a strong constitutive ermE*p promoter from Saccharopolyspora erythraea into bioinformatically identified cryptic clusters in the well studied strain S. albus J1074 led to the expression of an NRPS encoded blue pigment and two novel compounds 6-epi-alteramide A (13) and B, encoded by a hybrid type I PKS-NRPS cluster (Olano et al., 2014). In cases where putative transcription activators are identified in a cluster, induction of their constitutive expression has been shown to activate an entire cluster. For example, cloning out the large ATP-binding LuxR-type (LAL) regulatory gene and reintroducing it under control of a constitutive ermE*p led to identification of four novel, 51-membered, glycosylated macrolides stambomycins A–D (14) with activity against human cancer lines in Streptomyces ambofaciens ATCC23877 (Laureti et al., 2011). Not all regulatory elements are inducers of secondary metabolism, as was observed when mutagenic inactivation of the quorum-sensing regulator in Burkholderia thailandensis led to activation of a cryptic cluster encoding a novel polyketide metabolite, thailandamide lactone (Ishida et al., 2010).
Ochi and co-workers have worked on a more general, sequence-agnostic methods for cryptic cluster activation by exposure of bacteria of interest to antibiotics and selecting for drug-resistant mutants. With this approach, which they have termed “ribosome engineering”, they observed that mutations in the endogenous RNA polymerase and ribosomal protein S12 lead to strong activation of cryptic clusters and improve antibiotic expression in Streptomyces spp., myxobacteria, and fungi (reviewed in (Ochi and Hosaka, 2013)). This work led to the identification of 3,3’-neotrehalosadiamine, a novel aminosugar antibiotic, in B. subtilis and piperidamycin (15), a novel antibacterial class, in Streptomyces sp. 631689 (Hosaka et al., 2009; Inaoka et al., 2004).
Capturing Gene Clusters for Heterologous Expression in Model Hosts
To take advantage of many newly developed natural products discovery approaches it is often necessary to clone a gene cluster from its native host for subsequent introduction into a heterologous host. Secondary metabolite gene clusters vary in size from less than 10 kb to over 150 kb, with an average size around 50 kb (Starcevic et al., 2008) and thus cloning using standard “cut and paste” techniques can be challenging. If the sequence of the target gene cluster is known a variety of recombination-based techniques (e.g. transformation-assisted recombination (TAR) in the budding yeast Saccharomyces cerevisiae (Kim et al., 2010; Larionov et al., 1997), Red/ET recombination in E. coli (Bian etal., 2012; Fu et al., 2012; Muyrers et al., 2001), and oriT-directed capture (Chain et al., 2000; Kvitko et al., 2013)) can now used to directly clone intact gene clusters from genomic DNA. For metagenomic samples, which are significantly more complex than individual genomes and lack sequence data, metagenomic cosmid or fosmid libraries containing 30–40 kb inserts are first constructed and large gene clusters are assembled using TAR from overlapping metagenomic clones (Kim et al., 2010). Theoretically BAC libraries, capable of carrying inserts of up to 250 kb, can be used to circumvent this limitation; however, as mentioned above more efficient cloning methodologies are needed to generate useful BAC libraries from biosynthetically rich metagenomic source.
Host Modifications
At the host level, problems with heterologous expression can arise from issues associated with transcription and translation of the exogenous DNA and from insufficient availability of precursors. Most studies addressing translation and transcription issues have focused on E. coli as it is a rapidly growing and genetically tractable organism. Overexpression of an alternative sigma factor, σ54, in E. coli enabled expression of a heterologous oxytetracycline polyketide (16) encoded by a Type II PKS gene cluster (Stevens et al., 2013). In addition, E. coli strains expressing rare tRNAs have been engineered to alleviate translation problems (e.g. stalling, premature termination, frameshifts) due to the imbalance of available tRNAs caused by the differences in codon usage (Novy et al., 2001). In a similar vein, expression of chaperones to facilitate proper folding of biosynthetic enzymes has been beneficial for heterologous expression studies in E. coli (Hoertz et al., 2012; Mutka et al., 2006). Limited precursor availability can often be a result of the heterologous host not producing sufficient amount of the precursor or of precursor depletion by endogenous biosynthetic pathways. Precursor feeding has been used successfully to increase production of meridamycin (17), a 27-membered macrolide with potent neuroprotective properties, in S. lividans (Liu et al., 2009). Whereas, deletion of the competing pathways has generally led to a dramatic increase in the levels of heterologous expression of secondary metabolites in S. lividans (Pestov et al., 1999), S. avermitilis (Komatsu et al., 2010), and S. coelicolor (Gomez-Escribano and Bibb, 2011).
Refactoring of cloned gene clusters
At the gene cluster level, lack of recognition of promoters and ribosome binding sites (RBS) by the heterologous host frequently leads to low or no expression of the biosynthetic enzymes and a corresponding lack of small molecule expression. This challenge is most commonly overcome by replacing the native transcriptional regulatory elements within the biosynthetic gene cluster with ones known to function in the heterologous host. For small clusters, this can be accomplished by PCR amplification of individual genes and sub-cloning them under the control of such promoters, a strategy that yielded a family of novel tryptophan dimer cytotoxins erdasporines A–C from a metagenomic clone refactored for expression in E. coli (Chang et al., 2013). For larger clusters, Red/ET and TAR based tools can be used to replace promoters driving the expression of each polycistronic operon. Using Red/ET recombineering in E. coli, the myxochromide S biosynthetic gene cluster from myxobacteria Stigmatella aurantiaca was placed under the control of the toluic acid inducible Pm promoter suitable for use in pseudomonads. Transfer of this construct into Pseudomonas putida resulted in high expression level of myxochromide S, as well as two new derivatives that have not been previously observed (Wenzel et al., 2005). In two examples of promoter replacement in a multi-operon clusters, a powerful plug-and-play “DNA assembler” method based on homologous recombination in yeast (Shao et al., 2011; Shao et al., 2009) was used to activate and characterize cryptic clusters encoding four novel polycyclic tetramate macrolactams from S. griseus (Luo et al., 2013) and a daptomycin-like dichlorinated lipopeptide antibiotic taromycin A (18) from the marine actynomycete Saccharomonospora sp. CNQ-490 (Yamanaka et al., 2014).
Success of complete refactoring of biosynthetic gene clusters as a strategy for heterologous activation of cryptic metabolism prompted the development of new tools to simplify and accelerate this process. Gibson (Gibson et al., 2009) and Golden Gate (Engler et al., 2008) assembly techniques allow rapid assembly of multiple PCR generated fragments in a specific order determined by the user-designed overlapping sequences. These methods are particularly useful for assembling codon-optimized synthetic constructs into functional gene clusters. Seamless ligation cell extract (SLiCE) is an ex vivo recombineering method that allows both cloning and genetic manipulation of large DNA fragments using cell extracts from an E. coli strain expressing an optimized lambda prophage Red/ET system (Zhang et al., 2012). This method accelerates the process by eliminating multiple transformation and cell growth steps. Also recently introduced is the CRISPR/Cas system, which functions via an RNA-guided nuclease that, in isolation, can be programmed to target any sequence by providing a short (20–50 nucleotide) synthetic RNA sequence (Wiedenheft et al., 2012). It offers ease of use, high specificity, simplicity and speed for targeting virtually any DNA sequence. These characteristics have led to an immense amount of interest in developing a variety of genome editing tools based on CRISPR/Cas system, which are certain to revolutionize synthetic biology (Baker, 2014). Although these latter tools have not yet yielded novel molecules, they are very likely to turn out many new natural products in the coming years.
Even with appropriate transcriptional elements in place, the DNA sequence itself varies significantly among species and differences in GC content and codon usage can lead to decreased translation efficiency, early transcription termination, and self-resistance, if the final product is toxic to the host (Gustafsson et al., 2004). De novo synthesis with codon optimization and refactoring is possible, as the most aggressive approach to cryptic cluster activation, but has predominantly been used to increase yields of existing clinically important compounds by engineering their production in a heterologous host. In one such example, the gene cluster encoding production of the promising anticancer metabolites epothilone C and D (19) from the slow growing and genetically intractable myxobacterium Sorangium cellulosum was completely re-synthesized to optimize codon and promoters usage to allow production in E. coli (Mutka et al., 2006).
The Future of Metabiome Mining
Transformational changes in the field of DNA sequencing and molecular and synthetic biology (i.e. genome manipulation) are drastically altering the way we pursue the discovery of natural products. The ever increasing availability of sequenced genomes will undoubtedly continue to fuel targeted approaches to access cryptic clusters in cultured bacteria. Although many of the most important clinical therapeutic agents have been discovered in cultured bacteria, these organisms represent only the tip of the microbial diversity iceberg. Metagenomics is taking advantage of the sequencing revolution to bypass many of the traditional barriers to drug discovery by providing the means to profile and access the biosynthetic capacity of the environmental metabiome. Promising new technologies have the potential to extend the power of whole genome sequencing to metagenomes. Long-read nano-pore based, single molecule sequencing alleviates the problems inherent in metagenomic sequence assembly (Koren et al., 2013; Mavromatis et al., 2012), while additional new techniques can now obtain nearly complete genome sequence from a single cell (Fitzsimons et al., 2013; Lasken, 2012). At this point, a collection of heterologous expression strategies exists, that appears capable of inducing expression of effectively any gene cluster, given sufficient expended effort. The focus of heterologous expression studies has also clearly shifted from fine-tuning, aimed at increasing the production of industrially important metabolites, to discovery, aimed at the detection of cryptic metabolites. In the future, the challenge will be to develop more universally applicable heterologous expression strategies, which can streamline efforts to activate the diverse cryptic gene cluster now known to be present in the Earth’s microbiome.
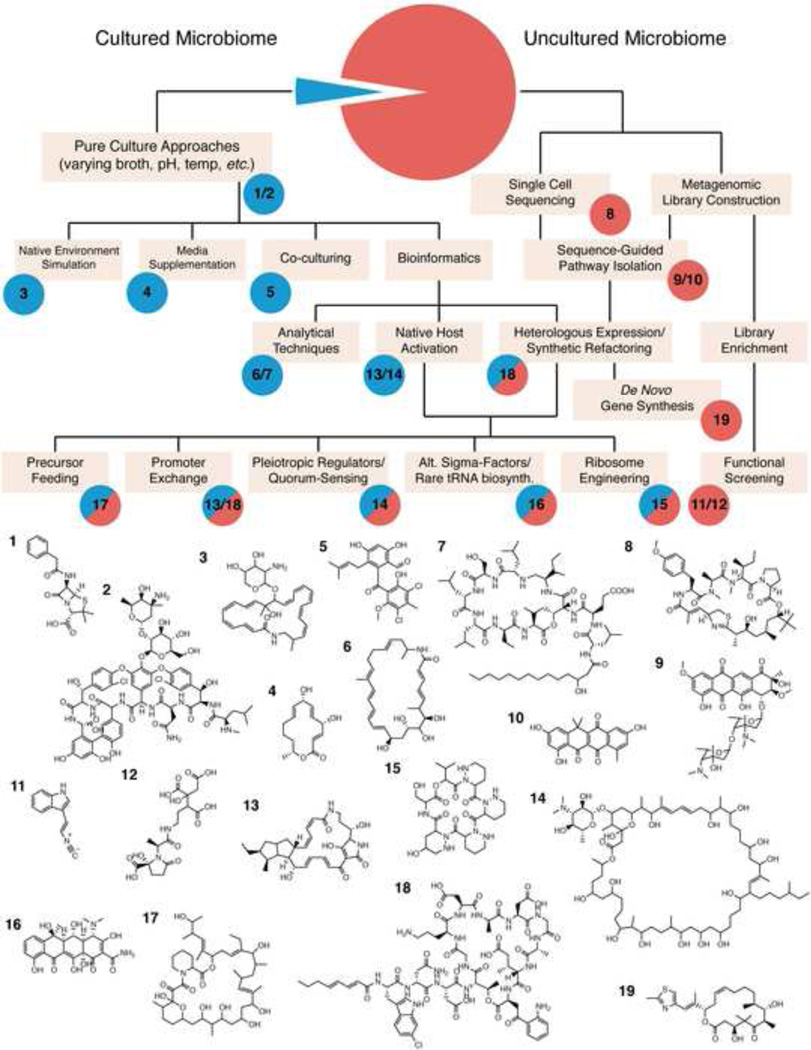
Figure 1.
Flow diagram of the approaches used for metabiome mining for natural product small molecules from cultured and uncultured bacteria and examples of molecules identified by various techniques. (1) Penicillin G; (2) Vancomycin; (3) NOVO4; (4) Mutolide; (5) Pestalone; (6) Salinilactam A; (7) Orfamide A; (8) Apratoxin A; (9) Arimetamycin A; (10) Tetarimycin A; (11) Isocyanide; (12) Vibrioferrin; (13) 6-epi-alteramide A; (14) Stambomycin A; (15) Piperidamycin A; (16) Oxytetracycline; (17) Meridamycin A; (18) Taromycin A; (19) EpothiloneC.
Figure 2.
Common approaches for activation of cryptic biosynthetic gene clusters.
Acknowledgements
This work was supported by National Institutes of Health (grant number GM077516). J.S.S. is supported by the Helmsley Postdoctoral Fellowship for Basic and Translational Research on Disorders of the Digestive System. S.F.B. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakvik T, Degnes KF, Dahlsrud R, Schmidt F, Dam R, Yu L, Völker U, Ellingsen TE, Valla S. A plasmid RK2-based broad-host-range cloning vector useful for transfer of metagenomic libraries to a variety of bacterial species. FEMS Microbiol. Lett. 2009;296:149–158. doi: 10.1111/j.1574-6968.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- Baker DD, Chu M, Oza U, Rajgarhia V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007;24:1225–1244. doi: 10.1039/b602241n. [DOI] [PubMed] [Google Scholar]
- Baker M. Gene editing at CRISPR speed. Nat. Biotechnol. 2014;32:309–312. doi: 10.1038/nbt.2863. [DOI] [PubMed] [Google Scholar]
- Ben-Dov E, Kramarsky-Winter E, Kushmaro A. An in situ method for cultivating microorganisms using a double encapsulation technique. FEMS Microbiol. Ecol. 2009;68:363–371. doi: 10.1111/j.1574-6941.2009.00682.x. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Bian X, Huang F, Stewart FA, Xia L, Zhang Y, Muller R. Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering. ChemBioChem. 2012;13:1946–1952. doi: 10.1002/cbic.201200310. [DOI] [PubMed] [Google Scholar]
- Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bode HB, Walker M, Zeeck A. Secondary metabolites by chemical screening, 41 - Structure and biosynthesis of mutolide, a novel macrolide from a UV mutant of the fungus F-24 ' 707. Eur. J. Org. Chem. 2000:1451–1456. [Google Scholar]
- Bollmann A, Palumbo AV, Lewis K, Epstein SS. Isolation and physiology of bacteria from contaminated subsurface sediments. Appl. Environ. Microbiol. 2010;76:7413–7419. doi: 10.1128/AEM.00376-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SF, Clardy J. Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J. Am. Chem. Soc. 2000;122:12903–12904. [Google Scholar]
- Brady SF, Clardy J. Palmitoylputrescine, an antibiotic isolated from the heterologous expression of DNA extracted from bromeliad tank water. J. Nat. Prod. 2004;67:1283–1286. doi: 10.1021/np0499766. [DOI] [PubMed] [Google Scholar]
- Brady SF, Clardy J. Cloning and heterologous expression of isocyanide biosynthetic genes from environmental DNA. Angew. Chem. 2005;44:7063–7065. doi: 10.1002/anie.200501941. [DOI] [PubMed] [Google Scholar]
- Britschgi TB, Giovannoni SJ. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 1991;57:1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. Microbial scout hypothesis and microbial discovery. Appl. Environ. Microbiol. 2012;78:3229–3233. doi: 10.1128/AEM.07308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PSG, Hernandez-Lucas I, Golding B, Finan TM. oriT-Directed Cloning of Defined Large Regions from Bacterial Genomes: Identification of the Sinorhizobium meliloti pExo Megaplasmid Replicator Region. J. Bacteriol. 2000;182:5486–5494. doi: 10.1128/jb.182.19.5486-5494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang FY, Brady SF. Discovery of indolotryptoline antiproliferative agents by homology-guided metagenomic screening. Pro. Natl. Acad. Sci. USA. 2013;110:2478–2483. doi: 10.1073/pnas.1218073110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang FY, Ternei MA, Calle PY, Brady SF. Discovery and synthetic refactoring of tryptophan dimergene clusters from the environment. J. Am. Chem. Soc. 2013;135:17906–17912. doi: 10.1021/ja408683p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlop-Powers Z, Banik JJ, Owen JG, Craig JW, Brady SF. Selective enrichment of environmental DNA libraries for genes encoding nonribosomal peptides and polyketides by phosphopantetheine transferase-dependent complementation of siderophore biosynthesis. ACS Chem. Biol. 2013;8:138–143. doi: 10.1021/cb3004918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlop-Powers Z, Owen JG, Reddy BV, Ternei MA, Brady SF. Chemical-biogeographic survey of secondary metabolism in soil. Pro. Natl. Acad. Sci. USA. 2014;111:3757–3762. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ. Plans as a source of anti-cancer agents. J. Ethnopharmacol. 2005;100:72–79. doi: 10.1016/j.jep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Craig JW, Chang F-Y, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl. Environ. Microbiol. 2010;76:1633–1641. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craney A, Ozimok C, Pimentel-Elardo SM, Capretta A, Nodwell JR. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 2012;19:1020–1027. doi: 10.1016/j.chembiol.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Cueto M, Jensen PR, Kauffman C, Fenical W, Lobkovsky E, Clardy J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 2001;64:1444–1446. doi: 10.1021/np0102713. [DOI] [PubMed] [Google Scholar]
- Dewick PM. Medicinal Natural Products: A Biosynthetic Approach. 3rd edn. Chichester, West Sussex, England ; New York NY, USA: John Wiley & Sons; 2009. [Google Scholar]
- Endo A. The discovery and development of HMG-CoA reductase inhibitors. J. Lipid Res. 1992;33:1569–1582. [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Chakraborty D, Dewell SB, Reddy BV, Brady SF. Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. J. Am. Chem. Soc. 2012;134:2981–2987. doi: 10.1021/ja207662w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari BC, Binnerup SJ, Gillings M, Ferrari BC, Binnerup SJ, Gillings M. Microcolony Cultivation on a Soil Substrate Membrane System Selects for Previously Uncultured Soil Bacteria. Appl. Environ. Microbiol. 2005;71 doi: 10.1128/AEM.71.12.8714-8720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch KM, Gurgui C, Heycke N, van der Sar Sa, Anderson Sa, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- Fitzsimons MS, Novotny M, Lo CC, Dichosa AE, Yee-Greenbaum JL, Snook JP, Gu W, Chertkov O, Davenport KW, McMurry K, et al. Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome. Genome Res. 2013;23:878–888. doi: 10.1101/gr.142208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to Their Use in the Isolation of B. Influenzae. Br. J. Exp. Pathol. 1929;10:226–236. [Google Scholar]
- Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- Fujita MJ, Kimura N, Sakai A, Ichikawa Y, Hanyu T, Otsuka M. Cloning and heterologous expression of the vibrioferrin biosynthetic gene cluster from a marine metagenomic library. Biosci. Biotechnol. Biochem. 2011;75:2283–2287. doi: 10.1271/bbb.110379. [DOI] [PubMed] [Google Scholar]
- Fujita MJ, Kimura N, Yokose H, Otsuka M. Heterologous production of bisucaberin using a biosynthetic gene cluster cloned from a deep sea metagenome. Mol. BioSyst. 2012;8:482–485. doi: 10.1039/c1mb05431g. [DOI] [PubMed] [Google Scholar]
- Garcia JAL, Fernández-Guerra A, Casamayor EO. A close relationship between primary nucleotides sequence structure and the composition of functional genes in the genome of prokaryotes. Mol. Phylogen. Evol. 2011;61:650–658. doi: 10.1016/j.ympev.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Gatte-Picchi D, Weiz A, Ishida K, Hertweck C, Dittmann E. Functional analysis of environmental DNA-derived microviridins provides new insights into the diversity of the tricyclic peptide family. Appl. Environ. Microbiol. 2014;80:1380–1387. doi: 10.1128/AEM.03502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 2014;21:509–518. doi: 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontang EA, Gaudencio SP, Fenical W, Jensen PR. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl. Environ. Microbiol. 2010;76:2487–2499. doi: 10.1128/AEM.02852-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MA, Till R, Smith MCM. Integration Site for Streptomyces Phage cpBT1 and Development of Site-Specific Integrating Vectors. J. Bacteriol. 2003;185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindberg RV, Ishoey T, Brinza D, Esquenazi E, Coates RC, Liu WT, Gerwick L, Dorrestein PC, Pevzner P, Lasken R, et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS One. 2011;6:e18565. doi: 10.1371/journal.pone.0018565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H, Stockwell VO, Henkels MD, Nowak-Thompson B, Loper JE, Gerwick WH. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 2007;14:53–63. doi: 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- Hoertz AJ, Hamburger JB, Gooden DM, Bednar MM, McCafferty DG. Studies on the biosynthesis of the lipodepsipeptide antibiotic Ramoplanin A2. Bioorg. Med. Chem. 2012;20:859–865. doi: 10.1016/j.bmc.2011.11.062. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Merrick MJ. Genetics of antibiotic production. Bacteriol. Rev. 1977;41:595–635. doi: 10.1128/br.41.3.595-635.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 2009;27:462–464. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- Inaoka T, Takahashi K, Yada H, Yoshida M, Ochi K. RNA polymerase mutation activates the production of a dormant antibiotic 3,3'-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 2004;279:3885–3892. doi: 10.1074/jbc.M309925200. [DOI] [PubMed] [Google Scholar]
- Ishida K, Lincke T, Behnken S, Hertweck C. Induced biosynthesis of cryptic polyketide metabolites in a Burkholderia thailandensis quorum sensing mutant. J. Am. Chem. Soc. 2010;132:13966–13968. doi: 10.1021/ja105003g. [DOI] [PubMed] [Google Scholar]
- Ji HF, Li HJ, Zhang HY. Natural products and drug discovery. EMBO Rep. 2009;10:194–200. doi: 10.1038/embor.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein T, Lewis K, Epstein SS. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- Kakirde KS, Wild J, Godiska R, Mead DA. Gram negative shuttle BAC vector for heterologous expression of metagenomic libraries. Gene. 2011;475:57–62. doi: 10.1016/j.gene.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallifidas D, Kang HS, Brady SF. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNAgene clusters. J. Am. Chem. Soc. 2012;134:19552–19555. doi: 10.1021/ja3093828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Zabinsky R, Bowerman S, Baker DR, Lidstrom ME, Chistoserdova L. Fluorescence in situ hybridization-flow cytometry-cell sorting-based method for separation and enrichment of type I and type II methanotroph populations. Appl. Environ. Microbiol. 2006;72:4293–4301. doi: 10.1128/AEM.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa A, Gagunashvili AN, Gulder TA, Morinaka BI, Daolio C, Godejohann M, Miao VP, Piel J, Andresson O. Metagenomic natural product discovery in lichen provides evidence for a family of biosynthetic pathways in diverse symbioses. Pro. Natl. Acad. Sci. USA. 2013;110:E3129–E3137. doi: 10.1073/pnas.1305867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HS, Brady SF. Arimetamycin A: improving clinically relevant families of natural products through sequence-guided screening of soil metagenomes. Angew. Chem. 2013;52:11063–11067. doi: 10.1002/anie.201305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K, Wang G, Okamoto S, Ochi K. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 2007;274:311–315. doi: 10.1111/j.1574-6968.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, Ziemert N, Gonzalez DJ, Duggan BM, Nizet V, Dorrestein PC, Moore BS. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Pro. Natl. Acad. Sci. USA. 2013;110:E4407–4416. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Feng Z, Bauer JD, Kallifidas D, Calle PY, Brady SF. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers. 2010;93:833–844. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Bauer JD, Brady SF. An environmental DNA-derived type II polyketide biosynthetic pathway encodes the biosynthesis of the pentacyclic polyketide erdacin. Angew. Chem. 2009;48:6257–6261. doi: 10.1002/anie.200901209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Pro. Natl. Acad. Sci. USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, McVey SD, Radune D, Bergman NH, Phillippy AM. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhstoss S, Rao RN. Analysis of the integration function of the streptomycete bacteriophage phi C31. J. Mol. Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- Kvitko BH, McMillan IA, Schweizer HP. An improved method for oriT-directed cloning and functionalization of large bacterial genomic regions. Appl. Environ. Microbiol. 2013;79:4869–4878. doi: 10.1128/AEM.00994-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. Genome streamlining and chemical defense in a coral reef symbiosis. Pro. Natl. Acad. Sci. USA. 2012;109:20655–20660. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer-Safer PR, Levine M, Ward DC. Immunological method for mapping genes on Drosophila polytene chromosomes. Pro. Natl. Acad. Sci. USA. 1982;79:4381–4385. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Solomon G, Barrett JC, Resnick MA. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Pro. Natl. Acad. Sci. USA. 1997;94:7384–7387. doi: 10.1073/pnas.94.14.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K, Rothfeder M, Ling LL, Moore C, Zhang Q, Peoples A. In: Novel macrolactam compounds. U.S.P.a.T. Office, editor. USA: NovoBiotic Pharmaceuticals; 2009. [Google Scholar]
- Li MH, Ung PM, Zajkowski J, Garneau-Tsodikova S, Sherman DH. Automated genome mining for natural products. BMC Bioinf. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Goodman RM, Handelsman J. Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Appl. Environ. Microbiol. 2008;74:3302–3305. doi: 10.1128/AEM.02630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HK, Chung EJ, Kim JC, Choi GJ, Jang KS, Chung YR, Cho KY, Lee SW. Characterization of a Forest Soil Metagenome Clone That Confers Indirubin and Indigo Production on Escherichia coli. Appl. Environ. Microbiol. 2005;71:7768–7777. doi: 10.1128/AEM.71.12.7768-7777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jiang H, Haltli B, Kulowski K, Muszynska E, Feng X, Summers M, Young M, Graziani E, Koehn F, et al. Rapid Cloning and Heterologous Expression of the Meridamycin Biosynthetic Gene Cluster Using a Versatile Escherichia coli-Streptomyces Artificial Chromosome Vector, pSBAC. J. Nat. Prod. 2009;72:389–395. doi: 10.1021/np8006149. [DOI] [PubMed] [Google Scholar]
- Liu J, Li J, Feng L, Cao H, Cui Z. An improved method for extracting bacteria from soil for high molecular weight DNA recovery and BAC library construction. J. Microbiol. 2010;48:728–733. doi: 10.1007/s12275-010-0139-1. [DOI] [PubMed] [Google Scholar]
- Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil Ia, Tiong CL, Minor C, August PR, Grossman TH, Loiacono Ka, Lynch Ba, Phillips T, Narula S, Sundaramoorthi R, et al. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotechnol. 2001;3:301–308. [PubMed] [Google Scholar]
- Martin MF, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, Clum A, Goodwin L, Woyke T, Lapidus A, Klenk HP, et al. The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS One. 2012;7:e48837. doi: 10.1371/journal.pone.0048837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer QC, Burton SG, Cowan Da. Subtractive hybridization magnetic bead capture: a new technique for the recovery of full-length ORFs from the metagenome. Biotechnol. J. 2007;2:36–40. doi: 10.1002/biot.200600156. [DOI] [PubMed] [Google Scholar]
- Moore JM, Bradshaw E, Seipke RF, Hutchings MI, McArthur M. Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. Methods Enzymol. 2012;517:367–385. doi: 10.1016/B978-0-12-404634-4.00018-8. [DOI] [PubMed] [Google Scholar]
- Morimoto S, Fujii T. A new approach to retrieve full lengths of functional genes from soil by PCR-DGGE and metagenome walking. Appl. Microbiol. Biotechnol. 2009;83:389–396. doi: 10.1007/s00253-009-1992-x. [DOI] [PubMed] [Google Scholar]
- Mutka SC, Carney JR, Liu Y, Kennedy J. Heterologous Production of Epothilone C and D in Escherichia coli. Biochemistry. 2006;45:1321–1330. doi: 10.1021/bi052075r. [DOI] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Stewart AF. Techniques: Recombinogenic engineering--new options for cloning and manipulating DNA. Trends Biochem. Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- NCBI. New NCBI Handbook chapters: Eukaryotic and prokaryotic genome annotation pipelines. In NCBI News. 2013 [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM, Snader KM. The influence of natural products upon drug discovery. Nat. Prod. Rep. 2000;17:215–234. doi: 10.1039/a902202c. [DOI] [PubMed] [Google Scholar]
- Novy R, Drott D, Yaeger K, Mierendorf R. Overcoming the codon bias of E. coli for enhanced protein expression. Novations. 2001;12:1–3. [Google Scholar]
- Ochi K, Hosaka T. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 2013;97:87–98. doi: 10.1007/s00253-012-4551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano C, Garcia I, Gonzalez A, Rodriguez M, Rozas D, Rubio J, Sanchez-Hidalgo M, Brana AF, Mendez C, Salas JA. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014;7:242–256. doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Onaka H, Mori Y, Igarashi Y, Furumai T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011;77:400–406. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen JG, Reddy BV, Ternei MA, Charlop-Powers Z, Calle PY, Kim JH, Brady SF. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Pro. Natl. Acad. Sci. USA. 2013 doi: 10.1073/pnas.1222159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen JG, Robins KJ, Parachin NS, Ackerley DF. A functional screen for recovery of 4'-phosphopantetheinyl transferase and associated natural product biosynthesis genes from metagenome libraries. Environ. Microbiol. 2012;14:1198–1209. doi: 10.1111/j.1462-2920.2012.02699.x. [DOI] [PubMed] [Google Scholar]
- Peoples A, Ling LL, Lewis K, Zhang Z. In: Novel Antibiotics. U.S.P.a.T. Office, editor. USA: NovoBiotic Pharmaceuticals; 2011. [Google Scholar]
- Pestov DG, Polonskaia M, Lau LF. Recombinant Polyketide Synthesis in Streptomyces: Engineering of Improved Host Strains. BioTechniques. 1999;26:106–110. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- Pop M. Genome assembly reborn: recent computational challenges. Briefings in Bioinf. 2009;10:354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Davo A, Villarreal-Gomez LJ, Forschner-Dancause S, Bull AT, Stach JE, Smith DC, Rowley DC, Jensen PR. Targeted search for actinomycetes from nearshore and deep-sea marine sediments. FEMS Microbiol. Ecol. 2013;84:510–518. doi: 10.1111/1574-6941.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Reddy BV, Milshteyn A, Charlop-Powers Z, Brady SF. eSNaPD: a versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chem. Biol. 2014 doi: 10.1016/j.chembiol.2014.06.007. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood Da, van Wezel GP. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbel L, Knappe TA, Linne U, Xie X, Marahiel MA. Erythrochelin-a hydroxamate-type siderophore predicted from the genome of Saccharopolyspora erythraea. FEBS J. 2010;277:663–676. doi: 10.1111/j.1742-4658.2009.07512.x. [DOI] [PubMed] [Google Scholar]
- Sakula A. Selman Waksman (1888–1973), discoverer of streptomycin: a centenary review. Br. J. Dis. Chest. 1988;82:137. doi: 10.1016/0007-0971(88)90005-8. [DOI] [PubMed] [Google Scholar]
- Schatz A, Bugie E, Waksman SA. Streptomycin, a Substance Exhibiting Antibiotic Activity Against Gram-Positive and Gram-Negative Bacteria. Exp. Biol. Med. 1944;55:66–69. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- Schatz A, Waksman SA. Effect of streptomycin upon Mycobacterium tuberculosis and related organisms. Proc. Soc. Exptl. Biol. & Med. 1944;57:244–248. [Google Scholar]
- Schroeckh V, Scherlach K, Nutzmann H-W, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of 53 archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Seow K-t, Meurer G, Gerlitz M, Wendt-pienkowski E, Hutchinson CR, Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA : a means to access and use genes from uncultured microorganisms. A Study of Iterative Type II Polyketide Synthases, Using Bacterial Genes Cloned from Soil DN. Microbiology. 1997;23:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Luo Y, Zhao H. Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol. BioSyst. 2011;7:1056–1059. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki RS. Shanidar IV, a Neanderthal Flower Burial in Northern Iraq. Science. 1975;190:880–881. [Google Scholar]
- Stahelin HF. The history of cyclosporin A (Sandimmune®) revisited: Another point of view. Experientia. 1996;52:5–13. doi: 10.1007/BF01922409. [DOI] [PubMed] [Google Scholar]
- Stahl DA, Lane DJ, Olsen GJ, Pace NR. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science. 1984;224:409–411. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- Starcevic A, Zucko J, Simunkovic J, Long PF, Cullum J, Hranueli D. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008;36:6882–6892. doi: 10.1093/nar/gkn685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DC, Conway KR, Pearce N, Villegas-Penaranda LR, Garza AG, Boddy CN. Alternative sigma factor over-expression enables heterologous expression of a type II polyketide biosynthetic pathway in Escherichia coli. PLoS One. 2013;8:e64858–e64858. doi: 10.1371/journal.pone.0064858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Hosaka T, Ochi K. Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2) J. Antibiot. 2010;63:477–481. doi: 10.1038/ja.2010.53. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Daae FL, Sandaa RA, Ovreas L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 1998;64:53–62. doi: 10.1016/s0168-1656(98)00103-5. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990a;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V, Salte K, Sorheim R, Goksoyr J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl. Environ. Microbiol. 1990b;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V, Sorheim R, Goksoyr J. Total bacterial diversity in soil and sediment communities--a review. J. Ind. Microbiol. 1996;17:170–178. [Google Scholar]
- Tyndall J. The Optical Deportment of the Atmosphere in Relation to the Phenomena of Putrefaction and Infection. Philos. Trans. R. Soc. Lond. 1876;766:27–74. doi: 10.1136/bmj.1.787.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Abe T, Ikemura T, Watanabe K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat. Biotechnol. 2005;23:88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Pro. Natl. Acad. Sci. USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. J. Antibiot. 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Andersen RJ, Davies J. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2000;2:2401–2404. doi: 10.1021/ol005860z. [DOI] [PubMed] [Google Scholar]
- Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller R, Weller JW, Ward DM. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl. Environ. Microbiol. 1991;57:1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel SC, Gross F, Zhang Y, Fu J, Stewart AF, Muller R. Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via red/ET recombineering. Chem. Biol. 2005;12:349–356. doi: 10.1016/j.chembiol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Wilson MC, Mori T, Ruckert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UAE, Heycke N, Schmitt S, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- Woodhouse JN, Fan L, Brown MV, Thomas T, Neilan BA. Deep sequencing of non-ribosomal peptide synthetases and polyketide synthases from the microbiomes of Australian marine sponges. ISME J. 2013;7:1842–1851. doi: 10.1038/ismej.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Pro. Natl. Acad. Sci. USA. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Straight PD, Hrvatin S, Dorrestein PC, Bumpus SB, Jao C, Kelleher NL, Kolter R, Walsh CT. Genome-wide high-throughput mining of natural-product biosynthetic gene clusters by phage display. Chem. Biol. 2007;14:303–312. doi: 10.1016/j.chembiol.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Yu W-H, Su S-C, Lee C-Y. A novel retrieval system for nearly complete microbial genomic fragments from soil samples. J. Microbiol. Methods. 2008;72:197–205. doi: 10.1016/j.mimet.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, He J, Yang M, Yen M, Yin J. Identifying natural product biosynthetic genes from a soil metagenome by using T7 phage selection. ChemBioChem. 2009;10:2599–2606. doi: 10.1002/cbic.200900297. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Werling U, Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Ishida K, Weiz A, Hertweck C, Dittmann E. Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Appl. Environ. Microbiol. 2010;76:3568–3574. doi: 10.1128/AEM.02858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012;7:e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann K, Engeser M, Blunt JW, Munro MHG, Piel J. Pederin-type pathways of uncultivated bacterial symbionts: analysis of o-methyltransferases and generation of a biosynthetic hybrid. J. Am. Chem. Soc. 2009;131:2780–2781. doi: 10.1021/ja808889k. [DOI] [PubMed] [Google Scholar]