Abstract
Nucleotide excision repair is a primary pathway in cells for coping with DNA interstrand cross-links (ICLs). Recently, C4′-oxidized (C4-AP) and C5′-oxidized abasic sites (DOB) that are produced following hydrogen atom abstraction from the DNA backbone were found to produce ICLs. Because some of the ICLs derived from C4-AP and DOB are too unstable to characterize in biochemical processes, chemically stable analogues were synthesized [Ghosh, S., and Greenberg, M. M. (2014) J. Org. Chem.79, 5948–5957]. UvrABC incision of DNA substrates containing stabilized analogues of the ICLs derived from C4-AP and DOB was examined. The incision pattern for the ICL related to the C4′-oxidized abasic site was typical for UvrABC substrates. UvrABC cleaved both strands of the substrate containing the C4-AP ICL analogue, but it was a poor substrate. UvrABC incised <30% of the C4-AP ICL analogue over an 8 h period, raising the possibility that this cross-link will be inefficiently repaired in cells. Furthermore, double-strand breaks were not detected upon incision of an internally labeled hairpin substrate containing the C4-AP ICL analogue. UvrABC incised the stabilized analogue of the DOB ICL more efficiently (∼20% in 1 h). Furthermore, the incision pattern was unique, and the cross-linked substrate was converted into a single product, a double-strand break. The template strand was exclusively incised on the template strand on the 3′-side of the cross-linked dA. Although the outcomes of the interaction between UvrABC and these two cross-linked substrates are different from one another, they provide additional examples of how seemingly simple lesions (C4-AP and DOB) can potentially exert significant deleterious effects on biochemical processes.
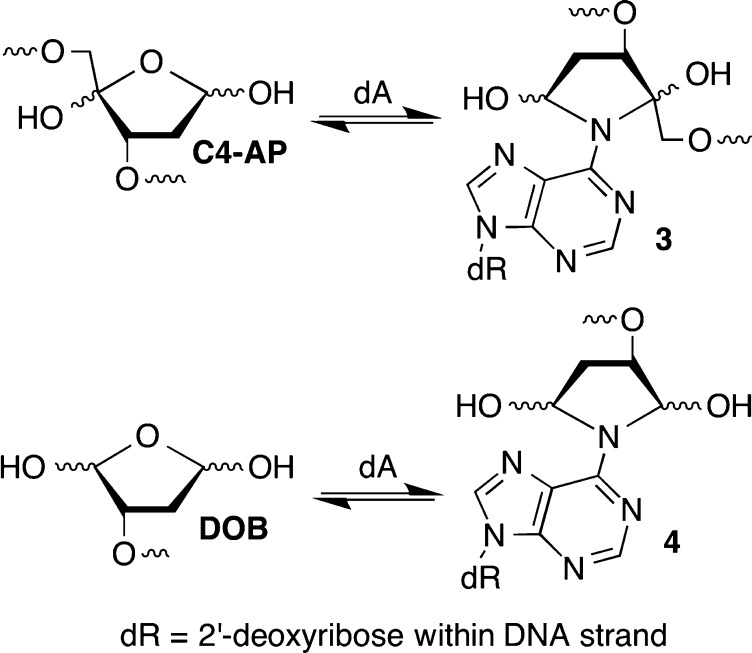
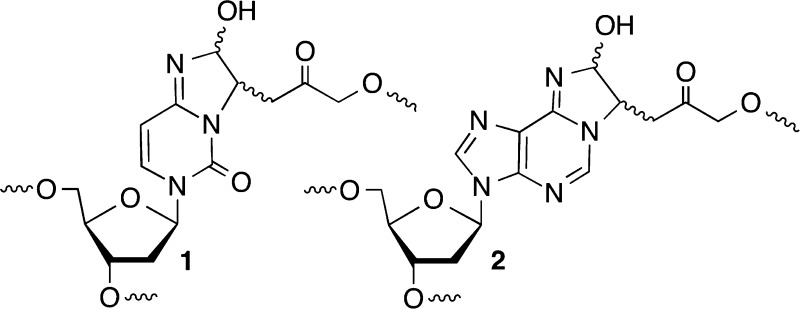
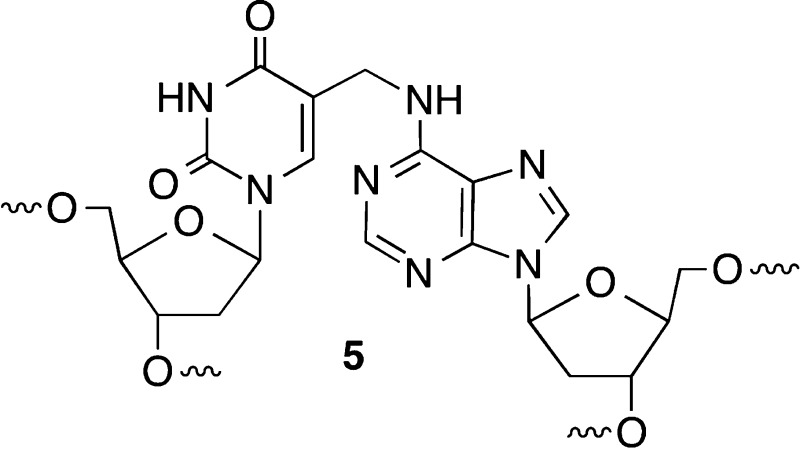
DNA interstrand cross-links are produced by a variety of exogenous electrophiles that react directly with two nucleotides in opposite strands.1−3 Some ICLs, such as those derived from acrolein, are in equilibrium with single-strand modification products, and biochemical studies of them are typically conducted with chemically stabilized cross-linked analogues.4,5 Recently, it was discovered that the AP, C4-AP, and DOB DNA lesions also form ICLs.6−12 C4-AP forms two types of ICLs; one of them in which the original strand containing the oxidized abasic site is cleaved (1 and 2) has been detected in cellular DNA (1).8 The DOB-containing strand in ICL 4 is also cleaved. In another ICL derived from C4-AP (3), both strands are intact.11 However, 3 as well as that produced from DOB (4) forms reversibly (Scheme 1). In ICLs 1–4 the oxidized abasic site is typically bonded to the nucleotide on the opposing strand that forms a base pair with the 3′-adjacent nucleotide. This relative positioning of the cross-linked nucleotides is consistent with the exclusive reactivity with dA and dC whose exocyclic amines lie in the major groove of DNA. The stabilities of ICLs involving AP lesions vary depending upon the native nucleotide to which it is cross-linked.6,7 Cross-links formed with dA are more stable than ICLs between AP and dG. ICLs are a dangerous form of DNA damage because they are absolute blocks of DNA replication and transcription. Although ICLs can be excised by other pathways, nucleotide excision repair is a primary pathway for their removal from DNA.13−16 In recent years, nucleotide excision repair of ICLs has produced unexpected and interesting observations. Investigations of 1 and two other ICLs revealed that NER actually misrepairs the lesions by converting them into the most dangerous form of DNA damage, double-strand breaks.17−19 These reports provided the impetus to examine the outcomes of interactions between the bacterial NER system, UvrABC with 3 and 4.
Scheme 1. Reversible Formation of ICLs from C4-AP and DOB.
UvrABC is present in prokaryotes and is still used to study DNA repair, despite its simplicity compared to the complexity of eukaryotic NER.20−23 UvrABC repair of psoralen cross-links occurs selectively on one strand or another but avoids dsb formation.24−26 Strand selection depends upon the DNA sequence, but how the NER system chooses one strand over another is not understood. Strand selection was of paramount importance during incision of 1, which is a complex (clustered) lesion.27,28 Complex (clustered) lesions contain two or more damaged sites within 1.5–2.0 turns of duplex DNA and are often more difficult to repair than isolated lesions.29−34 ICL 1 is an example of a clustered lesion because it also contains a strand break. NER incises the bottom strand of 1 approximately 15% of the time, producing a dsb.17 In addition, dsbs are produced ≥25% of the time when UvrABC acts on an ICL between dA and T (5).18 Finally, ICLs resulting from reaction of DNA with the antitumor agent mitomycin C are also transformed into dsbs by UvrABC.19 These observations raise the question of how general misrepair of ICLs is and led us to examine the NER of stabilized analogues of 3 and 4.
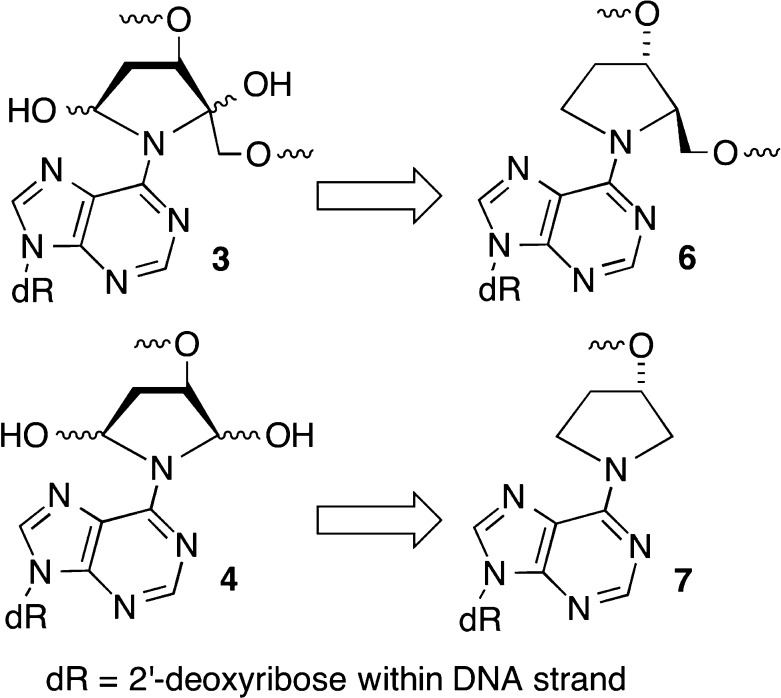
ICLs 3 and 4 form rapidly but reversibly from C4-AP and DOB, respectively.10−12 Cross-linking occurs preferentially with dA opposite a 3′-adjacent thymidine and not at all with dG. The half-life for reversion to the oxidized abasic sites is less than 4 and 12 h for ICLs 3 and 4, respectively. Although 3 and 4 were isolated by denaturing polyacrylamide gel electrophoresis (PAGE), they are too unstable to be used reliably in subsequent NER experiments. Consequently, a method for synthesizing oligonucleotides containing stabilized analogues [6 and 7 (Scheme 2)] was developed.35 The analogues retain the stereochemistry of the original lesions in natural DNA but lack the hydroxyl groups that are necessary for reversible cross-link formation.
Scheme 2. Stabilized Analogues of ICLs Derived from C4-AP (3) and DOB (4).
Materials and Methods
Materials and General Methods
Oligonucleotides were prepared on an Applied Biosystems Inc. 394 DNA synthesizer. Commercially available DNA synthesis reagents were obtained from Glen Research Inc. DNA substrates used in this study are presented in Chart 1. T4 polynucleotide kinase, terminal deoxynucleotide transferase, and T4 DNA ligase were obtained from New England Biolabs. [α-32P]dCTP, [γ-32P]ATP, and [α-32P]cordycepin 5′-triphosphate were purchased from PerkinElmer. Analysis of radiolabeled oligonucleotides was conducted using a Storm 840 phosphorimager and ImageQuant TL. UvrABC was obtained as previously described.36,37 C18-Sep-Pak cartridges were obtained from Waters. Experiments using radiolabeled oligonucleotides were analyzed following PAGE using a Storm 840 phosphorimager and Imagequant TL.
Chart 1. DNA Molecules Used in This Study.
UvrABC Reaction
The purified cross-linked DNA was resuspended in 100 mM NaCl and 10 mM potassium phosphate buffer (pH 7.2, 25 μL) and rehybridized by heating to 90 °C (2 min), cooling to room temperature over the course of 2 h, and storing at 4 °C overnight. UvrA, UvrB, and UvrC were freshly prepared from stock solutions and heated individually at 65 °C for 10 min before use. They were added sequentially to the reaction mixtures. The reaction buffer contains 50 mM Tris-HCl (pH 7.5), MgCl2 (10 mM), KCl (50 mM), dTT (5 mM), and ATP (1.0 mM). The cross-linked DNA (2.0 nM) was incubated with UvrA (20 nM), UvrB (100 nM), and UvrC (50 nM) in a total volume of 20 μL at 55 °C. The concentration of cross-linked DNA was based upon the specific activity of the initially labeled oligonucleotide, which was determined by counting the radioactivity (using a liquid scintillation counter) and measuring the concentration (A260). After 60 min, the reaction was quenched by precipitation with 5 M NH4OAc (5 μL), 1 μg/μL calf thymus DNA (5 μL), and cold ethanol (75 μL). The incision products were separated by 20% denaturing PAGE and visualized using the phosphorimager. For the time course reaction (total volume of 60 μL), aliquots (7 μL) were removed at the prescribed times and immediately quenched by addition of 90% formamide loading buffer (3 μL) and heating at 90 °C for 2 min.
Results
NER Substrate Design and Preparation
ICLs containing analogues 6 and 7 were designed on the basis of local sequences in which C4-AP and DOB, respectively, are known to produce cross-links. The lengths of the substrates (>50 bp) are also sufficient for UvrABC to bind and incise DNA. The cross-linked substrates were synthesized by a combination of solid-phase oligonucleotide synthesis and enzymatic ligation, as previously described.35 The enzyme ligation procedure also provided a convenient method for controlling which strand and which terminus were 32P-labeled. Because the UvrABC experiments are conducted at 55 °C, the stability of duplex 9 that contains the DOB analogue (7) was carefully examined. The melting temperature of the analogous un-cross-linked duplex was estimated to be ∼75 °C using a standard TM calculator (www.idtdna.com/analyzer/Applications/OligoAnalyzer/). Nonetheless, hybridization persistence was tested using 5′-32P-t-9 [please note that the strand labeled in each substrate is named “t” for the strand containing the abasic site analogue (top strand in Chart 1) or “b” for the strand containing the dA that is involved in the cross-link (bottom strand in Chart 1)] and subjecting this to the reaction conditions (without UvrABC) in the presence of 5 equiv of the unlabeled single-stranded 25mer (10) (see the Supporting Information). Aliquots were removed over the course of 60 min and analyzed by nondenaturing PAGE. There was no evidence of either the loss of 5′-32P-t-9 or the appearance of 5′-32P-10, indicating that the hybridized complex was stable to the UvrABC reaction conditions. UvrABC incision of 8 and 9 was calibrated using a duplex (12) containing a C5-fluoresceinylated thymidine derivative, which has previously been used as a benchmark for comparing enzyme activity.38 In addition, a hairpin substrate (11) containing 6 was also prepared and labeled either at the 5′-end of its top strand (5′-32P-t-11) or internally with 32P at the phosphate between dC39 and dG40. The integrity of 5′-32P-t-11 was confirmed by treatment with the restriction enzyme CviQI and comparing the migration of the product with that from CviQI treatment of 5′-32P-t-8, because both would generate the same radiolabeled product (see the Supporting Information). On the other hand, the integrity of internally labeled 11 was verified by treatment with the restriction enzymes Fnu4HI (generating a 44mer product) and HpyCH4V. These substrates facilitated determination of dsb formation (see the Supporting Information).
UvrABC Incision of an Analogue of the C4-AP ICL (6)
Unambiguous determination of the site(s) at which UvrABC incises an ICL comprised of 6 was conducted in four independent experiments in which each of the termini of 8 was 32P-labeled. Nucleotides in both strands of 8 were incised (Figure 1). A single nucleotide (dG22 and dC83) was incised on the 5′-side of the ICL in each strand. The 5′-incision site consisted of eight nucleotides from the cross-link in the “top” strand (that containing the abasic site analogue) and seven nucleotides in the strand containing the dA portion (“bottom” strand). The incision pattern on the 3′-side of 6 was more diffuse in both strands and closer to the cross-linked nucleotides. In the top strand of 8, the two nucleotides adjacent to the cross-linked abasic site (T92 and A93) were the major sites of incision. T94 was also incised but significantly less so than T92 and dA93 (Figure 1). The incision pattern on the 3′-side of the ICL of the bottom strand was also distributed over three nucleotides. However, the relative cleavage intensity at dA32, T33, and dA34 was more evenly distributed than in the top strand, and the incised nucleotides were further removed from the cross-linked nucleotide. Although the positions of incised nucleotides were shifted, the overall sizes of the unhooked oligonucleotides were very similar in both strands. Incision sites in the top strand spanned 10–12 nucleotides, whereas sites in the bottom strand consisted of 11–13 nucleotides.
Figure 1.
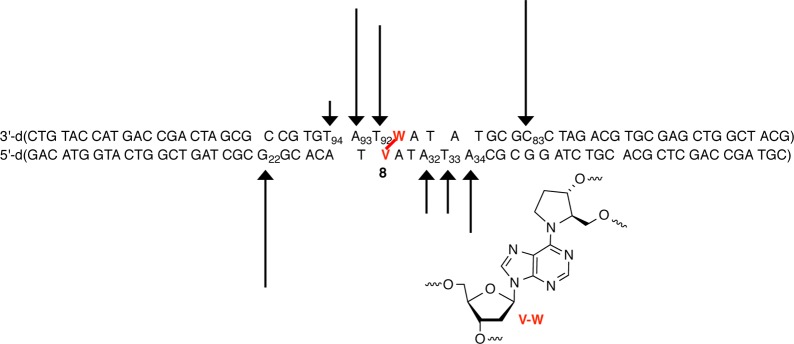
Histogram describing UvrABC incision of C4-AP ICL analogue 6 in 8. Data taken from four individual experiments in which each terminus of 8 was 32P-labeled.
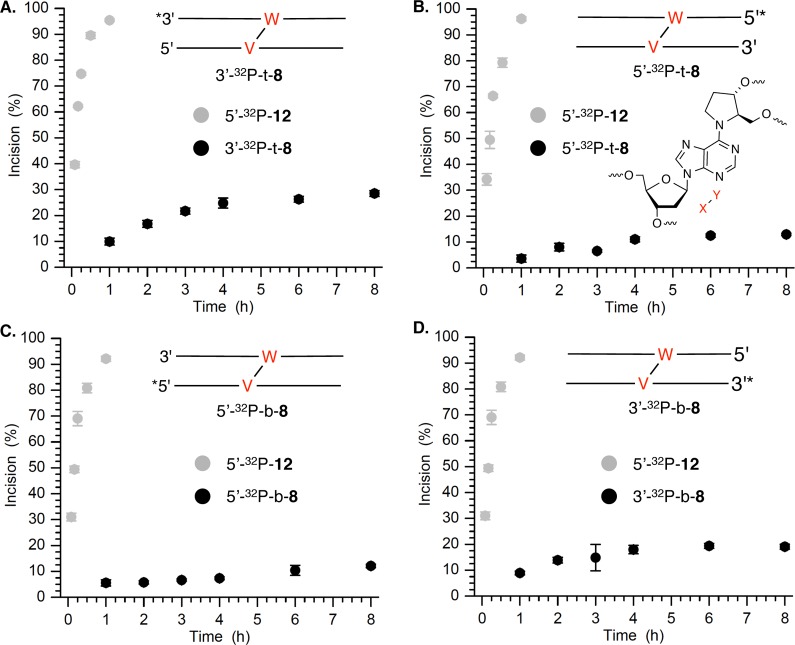
Although 6 was a substrate for UvrABC, it was incised significantly less efficiently by UvrABC than a duplex containing fluoresceinylated dT (12), which is often used as a basis for comparison (Figure 2) (see the Supporting Information). Greater than 90% of 5′-32P-12 was cleaved in 60 min. In contrast, the maximal cleavage observed in any region of 8 was less than 30% in 8 h. Incision on the labeled strand in the region between the ICL and the 32P label was quantified even though other cleaved regions could be detected. The overall extent of strand scission on the top strand was slightly greater than that on the bottom strand of 8. In addition, although cleavage on the 3′-side of the ICL was more diffuse in each strand than on the 5′-side, overall it was greater. Consequently, the extent of cleavage in the top strand on the 3′-side of the ICL (27–28%) was higher than in any other region of the substrate. Most notable is that the level of incision at T92–T94 was approximately twice that on the 5′-side of the ICL in the same strand at C83. In comparison, incision on either side of 6 in the bottom strand of 8 was much closer to 1:1, although as previously stated the total level of cleavage on the 3′-side of the ICL was consistently greater than on the 5′-side.
Figure 2.
Time dependence of UvrABC incision of C4-AP ICL analogue 6 in 32P-labeled 8 compared to that in 5′-32P-12. The total amount of incision at all nucleotides in the region between the 32P label and the cross-link is plotted: (A) 3′-32P-t-8, (B) 5′-32P-t-8, (C) 5′-32P-b-8, and (D) 3′-32P-b-8.
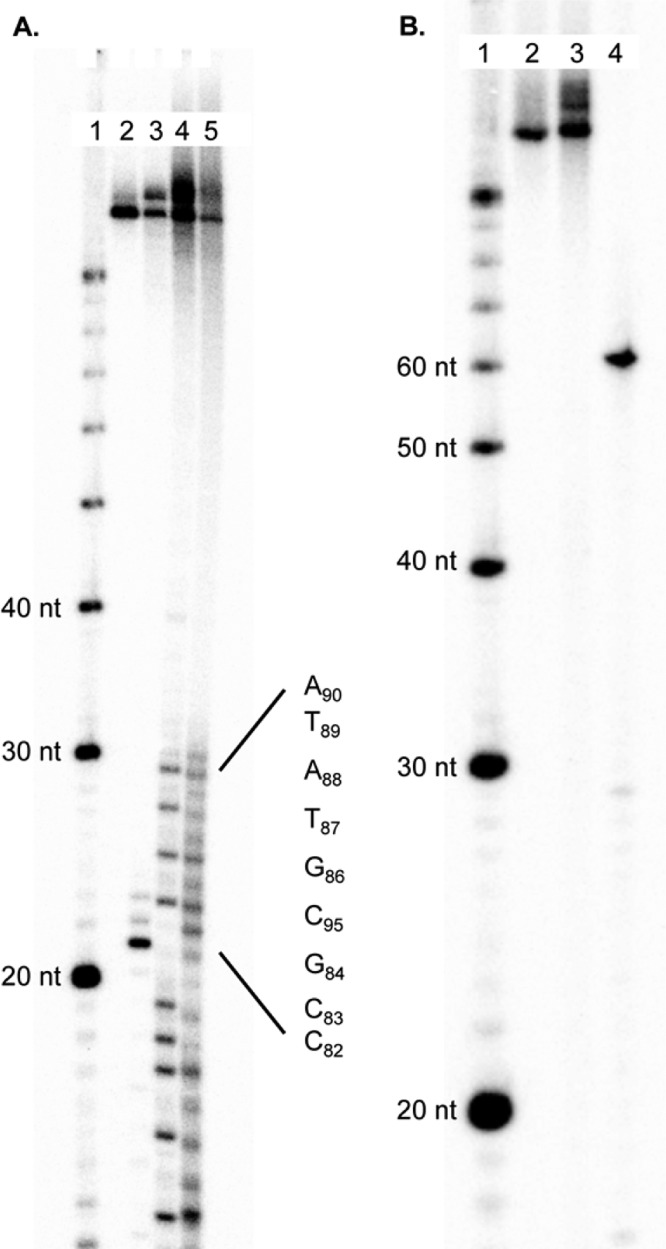
Because both strands of cross-linked 6 are incised by UvrABC, we determined whether 8 undergoes double-strand cleavage. Initial nondenaturing PAGE analysis of experiments with 5′-32P-t-8, 3′-32P-t-8, and 5′-32P-b-8 indicated ∼1–3% of the product migrated through the gel at the same rate as the anticipated dsb (see the Supporting Information). This would correspond to a ssb:dsb ratio of ∼10, which is slightly greater than that previously observed for 1.17 A control experiment was designed to determine whether the observed dsb product could be an artifact that results from the hybridization of products released from two independent single-strand incisions. On the basis of the incision sites determined for each strand in 8 (Figure 1), the anticipated dsb product (13) was prepared and 32P-labeled at the 5′-terminus of its shorter strand. This radiolabeled product was incubated under the UvrABC incision conditions (55 °C, 3 h) separately with 1 and 5 equiv of 15, which contains the identical complementary binding region as the unlabeled strand in 5′-32P-13 but is 13 nucleotides longer. The initial concentration of 5′-32P-13 (0.26 nM) was carefully chosen and based upon the amount of incision detected from the UvrABC reaction with 8. More than 9.8% of the longer, slower migrating exchange product (5′-32P-14) was observed following incubation with 1 equiv of 15 and 21.9% in the presence of 5 equiv (see the Supporting Information). Given the ambiguity introduced by this experiment, a hairpin substrate containing 6 (11) was prepared in which either the phosphate between dC39 and dG40 (Chart 1) or the 5′-terimus of the hairpin was 32P-labeled. Reaction of UvrABC with 5′-32P-11 yielded cleavage at a position and a yield that were the same as those of the analogously labeled 8, indicating that the slight modification of the substrate introduced in the form of the hairpin did not alter the recognition of the cross-link (6) by proteins (Figure 3A). Internally labeled 32P-11 was then subjected to UvrABC incision. If both strands of the internally labeled substrate were incised in the region between 6 and the hairpin loop at positions comparable to those in 8, the double-strand break would manifest itself in the form of a 54-nucleotide 32P-labeled product [16 (Figure 3B)]. UvrABC incision of 5′-32P-11 or internally labeled 32P-11 gave rise to products that migrate slightly more slowly than the cross-linked substrate. These products are believed to result from single-strand cleavage of 11 in the region between the hairpin loop and 6, as oligonucleotides that comigrated with these were also observed when ligation was incomplete (data not shown). Moreover, no product that migrated like the independently synthesized anticipated dsb product (5′-32P-16) was detected.
Figure 3.
UvrABC incision of hairpin DNA (11) containing C4-AP ICL analogue 6. (A) Incision of 5′-32P-11: lane 1, nucleotide size markers; lane 2, unreacted 5′-32P-11; lane 3, incised 5′-32P-11; lane 4, A + G reaction; lane 5, T + C reaction. (B) Incision of internally labeled 32P-11: lane 1, nucleotide size markers; lane 2, unreacted 11; lane 3, incised 11; lane 4, 5′-32P-16.
UvrABC Incision of an Analogue of the DOB ICL (7)
The structure of 7 (4) is similar to that of 1 in that the ICL is part of a complex lesion that contains a strand break in addition to a cross-link.17 Consequently, the choice of strand incised is important because cleavage of the strand containing the dA portion of the ICL (“bottom” strand) would give rise to a double-strand break. Indeed, treatment of 3′-32P-b-9 gives rise to double-strand breaks via strand scission at dA29–dG31 (Figure 4). Although the level of cleavage at dG30 was slightly greater than at dG31, the amount of incision at either of these purines was considerably higher than at dA29. After 1 h, the total amount of incision over these nucleotides ranged from ∼16 to 22%, while incision of the fluoresceinylated dT (5′-32P-12) varied from ∼80 and 87% (Figure 5). No other sites of strand scission were detected by denaturing PAGE analysis of 3′-32P-b-9. Analysis of 5′-32P-b-9 or 3′-32P-t-9 also revealed a single region of incision by UvrABC (see the Supporting Information). In neither instance does the migration of the cleavage product correlate with strand scission on the side of the cross-link between it and the terminus where the substrate is 32P-labeled. The migration of the cleavage product formed from both substrates coincides with strand scission just beyond the cross-link position, which is consistent with cleavage at dA29–dG31 detected using 3′-32P-b-9. The extent of strand scission measured was also similar in these substrates. Extending the reaction time for 3′-32P-t-9 with UvrABC to 8 h yielded more than 60% strand scission, but products corresponding to cleavage in the region of dA29–dG31 were still the only ones observed (Figure 4) (see the Supporting Information). Finally, cleavage in the noncovalently bound strand would be detectable only in the substrate in which it is labeled (5′-32P-t-9). Denaturing PAGE analysis of this substrate does not show any cleavage, confirming that UvrABC does not incise DNA containing 7 in this region (data not shown).
Figure 4.
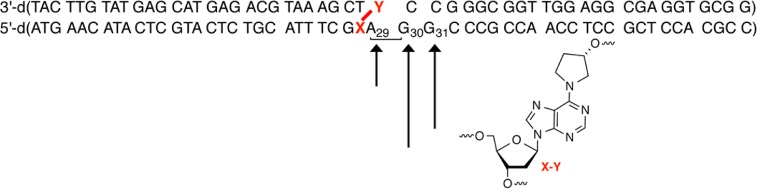
Histogram describing UvrABC incision of DOB ICL analogue 7 in 9. Data taken from four individual experiments in which one of the four termini of 9 was 32P-labeled.
Figure 5.
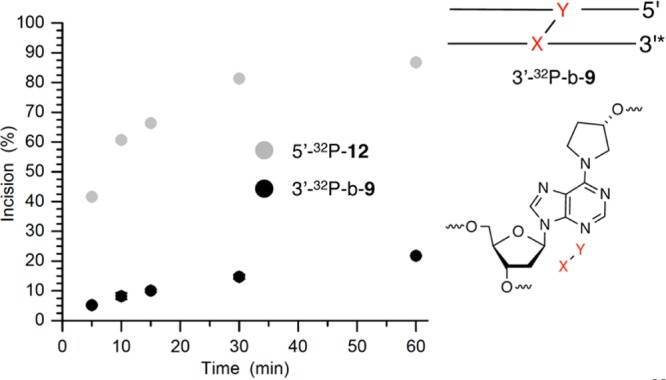
Time dependence of UvrABC incision of C4-AP ICL analogue 7 in 3′-32P-b-9 compared to that in 5′-32P-12.
Discussion
Nucleotide excision repair is a primary pathway for protecting the genome from interstrand cross-links. The mechanism(s) by which even the relatively simple bacterial NER system (e.g., UvrABC) recognizes its substrates is evolving but still not completely understood.13,39,40 Macroscopic distortion of the DNA duplex is certainly believed to be one structural feature that is recognized by the repair system. We characterized the UvrABC incision of two chemically stabilized analogues (Scheme 2) of interstrand cross-links produced by oxidatively damaged DNA (Scheme 1).
The C4-AP ICL analogue (6) is incised on both strands, which distinguishes it from psoralen-cross-linked DNA.24,41 Although the choice of incised strand varies depending upon DNA sequence, only one strand of psoralen-derived ICLs is cleaved by UvrABC. The sizes of the DNA fragments incised by UvrABC in 8 (10–13 nucleotides) are within the usual range.20 However, cleavage on the strand containing the cross-linked abasic site on the 3′-side of 6 occurs predominantly at the two adjacent nucleotides. This is one to two nucleotides closer to the damage site than one typically observes and is one difference between incision patterns on the abasic site-containing strand (“top”) and the “bottom” strand containing the dA that is cross-linked. Another difference between the interaction of the two strands in 8 with UvrABC is that incision on the two sides of the cross-link on the top strand are unequal. The level of cleavage on the 3′-side of the abasic site is ∼2-fold greater than on the 5′-side. Previously, it was shown that UvrABC incision of 1 was asymmetric.17 The inefficiency of incision is the most remarkable difference between 6 and other ICLs subjected to UvrABC. Although more than 90% of the standard UvrABC substrate, 12, is incised in 1 h, typically less than 10% of 8 is cleaved in this time frame. Even after reaction for 8 h, levels of cleavage of the strand in 8 that contains the abasic site and the opposite strand were less than 30 and 20%, respectively. Although it is not known at this time why 8 is cleaved so inefficiently, it suggests that DNA containing 6 is not significantly distorted. UvrABC incision of the C4-AP ICL analogue (6) is also different than that of other recently characterized cross-links, which have been observed to undergo double-strand scission. Utilization of internally 32P-labeled hairpin substrate containing 6 (11) unambiguously demonstrated that UvrABC does not generate dsbs in this molecule. Importantly, control experiments on ICL containing 6 revealed that products corresponding to dsbs can be produced under the reaction conditions from two independent cleavage events on complementary strands in separate molecules. This possibility may also explain the observed dsbs in other cross-linked substrates exposed to UvrABC.18,19
Incision of DOB cross-link analogue 7 in 9 was perhaps the most remarkable observation of any ICL reaction with UvrABC yet reported. UvrABC incision of 9 is not as efficient as the fluoresceinylated dT standard. However, DNA containing 7 is cleaved far more efficiently than cross-linked DNA containing the C4-AP analogue. We hypothesize that the cross-linked sugarlike components in 6 and 7 do not significantly perturb the duplex structure. However, 7 is accompanied by a strand break, which provides flexibility and gives rise to greater distortion that is recognized by UvrABC. The cleavage pattern of 9 on the 3′-side of the cross-linked dA (“bottom” strand) is similar to the corresponding location of the substrate containing the C4-AP ICL analogue in that it is distributed over three nucleotides adjacent to the cross-link. However, the region on the 3′-side of the cross-linked dA in 9 is the only one in the DNA containing the DOB ICL analogue (7) that is incised by UvrABC. Moreover, because DNA containing a cross-linked DOB already contains a nick, cleavage of the ICL on the bottom strand converts the substrate into a double-strand break. To the best of our knowledge, DOB ICL analogue 7 is the only cross-linked DNA that is exclusively transformed into a double-strand break, the most deleterious form of DNA damage.
Conclusions
Molecules that produce double-strand breaks and/or interstrand cross-links directly are of great interest as potential DNA-damaging therapeutic agents because these families of nucleic acid damage are difficult to repair.42−46 However, it is increasingly evident that DNA damage can be transformed into forms that are potentially more deleterious.47 For instance, it was discovered that seemingly simple abasic sites (AP, C4-AP, and DOB) spontaneously form interstrand cross-links.6,7,9−12 One type of cross-link was identified in cells (1), and its formation is catalyzed by the DNA sequence flanking it.8,11 In addition, UvrABC transforms cross-linked DNA containing 1 into a double-strand break ∼15% of the time.17
This work describes results for two chemically stabilized analogues of ICLs that add to a small but growing list of lesions that affect biochemical processes in unanticipated ways. The DOB lesion is produced by some of the most cytotoxic DNA-damaging agents.48 Only a short time ago it was demonstrated that DOB forms ICLs (4) in DNA.12 Exclusive conversion of the stabilized analogue (7) of 4 into a double-strand break by UvrABC is the first example of such selective misrepair. The interaction between UvrABC and the stabilized analogue (6) of the C4-AP ICL (3) is different but perhaps also significant. Unlike the other DNA ICLs whose interaction with UvrABC has been reported, 6 is a very poor substrate for this NER system. Given that ICLs are absolute blocks to replication and transcription, failure to repair an ICL such as 3 (6) could be very detrimental to a cell. Consequently, given the caveat that these studies were conducted with analogues of the actual lesions, the experiments suggest additional possibilities concerning the chemical bases for the cytotoxicity of DNA-damaging agents that produce simple lesions.49
Acknowledgments
We thank Professor Ben Van Houten (University of Pittsburgh Medical Center) for providing the plasmids for producing UvrA, UvrB, and UvrC.
Glossary
Abbreviations
- AP
apurinic/apyrimidinic site
- C4-AP
C4′-oxidized abasic site
- BER
base excision repair
- DOB
5′-(2-phosphoryl-1,4-dioxobutane)
- dsb
double-strand break
- ICL
interstrand cross-link
- NER
nucleotide excision repair.
Supporting Information Available
Procedures for preparing, characterizing, and radiolabeling UvrABC substrates; autoradiograms showing UvrABC incision of 8 and 9 containing various 32P-labeled termini; time course analysis of UvrABC incision of 32P-9; and an autoradiogram examining the stability of 32P-9 and 32P-13. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
We are grateful for the support of this research by National Institute of General Medical Sciences Grant GM-063028.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kozekov I. D.; Turesky R. J.; Alas G. R.; Harris C. M.; Harris T. M.; Rizzo C. J. (2010) Formation of deoxyguanosine cross-links from calf thymus DNA treated with acrolein and 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 23, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. P.; Cho Y.-J.; Huang H.; Kim H.-Y.; Kozekov I. D.; Kozekova A.; Wang H.; Minko I. G.; Lloyd R. S.; Harris T. M.; Rizzo C. J. (2008) Interstrand DNA cross-links induced by α,β-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res. 41, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozekov I. D.; Nechev L. V.; Moseley M. S.; Harris C. M.; Rizzo C. J.; Stone M. P.; Harris T. M. (2003) DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 125, 50–61. [DOI] [PubMed] [Google Scholar]
- Guainazzi A.; Scharer O. D. (2010) Using synthetic DNA interstrand crosslinks to elucidate repair pathways and identify new therapeutic targets for cancer chemotherapy. Cell. Mol. Life Sci. 67, 3683–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov T.; Guainazzi A.; Scharer O. D. (2009) Generation of DNA interstrand cross-links by post-synthetic reductive amination. Org. Lett. 11, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. E.; Johnson K. M.; Wang J.; Fekry M. I.; Wang Y.; Gates K. S. (2014) Interstrand DNA-DNA crosslink formation between adenine residues and abasic sites in duplex DNA. J. Am. Chem. Soc. 136, 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. M.; Price N. E.; Wang J.; Fekry M. I.; Dutta S.; Seiner D. R.; Wang Y.; Gates K. S. (2013) On the formation and properties of interstrand DNA,DNA cross-links forged by reaction of an abasic site with the opposing guanine residue of 5′-cap sequences in duplex DNA. J. Am. Chem. Soc. 135, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulus P.; Duroux B.; Bayle P.-A.; Favier A.; Cadet J.; Ravanat J.-L. (2007) Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc. Natl. Acad. Sci. U.S.A. 104, 14032–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczepanski J. T.; Hiemstra C. N.; Greenberg M. M. (2011) Probing DNA interstrand cross-link formation by an oxidized abasic site using nonnative nucleotides. Bioorg. Med. Chem. 19, 5788–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczepanski J. T.; Jacobs A. C.; Majumdar A.; Greenberg M. M. (2009) Scope and mechanism of interstrand cross-link formation by the C4′-oxidized abasic site. J. Am. Chem. Soc. 131, 11132–11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczepanski J. T.; Jacobs A. C.; Greenberg M. M. (2008) Self-promoted DNA interstrand cross-link formation by an abasic site. J. Am. Chem. Soc. 130, 9646–9647. [DOI] [PubMed] [Google Scholar]
- Guan L.; Greenberg M. M. (2009) DNA interstrand cross-link formation by the 1,4-dioxobutane abasic lesion. J. Am. Chem. Soc. 131, 15225–15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauson C.; Scharer O. D.; Niedernhofer L. (2013) Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspect. Biol. 5, a012732/012731–a012732/012725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. V.; Schärer O. D. (2010) Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ. Mol. Mutagen. 51, 552–566. [DOI] [PubMed] [Google Scholar]
- Räschle M.; Knipsheer P.; Enoiu M.; Angelov T.; Sun J.; Griffith J. D.; Ellenberger T. E.; Schärer O. D.; Walter J. C. (2008) Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. A.; Makarova A. V.; Samson L.; Thiesen K. E.; Dhar A.; Bessho T. (2012) Bypass of a psoralen DNA interstrand cross-link by DNA polymerases α β, and δ in vitro. Biochemistry 51, 8931–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczepanski J. T.; Jacobs A. C.; Van Houten B.; Greenberg M. M. (2009) Double strand break formation during nucleotide excision repair of a DNA interstrand cross-link. Biochemistry 48, 7565–7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X.; Ghosh A. K.; Van Houten B.; Greenberg M. M. (2010) Nucleotide excision repair of a DNA interstrand cross-link produces single- and double-strand breaks. Biochemistry 49, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M.-W.; Zheng Y.; Jasti V. P.; Champeil E.; Tomasz M.; Wang Y.; Basu A. K.; Tang M.-S. (2010) Repair of mitomycin C mono- and interstrand cross-linked DNA adducts by UvrABC: A new model. Nucleic Acids Res. 38, 6976–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio J. J.; Croteau D. L.; Van Houten B.; Kisker C. (2006) Prokaryotic nucleotide excision repair: The UvrABC system. Chem. Rev. 106, 233–252. [DOI] [PubMed] [Google Scholar]
- Noll D. M.; Mason T. M.; Miller P. S. (2006) Formation and repair of interstrand cross-links in DNA. Chem. Rev. 106, 277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.; Tang T.-S.; Friedberg E. C. (2010) Snapshot: Nucleotide excision repair. Cell 140, 754–4.e1. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., and Ellenberger T. (2006) DNA repair and mutagenesis, 2nd ed., American Society for Microbiology Press, Washington, DC. [Google Scholar]
- Van Houten B.; Gamper H.; Holbrook S. R.; Hearst J. E.; Sancar A. (1986) Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl. Acad. Sci. U.S.A. 83, 8077–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B.; Gamper H. B.; Hearst J. E.; Sancar A. (1988) Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J. Biol. Chem. 263, 16553–16560. [PubMed] [Google Scholar]
- Jones B. K.; Yeung A. T. (1990) DNA base composition determines the specificity of UvrABC endonuclease incision of a psoralen cross-link. J. Biol. Chem. 265, 3489–3496. [PubMed] [Google Scholar]
- Sage E.; Harrison L. (2011) Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat. Res. 711, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill P.; Wardman P. (2009) Radiation chemistry comes before radiation biology. Int. J. Radiat. Biol. 85, 9–25. [DOI] [PubMed] [Google Scholar]
- Eccles L. J.; Lomax M. E.; O’Neill P. (2010) Hierarchy of lesion processing governs the repair, double-strand break formation and mutability of three-lesion clustered DNA damage. Nucleic Acids Res. 38, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paap B.; Wilson D. M.; Sutherland B. M. (2008) Human abasic endonuclease action on multilesion abasic clusters: Implications for radiation-induced biological damage. Nucleic Acids Res. 36, 2717–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budworth H.; Matthewman G.; O’Neill P.; Dianov G. L. (2005) Repair of tandem base lesions in DNA by human cell extracts generates persisting single-strand breaks. J. Mol. Biol. 351, 1020–1029. [DOI] [PubMed] [Google Scholar]
- Lomax M. E.; Cunniffe S.; O’Neill P. (2004) Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry 43, 11017–11026. [DOI] [PubMed] [Google Scholar]
- Georgakilas A. G.; Bennett P. V.; Wilson D. M.; Sutherland B. M. (2004) Processing of bistranded abasic DNA clusters in γ-irradiated human hematopoietic cells. Nucleic Acids Res. 32, 5609–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland B. M.; Bennett P. V.; Sidorkina O.; Laval J. (2000) Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 97, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Greenberg M. M. (2014) Synthesis of cross-linked DNA containing oxidized abasic site analogues. J. Org. Chem. 79, 5948–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; DellaVecchia M. J.; Skorvaga M.; Croteau D. L.; Erie D. A.; Van Houten B. (2006) UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. J. Biol. Chem. 281, 15227–15237. [DOI] [PubMed] [Google Scholar]
- Croteau D. L.; DellaVecchia M. J.; Wang H.; Bienstock R. J.; Melton M. A.; Van Houten B. (2006) The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J. Biol. Chem. 281, 26370–26381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorvaga M.; Theis K.; Mandavilli B. S.; Kisker C.; Van Houten B. (2002) The β-hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J. Biol. Chem. 277, 1553–1559. [DOI] [PubMed] [Google Scholar]
- Pakotiprapha D.; Samuels M.; Shen K.; Hu J. H.; Jeruzalmi D. (2012) Structure and mechanism of the UvrA,UvrB DNA damage sensor. Nat. Struct. Mol. Biol. 19, 291–298. [DOI] [PubMed] [Google Scholar]
- Webster M. P. J.; Jukes R.; Zamfir V. S.; Kay C. W. M.; Bagnris C.; Barrett T. (2012) Crystal structure of the UvrB dimer: Insights into the nature and functioning of the UvraB damage engagement and UvrB,DNA complexes. Nucleic Acids Res. 40, 8743–8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. K.; Yeung A. T. (1988) Repair of 4,5′,8-trimethylpsoralen monoadducts and cross-links by the Escherichia coli UvrABC endonuclease. Proc. Natl. Acad. Sci. U.S.A. 85, 8410–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. D.; Ellestad G. A.; Borders D. B. (1991) Calicheamicins: Discovery, structure, chemistry, and interaction with DNA. Acc. Chem. Res. 24, 235–243. [Google Scholar]
- Dedon P. C.; Salzberg A. A.; Xu J. (1993) Exclusive production of bistranded DNA damage by calicheamicin. Biochemistry 32, 3617–3622. [DOI] [PubMed] [Google Scholar]
- Kennedy D. R.; Lu J.; Shen B.; Beerman T. A. (2007) Designer enediynes generate DNA breaks, interstrand cross-links, or both, with concomitant changes in the regulation of DNA damage responses. Proc. Natl. Acad. Sci. U.S.A. 104, 17632–17637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D. R.; Gawron L. S.; Ju J.; Liu W.; Shen B.; Beerman T. A. (2007) Single chemical modifications of the C-1027 enediyne core, a radiomimetic antitumor drug, affect both drug potency and the role of ataxia-telangiectasia mutated in cellular responses to DNA double-strand breaks. Cancer Res. 67, 773–781. [DOI] [PubMed] [Google Scholar]
- Colis L. C.; Woo C. M.; Hegan D. C.; Li Z.; Glazer P. M.; Herzon S. B. (2014) The cytotoxicity of (−)-lomaiviticin a arises from induction of double-strand breaks in DNA. Nat. Chem. 6, 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. M. (2014) Abasic and oxidized abasic site reactivity in DNA: Enzyme inhibition, crosslinking, and nucleosome catalyzed reactions. Acc. Chem. Res. 47, 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., and Goldberg I. H. (1999) DNA-damaging enediyne compounds. In Comprehensive natural products chemistry (Kool E. T., Ed.) pp 553–592, Elsevier, Amsterdam. [Google Scholar]
- Greenberg M. M. (2014) Looking beneath the surface to determine what makes DNA damage deleterious. Curr. Opin. Chem. Biol. 21, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.