ABSTRACT
Due to the increasing concerns about limited fossil resources and environmental problems, there has been much interest in developing biofuels from renewable biomass. Ethanol is currently used as a major biofuel, as it can be easily produced by existing fermentation technology, but it is not the best biofuel due to its low energy density, high vapor pressure, hygroscopy, and incompatibility with current infrastructure. Higher alcohols, including 1-propanol, 1-butanol, isobutanol, 2-methyl-1-butanol, and 3-methyl-1-butanol, which possess fuel properties more similar to those of petroleum-based fuel, have attracted particular interest as alternatives to ethanol. Since microorganisms isolated from nature do not allow production of these alcohols at high enough efficiencies, metabolic engineering has been employed to enhance their production. Here, we review recent advances in metabolic engineering of microorganisms for the production of higher alcohols.
INTRODUCTION
Increasing concerns on climate change and inevitable depletion of fossil resources are urging us to develop fuels and energy that are independent of fossil resources. Microbial production of biofuels from renewable biomass has been considered one of the solutions (1). Currently, ethanol is a major biofuel produced worldwide, mainly because it can be produced by fermentation technology that has been available for a long time. However, ethanol is not such a great biofuel due to its inferior fuel characteristics, such as low energy density, high vapor pressure, hygroscopy, and incompatibility with current infrastructure. Therefore, there has recently been much interest in producing advanced biofuels possessing fuel characteristics similar to those of petroleum-derived fuels, such as hydrocarbons and higher alcohols. In this paper, we review recent advances in the production of higher alcohols, with a focus on metabolic engineering strategies employed for the development of microbial strains efficiently producing them.
METABOLIC ENGINEERING STRATEGIES FOR THE PRODUCTION OF PRIMARY ALCOHOLS
Primary higher alcohols can be synthesized via either the fatty acid or amino acid pathway in microorganisms. In general, the use of fatty acid metabolism is advantageous for the production of linear-chain alcohols, while that of amino acid metabolism is suitable for branched-chain alcohols. In this section, we review the recent studies for the production of 1-butanol and extend the discussion to higher fatty alcohols. Finally, the strategies employed for the production of branched-chain alcohols are reviewed. Strategies for the production of higher alcohols as well as the metabolic pathways and key enzymes involved are shown in Fig. 1 and 2. Also, the results of recent studies on higher-alcohol production by various microorganisms are summarized in Table 1.
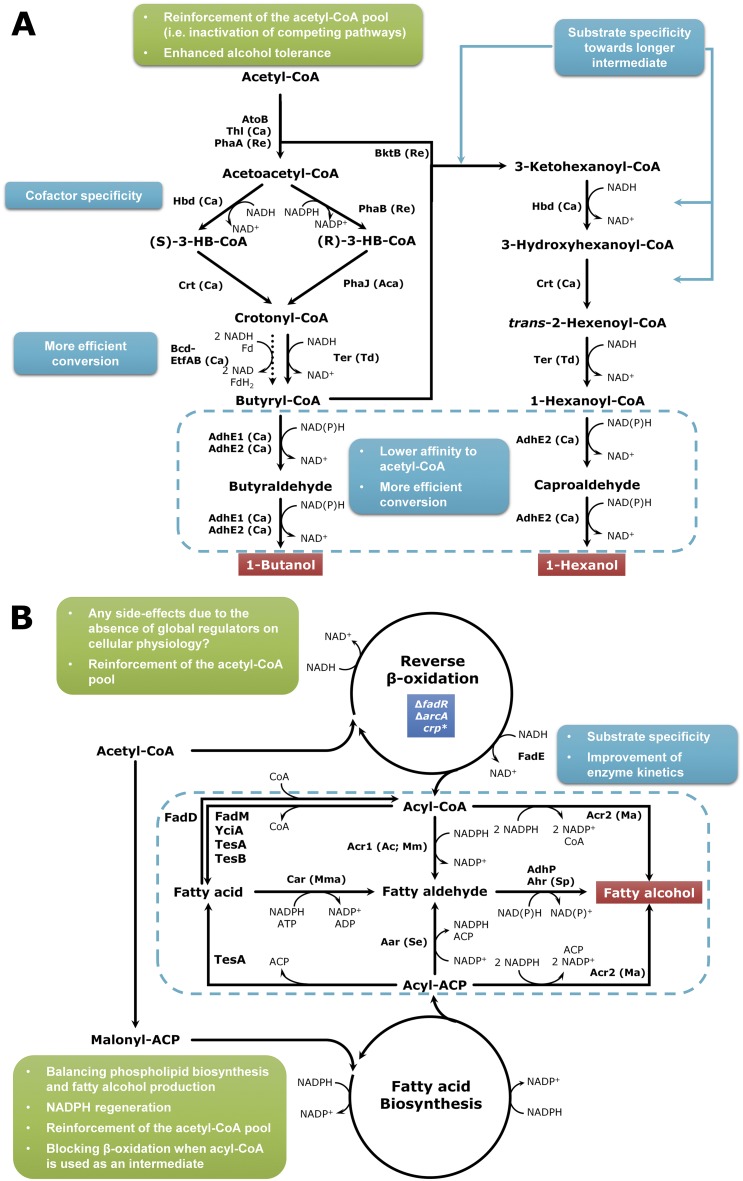
FIG 1 .
Strategies for the production of linear, primary alcohols. (A) Production of 1-butanol and 1-hexanol through the native or reconstructed clostridial pathway. The dotted arrow adjacent to Bcd-EtfAB indicates the weak activity of the Bcd enzyme in microbes other than clostridia. (B) Production of long-chain primary alcohols. In contrast to short-chain alcohols, long-chain fatty alcohols can be produced via various routes. The blue box in the reverse β-oxidation indicates the essential genetic manipulation to activate this pathway in the presence of glucose. The crp* gene encodes the mutant catabolite repressor protein for catabolite derepression. Points to be considered for further engineering in enzymatic and cellular levels are indicated in cyan and green boxes, respectively. For each reaction, the names of the corresponding enzymes used in the metabolic engineering studies are shown. The source of the enzyme was noted together with the enzyme, except for E. coli. The abbreviations of the species are as follows: Ac, Acinetobacter calcoaceticus; Aca, Aeromonas caviae; Ca, Clostridium acetobutylicum; Ch, Cuphea hookeriana; Ma, Marinobacter aqualeolei; Mm, Mus musculus; Mma, Mycobacterium marinum; Re, Ralstonia eutropha; Se, S. elongatus; Sp, Synechocystis sp. PCC 6803; Td, Treponema denticola; Uc, Umbellularia californica. See the main text for the abbreviations of the enzymes.
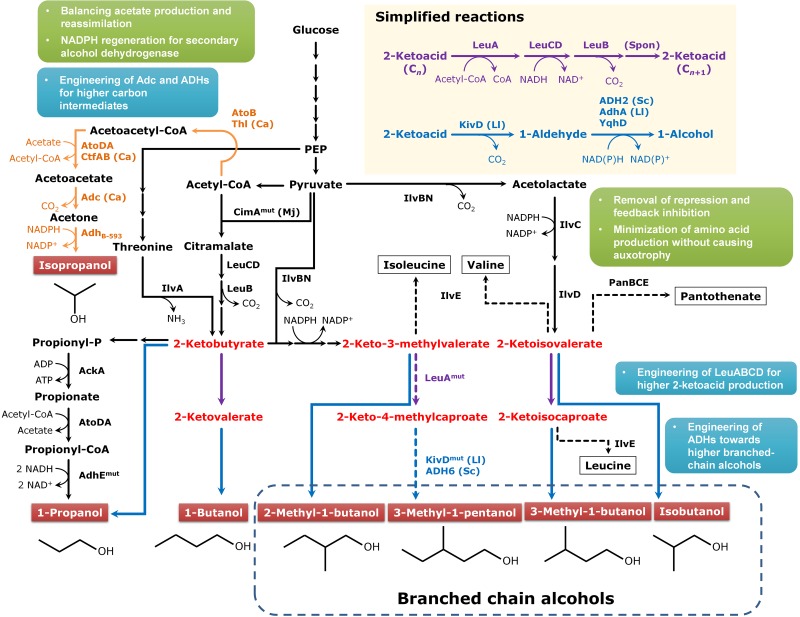
FIG 2 .
Production of branched-chain and secondary alcohols. Higher alcohols are shown in the red boxes, and the 2-ketoacid precursors are indicated in red text. The reactions in the isopropanol production pathway are shown with orange arrows. As in Fig. 1, points to be considered are indicated in cyan and green boxes. The source of the enzyme is noted together with the enzyme except E. coli, and follows that in Fig. 1. Additional abbreviations of the species are Ll, Lactococcus lactis; Sc, Saccharomyces cerevisiae. The enzymes shown are as follows: AckA, acetate kinase A and propionate kinase II; AdhEmut, aerobically functional alcohol dehydrogenase; IlvA, threonine dehydratase; IlvC, ketol-acid reductoisomerase; IlvD, dihydroxyacid dehydratase; IlvE, branched-chain amino-acid aminotransferase; IlvIH, acetolactate synthase I; IlvBN, acetolactate synthase III complex; KivD, 2-ketoacid decarboxylase; LeuA, 2-isopropylmalate synthase; LeuB, 3-isopropylmalate dehydrogenase; LeuCD, 3-isopropylmalate isomerase complex; YqhD, NADPH-dependent aldehyde reductase; AtoB and Thl, acetyl-CoA acetyltransferase; AtoDA, acetyl-CoA:acetoacetyl-CoA synthase; CtfAB, CoA transferase; Adc, acetoacetate decarboxylase; AdhB-593, primary/secondary alcohol dehydrogenase from C. beijerinckii B-593.
TABLE 1 .
Summary of microbial production of higher alcohols
| Product | Host | Genotypea (knockout; overexpression) |
Substrate | Medium | Titer | Cultivation | Comment | Reference |
|---|---|---|---|---|---|---|---|---|
| 1-Propanol | E. coli BW25113 F' | ΔilvA ΔilvB; LlkivD ScADH2 MjcimAmut leuABCD | Glucose | Defined | 2.78 g/liter | Shake flask | 1-Butanol, 0.39 g/liter | 40 |
| E. coli W3110 | ΔlacI ΔlysA ΔmetA ΔtdhA ΔiclR ΔilvIH ΔilvBN ΔrpoS thrAC1034T
lysCC1005T Pthr::Ptac Pppc::Ptrc ilvAC1139T,G1341T,C1351G,T1352C; thrABC MjcimA ackA adhEmut |
Glucose | Semidefined | 1.5 g/liter | Bioreactor | Aerotolerant AdhE; 20 g/liter initial glucose without feeding | 39 | |
| E. coli W3110 | ΔlacI ΔlysA ΔmetA ΔtdhA ΔiclR ΔilvIH ΔilvBN ΔrpoS thrAC1034T
lysCC1005T Pthr::Ptac Pppc::Ptrc ilvAC1139T,G1341T,C1351G,T1352C; thrABC MjcimA ackA adhEmut |
Glycerol | Semidefined | 10.3 g/liter | Bioreactor | Yield, 0.259 g/g; aerotolerant AdhE | 39 | |
| Isopropanol | E. coli ATCC 11303 | None; lacIq Cathl atoDA Caadc adhB-593 | Glucose | Semidefined | 5 g/literb | Shake baffled flask | Yield, 0.15 g/g; acetone accumulation when glucose is depleted | 61 |
| E. coli ATCC 11303 | None; lacIq Cathl atoDA Caadc adhB-593 | Glucose | Semidefined | 143 g/liter | Stirred flask | Yield, 0.23 g/g; with gas stripping; 240 h | 63 | |
| E. coli ATCC 11303 | None; Cathl atoDA Caadc adhB-593 Tfbgl-blc (fused) | Cellobiose | Semidefined | 4.1 g/liter | Shake flask | Yield, 0.08 g/g | 64 | |
| E. coli JM109 | None; Cathl CactfAB Caadc adhB-593 | Glucose | Complex | 13.6 g/liter | Shake baffled flask | Yield, 0.17 g/g; acetone yield, 0.03 g/g | 62 | |
| Alcohol mixture (IBE) | C. acetobutylicum ATCC 824 | Δbuk::ermC; adc ctfAB adhB-593 | Glucose | Semidefined | 20.4 g/liter | Bioreactor | Yield, 0.30 g/g; isopropanol, 4.4 g/liter; butanol, 14.1 g/liter; with gas stripping | 65 |
| C. acetobutylicum ATCC 824 | Δbuk::ermC; adc ctfAB adhB-593 | Glucose | Semidefined | 35 g/liter | Bioreactor | Yield, 0.26 g/g; isopropanol, 4.1 g/liter; butanol, 25.1 g/liter; with gas stripping | 65 | |
| C. acetobutylicum ATCC 824 | Δbuk ΔCA_C1502; adc ctfAB adhB-593 | Glucose | Semidefined | 20.4 g/liter | Bioreactor | Yield, 0.33 g/g; with gas stripping | 66 | |
| C. acetobutylicum Rh8 | None; adhB-593 | Glucose | Semidefined | 23.9 g/liter | Bioreactor | Random mutagenized strain; yield, 0.31 g/g; isopropanol, 7.6 g/liter; butanol, 15 g/liter | 67 | |
| C. acetobutylicum BKM19 | Δbuk::ermC; adhB-593 hydGB-593 | Glucose | Semidefined | 28.5 g/liter | Bioreactor | Obtained in a pilot-scale fermentation; random mutagenized strain; yield, 0.37 g/g; isopropanol, 3.5 g/liter; butanol, 15.4 g/liter; ethanol, 9.6 g/liter | 68 | |
| 1-Butanol | C. acetobutylicum ATCC 824 | Δpta Δbuk; adhED485G | Glucose | Semidefined | 18.9 g/liter | Bioreactor | Without in situ recovery; yield, 0.29 g/g; acetone, 1.5 g/liter | 8 |
| C. acetobutylicum ATCC 824 | Δpta Δbuk; adhED485G | Glucose | Semidefined | 130 g/liter | Bioreactor | Volumetric productivity, 1.32 g/liter/h; with in situ recovery; yield, 0.31 g/g | 8 | |
| C. acetobutylicum ATCC 824 | Δadc; Ecgsh adhE ctfAB thl hbd crt bcd | Glucose | Semidefined | 14.9 g/liter | Bioreactor | Yield, 0.34 g/g; 3.3 g/liter of ethanol | 9 | |
| C. tyrobutyricum ATCC 25755 | Δack; CaadhE2 | Glucose | Semidefined | 10 g/liter | Serum bottle; anaerobic | Yield, 0.27 g/g; 5.8 g/liter butyrate; manual pH control by NaOH | 16 | |
| C. tyrobutyricum ATCC 25755 | Δack; CaadhE2 | Mannitol | Semidefined | 16 g/liter | Serum bottle; anaerobic | Yield, 0.31 g/g; 1.0 g/liter butyrate; manual pH control by NaOH | 16 | |
| C. tyrobutyricum ATCC 25755 | None; CaadhE2 | Mannitol | Complex | 20.5 g/liter | Bioreactor; anaerobic | Yield, 0.33 g/g; productivity, 0.32 g/liter/h; 1.0 g/liter butyrate; manual pH control by NaOH | 17 | |
| E. coli DH1 | None; RephaAB AcaphaJ Tdter CaadhE2 aceEF lpd | Glucose | Complex | 3.4 g/liter | Shake flask | Shift to the anaerobic condition after induction | 18 | |
| E. coli DH1 | None; RephaA Cahbd Cacrt Tdter CaadhE2 aceEF lpd | Glucose | Complex | 4.7 g/liter | Shake flask | Yield, 0.28 g/g; shift to the anaerobic condition after induction | 18 | |
| E. coli BW25113/F' | ΔldhA ΔadhE ΔfrdBC Δpta; atoB Cahbd Cacrt CaadhE2 Cbfdh Tdter | Glucose | Complex | 15 g/liter | Bioreactor; anaerobic | Yield, 0.36 g/g; without gas stripping | 19 | |
| E. coli BW25113/F' | ΔldhA ΔadhE ΔfrdBC Δpta; atoB Cahbd Cacrt CaadhE2 Cbfdh Tdter | Glucose | Complex | 30 g/liter | Bioreactor; anaerobic; with gas stripping | Yield, 0.36 g/g; without gas stripping | 19 | |
| S. elongatus PCC 7942 | None; SclnphT7 Cahbd Cacrt Tdter Csbld EcyqhD | CO2 | Defined | 27 mg/literb | Static capped flask | Photosynthesis | 22 | |
| S. elongatus PCC 7942 | None; SclnphT7 RephaB AcaphaJ Tdter Csbld EcyqhD | CO2 | Defined | 29.9 mg/liter | Static capped flask | Photosynthesis | 22 | |
| E. coli MG1655 | fadR− crp* ΔarcA ΔadhE Δpta ΔfrdA ΔyqhD; atoC yqeF fucO | Glucose | Defined | 14 g/liter | Shake baffled flask | 24 | ||
| Isobutanol | E. coli BW25113 F' | ΔadhE ΔldhA ΔfrdBC Δfnr Δpta ΔpflB; BsalsS ilvCD LlkivD ScADH2 | Glucose | Semidefined | 22 g/liter | Shake flask | Yield, 0.35 g/g | 34 |
| E. coli BW25113 F' | ΔadhE ΔldhA ΔfrdBC Δfnr Δpta ΔpflB; BsalsS ilvCD LlkivD LladhA | Glucose | Semidefined | 50.9 g/liter | Bioreactor | With in situ gas stripping; volumetric productivity, 0.7 g/liter/h; yield, 0.29 g/g | 36 | |
| B. subtilis | Δldh; CgilvCD alsS LlkivD ScADH2 | Glucose | Complex | 2.62 g/liter | Bioreactor | 41 | ||
| B. subtilis | Δldh; CgilvCD alsS LlkivD ScADH2 | Glucose | Complex | 3.83 g/liter | Bioreactor | Auto-inducible 2-ketovalerate synthetic operon | 42 | |
| C. cellulolyticum ATCC 35319 | None; LlkivD EcyqhD BsalsS EcilvCD | Cellulose (Sigmacell type 50) | Defined | 660 mg/liter | Not specified | 7–9 days; strong expression of alsS might be deleterious | 43 | |
| C. glutamicum ATCC 13032 | Δpyc ΔldhA; BsalsS LlkivD ilvCD adhA | Glucose | Complex | 4.9 g/liter | Shake flask | Yield, 0.09 g/g | 44 | |
| C. glutamicum ATCC 13032 | ΔaceE Δpqo ΔilvE ΔldhA Δmdh; ilvBNCD EcpntAB LlkivD adhA | Glucose | Semidefined | 13 g/liter | Bioreactor | Yield, 0.20 g/g; volumetric productivity, 0.32 g/liter/h; shift to the anaerobic condition | 45 | |
| R. eutropha H16 | ΔphaCAB ΔilvE ΔbkdAB ΔaceE; adh(con) ilvBHCD LlkivD | Fructose | Defined | 270 mg/liter | Shake flask | Coproduced 40 mg/liter of 3-methyl-1-butanol | 46 | |
| R. eutropha H16 | ΔphaB2C2 ΔphaC1AB1; BsalsS ilvCD LlkivD EcyqhD | CO2 | Defined | 90 mg/liter | Bioreactor with electrodes | Coproduced 50 mg/liter of 3-methyl-1-butanol | 47 | |
| S. cerevisiae CEN.PK 2-1 C | None; ILV2 ILV5 ILV3 | Glucose | Complex | 4.12 mg/liter | Shake baffled flask | 48 | ||
| S. cerevisiae BY4741 | lpd1Δ; LlkivD ADH6 ILV2 ILV5c ILV3c ILV2C MAE1 | Glucose | Complex | 1.62 g/liter | Shake flask | Yield, 0.016 g/g | 49 | |
| S. cerevisiae S288C (MATa/α) | his3Δl1/HIS3 leu2Δ0/LEU2 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0; ILV2 ILV3 ADH7 ILV5 LlkivD | Glucose | Defined | 635 mg/liter | Shake tube | High-cell-density culture; yield, 6.4 mg/g; mitochondrial expression of ILV, KivD, ADH genes; 2-methyl-1-butanol, 118 mg/liter; 3-methyl-1-butanol, 95 mg/liter | 50 | |
| 2-Methyl- 1-butanol |
E. coli BW25113 F' | ΔmetA Δtdh; StyilvGM ilvCD CgilvA LlkivD ScADH2 thrABC | Defined | 1.25 g/liter | Shake baffled flask | Yield, 0.17 g/g; total alcohol, 3 g/liter | 37 | |
| S. elongatus PCC 7942 | None; LlkivD EcyqhD MjcimAmut leuBCD | CO2 | Defined | 177.5 mg/liter | Static flask | Photosynthesis; 12-day culture; isobutanol, 50 mg/liter; 1-propanol, 17.5 mg/liter | 70 | |
| 3-Methyl- 1-butanol |
E. coli BW25113 F' | None; BsalsS ilvCD LlkivD ScADH2 leuAG462D leuBCD | Glucose | Defined | 9.5 g/liter | Shake flask | Random mutagenized strain; two-phase culture with oleoyl alcohol; yield, 0.11 g/g; total alcohol, 12.5 g/liter | 38 |
| 1-Hexanol | E. coli BW25113 F' | ΔldhA ΔadhE ΔfrdBC Δpta; atoB RebktB Cahbd Cacrt CaadhE2 Cbfdh Tdter | Glucose | Complex | 47 mg/liter | Sealed test tube with shaking | Anaerobic; butanol, 5.1 g/liter | 23 |
| 3-Methyl- 1-pentanol |
E. coli ATCC 98082 | ΔilvE ΔtyrB; ilvGMCD tdcB LlkivDV461A,F381L ScADH6 leuAG462D,S139G leuBCD | Glucose | Semidefined | 793.5 mg/liter | Shake flask | Enzyme evolution | 52 |
| Fatty alcohols | E. coli MG1655 | fadR− ΔarcA Δcrp ΔadhE Δpta ΔfrdA ΔfucO ΔyqhD ΔfadD; crp* atoC(con) fadBA yiaY | Glucose | Defined | 0.33 g/liter | Shake baffled flask | Yield, 0.08 g/g; mixture of 1-hexanol, 1-octanol, and 1-decanol | 24 |
| S. cerevisiae BY4742 (MATα) | his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 acc1::PTEF1-ACC1 fas1::PTEF1-FAS1 fas2::PTEF1-FAS2; ACC1 FAS1 FAS2 MmFAR1 MalMAE | Glucose and galactose | Defined | 98 mg/liter | Shake flask | 26 | ||
| E. coli DH1 | fadE; ‘tesA fadD Acacr1 | Glucose | Defined | 60 mg/liter | Shake baffled flask | 27 | ||
| E. coli MG1655 | ΔaraBAD ΔfadE ΔfadAB ΔackA Δpta; UcfatB fadD Maacr2 (Maqu_2507) | Glucose | Defined | 1.65 g/liter | Bioreactor | Yield, 0.134 g/g; mainly C12 and C14 alcohols | 29 | |
| E. coli BL21 (DE3) | ΔfadE; fadD Maacr2 (Maqu_2220) ‘tesA | Glucose | Defined | 1.73 g/liter | Bioreactor | Yield, 28.3 mg/g | 30 | |
| E. coli BL21 (DE3) | None; TPC Mmacar Bssfp Spahr | Glucose | Defined | 360 mg/liter | Test tube with shaking | 31 | ||
| E. coli BL21 (DE3) | ΔfadE; Seaar | Glycerol | Defined | 0.75 g/liter | Bioreactor | Yield, 0.02 g/g | 33 |
For heterologous genes, the abbreviation of the species is followed by the gene name (e.g., ScADH2, the ADH2 gene from S. cerevisiae). The abbreviations of the species are as follows: Ac, A. calcoaceticus; Aca, A. caviae; Ca, C. acetobutylicum; Cb, Candida boidinii; Cs, Clostridium saccharoperbutylacetonicum N1-4; Ll, L. lactis; Ma, M. aquaeolei; Mal, Mortierella alpine; Mj, Methanococcus jannaschii; Mm, M. musculus; Mma, M. marinum; Sc, S. cerevisiae; Scl, Streptomyces sp. strain CL190; Se, Synechococcus elongatus PCC 7942; Sp, Synechocystis sp. PCC 6803; Td, Treponema denticola; Tf, Thermobifida fusca YX, Uc, Umbellularia californica. Other abbreviations are as follows: crp*, a cyclic AMP-independent mutant crp gene; ‘tesA, a leaderless tesA gene; gene(con), modified ITS for constitutive expression; adhB-593, the primary/secondary alcohol dehydrogenase gene from C. beijerinckii NRRL B-593; hydGB-593, a putative gene encoding an electron transfer protein from C. beijerinckii NRRL B-593.
These values were estimated from the figures in the original references, as the values were not described in the text.
1-Butanol.
1-Butanol can be best produced by the fermentation of clostridia (Fig. 1A) (see references 2–4 for the details on clostridial butanol fermentation). Due to difficulties in genetic manipulation and complex metabolic regulations in clostridia, metabolic engineering of clostridia has been rather difficult and has advanced only recently. The metabolic engineering strategies have been focused mainly on achieving an objective of increasing 1-butanol production and selectivity through the reduced formation of acetate, butyrate, and acetone. However, several previous studies suggested that elimination or reduction of the acetone flux is not sufficient to increase 1-butanol production; this resulted in accumulation of acids and reduced production of 1-butanol (5–7).
Jang et al. (8) successfully engineered Clostridium acetobutylicum ATCC 824 for the production of 1-butanol to high titer with high selectivity by reinforcing the direct 1-butanol biosynthetic pathway (hot channel). First, four genes (pta, ack, ptb, and buk) involved in production of short-chain fatty acid were individually or combinatorially knocked out. Among them, the pta/buk double knockout mutant produced the highest titer (16 g/liter) of 1-butanol. 1-Butanol production was further reinforced by the overexpression of a variant adhE1 gene, which was engineered to utilize NADPH as a cofactor (8). The resulting strain produced over 18.9 g/liter of 1-butanol in batch fermentation. Also, 585.3 g of butanol was produced from 1,861.9 g of glucose by fed-batch culture of this engineered strain with in situ recovery. Hou et al. (9) also reported enhanced 1-butanol production in C. acetobutylicum. The adc gene was replaced with the glutathione-encoding genes from Escherichia coli, which was previously suggested to increase 1-butanol production in C. acetobutylicum (10). Instead of deleting the acid production pathway, the whole 1-butanol-forming pathway (from thl to adhE1 genes) was amplified to produce 14.9 and 3.3 g/liter of 1-butanol and ethanol, respectively (9).
C. acetobutylicum has a pSOL1 megaplasmid harboring genes involved in solventogenesis, and the loss of this plasmid results in so-called strain degeneration incapable of producing solvents (11). Several clostridial species produce high titers of butyric acid. It was thus thought that 1-butanol can be produced in butyric acid-producing bacteria by introducing aldehyde and alcohol dehydrogenases. However, the overexpression of aldehyde and alcohol dehydrogenases (or bifunctional aldehyde/alcohol dehydrogenases) in degenerate C. acetobutylicum variants did not typically result in a higher titer of 1-butanol, as the butyric acid production was still very active (12–15). Recently, production of 1-butanol by engineering one of the best butyric acid producers, Clostridium tyrobutyricum, has been reported (16, 17). In one study (16), the ack and ptb single mutants of C. tyrobutyricum were employed for 1-butanol production. When the adhE2 gene from C. acetobutylicum was overexpressed in the C. tyrobutyricum ack mutant, about 10 g/liter of 1-butanol was produced. The use of mannitol, a more reduced carbon source than glucose, further increased the 1-butanol titer to 16 g/liter (16). Later, different replicons were examined to overexpress the C. acetobutylicum adhE2 gene in C. tyrobutyricum (17), and the best strain, without any gene knockout, was able to produce 21 g/liter of 1-butanol and 6 g/liter of butyric acid using mannitol as the sole carbon source.
Reconstruction of the clostridial 1-butanol production pathway in other microorganisms has also been reported. However, such engineered microorganisms equipped with clostridial genes did not result in sufficient 1-butanol production, probably due to the poor activity of butyryl coenzyme A (butyryl-CoA) dehydrogenase (Bcd). This bottleneck could be overcome by employing an alternative enzyme, trans-enoyl-CoA reductase (Ter) (18, 19). Shen et al. (19) deleted anaerobic fermentation genes in E. coli, including fumarate reductase, lactate dehydrogenase, and endogenous aldehyde/alcohol dehydrogenase, and reconstructed a chimeric pathway by overexpressing the atoB gene from E. coli, the hbd (encoding 3-hydroxybutyryl [HB]–CoA dehydrogenase), crt (3-hydroxybutyryl–CoA dehydratase), and adhE2 (bifunctional aldehyde/alcohol dehydrogenase) genes from C. acetobutylicum, and the ter gene from Treponema denticola. The formate production during anaerobic fermentation of engineered E. coli was reduced by introducing a fungal formate dehydrogenase, converting formate to CO2 and NADH (20). The resulting strain produced 1-butanol up to 15 g/liter in batch fermentation, and the yield reached up to 88% of the theoretical maximum.
In the case of redox enzymes, its cofactor preference as well as catalytic activity can affect 1-butanol production. Typically, NADH is the preferential electron donor, but some organisms are capable of efficiently generating NADPH (e.g., photosynthesis in cyanobacteria). In this case, the use of NADPH-dependent enzymes can be beneficial for 1-butanol production. A good alternative of clostridial NADH-dependent Hbd enzyme is the NADPH-dependent PhaB from bacteria producing polyhydroxyalkanoates (Fig. 1A). Unlike Hbd, PhaB produces the R form of 3-hydroxybutyryl–CoA instead of the S form (21). Since Crt does not accept the R form of 3-HB–CoA, it needs to be replaced with the R-form-specific dehydratase PhaJ (18). The use of PhaB-PhaJ as well as NADPH-specific aldehyde and alcohol dehydrogenases was successfully employed for the enhanced 1-butanol production from CO2 in cyanobacteria (22).
Fatty alcohols.
Fatty alcohols can be produced from acyl-CoAs. One strategy is to extend the clostridial pathway (Fig. 1A). The enzyme Ter has a broad substrate specificity, and it can be used to produce higher alcohols. Dekishima et al. (23) demonstrated such possibility by producing 1-hexanol in E. coli. They confirmed in vitro that Ter was able to convert 1-hexenoyl–CoA to 1-hexanoyl–CoA. Unlike the case of the 1-butanol production, the bktB gene from Ralstonia eutropha, encoding a β-ketothiolase, was additionally overexpressed since the endogenous AtoB could not condense acetyl-CoA and higher acyl-CoA. It was also demonstrated in vitro that AdhE2 from C. acetobutylicum has an activity toward octanoyl-CoA, even though 1-octanol was not produced. These results suggest that higher alcohols might be produced in the future after improving the substrate specificities of the other enzymes involved in acyl-CoA synthesis.
Another notable strategy is the use of the endogenous β-oxidation pathway in microorganisms (Fig. 1B), which was demonstrated in E. coli (24). The key metabolic engineering strategy employed was the elimination of regulatory mechanisms that repress the genes involved in β-oxidation. Through testing of various enzymes involved in initiation (condensation of acetyl- and acyl-CoAs) and termination (conversion of acyl-CoAs into fatty acids or alcohols), the reversal of the β-oxidation cycle combined with endogenous dehydrogenases and thioesterases was established to produce higher alcohols. Even though 1-butanol was preferentially produced (up to 14 g/liter), a mixture of higher alcohols (~C10; 0.33 g/liter) could also be produced by employing a different alcohol dehydrogenase. However, this strategy depended on the derepression of the genes, which made it difficult to fine-tune the pathway to control the chain length and to increase productivity. In a later study, the key enzymes involved in the β-oxidation pathway were characterized in vitro, and their various combinations were assembled and examined (25). Even though only fatty acid production was examined, it would be possible to produce higher alcohols by the introduction of aldehyde and alcohol dehydrogenases. In another study, Runguphan and Keasling (26) were able to produce fatty alcohols in Saccharomyces cerevisiae by engineering the triacylglyceride (TAG) biosynthetic pathway and blocking the β-oxidation pathway. Introduction of an acyl-CoA reductase from Mus musculus into the engineered strain resulted in the production of ca. 100 mg/liter of fatty alcohols.
Higher alcohols can also be produced via the fatty acid biosynthetic pathway, and various strategies have been reported (Fig. 1B). Fatty acyl-acyl carrier proteins (acyl-ACPs) can be converted into free fatty acids by thioesterase, and the resulting acids can be further converted to fatty acyl-CoAs. Then, acyl-CoA reductase (ACR) and aldehyde reductase (AHR) can convert fatty acyl-CoAs into fatty alcohols (Fig. 1B). Steen et al. (27) first demonstrated this strategy in E. coli using the acr1 gene, encoding ACR, from Acinetobacter calcoaceticus BD413. In flask cultures, it was possible to produce up to 60 mg/liter of total fatty alcohols (C12 to C16) when combined with the overexpression of the tesA and fadD genes to enhance the generation of free fatty acids and their conversion into acyl-CoAs, respectively, and deleting the fadE gene to block the degradation of acyl-CoAs via the β-oxidation pathway. The chain lengths of fatty alcohols were dependent mainly on the specificity of thioesterase (27). More recently, another type of ACR (Acr2) has been characterized (28). This ACR is able to directly convert fatty acyl-CoA, and acyl-ACP at a lower efficiency, into fatty alcohol. Youngquist et al. (29) found that the acyl-CoA reductase from Marinobacter aquaeolei VT8 (Acr2 encoded by Maqu_2507; Fig. 1B) converted acyl-CoA into fatty alcohol more rapidly than Acr1. After the optimization of fadD and acr2 expression, they were able to produce fatty alcohols up to 1.65 g/liter by fermentation with pH and dissolved oxygen (DO) control (29). Liu et al. (30) used another acr2 gene (Maqu_2220) from M. aquaeolei VT8, which resulted in 2-fold-increased production of fatty alcohols (ca. 650 mg/liter) compared to that obtained by using Maqu_2507 in shaking flask cultures. However, in a bioreactor experiment, fatty alcohols were produced to 1.73 g/liter (30), which is only slightly higher than that obtained by Youngquist et al. (29).
Conversion of acyl-ACPs to acyl-CoAs requires optimal control of both ACP and CoA pools in order to achieve enhanced production of fatty alcohols. To avoid such difficulty, acyl-CoA-independent production of fatty alcohols has been examined. Free fatty acids can be directly converted to fatty aldehydes by carboxylic acid reductase (Car) (Fig. 1B), which directly converts free fatty acids to fatty aldehydes using both NADPH and ATP. Akhtar et al. (31) characterized the kinetic properties of the Car from Mycobacterium marinum. When the car gene was overexpressed in E. coli together with the endogenous tesA gene for free fatty acid production, the Bacillus subtilis sfp gene (encoding a phosphopantetheinyl transferase) for the activation of Car (32), and also the endogenous yjgB gene, the product of which shares homology with the aldehyde reductase from Synechocystis sp. PCC 6803, about 360 mg/liter of fatty alcohols could be produced (31). The use of Car might be advantageous in that this enzyme does not depend on the intracellular CoA pool, and the reaction is more exergonic than that of cyanobacterial acyl-ACP reductases (Aar) (Fig. 1B) due to the coupling of ATP hydrolysis. Nonetheless, Liu et al. (33) produced ca. 0.75 g/liter of fatty alcohols in E. coli, exceeding the titer achieved by Akhtar et al. (31), by the overexpression of the Synechococcus elongatus aldehyde reductase gene alone. In that study, it was demonstrated in vitro that endogenous acetaldehyde dehydrogenase (AdhP) was the key enzyme for the reduction of fatty aldehydes to fatty alcohols, although it was downregulated in fatty alcohol-producing strains (33).
Branched-chain alcohols.
2-Ketoacids, which are metabolic intermediates of amino acid metabolism, can be used for the production of various branched-chain alcohols, including isobutanol (34–36), 2-methyl-1-butanol (37), and 3-methyl-1-butanol (38), as well as linear alcohols, including 1-propanol and 1-butanol (39, 40). The metabolic pathways designed for the production of various higher alcohols from the 2-ketoacid pathway are presented in Fig. 2. The key enzyme in this pathway is 2-ketoacid decarboxylase from Lactococcus lactis (KDC; encoded by the kivD gene), which shows a broad substrate specificity toward various 2-ketoacids (34). This strategy has been employed in a wide range of microorganisms, including Bacillus subtilis (41, 42), Clostridium cellulolyticum (43), Corynebacterium glutamicum (44, 45), Ralstonia eutropha (46, 47), and S. cerevisiae (48–50), in addition to E. coli. Among them, C. glutamicum has a great potential for the production of various amino acids and thus is advantageous for producing various alcohols from 2-ketoacids. Blombach et al. (45) have reported high-titer production of isobutanol from glucose by engineered C. glutamicum. In this study, an engineered C. glutamicum strain capable of producing a high titer of 2-ketoisovalerate (51) was employed as the base strain; in this strain, the ilvE, aceE, and pqo genes encoding transaminase B, pyruvate dehydrogenase subunit E1, and pyruvate:quinone oxidoreductase, respectively, were deleted, and the ilvBNCD genes were overexpressed to reinforce the carbon flux toward 2-ketoisovalerate (51). The key engineering strategies for isobutanol production include the inactivation of lactate and malate dehydrogenases, the use of endogenous alcohol dehydrogenase instead of S. cerevisiae ADH2, and the expression of the E. coli transhydrogenase (45). Interestingly, even though transhydrogenase PntAB from E. coli was overexpressed, inactivation of malic enzyme resulted in severe reduction of the production yield, indicating that C. glutamicum depends mainly on NADPH generation through the malic enzyme. The final strain was able to produce ca. 13 g/liter of isobutanol (45).
Similar to fatty acid biosynthesis, the chain length of 2-ketoacids can be extended by using the leuABCD genes; using this pathway, one carbon is added every cycle. As a proof-of-concept example (52), protein engineering was performed on KDC from L. lactis and LeuA (2-isopropylmalate synthase) from E. coli for the production of 3-methyl-1-pentanol (3MP). Based on the computational prediction of enzyme structures, the binding pockets of KDC and LeuA were modified, and their activities were examined. One combination, KDC V461A/F381L and LeuA G462D/S139G, resulted in a dramatic increase in 3MP production compared to that obtained with the wild-type enzymes (793.5 versus 6.5 mg/liter).
Another notable study is the use of an autolithotrophic bacterium such as R. eutropha. However, the low solubility of H2 in a growth medium acts as a major barrier for the efficient production of alcohols. Thus, Li et al. (47) directly supplied electrons to R. eutropha using electrodes, instead of using H2 as an electron donor. To produce higher alcohols from only CO2, an engineered R. eutropha strain was constructed and examined in a specially designed bioreactor. It was found that NO and O2− produced during the electric current flow inhibited cell growth, which was solved by shielding the anode with a ceramic cup, partly blocking their diffusion. Finally, the engineered R. eutropha strain produced ca. 90 and 50 mg/liter of isobutanol and 3-methyl-1-butanol, respectively, from only CO2 and electric current.
There had also been a report on higher-alcohol production by KDC-independent pathways (Fig. 2). Recently, there has been a report on the production of more than 10 g/liter of 1-propanol by engineering the threonine degradation pathway through 2-ketobutyrate to propionate and then to 1-propanol in E. coli (39). It was achieved by redirecting the carbon flux toward 2-ketobutyrate by the overexpression of the feedback-resistant threonine dehydratase gene (ilvA) and deletion of competing metabolic pathway genes (ilvI, ilvH, ilvB, and ilvN) followed by the overexpression of citramalate synthase (cimA) and mutant alcohol/aldehyde dehydrogenase (adhEmut) genes. In this study, the E. coli acetate kinase/propionate kinase II (ackA), acetyl-CoA:acetoacetyl-CoA synthase (atoDA), and an aerobically functional mutant alcohol/aldehyde dehydrogenase (adhEmut) were employed instead of KDCs and ADHs for converting 2-ketobutyrate to 1-propanol. Due to the clear feasibility and advantages of redirecting fluxes to the desired metabolites, the 2-ketoacid pathway will serve as an important platform for the production of biofuels and chemicals.
METABOLIC ENGINEERING STRATEGIES FOR THE PRODUCTION OF SECONDARY ALCOHOLS
Secondary alcohols have chemical properties different from those of primary alcohols due to the position of the hydroxyl group. The secondary alcohols of short-chain length can dissolve polar and nonpolar chemicals and thus are used as solvents in various industrial applications. Secondary alcohols are petrochemically synthesized by hydration of alkenes or oxidation of ketones. Biological secondary alcohol production depends on the reduction of ketones by secondary alcohol dehydrogenases (53–58). Acetone is a representative ketone produced as a major metabolite by clostridia, and biosynthesis of 2-butanone (59) and 2-pentanone (60) has only recently been reported. Although only the production of isopropanol will be described in this paper (see Fig. 2), it will be possible to produce other higher secondary alcohols by employing novel biosynthetic routes toward higher ketones and engineering secondary alcohol dehydrogenases.
The first isopropanol production in microorganisms other than clostridia was reported using engineered E. coli strains. Hanai et al. (61) produced ca. 5 g/liter of isopropanol by an engineered E. coli B strain after overexpressing the thl and adc genes from C. acetobutylicum ATCC 824 and the endogenous atoDA genes encoding CoA transferase. Even though their result suggested that the atoDA genes were better than C. acetobutylicum ctfAB genes, Jojima et al. (62) produced about 13.6 g/liter of isopropanol using the C. acetobutylicum ctfAB genes in E. coli JM109. The fed-batch fermentation of the final strain developed by Hanai et al. (61) coupled with in situ recovery by gas stripping allowed production of 143 g/liter of isopropanol with a yield of 0.23 g/g glucose in 240 h (63). In a recent study, this strain was further engineered to utilize cellobiose, and about 4.1 g/liter of isopropanol was produced from 50 g/liter of cellobiose in shaking-flask cultivation (64). Even though isopropanol could be produced to a high titer by this engineered E. coli strain, incomplete conversion of acetone might cause a problem in downstream processes. To solve this problem, the expression levels of heterologous genes and the redox balance, in particular that of NADPH required as the cofactor of the secondary alcohol dehydrogenase, should be optimized.
Isopropanol can be used as a fuel additive since it has a higher octane rate (118) than 1-butanol. In this context, an interesting idea was generated to convert acetone into isopropanol in the acetone-butanol-ethanol (ABE) fermentation of clostridia; by doing so, isopropanol-butanol-ethanol (IBE) fuel mixture can be produced by one-step fermentation. In a C. acetobutylicum PJC4BK buk-inactivated strain, Lee et al. (65) overexpressed the primary/secondary alcohol dehydrogenase from Clostridium beijerinckii NRRL B-593 (encoded by adhB-593) and endogenous acetone-producing enzymes. The engineered C. acetobutylicum PJC4BK strain was able to produce 20.4 g/liter of IBE mixture. Fed-batch fermentation coupled with in situ gas stripping allowed production of ca. 35 g/liter of IBE mixture (4.1, 6.3, and 25.1 g/liter of isopropanol, ethanol, and 1-butanol, respectively) from 133 g/liter of glucose. A similar result was obtained by an independent study employing different promoters in gene overexpression (66). In another study, the adhB-593 gene was overexpressed in a 1-butanol-tolerant mutant C. acetobutylicum Rh8 strain (67). The resulting strain produced 23.9 g/liter of IBE mixture containing 7.6 and 15 g/liter of isopropanol and 1-butanol, respectively. Jang et al. (68) also reported the use of a C. acetobutylicum mutant that overproduces butanol and ethanol. The engineered C. acetobutylicum BKM19 strain with the introduced adhB-593 and hydG genes was capable of producing butanol and ethanol to higher titers (69). The resulting strain produced 27.9 g/liter of IBE mixture containing 3.6, 14.8, and 9.5 g/liter of isopropanol, 1-butanol, and ethanol, respectively, in pilot-scale batch fermentation. These results suggest that isopropanol with or without other alcohols can be efficiently produced by metabolic engineering of E. coli or C. acetobutylicum.
CONCLUSION
Limited fossil fuel resources and increasing environmental concerns have been urging us to develop platform technologies for the sustainable and economical production of alternative fuels. In order to develop economically competitive bioprocesses for their production, the metabolic pathways need to be optimally engineered by designing the best pathways to increase the metabolic flux toward the desired product, improving the kinetics and substrate specificities of the enzymes involved, and balancing the cofactors and redox. As described above, several higher alcohols can be efficiently produced by employing metabolically engineered microorganisms. It is expected that more successful examples of microbial higher-alcohol production will appear through the strain development integrated with bioprocess engineering. Metabolic engineering will keep playing a key role in developing such economically competitive bioprocesses.
ACKNOWLEDGMENTS
This work was supported by the Advanced Biomass Research and Development Center of Korea (NRF-2010-0029799) through the Global Frontier Research Program of the Ministry of Science, ICT, and Future Planning through the National Research Foundation.
Footnotes
Citation Choi YJ, Lee J, Jang Y-S, Lee SY. 2014. Metabolic engineering of microorganisms for the production of higher alcohols. mBio 5(5):01524-14. doi:10.1128/mBio.01524-14.
REFERENCES
- 1. Tracy BP. 2012. Improving butanol fermentation to enter the advanced biofuel market. mBio 3(6):e00518-12. 10.1128/mBio.00518-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jang YS, Lee J, Malaviya A, Seung DY, Cho JH, Lee SY. 2012. Butanol production from renewable biomass: rediscovery of metabolic pathways and metabolic engineering. Biotechnol. J 7:186–198. 10.1002/biot.201100059 [DOI] [PubMed] [Google Scholar]
- 3. Dürre P. 2011. Fermentative production of butanol—the academic perspective. Curr. Opin. Biotechnol. 22:331–336. 10.1016/j.copbio.2011.04.010 [DOI] [PubMed] [Google Scholar]
- 4. Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. 2008. Fermentative butanol production by Clostridia. Biotechnol. Bioeng. 101:209–228. 10.1002/bit.22003 [DOI] [PubMed] [Google Scholar]
- 5. Tummala SB, Welker NE, Papoutsakis ET. 2003. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J. Bacteriol. 185:1923–1934. 10.1128/JB.185.6.1923-1934.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang Y, Xu C, Dong F, Yang Y, Jiang W, Yang S. 2009. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 11:284–291. 10.1016/j.ymben.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 7. Han B, Gopalan V, Ezeji TC. 2011. Acetone production in solventogenic Clostridium species: new insights from non-enzymatic decarboxylation of acetoacetate. Appl. Microbiol. Biotechnol. 91:565–576. 10.1007/s00253-011-3276-5 [DOI] [PubMed] [Google Scholar]
- 8. Jang YS, Lee JY, Lee J, Park JH, Im JA, Eom MH, Lee J, Lee SH, Song H, Cho JH, Seung DY, Lee SY. 2012. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. mBio 3(5):e00314-12. 10.1128/mBio.00314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hou X, Peng W, Xiong L, Huang C, Chen X, Chen X, Zhang W. 2013. Engineering Clostridium acetobutylicum for alcohol production. J. Biotechnol. 166:25–33. 10.1016/j.jbiotec.2013.04.013 [DOI] [PubMed] [Google Scholar]
- 10. Zhu L, Dong H, Zhang Y, Li Y. 2011. Engineering the robustness of Clostridium acetobutylicum by introducing glutathione biosynthetic capability. Metab. Eng. 13:426–434. 10.1016/j.ymben.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 11. Cornillot E, Nair RV, Papoutsakis ET, Soucaille P. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nair RV, Papoutsakis ET. 1994. Expression of plasmid-encoded aad in Clostridium acetobutylicum M5 restores vigorous butanol production. J. Bacteriol. 176:5843–5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sillers R, Chow A, Tracy B, Papoutsakis ET. 2008. Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab. Eng. 10:321–332. 10.1016/j.ymben.2008.07.005 [DOI] [PubMed] [Google Scholar]
- 14. Lee JY, Jang YS, Lee J, Papoutsakis ET, Lee SY. 2009. Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnol. J. 4:1432–1440. 10.1002/biot.200900142 [DOI] [PubMed] [Google Scholar]
- 15. Fontaine L, Meynial-Salles I, Girbal L, Yang X, Croux C, Soucaille P. 2002. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:821–830. 10.1128/JB.184.3.821-830.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu M, Zhang Y, Tang IC, Yang ST. 2011. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production. Metab. Eng. 13:373–382. 10.1016/j.ymben.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 17. Yu M, Du Y, Jiang W, Chang WL, Yang ST, Tang IC. 2012. Effects of different replicons in conjugative plasmids on transformation efficiency, plasmid stability, gene expression and n-butanol biosynthesis in Clostridium tyrobutyricum. Appl. Microbiol. Biotechnol. 93:881–889. 10.1007/s00253-011-3736-y [DOI] [PubMed] [Google Scholar]
- 18. Bond-Watts BB, Bellerose RJ, Chang MC. 2011. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 7:222–227. 10.1038/nchembio.537 [DOI] [PubMed] [Google Scholar]
- 19. Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC. 2011. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 77:2905–2915. 10.1128/AEM.03034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berríos-Rivera SJ, Bennett GN, San KY. 2002. Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD(+)-dependent formate dehydrogenase. Metab. Eng. 4:217–229. 10.1006/mben.2002.0227 [DOI] [PubMed] [Google Scholar]
- 21. Lee SY. 1996. Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 49:1–14. [DOI] [PubMed] [Google Scholar]
- 22. Lan EI, Liao JC. 2012. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 109:6018–6023. 10.1073/pnas.1200074109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dekishima Y, Lan EI, Shen CR, Cho KM, Liao JC. 2011. Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J. Am. Chem. Soc. 133:11399–11401. 10.1021/ja203814d [DOI] [PubMed] [Google Scholar]
- 24. Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. 2011. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476:355–359. 10.1038/nature10333 [DOI] [PubMed] [Google Scholar]
- 25. Clomburg JM, Vick JE, Blankschien MD, Rodriguez-Moya M, Gonzalez R. 2012. A synthetic biology approach to engineer a functional reversal of the beta-oxidation cycle. ACS Synth. Biol. 1:541–554. 10.1021/sb3000782 [DOI] [PubMed] [Google Scholar]
- 26. Runguphan W, Keasling JD. 2014. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 21:103–113. 10.1016/j.ymben.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 27. Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562. 10.1038/nature08721 [DOI] [PubMed] [Google Scholar]
- 28. Hofvander P, Doan TT, Hamberg M. 2011. A prokaryotic acyl-CoA reductase performing reduction of fatty acyl-CoA to fatty alcohol. FEBS Lett. 585:3538–3543. 10.1016/j.febslet.2011.10.016 [DOI] [PubMed] [Google Scholar]
- 29. Youngquist JT, Schumacher MH, Rose JP, Raines TC, Politz MC, Copeland MF, Pfleger BF. 2013. Production of medium chain length fatty alcohols from glucose in Escherichia coli. Metab. Eng. 20:177–186. 10.1016/j.ymben.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu A, Tan X, Yao L, Lu X. 2013. Fatty alcohol production in engineered E. coli expressing Marinobacter fatty acyl-CoA reductases. Appl. Microbiol. Biotechnol. 97:7061–7071. 10.1007/s00253-013-5027-2 [DOI] [PubMed] [Google Scholar]
- 31. Akhtar MK, Turner NJ, Jones PR. 2013. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. U. S. A. 110:87–92. 10.1073/pnas.1216516110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Venkitasubramanian P, Daniels L, Rosazza JP. 2007. Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme. J. Biol. Chem. 282:478–485. 10.1074/jbc.M607980200 [DOI] [PubMed] [Google Scholar]
- 33. Liu R, Zhu F, Lu L, Fu A, Lu J, Deng Z, Liu T. 2014. Metabolic engineering of fatty acyl-ACP reductase-dependent pathway to improve fatty alcohol production in Escherichia coli. Metab. Eng. 22:10–21. 10.1016/j.ymben.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 34. Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. 10.1038/nature06450 [DOI] [PubMed] [Google Scholar]
- 35. Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC. 2010. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 85:651–657. 10.1007/s00253-009-2085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baez A, Cho KM, Liao JC. 2011. High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 90:1681–1690. 10.1007/s00253-011-3173-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cann AF, Liao JC. 2008. Production of 2-methyl-1-butanol in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 81:89–98. 10.1007/s00253-008-1631-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Connor MR, Cann AF, Liao JC. 2010. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Appl. Microbiol. Biotechnol. 86:1155–1164. 10.1007/s00253-009-2401-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi YJ, Park JH, Kim TY, Lee SY. 2012. Metabolic engineering of Escherichia coli for the production of 1-propanol. Metab. Eng. 14:477–486. 10.1016/j.ymben.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 40. Atsumi S, Liao JC. 2008. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl. Environ. Microbiol. 74:7802–7808. 10.1128/AEM.02046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li S, Wen J, Jia X. 2011. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl. Microbiol. Biotechnol. 91:577–589. 10.1007/s00253-011-3280-9 [DOI] [PubMed] [Google Scholar]
- 42. Li S, Jia X, Wen J. 2012. Improved 2-methyl-1-propanol production in an engineered Bacillus subtilis by constructing inducible pathways. Biotechnol. Lett. 34:2253–2258. 10.1007/s10529-012-1041-1 [DOI] [PubMed] [Google Scholar]
- 43. Higashide W, Li Y, Yang Y, Liao JC. 2011. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl. Environ. Microbiol. 77:2727–2733. 10.1128/AEM.02454-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith KM, Cho KM, Liao JC. 2010. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87:1045–1055. 10.1007/s00253-010-2522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77:3300–3310. 10.1128/AEM.02972-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu J, Brigham CJ, Gai CS, Sinskey AJ. 2012. Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha. Appl. Microbiol. Biotechnol. 96:283–297. 10.1007/s00253-012-4320-9 [DOI] [PubMed] [Google Scholar]
- 47. Li H, Opgenorth PH, Wernick DG, Rogers S, Wu TY, Higashide W, Malati P, Huo YX, Cho KM, Liao JC. 2012. Integrated electromicrobial conversion of CO2 to higher alcohols. Science 335:1596. 10.1126/science.1217643 [DOI] [PubMed] [Google Scholar]
- 48. Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K. 2011. Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol. Biofuels 4:21. 10.1186/1754-6834-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matsuda F, Ishii J, Kondo T, Ida K, Tezuka H, Kondo A. 2013. Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance. Microb. Cell. Fact. 12:119. 10.1186/1475-2859-12-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Avalos JL, Fink GR, Stephanopoulos G. 2013. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat. Biotechnol. 31:335–341. 10.1038/nbt.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krause FS, Blombach B, Eikmanns BJ. 2010. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl. Environ. Microbiol. 76:8053–8061. 10.1128/AEM.01710-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang K, Sawaya MR, Eisenberg DS, Liao JC. 2008. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. U. S. A. 105:20653–20658. 10.1073/pnas.0807157106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. George HA, Johnson JL, Moore WE, Holdeman LV, Chen JS. 1983. Acetone, isopropanol, and butanol production by Clostridium beijerinckii (syn. Clostridium butylicum) and Clostridium aurantibutyricum. Appl. Environ. Microbiol. 45:1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ismaiel AA, Zhu CX, Colby GD, Chen JS. 1993. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J. Bacteriol. 175:5097–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen JS. 1995. Alcohol dehydrogenase: multiplicity and relatedness in the solvent-producing clostridia. FEMS Microbiol. Rev. 17:263–273. 10.1111/j.1574-6976.1995.tb00210.x [DOI] [PubMed] [Google Scholar]
- 56. Chen J-S, Hiu SF. 1986. Acetone-butanol-isopropanol production by Clostridium beijerinckii (synonym, Clostridium butylicum). Biotechnol. Lett. 8:371–376. 10.1007/BF01040869 [DOI] [Google Scholar]
- 57. Hiu SF, Zhu CX, Yan RT, Chen JS. 1987. Butanol-ethanol dehydrogenase and butanol-ethanol-isopropanol dehydrogenase: different alcohol dehydrogenases in two strains of Clostridium beijerinckii (Clostridium butylicum). Appl. Environ. Microbiol. 53:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoneda H, Tantillo DJ, Atsumi S. 2014. Biological production of 2-butanone in Escherichia coli. ChemSusChem 7:92–95. 10.1002/cssc.201300853 [DOI] [PubMed] [Google Scholar]
- 60. Lan EI, Dekishima Y, Chuang DS, Liao JC. 2013. Metabolic engineering of 2-pentanone synthesis in Escherichia coli. AIChE J. 59:3167–3175. 10.1002/aic.14086 [DOI] [Google Scholar]
- 61. Hanai T, Atsumi S, Liao JC. 2007. Engineered synthetic pathway for isopropanol production in Escherichia coli. Appl. Environ. Microbiol. 73:7814–7818. 10.1128/AEM.01140-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jojima T, Inui M, Yukawa H. 2008. Production of isopropanol by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:1219–1224. 10.1007/s00253-007-1246-8 [DOI] [PubMed] [Google Scholar]
- 63. Inokuma K, Liao JC, Okamoto M, Hanai T. 2010. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 110:696–701. 10.1016/j.jbiosc.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 64. Soma Y, Inokuma K, Tanaka T, Ogino C, Kondo A, Okamoto M, Hanai T. 2012. Direct isopropanol production from cellobiose by engineered Escherichia coli using a synthetic pathway and a cell surface display system. J. Biosci. Bioeng. 114:80–85. 10.1016/j.jbiosc.2012.02.019 [DOI] [PubMed] [Google Scholar]
- 65. Lee J, Jang YS, Choi SJ, Im JA, Song H, Cho JH, Seung DY, Papoutsakis ET, Bennett GN, Lee SY. 2012. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for isopropanol-butanol-ethanol fermentation. Appl. Environ. Microbiol. 78:1416–1423. 10.1128/AEM.06382-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dusséaux S, Croux C, Soucaille P, Meynial-Salles I. 2013. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for the high-yield production of a biofuel composed of an isopropanol/butanol/ethanol mixture. Metab. Eng. 18:1–8. 10.1016/j.ymben.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 67. Dai Z, Dong H, Zhu Y, Zhang Y, Li Y, Ma Y. 2012. Introducing a single secondary alcohol dehydrogenase into butanol-tolerant Clostridium acetobutylicum Rh8 switches ABE fermentation to high level IBE fermentation. Biotechnol. Biofuels 5:44. 10.1186/1754-6834-5-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jang YS, Malaviya A, Lee J, Im JA, Lee SY, Lee J, Eom MH, Cho JH, Seung DY. 2013. Metabolic engineering of Clostridium acetobutylicum for the enhanced production of isopropanol-butanol-ethanol fuel mixture. Biotechnol. Prog. 29:1083–1088. 10.1002/btpr.1733 [DOI] [PubMed] [Google Scholar]
- 69. Jang YS, Malaviya A, Lee SY. 2013. Acetone-butanol-ethanol production with high productivity using Clostridium acetobutylicum BKM19. Biotechnol. Bioeng. 110:1646–1653. 10.1002/bit.24843 [DOI] [PubMed] [Google Scholar]
- 70. Shen CR, Liao JC. 2012. Photosynthetic production of 2-methyl-1-butanol from CO2 in cyanobacterium Synechococcus elongatus PCC7942 and characterization of the native acetohydroxyacid synthase. Energy Environ. Sci. 5:9574–9583. 10.1039/c2ee23148d [DOI] [Google Scholar]