ZNF750 controls epithelial homeostasis by inhibiting progenitor genes while inducing differentiation genes. Here, Boxer et al. characterized ZNF750 as a transcription factor that binds both the progenitor and differentiation genes that it controls at a CCNNAGGC DNA motif. ZNF750 controls differentiation in concert with RCOR1 and CTBP1/2 by acting with either KDM1A to repress progenitor genes or KLF4 to induce differentiation genes.
Keywords: stem cell, differentiation, ZNF750, KLF4, chromatin regulator
Abstract
ZNF750 controls epithelial homeostasis by inhibiting progenitor genes while inducing differentiation genes, a role underscored by pathogenic ZNF750 mutations in cancer and psoriasis. How ZNF750 accomplishes these dual gene regulatory impacts is unknown. Here, we characterized ZNF750 as a transcription factor that binds both the progenitor and differentiation genes that it controls at a CCNNAGGC DNA motif. ZNF750 interacts with the pluripotency transcription factor KLF4 and chromatin regulators RCOR1, KDM1A, and CTBP1/2 through conserved PLNLS sequences. ChIP-seq (chromatin immunoprecipitation [ChIP] followed by high-throughput sequencing) and gene depletion revealed that KLF4 colocalizes ∼10 base pairs from ZNF750 at differentiation target genes to facilitate their activation but is unnecessary for ZNF750-mediated progenitor gene repression. In contrast, KDM1A colocalizes with ZNF750 at progenitor genes and facilitates their repression but is unnecessary for ZNF750-driven differentiation. ZNF750 thus controls differentiation in concert with RCOR1 and CTBP1/2 by acting with either KDM1A to repress progenitor genes or KLF4 to induce differentiation genes.
Somatic tissue differentiation commonly requires the coordinated and simultaneous repression of progenitor genes mediating the proliferative cell program in concert with the activation of tissue-specific differentiation genes. Epidermal tissue contains a basal layer of self-renewing progenitor keratinocytes that migrate outward as they undergo cell cycle arrest and subsequently differentiate to form the epidermal permeability barrier. Epidermal differentiation involves both the repression of progenitor genes involved in cell proliferation and adhesion to the underlying basement membrane as well as the induction of epidermis-specific differentiation genes involved in barrier formation. This process is tightly regulated, and disruption of epidermal differentiation characterizes epidermal cancers and common skin disorders, such as psoriasis and chronic wounds. Recent studies have identified regulators of epidermal differentiation, including the transcription factors KLF4 (Segre et al. 1999; Patel et al. 2006), OVOL1 (Teng et al. 2007), and GRHL3 (Yu et al. 2006; Hopkin et al. 2012) and the chromatin regulators JMJD3 (Sen et al. 2008) and BRG1/BRM (Indra et al. 2005; Bao et al. 2013). Transcription factors often interact with other transcription factors and chromatin regulators to enable specific, context-dependent gene activation or repression. For example, in epidermal differentiation, the transcription factor GRHL3 interacts with the Trithorax epigenetic regulators MLL2 and WDR5 to activate epidermal differentiation genes (Hopkin et al. 2012). However, few other transcription factor complexes that regulate epidermal differentiation have been characterized. It is likely that additional transcription factors and chromatin regulators interact to activate epidermal differentiation genes and repress epidermal progenitor genes.
We recently found that ZNF750 is an essential regulator of epidermal differentiation (Sen et al. 2012). ZNF750 is highly up-regulated during epidermal differentiation by the epidermal master regulator p63, and its expression is necessary for the two types of global gene expression changes required for progenitor differentiation; namely, the induction of epidermal differentiation genes and the repression of epidermal progenitor genes (Sen et al. 2012). Ectopic expression of ZNF750 in progenitor keratinocytes induces differentiation through an evolutionarily conserved atypical C2H2 zinc finger motif (Sen et al. 2012). ZNF750-driven epidermal differentiation occurs at least partially through the induction of the transcription factor KLF4 (Sen et al. 2012). However, the mechanism by which ZNF750 regulates epidermal differentiation, including its direct target genes and the proteins it interacts with to control these genes, remains unknown.
Recent work implicates mutations in ZNF750, whose expression is largely confined to stratified epithelial tissue, in the pathogenesis of multiple human diseases characterized by disrupted epithelial homeostasis. Monogenic sebopsoriasis, a disease characterized by incomplete epidermal barrier formation and a failure to suppress the proliferative progenitor program, was found to be caused by a frameshift mutation in ZNF750 that resulted in a truncated protein prior to the C2H2 motif (Birnbaum et al. 2006). Genome-wide association studies of psoriasis, which display similar abnormalities in epidermal homeostasis, identified recurrent variants in the ZNF750 promoter region that resulted in decreased ZNF750 expression (Yang et al. 2008; Birnbaum et al. 2011). Additionally, a recent study of esophageal squamous cell carcinoma identified missense and truncating mutations in ZNF750 and decreased expression of ZNF750 in tumors compared with normal tissue, suggesting that ZNF750 may function as a tumor suppressor in this context (Lin et al. 2014). Specifically, ZNF750 loss is associated with impaired differentiation as well as a failure to fully repress the proliferative genetic program, both of which are key hallmarks of cancer (Lin et al. 2014). An understanding of the mechanism by which ZNF750 represses the progenitor gene expression program while activating differentiation may yield insight into the role of ZNF750 in both normal homeostasis and disease.
Here, we characterize ZNF750 as a DNA-binding transcription factor that binds both the progenitor and differentiation genes that it controls at a unique CCNNAGGC DNA motif. ZNF750-interacting proteins were identified by ZNF750 purification followed by mass spectrometry and were found to include the KLF4 transcription factor as well as the RCOR1 (CoREST), KDM1A (LSD1), and CTBP1 and CTBP2 (CTBP1/2) chromatin regulators. Two evolutionarily conserved PLNLS motifs in ZNF750 were identified as required for these newly identified ZNF750 protein–protein interactions. ChIP-seq (chromatin immunoprecipitation [ChIP] followed by high-throughput sequencing) and gene depletion of ZNF750-interacting proteins identified that KLF4 cobound and was required to activate ZNF750 differentiation gene targets. In contrast, KDM1A was found to cobind and help repress ZNF750-regulated progenitor genes, while RCOR1 and CTBP1/2 helped regulate both categories of ZNF750 targets. ZNF750 was required for full target gene binding by CTBP1/2, RCOR1, and KDM1A but not by KLF4, consistent with the latter’s status as a pioneer transcription factor. These data indicate that ZNF750 controls epidermal homeostasis in concert with RCOR1 and CTBP1/2 by acting with either KLF4 to induce differentiation genes or KDM1A to repress progenitor genes.
Results
ZNF750 binds both differentiation genes that it activates and progenitor genes that it represses
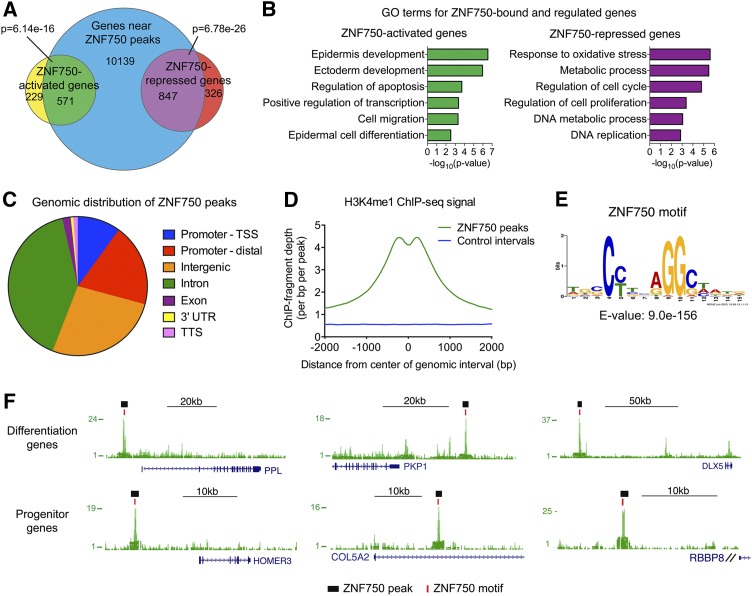
Given the conserved atypical C2H2 zinc finger motif in ZNF750 and the previously described binding of ZNF750 to the KLF4 genomic locus (Sen et al. 2012), we explored the genomic binding sites of ZNF750. We performed ChIP-seq on endogenous ZNF750 in differentiating epidermal keratinocytes, the main context in which ZNF750 is expressed (Supplemental Table S1). To search for potential direct target genes of ZNF750, we assigned ZNF750 ChIP-seq peaks to nearby genes using GREAT and assessed them with respect to ZNF750-regulated genes identified by global profiling (Sen et al. 2012). We observed a statistically significant overlap between ZNF750-binding sites in the genome with both ZNF750-activated and ZNF750-repressed genes, suggesting that ZNF750 may both directly activate and repress transcription (Fig. 1A). Genes that were ZNF750-bound and ZNF750-activated displayed gene ontology analysis enrichment for terms related to epidermal differentiation. In contrast, ZNF750-bound and ZNF750-repressed genes were enriched for terms relevant to cellular proliferation (Fig. 1B; Supplemental Fig. S1A). ZNF750 ChIP-seq peaks are located in proximal and distal regulatory regions, intergenic regions, and introns (Fig. 1C; Supplemental Fig. S1B) and are enriched with the enhancer-associated histone modification H3K4me1 (Fig. 1D; The ENCODE Project Consortium 2012). A CCNNAGGC motif was enriched in the centers of ZNF750 ChIP-seq peaks (E-value = 9 × 10−156) (Fig. 1E). This motif was found in ∼90% of ZNF750-binding peaks proximal to both the differentiation and progenitor genes (Fig. 1F; Supplemental Fig. S1C). ZNF750 therefore binds both the epidermal differentiation genes that it activates as well as epidermal progenitor genes that it represses.
Figure 1.
ZNF750 genomic binding. (A) Overlap of genes associated with ZNF750 ChIP-seq peaks by GREAT with ZNF750-activated and ZNF750-repressed genes. P-values determined by Fisher’s exact test. (B) Gene ontology (GO) terms for putative direct ZNF750-activated or ZNF750-repressed genes. (C) Distribution of ZNF750 peaks across the genome. (TSS) Transcription start site; (TTS) transcription termination site; (Promoter-TSS) +1 to −5 kb of TSS and 5′ untranslated region (UTR); (Promoter-distal) −5 to −100 kb of TSS. (D) Histogram of keratinocyte H3K4me1 ChIP-seq fragment depth (The ENCODE Project Consortium 2012) in ZNF750 peaks compared with size-matched control genomic intervals. (E) ZNF750 sequence-specific DNA motif identified in ChIP-seq peaks. (F) Representative ZNF750 ChIP-seq peaks near differentiation or progenitor genes. Black bars denote MACS-called peaks, and red bars denote ZNF750 motifs. See also Supplemental Figure S1 and Supplemental Table S1.
Identification of functional ZNF750-interacting proteins
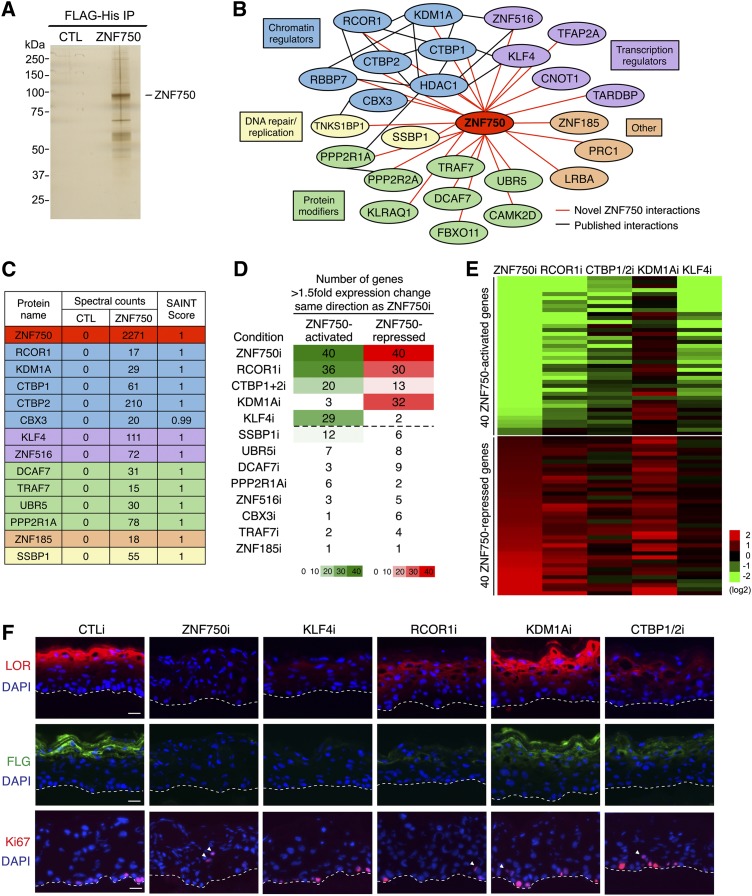
To understand the basis for the contrasting activating and repressive gene regulatory impacts of ZNF750 in epidermal differentiation, we sought to identify the nuclear proteins that interact with ZNF750. Functionally active, C-terminally tagged ZNF750, with dual sequences added for tandem affinity purification via the Flag epitope and hexahistidine (HIS) tags, was purified from differentiating keratinocytes along with a Flag-HIS tag-only control (Fig. 2A) and then subjected to mass spectrometry. Proteins with no spectral counts in the tag-only control, ≥10 spectral counts with purified ZNF750, a SAINT score of ≥0.9 (Choi et al. 2011), and absence in common background data sets (Trinkle-Mulcahy et al. 2008; Mellacheruvu et al. 2013) were used to generate a network of novel ZNF750-interacting proteins (Fig. 2B). Functional categorization revealed that the majority of ZNF750-interacting proteins are transcription factors, chromatin regulators, and protein modifiers (Fig. 2B). Interestingly, a number of ZNF750-interacting proteins have been previously observed to interact with each other, suggesting that ZNF750 might interface with several known protein complexes (Fig. 2B). In particular, several chromatin regulators were identified that are components of chromatin regulator complexes, including KDM1A, RCOR1, and CTBP1/2 (You et al. 2001; Shi et al. 2003). These chromatin regulators have been previously observed to interact with a number of other transcription factors, some of which were also among ZNF750-interacting proteins, including ZNF516 (Lee et al. 2005) and KLF4 (Liu et al. 2009).
Figure 2.
Identification of functional ZNF750-interacting proteins. (A) Silver stain gel of Flag-HIS ZNF750 or tag-only control (CTL) tandem affinity-purified from differentiated keratinocytes. (B) Network of ZNF750-interacting proteins identified by mass spectrometry, grouped by functional category. Red lines indicate novel ZNF750 interactions, and black lines indicate published interactions. (C) Table of spectral counts and SAINT scores for 13 ZNF750-interacting proteins tested for phenotypic overlap with ZNF750 depletion. (D) Heat map of gene expression changes with depletion of ZNF750-interacting proteins by siRNA in keratinocytes induced to differentiate for 3 d with calcium, shown as number of genes with >1.5-fold expression change by qPCR in the same direction as ZNF750 depletion. Proteins above the dotted line were considered for further analysis. (E) Heat map of gene expression changes with depletion of ZNF750, RCOR1, CTBP1/2, KDM1A, or KLF4 in differentiated keratinocytes by qPCR. (F) Effects of depletion of ZNF750-interacting proteins on differentiation proteins loricrin (LOR; red) and filaggrin (FLG; green) and proliferation (Ki67; red) in regenerated organotypic epidermal tissue. Epidermal basement membrane is noted with dashed lines, and white arrowheads denote suprabasal Ki67-positive cells. Bar, 25μm. See also Supplemental Figure S2.
We next determined which ZNF750-interacting proteins are important for ZNF750 function in epidermal differentiation. Thirteen top ZNF750-interacting proteins were selected based on the number of spectral counts, SAINT score, known interactions with other ZNF750-interacting proteins, and known biological importance (Fig. 2C). To determine whether these proteins are important for ZNF750 function, we depleted each protein in keratinocytes undergoing calcium-induced differentiation for 3 d in vitro and assayed the expression of a panel of 40 known ZNF750-activated genes and 40 known ZNF750-repressed genes (Sen et al. 2012) by quantitative PCR (qPCR) (Fig. 2D; Supplemental Fig. S2A,B). Among the 13 proteins tested, depletion of RCOR1, CTBP1/2, KDM1A, and KLF4 showed the strongest phenotypic overlap with ZNF750 depletion, with each regulating the expression of between 38% and 82.5% of analyzed ZNF750 targets (Fig. 2D,E). Interestingly, KLF4 depletion selectively impaired the induction of ZNF750-activated genes, while KDM1A loss selectively impaired the inhibition of ZNF750-repressed genes. RCOR1 and CTBP1/2 depletion affected the differentiation-mediated regulation of both categories of ZNF750 target genes (Fig. 2D,E). To confirm the functional importance of ZNF750-interacting proteins in three-dimensional tissue, we depleted ZNF750-interacting proteins in regenerated organotypic epidermis. Depletion of ZNF750, KLF4, RCOR1, and CTBP1/2, but not KDM1A, resulted in a decrease in expression of terminal differentiation proteins loricrin (LOR) and filaggrin (FLG) (Fig. 2F). Depletion of ZNF750, RCOR1, KDM1A, and CTBP1/2 caused an increase in the percentage of proliferative cells in the normally differentiated suprabasal layer (Fig. 2F; Supplemental Fig. S2C). Functional ZNF750-interacting proteins thus include KLF4, which is required for differentiation gene activation, and KDM1A, which is required for progenitor gene repression, as well as RCOR1 and CTBP1/2, which are required for both effects.
ZNF750 interacts directly with KLF4, RCOR1, KDM1A, and CTBP1/2
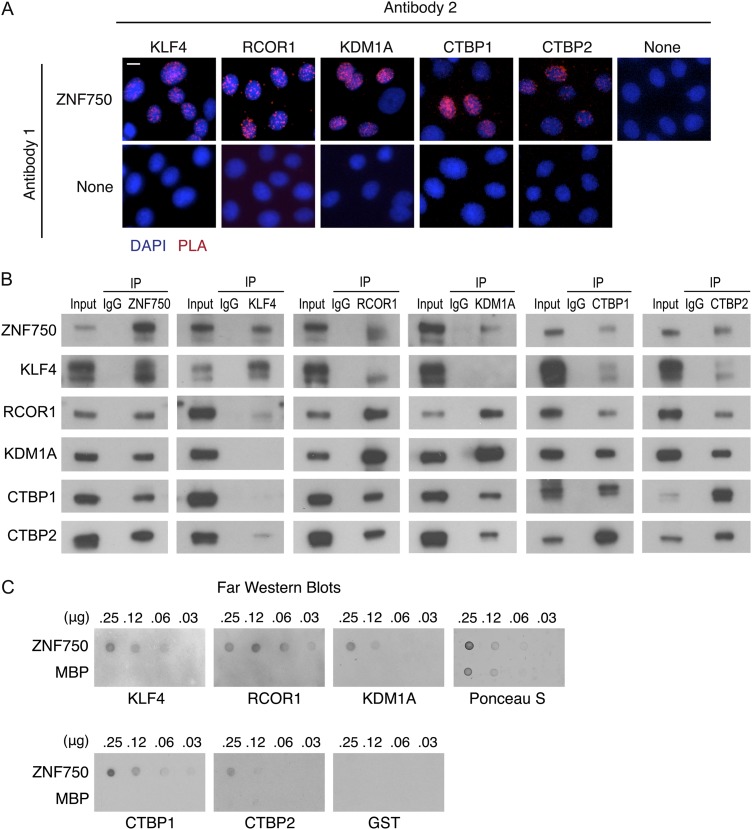
The functional impacts on ZNF750 target genes by the proteins copurifying with ZNF750 stimulated the assessment of the nature of these interactions. First, to confirm that ZNF750 is in close proximity with KLF4, RCOR1, KDM1A, and CTBP1/2 in differentiating keratinocytes, proximity ligation analysis (PLA), a method for visualizing individual protein interactions in cells, was performed. Nuclear PLA signal was observed between ZNF750 and KLF4, RCOR1, KDM1A, and CTBP1/2 but not with the single-antibody controls (Fig. 3A), consistent with proximity of each of these proteins to ZNF750 within the nucleus. To further confirm the interactions among ZNF750, KLF4, RCOR1, KDM1A, and CTBP1/2, two-way coimmunoprecipitations (co-IPs) were performed among the six endogenous proteins in differentiating keratinocytes. These two-way co-IPs confirmed the interactions of ZNF750 with KLF4, RCOR1, KDM1A, and CTBP1/2 (Fig. 3B; Supplemental Fig. S3A). Interactions among RCOR1, KDM1A, CTBP1, and CTBP2 were also observed (Fig. 3B), as previously described in other cell types (You et al. 2001; Shi et al. 2003; Lee et al. 2005). Interestingly, KLF4 interacted with RCOR1, CTBP1, and CTBP2 but not with KDM1A (Fig. 3B), suggesting that KLF4 and KDM1A may be part of different ZNF750-interacting protein complexes. To determine whether these proteins interact directly with ZNF750, we performed far-Western blotting with purified recombinant proteins. ZNF750, but not the MBP-negative control, bound directly to KLF4, RCOR1, KDM1A, CTBP1/2 but not to a GST-negative control (Fig. 3C). Thus, ZNF750 interacts directly with KLF4, RCOR1, KDM1A, and CTBP1/2.
Figure 3.
ZNF750 interacts with KLF4, RCOR1, KDM1A, CTBP1, and CTBP2. (A) PLA in keratinocytes with antibody recognizing ZNF750 or no antibody control in combination with KLF4, RCOR1, KDM1A, CTBP1, or CTBP2 antibodies. Bar, 5 μm. (B) Western blots of co-IPs among ZNF750, KLF4, RCOR1, KDM1A, CTBP1, and CTBP2. Inputs are 1%. (C) Far-Western analysis with recombinant ZNF750 or MBP-negative control on membrane, probed with recombinant proteins and antibodies to KLF4, RCOR1, KDM1A, CTBP1, and CTBP2 or GST as negative control. Ponceau S was used as a loading control. See also Supplemental Figure S3.
Given the distinct gene regulatory outcomes of KLF4 and KDM1A depletion and the binding of KLF4 and KDM1A to ZNF750 but not to each other, we explored whether KLF4 and KDM1A compete for binding to ZNF750. We performed ZNF750 immunoprecipitations with KLF4 or KDM1A depletion and found that KDM1A binding to ZNF750 increased over twofold with depletion of KLF4, and KLF4 binding to ZNF750 increased with depletion of KDM1A (Supplemental Fig. S3B). Far-Western blot analysis indicated that KDM1A binding to ZNF750 decreased with addition of KLF4 but not an MBP-negative control and that KLF4 binding to ZNF750 decreased slightly with addition of KDM1A as well. (Supplemental Fig. S3C). This suggests that KLF4 and KDM1A each bind independently to ZNF750 and that they may compete with each other for binding to ZNF750.
Conserved ZNF750 C2H2 and PLNLS motifs in DNA-binding and protein interactions
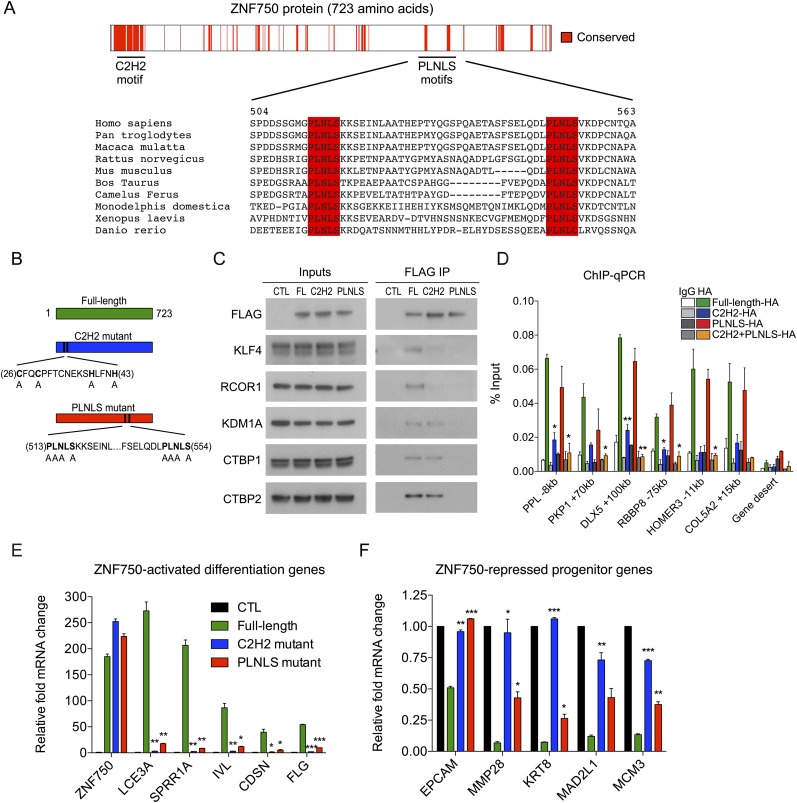
ZNF750 appeared during vertebrate evolution with the emergence of stratified epithelium. To discover the portions of ZNF750 that facilitate its function, including its DNA binding and its protein–protein interactions, we analyzed the evolutionary sequence conservation of the ZNF750 protein. In addition to the previously identified, evolutionarily conserved atypical C2H2 zinc finger motif that is required for ZNF750 function (Sen et al. 2012), we observed two highly conserved PLNLS sequences in the ZNF750 C terminus (Fig. 4A). To explore the role of the C2H2 and PLNLS motifs in ZNF750 protein–protein interactions, we mutagenized the C2H2 and PLNLS motifs by alanine substitution (Fig. 4B) and performed co-IPs with full-length, C2H2 mutant, or PLNLS mutant ZNF750 from differentiated keratinocytes. Disruption of the PLNLS motif abolished the interactions of ZNF750 with KLF4, RCOR1, KDM1A, and CTBP1/2 (Fig. 4C; Supplemental Fig. S4A). Mutagenesis of the C2H2 motif partially reduced ZNF750 interactions with KLF4 and RCOR1 but not with KDM1A or CTBP1/2 (Fig. 4C). ChIP-qPCR with the ZNF750 mutants revealed a significant decrease in DNA binding with mutagenesis of the C2H2 motif (Fig. 4D). Disruption of the PLNLS motif also partially decreased DNA binding, and mutation of both the C2H2 and PLNLS motifs resulted in a further decrease in DNA binding than the C2H2 mutant alone. (Fig. 4D; Supplemental Fig. S4B). The PLNLS motifs are thus required for ZNF750 binding to KLF4, RCOR1, KDM1A, and CTBP1/2, and the C2H2 motif is partially required for ZNF750 binding to KLF4 and RCOR1, while both motifs are required for full ZNF750 binding to its target genes.
Figure 4.
Domains of ZNF750 required for protein–protein interactions and DNA binding. (A) Evolutionarily conserved C2H2 and PLNLS motifs in ZNF750 protein. (B) Diagram of mutations made in C2H2 and PLNLS motifs. (C) Inputs and Flag immunoprecipitation (IP) of tag-only control (CTL), full-length (FL), C2H2 mutant, or PLNLS mutant ZNF750 from keratinocytes, with Western blot for Flag, KLF4, RCOR1, KDM1A, CTBP1, or CTBP2. (D) ChIP-qPCR at ZNF750 target genes with HA-tagged full-length, C2H2 mutant, PLNLS mutant, or C2H2 PLNLS double-mutant ZNF750, compared with IgG controls. t-test: (*) P < 0.05; (**) P < 0.01, compared with full-length. Differentiation gene (E) or progenitor gene (F) expression changes with ectopic expression of full-length, C2H2 mutant, or PLNLS mutant ZNF750 in undifferentiated keratinocytes compared with tag-only control (CTL). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, compared with full-length) See also Supplemental Figure S4.
The observed disruption of ZNF750 binding to its interacting proteins as well as to its genomic targets suggested that both the C2H2 and PLNLS motifs would be important functionally in ZNF750 target gene regulation. To examine this possibility, we expressed full-length, C2H2 mutant, or PLNLS mutant ZNF750 in undifferentiated keratinocytes, where ZNF750 is not normally expressed. While full-length ZNF750 induced differentiation gene targets and repressed progenitor target gene expression as expected, both the C2H2 and PLNLS mutants failed to fully achieve this (Fig. 4E,F; Supplemental Fig. S4C). The effects of mutation of C2H2 and PLNLS motifs were also confirmed in regenerated organotypic epidermal tissue, where full-length ZNF750 induced ectopic late differentiation protein expression into the lower epidermis, while the C2H2 and PLNLS mutants did not (Supplemental Fig. S4D). Mutation of the C2H2 and PLNLS motifs also rescued the impaired clonogenic growth induced by full-length ZNF750 expression (Supplemental Fig. S4E). Thus, both the C2H2 and PLNLS motifs of ZNF750 are functionally required for ZNF750’s dual activating and repressive gene regulatory impacts during epidermal differentiation.
Genomic colocalization of ZNF750, KLF4, RCOR1, and KDM1A at ZNF750 target genes
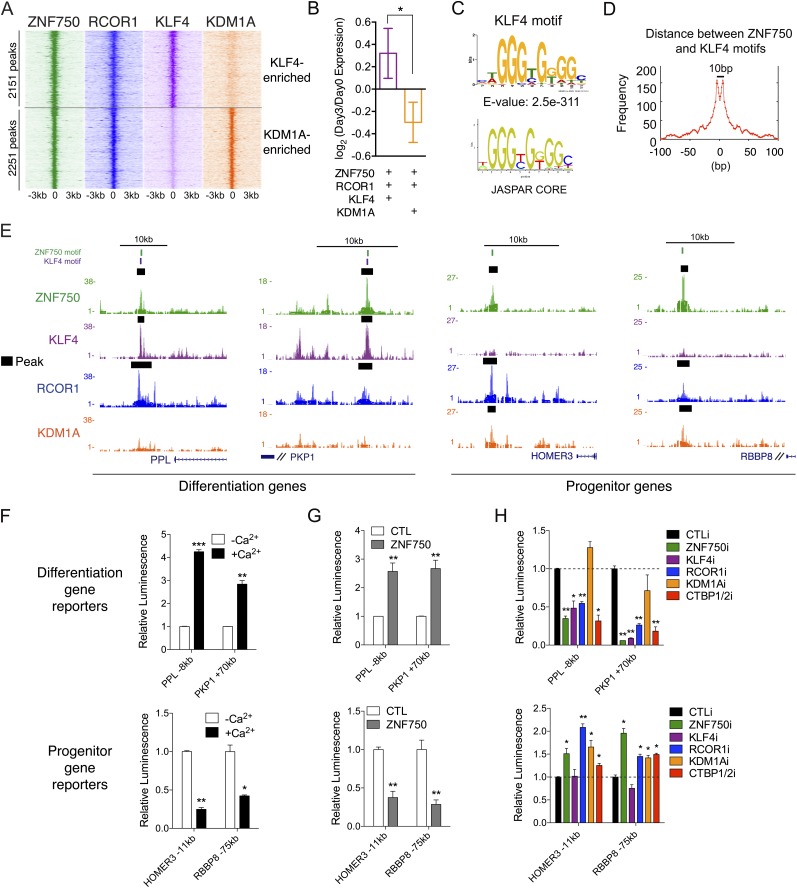
We next explored whether ZNF750 colocalizes with its interacting proteins with respect to genomic binding. ChIP-seq was performed in differentiating epidermal cells for ZNF750-interacting proteins known to directly associate with chromatin; namely, KLF4, RCOR1, and KDM1A. Analyses of these data revealed a striking overlap in regions of genomic binding between ZNF750 and KLF4, RCOR1, and KDM1A (Fig. 5A; Supplemental Tables S2–S4). Of interest, the gene set displaying KLF4 and RCOR1 cobinding with ZNF750 in the setting of relatively less KDM1A binding was enriched for ZNF750-induced differentiation genes. In contrast, the gene subset characterized by KDM1A and RCOR1 cobinding with ZNF750 in the context of relatively diminished KLF4 binding was enriched for ZNF750-repressed progenitor genes (Fig. 5A,B; Supplemental Fig. S5A; Supplemental Tables S5, S6; Kretz et al. 2012; Sen et al. 2012). De novo motif discovery identified the canonical KLF4 DNA motif (Chen et al. 2008) in KLF4 ChIP-seq peaks, supporting the accuracy of KLF4 ChIP-seq data in this setting (Fig. 5C). At sites of colocalized genomic binding, ZNF750 and KLF4 motifs are frequently 10 base pairs (bp) apart, consistent with a close localization at potential genomic sites of gene coregulation by these two physically interacting transcription factors (Fig. 5D). Examples of ChIP-seq tracks at two differentiation genes, PPL and PKP1, demonstrate colocalization of ZNF750, KLF4, and RCOR1 at ZNF750 and KLF4 motifs (Fig. 5E). ChIP-seq tracks near two progenitor genes, HOMER3 and RBBP8, show colocalization of ZNF750, RCOR1, and KDM1A at the ZNF750 motif (Fig. 5E). This is consistent with a ZNF750 differentiation gene-activating functionality involving KLF4 and a distinct ZNF750 progenitor gene-repressive functionality involving KDM1A.
Figure 5.
ZNF750 activates or represses transcription with different interacting proteins. (A) Heat map of combined ZNF750, KLF4, RCOR1, and KDM1A ChIP-seq peaks, selected for KLF4 or KDM1A enrichment in ZNF750- and RCOR1-binding regions. Heat map shows ChIP-seq signal over 6-kb regions around peak center. (B) Log2 fold change during keratinocyte differentiation of genes near the top 500 ZNF750, KLF4, and RCOR1 peaks or the ZNF750, RCOR1, and KDM1A peaks. (*) P < 0.05. (C) Sequence-specific motif identified in KLF4 ChIP-seq peaks, compared with canonical KLF4 motif. (D) Frequency of distances between pairs of ZNF750 and KLF4 motifs in ChIP-seq peaks. (E) Example ZNF750, KLF4, RCOR1, and KDM1A ChIP-seq tracks near two differentiation genes (left) and two progenitor genes (right), showing locations of motifs for ZNF750 (green) and KLF4 (purple). (Black bars) MACS-called peaks. (F) Change in luciferase activity of reporters near differentiation genes (top) or progenitor genes (bottom) during calcium-induced keratinocyte differentiation. t-test: (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, compared with –Ca2+. (G) Change in luciferase activity of each reporter with ectopic ZNF750 expression in progenitor keratinocytes compared with empty vector control (CTL). (**) P < 0.01, compared with CTL. (H) Luciferase activity of reporters with control knockdown (CTLi) compared with depletion of ZNF750, KLF4, RCOR1, KDM1A, or CTBP1/2. (*) P < 0.05; (**) P < 0.01, compared with CTLi. See also Supplemental Figure S5 and Supplemental Tables S2–S6.
ZNF750 activates and represses genes with differing combinations of interacting proteins
We next used reporter assays to test whether ZNF750 and its interacting proteins are involved in gene activation or repression at specific sites. Reporters at ZNF750-binding sites near differentiation genes showed increased reporter activity during calcium-induced keratinocyte differentiation, and reporters at ZNF750-binding sites near progenitor genes showed decreased reporter activity during differentiation, consistent with the expression changes of these genes during differentiation (Fig. 5F). Additionally, ectopic expression of ZNF750 in progenitor keratinocytes resulted in increased activity of differentiation gene reporters and decreased activity of progenitor gene reporters, confirming that ZNF750 can either activate or repress transcription, depending on the context (Fig. 5G).
To assess the role of ZNF750-interacting proteins in transcriptional regulation of ZNF750-dependent reporters, we depleted the expression of ZNF750, KLF4, RCOR1, KDM1A, or CTBP1/2 in differentiated keratinocytes and assessed the changes in activity of each reporter. Depletion of ZNF750, KLF4, RCOR1, or CTBP1/2, but not KDM1A, resulted in decreased activity of the differentiation gene reporters, whereas depletion of ZNF750, RCOR1, KDM1A, or CTBP1/2, but not KLF4, resulted in increased activity of progenitor gene reporters (Fig. 5H). Additionally, mutation of the ZNF750 and KLF4 motifs in the differentiation gene reporters resulted in decreased activity, while mutation of the ZNF750 motif in progenitor gene reporters resulted in increased activity (Supplemental Fig. S5B,C), confirming that ZNF750-dependent gene activation or repression occurs directly through these two motifs. To further explore the chromatin context of ZNF750-dependent gene activation or repression, we performed ChIP-qPCR for the active histone modifications H3K27ac and H3K4me1 at ZNF750-binding sites with depletion of ZNF750 and interacting proteins. Depletion of ZNF750, KLF4, RCOR1, or CTBP1/2, but not KDM1A, resulted in a significant decrease in H3K27ac at differentiation genes (Supplemental Fig. S5D). Depletion of ZNF750, RCOR1, KDM1A, or CTBP1/2 resulted in an increase in H3K4me1 at progenitor genes (Supplemental Fig. S5D), consistent with gene repression mediated by KDM1A H3K4me1/2 demethylase activity. Thus, ZNF750-driven differentiation gene activation requires KLF4, RCOR1, and CTBP1/2, and ZNF750-mediated progenitor gene repression requires RCOR1, KDM1A, and CTBP1/2.
ZNF750 recruits chromatin regulators to specific sites
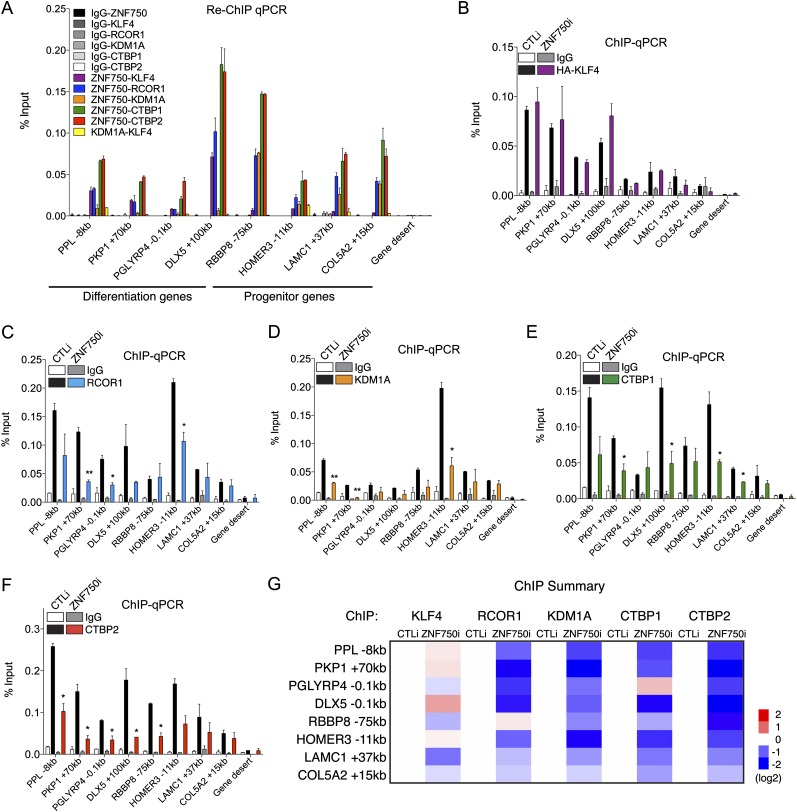
To confirm that ZNF750 binds with different sets of proteins to specific sites and to explore whether CTBP1/2 are also present at these sites, we performed re-ChIP-qPCR with ZNF750 and each interacting protein (Fig. 6A). Consistent with the ChIP-seq signal at these specific sites, ZNF750 re-ChIP with KLF4 gave enriched signal at differentiation genes, and ZNF750 re-ChIP with KDM1A gave enriched signal at progenitor genes (Fig. 6A). KLF4 re-ChIP with KDM1A did not give enriched signal at any of the sites tested (Fig. 6A). ZNF750 re-ChIP with RCOR1 and CTBP1/2 gave enriched signal at sites near both differentiation and progenitor genes, confirming that these proteins reside at both ZNF750-activating and ZNF750-repressing genomic contexts (Fig. 6A).
Figure 6.
ZNF750 facilitates chromatin regulator association with target genes. (A) Re-ChIP-qPCR with ZNF750 and each interacting protein to sites near four differentiation genes and four progenitor genes compared with gene desert-negative control. (B–F) ChIP-qPCR with control or ZNF750 depletion for KLF4 (B), RCOR1 (C), KDM1A (D), CTBP1 (E), or CTBP2 (F). t-test: (*) P < 0.05; (**) P < 0.01, compared with CTLi. (G) Heat map summarizing ChIP-qPCR data from B–F. See also Supplemental Figure S6.
While ZNF750 expression is highly up-regulated during epidermal differentiation, the levels of ZNF750-interacting proteins are much more stable during epidermal differentiation (Supplemental Fig. S6A,B; Kretz et al. 2012), suggesting that ZNF750 might recruit these proteins to specific sites on DNA upon induction of expression. To investigate whether ZNF750-interacting proteins require ZNF750 to bind specific sites on DNA, we performed ChIP-qPCR on each ZNF750-interacting protein in the context of ZNF750 depletion in differentiated keratinocytes. KLF4 genomic binding was not reduced with ZNF750 depletion, consistent with KLF4 binding to DNA independently of ZNF750 through its own DNA motif (Fig. 6B) and with the known role of KLF4 as a pioneer transcription factor that can directly bind closed chromatin (Soufi et al. 2012). However, ZNF750 depletion decreased target gene binding by RCOR1, KDM1A, and CTBP1/2 at most of the sites tested (Fig. 6C–G) despite the fact that these proteins were expressed at the same levels with control or ZNF750 depletion (Supplemental Fig. S6C). Thus, ZNF750 is dispensable for the association of KLF4 with its target genes but is required for full target gene binding by the chromatin regulators RCOR1, KDM1A, and CTBP1/2.
Discussion
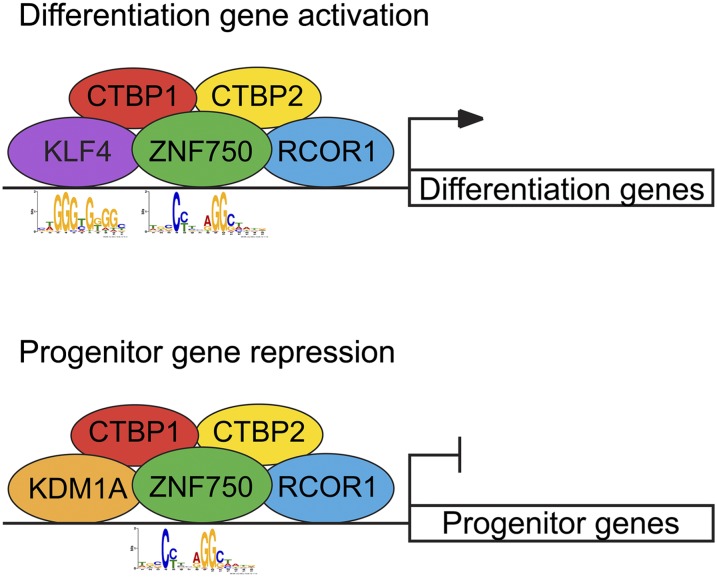
Here, we found that ZNF750 acts with RCOR1 and CTBP1/2 to exert dual gene regulatory impacts; namely, by activating differentiation gene expression in concert with KLF4 and repressing progenitor gene expression in association with KDM1A (Fig. 7). In doing so, ZNF750 functions as a DNA sequence-specific transcription factor essential for the switch from the self-renewal genetic program to the terminal differentiation program in epidermis. Two evolutionarily conserved PLNLS motifs facilitate ZNF750 protein–protein interactions and function in this context. These PLNLS motifs, together with the atypical C2H2 zinc finger motif, facilitate ZNF750 target gene binding at sites containing the CCNNAGGC DNA motif. This supports a model (Fig. 7) in which ZNF750 engages progenitor gene repression and differentiation gene activation via association with these pre-existing proteins, which require the presence of ZNF750 to exert their full gene regulatory effects. In this regard, although KLF4 target gene binding occurs independently of ZNF750, in spite of the close proximity of the two proteins and their motifs on DNA, KLF4 requires ZNF750 for activation of their shared differentiation gene targets. In contrast, the chromatin regulators identified here all depend on ZNF750 for full target gene binding as well as target gene regulation, indicating that ZNF750 can regulate transcription by both synergizing with factors already present on DNA and enhancing the recruitment of additional factors.
Figure 7.
Model of dual ZNF750 gene regulatory impacts in epidermal differentiation. ZNF750 physically interacts with KLF4, RCOR1, and CTBP1/2 to bind and activate epidermal differentiation genes. ZNF750 binds in close proximity to KLF4 DNA binding at differentiation genes. At progenitor genes, ZNF750 interacts with KDM1A, RCOR1, and CTBP1/2 to repress gene expression.
The massive up-regulation of ZNF750 during epidermal differentiation contrasts with the relatively stable protein levels of its functionally active interacting partners. Taken together with the tight restriction of ZNF750 expression to differentiating stratified epithelial cells, these observations suggest that ZNF750 acts dominantly to mobilize its presynthesized interacting proteins to effect the switch from the epidermal progenitor to differentiating cell state. Consistent with this is our recent observation that ZNF750 expression alone, without any additional factors, is sufficient to engage terminal differentiation gene induction in epidermal progenitors both in vitro and in tissue (Sen et al. 2012). ZNF750 thus may represent a tissue-specific transcription factor that can engage with both more globally active transcription factors, such as KLF4, and general chromatin regulators, such as KDM1A, RCOR1, and CTBP1/2, as observed here, to achieve a tissue-specifying gene expression output.
ZNF750’s function as either a transcriptional activator of differentiation genes or a repressor of progenitor genes depends on its interacting proteins and genomic context. Other transcription factors have also been shown to either activate or repress transcription, depending on context. For example, the transcription factor p53 activates or represses transcription depending on its interactions with different sets of chromatin modifiers or transcription factors that act as coactivators or corepressors as well as variations in the specific DNA sequence to which it binds (for review, see Rinn and Huarte 2011). Here, we found that the main defining factor for ZNF750-mediated transcriptional activation is colocalization with KLF4 near epidermal differentiation genes. This is consistent with the observation that the primary phenotype of KLF4 knockout mice is impaired epidermal differentiation (Segre et al. 1999). This is also consistent with the observed capacity of KLF4 to promote epidermal differentiation gene expression (Patel et al. 2006) and with its function as a transcriptional activator through its interactions with the coactivators p300 and CBP (Geiman et al. 2000; Evans et al. 2007). The main defining factor for ZNF750-mediated transcriptional repression, in contrast, is colocalization with KDM1A near progenitor genes. This is consistent with the known role of KDM1A in transcriptional repression through its H3K4me1/2 demethylase activity (Shi et al. 2004) and the observed increase in H3K4me1 with KDM1A depletion at ZNF750-binding sites. Of interest, in spite of their mutual association with ZNF750, we observed that KLF4 and KDM1A do not themselves associate with each other by co-IP and appear to compete for binding to ZNF750, consistent with their action with ZNF750 in separate contexts.
Interestingly, we found that RCOR1 and CTBP1/2 are part of both the ZNF750-activating and ZNF750-repressing complexes. RCOR1 and CTBP1/2 have known roles in transcriptional repression in a complex with KDM1A (Shi et al. 2003; Lee et al. 2005). Roles for RCOR1 and CTBP1/2 in transcriptional activation have also been described, but the mechanism is not well understood (Fang et al. 2006; Jin et al. 2007; Abrajano et al. 2010; Paliwal et al. 2012). Transcriptional activation or repression by RCOR1 and CTBP1/2 likely depends on the specific transcription factors and chromatin regulators with which they are associated, as shown here. We found that ZNF750 is required for full association of RCOR1, KDM1A, and CTBP1/2 with specific genomic sites, consistent with the previously described recruitment of these general chromatin regulators to specific sites by other transcription factors (Lin et al. 2010; Paliwal et al. 2012). It is possible that additional ZNF750-interacting proteins identified by mass spectrometry may also be involved in distinguishing the ZNF750-activating and ZNF750-repressing complexes.
KLF4 is important for a wide variety of processes in different tissues. In addition to being essential for epidermal differentiation and barrier formation (Segre et al. 1999), KLF4 is important for cell differentiation in a number of other cell types, including monocytes (Feinberg et al. 2007), vascular smooth muscle cells (Li et al. 2010), and goblet cells in the colon (Katz et al. 2002). KLF4 is also one of the four factors that can induce pluripotent stem cells (Takahashi and Yamanaka 2006) and plays a role in embryonic stem cell self-renewal (Jiang et al. 2008). It is currently unclear how KLF4 is capable of regulating such diverse processes, but one possibility is that KLF4 interacts with different tissue-specific factors in different contexts that specify KLF4 functions. It is possible that the interaction of KLF4 with ZNF750, which is expressed exclusively in stratified epithelial tissues, specifies KLF4 as an epidermal differentiation factor in that context. Interestingly, we previously identified KLF4 as a direct target gene of ZNF750 (Sen et al. 2012). In this study, ChIP-seq analysis revealed that both ZNF750 and KLF4 bind to several locations in the KLF4 and ZNF750 genomic loci (Supplemental Tables S1, S2), consistent with a differentiation-driving autoregulatory positive feedback loop. Autoregulatory positive feedback loops have been described as a feature of master tissue-specific transcription factors, such as OCT4, SOX2, and NANOG in embryonic stem cells (Boyer et al. 2005). The ZNF750–KLF4 positive feedback loop likely serves as a mechanism for the rapid induction and stable maintenance of gene expression during epidermal differentiation.
We identify two highly conserved PLNLS sequences in ZNF750 that are required for ZNF750 protein–protein interactions with KLF4, RCOR1, KDM1A, and CTBP1/2. The PLNLS sequence and other similar sequences are found in several other transcription factors, including ZEB1 (Postigo and Dean 1999), ZNF217 (Quinlan et al. 2006), and KLF4 (Liu et al. 2009), where this sequence facilitates protein–protein interactions with CTBP1/2. The finding that ZNF750 protein–protein interactions are all dependent on the two PLNLS sequences is consistent with KLF4 and KDM1A binding independently to and competing for binding to ZNF750. This competition is likely involved in distinguishing the ZNF750-activating and ZNF750-repressing complexes. A recent study identified several mutations in ZNF750 in esophageal squamous cell carcinoma and suggested a role for ZNF750 as a tumor suppressor (Lin et al. 2014). This work identified several missense mutations within the C2H2 motif, which, as we show, would disrupt ZNF750 DNA binding and function. Interestingly, most of the identified mutations are truncating mutations that occur after the C2H2 motif but before the PLNLS motif. These truncated mutant forms of ZNF750 would be unable to engage in protein–protein interactions and unable to induce differentiation and suppress proliferation. Thus, the ZNF750 protein–protein interactions and genomic binding sites identified here offer insight into both normal epidermal tissue differentiation and the suppression of epithelial tumorigenesis.
Materials and methods
Lentiviral constructs, siRNA sequences, and primer sequences are in the Supplemental Material.
Cell culture
Primary human keratinocytes were isolated from fresh, surgically discarded neonatal foreskin. Keratinocytes were grown in KCSFM and Medium 154 (Life Technologies). Keratinocytes were induced to differentiate by addition of 1.2 mM calcium for 3 d in full confluence. 293T cells were grown in DMEM with 10% fetal bovine serum. For regenerated organotypic epidermal tissue, 1 million genetically modified keratinocytes were seeded onto devitalized human dermis for 4 d and then harvested for RNA or protein expression.
Gene transfer and knockdown
293T cells were transfected with 8 μg of lentiviral expression construct, 6 μg of pCMVΔ8.91, and 2 μg of pUC MD.G. Transfections were done in 10-cm plates using Lipofectamine 2000 (Life Technologies). Viral supernatant was collected 72 h after transfection and concentrated using a Lenti-X concentrator (Clontech). Keratinocytes were transduced overnight with virus and 5 μg/mL polybrene and selected with 1 μg/mL puromycin. For knockdown with siRNA, 1 million keratinocytes were electroporated with 1 nmol of siRNA using Amaxa nucleofection reagents.
qRT–PCR expression analysis
Total RNA was extracted using the RNeasy plus kit (Qiagen). One microgram of RNA was reverse-transcribed using iSCRIPT cDNA synthesis kit (Bio-Rad). qPCR was performed using the Roche480 LightCycler with Maxima SYBR Green master mix (Fermentas). Samples were run in duplicate and normalized to 18S RNA.
Cell and nucleus lysis for immunoprecipitation
Keratinocytes were harvested by trypsinization, and subsequent steps were performed at 4°C. Cells were washed in PBS and resuspended in five cell pellet volumes of hypotonic buffer (10 mM HEPES at pH 7.6, 1.5 mM MgCl2, 10 mM KCl, 1× protease inhibitor cocktail [PIC] [Roche]). Cells were lysed by addition of an equal volume of hypotonic buffer with 0.4% NP-40 for 2 min. Nuclei were pelleted by centrifugation at 4000 rpm and lysed in three cell pellet volumes of nucleus lysis buffer (50 mM Tris at pH 7.6, 0.05% igepal, 10% glycerol, 2 mM MgCl2, 250 mM NaCl, PIC). Nuclei were sheared with a 27.5-gauge needle, and lysis proceeded for 30 min. Insoluble material was removed by centrifugation at 13,000 rpm for 10 min, and nuclear supernatant was used for immunoprecipitation.
Flag-HIS purification and mass spectrometry
All steps were performed at 4°C unless otherwise indicated. Flag M2 beads (Sigma) were added to 15 mg of nuclear supernatant for 16 h. Flag beads were washed four times in wash buffer (50 mM Tris at pH 7.5, 5% glycerol, 0.05% igepal, 150 mM NaCl, PIC), and protein was eluted off beads with 500 μg/mL Flag peptide for 30 min. Flag eluate was precleared with Protein G sepharose beads (GE Healthcare) for 10 min. HIS beads (Life Technologies) were added to precleared supernatant with 10 mM imidazole for 30 min. Beads were washed twice with wash buffer plus 25 mM imidazole. Protein was eluted off beads by boiling in LDS sample buffer (Life Technologies). Ten percent of sample was resolved with 4%–12% SDS-PAGE (Life Technologies) for silver stain (Pierce), and the remaining material was stained with colloidal blue (Life Technologies), cut into 10 size fractions, and analyzed by the Harvard Mass Spectrometry Facility by microcapillary reverse-phase high-pressure liquid chromatography (HPLC) nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Thermo LTQ-Orbitrap mass spectrometer.
Immunoprecipitation
All steps were performed at 4°C. Two micrograms of antibody was added to 1 mg of nuclear supernatant for 1 h. Twenty microliters of Protein G beads was added for 1 h. Beads were washed four times in wash buffer, and protein was eluted off beads by boiling in LDS sample buffer.
Antibodies
Antibodies used for immunoprecipitation, PLA, immunofluorescence, Western blot, far-Western blot, and ChIP were ZNF750 (Sigma, HPA023012), KLF4 (R&D Systems, AF3640; Sigma, HPA002926), RCOR1 (Santa Cruz Biotechnology, 30189 and 376567), KDM1A (Bethyl Laboratories, A300-215A), CTBP1 (BD Bioscience, 612042), CTBP2 (BD Bioscience, 612044), HA tag (Abcam, ab9110; Covance, MMS-101P), Flag tag (Sigma, F1804), IgG (Santa Cruz Biotechnology, 2027), LOR (Covance, PRB-145P), FLG (Santa Cruz Biotechnology, 66192), Ki67 (Thermo Scientific, SP6), H3K27ac (Abcam, ab4729), H3K4me1 (Abcam, ab8895), and GST (Cell Signaling).
Western blotting
Immunoprecipitation samples or 25 μg of cell lysates were resolved on 4%–12% SDS-PAGE and transferred to PVDF membranes. Membranes were incubated in primary antibodies overnight at 4°C and in secondary antibodies (Santa Cruz or Rockland) for 45 min at room temperature.
Far-Western blotting
The following recombinant proteins were purchased or purified: ZNF750 (Origene), MBP (Prospec), KLF4 (Origene), RCOR1 (Abnova), KDM1A (Origene), CTBP1 (Origene), CTBP2 (Origene), and GST (purified with Pierce GST spin purification kit). ZNF750 and MBP were spotted on nitrocellulose membranes. Recombinant proteins KLF4, RCOR1, KDM1A, CTBP1, CTBP2, or GST were added to membrane at 1 μg/mL overnight at 4°C. Membranes were incubated in primary antibodies overnight at 4°C and in secondary antibodies for 45 min at room temperature. Ponceau S (Sigma) was used for loading control.
Immunofluorescence and PLA
Keratinocytes or 7-μm sections of epidermal tissue were fixed with 4% formaldehyde for 15 min followed by blocking in PBS with 2.5% normal goat serum, 0.3% Triton X-100, and 2% bovine serum albumin for 30 min. Primary antibodies were added overnight at 4°C, and secondary antibodies were added for 1 h. PLA was performed using Duolink with PLA probe anti-mouse plus and PLA probe anti-rabbit minus (Olink Bioscience). Due to antibody limitations, HA-tagged ZNF750 with HA antibody was used in place of endogenous ZNF750 with KLF4 and KDM1A PLA.
ChIP assays
Keratinocytes were cross-linked with 1% formaldehyde for 10 min. Fifty million keratinocytes were used for ChIP-seq, 5 million keratinocytes were used for ChIP-qPCR, and 10 million keratinocytes were used for re-ChIP-qPCR. All steps were performed at 4°C unless otherwise indicated. Cells were lysed in swelling buffer (100 mM Tris at pH 7.5, 10 mM KOAc, 15 mM MgOAc, 1% igepal, PIC) for 10 min followed by Dounce homogenization. Nuclei were pelleted at 4000 rpm for 5 min and lysed in modified RIPA buffer (PBS with 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, PIC) for 10 min. Chromatin was sonicated to 150–300 bp using a Diagenode bioruptor. Insoluble material was removed by centrifugation at 13,000 rpm for 10 min followed by preclear with StaphA cells for 15 min. One microgram of primary antibody/5 million keratinocytes was incubated with precleared chromatin for 16 h. For goat or mouse primary antibodies, rabbit anti-goat or rabbit anti-mouse secondary antibodies (Millipore) were added for 1 h. Ten microliters of StaphA cells/5 million keratinocytes was added for 15 min at room temperature. StaphA cells were washed twice with dialysis buffer (50 mM Tris at pH 8, 2 mM EDTA, 0.2% sarkosyl) and four times with immunoprecipitation wash buffer (100 mM Tris at pH 8, 500 mM LiCl, 1% NP-40, 1% sodium deoxycholate). Chromatin was eluted off StaphA cells in 1% SDS and 50 mM NaHCO3. For re-ChIP, eluted chromatin was diluted 10-fold in modified RIPA without SDS, and second primary antibody was added overnight. For ChIP-seq and ChIP-qPCR, 200 mM NaCl was added, and cross-links were reversed for 16 h at 67°C. DNA was purified using PCR purification kit (Qiagen), eluting in 50 μL of water. One microliter of ChIP DNA was used for qPCR. For ChIP-seq, libraries were prepared using the Mondrian (NuGen) and size-selected using Pippin Prep (Sage Science). Libraries were sequenced with 1 × 50 base pair reads on the Illumina HiSeq2500.
ChIP-seq data analysis
Sequencing reads were mapped to the hg19 genome using Bowtie, and PCR duplicates were removed, resulting in ∼20 million reads per sample. Peaks were called using MACS with a P-value cutoff of 1 × 10−5 and fold enrichment ≥10. MEME was used for de novo motif discovery. Peaks were assigned to genes using GREAT with basal plus 200 kb maximal extension. HOMER was used for genomic distribution analysis and quantification of ChIP-seq fragment depth over genomic intervals for H3K4me1 analysis and peak heat map. For comparing ChIP-seq data sets, KLF4, RCOR1, or KDM1A ChIP-seq signal in ZNF750 peaks was considered enriched if peak was called or summit height was ≥5 and reads per base pair were ≥0.004, similar to as previously described (Bardet et al. 2012).
Reporter assays
The lentiviral constructs for reporter assays were made by cloning regions of interest into pGreenFire (System Biosciences). Firefly luciferase activity was measured using the dual-luciferase reporter assay system from Promega. Luminescence was normalized to protein concentration and proviral integrants as previously described (Sen et al. 2012).
Clonogenic assays
Clonogenic assays were performed as previously described (Bao et al. 2013) with minor modifications. Mouse fibroblast 3T3 cells were treated with 15 μg/mL Mitomycin C (Sigma) in DMEM for 2 h and then plated at 8 × 105 cells per well in six-well plates. After 24 h, 500 keratinocytes were seeded onto 3T3s. After 7–10 d, 3T3 cells were removed with PBS, and keratinocytes were fixed with 1:1 methanol:acetone for 4 min and stained with crystal violet.
Accession numbers
ChIP-seq data have been deposited with Gene Expression Omnibus accession code GSE57702.
Acknowledgments
We thank J. Wysocka, A. Oro, J. Crabtree, H. Chang, and M. Fuller as well as A. Ungewickell, V. Lopez-Pajares, B. Zarnegar, B. Sun, A. Rubin, and X. Bao for presubmission review. We thank A. Bhaduri and K. Qu for helpful discussions and assistance with data analysis, S. Tao for technical support, and L. Morcom and P. Bernstein for administrative assistance. This work was supported by the U.S. Department of Veterans Affairs Office of Research and Development, National Institute of Arthritis and Musculoskeletal and Skin Diseases National Institutes of Health grant R01 AR45192 (to P.A.K), and National Science Foundation Graduate Research Fellowship grant DGE-0645962 (to L.D.B.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.246579.114.
References
- Abrajano JJ, Qureshi IA, Gokhan S, Molero AE, Zheng D, Bergman A, Mehler MF. 2010. Corepressor for element-1-silencing transcription factor preferentially mediates gene networks underlying neural stem cell fate decisions. Proc Natl Acad Sci 107: 16685–16690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, Khavari PA. 2013. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 12: 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet AF, He Q, Zeitlinger J, Stark A. 2012. A computational pipeline for comparative ChIP-seq analyses. Nat Protoc 7: 45–61 [DOI] [PubMed] [Google Scholar]
- Birnbaum RY, Zvulunov A, Hallel-Halevy D, Cagnano E, Finer G, Ofir R, Geiger D, Silberstein E, Feferman Y, Birk OS. 2006. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat Genet 38: 749–751 [DOI] [PubMed] [Google Scholar]
- Birnbaum RY, Hayashi G, Cohen I, Poon A, Chen H, Lam ET, Kwok P-Y, Birk OS, Liao W. 2011. Association analysis identifies ZNF750 regulatory variants in psoriasis. BMC Med Genet 12: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin Z-Y, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras A-C, Nesvizhskii AI. 2011. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods 8: 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. 2007. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem 282: 33994–34002 [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. 2006. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 25: 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, et al. 2007. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J 26: 4138–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman DE, Ton-That H, Johnson JM, Yang VW. 2000. Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein–protein interaction. Nucleic Acids Res 28: 1106–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, Baldi P, Andersen B. 2012. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet 8: e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Dupé V, Bornert J-M, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D. 2005. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development 132: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Jiang J, Chan Y-S, Loh Y-H, Cai J, Tong G-Q, Lim C-A, Robson P, Zhong S, Ng H-H. 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 10: 353–360 [DOI] [PubMed] [Google Scholar]
- Jin W, Scotto KW, Hait WN, Yang J-M. 2007. Involvement of CtBP1 in the transcriptional activation of the MDR1 gene in human multidrug resistant cancer cells. Biochem Pharmacol 74: 851–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129: 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GXY, Chow J, Kim GE, et al. 2012. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 26: 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437: 432–435 [DOI] [PubMed] [Google Scholar]
- Li HX, Han M, Bernier M, Zheng B, Sun SG, Su M, Zhang R, Fu JR, Wen JK. 2010. Kruppel-like Factor 4 promotes differentiation by transforming growth factor- receptor-mediated Smad and p38 MAPK signaling in vascular smooth muscle cells. J Biol Chem 285: 17846–17856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi Y-I, Evers BM, Zhou BP. 2010. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 29: 1803–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D-C, Hao J-J, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding L-W, et al. 2014. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 46: 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zheng H, Ai W. 2009. C-terminal binding proteins (CtBPs) attenuate KLF4-mediated transcriptional activation. FEBS Lett 583: 3127–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. 2013. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10: 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal S, Ho N, Parker D, Grossman SR. 2012. CtBP2 promotes human cancer cell migration by transcriptional activation of Tiam1. Genes Cancer 3: 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. 2006. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc Natl Acad Sci 103: 18668–18673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Dean DC. 1999. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci 96: 6683–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan KGR, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, Bolognesi M, Crossley M. 2006. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol 26: 8159–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Huarte M. 2011. To repress or not to repress: this is the guardian’s question. Trends Cell Biol 21: 344–353 [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 22: 356–360 [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. 2008. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. 2012. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell 22: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J-I, Sui G, Affar EB, Whetstine JR, Lan F, Ogawa H, Luke MP-S, Nakatani Y, Shi Y. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Teng A, Nair M, Wells J, Segre JA, Dai X. 2007. Strain-dependent perinatal lethality of Ovol1-deficient mice and identification of Ovol2 as a downstream target of Ovol1 in skin epidermis. Biochimica et Biophysica Acta 1772: 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert F-M, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. 2008. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol 183: 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C-F, Hwu W-L, Yang L-C, Chung W-H, Chien Y-H, Hung C-F, Chen H-C, Tsai P-J, Fann CSJ, Liao F, et al. 2008. A promoter sequence variant of ZNF750 is linked with familial psoriasis. J Invest Dermatol 128: 1662–1668 [DOI] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. 2001. CoREST is an integral component of the CoREST–human histone deacetylase complex. Proc Natl Acad Sci 98: 1454–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, et al. 2006. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol 299: 122–136 [DOI] [PubMed] [Google Scholar]