ABSTRACT
The plant pathogen Pseudomonas syringae pv. syringae B728a grows and survives on leaf surfaces and in the leaf apoplast of its host, bean (Phaseolus vulgaris). To understand the contribution of distinct regulators to B728a fitness and pathogenicity, we performed a transcriptome analysis of strain B728a and nine regulatory mutants recovered from the surfaces and interior of leaves and exposed to environmental stresses in culture. The quorum-sensing regulators AhlR and AefR influenced few genes in planta or in vitro. In contrast, GacS and a downstream regulator, SalA, formed a large regulatory network that included a branch that regulated diverse traits and was independent of plant-specific environmental signals and a plant signal-dependent branch that positively regulated secondary metabolite genes and negatively regulated the type III secretion system. SalA functioned as a central regulator of iron status based on its reciprocal regulation of pyoverdine and achromobactin genes and also sulfur uptake, suggesting a role in the iron-sulfur balance. RetS functioned almost exclusively to repress secondary metabolite genes when the cells were not on leaves. Among the sigma factors examined, AlgU influenced many more genes than RpoS, and most AlgU-regulated genes depended on RpoN. RpoN differentially impacted many AlgU- and GacS-activated genes in cells recovered from apoplastic versus epiphytic sites, suggesting differences in environmental signals or bacterial stress status in these two habitats. Collectively, our findings illustrate a central role for GacS, SalA, RpoN, and AlgU in global regulation in B728a in planta and a high level of plasticity in these regulators’ responses to distinct environmental signals.
IMPORTANCE
Leaves harbor abundant microorganisms, all of which must withstand challenges such as active plant defenses and a highly dynamic environment. Some of these microbes can influence plant health. Despite knowledge of individual regulators that affect the fitness or pathogenicity of foliar pathogens, our understanding of the relative importance of various global regulators to leaf colonization is limited. Pseudomonas syringae strain B728a is a plant pathogen and a good colonist of both the surfaces and interior of leaves. This study used global transcript profiles of strain B728a to investigate the complex regulatory network of putative quorum-sensing regulators, two-component regulators, and sigma factors in cells colonizing the leaf surface and leaf interior under stressful in vitro conditions. The results highlighted the value of evaluating these networks in planta due to the impact of leaf-specific environmental signals and suggested signal differences that may enable cells to differentiate surface versus interior leaf habitats.
INTRODUCTION
Pseudomonas syringae has been found in diverse habitats (1), with extensive multiplication occurring primarily during its association with plants. Many pathovars of P. syringae cause foliar diseases that manifest as necrotic spots. The P. syringae pv. syringae B728a strain, which causes bacterial spot of bean (Phaseolus vulgaris L.), exhibits particularly pronounced growth on leaf surfaces, designated epiphytic sites, as well as in the intercellular spaces of leaves, known as the apoplast. In a recent study, we demonstrated differences in the global transcript profiles of B728a cells during growth in epiphytic versus apoplastic sites, suggesting that P. syringae encounters distinct environments in these two plant habitats (2). The transcript profiles suggested that B728a cells experience a spatially heterogeneous nutritional habitat in epiphytic sites, thus requiring active motility and chemotaxis for relocation, compared to a nutritionally uniform environment in the apoplast where these traits were repressed. They also indicated habitat-specific differences in strategies for coping with plant defenses and water limitation. This habitat specificity of the global transcript profiles implies the involvement of distinct regulatory networks in each leaf habitat.
Like most pseudomonads, the genome of P. syringae B728a encodes a large number of transcriptional regulators consistent with the need to adapt to diverse environments during its life cycle. Here, our goal was to begin to identify the major regulatory networks that operate in strain B728a during colonization of the surfaces and apoplast of leaves, with the hope of better understanding their relative importance during the epiphytic and pathogenic stages of the P. syringae life cycle. The portfolio of genes controlled by bacterial regulators, such as quorum-sensing regulators, sigma factors, and two-component systems, is often characterized in a single environment, despite the possible influence of environmental signals on regulon content. The two-component activator GacS/GacA is among the most widely studied of the global regulators in Pseudomonas spp. GacS was discovered in strain B728a based on its role in inducing disease (3), with GacA later identified as a regulator of secreted secondary metabolites and enzymes in Pseudomonas protegens (4). GacS/GacA commonly contributes to the virulence and biocontrol activities of Pseudomonas spp., and in some strains, to motility and the oxidative stress response. The environmental signals activating GacS are not known. The regulatory mechanism involves GacS-mediated phosphorylation of GacA, enabling GacA-mediated transcriptional activation of genes encoding small RNAs (sRNAs), which bind to translation repressors and relieve repression of translational initiation (5). In strain B728a, GacS/GacA regulates production of the phytotoxins syringomycin and syringopeptin, a quorum signal molecule, and the exopolysaccharide (EPS) alginate, swarming motility, and the transcriptional activator SalA (6–8). SalA activates genes for syringomycin production and lesion formation in other P. syringae pv. syringae strains as well (9, 10). GacS/GacA- and SalA-regulated genes in P. syringae have been examined in culture but not yet in planta, despite the fact that plant products enhance the expression of the SalA-regulated syringomycin and syringopeptin genes (11, 12). Recently, the hybrid two-component sensor-kinase RetS was found to regulate EPS production and the type VI secretion system (T6SS) in a manner opposite that of GacS/GacA in B728a (13), suggesting that it is reciprocal to GacS/GacA in its regulatory role.
Among the 15 sigma factors in strain B728a, 10 are extracytoplasmic function sigma (ECF-σ) factors. The best-characterized ECF-σ factor in P. syringae, HrpL, has a central role in virulence due to its activation of genes encoding the type III secretion system (T3SS) and effector proteins that help suppress the plant’s defenses during infection. HrpL-regulated genes have been widely studied in P. syringae, including in strain B728a (14), and clearly contribute to growth in the apoplast and in epiphytic sites (15). Among the remaining ECF-σ factors, several are involved in responding to iron limitation (16, 17), but only AlgU, also known as AlgT, has been shown to contribute to P. syringae fitness on leaves (18). AlgU was discovered as an activator of alginate biosynthetic genes (19) but regulates a broad range of genes in response to cell envelope stress (20). The finding that B728a cells are exposed to envelope stress in the form of water limitation in both epiphytic and apoplastic sites (2) suggests a major role for AlgU in the regulatory networks active on leaves. Like AlgU, the sigma factor RpoS has a role in bacterial stress tolerance, including in pseudomonads (21), although its role in the stress tolerance and in planta fitness of strain B728a has not been examined. A final sigma factor, RpoN, is distinct from other sigma factors in that it requires one or more transcriptional activators for regulation, whereas other sigma factors may or may not involve such activators. RpoN was identified based on its activity during nitrogen starvation, but a preliminary analysis indicated that in B728a, as in other bacteria, RpoN regulates genes with diverse functions (22). Moreover, these functions are likely relevant to growth in planta based on RpoN regulation of the phytotoxin coronatine (23) and HrpL, and thus the T3SS (24), in other P. syringae pathovars.
Quorum regulation also has the potential to be a major form of regulation in planta. The perception of diffusible signal molecules is, in fact, enhanced on leaf surfaces due to restricted diffusion, as evidenced by the requirement for only a small quorum size for signal-mediated activation (25). P. syringae B728a uses the LuxI/LuxR homologs designated AhlI/AhlR to produce and respond to the signal molecule 3-oxo-hexanoyl-homoserine lactone (3-oxo-C6-HSL) (26). Loss of 3-oxo-C6-HSL synthesis was associated with reduced epiphytic fitness as well as reduced alginate production and increased swarming in culture (26, 27). Production of 3-oxo-C6-HSL was enhanced by the transcriptional activator AefR in B728a and other P. syringae strains (26, 28–30). Despite the evidence for quorum-sensing regulation in strain B728a and for regulation of large numbers of genes by many bacterial quorum-sensing regulators, we do not yet have a comprehensive picture of the breadth of genes and traits subject to this regulation in B728a in culture or in planta.
Here, we performed a global transcriptome analysis of P. syringae pv. syringae B728a and nine B728a mutants deficient in the regulators that we predicted to have the greatest role in growth in planta, namely, GacS, SalA, RetS, HrpL, AlgU, RpoS, RpoN, AhlR, and AefR. We compared the transcriptomes of cells recovered from epiphytic sites and apoplastic sites to better understand their relative contributions at these distinct leaf sites. Last, we compared the transcriptomes of these strains when unstressed or exposed to four environmental stresses predicted to occur in the phyllosphere, thus elucidating the roles of environmental signals on the genes influenced by each regulator.
RESULTS AND DISCUSSION
The sizes of the regulons indicated that RpoN, GacS, SalA, and AlgU influenced the greatest number of genes in planta among the regulators examined.
We constructed nonpolar deletion mutations in each of the target regulatory genes ahlR, aefR, salA, rpoS, algU, hrpL, and rpoN and used nonpolar deletion mutations in retS and gacS that were described previously (13). We examined the global gene expression profiles of P. syringae B728a and these nine mutants under seven environmental conditions: on bean leaf surfaces after incubation for 24 h under moist conditions and 48 h under dry conditions (i); in the leaf apoplast after a 48-h incubation (ii); in log-phase cells grown in a basal medium (iii); in log-phase cells grown in a basal medium followed by 15 min of exposure to osmotic stress (iv) or oxidative stress (v) or followed by 2 h of starvation for nitrogen (vi) or iron (vii), as described previously (2). Each treatment was performed using two biological replicates. P. syringae B728a and ∆ahlR and ∆aefR mutants were examined in one laboratory, designated lab I. P. syringae B728a and ∆retS, ∆gacS, and ∆salA mutants were examined in lab II, and strain B728a and ∆rpoS, ∆algU, ∆hrpL, and ∆rpoN mutants were examined in lab III. Transcriptome data were generated based on hybridization of cDNA derived from total RNA to a B728a microarray, with subsequent calculation of robust estimated mean values for the fluorescence intensity measurements for each open reading frame (ORF) on the array, which was then subjected to linear models for microarray data analysis, as described previously (2).
For each regulator, the set of genes that significantly differed in transcript levels between the regulatory mutant and the wild type was identified in each of the environmental conditions tested. The criterion used for differential expression was a false discovery rate of 1% (i.e., q value of <0.01). Each regulon, defined as a collection of genes that were directly or indirectly affected by a given regulator, was estimated by the union of the sets of identified differentially expressed genes from each of the seven environmental conditions. The regulon sizes, shown as the total number of unique genes (Table 1), ranged from only 9 genes for AhlR to 3,635 genes, or 71% of the P. syringae B728a ORFs examined for RpoN. The magnitude of the differences in the sizes of these regulons was not expected, with the greatest surprise being the small sizes of the regulons influenced by AhlR, AefR, and HrpL; these findings will be discussed below. The minimal regulon, i.e., the genes that were differentially expressed under every condition, was very small for each regulator, suggesting a major impact of environmental conditions on the composition of these regulons.
TABLE 1 .
Number of genes that differed in transcript levels between mutants lacking individual regulators and the wild type under seven environmental conditionsa
| Regulator | No. of genes that differed under environmental conditionb: |
Total unique genesc |
Minimal regulond |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Basal | NaCl | H2O2 | Low Fe | Low N | Epi | Apo | |||
| AhlR | 1 | 3 | 3 | 7 | 0 | 5 | 9 | 9 | 0 |
| AefR | 5 | 9 | 7 | 9 | 6 | 8 | 20 | 29 | 3 |
| GacS | 320 | 311 | 920 | 9 | 1,643 | 360 | 28 | 2,305 | 3 |
| SalA | 243 | 273 | 307 | 134 | 633 | 234 | 22 | 990 | 4 |
| RetS | 33 | 12 | 12 | 0 | 1 | 14 | 0 | 43 | 0 |
| RpoS | 1 | 3 | 3 | 44 | 162 | 104 | 2 | 213 | 0 |
| HrpL | 3 | 3 | 3 | 5 | 14 | 8 | 0 | 16 | 0 |
| AlgU | 63 | 810 | 19 | 12 | 29 | 272 | 34 | 866 | 6 |
| RpoN | 1,344 | 2,171 | 900 | 1,248 | 801 | 733 | 22 | 3,635 | 9 |
The regulatory genes that were deleted in the mutants were excluded from the gene counts; differences were determined based on a q value of<0.01.
The number of genes that differed in transcript levels between mutants lacking individual regulators and the wild type under environmental conditions. The conditions included the basal medium (Basal), osmotic stress (NaCl), oxidative stress (H2O2), iron starvation (Low Fe), nitrogen starvation (Low N), epiphytic sites (Epi), and apoplastic sites (Apo).
The total number of genes influenced by the regulator in at least one environmental condition.
The total number of genes that exhibited significant differences in every one of the environmental conditions.
The subset of each regulon that was differentially influenced by the environmental conditions was also determined by using a contrast analysis to identify the genes for which the difference in transcript levels between the mutant and the wild type in a given environmental condition was distinct from that same difference in the basal medium (see Table S1 in the supplemental material). In contrast to identifying genes that differed between a mutant and a wild type under a given condition (Table 1), this conservative approach minimized the inclusion of genes for which the differential expression in a given environmental condition was due to random variation in gene expression by either the mutant or the wild type. Here, we considered the size of the regulon to be the sum of these genes and those differing in transcript abundance between the mutant and the wild type in the basal medium (Table S1). Greater than 90% of the genes in the RpoS and AlgU regulons exhibited differential expression in a single condition, namely, nitrogen starvation and osmotic stress, respectively, whereas <60% of the genes in the GacS, SalA, and RpoN regulons were differentially expressed under any single condition; this finding indicates that, among the regulons comprised of at least 15 genes, the GacS, SalA, and RpoN regulons were especially impacted by multiple environmental conditions. Moreover, although environmental regulation of individual genes could be overlooked if the genes were not expressed in the wild type in a given environment, with the exception of the SalA-regulated genes under low-Fe conditions, >80% of the genes in the GacS and SalA regulons were expressed at detectable levels in strain B728a under all of the conditions tested, supporting the conclusion that the composition of these regulons is strongly influenced by the environment.
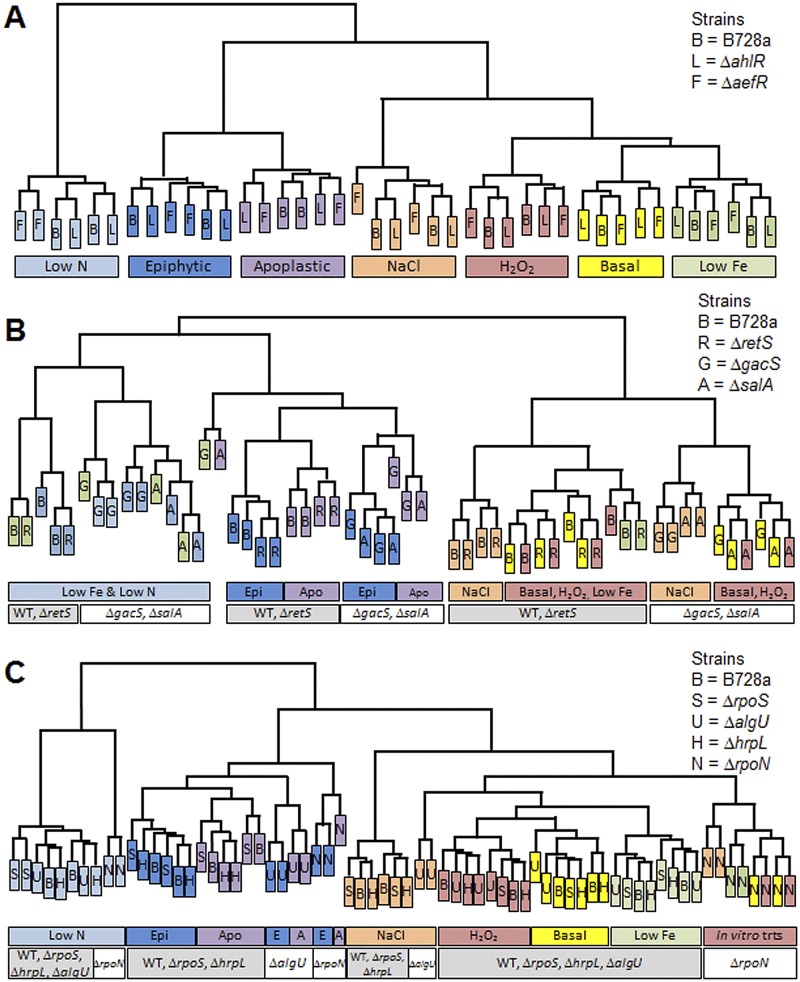
We performed hierarchical clustering, as described in Materials and Methods, to evaluate the similarity in global transcript profiles among the samples examined in each lab. As seen previously (2), the transcript profiles for the wild-type B728a data combined across the labs showed that N starvation, growth in planta, and osmotic stress had the largest impacts on the transcriptome profiles (Fig. 1A). The ∆ahlR and ∆aefR mutants were similar to strain B728a in their responses to these environmental conditions (Fig. 1A). The data from the second lab showed that the ΔretS mutant clustered with the wild type (Fig. 1B), indicating that, like AhlR and AefR, RetS has a minimal impact on the transcriptome under these conditions. In contrast, the transcriptome profiles of ∆gacS and ∆salA mutants diverged from that of the wild type under every environmental condition, including both in planta treatments (Fig. 1B), illustrating a major role for these two regulators under every condition tested. The data from the third lab showed that the transcriptome profiles of the ΔrpoN mutant diverged from those of the wild type in all of the treatments (Fig. 1C), illustrating its key global role in sensing the environment, whereas the ΔalgU mutant diverged only in response to osmotic stress and the in planta treatments (Fig. 1C), confirming the specificity of the AlgU response to hyperosmotic stress, as indicated in Table 1 and Table S1 in the supplemental material. The global transcriptome profiles for all of the mutants were distinct for cells recovered from epiphytic versus apoplastic sites, as they were for the wild type (2), consistent with the distinct nature of these habitats and the potential for distinct roles for these regulators in each habitat.
FIG 1 .
The regulators varied in their impact on the global transcriptome in an environmental context-dependent manner. (A to C) Hierarchical clustering was performed on the 500 genes that had the lowest P values in an F test for the combined effect among all of the samples within the data set from lab I (A), lab II (B), and lab III (C). The letters indicate the strains, as shown in the keys in the panels, and the colors indicate the in vitro treatments, including the basal medium (Basal), osmotic stress (NaCl), oxidative stress (H2O2), iron starvation (Low Fe), and nitrogen starvation (Low N), and the in planta treatments, including cells from epiphytic (Epi) and apoplastic (Apo) sites. The analysis was performed using the fluorescence intensities for each of the biological replicates, and the replicates are similarly labeled; values that are missing are described in the legend to Fig. S1. WT, wild-type strain B728a.
AefR and AhlR form a regulatory network comprised of surprisingly few genes.
In Pseudomonas aeruginosa, Sinorhizobium meliloti, and Burkholderia spp., quorum-sensing regulons of N-(β-ketocaproyl)-l-homoserine lactone (AHL)-dependent LuxR homologs were comprised of >300 genes (31–34), although considerable diversity can exist among strains (35). Based on transcript levels in the ΔahlR mutant, which was confirmed to exhibit reduced production of the 3-oxo-C6-HSL quorum-sensing signal (R. A. Scott and S. E. Lindow, unpublished data), AhlR regulated only nine genes under the conditions tested (Table 1), and all of these genes were located near the ahlR locus (Fig. 2). The basal medium in these experiments was amended with 3-oxo-C6-HSL (10 µM) to maximize differences between the wild type and the ΔahlR mutant. The decreased expression of the AhlR regulon genes in the ΔahlR mutant compared to P. syringae B728a supports a role for AhlR as a positive regulator of these few genes (Fig. 2). The attenuated decrease in the transcript levels of the AhlR regulon genes in epiphytic sites compared to the transcript levels in the basal medium could be due to the presence of lower concentrations of quorum-sensing signals on the leaf surface than in the medium. Similarly, the larger decreases in transcript levels for the ∆ahlR mutant in the apoplastic sites could be associated with high cell densities in the apoplast. Our results do not provide mechanistic insights into the quorum-sensing regulation previously reported for alginate production and swarming motility in strain B728a (27). P. syringae pv. tomato DC3000 has orthologs of ahlI and ahlR that show a convergent orientation, as in B728a, and produces antisense transcripts of the ahlR ortholog (36). The convergent orientation of ahlI and ahlR in strain B728a (Fig. 2) and the induction of ahlI by 3-oxo-C6-HSL (26) suggest that ahlR transcript levels may be subject to quorum sensing-mediated negative feedback, which could strongly attenuate AhlR regulon expression.
FIG 2 .
Deleting ahlR or aefR influenced the transcript levels of the genes neighboring the ahlI-ahlR locus. The numbers above the arrows are the Psyr locus numbers, and those below are the fold change values in transcript abundance for the indicated mutant relative to the wild-type strain B728a (WT) in cells from each of three environmental conditions. The predicted binding site for AhlR complexed with 3-oxo-C6-HSL is shown. The predicted functions of these genes are shown in Table S2 in the supplemental material.
AefR was a positive regulator of genes in the AhlR regulon, consistent with its identification as a positive regulator of 3-oxo-C6-HSL production (26); this regulation was observed only in planta (Fig. 2). AefR also regulated 20 genes beyond those in the AhlR regulon (Table 1), among which was a notable repression of two efflux systems, MexEF-OprN (Psyr_2967 to -2969) and MexAB-OprM (Psyr_4007 to -4009), for which the transcripts were increased an average of 11- and 3-fold, respectively, across the seven treatments in the ∆aefR mutant relative to the wild type (see Table S2 in the supplemental material). These efflux pumps were identified in P. aeruginosa as RND (resistance-nodulation-cell division) family multidrug efflux pumps. MexAB-OprM in strain B728a is known to contribute to fitness on leaves and tolerance to antibacterial compounds (37). Although this pump effectively exports phenolic antimicrobial compounds like those associated with plant defenses (37), the role of AefR in repressing these genes is not consistent with a role for AefR in evasion of the host immune response. The AhlR-dependent subset of the AefR regulon was regulated by GacS (Table S2), as suggested previously (26), and moreover in a SalA-dependent manner (Fig. 3), but GacS/SalA did not regulate AefR repression of these efflux systems.
FIG 3 .
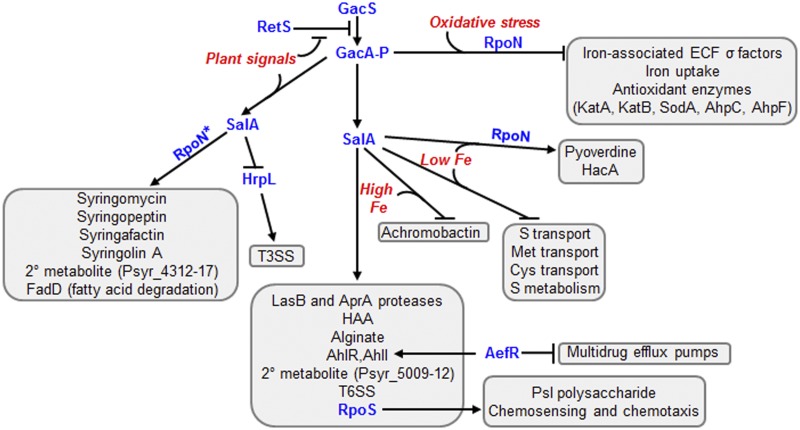
The GacS, SalA, and RetS regulatory networks in P. syringae B728a impacted diverse traits in an environmental signal-dependent manner. The regulators examined in this study are shown in blue type, regulated genes or traits are in black type, and relevant environmental signals are in red t ype. Not all traits identified in these networks are shown, and the arrows may indicate direct or indirect regulation. RpoN contributed to the coactivation or corepression of traits, as indicated. RpoN* indicates a contribution of RpoN to positive regulation in epiphytic sites and the in vitro treatments and to weaker positive regulation or even negative regulation in the apoplast. 2°, secondary.
GacS activated only one Rsm sRNA family member, RsmY.
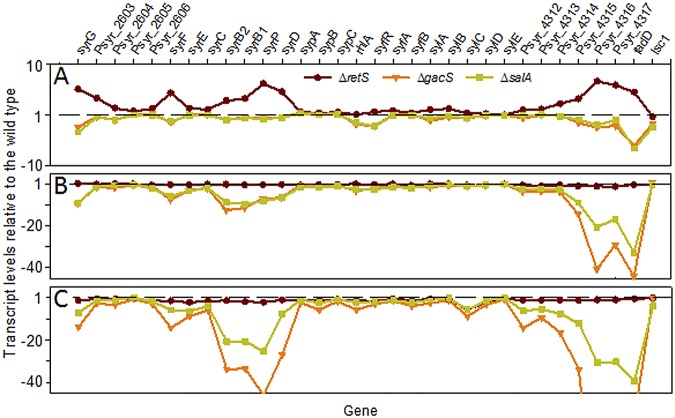
The majority of differentially expressed genes in the ΔgacS and ΔsalA mutants showed decreased expression, indicating that GacS and SalA are primarily positive regulators under all of the conditions tested. For example, 74% of the 320 differentially expressed genes in the ΔgacS mutant in the basal medium and 85% of the 243 differentially expressed genes in the ΔsalA mutant in the basal medium (Table 1) exhibited decreased expression compared to the wild type. To identify the P. syringae B728a small RNAs (sRNAs) that may be involved in GacS and SalA signal transduction, we included in the microarray design candidate sRNA genes predicted using the bioinformatic tool SIPHT (38). Out of 29 candidate sRNAs, one exhibited significantly reduced expression in the ΔgacS mutant under most of the conditions examined. This gene was identified as rsmY (Table 2) based on synteny and a 98% identity with rsmY in P. syringae pv. tomato DC3000 (39). Although the reduction of the rsmY transcripts in the ΔsalA and ΔgacS mutants suggests that, as an upstream activator of SalA (7), GacS may regulate rsmY via SalA, the wealth of studies showing direct GacS regulation of Rsm family genes in pseudomonads supports that GacS regulates rsmY directly; the ability of SalA to enhance rsmY transcript levels may be via feedback activation.
TABLE 2 .
Fold changes in the transcript abundance of selected genes due to the deletion of gacS, salA, or rpoN
| Locus | Gene | Function | Fold change in transcript abundancea |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Basal |
Epiphytic sites |
Apoplastic sites |
|||||||||
| gacS | salA | rpoN | gacS | salA | rpoN | gacS | salA | rpoN | |||
| rsmYb,c | rsmY | Regulation | −6.3 | −4.3 | 1.3 | −6.0 | −4.7 | 1.4 | −19.5 | −12.1 | 1.1 |
| rsmZc | rsmZ | Regulation | −1.5 | −1.2 | −1.1 | −1.5 | 1.2 | 1.2 | −2.0 | 2.0 | −7.5 |
| sRNA_P16 | rgsA | Regulation | 1.0 | 1.0 | −1.6 | −3.1 | −2.5 | 3.2 | −1.6 | −1.1 | −1.2 |
| Psyr_2601 | salA | Regulation | −2.2 | D | 9.1 | −9.0 | D | −1.8 | −7.6 | D | 1.6 |
| Psyr_2607 | syrF | Regulation | −2.4 | −2.3 | 44.5 | −8.5 | −6.7 | −4.7 | −15.2 | −6.8 | 2.7 |
| Psyr_2611 | syrB1 | Syringomycin synthesis | −1.5 | −1.6 | 54.7 | −12.5 | −10.5 | −5.1 | −34.5 | −21.7 | 2.8 |
| Psyr_2615 | sypB | Syringopeptin synthesis | 1.1 | 1.0 | 4.7 | −1.9 | −2.7 | −1.7 | −6.8 | −3.5 | 4.0 |
| Psyr_3041 | lasB | Protease (elastin) | −3.0 | −3.0 | 1.2 | −4.7 | −5.7 | −2.7 | −5.5 | −10.0 | −1.8 |
| Psyr_3163 | aprA | Alkaline protease | −29.5 | −22.9 | 2.6 | −28.9 | −26.8 | −1.7 | −36.1 | −41.5 | 2.8 |
| Psyr_3129 | rhlA | HAA synthesis | −3.0 | −2.5 | 15.8 | −4.3 | −3.6 | 1.6 | −6.9 | −3.3 | 1.9 |
| Psyr_2576 | syfA | Syringafactin synthesis | 1.0 | −1.1 | 3.9 | −2.0 | −2.6 | −1.2 | −2.5 | −2.3 | 6.6 |
| Psyr_2577 | syfB | Syringafactin synthesis | 1.1 | −1.2 | 1.8 | −2.5 | −2.9 | −1.2 | −5.0 | −3.4 | 6.6 |
| Psyr_2575 | syfR | Regulation | −3.2 | −3.3 | 6.4 | −3.0 | −3.6 | −1.2 | −4.1 | −2.9 | 1.6 |
| Psyr_1621 | ahlI | 3-oxo-C6-HSL synthase | −6.2 | −6.5 | 1.2 | −2.9 | −2.5 | −1.1 | −5.6 | −4.3 | −3.1 |
| Psyr_1622 | ahlR | Quorum regulator | −4.1 | −4.2 | −1.2 | −4.3 | −2.7 | 1.5 | −4.1 | −2.7 | −1.6 |
| Psyr_1059 | algE | Alginate synthesis | −3.7 | −1.8 | −3.0 | 1.3 | 1.0 | −3.6 | −1.8 | −2.7 | 4.5 |
| Psyr_3301 | pslA | Psl synthesis | −1.6 | −2.3 | 1.1 | −3.3 | −3.1 | −1.7 | −3.6 | −2.4 | 3.7 |
| Psyr_0754 | lsc1 | Levansucrase | −2.7 | −3.4 | −1.8 | 1.8 | −1.2 | −1.6 | −1.4 | −5.1 | 1.3 |
| Psyr_1704 | sylC | Syringolin A synthesis | −1.7 | −1.6 | 6.4 | −2.1 | −2.0 | −1.2 | −9.8 | −6.7 | 2.8 |
| Psyr_5009 | Secondary metabolite synthesis | −5.3 | −4.2 | 2.1 | −3.9 | −3.4 | −1.2 | −4.4 | −2.1 | 1.4 | |
| Psyr_4314 | Secondary metabolite synthesis | −1.4 | −1.3 | 4.0 | −4.8 | −3.7 | −3.9 | −17.7 | −8.7 | 2.5 | |
| Psyr_1374 | rpoS | Sigma factor | −8.7 | −7.0 | 2.3 | −7.3 | −6.1 | 1.6 | −8.8 | −10.8 | 2.5 |
| Psyr_0749 | fadD | Fatty acid metabolism | −7.1 | −7.6 | −1.2 | −45.1 | −33.8 | −2.4 | −57.1 | −40.3 | 1.1 |
| Psyr_3185 | cspD | Cold shock protein | −3.6 | −1.7 | 2.6 | −4.5 | −2.7 | 2.6 | −3.5 | −1.9 | 1.1 |
| Psyr_1217 | hrpL | T3SS regulation | −1.6 | −1.8 | 1.0 | 2.4 | 2.0 | −1.2 | 2.9 | 4.2 | −3.7 |
| Psyr_1192 | hrpA2 | T3SS pilus | −1.6 | −1.9 | −1.1 | 4.2 | 2.9 | −2.6 | 12.7 | 4.5 | −2.0 |
| Psyr_4919 | avrPto1 | T3SS effector protein | −1.1 | −2.1 | −3.0 | 10.4 | 5.2 | −8.7 | 28.1 | 8.6 | −9.6 |
| Psyr_2629 | ppkA | T6SS-associated kinase | −6.3 | −8.2 | 1.0 | −5.0 | −3.6 | −2.2 | −2.3 | −1.4 | −1.2 |
Values shown are the fold change in transcript abundance in the mutants lacking gacS, salA, or rpoN relative to transcript abundance in the wild type. The values in boldface type exhibited a significant change in transcript abundance (q value of <0.01); few genes showed significant changes in the cells from apoplastic sites due to the high variation among replicate samples in the lab II data set (see Fig. S1 in the supplemental material). For results in all of the environmental conditions evaluated, see Table S2. D, the gene was deleted in this mutant.
rsmY also showed reduced transcript abundance in the ∆gacS and ∆salA mutants in most of the stress conditions in vitro (Table S2).
Comparisons to sRNAs in other pseudomonads indicated that each of these sRNAs was smaller than those predicted using SIPHT (38), with rsmY, rsmZ, and rgsA encoded as orthologs of PSPTO_5647, PSPTO_5652, and PSPTO_5600, respectively.
We identified rsmZ based on synteny and 93% identity with P. syringae pv. tomato DC3000 rsmZ (39); however, unlike in the P. protegens, P. aeruginosa, and P. syringae pv. tomato strains examined (9, 40, 41), rsmZ in strain B728a did not show altered regulation upon loss of GacS (Table 2). Three of the candidate sRNAs that contain part or all of the rsmX3, rsmX4, and rsmX5 genes, which were predicted to be regulated by GacS/GacA (39), also did not show altered regulation in the ΔgacS mutant except in the low-N conditions (see Table S2 in the supplemental material). However, an sRNA with 68% identity to the GacA-regulated sRNA RgsA in P. aeruginosa PAO1, which does not function by sequestration of the translational inhibitor RsmA (42), was positively regulated by GacS, with this regulation specific to epiphytic sites (Table 2 and Table S2). Collectively, these results indicate that GacS/GacA regulation in strain B728a employs primarily a single member of the Rsm family of protein-binding, noncoding RNAs, namely, RsmY, with possible additional involvement of RgsA on leaves. Although RsmY-mediated regulation is primarily at the posttranscriptional level via inhibition of translational initiation, this inhibition is associated with reduced transcript stability and thus can be detected based on transcript abundance (43).
GacS and SalA activated genes that are consistent with the known phenotypes of P. syringae B728a gacS and salA mutants.
The GacS/GacA and SalA regulons have not yet been characterized in P. syringae B728a despite the fact that gacS and salA were originally identified in this strain. Although clear orthologs of SalA are absent in most pseudomonads, including other P. syringae strains, our data indicate a major role for SalA in the GacS/GacA regulatory pathway in strain B728a. SalA affected about a third of the 2,305 genes affected by GacS. Loss of gacS reduced salA transcripts 2- to 3-fold in most of the in vitro conditions but reduced salA transcripts 8- to 9-fold in planta (Table 2; see Table S2 in the supplemental material). This finding supports GacS regulation of SalA under all conditions and suggests that GacS activation of salA is enhanced by a plant-specific environmental signal, hereafter referred to as a plant signal (Fig. 3). Similarly, the plant signal-mediated enhancement of syringomycin production and syr or syp gene expression reported previously (10, 44, 45) occurs upstream of SalA in the regulatory hierarchy (Fig. 3) based on the similar impact of the in planta conditions on salA and syrF (Table 2), with SyrF functioning as a SalA-activated transcriptional regulator of syringomycin synthesis (12) (Table 2).
GacS is known to regulate extracellular protease production in strain B728a (46), and here we demonstrated that two proteases, LasB and AprA, out of 18 were regulated by GacS (Table 2). The lasB gene encodes elastase, which is a major secreted protease in P. aeruginosa and has a broad substrate specificity; the target substrates of LasB during P. syringae growth in planta are not clear. In contrast, the aprA gene encodes a secreted alkaline protease in P. aeruginosa that degrades monomers of flagellin and thus can minimize activation of flagellin monomer-induced defenses, including stomatal closure in plants (47). GacS and SalA also activated the other genes in the apr gene cluster, including the AprA secretion system encoded by aprFED (Psyr_3159 to Psyr_3161) and a periplasmic inhibitor of AprA activity encoded by aprI (Psyr_3162), which may protect intracellular flagellin monomers for flagellar assembly (47). The effects of the loss of gacS and salA on these protease genes were similar in vitro and in planta and thus were part of a plant signal-independent subset of the GacS/SalA regulon (Fig. 3), as shown for other genes using microarray analysis of SalA-targeted genes (10).
GacS regulation of swarming but not swimming motility in strain B728a (6) suggested GacS-mediated regulation of surfactant production, as observed in other pseudomonads (48, 49). Our data showed that genes for both of the surfactants synthesized by P. syringae, 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAA) and syringafactin (50), were expressed in planta (2) and were positively regulated by GacS and SalA in planta (Table 2). Whereas an HAA biosynthetic gene, rhlA, was also regulated by GacS/SalA in culture, the syringafactin genes syfAB were not, consistent with a requirement for surface signals for syfA expression (51). GacS and SalA were not involved in regulating flagellar synthesis and motility genes directly but were involved in inducing a large number of chemosensing and chemotaxis genes. This category was among the functional categories identified in which the representation of differentially expressed genes was greater than their representation among all of the B728a genes (see Table S3 in the supplemental material).
GacS regulated the 3-oxo-C6-HSL synthase gene ahlI and did so in all of the conditions tested (Table 2; see Table S2 in the supplemental material), as expected (7, 26). In contrast to previous findings (7), however, this regulation was mediated through SalA based on the decreased ahlI transcript levels in the ΔsalA mutant (Table 2). GacS also regulated biosynthetic genes for alginate production, as expected (8), and also for the two EPSs Psl and levan (Table 2 and Table S2). GacS and SalA contributed only modestly to the activation of the alginate genes in planta, but with the exception of the iron-limited conditions, contributed greatly to their regulation in vitro. The lack of regulation in the iron-limited conditions is consistent with a requirement for high iron availability for GacS activation of some genes, as shown for the P. syringae pv. tabaci virulence genes (52). The Psl genes generally were not induced in planta compared to in culture (2), but their steady-state expression in the epiphytic and apoplastic sites as well as in the in vitro conditions depended on the presence of SalA as well as GacS, as shown previously (13). Last, GacS and SalA regulated the expression of one of two levansucrases in strain B728a, lsc1 (Table 2), with SalA, in particular, at least partially responsible for the previously recognized apoplast specificity of lsc1 induction in planta (Table 2) (2).
Consistent with GacS/GacA as a master regulator of secondary metabolism genes in pseudomonads (53), GacS regulated genes for the biosynthesis of syringomycin, syringopeptin, syringafactin, HAA, syringolin A, which is a peptide derived from a mixed nonribosomal peptide synthase (NRPS) and polyketide synthase, and two additional operons encoding NRPSs. Most of these required a plant-associated signal for maximal activation by GacS and showed higher activation in the apoplast than in epiphytic sites (Table 2 and Fig. 3; see Table S2 in the supplemental material). Expression of the NRPS operon Psyr_5009 to -5012, which includes genes originally misidentified as mangotoxin synthase genes (54), depended on GacS and SalA for expression under all of the conditions tested, including in planta (Table 2 and Table S2). In contrast, the NRPS operon Psyr_4312 to -4317 depended on GacS and SalA for expression primarily in planta (Table 2), thus indicating an additional secondary metabolite in the GacS regulon that may be relevant to B728a-host interactions.
Among individual genes, we found evidence that GacS and SalA activated genes for the sigma factor RpoS, the fatty acid degradation protein FadD (Table 2), as was observed in P. syringae DC3000 (9), and one of five P. syringae B728a cold shock proteins, CspD, as was found in P. protegens Pf-5 (43). Another functional category with a high number of genes that are positively regulated by GacS or SalA under multiple environmental conditions is the category of hypothetical proteins (see Table S3 in the supplemental material). Of the 1,216 hypothetical proteins, GacS positively regulated ~30% in at least one environmental condition and 12% in two or more conditions.
The GacS-SalA pathway reciprocally regulates genes for the type III and type VI secretion systems.
GacS and SalA negatively regulated hrpL, genes for components of the type III secretion pilus, and several type III secreted effector proteins in P. syringae B728a during its association with bean leaves (Tables 2 and 3). This contrasts sharply with the known role of GacA and SalA as positive regulators of hrpL and HrpL-regulated genes in P. syringae pv. tomato DC3000 (9) and P. syringae pv. syringae B301D (11) and with the reduced pathogenicity and hypersensitive reaction (HR) induction of a DC3000 gacA mutant (9). Our findings are consistent with the reduced HR induction of several P. syringae strains, but not B728a, upon overexpression of the GacS/GacA-mediated translational regulator RsmA (55). They are also consistent with the finding that gacS and gacA mutants of strain B728a are not altered in HR induction (3, 9). The T3SS genes in strain B728a were expressed at only low levels in planta in these experiments, leading us to speculate that these genes were induced only transiently or in only a subset of the cells (2, 56). A plausible model is that the GacS/SalA pathway in strain B728a is involved in minimizing the expression of the T3SS genes in the absence of key plant signals and that these key plant signals are spatially or temporally heterogeneous in the phyllosphere. The involvement of GacS and SalA in positive regulation of phytotoxins but negative regulation of the T3SS illustrates the complexity of factors regulating virulence traits on leaves.
TABLE 3 .
Fold changes in the transcript abundance of selected type III secretion system-related genes due to the deletion of hrpL, rpoN, or gacS
| Locus | Gene | CLa | REb | Fold change in transcript abundancec |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N starvation |
Epiphytic sites |
Apoplastic sites |
||||||||||
| hrpL | rpoN | gacS | hrpL | rpoN | gacS | hrpL | rpoN | gacS | ||||
| Psyr_1197 | hrpE | C | 48 | −1.6 | −1.4 | 1.1 | −1.1 | 1.2 | 1.4 | −1.2 | 1.1 | 1.3 |
| Psyr_1210 | hrcQa | C | 54 | −1.6 | −1.6 | 1.4 | 1.1 | 1.6 | 1.4 | −1.2 | 1.1 | 1.1 |
| Psyr_1206 | hrcT | I | 60 | 1.0 | −1.2 | −1.3 | 1.0 | 1.3 | 1.1 | −2.6 | −2.9 | 1.3 |
| Psyr_1215 | hrcV | I | 60 | −1.3 | −1.5 | 1.2 | 1.1 | 1.5 | 1.3 | −2.0 | −1.3 | 1.8 |
| Psyr_1205 | hrcU | I | 65 | −1.0 | −1.2 | −1.5 | 1.1 | 1.7 | 1.2 | −2.9 | −3.0 | 1.3 |
| Psyr_1213 | hrcN | C | 82 | −1.8 | −1.3 | 1.4 | 1.1 | 1.8 | 1.5 | −1.2 | −1.1 | 1.1 |
| Psyr_1208 | hrcR | I | 84 | −1.3 | −1.5 | −1.1 | −1.3 | 1.1 | 1.7 | −2.2 | −2.0 | 1.4 |
| Psyr_1209 | hrcQb | C | 85 | −2.3 | −2.0 | 1.4 | −1.1 | 1.3 | 1.7 | −1.7 | −1.4 | 1.8 |
| Psyr_1207 | hrcS | I | 114 | −1.4 | −1.6 | −1.7 | −1.2 | 1.0 | 1.6 | −5.1 | −6.5 | 3.2 |
| Psyr_1199 | hrpG | C | 135 | −7.8 | −8.9 | 1.4 | −1.5 | 1.1 | 2.6 | −1.3 | 1.0 | 3.3 |
| Psyr_1195 | hrcJ | P | 178 | −5.6 | −3.9 | 1.2 | −1.6 | −1.3 | 3.3 | −1.8 | −1.5 | 4.7 |
| Psyr_1200 | hrcC | O | 237 | −3.9 | −4.8 | −1.1 | −1.7 | −1.1 | 2.4 | −2.1 | −2.2 | 3.2 |
| Psyr_1183 | hopAA1 | E | 312 | −9.7 | −10.8 | −1.1 | −2.0 | −1.4 | 2.2 | −2.8 | −2.9 | 3.2 |
| Psyr_1184 | hrpW1 | E | 335 | −14.2 | −14.9 | 1.4 | −2.1 | −2.2 | 4.2 | −4.7 | −4.6 | 11.0 |
| Psyr_1188 | avrE1 | E | 357 | −9.8 | −10.3 | −3.9 | −1.1 | 1.0 | 1.1 | −3.4 | −3.8 | 1.5 |
| Psyr_4326 | hopI1 | E | 554 | −5.0 | −3.9 | −1.3 | −1.1 | 1.1 | 1.7 | −2.6 | −2.5 | 2.8 |
| Psyr_1017 | hopJ1 | E | 826 | −1.3 | −1.1 | 2.7 | −1.2 | 1.3 | 1.5 | 2.6 | 2.8 | 1.2 |
| Psyr_1192 | hrpA2 | E | 1719 | −27.0 | −19.8 | 1.3 | −4.5 | −2.5 | 4.2 | −3.6 | −2.0 | 12.7 |
| Psyr_4919 | avrPto1 | E | 1728 | −17.7 | −21.4 | 1.1 | −6.2 | −8.7 | 10.4 | −3.4 | −9.6 | 28.1 |
| Psyr_1193 | hrpZ1 | E | 2791 | −24.7 | −14.3 | −1.1 | −7.8 | −6.6 | 4.2 | −3.7 | −3.8 | 10.2 |
The cellular location (CL) indicated as follows: C, cytoplasmic; I, inner membrane; P, periplasmic; O, outer membrane; and E, extracellular.
RE, relative expression. The average expression based on fluorescence intensity of each gene in strain B728a in the basal medium in the data sets from the three laboratories, where the minimum and maximum levels were 42 and 55,935, respectively, across all of the genes examined.
Values shown are the fold change in transcript abundance of the mutants lacking hrpL, rpoN, or gacS relative to transcript abundance in strain B728a, as described in Table 2, footnote a.
GacS and SalA positively regulated the genes encoding the T6SS in strain B728a. Positive regulation by GacS was shown previously for two T6SS-associated genes, icmF and hcp (13). Positive regulation by GacS and SalA was shown here for almost all of the T6SS-associated genes and under most conditions (see Table S2 in the supplemental material). GacS/SalA activation was particularly large for genes in the tagR1-tagS1-tagT1-ppkA transmembrane signaling operon (57) and for most T6SS genes in cells exposed to oxidative stress (Table 2 and Table S2). This enhanced activation by oxidative stress supports the reported regulatory coupling between the oxidative stress response and cellular proteins critical to T6SS function in P. aeruginosa (58). P. aeruginosa also shows reciprocal regulation of the T3SS and T6SS by GacS/GacA (59). The finding that the GacS/SalA pathway induced the T6SS genes irrespective of a plant signal is consistent with the mounting evidence that the primary role of the T6SS in bacteria, including P. syringae, is to kill other microbes, regardless of their interactions with plants (60, 61).
SalA employs distinct strategies to regulate the synthesis of two siderophores.
Strain B728a produces the siderophores pyoverdine and achromobactin. Pyoverdine is common to all fluorescent pseudomonads, whereas achromobactin is restricted primarily to P. syringae and other phytopathogens. As expected, the biosynthetic genes for these siderophores were induced in strain B728a by iron starvation (2). The transcriptome data indicate that GacS and SalA regulate the genes encoding these siderophores differently, with SalA having a particularly dominant role (Fig. 3 and 4A). The dramatic reduction of pyoverdine synthesis genes in iron-limited ΔsalA cells suggests that SalA functions as a major activator of pyoverdine synthesis in response to iron limitation. In contrast, the increase in the expression of achromobactin synthesis and transport genes in the ΔsalA mutant in iron-replete conditions suggests that SalA functions to repress achromobactin genes when iron is available. The effect of SalA on these genes was much greater than that of GacS, suggesting the responsiveness of SalA to iron may be, at least in part, independent of GacS (Fig. 4A).
FIG 4 .
(A to C) SalA dominated the regulation of genes involved in siderophore biosynthesis (A) and iron-sulfur homeostasis (B), whereas GacS independently regulated genes involved in an oxidative stress response (C). The heat maps display the fold change in gene expression in the gacS and salA deletion mutants relative to gene expression in strain B728a. All genes that were altered >10-fold are shown with the same intensity as those altered 10-fold. Numbers to the left are the Psyr locus number, and treatments are shown on the top, with abbreviations as defined in the legend to Fig. 1. σECF, extracytoplasmic function sigma factor.
The gacS and salA mutations generally did not result in altered expression of the siderophore biosynthetic genes in planta. This is consistent with a lack of evidence for iron limitation by most B728a cells in planta, as determined using pyoverdine gene-based biosensors (62). This finding suggests not only the absence of GacS- or SalA-mediated pyoverdine induction but also suggests that SalA is not a major contributor to the suppression of the achromobactin genes in planta. The finding that the achromobactin genes were induced up to 5-fold in B728a cells in epiphytic and apoplastic sites relative to the levels of expression in the basal medium, whereas the pyoverdine genes were not (2) supports the possibility that the leaf habitat has a moderate level of iron. That is, the iron levels were too high to allow SalA-mediated activation of pyoverdine but too low to promote full SalA-mediated repression of the achromobactin genes, thus suggesting that the low-affinity siderophore achromobactin is the primary siderophore produced in planta.
SalA and GacS independently regulate processes involved in iron-sulfur chemistry and oxidative stress tolerance, respectively.
About half of the 61 genes identified as sulfur metabolism and transport genes were increased in expression in the ΔsalA mutant under iron- and N-limited conditions (Fig. 4B). These genes included many involved in the transport and metabolism of sulfate and sulfonate. A major use for sulfur in cells is as a component of iron-sulfur proteins. The dramatic suppression by SalA in the iron-limited conditions of 22 genes involved in sulfur, sulfonate, and methionine transport (an average of 14-fold for the transporter genes shown in Fig. 4B), combined with the central role of SalA in balancing the expression of the pyoverdine and achromobactin biosynthetic genes in response to iron, supports a role for SalA as a central regulator that coordinates the stoichiometric uptake of iron and sulfur compounds for iron-sulfur cluster synthesis in strain B728a. The transcriptome of a P. protegens gacA mutant identified regulation of iron homeostasis as one of the dominant roles of GacA (43), although it also showed GacA-mediated positive regulation of sulfate and sulfonate transporters compared to SalA-mediated negative regulation in strain B728a. The B728a strain has 19 TonB-dependent receptors (63), at least 9 of which may be involved in iron uptake based on their induction in our iron-limited medium, and six TonB-ExbB-ExbD uptake systems, only one of which was induced in our iron-limited medium (2). SalA, but not GacS, strongly repressed one of the TonB-dependent receptors, Psyr_4826, and one of the six operons encoding a TonB uptake system, Psyr_2287 to −2289, under iron-limited conditions (Fig. 4B).
A global transcriptome analysis of a P. protegens gacA mutant discovered that GacA negatively regulated 18 of 20 ECF-σ factors and positively regulated genes involved in oxidative stress tolerance (43). In strain B728a, GacS negatively regulated four of its 10 ECF-σ factors, and this regulation was specific to cells exposed to oxidative stress (Fig. 4C). The ECF-σ factors that showed increased transcripts in the ∆gacS mutant function in iron homeostasis; these factors included two FecI-type regulators, Ecf5 (Psyr_1040) and Ecf6 (Psyr_4731) (16), the pyoverdine regulator PvdS (Psyr_1943), and the achromobactin regulator AcsS (Psyr_2580). Furthermore, 8 of 17 antioxidant enzymes in strain B728a exhibited increased transcript levels in the ∆gacS mutant and did so only in cells exposed to oxidative stress (Fig. 4C); GacS did not mediate full suppression of these genes, however, as transcripts for five of these antioxidant enzymes were increased in B728a cells exposed to oxidative stress (2). GacS also mediated the suppression of an operon for a polyamine transporter (Psyr_4613 to −4615), an iron permease (Psyr_3367), and an ortholog of PhuR (Psyr_1105), which contributes to the uptake of heme. None of these GacS-regulated genes, except for a peroxidase, was influenced by the loss of salA, indicating that they were part of a SalA-independent branch of the GacS regulon (Fig. 3). Altogether, these results support a model in which GacS helps reduce iron uptake in the face of oxidative stress, probably to minimize stress resulting from the iron-mediated generation of reactive oxygen species via the Fenton reaction. This reduced iron uptake would result from reductions in iron and heme uptake systems, reduced sigma factor-mediated activation of siderophore synthesis, and potential restrictions to siderophore synthesis due to reduced polyamine uptake, as polyamines can function as the substrates in NRPS-independent siderophore synthesis pathways (64). Achromobactin synthesis is NRPS independent and conceivably could be influenced by polyamines via the 2,4-diaminobutyrate precursor pool (65). Overall, our results with GacS and SalA demonstrate that that their roles in the regulatory hierarchy and the responses that they activate strongly depend on environmental signals, as illustrated by the differential effects of plant-associated signals, iron availability, and oxidative stress on the GacS and SalA regulons.
RetS negatively regulates a subset of the GacS regulon, including biosynthetic genes for phytotoxins and other secondary metabolites.
RetS is a hybrid sensor kinase that was discovered in P. aeruginosa and was shown to act inversely to GacS (66). In particular, RetS interacts directly with GacS, forming an inactive RetS-GacS heterodimer and thus blocking the formation of a GacS homodimer that is required for GacA phosphorylation (67). In this manner, RetS blocks GacS-mediated gene activation. Here, loss of RetS was associated with increased transcripts in at least one condition for 41 of the 43 genes in the RetS regulon (Table 1), illustrating that RetS functions primarily as a negative regulator but influences a relatively small set of genes. Although half of the RetS regulon was comprised of genes encoding hypothetical proteins, the other half was comprised of the subset of the GacS/SalA regulon that showed enhanced transcript levels in the presence of plant signals (Fig. 3). This subset included the biosynthetic genes for syringomycin, syringopeptin, syringafactin, syringolin A, and the secondary metabolite encoded by Psyr_4312 to −4317, and the fadD operon (Fig. 3). As would be predicted for a regulator that blocks GacS-mediated activation, these genes exhibited reciprocal regulation by RetS and GacS (Fig. 5A), as was shown with a few target genes in strain B728a (13). Interestingly, plant-associated signals not only enhanced GacS and SalA activation of these genes but also suppressed the RetS regulation (Fig. 5B and C).
FIG 5 .
(A to C) RetS repression of selected genes in the basal medium (A) was reciprocal to GacS and SalA activation of these genes in epiphytic sites (B) and apoplastic sites (C). The transcript levels in the ∆retS, ∆gacS, and ∆salA mutants relative to those in the wild-type strain B728a are shown. The dashed line indicates a value of 1 for reference.
The genes for the type VI secretion system and alginate production were not differentially expressed in the ΔretS mutant, although they were regulated by RetS in strain B728a in a previous study that used complex media for assessing gene expression (13). The absence of evidence for RetS regulation of T6SS or alginate genes in our in vitro conditions may be due to a requirement for additional factors for RetS repression, although we did not find evidence for such factors in planta. RetS is widely recognized as having a critical role in P. aeruginosa pathology by helping to mediate a switch between acute and chronic infections. In contrast, our results indicate that RetS does not have a significant regulatory role in P. syringae during its growth on or in bean plants, at least under the conditions tested. The sensitivity of RetS regulation to environmental context, however, suggests that RetS may have a role in P. syringae ecology under environmental conditions that have not yet been examined.
RpoS regulates genes for chemosensing and chemotaxis and Psl polysaccharide synthesis but few genes involved in stress tolerance.
RpoS regulation was detected primarily in cells limited for N or recovered from leaves (Table 1). We found no evidence for RpoS regulation of genes involved in environmental stress tolerance in P. syringae based on the near-absence of RpoS-regulated genes when osmotic or oxidative stress was imposed in vitro. RpoS contributes to the tolerance of a P. protegens strain to stresses, including osmotic and oxidative stresses, desiccation, UV irradiation, freezing, and starvation (21), but it has a much smaller influence (68) or no influence (69) on the stress tolerance of two P. fluorescens strains. This illustrates the potential for differences among pseudomonad strains in the role of RpoS and precedence for little to no role for RpoS in stress tolerance in pseudomonads.
The rpoS gene was subject to strong positive activation by GacS and SalA under all of the conditions examined (Table 2; see Table S2 in the supplemental material), indicating that RpoS is in the GacS/SalA regulatory hierarchy, as in P. syringae DC3000 (9). Approximately 75% of the RpoS-regulated genes were also regulated by GacS, similar to in P. protegens (70). The GacS/SalA/RpoS regulatory pathway positively regulated 25% of the genes involved in chemosensing and chemotaxis in epiphytic sites or when N was limited, as supported by a test for the functional categories with overrepresentation of differentially expressed genes in the ∆rpoS, ∆gacS, and ∆salA mutants (Table S3). Moreover, the GacS/SalA/RpoS pathway appeared to be a major pathway for the positive regulation of the Psl polysaccharide biosynthetic genes under N-limited conditions as well as in planta, although this regulatory pathway did not result in elevated psl gene transcripts in planta (2).
The HrpL regulon may be subject to repression by a negative regulatory element.
The HrpL regulon in P. syringae strains contains T3SS genes (14), which in strain B728a includes approximately 50 genes for the structural components of the T3SS and 22 confirmed effectors (71). We selected the basal medium HMM (see Materials and Methods) for this study based, in part, on its potential to enable expression of T3SS genes, as first demonstrated in P. syringae pv. glycinea (72). However, the T3SS genes in strain B728a generally exhibited low expression in this medium (Table 3), with almost 40% of the T3SS genes falling in the bottom 15% of the genes when ranked by their expression levels in the basal medium. A majority of these genes was induced in the wild type under N starvation (2), but even under these inducing conditions, HrpL activation of many genes was too low in the wild type to detect a significant decrease in expression upon deletion of hrpL. Interestingly, the genes with the lowest level of expression and lowest HrpL activation in vitro were those that encoded the proteins for the cytoplasmic and inner membrane components of the type III secretion pilus (Table 3). In contrast, the genes that encoded periplasmic, outer membrane, and extracellular components were expressed at higher levels in the wild type and were activated by HrpL up to 12-fold, on average, under N starvation (Table 3). These genes exhibited parallel positive regulation by RpoN in the N-limited conditions, as expected, since the expression of T3SS-related genes requires RpoN (73).
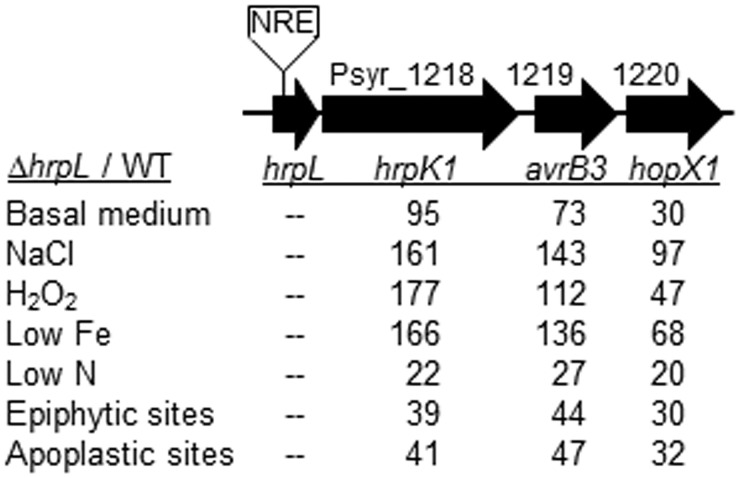
Although many T3SS genes exhibited reduced transcript levels in the hrpL mutant under the inducing conditions in vitro, consistent with positive HrpL regulation, the three effector genes downstream of hrpL, Psyr_1218 to −1220, exhibited dramatic increases in their transcript levels upon deletion of hrpL (Fig. 6). Although HrpL could function as a negative autoregulator, this seems unlikely given the specificity of the change to these three genes. Alternatively, a negative regulatory element may be located within the deleted region of the hrpL coding sequence. Such a negative regulatory element could silence hrpL under noninducing conditions, as supported by the noticeably large impact of the hrpL deletion under the in vitro conditions other than N starvation (Fig. 6). To the best of our knowledge, this is the first report of a possible negative regulatory element in hrpL.
FIG 6 .
The effect of deleting hrpL on the expression of its downstream genes suggested the presence of a negative regulatory element (NRE) in hrpL. The Psyr locus numbers and gene names are shown above and below the arrows, respectively. The fold change values in transcript abundance for the ∆hrpL mutant relative to the wild-type strain B728a (WT) are shown for cells from each environmental condition.
The data in Table 3 illustrate active HrpL regulation in planta, with HrpL activation generally greater in the apoplast than on the leaf surface, although we observed yet further activation by N starvation. Our ability to identify significant differences between the hrpL mutant and the wild type in the apoplast was limited by the high variability among samples (see Fig. S1 in the supplemental material); however, 55% of the T3SS genes showed at least a 2-fold reduction in transcript levels in the hrpL mutant for cells from the apoplast compared to 14% for cells from the leaf surface. The changes in the T3SS transcripts in the hrpL mutant in planta were generally reciprocal to the changes in the gacS and salA mutants, indicating GacS/SalA-mediated suppression of the T3SS genes in planta, but GacS/SalA regulation of these genes was not observed in the only in vitro conditions to induce hrpL, namely, the N-limited conditions (Table 3), demonstrating that this suppression requires a plant signal (Fig. 3). Interestingly, the gene for the HopJ1 effector was uniquely subject to negative regulation by HrpL in the apoplast (Table 3). In support of our evidence for HrpL activation of at least some T3SS genes in epiphytic sites, Lee et al. (15) showed that strain B728a expressed a heterologous effector gene, avrPto, on leaf surfaces. Collectively, our results support the possibility of a negative regulatory element involved in T3SS regulation, and for HrpL activation of some T3SS genes on leaf surfaces, which is a trait that may be unique to B728a and contribute to its uniquely robust epiphytic fitness (15).
RpoN and AlgU are major regulators in planta, with RpoN influencing the majority of genes that are regulated by AlgU, GacS, and SalA in planta.
Among the conditions tested, AlgU affected the greatest number of genes under the osmotic stress conditions, consistent with its role in responding to envelope stress (20). RpoN, known for its role in N metabolism, affected more than twice as many genes under osmotic stress as under N starvation (Table 1), as well as activated algU (see Table S2 in the supplemental material). RpoN coregulated 71% of the 810 AlgU-regulated genes under osmotic stress and contributed to the activation of 88% of the AlgU-activated genes under osmotic stress, but it had an even broader impact than AlgU under osmotic stress based on its contribution to the activation of an additional 623 AlgU-independent genes.
Both AlgU and RpoN impacted a large number of genes in planta (Table 1; see Fig. S2 in the supplemental material). The importance of these regulators to bacterial growth in planta was illustrated by the dramatically reduced growth of epiphytic and apoplastic populations of the algU and rpoN mutants relative to the wild type (Fig. S3). AlgU induced genes in planta that also responded to osmotic stress, including genes for compatible solute synthesis, osmoprotectant transporters, the known osmoresponsive protein OsmC, the LasB protease, and an sRNA, sRNA_42 (Table S2). AlgU also induced genes that responded to oxidative stress, including genes for the antioxidant enzymes KatE, SodC, and CpoF. AlgU activates genes in response to conditions that induce misfolded proteins in the periplasm; therefore, the finding that only a third of the genes that were differentially expressed in epiphytic sites were differentially expressed in cells exposed to 0.2 M NaCl suggests that cells were exposed to less envelope stress on leaves than at this osmolarity in culture. The percentage of genes regulated in epiphytic sites that were also regulated in osmotic-stressed cells was 90 for AlgU but only 56 for RpoN, suggesting that the leaf surface-responsive AlgU-regulated genes responded to envelope stress in planta, whereas the RpoN-regulated genes responded to a broader array of plant signals.
RpoN strongly activated genes involved in N metabolism, as expected, including genes for the nitrogen regulator NtrBC, nitrate reductase, an ammonium transporter, glutamine synthetases, and enzymes for urea utilization (see Table S2 in the supplemental material). The specificity of this regulation to ∆rpoN cells subjected to N starvation but not to the in planta habitats confirms our previous conclusion that cells in the apoplast or epiphytic sites were not strongly starved for N in planta (2). RpoN also strongly activated the vast majority of genes involved in flagellar synthesis and motility, as expected based on the critical role of RpoN to most flagellar synthesis genes in P. aeruginosa (74). The greater impact of the rpoN deletion on the transcript levels of these genes in cells in epiphytic sites than in the apoplast also confirms our previous conclusion that, under moist conditions favorable to motility, motility is more strongly favored by B728a cells on leaf surfaces than by cells in the apoplast (Table S2) (2).
Regulation of alginate biosynthesis is complex, but although it was subject to regulation by AlgU, RpoN, GacS, and SalA under osmotic stress conditions in culture, AlgU was the primary activator among these regulators in planta (see Table S2 in the supplemental material). This was also true for a large number of the core T6SS genes. In contrast to many osmotically regulated genes that were coactivated by RpoN and AlgU, genes for alginate biosynthesis and the T6SS were subject to negative regulation by RpoN in the apoplast based on their increased transcript levels in ∆rpoN cells from the apoplast but not from epiphytic sites (Fig. 7A and Fig. S4). In P. aeruginosa, RpoN-mediated negative regulation of algD, the first gene in the alginate biosynthetic operon, was previously found to result from RpoN interference with AlgU binding, which prevented the formation of an open complex for transcription (75); this negative regulation occurred when RpoN bound in the absence of an activator protein. Thus, the apoplast specificity of the AlgU activation and RpoN repression of the alginate and T6SS genes (Fig. 7A) is consistent with competitive inhibition from RpoN for promoter binding and a lack of relevant activator proteins in the apoplast.
FIG 7 .
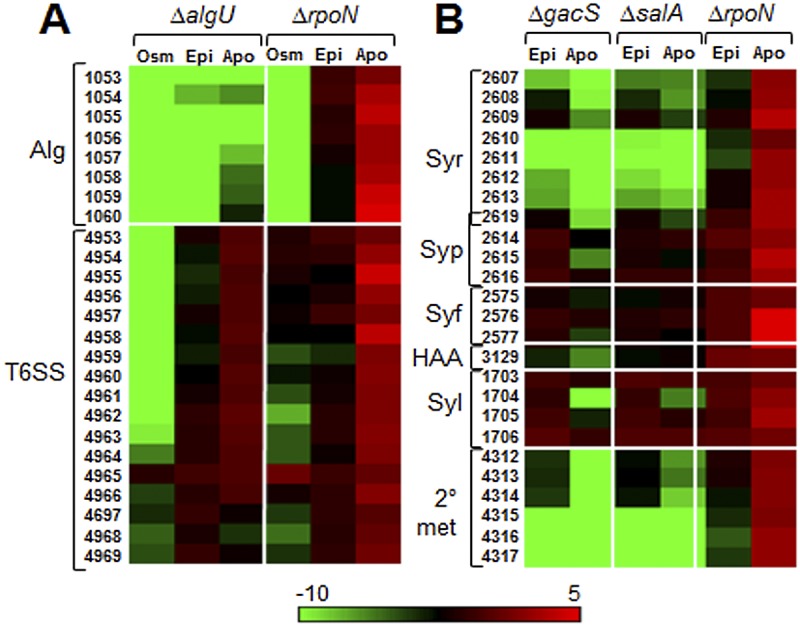
(A and B) RpoN differentially impacted AlgU-activated genes (A) and GacS- and SalA-regulated genes (B) in cells from epiphytic sites versus apoplastic sites. The heat maps display the fold change in gene expression in the indicated mutants relative to gene expression in strain B728a. All genes that showed a >10-fold decrease or >5-fold increase in transcript abundance are shown with the same intensity as those decreased 10- and increased 5-fold, respectively. The numbers to the left are the Psyr locus number, and treatments are shown on the top. Genes that are involved in the synthesis of alginate (Alg), the type VI secretion system (T6SS), syringomycin (Syr), syringopeptin (Syp), syringafactin (Syf), the surfactant HAA, syringolin (Syl), and a secondary metabolite (2° met) are shown. Osm, osmotic stress; Epi, epiphytic sites; Apo, apoplastic sites.
RpoN contributed to the regulation of most of the genes activated by GacS and SalA in planta. Transcript levels of these genes were greatly increased in the rpoN mutant in the basal medium and other in vitro treatments (Table 2; see Table S2 in the supplemental material), indicating negative regulation; this is likely due to RpoN binding in the absence of transcriptional regulators that are activated by the GacS/SalA pathway in planta. The gacS gene itself was subject to slight negative regulation by RpoN under most of the in vitro and in planta conditions (Table S2), as reported in P. aeruginosa (76). Similar to the alginate and T6SS genes, many of the GacS/SalA-activated genes were subject to apoplast-specific, RpoN-mediated repression (Fig. 7B); moreover, GacS/SalA activation of these genes was greater in the apoplast. A plausible model for this association between high-level GacS/SalA activation and increased transcript abundance upon deletion of rpoN is interference between RpoN and the GacS/SalA-regulated sigma factor RpoS. Such RpoS-RpoN antagonism was observed in Escherichia coli (77). We predict that the interference by RpoS in RpoN activation is relatively small based on the absence of significant changes in transcript levels of GacS-regulated genes in the ∆rpoS mutant but that the impact on RpoN activation was large enough in the apoplast to detect. Collectively, these results demonstrate major roles for RpoN and AlgU in cells associated with leaves and suggest a greater impact of RpoN interference in AlgU- and GacS/SalA-mediated gene activation in the apoplast than in epiphytic sites.
Conclusions.
This is the first global transcriptome study investigating the complex regulatory network of putative quorum-sensing regulators (AhlR and AefR), global regulators (GacS, SalA, and RetS), and sigma factors (RpoS, HrpL, AlgU, and RpoN) in P. syringae, and particularly in the context of stressful environmental and in planta conditions. One of our first surprises was that the loss of the only known quorum regulator in P. syringae B728a, AhlR, and an associated regulator, AefR, influenced the expression of only a small number of genes. Although quorum-sensing regulation in most pathogens, including P. aeruginosa, generally impacts large gene sets, our evidence suggests that for strain B728a, AHL-based quorum-sensing regulation plays a relatively minor role in gene regulation on leaves.
The GacS/SalA regulatory network includes distinct branches that are separable based on their dependence on coactivation by plant signals (Fig. 3). We expanded the inventory of plant signal-dependent traits from primarily phytotoxins to other secondary metabolites, including surfactants, syringolin A, and an as-yet uncharacterized secondary metabolite. We also discovered unexpected negative regulation of the T3SS by GacS, which in the context of previous studies indicates that P. syringae pathovars differ in how they regulate this dominant virulence trait. Our results support the emerging model in P. aeruginosa and P. syringae that RetS regulation is reciprocal to that of GacS, but only for a small number of genes, and that the primary function of RetS in strain B728a is to prevent the expression of genes for secondary metabolite synthesis in the absence of the plant (Fig. 5). The need for such tight regulation may reflect a benefit of these metabolites primarily in plants, as well as a centrality of these secondary metabolites to B728a-plant interactions, as evidenced in the proposal that production of syringomycin and syringopeptin may decrease the dependence of this pathovar of P. syringae on its effector genes for virulence (78). Collectively, our data confirm a role for GacS in oxidative stress tolerance, as suggested in P. protegens, and also highlight a role for SalA as a major regulator of both iron homeostasis and sulfur transport, presumably to ensure an iron-sulfur balance for iron-sulfur cluster synthesis.
With the exception of HrpL, the size of the gene sets altered by the loss of the sigma factor genes was paralleled by the change in population sizes during leaf colonization (see Fig. S3 in the supplemental material). In particular, RpoS influenced only a small number of genes, and the growth of the ∆rpoS mutant was similar to the growth of the wild type. As observed with Pseudomonas fluorescens A506 (69), RpoS was not a major regulator of environmental stress tolerance in strain B728a. In contrast, AlgU regulated a large number of genes involved in mediating tolerance to water limitation and oxidative stress, and the ∆algU mutant established populations that were approximately 10-fold lower than those of the wild type on and in leaves (Fig. S3). Moreover, RpoN coregulated much of the AlgU regulon and a wide inventory of additional genes (Fig. S4), and the ∆rpoN mutant was severely impaired in its ability to establish epiphytic or apoplastic populations.
Last, although the number of genes that were differentially expressed in the ∆hrpL mutant in these studies was low, HrpL has a large impact on the growth of strain B728a in planta (see Fig. S3 in the supplemental material), as expected based on its known role in regulating the T3SS. Lee et al. (15) showed that only a fraction of B728a cells actually express hrpL-regulated genes in planta. Thus, it is tempting to speculate that the two major regulatory systems in strain B728a, GacS/SalA and RpoN, influence distinct subpopulations of B728a cells by reciprocally influencing their expression of HrpL. We propose that the T3SS is suppressed in many cells due to the GacS response to particular plant signals but that it is activated in a subpopulation of cells due to the RpoN response to particular environmental signals.
Collectively, these data demonstrate the activity of multiple regulatory networks in strain B728a in planta. The major impact of physical or chemical signals in the leaf environment on the activity of the GacS/SalA-, AlgU-, and RpoN-driven regulatory networks illustrates the importance of evaluating these networks in planta. This multifactorial approach of examining regulatory mutants in several environments, including leaf habitats, provided evidence that at least some of these signals differ at least in intensity in epiphytic versus apoplastic sites, thus allowing P. syringae to fine-tune its expression of traits to best exploit each habitat.
MATERIALS AND METHODS
Bacterial strains, mutants, and growth conditions.
P. syringae pv. syringae strain B728a and its derivatives were grown in King’s B medium containing rifampin (50 µg/ml) or in HMM medium. HMM medium contained l-glutamine (10 mM), FeCl3 (10 µM), and N-(β-ketocaproyl)-l-homoserine lactone (AHL) (10 µM), in addition to nutrients and salts, as previously described (2), to help maximize phenotypic differences between strain B728a and selected mutants. Mutants were constructed that contained unmarked deletions of the following genes: ahlR (Psyr_1622), aefR (Psyr_3324), salA (Psyr_2601), algU (Psyr_3958), hrpL (Psyr_1217), rpoN (Psyr_4147), and rpoS (Psyr_1374), using the primers shown in Table S4 in the supplemental material. The ahlR and aefR mutants were constructed by amplifying the loci and flanking regions using primer set A, cloning the resulting fragment into pENTR/D (Invitrogen Corp.), amplifying the plasmid with primers to omit ahlR or aefR and introduce a KpnI site using primer set B, marking the deletion site via introduction of a kanamycin (kan) cassette surrounded by FLP recombination target (FRT) sites and KpnI sites (generated using primer set C), and incorporating this plasmid construct into the integration vector pTOK2T (79). The salA mutant was constructed as described previously (17). For the remaining mutants, marked deletions were generated using splicing by overlap extension PCR and a kan cassette surrounded by FRT sites and the integration vector pTOK2T, as described previously (79). After introduction of the pTOK2T constructs to generate marked mutants, the kan cassette was excised from each mutant by introducing pFlp2 (80), which was later cured via two passages on King’s B agar without selection for the ahlR and aefR mutants, or using sucrose (20%) counterselection for the other mutants. The deletions were confirmed by PCR and DNA sequencing. Mutants containing unmarked deletions in gacS (Psyr_3698) and retS (Psyr_4408) were constructed previously (13).
Exposure of bacteria to in vitro and in planta treatments prior to RNA extraction.
Strain B728a and the nine mutants were exposed to seven treatments in a multifactorial design that included exposure of each strain to each of the treatments. Three separate laboratories performed the treatments; they were located at the University of California at Berkeley, Texas A&M University, and Iowa State University and were designated laboratories I, II, and III, respectively. Lab I performed treatments on strain B728a and the ∆ahlR and ∆aefR mutants. Lab II performed treatments on strain B728a and the ∆gacS, ∆retS, and ∆salA mutants. Lab III performed treatments on strain B728a and the ∆algU, ∆hrpL, ∆rpoN, and ∆rpoS mutants. The treatments were performed by the method of Yu et al. (2). Briefly, log-phase cells grown in HMM medium were washed, resuspended in HMM medium lacking the components l-glutamine, FeCl3, AHL, and (NH4)2SO4, designated HMM-FeN medium, then diluted to 2.5 × 108 CFU/ml with either (i) HMM medium, (ii) HMM medium with NaCl to a final concentration of 0.23 M, (iii) HMM medium with H2O2 to a final concentration of 0.5 mM, (iv) HMM medium lacking FeCl3 but with N,N′-di(2-hydroxybenzyl) ethylenediamine-N,N′-diacetic acid monohydrochloride hydrate (HBED) (Strem Chemicals Inc., Newburyport, MA) to a final concentration of 100 µM, or (v) HMM medium lacking both l-glutamine and (NH4)2SO4; these were designated the basal medium, osmotic stress, oxidative stress, iron starvation, and nitrogen starvation treatments, respectively, with the latter four collectively referred to as the in vitro treatments. The cells were immediately diluted with an RNA stabilizing agent (RNAprotect bacterial reagent; Qiagen Inc., Valencia, CA) following exposure to the basal medium and in vitro treatments. In addition, the bacterial strains were introduced onto the adaxial and abaxial surfaces of leaves of bean plants (Phaseolus vulgaris L. cultivar Bush Blue Lake 274) and were recovered from these surfaces via sonication in an acidic phenol RNA-stabilizing solution after 24 h of growth under moist conditions followed by 48 h of growth without supplemental humidification; these cells were designated epiphytic cells. Due to limitations in growth space capacity, all of the epiphytic treatments were performed using the facilities of lab I, with cultures provided from the other laboratories and pelleted epiphytic cells returned to those labs for RNA extraction and analysis. Last, the bacterial strains were introduced by vacuum infiltration into bean leaves and were recovered after 48 h of growth by cutting the leaves during submersion in the RNA stabilizing solution, sonicating, and removing the plant tissues via filtration; these cells were designated apoplastic cells. These treatments were performed exactly by the method of Yu et al. (2), with the exception that the inoculum densities for recovering epiphytic cells varied by strain, with the densities (CFU/ml) as follows: ΔrpoN mutant (109), ΔhrpL mutant (108), ΔalgU mutant (107), ΔrpoS mutant (107), and strain B728a and the remaining mutants (106). The inoculum densities for recovering apoplastic cells were the same as for the epiphytic cells, with the exception of the ΔrpoS mutant, for which 106 cells/ml were used. The higher inoculum densities were to compensate for the reduced growth of the mutants in planta (see Fig. S3 in the supplemental material) and to ensure sufficient RNA recovery.
RNA extraction, microarray design, and hybridization.
We used a previously described experimental design (2) to generate two biological replicates for each treatment, with each replicate for the in vitro treatments containing RNA pooled from four independent cultures. RNA was purified using the Qiagen RNeasy minikit, DNA was removed using on-column DNase I digestion and a subsequent DNase I treatment, RNA integrity was assessed using an Agilent 2100 bioanalyzer, and the RNA samples from each laboratory were sent to Roche NimbleGen Inc. (Reykjavík, Iceland) for conversion into cDNA, labeling with U-CYA-3 fluorophore, and hybridization to a P. syringae B728a open reading frame (ORF)-based microarray, which was constructed based on RefSeqNC_007005.1 and was described in the National Center for Biotechnology Information’s Gene Expression Omnibus (accession no. GSE42544). The 16 putative sRNAs designated PIG (plant-inducible genes) due to their identification in a promoter-trapping assay were found to be encoded, or initiated, within the coding regions of annotated genes and thus were excluded from the analysis.
Microarray data analysis.
The fluorescence intensity for each probe was measured and subjected to robust multiarray averaging, which included adjustment for the background intensity, log2 transformation, quantile normalization, and median polishing (81). For each feature (ORF or small RNA [sRNA]) on the array, a robust estimated mean value was determined, and a linear model analysis of the resulting data was conducted. Each linear model included fixed effects for replications, treatments, strains, and treatment-by-strain interactions, as well as a fixed intercept parameter and one random error effect for each observation. Linear models for microarray data analysis (LIMMA) (82) were applied to share information across genes when estimating error variances. Application of LIMMA was performed separately for distinct groups of treatments that exhibited similar variability among replicates. Variance estimates obtained from the LIMMA analyses were used to calculate Welch t statistics and corresponding P values among all pairwise treatment comparisons of interest. For each comparison of interest, q values were estimated from the corresponding distribution of P values, as described previously (83). Features exhibiting a P value of <0.05 and a q value of <0.01 (i.e., an estimated false discovery rate of <1%) were identified as differentially expressed.
Hierarchical clustering.
Hierarchical clustering was performed to evaluate the similarity in global transcript profiles among the samples examined in each lab according to the following protocol. For each of the three labs, a dendrogram was generated using the fluorescent intensities for each of the biological replicates. The data for each gene were subjected to an analysis of variance (ANOVA) across the seven treatments. The observed intensities from the 500 genes that exhibited the lowest P values in an F test for the combined effect of all treatments from this ANOVA were used to perform hierarchical clustering among all the samples. The observed intensities for each gene were divided by the gene’s estimated standard deviation, so that each gene would contribute roughly equally to the distance metric during clustering. The hierarchical clustering was performed by the “hclust” function in R using Manhattan distance in a “bottom-up” approach.
Analysis of gene representation in functional categories.
The P. syringae B728a genes were assigned to 63 functional categories (as given in Table S2 from reference 2). For each functional category, we formed a 2 × 2 contingency table reporting the number of differentially expressed and non-differentially-expressed genes included in the given category and did the same for the differentially expressed and non-differentially-expressed genes in all of the categories except the given category. We then performed a Fisher’s exact test to evaluate overrepresentation of the differentially expressed genes. We performed this analysis separately for the differentially induced genes and the differentially repressed genes. The q values were generated from the resulting P values, as described above. Heat maps were generated using Matrix2png version 1.2.1 (84).
Microarray data accession number.
The expression data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible in GEO under accession no. GSE59273.
SUPPLEMENTAL MATERIAL
Relative variation in transcript levels among each of the strain-treatment combinations based on median absolute residuals computed from our linear model analyses. Download
Loss of algU and rpoN was correlated with decreases in the abundance of the transcripts induced by both osmotic stress and growth in planta. Download
(A and B) Mutants lacking algU, hrpL, and rpoN exhibited large reductions in growth in epiphytic sites (A) and apoplastic sites (B)of bean leaves. Download
RpoN activated genes involved in N metabolism, coregulated much of the AlgU regulon, and regulated many traits in a plant signal-dependent manner. Download
Number of genes for which the difference in transcript abundance between the mutant and the wild type in a given environmental condition differed from that in the basal medium
Effects of environmental conditions on the responses of selected genes to the loss of target regulators
The functional categories in which the representation of differentially-expressed genes was greater than their representation among all of the strain B728a genes
Primers used in this study to construct deletion mutants of the indicated target genes
ACKNOWLEDGMENTS
This project was supported by the National Research Initiative Competitive Grants Program grant 2008-35600-18766 from the U.S. Department of Agriculture National Institute of Food and Agriculture.
We thank David Morgan and Jia Wang for help with bacterial recovery from leaf surfaces.
Any mention of commercial products is for information only; it does not imply recommendation for endorsement by the National Institute of Standards and Technology.
Footnotes
Citation Yu X, Lund SP, Greenwald JW, Records AH, Scott RA, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2014. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 5(5):e01683-14. doi:10.1128/mBio.01683-14.
REFERENCES
- 1. Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffiere A, Yan SC, Dominguez H, Thompson BM. 2008. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2:321–334. 10.1038/ismej.2007.113 [DOI] [PubMed] [Google Scholar]
- 2. Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2013. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. U. S. A. 110:E425–E434. 10.1073/pnas.1221892110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Willis DK, Hrabak EM, Rich JJ, Barta TM, Lindow SE, Panopoulos NJ. 1990. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol. Plant Microbe Interact. 3:149–156. 10.1094/MPMI-3-149 [DOI] [Google Scholar]
- 4. Laville J, Voisard C, Keel C, Maurhofer M, Défago G, Haas D. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. U. S. A. 89:1562–1566. 10.1073/pnas.89.5.1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lapouge K, Schubert M, Allain FH-T, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67:241–253. 10.1111/j.1365-2958.2007.06042.x [DOI] [PubMed] [Google Scholar]
- 6. Kinscherf TG, Willis DK. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kitten T, Kinscherf TG, McEvoy JL, Willis DK. 1998. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28:917–929. 10.1046/j.1365-2958.1998.00842.x [DOI] [PubMed] [Google Scholar]
- 8. Willis DK, Holmstadt JJ, Kinscherf TG. 2001. Genetic evidence that loss of virulence associated with gacS or gacA mutations in Pseudomonas syringae B728a does not result from effects on alginate production. Appl. Environ. Microbiol. 67:1400–1403. 10.1128/AEM.67.3.1400-1403.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatterjee A, Cui YY, Yang HL, Collmer A, Alfano JR, Chatterjee AK. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant Microbe Interact. 16:1106–1117. 10.1094/MPMI.2003.16.12.1106 [DOI] [PubMed] [Google Scholar]
- 10. Wang N, Lu SE, Records AR, Gross DC. 2006. Characterization of the transcriptional activators SalA and SyrF, which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 188:3290–3298. 10.1128/JB.188.9.3290-3298.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu SE, Wang N, Wang JL, Chen ZJ, Gross DC. 2005. Oligonucleotide microarray analysis of the SalA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae. Mol. Plant Microbe Interact. 18:324–333. 10.1094/MPMI-18-0324 [DOI] [PubMed] [Google Scholar]
- 12. Lu SE, Scholz-Schroeder BK, Gross DC. 2002. Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant Microbe Interact. 15:43–53. 10.1094/MPMI.2002.15.1.43 [DOI] [PubMed] [Google Scholar]
- 13. Records AR, Gross DC. 2010. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J. Bacteriol. 192:3584–3596. 10.1128/JB.00114-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. 2006. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant Microbe Interact. 19:1151–1158. 10.1094/MPMI-19-1151 [DOI] [PubMed] [Google Scholar]
- 15. Lee J, Teitzel GM, Munkvold K, del Pozo O, Martin GB, Michelmore RW, Greenberg JT. 2012. Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol. 158:1803–1818. 10.1104/pp.111.190686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thakur PB, Vaughn-Diaz VL, Greenwald JW, Gross DC. 2013. Characterization of five ECF sigma factors in the genome of Pseudomonas syringae pv. syringae B728a. PLoS One 8:e58846. 10.1371/journal.pone.0058846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greenwald JW, Greenwald CJ, Philmus BJ, Begley TP, Gross DC. 2012. RNA-seq analysis reveals that an ECF sigma factor, AcsS, regulates achromobactin biosynthesis in Pseudomonas syringae pv. syringae B728a. PLoS One 7:e34804. 10.1371/journal.pone.0034804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu J, Penaloza-Vazquez A, Chakrabarty AM, Bender CL. 1999. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 33:712–720. 10.1046/j.1365-2958.1999.01516.x [DOI] [PubMed] [Google Scholar]
- 19. Martin DW, Holloway BW, Deretic V. 1993. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 175:1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wood LF, Ohman DE. 2012. Identification of genes in the σ22 regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. mBio 3(3):e00094-12. 10.1128/mBio.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stockwell VO, Loper JE. 2005. The sigma factor RpoS is required for stress tolerance and environmental fitness of Pseudomonas fluorescens Pf-5. Microbiology 151:3001–3009. 10.1099/mic.0.28077-0 [DOI] [PubMed] [Google Scholar]
- 22. Lorge A, Gross D. 2008. Characterization of the RpoN global regulatory gene of Pseudomonas syringae pv. syringae B728a and its impact on the plant-pathogen interaction. Phytopathology 98:S94–S94 [Google Scholar]
- 23. Alarcón-Chaidez FJ, Keith L, Zhao YF, Bender CL. 2003. RpoN (σ54) is required for plasmid-encoded coronatine biosynthesis in Pseudomonas syringae. Plasmid 49:106–117. 10.1016/S0147-619X(02)00155-5 [DOI] [PubMed] [Google Scholar]
- 24. Hendrickson EL, Guevera P, Ausubel FM. 2000. The alternative sigma factor RpoN is required for Hrp activity in Pseudomonas syringae pv. maculicola and acts at the level of hrpL transcription. J. Bacteriol. 182:3508–3516. 10.1128/JB.182.12.3508-3516.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dulla G, Lindow SE. 2008. Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proc. Natl. Acad. Sci. U. S. A. 105:3082–3087. 10.1073/pnas.0711723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quinones B, Pujol CJ, Lindow SE. 2004. Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 17:521–531. 10.1094/MPMI.2004.17.5.521 [DOI] [PubMed] [Google Scholar]
- 27. Quinones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 18:682–693. 10.1094/MPMI-18-0682 [DOI] [PubMed] [Google Scholar]
- 28. Chatterjee A, Cui Y, Hasegawa H, Chatterjee AK. 2007. PsrA, the Pseudomonas sigma regulator, controls regulators of epiphytic fitness, quorum-sensing signals, and plant interactions in Pseudomonas syringae pv. tomato strain DC3000. Appl. Environ. Microbiol. 73:3684–3694. 10.1128/AEM.02445-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng X, Xiao YM, Lan LF, Zhou JM, Tang XY. 2009. Pseudomonas syringae pv. phaseolicola mutants compromised for type III secretion system gene induction. Mol. Plant Microbe Interact. 22:964–976. 10.1094/MPMI-22-8-0964 [DOI] [PubMed] [Google Scholar]
- 30. Kawakita Y, Taguchi F, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. 2012. Characterization of each aefR and mexT mutant in Pseudomonas syringae pv. tabaci 6605. Mol. Genet. Genomics 287:473–484. 10.1007/s00438-012-0693-9 [DOI] [PubMed] [Google Scholar]
- 31. Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066–2079. 10.1128/JB.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. 2006. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188:3365–3370. 10.1128/JB.188.9.3365-3370.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gurich N, González JE. 2009. Role of quorum sensing in Sinorhizobium meliloti-alfalfa symbiosis. J. Bacteriol. 191:4372–4382. 10.1128/JB.00376-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coutinho BG, Mitter B, Talbi C, Sessitsch A, Bedmar EJ, Halliday N, James EK, Cámara M, Venturi V. 2013. Regulon studies and in planta role of the BraI/R quorum-sensing system in the plant-beneficial Burkholderia cluster. Appl. Environ. Microbiol. 79:4421–4432. 10.1128/AEM.00635-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chugani S, Kim BS, Phattarasukol S, Brittnacher MJ, Choi SH, Harwood CS, Greenberg EP. 2012. Strain-dependent diversity in the Pseudomonas aeruginosa quorum-sensing regulon. Proc. Natl. Acad. Sci. U. S. A. 109:E2823–E2831. 10.1073/pnas.1214128109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo SJ, Khrebtukova I, Yang Y, Thannhauser T, Butcher BG, Cartinhour S, Schneider DJ. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J. Bacteriol. 192:2359–2372. 10.1128/JB.01445-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stoitsova SO, Braun Y, Ullrich MS, Weingart H. 2008. Characterization of the RND-type multidrug efflux pump MexAB-OprM of the plant pathogen Pseudomonas syringae. Appl. Environ. Microbiol. 74:3387–3393. 10.1128/AEM.02866-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livny J, Teonadi H, Livny M, Waldor MK. 2008. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS One 3:e3197. 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moll S, Schneider DJ, Stodghill P, Myers CR, Cartinhour SW, Filiatrault MJ. 2010. Construction of an rsmX co-variance model and identification of five rsmX non-coding RNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol. 7:508–516. 10.4161/rna.7.5.12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heeb S, Blumer C, Haas D. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens Chao. J. Bacteriol. 184:1046–1056. 10.1128/jb.184.4.1046-1056.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73:434–445. 10.1111/j.1365-2958.2009.06782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. González N, Heeb S, Valverde C, Kay E, Reimmann C, Junier T, Haas D. 2008. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics 9:167. 10.1186/1471-2164-9-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hassan KA, Johnson A, Shaffer BT, Ren QH, Kidarsa TA, Elbourne LDH, Hartney S, Duboy R, Goebel NC, Zabriskie TM, Paulsen IT, Loper JE. 2010. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 12:899–915. 10.1111/j.1462-2920.2009.02134.x [DOI] [PubMed] [Google Scholar]
- 44. Rich JJ, Kinscherf TG, Kitten T, Willis DK. 1994. Genetic evidence that the gacA gene encodes the cognate response regulator for the LemA sensor in Pseudomonas syringae. J. Bacteriol. 176:7468–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mo YY, Gross DC. 1991. Plant signal molecules activate the syrB gene, which is required for syringomycin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 173:5784–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hrabak EM, Willis DK. 1993. Involvement of the lemA gene in production of syringomycin and protease by Pseudomonas syringae pv. syringae. Mol. Plant Microbe Interact. 6:368–375. 10.1094/MPMI-6-368 [DOI] [Google Scholar]
- 47. Bardoel BW, van der Ent S, Pel MJC, Tommassen J, Pieterse J, van Kessel KPM, van Strijp JAG. 2011. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog. 7:e1002206. 10.1371/journal.ppat.1002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Bruijn I, de Kock MJD, Yang M, de Waard P, van Beek TA, Raaijmakers JM. 2007. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 63:417–428. 10.1111/j.1365-2958.2006.05525.x [DOI] [PubMed] [Google Scholar]
- 49. Koch B, Nielsen TH, Sørensen D, Andersen JB, Christophersen C, Molin S, Givskov M, Sørensen J, Nybroe O. 2002. Lipopeptide production in Pseudomonas sp. strain DSS73 is regulated by components of sugar beet seed exudate via the Gac two-component regulatory system. Appl. Environ. Microbiol. 68:4509–4516. 10.1128/AEM.68.9.4509-4516.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Burch AY, Shimada BK, Mullin SWA, Dunlap CA, Bowman MJ, Lindow SE. 2012. Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. J. Bacteriol. 194:1287–1298. 10.1128/JB.06058-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Burch AY, Browne PJ, Dunlap CA, Price NP, Lindow SE. 2011. Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Environ. Microbiol. 13:2681–2691. 10.1111/j.1462-2920.2011.02534.x [DOI] [PubMed] [Google Scholar]
- 52. Cha JY, Lee DG, Lee JS, Oh JI, Baik HS. 2012. GacA directly regulates expression of several virulence genes in Pseudomonas syringae pv. tabaci 11528. Biochem. Biophys. Res. Commun. 417:665–672. 10.1016/j.bbrc.2011.11.124 [DOI] [PubMed] [Google Scholar]
- 53. Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, Haas D. 2007. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol. Microbiol. 66:341–356. 10.1111/j.1365-2958.2007.05909.x [DOI] [PubMed] [Google Scholar]
- 54. Arrebola E, Carrión VJ, Cazorla FM, Pérez-García A, Murillo J, de Vicente A. 2012. Characterisation of the mgo operon in Pseudomonas syringae pv. syringae UMAF0158 that is required for mangotoxin production. BMC Microbiol. 12:10. 10.1186/1471-2180-12-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kong HS, Roberts DP, Patterson CD, Kuehne SA, Heeb S, Lakshman DK, Lydon J. 2012. Effect of overexpressing rsmA from Pseudomonas aeruginosa on virulence of select phytotoxin-producing strains of P. syringae. Phytopathology 102:575–587. 10.1094/PHYTO-09-11-0267 [DOI] [PubMed] [Google Scholar]
- 56. Zeng Q, Laiosa MD, Steeber DA, Biddle EM, Peng Q, Yang CH. 2012. Cell individuality: the bistable gene expression of the type III secretion system in Dickeya dadantii 3937. Mol. Plant Microbe Interact. 25:37–47. 10.1094/MPMI-05-11-0105 [DOI] [PubMed] [Google Scholar]
- 57. Casabona MG, Silverman JM, Sall KM, Boyer F, Couté Y, Poirel J, Grunwald D, Mougous JD, Elsen S, Attree I. 2013. An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ. Microbiol. 15:471–486. 10.1111/j.1462-2920.2012.02816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goldová J, Ulrych A, Hercík K, Branny P. 2011. A eukaryotic-type signalling system of Pseudomonas aeruginosa contributes to oxidative stress resistance, intracellular survival and virulence. BMC Genomics 12:437. 10.1186/1471-2164-12-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gooderham WJ, Hancock REW. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33:279–294. 10.1111/j.1574-6976.2008.00135.x [DOI] [PubMed] [Google Scholar]
- 60. Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. 10.1016/j.cell.2013.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Haapalainen M, Mosorin H, Dorati F, Wu RF, Roine E, Taira S, Nissinen R, Mattinen L, Jackson R, Pirhonen M, Lin NC. 2012. Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 194:4810–4822. 10.1128/JB.00611-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Joyner DC, Lindow SE. 2000. Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology 146:2435–2445 [DOI] [PubMed] [Google Scholar]
- 63. Cornelis P, Bodilis J. 2009. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Rep. 1:256–262. 10.1111/j.1758-2229.2009.00041.x [DOI] [PubMed] [Google Scholar]
- 64. Burrell M, Hanfrey CC, Kinch LN, Elliott KA, Michael AJ. 2012. Evolution of a novel lysine decarboxylase in siderophore biosynthesis. Mol. Microbiol. 86:485–499. 10.1111/j.1365-2958.2012.08208.x [DOI] [PubMed] [Google Scholar]
- 65. Berti AD, Thomas MG. 2009. Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. J. Bacteriol. 191:4594–4604. 10.1128/JB.00457-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754. 10.1016/j.devcel.2004.08.020 [DOI] [PubMed] [Google Scholar]
- 67. Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23:249–259. 10.1101/gad.1739009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stockwell VO, Hockett K, Loper JE. 2009. Role of RpoS in stress tolerance and environmental fitness of the phyllosphere bacterium Pseudomonas fluorescens strain 122. Phytopathology 99:689–695. 10.1094/PHYTO-99-6-0689 [DOI] [PubMed] [Google Scholar]
- 69. Hagen MJ, Stockwell VO, Whistler CA, Johnson KB, Loper JE. 2009. Stress tolerance and environmental fitness of Pseudomonas fluorescens A506, which has a mutation in RpoS. Phytopathology 99:679–688. 10.1094/PHYTO-99-6-0679 [DOI] [PubMed] [Google Scholar]
- 70. Kidarsa TA, Shaffer BT, Goebel NC, Roberts DP, Buyer JS, Johnson A, Kobayashi DY, Zabriskie TM, Paulsen I, Loper JE. 2013. Genes expressed by the biological control bacterium Pseudomonas protegens Pf-5 on seed surfaces under the control of the global regulators GacA and RpoS. Environ. Microbiol. 15:716–735. 10.1111/1462-2920.12066 [DOI] [PubMed] [Google Scholar]
- 71. Vinatzer BA, Teitzel GM, Lee M-W, Jelenska J, Hotton S, Fairfax K, Jenrette J, Greenberg JT. 2006. The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62:26–44. 10.1111/j.1365-2958.2006.05350.x [DOI] [PubMed] [Google Scholar]
- 72. Huynh TV, Dahlbeck D, Staskawicz BJ. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374–1377. 10.1126/science.2781284 [DOI] [PubMed] [Google Scholar]
- 73. Tang XY, Xiao YM, Zhou JM. 2006. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant Microbe Interact. 19:1159–1166. 10.1094/MPMI-19-1159 [DOI] [PubMed] [Google Scholar]
- 74. Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809–824. 10.1046/j.1365-2958.2003.03740.x [DOI] [PubMed] [Google Scholar]
- 75. Boucher JC, Schurr MJ, Deretic V. 2000. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol. Microbiol. 36:341–351. 10.1046/j.1365-2958.2000.01846.x [DOI] [PubMed] [Google Scholar]
- 76. Heurlier K, Dénervaud V, Pessi G, Reimmann C, Haas D. 2003. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:2227–2235. 10.1128/JB.185.7.2227-2235.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dong T, Yu R, Schellhorn H. 2011. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 79:375–386. 10.1111/j.1365-2958.2010.07449.x [DOI] [PubMed] [Google Scholar]
- 78. Lindeberg M, Cunnac S, Collmer A. 2012. Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20:199–208. 10.1016/j.tim.2012.01.003 [DOI] [PubMed] [Google Scholar]
- 79. Chen CL, Malek AA, Wargo MJ, Hogan DA, Beattie GA. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29–45. 10.1111/j.1365-2958.2009.06962.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- 81. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 82. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 83. Nettleton D, Hwang JTG, Caldo RA, Wise RP. 2006. Estimating the number of true null hypotheses from a histogram of p values. J. Agric. Biol. Environ. Stat. 11:337–356. 10.1198/108571106X129135 [DOI] [Google Scholar]
- 84. Pavlidis P, Noble WS. 2003. Matrix2png: a utility for visualizing matrix data. Bioinformatics 19:295–296. 10.1093/bioinformatics/19.2.295 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative variation in transcript levels among each of the strain-treatment combinations based on median absolute residuals computed from our linear model analyses. Download
Loss of algU and rpoN was correlated with decreases in the abundance of the transcripts induced by both osmotic stress and growth in planta. Download
(A and B) Mutants lacking algU, hrpL, and rpoN exhibited large reductions in growth in epiphytic sites (A) and apoplastic sites (B)of bean leaves. Download
RpoN activated genes involved in N metabolism, coregulated much of the AlgU regulon, and regulated many traits in a plant signal-dependent manner. Download
Number of genes for which the difference in transcript abundance between the mutant and the wild type in a given environmental condition differed from that in the basal medium
Effects of environmental conditions on the responses of selected genes to the loss of target regulators
The functional categories in which the representation of differentially-expressed genes was greater than their representation among all of the strain B728a genes
Primers used in this study to construct deletion mutants of the indicated target genes