Abstract
Human mesenchymal stromal cells (hMSCs) show great potential for clinical and experimental use due to their capacity to self-renew and differentiate into multiple mesenchymal lineages. However, disadvantages of primary cultures of hMSCs are the limited in vitro lifespan, and the variable properties of cells from different donors and over time in culture. In this article, we describe the generation of a telomerase-immortalized nontumorigenic human bone marrow-derived stromal mesenchymal cell line, and its detailed characterization after long-term culturing (up to 155 population doublings). The resulting cell line, iMSC#3, maintained a fibroblast-like phenotype comparable to early passages of primary hMSCs, and showed no major differences from hMSCs regarding surface marker expression. Furthermore, iMSC#3 had a normal karyotype, and high-resolution array comparative genomic hybridization confirmed normal copy numbers. The gene expression profiles of immortalized and primary hMSCs were also similar, whereas the corresponding DNA methylation profiles were more diverse. The cells also had proliferation characteristics comparable to primary hMSCs and maintained the capacity to differentiate into osteoblasts and adipocytes. A detailed characterization of the mRNA and microRNA transcriptomes during adipocyte differentiation also showed that the iMSC#3 recapitulates this process at the molecular level. In summary, the immortalized mesenchymal cells represent a valuable model system that can be used for studies of candidate genes and their role in differentiation or oncogenic transformation, and basic studies of mesenchymal biology.
Introduction
Human mesenchymal stromal cells (hMSCs) have great potential for clinical and experimental use. The hMSCs are characterized by the fibroblastic spindle shape and expression of phenotypic markers such as CD44 molecule (CD44), 5′-nucleotidase ecto (NT5E, also known as CD73), Thy-1 cell surface antigen (THY1, also known as CD90), and endoglin (ENG, also known as CD105), but are negative for protein tyrosine phosphatase, receptor type C (PTPRC, also known as CD45) and CD34 molecule (CD34) [1]. Furthermore, hMSCs have the capability to self-renew in vivo and to differentiate into cells of mesodermal lineage, including adipocytes, osteoblasts, and chondrocytes. Studies also suggest that hMSCs under certain circumstances can differentiate into ecto- and endodermal lineages [2,3]. The multi-lineage differentiation potential and the relative easiness of obtaining the cells from bone marrow [4] and adipose tissue [5] have made these cells subject of great interest for tissue engineering purposes. It also has been shown that hMSCs display immunosuppressive effects both in vitro and in vivo (reviewed in Ghannam et al. [6]), and enhance engraftment of hematopoietic stem cells during bone marrow transplantation [7]. In addition, hMSCs are important for tissue repair, as demonstrated in the ischemia model of the brain [8]. Since MSCs migrate toward inflamed ischemic and damaged regions, they have become attractive vehicles for gene therapy approaches [9], but may also contribute to cancer development [10].
Besides clinical applications, hMSCs are valuable model systems for studying candidate oncogenes found in tumors of mesenchymal origin. For instance, it has been shown that introduction of a Ewing sarcoma specific fusion protein into hMSCs resulted in malignant transformation, whereas this was not observed in fibroblasts [11]. hMSCs are also frequently used as model systems to elucidate the molecular mechanisms controlling cell fate determination and differentiation [12]. A major drawback of adult human bone marrow-derived MSCs is their limited lifespan in vitro, as after a certain number of cell divisions they enter senescence, which among other mechanisms involves a progressive shortening of the telomeres due to the lack of telomerase activity [13]. Several studies have also demonstrated that long-term culturing of hMSCs affects their capacity to proliferate and differentiate, limiting their use in therapeutic applications and basic research. Immortalization can be achieved by several methods, including introduction of telomerase reverse transcriptase (TERT), human papillomavirus-16 E6 and E7 genes or simian virus 40 large antigen [14–18]. Previous studies have demonstrated that the phenotype of hMSCs is maintained after immortalization [19–21] and their differentiation potential has even been reported to improve thereafter [21].
We here report the generation and detailed characterization of a nontumorigenic bone marrow-derived stromal cell line, immortalized by retroviral transduction of TERT. The resulting cell line iMSC#3 has a reproducible and consistent phenotype, while maintaining important properties of primary hMSCs.
Materials and Methods
Cell culture
Human bone marrow was collected from the hip of a healthy male donor after written informed consent. The informed consent and sample collection were approved by the Regional Ethical Committee for Southern Norway (S-90128). Mononuclear cells were isolated using standard density centrifugation, transferred to a culture flask (Nunc) and incubated overnight. Nonadherent cells were washed away and the adherent cells were cultured in minimum essential medium alpha medium (Life Technologies), supplemented with 10% heat inactivated fetal calf serum (Integro), GlutaMAX (2 mM), penicillin (100 U/mL), and streptomycin (100 μg/mL) (all from Life Technologies) at 37°C and 5% CO2. Cells were grown in T175 culture flasks (Nunc) and passaged every 2 weeks. Cells from passage number 2 were used for immortalization. Population doublings (PDs) of the cells were calculated as follows: PD=Log (cells harvested/cells seeded)/Log (2). The hMSC-Tert20 cells were provided by M Kassem, Department of Endocrinology and Metabolism, University Hospital of Odense, Denmark [22]. hMSC-Tert20 and primary hMSCs from a female and a male donor (∼50 and 25 years old, respectively) were cultured as previously described [22]. The established iMSC#3 cell line was DNA typed using the PowerPlex 16 HS System (Promega) (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd), and tested negative for hepatitis B virus, hepatitis C virus, human immunodeficiency virus-1, human immunodeficiency virus-2, human T-cell lymphotropic virus I/II, cytomegalovirus, Epstein-Barr Virus, herpes simplex virus-1, and Herpes simplex virus-2 by GlobalStem (GlobalStem) (Supplementary Table S2). The iMSC#3 cells were also routinely checked for Mycoplasma contamination, using standard polymerase chain reaction (PCR)-based methods. Unless otherwise stated, iMSC#3 cells at approximately passage 35 (155 PDs) were used. Experiments with primary hMSCs and the telomerase-transduced iMSC#3 and −9 clones and the hMSC-Tert20 were all performed while the cells were in proliferation phase.
Production of retroviruses and transduction
The pBabe-puro-hTERT vector (Addgene plasmid 1771) used for immortalization was a gift from RA Weinberg [23], The Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute of Technology, Cambridge, Boston. The retrovirus was produced using Phoenix-Ampho cells [24]. Phoenix cells were plated in 100 mm Petri-dishes (Nunc) at a density of 6×106 per plate in 10 mL Iscove's modified Dulbecco's medium (Life Technologies), supplemented with 10% fetal calf serum (Integro), penicillin (100 U/mL), and streptomycin (100 μg/mL) (Life Technologies). Cells were incubated overnight and transfected using calcium chloride. Briefly, 20 μg plasmid DNA was mixed with 36 μL of calcium chloride (2.0 M) and the volume was adjusted to 300 μL with Milli-Q water. After 10 min, the mixture was added to 300 μL 2× HEPES buffered saline, while bubbling air. The final mixture was incubated for 30 min and added one drop at the time to the Phoenix-Ampho cells and incubated overnight before the medium was replaced. Selection with puromycin (Life Technologies) at a final concentration of 2 μg/mL was started 2 days post-transfection and resistant cells were expanded for virus production. To produce retroviruses, 6×106 cells were seeded per 100 mm Petri-dish (Nunc) in medium without puromycin (Life Technologies). After an overnight incubation, the medium was replaced and the cells were cultured for another day. The viruses, present in the medium, were harvested and passed through a standard 0.45 μm filter.
To immortalize hMSCs, 2×105 cells were plated in six-well plates (Nunc) and incubated overnight. DOTAP Liposomal Transfection Reagent (Roche Applied Science) to a final concentration of 10 μg/mL was then added to 2 mL of 100× diluted pBabe-puro-hTERT viral supernatant, mixed, and incubated on ice for 10 min. The medium was removed, replaced by the viral supernatant, and the cells were transduced for 6 h. Selection using puromycin (Life Technologies) at a final concentration of 2 μg/mL was initiated 2 days post-transduction. After 2 weeks, the cells were transferred to Petri-dishes (Nunc) for clonal selection. After 2 additional weeks nine colonies were picked using cloning disks (Sigma-Aldrich). The telomerase-transduced hMSC clones (iMSC#1–9) were expanded for further characterization.
Determination of surface markers
iMSC#3 cells were harvested and resuspended in cold fluorescence-activated cell sorting staining buffer consisting of 1× phosphate-buffered saline (PBS) with 2% fetal bovine serum and 0.1% sodium azide. About 1×106 cells were incubated in the presence of 0.5–1 μg of CD44-PE, CD90-PE, CD73-PE (BD Biosciences), and CD105-APC (eBioscience), or their respective isotype controls for 30 min at 4°C. Flow cytometry was performed on a BD LSRII Flow Cytometer (BD Biosciences), and offline data analysis was performed with the Flowjo software (Tree Star).
Cloning of ectopic TERT integration sites
The genomic integration site for TERT was identified using inverse PCR (IPCR) [25]. The primers used (Supplementary Table S3) were designed to give products that cover the viral/chromosomal junction between the 5′-long-terminal repeat (LTR) of the pBabe-puro-hTERT DNA and genomic DNA, isolated from transduced cells using the High Pure PCR Template Preparation Kit (Roche Applied Science). Four hundred nanograms of DNA was digested with 20 U of HindIII or EcoRI (New England Biolabs) for 6 h and purified using the Qiaquick PCR Purification Kit (Qiagen). One hundred nanograms of purified DNA was circularized by re-ligation overnight at 16°C, using 400 U of T4 DNA Ligase (New England Biolabs) in a total volume of 100 μL. Ten microliters of the ligation mix was used as template for the primary PCR in a final volume of 50 μL, using 5 U of a Pfu polymerase mixed with dUTPase (provided by Dave Warren at the Norwegian Radium Hospital). Denaturation was carried out at 94°C for 2 min, followed by 30 cycles at 94°C for 30 s, 54°C for 25 s, and 72°C for 5 min, followed by a final step of 72°C for 7 min. Nested PCR was performed using 10 μL of the primary PCR as template and the same conditions as for the primary IPCR. The opposite junction between the 3′-LTR and chromosomal DNA was amplified using standard PCR. The resulting PCR products were tailed with adenosine and cloned into the pGEM-T Easy vector (Promega), before being sequenced and mapped to the human genome (human reference genome assembly GRCh37/hg19).
Karyotyping and high-resolution array comparative genomic hybridization
Karyotype analysis of cells after ∼50 and 155 PDs was performed using 3×25 nuclei. Cells were arrested in metaphase with 0.04 μg/mL demecolcine and harvested. The chromosomes were G-banded using standard techniques. Image acquisition of metaphase cells and karyotyping were performed using CytoVision System (Applied Imaging). The karyotypic description was according to the ISCN (2009) guidelines [26].
High-resolution array comparative genomic hybridization (array CGH) was performed using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix), as previously described [27]. DNA was isolated from iMSC#3 using the Wizard Genomic DNA Purification Kit (Promega). Experimental quality was assessed in Genotyping Console v3.0.1 (Affymetrix) using the contrast quality control algorithm with a minimal call rate of >86%. DNA copy number analysis was performed in Nexus (BioDiscovery), using the SNP-FASST2 segmentation algorithm (threshold of 0.7 for high copy gain, 0.2 for gain, −0.2 for loss, and −1.0 for homozygous loss). The array CGH dataset has been deposited in the GEO data repository (www.ncbi.nlm.nih.gov/geo/, accession No. GSE47646).
Isolation of total RNA
Total RNA was isolated using TRIzol solution (Life Technologies). For quantification of adipocyte markers using quantitative real-time PCR (qPCR), the total RNA was treated with amplification grade DNase I (Life Technologies). For the global expression profiling, the RNA was further purified with an RNeasy mini column (Qiagen). The purity and quantity of the extracted RNA was measured using the NanoDrop ND1000 spectrophotometer (Nanodrop Technologies), and the RNA integrity was evaluated using the Agilent 2100 Bioanalyzer and the RNA nano6000 kit (Agilent Technologies).
Genome-wide expression and DNA methylation profiling and analyses
Expression profiling was performed using the Illumina HumanWG-6 v2 Expression BeadChip (adipocyte differentiation) and Illumina HumanHT12 v4 Expression BeadChip (comparison of iMSC#3 and primary hMSCs), both provided by Illumina. Data extraction and initial quality control of the bead summary raw data were performed essentially as previously described [27]. Expression data were quantile normalized in GeneSpring GX10 (Agilent) for Human WG-6 beadarrays or in GenomeStudio v2011.1 (Illumina) for HumanHT12 v4 beadarrays, and visualized in R using the Bioconductor package Geneplotter. The mRNA expression dataset has been deposited in the GEO data repository (www.ncbi.nlm.nih.gov/geo/, accession No. GSE47647 and GSE47648, respectively). For the Illumina HumanHT12 v4 Expression BeadChip, differentially expressed genes were identified by calculating the ratio of intensities for each microarray probe and calling differentially expressed probes with a fold-change ≥2.
DNA methylation profiling of cells was performed using the Illumina HumanMethylation450 BeadChip (Illumina). One microgram of total genomic DNA was bisulfite converted using the Zymo EZ DNA Methylation Kit (Zymo Research). Labeling, hybridization, and scanning were performed following Illumina's protocol, and data extraction and quality control were done in GenomeStudio v2011.1 and the Methylation module v1.9 (both provided by Illumina). For each sample, tab separated text files with Beta values for each probe was exported and visualized as density scatter plots using Geneplotter. The DNA methylation dataset has been deposited in the GEO data repository (www.ncbi.nlm.nih.gov/geo/, accession No. GSE47649). The thresholds used to define probes showing hypermethylation and hypomethylation were deltaBeta (difference between iMSC#3 and primary hMSC cultures) >0.3 and <−0.3, respectively.
Ingenuity pathway analyses of differentially expressed genes
Functional enrichment analysis of differentially expressed genes was performed in Ingenuity Pathway Analysis (IPA) (Ingenuity Systems). Differentially expressed genes from the genome-wide expression comparisons were uploaded into IPA as Illumina probes with the corresponding gene expression ratios. The analysis was conducted using default settings, and reported pathways, canonical pathways, biological functions, and upstream regulators.
Determination of tumorigenicity
To determine whether the iMSC#3 cells were tumorigenic, 2 and 1×106 cells at p16 (50 PDs) and p35 (155 PDs) in 100 μL pf serum-free medium were subcutaneously injected into the flank of immunodeficient athymic mice (n=3), and observed for 6 months for tumor formation. The tumorigenic and immortalized bone marrow-derived stromal cell line hMSC-Tert20 was used as positive control [28]. All animal experiments were performed according to protocols approved by the National Animal Research Authority in compliance with the European Convention of the Protection of Vertebrates Used for Scientific Purposes (approval ID1499 and 3275; www.fdu.no/).
Differentiation to osteoblasts and adipocytes
For osteoblast differentiation, cells were seeded in six-well plates (Nunc) at a density of 288,000 cells per well. After incubation overnight, the medium was replaced by a differentiation medium containing 3.5 mM β-glycerol phosphate (Sigma-Aldrich), 10 nM dexamethasone (Decadron), and 67 μM ascorbic acid 2-phosphate (Sigma-Aldrich), which was replaced twice a week for 28 days. The degree of differentiation was determined by staining for alkaline phosphatase (ALPL) activity and calcium deposition as previously described [29]. Wells were washed with PBS and the cells were fixed with 4% formalin. After washing with water, 300 μL substrate solution for ALPL was added consisting of 0.2 mg/mL α-naphtyl-1-phosphate, 0.01% MgSO4, and 0.6 mg/mL fastblue RR in 0.1 M Tris, pH=8.9. After incubation for 15 min, the reaction was stopped by washing with PBS. Subsequently, the purple reaction product was extracted with 300 μL 0.05 M sodium hydroxide in ethanol and absorbance was measured at 540 nm using a Victor2 1420 Multilabel Counter (PerkinElmer). Calcium deposition was determined by adding 300 μL 40 mM Alizarin Red S (Alizarin R) from Sigma-Aldrich, pH=4.1 adjusted with 1% ammonium hydroxide, to the wells and incubated for 2 h [30]. Unbound dye was washed away with water. To extract the bound dye, 300 μL of 10% acetic acid was added to the wells and incubated for 30 min with agitation. Cells were scraped and transferred with the acetic acid to 1.5 mL tubes and incubated for 10 min at 85°C. The tubes were cooled on ice, centrifuged for 5 min at 13,000 rounds/min, 200 μL supernatant was transferred to another tube, and the pH was adjusted with 22.5 μL 10% ammonium hydroxide. The absorbance was measured at 405 nm against a calibration curve. To correct for the amount of cells, the formalin-fixed cells were incubated with 500 μL 50% trichloroacetic acid for 1 h, washed with water, and subsequently stained with 300 μL 0.4% sulforhodamine B (Sigma-Aldrich) in 1% acetic acid. Unbound dye was washed away with 1% acetic acid. The bound dye was solubilized with 300 μL 10 mM Tris and the absorbance was measured at 540 nm. Adipocyte differentiation was performed as described previously [22].
High-throughput sequencing of microRNAs
Small RNA sequencing libraries were prepared using the IlluminaTruSeq Small RNA Sample Preparation protocol (Illumina). One microgram of total RNA was ligated to 3′- and 5′-RNA adapters. The ligated products were reverse transcribed to generate cDNA libraries for each sample, and subsequently PCR amplified, where unique index sequences were incorporated. Small RNA libraries were pooled and 32 bases were sequenced for each cDNA molecule using an Illumina Genome Analyzer IIx (Illumina). Indexes were sequenced to identify the source of each read. Real-time analysis, base calling, and filtering of low quality reads were done by Illumina's software packages (SCS2.9/RTA1.9 and Off-line Basecaller v1.9). Novoalign (V2.08.01 Novocraft 2010; www.novocraft.com) was used to remove remaining adapter sequences and map the reads to the reference human genome (human reference genome assembly GRCh37/hg19). All reads mapping to 10 or more genomic regions were excluded from further analysis. The miRBase database release 18 was used to identify miRNAs.
To calculate the read count for each miRNA, the reads that mapped uniquely within a mature miRNA sequence with a maximum of one mismatch were considered as hits. Reads mapping to more than one mature miRNA sequence were assigned according to the frequency of uniquely mapped reads found for these miRNAs. That means when two miRNAs shared a given number of multiple mapped reads, we identified the ratio of unique reads between these two miRNAs. This ratio was applied to divide the number of multiple mapped reads and assign them. If multiple hits were found to be perfectly mapped to one genomic region and mapped with mismatch to another one, only the perfect matches were considered.
For normalization we calculated the sample-specific normalization factor by dividing the total number of reads mapped to annotated miRNAs with 1 million. The normalized expression values were generated by dividing the read count of each miRNA with the according sample-specific normalization factor. After normalization, all miRNAs with read counts less than 10 across all samples were considered as background and set to 0. The raw and normalized read counts for the miRNA sequencing dataset has been deposited in the GEO data repository (www.ncbi.nlm.nih.gov/sra/, accession No. SRP028157).
Production of lentiviruses and transduction
The constructs used to produce lentiviruses were generated by standard Gateway cloning (Life Technologies). Enhanced green fluorescent protein (EGFP) was amplified from pEGFP-C3 (Clontech), whereas a 371 base pair long fragment consisting of the mir-155 precursor sequence with flanking regions was amplified from iMSC#3 genomic DNA (Supplementary Table S3 for primer sequences). After recombination into the donor vector pDONR 221 (Life Technologies), the inserts were transferred to the pLenti CMV/TO Neo DEST (685-3) (Addgene plasmid 17292) [31]. The lentiviruses were produced essentially as described for the ViraPower Lentiviral Expression System (Life Technologies).
Transient transfection of miRNA mimics
The iMSC#3 cells were transiently transfected with hsa-miR-155 Pre-miR miRNA Precursor or Negative control #1 using Lipofectamine 2000 (all from Life Technologies), as previously described [22]. Adipocyte differentiation was induced 24 h post-transfection.
Quantitative real-time PCR
The High Capacity cDNA Archive Kit (Life Technologies) was used to synthesize cDNA. TaqMan Gene Expression Master Mix and various TaqMan Gene Expression Assays (Life Technologies; Supplementary Table S4) were used to perform the qPCR. The TATA box binding protein (TBP) was used to normalize the expression data. The miRNA qPCR was performed as previously described [22]. The error bars in all qPCR experiments show the standard deviation of technical replicates, calculated as previously described [32].
Results
Characterization of morphology, proliferation rates, and surface markers of transduced cells
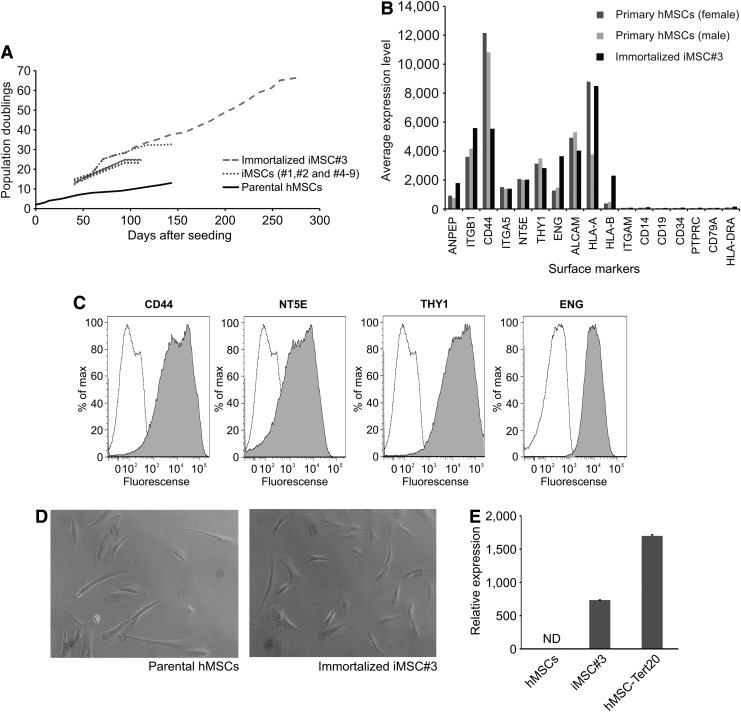
Primary stromal cells derived from the bone marrow of a healthy donor were transduced at passage two (p2) with pBabe-puro-hTERT. After transduction, nine clones (referred to as iMSC#1–9) were selected and expanded. The doubling time of selected clones varied from 97 h for clone iMSC#8 to 140 h for clone iMSC#9. The proliferation rate of iMSC#3 remained largely the same with a doubling time of about 100 h, which was somewhat lower than the doubling time of 120 h of the parental primary hMSCs (Fig. 1A). The parental primary hMSCs ceased to proliferate after about 15 PDs, at which time the clones iMSC#2, #4, #5, #6, #7, and #8 had gone through about 25 PDs, whereas the iMSC#1 clone was lost within 1 week. The iMSC#9 clone ceased to proliferate after 32 PDs and only the iMSC#3 clone continued to divide and has at the present time been in culture for a prolonged period of more than 200 PDs (Fig. 1A). The cells had an expression pattern of markers consistent with multipotent MSCs as defined by Dominici et al. [33] (Fig. 1B, C). Several additional markers associated with hMSCs were also abundantly expressed at the transcript level, including alanyl aminopeptidase (ANPEP, ie, CD13), integrin beta 1 (ITGB1, ie, CD29), integrin alpha 5 (ITGA5, ie, CD49e), activated leukocyte cell adhesion molecule (ALCAM, ie, CD166), and major histocompatibility complex, class I, A and B (HLA-A and B). As determined by flow cytometry, the iMSC#3 cells were positive for CD44, NT5E, THY1, and ENG (Fig. 1C), and, like primary hMSCs, were positive for CD90, and were negative for the hematopoietic markers CD34, PTPRC, and v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT, also known as CD117) (Supplementary Fig. S1A). They also maintained the same cellular morphology as the primary hMSCs (Fig. 1D). Staining for β-galactosidase, a marker of senescence, showed a high percentage of positive cells with a flattened morphology in the parental primary cells and the iMSC#9 clone at senescence, whereas iMSC#3 maintained a normal spindle-shaped morphology and contained only a limited number of positive cells (Supplementary Fig. S1B). Gene expression profiling showed high transcription of the corresponding genes.
FIG. 1.
Analyses of morphology, proliferation rates, surface markers, and ectopic expression of TERT. (A) Comparison of proliferation rates of parental hMSCs and telomerase-transduced hMSCs (iMSC#1–9). All clones except iMSC#3 ceased to grow after ∼25 to 32 PDs. (B) mRNA expression levels of the markers defining multipotent mesenchymal stroma cells according to Dominici et al. [33]. (C) Flow cytometry analysis of the mesenchymal surface markers CD44, NT5E (ie, CD73), THY1 (ie, CD90), and ENG (ie, CD105) in iMSC#3 (gray histograms). The respective isotype controls are indicated by white histograms. (D) Comparison of the cellular morphology of parental hMSCs after 6 PDs and immortalized mesenchymal bone marrow-derived stromal cells, iMSC#3, after 155 PDs. (E) The expression level of the human telomerase reverse transcriptase (TERT) in primary hMSCs (5 PDs), iMSC#3 cells after 155 PDs and hMSC-Tert20 cells determined by quantitative real-time PCR, and compared to the endogenous level in mononuclear cells (set to 1). TBP was used to normalize the data. Error bars represent standard deviations of technical replicates. ND, not detected. hMSCs, human mesenchymal stromal cells; PD, population doubling; TBP, TATA box binding protein; TERT, telomerase reverse transcriptase.
Expression of ectopic TERT and characterization of integration site
qPCR was used to investigate whether iMSC#3 cells had increased levels of TERT, and to determine whether the expression level was maintained after long-term culturing. Prior to retroviral transduction, TERT expression was not detected in primary hMSCs. Thus, mononuclear cells from the bone marrow of early stage breast cancer patients were used to set the base line, as these cells have low endogenous levels of TERT. Although not as high as in the positive control hMSC-Tert20, the expression of TERT in iMSC#3 was ∼750-fold higher than in the mononuclear cells (Fig. 1E).
A single chromosomal integration site of the TERT proviral DNA was mapped to chromosome segment 1q42.3 (position 235115596-235115597) using IPCR. Except for the expected short target-site duplication at the insertion site, no other changes were observed. The ectopic TERT gene was located in an exon of a putative gene of unknown function (ENSG00000258082). Moreover, the nearest protein-coding genes were located ∼370 kb upstream (interferon regulatory factor 2 binding protein 2, IRF2BP2) and ∼157 kb downstream (translocase of outer mitochondrial membrane 20 homolog, TOMM20) from the integration site.
Karyotype analysis and high-resolution mapping of genetic alterations
To determine whether retroviral transduction and long-term culturing of iMSC#3 cells resulted in any chromosomal aberrations, karyotype analysis after 50 and 155 PDs was performed. No chromosomal aberrations were discovered (Supplementary Fig. S2), and high-resolution mapping using array CGH confirmed that the cells in general had a normal copy number. Supplementary Figure S3 shows a summary of the identified copy number variations (CNV) and allelic events, and detailed information is given in Supplementary Table S5. Some narrow regions with deviations were identified, but most of these were associated with known regions of normal CNV. Since we do not have normal material from the donor, we do not know whether any of these are in fact mutations. Eight small regions with copy number aberration showed less than 25% overlap with known regions of CNV, two with increased copy number (located in 7q11.22 and 9p24.1) and six with decreased copy number (located in 3q13.31, 7q34, 9p24.1 10q21.3, 11p14.1-p13, and Xq23). Six of these small regions overlapped with one or more genes (Supplementary Table S5).
High-throughput sequencing of miRNAs
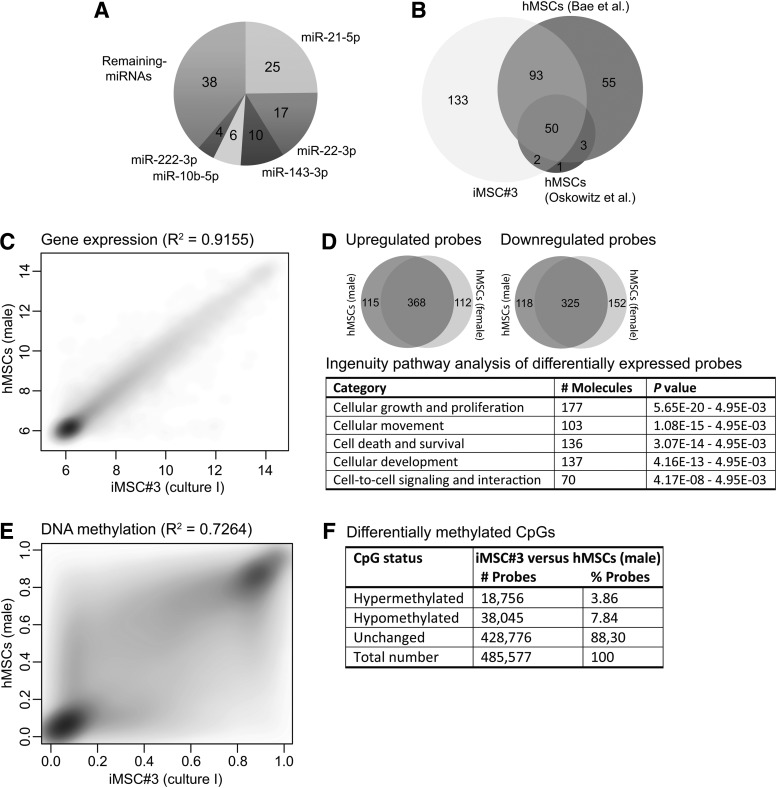
High-throughput sequencing of small RNAs was used to determine the miRNA expression profile of iMSC#3 cells. Expression of total of 278 miRNAs was detected (≥10 reads) (Supplementary Table S6) and the five most abundant miRNAs were miR-21-5p, miR-22-3p, miR-143-3p, miR-10b-5p, and miR-222-3p, accounting for 62% of the total reads (Fig. 2A). A comparison between the miRNAs found in iMSC#3 and the miRNAs reported to be expressed in bone marrow-derived hMSCs by Oskowitz et al. [34] and Bae et al. [35] was performed (Fig. 2B). Interestingly, a set of 50 miRNAs, including the five most abundant miRNAs in iMSC#3, was common for all three studies, whereas 133 miRNAs were only expressed in iMSC#3 (Supplementary Table S7).
FIG. 2.
Analyses of genome-wide gene expression and DNA methylation of iMSC#3 and primary hMSCs. (A) Shows the relative distribution of the five most abundantly expressed miRNAs to the complete population of miRNAs in iMSC#3 (numbers within the chart depict the percentages). (B) Venn diagram showing the overlaps of 278 miRNAs identified in undifferentiated iMSC#3 by high-throughput sequencing and in primary hMSCs in two other studies [34,35]. (C) Density scatter plot of genome-wide Log2-transformed gene expression signal intensities (depicted on the axis) between iMSC#3 and primary hMSCs (male donor). Coefficient of determination R2=0.92. (D) Venn diagrams showing the number of ≥2-fold differentially expressed probes in iMSC#3 compared with primary hMSCs from either male or female donor, or both. The table shows the top molecular and cellular functions associated with the common 693 differentially expressed probes identified by Ingenuity Pathway Analysis. (E) Genome-wide DNA methylation profiling of iMSC#3 and primary hMSCs (male donor). The density scatter plots show the average beta values (depicted on the axis) for each CpG site, which range from 0 (unmethylated) to 1 (fully methylated). Coefficient of determination R2=0.73. (F) Distribution and number of hyper- and hypomethylated CpGs sites in iMSC#3 compared with the primary hMSCs (male donor). The thresholds for hyper- and hypomethylation were deltaBeta >0.3 and <−0.3, respectively.
Gene expression and DNA methylation profiling of iMSC#3 cells and primary hMSCs
To investigate the genome-wide effects of ectopic expression of TERT and long-term culturing of iMSC#3, comparative gene expression and DNA methylation profiling of iMSC#3 after ∼155 PDs and primary hMSCs from two different donors were performed.
The expression profiles of the two donors were highly similar with a coefficient of determination (R2) of 0.98, whereas the R2 between iMSC#3 and the female and male donor were 0.93 and 0.92, respectively (Fig. 2C and Supplementary Fig. S4). This showed that their expression profiles were overall similar. Nevertheless, 693 probes were ≥2-fold differentially expressed in iMSC#3 compared with both of the primary hMSC cultures, of which 368 were upregulated and 325 were downregulated (Fig. 2D and Supplementary Table S8). An IPA of differentially expressed probes indicated that the top molecular and cellular functions affected were cellular growth and proliferation, cellular movement and development, cell death and survival, and cell-to-cell signaling and interactions (Fig. 2D). The relative expression levels of the 50 most differentially expressed genes are shown in Supplementary Fig. S5. Interestingly, most of the upregulated genes were represented by cancer/testis antigens in the G- and melanoma antigens family (GAGE and MAGE, respectively).
As for the expression profiles, the genome-wide DNA methylation profiles of the two primary hMSC cultures were highly similar (R2=0.96) (Supplementary Fig. S6), whereas less similarity was found between iMSC#3 and the hMSCs from the female and male donor (both R2=0.73) (Fig. 2E and Supplementary Fig. S6). However, 88% of the CpGs in iMSC#3 were unchanged (ie,<30% differentially methylated) compared with the primary hMSCs, whereas ∼4% and 8% were hyper- and hypomethylated (ie, >30% differentially methylated), respectively (Fig. 2F).
Tumorigenicity
To determine whether the iMSC#3 cells were tumorigenic, they were subcutaneously injected into immunodeficient athymic mice (data not shown). No tumor formation was observed after 6 months with cells after 50 and 155 PDs, whereas the immortalized bone marrow-derived stromal cell line hMSC-Tert20 [14] formed large tumors within much shorter time.
Differentiation potential
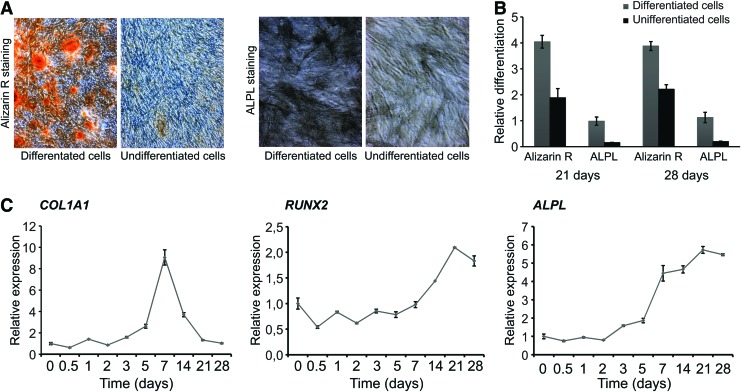
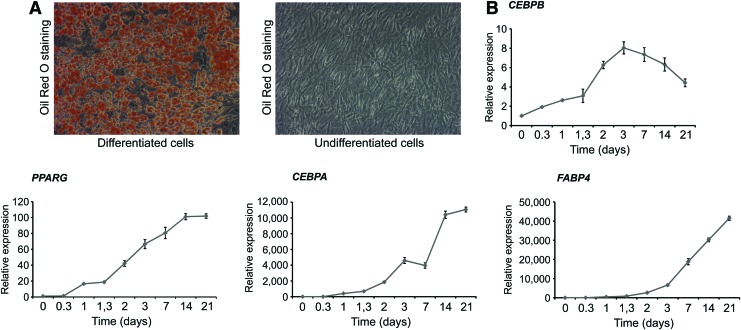
To examine the differentiation potential of iMSC#3 after long-term culturing (∼155 PDs), immortalized cells were exposed to differentiation medium that induced either osteogenesis or adipogenesis. Osteoblast differentiation was detected by staining and quantification of calcium deposition by Alizarin Red and by ALPL activity (Fig. 3A, B), which were increased after 21 days and remained elevated until the experiment was ended 7 days later. In addition, the transcript levels of collagen type I alpha 1 (COL1A1), runt-related transcription factor 2 (RUNX2), and ALPL were consistently increased, although expression of COL1A1 peaked at day 7 (Fig. 3C). The increase of COL1A1 at day 7 of expansion of hMSC has been reported previously (GEO ID: 28591335), and also its induction by dexamethasone in rodent cells (GEO ID: 28591335 and 28582513). Exposure to adipocyte differentiation medium showed the expected change toward an adipocyte phenotype by accumulation of triglycerides as shown by Oil Red O staining 21 days after induction (Fig. 4A). In addition, the expression patterns of four genes that are known to participate in adipogenesis were investigated using qPCR (Fig. 4B). CCAAT/enhancer-binding protein alpha (CEBPA), CCAAT/enhancer-binding protein beta (CEBPB), peroxisome proliferator-activated receptor gamma (PPARG), and fatty acid binding protein 4 (FABP4) all showed the expected temporal expression during adipocyte differentiation. Single osteogenic and adipogenic differentiation experiments were also performed with the iMSC#9 clone and showed a lower degree of differentiation both for osteogenic and the adipogenic lineage (data not shown). This might be related to the growth stage of the cells since shortly after the first experiments the cells ceased to grow. As demonstrated by Wu et al., the iMSC#3 cells can also be differentiated toward chondrocytes [36].
FIG. 3.
Osteoblast differentiation of iMSC#3. (A) iMSC#3 cells was differentiated for 28 days before the cells were fixated and stained with Alizarin Red S (Alizarin R) and ALPL. (B) Quantification of Alizarin Red staining and ALPL activity 21 and 28 days after induction of osteogenesis, normalized to total protein using sulforhodamine B staining. (C) The expression patterns of COL1A1, RUNX2, and ALPL genes during osteogenesis determined by quantitative real-time PCR at the indicated time points, and compared to the level at day 0 (set to 1). TBP was used to normalize the data. Error bars represent standard deviations of technical replicates. ALPL, alkaline phosphatase; COL1A1, collagen type I alpha 1; RUNX2, runt-related transcription factor 2. Color images available online at www.liebertpub.com/scd
FIG. 4.
Adipocyte differentiation of iMSC#3. (A) iMSC#3 cells were differentiated for 21 days before the cells were fixated and stained with Oil Red O at day 21. (B) Gene expression patterns of adipocyte marker genes during differentiation. The expression of the adipocyte markers CEBPB, PPARG, CEBPA, and FABP4 was investigated using quantitative real-time PCR at the indicated time points, and compared to the level at day 0 (set to 1). TBP was used to normalize the data. Error bars represent standard deviations of technical replicates. CEBPA, CCAAT/enhancer-binding protein alpha; CEBPB, CCAAT/enhancer-binding protein beta; FABP4, fatty acid binding protein 4; PPARG, peroxisome proliferator-activated receptor gamma. Color images available online at www.liebertpub.com/scd
Genome-wide identification of genes involved in adipocyte differentiation
To investigate the molecular basis of the adipocyte differentiation potential of iMSC#3, a comprehensive genome-wide profiling of mRNAs at six time points during adipogenesis was performed. A total of 594 probes were differentially expressed (≥5-fold) between undifferentiated iMSC#3 cells at day 0 and differentiated cells at day 21, of which 348 were upregulated and 246 were downregulated (Supplementary Table S9). As expected, many of these were known to be involved in adipogenesis, including PPARG, CEBPA, and FABP4, whose inductions were validated using qPCR (Fig. 4B). To further characterize the changes during adipocyte differentiation, Ingenuity Pathway Analyses of the differentially expressed genes were performed. Numerous biological processes associated with adipogenesis and mature adipocytes were identified. These included activation of retinoid X receptor and its dimerization partner, biosynthesis, activation, and oxidation of various fatty acids and di/triacylglycerols, biosynthesis of cholesterol, insulin signaling, PPARG signaling, and processes implicated in cell cycle regulation and control. All identified Ingenuity pathways are listed in Supplementary Table S10.
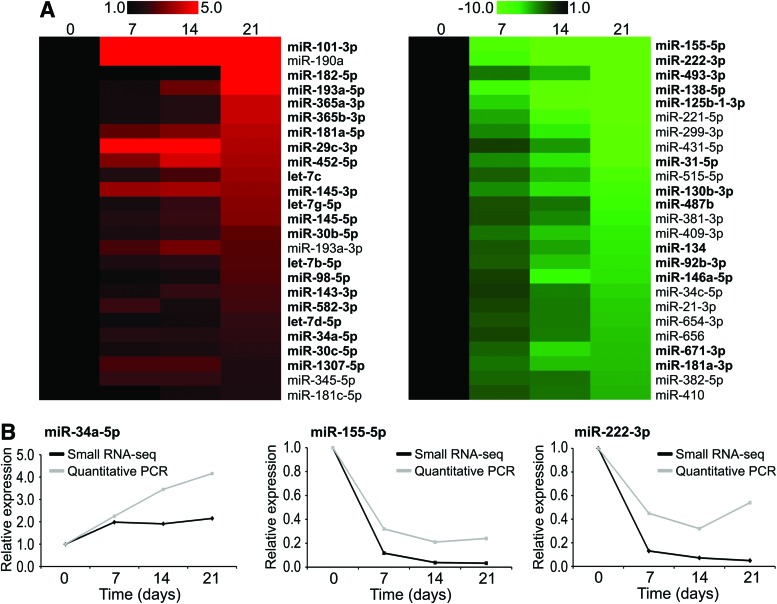
As we have previously shown that miRNAs play a critical role in differentiation of hMSCs into adipocytes [22], we wanted to determine how iMSC#3 performed in this respect. To investigate this, expression profiles of miRNAs at three time points during adipogenesis were generated by high-throughput sequencing (Supplementary Table S11). After 21 days, 22 and 78 miRNAs were ≥2-fold up- and downregulated, respectively, compared with undifferentiated cells at day 0. A comparison of the 50 most differentially expressed miRNAs, showed that more than 30 of these have been identified in previous adipocyte-related studies, also across species (Fig. 5A and Supplementary Table S12). The expression level of three miRNAs, that is, miR-34a-5p, miR-155-5p, and miR-222-3p, was validated by qPCR (Fig. 5B).
FIG. 5.
High-throughput sequencing of miRNAs during adipogenesis of iMSC#3. (A) Twenty-five most up- and downregulated miRNAs during adipogenesis of iMSC#3 as determined by high-throughput sequencing. miRNAs in bold have previously been shown to be differentially expressed in either different adipose tissues or during adipogenesis. The numbers above the heat map depicts the days after induction of differentiation (day 0–21), and the color scale indicates fold-change expression relative to undifferentiated cells at day 0. (B) Validation of high-throughput sequencing (small RNA-seq) data. The expression level of miR-34a-5p, miR-155-5p, and miR-222-3p was determined by quantitative real-time PCR (quantitative PCR). RNU44 was used to normalize the data. Color images available online at www.liebertpub.com/scd
iMSC#3 as a model system for functional gene studies
The iMSC#3 cells also provide a flexible model system to study candidate genes as they can be easily transfected and transduced. We took advantage of our recent discovery that adipogenesis is suppressed by ectopic expression of miR-155-5p in primary hMSCs [22] and used miR-155-5p as a proof of principle to demonstrate that the immortalized iMSC#3 cells can be transiently transfected using standard procedures. Consistent with our previous results, ectopic expression of synthetic miR-155-5p inhibited differentiation, observed as reduced accumulation of triglycerides and transcriptional inhibition of CEBPA, PPARG, and FABP4 (Supplementary Fig. S7). Furthermore, iMSC#3 can also be used to study stable gene expression. This was demonstrated by transducing the cells using lentiviruses that expressed miR-155-5p or EGFP. Stably transduced cells with ∼25-fold increased expression of miR-155-5p and a homogenous population of EGFP-expressing cells were obtained after selection with G418 for 14 days (Supplementary Fig. S8).
Discussion
This article describes the establishment and extensive characterization of the immortalized human bone marrow-derived stromal cell line iMSC#3, which has been in culture for many years. During this extended period, the cells maintained their morphology, differentiation potential, and proliferation rate and did not undergo malignant transformation. This is in accordance with results obtained by others using TERT transduction as a way of extending the lifespan of cells with stem cell-like properties [37].
The integration site of a single copy of the provirus encoding the ectopic TERT was identified. The integration event did not lead to insertional mutagenesis of any known genes, and the ectopic expression of TERT was maintained during continuous culturing of iMSC#3. Prolonged culture of the iMSC#3 cells did not result in chromosomal aberrations, as shown by the normal karyotype. In addition, high-resolution array CGH of the same cells confirmed that they in general had normal copy number. Also, the transcriptome of the iMSC#3 cells resembled that of primary hMSCs, which is consistent with results from a similar study where immortalization of hMSCs was achieved by tumor protein p53 (TP53) knockdown and ectopic expression of TERT [21]. To identify biological processes that might be changed in iMSC#3, enrichment analysis of the differentially expressed genes was performed. This analysis suggested that not only molecular and cellular functions such as cellular growth, proliferation, movement, and development, but also cell death and survival, and cell signaling and interactions were affected. Interestingly, a large number of cancer/testis antigens in the GAGE and MAGE-family were among the most highly upregulated genes in iMSC#3 compared with the primary hMSCs. The transcripts encoded by these genes are normally not found in somatic tissues, but are frequently expressed in embryonic tissues and various cancers (reviewed in Simpson et al. [38]). The GAGE antigens are not well described, but they have been shown to confer resistance to apoptosis [39], and they might be involved in reorganization of chromatin [40]. The MAGE antigens have been more studied, and MAGE2C, which was increased 15-fold in IMSC#3, has been demonstrated to promote both cell proliferation and viability of mast cells [41]. Therefore, these genes might play a role in mediating cell survival of iMSC#3, which is in agreement with other studies where it has been reported that immortalization of hMSCs might be dependent on additional factors besides increased telomerase activity per se [21,42,43]. Comparison with other selected clones to add insight in what factors represent variation in cellular phenotypes, and whether some might have contributed to the process of immortalization was not possible, since only one clone continued to grow. Despite the observed changes in gene expression, only small changes were observed in doubling time between iMSC#3 and parental primary hMCSs, and the iMSC#3 cells did not form tumors when subcutaneously injected in immunodeficient mice.
Long-term culturing of MSCs has also been demonstrated to induce epigenetic modifications such as DNA methylation [44,45]. Although a detailed comparison of the DNA methylation profiles of iMSC#3 and the hMSCs was beyond the scope of the present study, we identified that 12% of the CpGs that were more than 30% differentially methylated in iMSC#3 cells, were either hyper- or hypomethylated. For instance, the aforementioned upregulation of MAGE2C was associated with hypomethylation. Furthermore, we found expression of 278 miRNAs in iMSC#3 and 50 of these, including the five most abundantly expressed miRNAs miR-21-5p, miR-22-3p, miR-143-3p, miR-10b-5p, and miR-222-3p, were found to be present in hMSCs in two other studies [34]. Some of these might have a function in maintaining the undifferentiated state or mesenchymal phenotype of hMSCs. An interesting candidate is the highly abundant miR-21-5p, which has been shown to promote development of the mesenchymal phenotype of epicardial mesothelial cells [46].
We show that the capacity to differentiate into the adipogenic and osteogenic lineage was maintained in iMSC#3 after immortalization, which is in line with previous studies [14–17]. In addition, a genome-wide profiling of mRNAs during adipogenesis, followed by IPA showed that numerous highly relevant biological processes were affected. Identification of differentially expressed miRNAs during adipogenesis further showed that iMSC#3 recapitulate this process at the molecular level, as several known adipocyte-promoting miRNAs, including miR-143-3p, were induced [47]. In addition to the data presented here, differentiation of iMSC#3 cells into adipocytes and osteoblasts was also previously demonstrated in a microfluidic device [48]. The chondrogenic potential of iMSC#3 appears to be relatively low, but the iMSC#3 cells can stimulate chondrocyte differentiation of primary chondrocytes in cocultures in a similar manner as primary hMSCs [36]. The iMSC#3 cells also have been shown to support the proliferation and development of human B cell precursor cells in a coculture system to an equal or higher extent than primary hMSCs [49]. These other studies thus confirm further the MSC phenotypes of iMSC#3. Although the cell line fulfilled our objectives, it was a single successful event out of the clones obtained, and we do not know how reproducible our procedures are, or whether the inability of the other clones to grow was the result of additional events extinguishing TERT, or on the contrary, iMSC#3 was the result of some second mutation facilitating its continuous growth. Since other adipocyte cloning studies have shown the presence of subclones with different balance between growth, and differentiation propensity toward certain cellular phenotypes, it may well be that the isolation and characterization of further clones could provide new cell lines, for example, with better ability to differentiate to cartilage.
In summary, the immortalized nontumorigenic iMSC#3 cell line maintains the main characteristics found in primary hMSCs, thus making this a valuable model for studying basic mesenchymal biology and differentiation, and functional studies of candidate genes involved in mesenchymal tumors. Furthermore, the comprehensive phenotypic and molecular characterization of iMSC#3 provides a framework to increase current knowledge of mesenchymal biology.
Supplementary Material
Acknowledgments
We thank Siri Tveito and Russell Castro at the Department of Tumor Biology for providing mononuclear cells from early stage breast cancer patients and for performing flow cytometry, respectively. This work was supported by the Norwegian Center for Stem Cell Research, the Research Council of Norway (grant number 193056), and the Norwegian Cancer Society.
Author Disclosure Statement
The authors have no conflicts of interest.
References
- 1.Fibbe WE. and Noort WA. (2003). Mesenchymal stem cells and hematopoietic stem cell transplantation. Ann N Y Acad Sci 996:235–244 [DOI] [PubMed] [Google Scholar]
- 2.Greco SJ, Zhou C, Ye JH. and Rameshwar P. (2007). An interdisciplinary approach and characterization of neuronal cells transdifferentiated from human mesenchymal stem cells. Stem Cells Dev 16:811–826 [DOI] [PubMed] [Google Scholar]
- 3.Lee KD, Kuo TK, Whang-Peng J, Chung YF, Lin CT, et al. (2004). In vitro hepatic differentiation of human mesenchymal stem cells. Hepatology 40:1275–1284 [DOI] [PubMed] [Google Scholar]
- 4.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, et al. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147 [DOI] [PubMed] [Google Scholar]
- 5.Boquest AC, Shahdadfar A, Fronsdal K, Sigurjonsson O, Tunheim SH, et al. (2005). Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol Biol Cell 16:1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghannam S, Bouffi C, Djouad F, Jorgensen C. and Noel D. (2010). Immunosuppression by mesenchymal stem cells: mechanisms and clinical applications. Stem Cell Res Ther 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noort WA, Kruisselbrink AB, in't Anker PS, Kruger M, van Bezooijen RL, et al. (2002). Mesenchymal stem cells promote engraftment of human umbilical cord blood-derived CD34(+) cells in NOD/SCID mice. Exp Hematol 30:870–878 [DOI] [PubMed] [Google Scholar]
- 8.Honma T, Honmou O, Iihoshi S, Harada K, Houkin K, et al. (2006). Intravenous infusion of immortalized human mesenchymal stem cells protects against injury in a cerebral ischemia model in adult rat. Exp Neurol 199:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamada H, Kobune M, Nakamura K, Kawano Y, Kato K, et al. (2005). Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Sci 96:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, et al. (2004). Gastric cancer originating from bone marrow-derived cells. Science 306:1568–1571 [DOI] [PubMed] [Google Scholar]
- 11.Riggi N, Cironi L, Provero P, Suva ML, Kaloulis K, et al. (2005). Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 65:11459–11468 [DOI] [PubMed] [Google Scholar]
- 12.Liu TM, Martina M, Hutmacher DW, Hui JHP, Lee EH, et al. (2007). Identification of common pathways mediating differentiation of bone marrow- and adipose tissue-derived human mesenchymal stem cells into three mesenchymal lineages. Stem Cells 25:750–760 [DOI] [PubMed] [Google Scholar]
- 13.Baxter MA, Wynn RF, Jowitt SN, Wraith JE, Fairbairn LJ, et al. (2004). Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells 22:675–682 [DOI] [PubMed] [Google Scholar]
- 14.Simonsen JL, Rosada C, Serakinci N, Justesen J, Stenderup K, et al. (2002). Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol 20:592–596 [DOI] [PubMed] [Google Scholar]
- 15.Mihara K, Imai C, Coustan-Smith E, Dome JS, Dominici M, et al. (2003). Development and functional characterization of human bone marrow mesenchymal cells immortalized by enforced expression of telomerase. Br J Haematol 120:846–849 [DOI] [PubMed] [Google Scholar]
- 16.Kawano Y, Kobune M, Yamaguchi M, Nakamura K, Ito Y, et al. (2003). Ex vivo expansion of human umbilical cord hematopoietic progenitor cells using a coculture system with human telomerase catalytic subunit (hTERT)-transfected human stromal cells. Blood 101:532–540 [DOI] [PubMed] [Google Scholar]
- 17.Kang SK, Putnam L, Dufour J, Ylostalo J, Jung JS, et al. (2004). Expression of telomerase extends the lifespan and enhances osteogenic differentiation of adipose tissue-derived stromal cells. Stem Cells 22:1356–1372 [DOI] [PubMed] [Google Scholar]
- 18.Gong M, Bi Y, Jiang W, Zhang Y, Chen L, et al. (2011). Immortalized mesenchymal stem cells: an alternative to primary mesenchymal stem cells in neuronal differentiation and neuroregeneration associated studies. J Cell Biochem 18:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdallah BM, Haack-Sorensen M, Burns JS, Elsnab B, Jakob F, et al. (2005). Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene despite extensive proliferation. Biochem Biophys Res Commun 326:527–538 [DOI] [PubMed] [Google Scholar]
- 20.Gao K, Lu YR, Wei LL, Lu XF, Li SF, et al. (2008). Immortalization of mesenchymal stem cells from bone marrow of rhesus monkey by transfection with human telomerase reverse transcriptase gene. Transplant Proc 40:634–637 [DOI] [PubMed] [Google Scholar]
- 21.Liu TM, Ng WM, Tan HS, Vinitha D, Yang Z, et al. (2013). Molecular basis of immortalization of human mesenchymal stem cells by combination of p53 knockdown and human telomerase reverse transcriptase overexpression. Stem Cells Dev 22:268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skarn M, Namlos HM, Noordhuis P, Wang MY, Meza-Zepeda LA, et al. (2012). Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222. Stem Cells Dev 21:873–883 [DOI] [PubMed] [Google Scholar]
- 23.Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, et al. (1998). Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene 16:1217–1222 [DOI] [PubMed] [Google Scholar]
- 24.Kinsella TM. and Nolan GP. (1996). Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther 7:1405–1413 [DOI] [PubMed] [Google Scholar]
- 25.Ochman H, Gerber AS. and Hartl DL. (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaffer L, Slovak M. and Campbell L. (2009). ISCN 2009 an international system for human cytogenetic nomenclature. Hum Genet 126:603–604 [Google Scholar]
- 27.Kresse SH, Rydbeck H, Skårn M, Namløs HM, Barragan-Polania AH, et al. (2012). Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma. PLoS One 7:e48262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serakinci N, Guldberg P, Burns JS, Abdallah B, Schrodder H, et al. (2004). Adult human mesenchymal stem cell as a target for neoplastic transformation. Oncogene 23:5095–5098 [DOI] [PubMed] [Google Scholar]
- 29.in't Anker PS, Noort WA, Scherjon SA, Kleijburg-van der Keur C, Kruisselbrink AB, et al. (2003). Mesenchymal stem cells in human second-trimester bone marrow, liver, lung, and spleen exhibit a similar immunophenotype but a heterogeneous multilineage differentiation potential. Haematologica 88:845–852 [PubMed] [Google Scholar]
- 30.Gregory CA, Grady Gunn W, Peister A. and Prockop DJ. (2004). An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem 329:77–84 [DOI] [PubMed] [Google Scholar]
- 31.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, et al. (2009). A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4:e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM. and Kramer MF. (2001). High-Throughput Real-Time Quantitative Reverse Transcription PCR. Current Protocols in Molecular Biology. John Wiley & Sons, Inc., Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 33.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, et al. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8:315–317 [DOI] [PubMed] [Google Scholar]
- 34.Oskowitz AZ, Lu J, Penfornis P, Ylostalo J, McBride J, et al. (2008). Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci U S A 105:18372–18377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae S, Ahn JH, Park CW, Son HK, Kim KS, et al. (2009). Gene and microRNA expression signatures of human mesenchymal stromal cells in comparison to fibroblasts. Cell Tissue Res 335:565–573 [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Leijten JC, Georgi N, Post JN, van Blitterswijk CA, et al. (2011). Trophic effects of mesenchymal stem cells increase chondrocyte proliferation and matrix formation. Tissue Eng 17:1425–1436 [DOI] [PubMed] [Google Scholar]
- 37.Partridge KA. and Oreffo RO. (2004). Gene delivery in bone tissue engineering: progress and prospects using viral and nonviral strategies. Tissue Eng 10:295–307 [DOI] [PubMed] [Google Scholar]
- 38.Simpson AJG, Caballero OL, Jungbluth A, Chen Y-T. and Old LJ. (2005). Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 5:615–625 [DOI] [PubMed] [Google Scholar]
- 39.Kular RK, Yehiely F, Kotlo KU, Cilensek ZM, Bedi R, et al. (2009). GAGE, an antiapoptotic protein binds and modulates the expression of nucleophosmin/B23 and interferon regulatory factor 1. J Interferon Cytokine Res 29:645–655 [DOI] [PubMed] [Google Scholar]
- 40.Gjerstorff MF, Rösner HI, Pedersen CB, Greve KBV, Schmidt S, et al. (2012). GAGE cancer-germline antigens are recruited to the nuclear envelope by germ cell-less (GCL). PLoS One 7:e45819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang B, O'Herrin S, Wu J, Reagan-Shaw S, Ma Y, et al. (2007). Select cancer testes antigens of the MAGE-A, -B, and -C families are expressed in mast cell lines and promote cell viability in vitro and in vivo. J Invest Dermatol 127:267–275 [DOI] [PubMed] [Google Scholar]
- 42.Mori T, Kiyono T, Imabayashi H, Takeda Y, Tsuchiya K, et al. (2005). Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol Cell Biol 25:5183–5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto T, Aoyama T, Nakayama T, Nakamata T, Hosaka T, et al. (2002). Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem Biophys Res Commun 295:354–361 [DOI] [PubMed] [Google Scholar]
- 44.Bork S, Pfister S, Witt H, Horn P, Korn B, et al. (2010). DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 9:54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi MR, In YH, Park J, Park T, Jung KH, et al. (2012). Genome-scale DNA methylation pattern profiling of human bone marrow mesenchymal stem cells in long-term culture. Exp Mol Med 44:503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brønnum H, Andersen DC, Schneider M, Sandberg MB, Eskildsen T, et al. (2013). miR-21 promotes fibrogenic epithelial-to-mesenchymal transition of epicardial mesothelial cells involving programmed cell death 4 and sprouty-1. PLoS One 8:e56280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, et al. (2004). MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279:52361–52365 [DOI] [PubMed] [Google Scholar]
- 48.Tenstad E, Tourovskaia A, Folch A, Myklebost O. and Rian E. (2010). Extensive adipogenic and osteogenic differentiation of patterned human mesenchymal stem cells in a microfluidic device. Lab Chip 10:1401–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Døsen-Dahl G, Munthe E, Nygren MK, Stubberud H, Hystad ME, et al. (2008). Bone marrow stroma cells regulate TIEG1 expression in acute lymphoblastic leukemia cells: Role of TGFβ/BMP-6 and TIEG1 in chemotherapy escape. Int J Cancer 123:2759–2766 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.