Abstract
Septic shock is the most common cause of death in intensive care units due to an aggressive inflammatory response that leads to multiple organ failure. However, a lipopolysaccharide (LPS) tolerance phenomenon (a nonreaction to LPS), is also often described. Neither the inflammatory response nor the tolerance is completely understood. In this work, both of these responses were analyzed using microarrays in zebrafish. Fish that were 4 or 6 days postfertilization (dpf) and received a lethal dose (LD) of LPS exhibited 100% mortality in a few days. Their transcriptome profile, even at 4 dpf, resembled the profile in humans with severe sepsis. Moreover, we selected 4-dpf fish to set up a tolerance protocol: fish treated with a nonlethal concentration of Escherichia coli LPS exhibited complete protection against the LD of LPS. Most of the main inflammatory molecules described in mammals were represented in the zebrafish microarray experiments. Additionally and focusing on this tolerance response, the use of cyclodextrins may mobilize cholesterol reservoirs to decrease mortality after a LD dose of LPS. Therefore, it is possible that the use of the whole animal could provide some clues to enhance the understanding of the inflammatory/tolerance response and to guide drug discovery.
Introduction
In mammals, microbial products such as lipopolysaccharide (LPS) or endotoxin are potent inducers of inflammation. These components stimulate immune system cells after the components are recognized, usually by toll-like receptors (TLRs). TLRs are a family of closely related transmembrane proteins that initiate signaling cascades, leading to the enhanced transcription of cytokines and other proinflammatory mediators.1 Due to an excessive inflammatory response, some individuals develop a sepsis reaction, which can lead to death in the case of critically ill individuals; septic shock is the most common cause of death in intensive care units.2 The aggressive inflammatory response causes multiple organ failure rather than an inability to fight infection.2 However, a hyporesponsive or cell desensitization state called tolerance (TOL) develops after continued exposure to LPS and prevents excessive cell activation, limiting the proinflammatory responses of neutrophils.3 The pathogenesis of severe sepsis and septic shock is a complex interaction of inflammatory and anti-inflammatory responses, coagulation, apoptosis, and metabolic disorders. However, the molecular processes that lead to multiple organ failure remain unclear.4 The LPS tolerance consisting of a nonreaction to an LPS treatment is also a response that is not completely understood. A whole gene expression study is a useful approach to examine the characteristic signature gene profile that is associated with a particular process. In this case, knowledge of which genes are modulated might reveal novel insights into the conserved host responses to sepsis and LPS tolerance.
In the present work, we used zebrafish (Danio rerio) to increase our knowledge of the global transcriptome associated with sepsis and LPS tolerance. In the past few years, this species has been gaining importance as a model for many human diseases because it has important advantages: its fertilized embryos are transparent, fluorescent tools are available to study host–pathogen interactions, it is suitable for large-scale genetic analyses, and there is a clear temporal separation between the innate and adaptive immune systems.5–10 Moreover, we can investigate processes in vivo by using the complete organism and by studying the global response. Therefore, to determine the associated transcriptome profile and to identify the candidate genes associated with sepsis and tolerance, we studied the effects of a LPS treatment or the combination of this lethal treatment preceded by the administration of a sublethal concentration of LPS, which results in a tolerized state. The sepsis/inflammation/tolerance processes in zebrafish were analyzed by comparing them with the same responses in mammals to determine if there are similar gene expression profiles.
Materials and Methods
Animals
Wild type zebrafish embryos and larvae were obtained from our experimental facilities where zebrafish are cultured following established protocols11,12 (also see http://zfin.org/zf_info/zfbook/zfbk.html). Tg(mpx:GFP) fish were kindly provided by S. Renshaw (University of Sheffield). Fish care and challenge experiments were conducted according to the CSIC National Committee on Bioethics guidelines under approval number ID 01_09032012.
Experimental treatments
Zebrafish larvae that were 4 or 6 days postfertilization (dpf) were treated with LPS following a previously described protocol.13 Briefly, a concentration of 50 μg/mL of LPS from Escherichia coli serotype 0111:B4 (Sigma-Aldrich) was used as a sublethal dose treatment (SLD) (after confirming that it did not induce any mortalities after the treatment), and a concentration in water of 50 μg/mL of LPS from Pseudomonas aeruginosa (Sigma-Aldrich) was used as a lethal dose (LD) (after determining that all fish died within 2–5 days). The treatments were conducted in six-well plates that were maintained at 28°C.
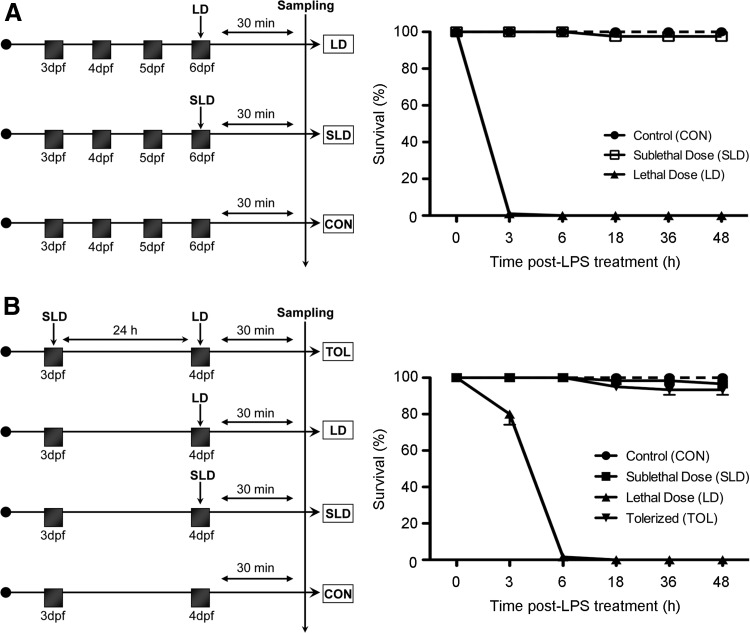
Combinations of sublethal concentrations of E. coli LPS and lethal concentrations of P. aeruginosa LPS were used to generate a tolerance state. The diagrams of the various experiments are shown in Figure 1A and B. After the different treatments, sampling was conducted after 30 min, and four biological replicates were collected for each treatment. Parallel experiments with the same stock of fish larvae were always conducted to follow the mortalities observed during each treatment.
FIG. 1.
Graphical representation of the different experiments conducted for sampling and further microarray hybridization. (A) Treatments applied to 6-dpf fish receiving a LD of LPS (LD), a SLD of LPS (SLD), and controls (CON). (B) Treatments applied to 4-dpf fish receiving a LD of LPS (LD), a SLD of LPS (SLD), SLD and LDs (TOL), and controls (CON). h, hours poststimulation; min, minutes; dpf, days postfertilization. Parallel experiments to follow the mortalities per treatment with the same stock of fish larvae used for sampling are shown in the plots both for (A) and (B). LPS, lipopolysaccharide.
Moreover, to determine the importance of the cholesterol metabolism in sepsis, different concentrations of alpha and beta cyclodextrins (Sigma-Aldrich) were added 24 h before the LD of LPS.
Neutrophil migration studies
Tg(mpx:GFP) zebrafish larvae with GFP fluorescent neutrophils were used to investigate the neutrophil migration to a wound. After larvae hatching (3 dpf), a SLD of LPS was administered as described above, and 24 h later, the zebrafish tails were cut using a razor. Four hours after the tail ablations, neutrophil migration to the wound was observed under an AZ100 microscope (Nikon) and photographed with a DS-Fi1 digital camera (Nikon). The same protocol was conducted using the 4-dpf larvae, except that a 30-min SLD treatment was used instead of a 24-h treatment for comparison. Controls were used in both cases with tail ablation, but without the LPS treatment. Finally, the relative fluorescence intensity in the tail was measured using ImageJ software.14
RNA isolation and cDNA transcription
Larvae (n=10–15) from each condition were pooled 30 min poststimulation in 500 μL of TRIzol reagent (Life Technologies) and preserved at −80°C until use. Total RNA isolation was conducted following the TRIzol manufacturer's specifications in combination with the RNeasy Mini Kit (Qiagen) for RNA purification after DNase I treatment. One microgram of total RNA was then used to obtain cDNA using the SuperScript III First-Strand Synthesis SuperMix for quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) (Life Technologies).
Microarray analyses
The 4×44K Zebrafish Gene Expression Microarray (V2, AMADID 019161) from Agilent Technologies was used. Briefly, this array is composed of a 60-mer oligonucleotide array containing 43,803 probes representing 23,207 genes. A total of six slides (21 microarrays) were hybridized. RNA quality was assessed with the Agilent 2100 Bioanalyzer and kept frozen at −80°C until all of the experiments could be hybridized and processed simultaneously. The labeling of 2 μg of RNA (∼50 μg/mL) and hybridizations were carried out at the Universidad Autónoma de Barcelona microarray facility, complying with the Minimum Information about a Microarray Experiment (MIAME) standards.15 The signal was captured, processed, and segmented using an Agilent G2565B scanner (Agilent Technologies) with the Agilent Feature Extraction Software (v9.5) protocol GE1-v5_95 using an extended dynamic range and preprocessing by the Agilent Feature Extraction v9.5.5.1.
The results for the fluorescence intensity data and quality annotations were imported into GeneSpring GX version 11.0.2 (Agilent Technologies). All of the control features (including the positive and negative controls and the landing lights) were excluded from the subsequent analyses. Normalization was then carried out by a percentile shift at the 75th percentile. Entities with an expression between the 20th and 100th percentile in the raw data were retained and used in the subsequent analyses. To assess genes for differential expression, the normalized log intensity ratios were analyzed with Student's t-test without multiple testing correction.
Differential modulation of gene expression after each treatment has been compared using Venn diagrams. Lists of modulated genes in each treatment compared with controls were selected and compared with other treatments or fish age using the Venny software program16 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
For enrichment analysis, the functional classes that were significantly different among the treatments were identified using the Blast2GO software program.17 Fisher's exact test with a term filter value of 0.05 and multiple testing correction have been used to compare the list of up- or down-modulated probes for each treatment separately (fold change >1.5). Following the true path rule, only the most specific terms obtained were presented.
For the interactome analysis, interactions and overlays of expression profiles were visualized using the Cytoscape (version 2.8.2.; www.systemsbiology.org). The interactome network was obtained from all interactions with a Final Bayesian Score >6. The interactome backbone contains 5,760 nodes (protein–protein and protein–DNA interactions) and 99,573 relationships between these proteins (interactions). The designation of protein properties was drawn from Alexeyenko et al. 201018 NCBI gene name attributes were used to unify the protein list and were imported through the Biomart plugin. The network for the zebrafish challenges was built from within the Danio_rerio_CS interactome. Topological analysis of individual and combined networks was performed with Network Analyzer, and jActiveModules 2.2 was used to analyze network characteristics.19,20 GO analyses were conducted with the Biological Network Gene Ontology (BinGO, version 2.0) plugin21 that was used for statistical evaluation of groups of proteins with respect to the current annotations available at the Gene Ontology Consortium (www.geneontology.org). GO overrepresentation was calculated using the hypergeometric test with Benjamini and Hochberg false discovery rate (FDR) multiple testing correction and significance (pFDR<0.05). The redundant GO classes were deleted to simplify the figure, and all GO classes for each treatment were included as Supplementary Tables S1 and S2 (Supplementary Data are available online at www.liebertpub.com/zeb). In addition, we conducted a complementary analysis with the ClusterMaker cytoscape plugin,22 using the MCL algorithm to search protein–protein interaction network modules derived from TAP/MAS (tandem affinity purification/mass spectrometry). This approach clustered the network into modules based on the purification enrichment (PE) score to indicate the strength of the node association given a fixed set of genes with high protein–protein affinity (interactome cluster nodes). The subcellular localization was determined by the Cytoscape plugin, Cerebral.23
Quantitative reverse transcriptase-polymerase chain reaction
Specific PCR primers were designed from the sequences of the selected probes (Supplementary Table S3) using the Primer3 program24 according to qRT-PCR restrictions. Oligo Analyzer (version 1.0.2) was used to check for dimer and hairpin formation, and the efficiency of each primer pair was also analyzed from the slope of the regression line of the quantification cycle versus the relative concentration of cDNA.25 A melting curve analysis was also performed to verify that only specific amplification occurred and that no primer dimers were amplified. If these conditions were not accomplished, new primer pairs were designed.
qRT-PCR was performed using the 7300 Real Time PCR System (Applied Biosystems). One microliter of fivefold diluted cDNA template was mixed with 0.5 μL of each primer (10 μM) and 12.5 μL of SYBR green PCR master mix (Applied Biosystems) in a final volume of 25 μL. The standard cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were performed as technical triplicates, and an analysis of melting curves was performed in each reaction. The relative expression levels of the genes were normalized using 18S ribosomal RNA (BX296557) expression as a housekeeping gene control, which was constitutively expressed and not affected by the treatments, following the Pfaffl method.25 Fold change units were calculated by dividing the normalized expression values of the stimulated tissues by the normalized expression values of the controls. For the biological replicates, the average relative level of expression from each replicate was considered a single point, and the mean and standard error were calculated.
Results and Discussion
LPS-induced mortalities and microarray validation results
The experiments conducted with parallel fish groups that received the same treatments than those used for analyzing the gene expression profile showed that fish (4 or 6 dpf) died after the LD of P. aeruginosa LPS with mortalities reaching 100% within a few hours posttreatment. However, no mortalities were recorded when the fish were treated with a low concentration of the E. coli LPS. These results were highly consistent, and for this reason, we used the P. aeruginosa LPS and the E. coli LPS as the lethal and SLDs, respectively (Fig. 1A). In the tolerance experiments, 4-dpf fish previously treated with a SLD of E. coli LPS did not die after the P. aeruginosa LD treatment, which allowed us to obtain the sample of tolerized fish according to Novoa et al.13 (Fig. 1B).
A group of four transcripts modulated in 4- and 6-dpf fish was selected to quantify their expression pattern and validate the transcriptome profile after microarray hybridization with the samples described above. Increased expression was confirmed by qRT-PCR for the four transcripts selected for each age (Supplementary Fig. S1A, B).
Transcriptome of zebrafish after severe (LD) LPS treatment
The number and magnitude of the response of differentially modulated genes after LPS treatment were both higher when the experiments were conducted in 6-dpf fish (25.7% of the modulated genes, based on a cut-off value of >1.5-fold change) than in 4-dpf fish (1.1% of modulated genes), indicating that maturation of the immune system occurs at these stages and that older fish respond by expressing a more complex repertoire of genes. Although the number of common modulated genes between 4- and 6-dpf LD-treated fish was low (only 36 genes) and the most modulated genes were not the same after the LPS treatment when the fish at the two ages were compared, there was an important up-modulation of proinflammatory genes such as interleukin 1, beta (IL1B), tumor necrosis factor-beta (TNFB), and TNF receptor family members that clearly confirm a strong inflammatory response in both cases (Table 1). The basis for the differences between the ages could be that at 4 dpf, not all the genes have started to be expressed or that the expression was too low to provide significant values based on the statistical analysis.6,26,27
Table 1.
Top25 Most Modulated Genes in Fish Treated with the Lethal and Sub-LD Compared with Control Fish (LD/Control); (SLD/Control)
| Description (4 dpf) | FC | Reg | Description (6 dpf) | FC | Reg |
|---|---|---|---|---|---|
| LD/control comparison | |||||
| Glutaminyl-peptide cyclotransferase-like | 8.3 | Up | Tumor necrosis factor-beta | 28.2 | Up |
| Interleukin 1, beta | 5.5 | Up | Cholesterol 25-hydroxylase | 15.6 | Up |
| Alpha globin type-2, transcript variant 1 | 5.3 | Down | E74-like factor 3 (ets domain transcription factor, epithelial-specific) | 12.9 | Up |
| Diablo, IAP-binding mitochondrial protein | 4.8 | Up | Zinc finger and BTB domain containing 25 | 9.7 | Up |
| Pyruvate dehydrogenase kinase, isoenzyme 2 | 4.1 | Up | Early growth response 2 | 9.5 | Up |
| Cyclin-dependent protein kinase 5 | 3.7 | Down | D-alanyl-D-alanine carboxypeptidase protein | 9.4 | Up |
| Patched 1 | 3.7 | Up | Pleckstrin homology domain containing, family F (with FYVE domain) member 1 | 8.6 | Up |
| Somatolactin beta | 3.3 | Up | FOS-like antigen 2 | 7.8 | Up |
| DNA fragmentation factor, alpha polypeptide | 3.3 | Up | FBJ murine osteosarcoma viral oncogene homolog B | 7.3 | Up |
| Centrosomal protein 70 | 2.9 | Up | Early growth response 3 | 7.0 | Up |
| Very large inducible GTPase 1 | 2.8 | Down | Coagulation factor IIIb | 6.8 | Up |
| Caspase b-like | 2.7 | Up | Interleukin 1, beta | 6.2 | Up |
| Polymeric immunoglobulin receptor | 2.7 | Up | Prostaglandin-endoperoxide synthase 2b | 6.0 | Up |
| Transmembrane protein 220 | 2.7 | Up | Alpha 1 type II procollagen | 5.2 | Up |
| Preimplantation protein 4-like | 2.6 | Up | Rho-related BTB domain containing 2a | 5.2 | Up |
| ST3 beta-galactoside alpha-2,3-sialyltransferase 1 | 2.4 | Up | CD276 molecule precursor | 5.1 | Up |
| Junctional adhesion molecule C precursor | 2.4 | Down | JUN B proto-oncogene | 4.9 | Up |
| Diaphanous homologue 2 | 2.4 | Up | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a | 4.4 | Up |
| Tumor necrosis factor receptor superfamily member 18 | 2.3 | Up | JUN dimerization protein 2 | 4.2 | Up |
| Interleukin 10 | 2.3 | Up | ANP32A protein | 4.1 | Up |
| Wingless-type MMTV integration site family, member 3A | 2.3 | Up | Glypican 6 | 4.1 | Up |
| Keratocan | 2.3 | Down | CCAAT/enhancer binding protein (C/EBP), beta | 4.0 | Up |
| Cytochrome P450 | 2.3 | Down | Somatolactin alpha | 4.0 | Up |
| Phosphatidylcholine-sterol acyltransferase precursor | 2.3 | Up | Prostaglandin E receptor 2b (subtype EP2) | 3.9 | Up |
| Zinc finger protein 347 | 2.3 | Up | Galactosidase, beta 1-like 2 | 3.6 | Up |
| SLD/control comparison | |||||
| Low density lipoprotein receptor | 3.4 | Up | Centrosomal protein 135 | 8.9 | Up |
| Tumor necrosis factor receptor superfamily, member 14 | 2.8 | Down | Glucose-6-phosphatase | 8.7 | Up |
| C1q-like adipose specific protein | 2.6 | Up | Zinc finger protein 37 homolog | 7.7 | Up |
| NADPH oxidase organizer 1 | 2.4 | Down | Solute carrier family 12 member 9-like | 7.3 | Up |
| Splicing factor 3a, subunit 1 | 2.4 | Up | Nipped-B homolog | 6.9 | Up |
| Macrophage expressed 1 | 2.3 | Down | Histamine receptor H2 | 6.9 | Up |
| Phosphoenolpyruvate synthase | 2.2 | Up | PRKC, apoptosis, WT1, regulator like | 5.9 | Up |
| ADP-ribosylation factor GTPase activating protein 1 | 2.2 | Up | Zinc finger protein 592 | 5.9 | Up |
| Integrin, alpha 2 | 2.1 | Up | Leucine-rich repeat-containing G protein-coupled receptor 4 | 5.7 | Up |
| Synaptosomal-associated protein 23 | 2.1 | Down | PR domain containing 1c | 5.7 | Up |
| Zinc finger protein 782 | 2.0 | Down | EF-hand calcium-binding domain-containing protein 4B | 5.3 | Up |
| Sterile alpha and TIR motif containing 1 | 5.2 | Up | |||
| Gamma-glutamyltransferase family | 5.2 | Up | |||
| Ataxin 2 | 5.2 | Up | |||
| Notum 3 | 5.1 | Up | |||
| Oxytocin receptor | 5.1 | Up | |||
| Glucagon receptor | 5.1 | Up | |||
| 40S ribosomal protein S21 | 5.1 | Up | |||
| Hyaluronan and proteoglycan link protein 1b | 5.1 | Up | |||
| Zinc finger protein 271 | 4.9 | Up | |||
| Activating molecule in beclin-1-regulated autophagy | 4.8 | Up | |||
| Glutamate receptor, ionotrophic, AMPA 3b | 4.7 | Up | |||
| Adenosine A2b receptor | 4.7 | Up | |||
| PE-PGRS family protein | 4.7 | Up | |||
| Transcription termination factor, RNA polymerase II | 4.7 | Up | |||
dpf, days postfertilization; FC, fold change; LD, lethal dose; Reg, regulation; SLD, sublethal dose.
Many of the differentially modulated transcripts were associated with biological processes that occur during the inflammatory and anti-inflammatory cascades in mammals, including macrophage and neutrophil activation programs, the mitogen-activated protein kinase signaling network, apoptosis, ROS activity, lipid metabolism, and coagulation (Table 1). This can also be observed in the enrichment analysis of Gene Ontology (GO) terms carried out with the microarray data (Supplementary Table S4): for the genes that were up-modulated in the 6-dpf fish, there was a significant enrichment of GO terms such as immune response, G-protein coupled receptor signaling, detection of stimulus, etc. In contrast, the genes that were down-modulated in LD-treated fish were enriched in gene categories that are related to normal development of the fish, suggesting a strong deviation from the normal gene expression pattern occurred in response to the excessive inflammation.
In addition to IL1B or TNFB, other proinflammatory genes were modulated after the LPS treatment: the CCAAT/enhancer binding protein (C/EBP), beta (CEBPB) (at 6 dpf), which is known as a key modulator in inflammatory diseases28–30; the glutaminyl-peptide cyclotransferase-like (QPCTL) was the most up-modulated gene in the 4-dpf LD-exposed fish (fold change of 8.3) and have been reported to be involved in several inflammatory processes, such as Alzheimer's disease31 or monocyte migration.32,33 The elevated expression levels of all these molecules explain the exaggerated inflammation response that characterizes septic shock. Moreover, the CD276 molecule precursor (B7-H3), which was up-modulated under LD conditions at 6 dpf, functions as both a costimulator of proinflammatory cytokine release through NFKB34 and a co-inhibitor to control the exuberant immune responses.35 The up-modulation of other genes that limit the inflammatory process was also observed: IL10 (at 4 dpf), the inhibitor of NFKB (NFKBIA, the nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha) and the early growth response 2 and 3 transcription factors (EGR2, 3) (at 6 dpf); these are essential for balancing and successfully controlling inflammation.36
During the final process that leads to organ failure due to oxidative stress, tissue-damaging enzymes and apoptosis, several proteins were found to be up-modulated in the zebrafish transcriptome: the pyruvate dehydrogenase kinase isoenzyme 2 (PDK2) was up-modulated at 4 dpf and increases the concentration of lactate in mammals. Lactate concentration is a measure of tissue hypoxia and is sufficient for the diagnosis of septic shock.37 Three apoptosis-related proteins were also up-modulated in 4-dpf fish: diablo, IAP-binding mitochondrial protein (DIABLO); DNA fragmentation factor, alpha polypeptide (DFFA); and caspase b-like and one in 6-dpf fish: a protein with homology to pleckstrin (PLEKHF1) that was reported to be an important intermediate in the activation pathways of inflammatory reactions.38
In the pathogenesis of sepsis, inflammation and coagulation play pivotal roles. An up-modulation of coagulation factor IIIb (F3b) was observed at 6 dpf during the LD treatment. Moreover, the ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (ST3GAL1) was up-modulated at 4 dpf. Sialylation/desialylation is a process that seems to be involved in the clearance of glycoproteins decorated with sialic acid to avoid disseminated intravascular coagulation, a feared complication of sepsis.39
It is worth emphasizing that an important activation of lipid-mediated signaling was observed; this activation is similar to the one observed in mammals, which is also a critical step in the mammalian inflammation cascade.40–43 Prostaglandin-endoperoxide synthase 2 (PTGS2) and prostaglandin E receptor 2b (subtype EP2) (PTGER2) were up-modulated at 6 dpf. Phosphatidylcholine-sterol acyltransferase or lecithin-cholesterol acyltransferase (LCAT) was up-modulated at 4 dpf, and it is a key enzyme in the intravascular metabolism of HDL cholesterol.44 Importantly, cholesterol 25-hydroxylase (CH25H) was significantly up-modulated at 6 dpf, and it is thought to play a role in the inflammatory response because its expression is induced rapidly, selectively, and robustly by the TLR ligands poly I:C and LPS.45–48
Transcriptome after mild (SLD) LPS treatment
The inflammatory response observed for this LPS treatment (Table 1) was not as clear as that for the LD described above. The percentage of modulated genes at 4 dpf (0.3%) and 6 dpf (14.5%) was lower than for the LD. However, there was a specific modulation of several genes related to chemokines and G protein-coupled receptor signaling that appeared to be modulated only in response to the E. coli LPS (sublethal treatment): the integrin, alpha 2 (ITGA2) is crucial for macrophage and neutrophil migration to host tissues49; and the adenosine A2b receptor (ADORA2B), which is responsible for the movement of macrophages through a gradient of the chemoattractant C5a.50–55 The up-modulated ADP-ribosylation factor GTPase activating protein 1 (ARFGAP1) also seems to significantly modify the transport of G protein-coupled receptors,56 including chemokine (C-X-C motif) receptor 4 (CXCR4), which has been related to TLR4 and LPS tolerance.13,57 This response was also observed in the enrichment of GO terms in the comparison SLD-treated fish against controls (Supplementary Table S4).
There were also some modulated genes with important anti-inflammatory properties in 6-dpf fish: the histamine receptor H2 (HRH2), that acts as a suppressor of antigen presentation capacity and enhances IL10 production58–63 and the toll IL1 receptor domain-containing adaptor (SARM1) that down-modulates NFKB and IRF3-mediated TLR3 and TLR4 signaling.64
Similar to the LD treatment, some proteins related to tissue damage and apoptosis were also modulated in zebrafish in response to the SLD treatment, but the inflammatory effects did not seem to be as apparent as for the lethal treatment.
Genes involved in glucose homeostasis, such as glucose-6-phosphatase (G6P) or the glucagon receptor (GCGR), were highly up-modulated in SLD-treated fish. This is consistent with reports in mammals because insulin regulates the inflammatory response either directly or indirectly.65–72
As in the case of the lethal LPS treatment, the lipid metabolism-related genes were affected: the low-density lipoprotein receptor (LDLR) was the most up-modulated gene at 4 dpf. Because LPS can be bound by triglyceride-rich lipoproteins (TRL) that may be internalized through the LDLR pathway, the internalization of lipoprotein bound endotoxin (TRL-LPS) could attenuate the systemic inflammatory response.73 Moreover, LDLR dysfunction leads to the accumulation of cholesterol-rich LDL (low-density lipoproteins) in plasma and premature atherosclerosis.74 This fact again suggests the key role of cholesterol during the inflammation process.
Transcriptome in LPS tolerance
Although the transcriptomic response to LPS was more complex for 6-dpf fish, the treatment with a previous sublethal concentration of LPS induced a complete protection against a LD of LPS in 4-dpf fish. For this reason, we used 4-dpf zebrafish larvae to further determine the global transcriptome in a tolerized state against LPS (Table 2). For this sample, the percentage of modulated genes was low (0.99% and 2.2% for TOL/CON and LD/TOL comparisons, respectively), most likely corresponding to an immature development of the organism.
Table 2.
Top25 Most Modulated Genes in Fish Treated With the Sublethal and Lethal Dose Compared With Control Fish (TOL/Control) and in Fish Treated With the Lethal Dose Compared to Fish Treated With the Sublethal and Lethal Dose (LD/Tol)
| Description (4 dpf) | FC | Reg |
|---|---|---|
| TOL/control comparison | ||
| Somatolactin beta | 6.6 | Up |
| Hatching enzyme 1 (he1a) | 4.0 | Down |
| FBJ murine osteosarcome viral oncogene homolog B | 3.6 | Up |
| Complement C3-H1 | 3.2 | Down |
| Splicing factor 3a, subunit 1 | 3.2 | Up |
| Transmembrane protein 144 | 3.1 | Up |
| Very large inducible GTPase 1 | 2.8 | Down |
| Transient receptor potential cation channel, subfamily V, member 6 | 2.8 | Up |
| C1q-like adipose specific protein | 2.7 | Up |
| Heat shock protein, alpha-crystallin-related, 9 | 2.5 | Up |
| Ghrelin/obestatin prepropeptide | 2.4 | Up |
| Siaz-interacting nuclear protein | 2.4 | Up |
| Cylicin-1 | 2.4 | Up |
| All-trans-13,14-dihydroretinol saturase | 2.3 | Up |
| Apolipoprotein B (including Ag(x) antigen) | 2.3 | Down |
| Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | 2.3 | Down |
| Prominin-like 1 | 2.2 | Up |
| Prickle-like 1 | 2.2 | Down |
| Phosphoenolpyruvate synthase | 2.2 | Up |
| Complement factor B | 2.1 | Down |
| Prostaglandin-endoperoxide synthase 2b | 2.1 | Up |
| DnaJ (Hsp40) homolog, subfamily B, member 12 | 2.1 | Down |
| UDP glucuronosyltransferase 2 family, polypeptide A3 | 2.1 | Up |
| Peptidyl-tRNA hydrolase | 2.0 | Down |
| Zona pellucida glycoprotein 3a | 2.0 | Down |
| LD/TOL comparison | ||
| Glutaminyl-peptide cyclotransferase-like | 7.3 | Up |
| Interleukin 1, beta | 6.8 | Up |
| Trypsinogen | 5.6 | Up |
| Stanniocalcin 1 | 5.1 | Up |
| Cholesterol 25-hydroxylase | 4.6 | Up |
| S100 calcium binding protein T | 4.3 | Down |
| Histone cluster 3, H3c | 3.9 | Up |
| Wingless-type MMTV integration site family, member 2Bb | 3.8 | Up |
| Ubiquitin family | 3.7 | Up |
| Glutamate receptor-associated protein 1 | 3.7 | Up |
| Diablo, IAP-binding mitochondrial protein | 3.7 | Up |
| Transcription elongation factor B (SIII), polypeptide 3 | 3.6 | Up |
| Centrosomal protein 70 | 3.4 | Up |
| Alpha-tectorin | 3.2 | Down |
| Insulin-responsive sequence DNA-binding protein 1 | 3.1 | Down |
| Monooxygenase, DBH-like 1 | 3.1 | Up |
| Patched 1 | 3.1 | Up |
| Glutathione peroxidase 7 | 3.0 | Down |
| Phosphatidylcholine-sterol acyltransferase precursor | 2.9 | Up |
| Histone cluster 4, H4 | 2.9 | Up |
| Tumor necrosis factor receptor superfamily member 18 | 2.7 | Up |
| Membrane bound O-acyltransferase domain containing protein 1 | 2.7 | Up |
| FBJ murine osteosarcome viral oncogene homolog B | 2.7 | Down |
| Epidermal growth factor receptor | 2.6 | Up |
| Leucine zipper and W2 domains 1b | 2.6 | Up |
TOL, tolerized.
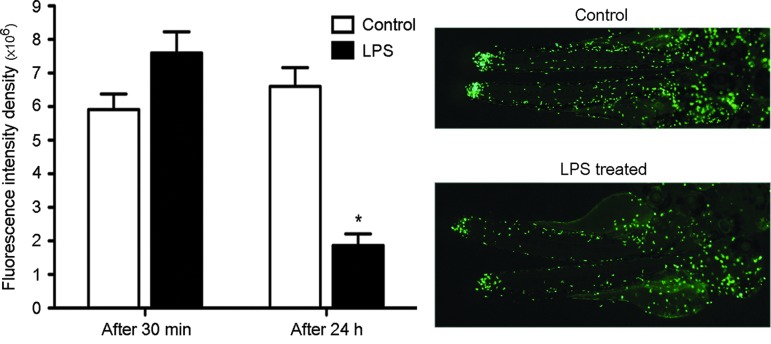
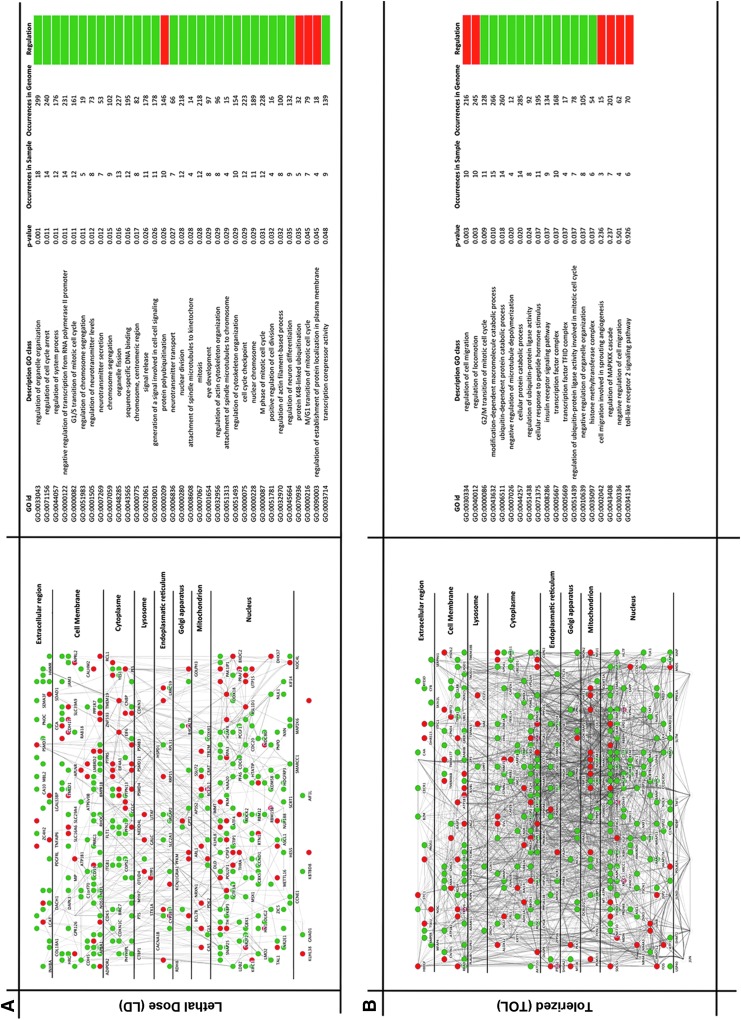
In general, we determined that many of the proinflammatory genes that were up-modulated during the lethal treatment (Table 1) were down-modulated in the tolerance state (Table 2), most likely to avoid the excessive inflammation associated with the mortality of fish treated with the lethal concentration of LPS. This was also confirmed in the GO enrichment analysis where only the immune response term was up-modulated in LD-treated fish (LD/TOL comparison) reflecting a generalized defense response that most likely leads to fish death due to an exaggerated inflammation response (Supplementary Table S4). The reduced capacity to respond to LPS activation after an initial exposure to this stimulus has been previously described in mammals and indicates the development of a hyporeactive state.3,75–79 This reduced response to the LD of LPS could be visualized by using the zebrafish Tg(mpx:GFP) transgenic line with GFP fluorescent neutrophils.80 The neutrophil migration to an injury induced in the tail and, therefore, the inflammatory reaction was significantly reduced after 24 h of the SLD of LPS treatment (Fig. 2). Despite this reduced capacity to respond to LPS in tolerized fish, the interactome analysis displayed a clearly different response between the LD- and TOL-treated fish. Whereas the down-modulated genes in LD-treated 4-dpf fish are enriched in gene categories that are mainly related to the normal development and differentiation of the fish, this inhibition was not observed for TOL-treated fish, with a modulation of cell migration as described above and a much more complex interactome scenario that requires further study (Fig. 3A, B).
FIG. 2.
Neutrophil migration after LPS treatment and tail ablation. Pictures show the neutrophil migration to the wound 4 h after tail ablation in mpx:GFP zebrafish larvae treated with a SLD of LPS during 30 min (4-dpf larvae) or 24 h (3-dpf larvae). Controls were used in both cases with tail ablation, but without the LPS treatment. Bars represent the relative fluorescence intensity density on the tail measured using ImageJ software (*p<0.05).
FIG. 3.
Interactome mapping of 4-dpf zebrafish larvae treated with (A) lethal dose (LD) and (B) tolerized (TOL). Left panel: interactome modules of mRNAs expressed in larvae based on the MCL algorithm (ClusterMaker plugin). The over-represented pathways are visualized in Cytoscape with the Cerebral plugin. The larger network shows the pathway according to subcellular localization. Right panel: Gene ontology analysis (BinGO plugin) of each interactome module of over-expressed GO categories (p<0.01); the color scale bar indicates relative abundance (high-low) of GO categories in each treatment. The redundant GO classes were deleted. The colors in the main view for each gene expression condition (two, in this case) were represented according to the quantitative data provided (green; down-modulated, red; up-modulated). Sample size, n=532 in LD, n=481 in TOL. BinGO, Biological Network Gene Ontology.
Interestingly, several genes with anti-inflammatory or potential anti-inflammatory properties were up-modulated in the zebrafish transcriptome: ghrelin/obestatin prepropeptide (GHRL)81 or S100 calcium binding protein T (S100T). Although the role of this protein in zebrafish is still unknown, S100 proteins are endogenous activators of innate immune responses in mammals, and some of them have proinflammatory properties and are associated with different inflammatory diseases, such as inflammatory bowel disease.82–84
Several genes involved in the relief of oxidative stress as a compensatory mechanism were up-modulated in tolerized zebrafish as described for mammals85,86; these include glutathione peroxidase 7 (GPX7), which mitigates the organ dysfunction during chronic inflammation, and phosphoenolpyruvate synthase, which is an essential enzyme when pyruvate and lactate are used as a carbon source. The up-modulation in tolerized fish suggests a mechanism for avoiding the excess of lactate and hypoxia that induces the organ failure associated with sepsis. In addition to being a metabolic intermediate, pyruvate is an effective scavenger of ROS, playing a role as an anti-inflammatory molecule in the last stages of the inflammation process.87
As already described above for the SLD treatment, the carbohydrate metabolism seemed to be substantially altered in the tolerized zebrafish as part of the metabolic syndrome that it is concurrent with most of the inflammatory diseases described in mammals. The insulin-responsive sequence DNA-binding protein 1 (IRE-BP1) that was up-modulated in tolerized fish compared with LD-treated fish appears to be a downstream effector of insulin-induced phosphoinositide-3-kinase (PI3K) signaling and mediates the action of insulin on multiple target genes.88,89 Moreover, IRE-BP1 expression also increased the mRNA levels of a number of genes involved in fatty acid homeostasis.90
Regarding the lipid mediators role, some of the molecules that were up-modulated in LD-treated fish were down-modulated for the tolerance treatment as described above for the main proinflammatory genes. This is the case for LCAT and CH25H (both involved in cholesterol metabolism). Interestingly, C1q-like adipose specific protein or adiponectin only appeared to be up-modulated in tolerized fish compared with the controls. Several studies have shown that adiponectin attenuates the production of inflammatory cytokines through LPS-induced macrophages in vitro.91 Moreover, Benoit and Tenner92 recently reported that C1q prevents β-amyloid-induced neuronal death in vitro and induces an up-modulation of genes associated with cholesterol metabolism.
In summary, we can highlight that transcriptomic analysis in zebrafish is very suitable for the study of the inflammatory response. The results obtained with the whole animal are also commonly observed in mammals using specific tissues with the same approach. This includes the following: an early inflammatory response with up-modulation of cytokines, chemokines, and proinflammatory mediators; neutrophil, complement and coagulation activation; apoptosis; and oxidative stress events. The early inflammatory response is then followed by the transition to an anti-inflammatory state with increasing abundance of proteins that limit the inflammatory response, such as proteins responsible for preventing tissue injury.93–101
Involvement of cholesterol in sepsis
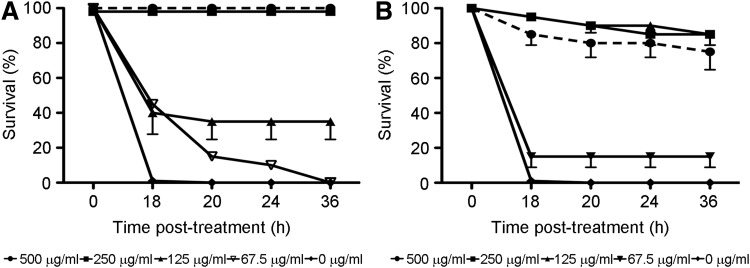
Considering that some of the genes involved in lipid metabolism were the most modulated after LPS treatment, we sought to determine whether cholesterol metabolism was a key element in the response observed in LPS-treated fish and whether our zebrafish model was suitable for this study. To accomplish these objectives, we used cyclodextrins because it is well-known that they remove cholesterol from lipid rafts, which are targets in several diseases, including chronic inflammation, sepsis and septic shock, Alzheimer's disease, and atherosclerosis.102–104 The addition of different concentrations of alpha cyclodextrins before the LD treatment reduced the mortality to 0% at the higher concentrations (250 and 500 μg/mL) achieving a complete tolerance status to a LPS LD. For the beta cyclodextrins, a reduction in mortality was also observed for the concentrations of 125, 250, and 500 μg/mL (Fig. 4A, B). To the best of our knowledge, this is the first time that a tolerance status with 100% of survival rate was achieved using zebrafish as a model. Furthermore, as far as we know, this is the first report on the use of cyclodextrins based on their anti-inflammatory properties instead of their ability to increase the bioavailability of several drug–cyclodextrin complexes, which is well documented.105–107 These results, therefore, demonstrated the importance of lipid mediators in the sepsis process.
FIG. 4.
Representative experiment conducted to follow mortalities after alpha cyclodextrin (A) and beta cyclodextrin (B) treatment. Different concentrations (67.5, 125, 250, 500 μg/mL) of alpha and beta cyclodextrins were added to zebrafish larvae 24 h before LD treatment for mortality registration.
In conclusion, our results showed that the inflammatory/tolerance response could be clearly described in a whole organism model of zebrafish. Our results suggest a fine-tuning of gene regulation similar to that described in humans. The modulation was different in larvae administered a LPS LD (resulting in a strong inflammation response, similar to a sepsis process) from the larvae administered the milder treatment (SLD). Importantly, the mammalian inflammation models that have been proposed in the literature do not seem to be completely appropriate for the discovery of target genes to resolve septic shock. However, it is possible that the use of this simple zebrafish model, which takes the animal as a whole into account, could provide us with some conserved insight to aid in our understanding of the inflammatory/tolerance response and drug discovery.
Supplementary Material
Acknowledgments
This work was supported by grants CSD2007-00002 Aquagenomics of the Consolider-Ingenio 2010 program and AGL2011-28921-C03 IMTRA-VAC from the Spanish Ministerio de Ciencia e Innovación (MICINN) and grant 201230E057 Proyecto Intramural Especial, PIE from Agencia Estatal Consejo Superior de Investigaciones Científicas (CSIC). This work was also partially supported by grant 289202 FishForPharma from the European Union. The authors wish to thank Dr. Steve Renshaw (University of Sheffield) for the zebrafish transgenic line Tg(mpx:GFP) and Rubén Chamorro for his technical assistance.
Disclosure Statement
No competing financial interests exist.
References
- 1.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal 2001;13:85–94 [DOI] [PubMed] [Google Scholar]
- 2.Angus DC, Wax RS. Epidemiology of sepsis: an update. Crit Care Med 2001;29(Suppl 7):S109–S116 [DOI] [PubMed] [Google Scholar]
- 3.Parker LC, Jones EC, Prince LR, Dower SK, Whyte MK, Sabroe I. Endotoxin tolerance induces selective alterations in neutrophil function. J Leukoc Biol 2005;78:1301–1305 [DOI] [PubMed] [Google Scholar]
- 4.Marshall JC. Inflammation, coagulopathy, and the pathogenesis of multiple organ dysfunction syndrome. Crit Care Med 2001;29:S99–S106 [DOI] [PubMed] [Google Scholar]
- 5.Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 2002;17:693–702 [DOI] [PubMed] [Google Scholar]
- 6.Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol 2004;28:9–28 [DOI] [PubMed] [Google Scholar]
- 7.Redd MJ, Kelly G, Dunn G, Way M, Martin P. Imaging macrophage chemotaxis in vivo: studies of microtubule function in zebrafish wound inflammation. Cell Motil Cytoskeleton 2006;63:415–422 [DOI] [PubMed] [Google Scholar]
- 8.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet 2007;8:353–367 [DOI] [PubMed] [Google Scholar]
- 9.Trede NS, Ota T, Kawasaki H, Paw BH, Katz T, Demarest B, et al. Zebrafish mutants with disrupted early T-cell and thymus development identified in early pressure screen. Dev Dyn 2008;237:2575–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockhammer OW, Zakrzewska A, Hegedûs Z, Spaink HP, Meijer AH. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J Immunol 2009;182:5641–5653 [DOI] [PubMed] [Google Scholar]
- 11.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene: University of Oregon Press, 2000 [Google Scholar]
- 12.Nusslein-Volhard C, Dahm R. Zebrafish, a Practical Approach. Oxford: Oxford University Press, 2002 [Google Scholar]
- 13.Novoa B, Bowman TV, Zon L, Figueras A. LPS response and tolerance in the zebrafish (Danio rerio). Fish Shellfish Immunol 2009;26:326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophoton Int 2004;11:36–42 [Google Scholar]
- 15.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 2001;29:365–371 [DOI] [PubMed] [Google Scholar]
- 16.Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007. Available at http://bioinfogp.cnb.csic.es/tools/venny/index.html
- 17.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005;21:3674–3676 [DOI] [PubMed] [Google Scholar]
- 18.Alexeyenko A, Wassenberg DM, Lobenhofer EK, Yen J, Linney E, Sonnhammer EL, et al. Dynamic zebrafish interactome reveals transcriptional mechanisms of dioxin toxicity. PLoS One 2010;5:e10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics (Oxford, England) 2010;26:2927–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics (Oxford, England) 2005;21:3448–3449 [DOI] [PubMed] [Google Scholar]
- 22.Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, et al. ClusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 2011;12:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barsky A, Gardy JL, Hancock RE, Munzner T. Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics 2007;23:1040–1042 [DOI] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky H. Primer3 on the www for general users and for biologist programmers. Methods Mol Biol 2000;132:365–386 [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willett CE, Cortes A, Zuasti A, Zapata AG. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev Dyn 1999;214:323–336 [DOI] [PubMed] [Google Scholar]
- 27.Trede NS, Zapata A, Zon LI. Fishing for lymphoid genes. Trends Immunol 2001;22:302–307 [DOI] [PubMed] [Google Scholar]
- 28.Hasselgren PO, Menconi MJ, Fareed MU, Yang H, Wei W, Evenson A. Novel aspects on the regulation of muscle wasting in sepsis. Int J Biochem Cell Biol 2005;37:2156–2168 [DOI] [PubMed] [Google Scholar]
- 29.Hung CC, Liu X, Kwon MY, Kang YH, Chung SW, Perrella MA. Regulation of heme oxygenase-1 gene by peptidoglycan involves the interaction of Elk-1 and C/EBPalpha to increase expression. Am J Physiol Lung Cell Mol Physiol 2010;298:L870–L879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua F, Wang J, Ishrat T, Wei W, Atif F, Sayeed I, et al. Genomic profile of Toll-like receptor pathways in traumatically brain-injured mice: effect of exogenous progesterone. J Neuroinflamm 2011;8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schilling S, Zeitschel U, Hoffmann T, Heiser U, Francke M, Kehlen A, et al. Glutaminyl cyclase inhibition attenuates pyroglutamate Abeta and Alzheimer's disease-like pathology. Nat Med 2008;14:1106–1111 [DOI] [PubMed] [Google Scholar]
- 32.Cynis H, Hoffmann T, Friedrich D, Kehlen A, Gans K, Kleinschmidt M, et al. The isoenzyme of glutaminyl cyclase is an important regulator of monocyte infiltration under inflammatory conditions. EMBO Mol Med 2011;3:545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YL, Huang KF, Kuo WC, Lo YC, Lee YM, Wang AH. Inhibition of glutaminyl cyclase attenuates cell migration modulated by monocyte chemoattractant proteins. Biochem J 2012;442:403–412 [DOI] [PubMed] [Google Scholar]
- 34.Zhang G, Wang J, Kelly J, Gu G, Hou J, Zhou Y, et al. B7-H3 Augments the inflammatory response and is associated with human sepsis. J Immunol 2010;185:3677–3684 [DOI] [PubMed] [Google Scholar]
- 35.Fassbender M, Gerlitzki B, Ullrich N, Lupp C, Klein M, Radsak MP, et al. Cyclic adenosine monophosphate and IL-10 coordinately contribute to nTreg cell-mediated suppression of dendritic cell activation. Cell Immunol 2010;265:91–96 [DOI] [PubMed] [Google Scholar]
- 36.Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity 2012;37:685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen HB, Loomba M, Yang JJ, Jacobsen G, Shah K, Otero RM, et al. Early lactate clearance is associated with biomarkers of inflammation, coagulation, apoptosis, organ dysfunction and mortality in severe sepsis and septic shock. J Inflamm (Lond) 2010;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Y, Kantarci A, Badwey JA, Hasturk H, Malabanan A, Van Dyke TE. Phosphorylation of pleckstrin increases proinflammatory cytokine secretion by mononuclear phagocytes in diabetes mellitus. J Immunol 2007;179:647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grewal PK, Uchiyama S, Ditto D, Varki N, Le DT, Nizet V, et al. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Med 2008;14:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decker T. Sepsis: avoiding its deadly toll. J Clin Invest 2004;113:1387–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makowski L, Hotamisligil GS. Fatty acid binding proteins-the evolutionary crossroads of inflammatory and metabolic responses. J Nutr 2004;134:2464S–2468S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aziz M, Jacob A, Yang WL, Matsuda A, Wang P. Current trends in inflammatory and immunomodulatory mediators in sepsis. J Leukoc Biol 2013;93:329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das UN. Is sepsis a pro-resolution deficiency disorder? Med Hypotheses 2013;80:297–299 [DOI] [PubMed] [Google Scholar]
- 44.Li L, Hossain MA, Sadat S, Hager L, Liu L, Tam L, et al. Lecithin cholesterol acyltransferase null mice are protected from diet-induced obesity and insulin resistance in a gender-specific manner through multiple pathways. J Biol Chem 2011;286:17809–17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauman DR, Bitmansour AD, McDonald JG, Thompson BM, Liang G, Russell DW. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc Natl Acad Sci U S A 2009;106:16764–16769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diczfalusy U, Olofsson KE, Carlsson AM, Gong M, Golenbock DT, Rooyackers O, et al. Marked upregulation of cholesterol 25-hydroxylase expression by lipopolysaccharide. J Lipid Res 2009;50:2258–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park K, Scott AL. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J Leukoc Biol 88;1081–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou T, Garifulin O, Berland R, Boyartchuk VL. Listeria monocytogenes infection induces pro-survival metabolic signaling in macrophages. Infect Immun 2011;79:1526–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strassheim D, Park JS, Abraham E. Sepsis: current concepts in intracellular signaling. Int J Biochem Cell Biol 2002;34:1527–1533 [DOI] [PubMed] [Google Scholar]
- 50.Haskó G, Linden J, Cronstein B, Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov 2008;7:759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Csóka B, Németh ZH, Rosenberger P, Eltzschig HK, Spolarics Z, Pacher P, et al. A2B adenosine receptors protect against sepsis-induced mortality by dampening excessive inflammation. J Immunol 2010;185:542–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kronlage M, Song J, Sorokin L, Isfort K, Schwerdtle T, Leipziger J, et al. Autocrine purinergic receptor signaling is essential for macrophage chemotaxis. Sci Signal 2010;3:ra55. [DOI] [PubMed] [Google Scholar]
- 53.Aherne CM, Kewley EM, Eltzschig HK. The resurgence of A2B adenosine receptor signaling. Biochim Biophys Acta Biomembr 2011;1808:1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrero ME. Purinoceptors in inflammation: potential as anti-inflammatory therapeutic targets. Front Biosci 2011;17:2172–2186 [DOI] [PubMed] [Google Scholar]
- 55.Ramakers BP, Riksen NP, van der Hoeven JG, Smits P, Pickkers P. Modulation of innate immunity by adenosine receptor stimulation. Shock 2011;36:208–215 [DOI] [PubMed] [Google Scholar]
- 56.Dong C, Zhang X, Zhou F, Dou H, Duvernay MT, Zhang P, et al. ADP-ribosylation factors modulate the cell surface transport of G protein-coupled receptors. J Pharmacol Exp Ther 2010;333:174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novoa B, Figueras A. Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases. Adv Exp Med Biol 2012;946:253–275 [DOI] [PubMed] [Google Scholar]
- 58.van der Pouw Kraan TC, Snijders A, Boeije LC, de Groot ER, Alewijnse AE, Leurs R, et al. Histamine inhibits the production of interleukin-12 through interaction with H2 receptors. J Clin Invest 1998;102:1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caron G, Delneste Y, Roelandts E, Duez C, Herbault N, Magistrelli G, et al. Histamine induces CD86 expression and chemokine production by human immature dendritic cells. J Immunol 2001;166:6000–6006 [DOI] [PubMed] [Google Scholar]
- 60.Mazzoni A, Young HA, Spitzer JH, Visintin A, Segal DM. Histamine regulates cytokine production in maturing dendritic cells, resulting in altered T cell polarization. J Clin Invest 2001;108:1865–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morichika T, Takahashi HK, Iwagaki H, Yoshino T, Tamura R, Yokoyama M, et al. Histamine inhibits lipopolysaccharide-induced tumor necrosis factor-alpha production in an intercellular adhesion molecule-1- and B7.1-dependent manner. J Pharmacol Exp Ther 2003;304:624–633 [DOI] [PubMed] [Google Scholar]
- 62.Takahashi HK, Iwagaki H, Mori S, Yoshino T, Tanaka N, Nishibori M. Histamine inhibits lipopolysaccharide-induced interleukin (IL)-18 production in human monocytes. Clin Immunol 2004;112:30–34 [DOI] [PubMed] [Google Scholar]
- 63.Jutel M, Akdis M, Akdis CA. Histamine, histamine receptors and their role in immune pathology. Clin Exp Allergy 2009;39:1786–1800 [DOI] [PubMed] [Google Scholar]
- 64.Peng J, Yuan Q, Lin B, Panneerselvam P, Wang X, Luan XL, et al. SARM inhibits both TRIF- and MyD88-mediated AP-1 activation. Eur J Immunol 2010;40:1738–1747 [DOI] [PubMed] [Google Scholar]
- 65.Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab 2003;284:E671–E678 [DOI] [PubMed] [Google Scholar]
- 66.Jeschke MG, Klein D, Bolder U, Einspanier R. Insulin attenuates the systemic inflammatory response in endotoxemic rats. Endocrinology 2004;145:4084–4093 [DOI] [PubMed] [Google Scholar]
- 67.Russell JA. Management of sepsis. N Engl J Med 2006;355:1699–1713 [DOI] [PubMed] [Google Scholar]
- 68.Shapiro NI, Howell MD, Talmor D, Lahey D, Ngo L, Buras J, et al. Implementation and outcomes of the multiple urgent sepsis therapies (MUST) protocol. Crit Care Med 2006;34:1025–1032 [DOI] [PubMed] [Google Scholar]
- 69.Sato Y, Nishio Y, Sekine O, Kodama K, Nagai Y, Nakamura T, et al. Increased expression of CCAAT/enhancer binding protein-beta and -delta and monocyte chemoattractant protein-1 genes in aortas from hyperinsulinaemic rats. Diabetologia 2007;50:481–489 [DOI] [PubMed] [Google Scholar]
- 70.Authier F, Desbuquois B. Glucagon receptors. Cell Mol Life Sci 2008;65:1880–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martins JO, Ferracini M, Ravanelli N, Landgraf RG, Jancar S. Insulin inhibits LPS-induced signaling pathways in alveolar macrophages. Cell Physiol Biochem 2008;21:297–304 [DOI] [PubMed] [Google Scholar]
- 72.Martins JO, Ferracini M, Ravanelli N, Landgraf RG, Jancar S. Insulin suppresses LPS-induced iNOS and COX-2 expression and NF-kappaB activation in alveolar macrophages. Cell Physiol Biochem 2008;22:279–286 [DOI] [PubMed] [Google Scholar]
- 73.Spitzer AL, Harris HW. Statins attenuate sepsis. Surgery 2006;139:283–287 [DOI] [PubMed] [Google Scholar]
- 74.Schuster H. High risk/high priority: familial hypercholesterolemia—a paradigm for molecular medicine. Atheroscler Suppl 2002;2:27–30; discussion 30–32. [DOI] [PubMed] [Google Scholar]
- 75.Tominaga K, Saito S, Matsuura M, Nakano M. Lipopolysaccharide tolerance in murine peritoneal macrophages induces downregulation of the lipopolysaccharide signal transduction pathway through mitogen-activated protein kinase and nuclear factor-κ B cascades, but not lipopolysaccharide-incorporation steps. Biochim Biophys Acta 1999;1450:130–144 [DOI] [PubMed] [Google Scholar]
- 76.Medvedev AE, Kopydlowski KM, Vogel SN. Inhibition of lipopolysaccharide-induced signal transduction in endotoxin-tolerized mouse macrophages: dysregulation of cytokine, chemokine, and Toll-like receptor 2 and 4 gene expression. J Immunol 2000;164:5564–5574 [DOI] [PubMed] [Google Scholar]
- 77.Medvedev AE, Lentschat A, Wahl LM, Golenbock DT, Vogel SN. Dysregulation of LPS-induced Toll-like receptor 4-MyD88 complex formation and IL-1 receptor-associated kinase 1 activation in endotoxin-tolerant cells. J Immunol 2002;169:5209–5216 [DOI] [PubMed] [Google Scholar]
- 78.Dobrovolskaia MA, Vogel SN. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microbes Infect 2002;4:903–914 [DOI] [PubMed] [Google Scholar]
- 79.Dobrovolskaia MA, Medvedev AE, Thomas KE, Cuesta N, Toshchakov V, Ren T, et al. Induction of in vitro reprogramming by Toll-like receptor (TLR)2 and TLR4 agonists in murine macrophages: effects of TLR “homotolerance” versus “heterotolerance” on NF-κ B signaling pathway components. J Immunol 2003;170:508–519 [DOI] [PubMed] [Google Scholar]
- 80.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood 2006;108:3976–3978 [DOI] [PubMed] [Google Scholar]
- 81.Baatar D, Patel K, Taub DD. The effects of ghrelin on inflammation and the immune system. Mol Cell Endocrinol 2011;340:44–58 [DOI] [PubMed] [Google Scholar]
- 82.Foell D, Wittkowski H, Ren Z, Turton J, Pang G, Daebritz J, et al. Phagocyte-specific S100 proteins are released from affected mucosa and promote immune responses during inflammatory bowel disease. J Pathol 2008;216:83–192 [DOI] [PubMed] [Google Scholar]
- 83.Manolakis AC, Kapsoritakis AN, Georgoulias P, Tzavara C, Valotassiou V, Kapsoritaki A, et al. Moderate performance of serum S100A12, in distinguishing inflammatory bowel disease from irritable bowel syndrome. BMC Gastroenterol 2010;10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O'Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood 2010;116:1136–1146 [DOI] [PubMed] [Google Scholar]
- 85.Shen SS, Callaghan D, Juzwik C, Xiong H, Huang P, Zhang W. ABCG2 reduces ROS-mediated toxicity and inflammation: a potential role in Alzheimer's disease. J Neurochem 2010;114:1590–1604 [DOI] [PubMed] [Google Scholar]
- 86.Olson N, Hristova M, Heintz NH, Lounsbury KM, Van Der Vliet A. Activation of hypoxia-inducible factor-1 protects airway epithelium against oxidant-induced barrier dysfunction. Am J Physiol Lung Cell Mol Physiol 2011;301:L993–L1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kao KK, Fink MP. The biochemical basis for the anti-inflammatory and cytoprotective actions of ethyl pyruvate and related compounds. Biochem Pharmacol 2010;80:151–159 [DOI] [PubMed] [Google Scholar]
- 88.Villafuerte BC, Kaytor EN. An insulin-response element-binding protein that ameliorates hyperglycemia in diabetes. J Biol Chem 2005;280:20010–20020 [DOI] [PubMed] [Google Scholar]
- 89.Villafuerte BC, Barati MT, Song Y, Moore JP, Epstein PN, Portillo J. Transgenic expression of insulin-response element binding protein-1 in beta-cells reproduces type 2 diabetes. Endocrinology 2009;150:2611–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakamura MT, Nara TY. Gene regulation of mammalian desaturases. Biochem Soc Trans 2002;30:1076–1079 [DOI] [PubMed] [Google Scholar]
- 91.Peake PW, Shen Y, Campbell LV, Charlesworth JA. Human adiponectin binds to bacterial lipopolysaccharide. Biochem Biophys Res Commun 2006;341:108–115 [DOI] [PubMed] [Google Scholar]
- 92.Benoit ME, Tenner AJ. Complement protein C1q-mediated neuroprotection is correlated with regulation of neuronal gene and microRNA expression. J Neurosci 2011;31:3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chinnaiyan AM, Huber-Lang M, Kumar-Sinha C, Barrette TR, Shankar-Sinha S, Sarma VJ, et al. Molecular signatures of sepsis: multiorgan gene expression profiles of systemic inflammation. Am J Pathol 2001;159:1199–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature 2005;437:1032–1037 [DOI] [PubMed] [Google Scholar]
- 95.Pachot A, Lepape A, Vey S, Bienvenu J, Mougin B, Monneret G. Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol Lett 2006;106:63–71 [DOI] [PubMed] [Google Scholar]
- 96.Johnson SB, Lissauer M, Bochicchio GV, Moore R, Cross AS, Scalea TM. Gene expression profiles differentiate between sterile SIRS and early sepsis. Ann Surg 2007;245:611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu H, Tang Y, Ivanciu L, Centola M, Lupu C, Taylor FB Jr., et al. Temporal dynamics of gene expression in the lung in a baboon model of E. coli sepsis. BMC Genomics 2007;8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhatty M, Fan R, Muir WM, Pruett SB, Nanduri B. Transcriptomic analysis of peritoneal cells in a mouse model of sepsis: confirmatory and novel results in early and late sepsis. BMC Genomics 2012;13:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lambeck S, Weber M, Gonnert FA, Mrowka R, Bauer M. Comparison of sepsis-induced transcriptomic changes in a murine model to clinical blood samples identifies common response patterns. Front Microbiol 2012;3:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maslove DM, Tang BM, McLean AS. Identification of sepsis subtypes in critically ill adults using gene expression profiling. Crit Care 2012;16:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Starr ME, Hu Y, Stromberg AJ, Carmical JR, Wood TG, Evers BM, et al. Gene expression profile of mouse white adipose tissue during inflammatory stress: age-dependent upregulation of major procoagulant factors. Aging Cell 2013;12:194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Simons K, Ikonen E. How cells handle cholesterol. Science 2000;290:1721–1726 [DOI] [PubMed] [Google Scholar]
- 103.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 2000;1:31–39 [DOI] [PubMed] [Google Scholar]
- 104.Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. J Clin Invest 2002;110:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Laza-Knoerr AL, Gref R, Couvreur P. Cyclodextrins for drug delivery. J Drug Target 2010;18:645–656 [DOI] [PubMed] [Google Scholar]
- 106.Mura P, Corti G, Cirri M, Maestrelli F, Mennini N, Bragagni M. Development of mucoadhesive films for buccal administration of flufenamic acid: effect of cyclodextrin complexation. J Pharm Sci 2010;99:3019–3029 [DOI] [PubMed] [Google Scholar]
- 107.Danciu C, Soica C, Csanyi E, Ambrus R, Feflea S, Peev C, et al. Changes in the anti-inflammatory activity of soy isoflavonoid genistein versus genistein incorporated in two types of cyclodextrin derivatives. Chem Cent J 2012;6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.