Abstract
Background
The concept of adaptive evolution implies underlying genetic mutations conferring a selective advantage to an organism under particular environmental conditions. Thus, a flow cytometry-based strategy was used to study the adaptive evolution in Chlamydomonas reinhardtii wild-type strain CC124 and starchless mutant sta6-1 cells, with respect to lipid metabolism under nitrogen-(N) depleted and -replete conditions.
Results
The successive sorting and regeneration of the top 25,000 high-lipid content cells of CC124 and sta6-1, combined with nitrogen starvation, led to the generation of a new population with an improved lipid content when compared to the original populations (approximately 175% and 50% lipid increase in sta6-1 and CC124, respectively). During the adaptive evolution period, the major fatty acid components observed in cells were C16:0, C16:1, C18:0, and C18:1-3, and elemental analysis revealed that cellular carbon to nitrogen ratio increased at the end of adaptive evolution period In order to gain an insight into highly stimulated intracellular lipid accumulation in CC124 and sta6-1 resulting from the adaptive evolution, proteomics analyses of newly generated artificial high-lipid content populations were performed. Functional classifications showed the heightened regulation of the major chlorophyll enzymes, and the enzymes involved in carbon fixation and uptake, including chlorophyll-ab-binding proteins and Rubisco activase. The key control protein (periplasmic L-amino acid oxidase (LAO1)) of carbon-nitrogen integration was specifically overexpressed. Glutathione-S-transferases and esterase, the enzymes involved in lipid-metabolism and lipid-body associated proteins, were also induced during adaptive evolution.
Conclusions
Adaptive evolution results demonstrate the potential role of photosynthesis in terms of carbon partitioning, flux, and fixation and carbon-nitrogen metabolism during lipid accumulation in microalgae. This strategy can be used as a new tool to develop C. reinhardtii strains and other microalgal strains with desired phenotypes such as high lipid accumulation.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-014-0117-7) contains supplementary material, which is available to authorized users.
Keywords: Chlamydomonas reinhardtii, Adaptive evolution, Flow cytometry, Proteomics
Background
Chlamydomonas reinhardtii, a unicellular green alga, has been investigated intensely as a robust model system for the formation of intracellular lipid bodies that contain triacylglycerol (TAG) [1-3]. The intracellular lipid body formation in C. reinhardtii depends on several factors including stress conditions such as nutrient starvation, temperature, salinity, and light intensity [4]. In the past few years, researchers have investigated the intracellular lipid accumulation in microalgae under different stress conditions [1,5]. However, the molecular mechanisms of lipid accumulation in relation with carbon and nitrogen metabolisms, and cell division remain poorly understood in microalgae. Detailed studies on the molecular mechanism of lipid accumulation in microalgae under stress conditions should facilitate improvements in the lipid productivity, cultivation processes, and strain development for biofuels production [4]. The cellular physiology of C. reinhardtii changes depending on nitrogen availability [6]. Under nitrogen-depleted conditions, neutral lipids and starch will accumulate to high levels to serve as a primary form of energy storage [4].
Among several C. reinhardtii strains, CC124 wild-type and sta6-1 mutants are broadly studied. The starchless sta6-1 mutants of C. reinhardtii have been reported to produce a higher level of TAG than the wild-type under nitrogen-depleted conditions [5]. The sta6-1 mutants are deficient of a central starch synthesis enzyme (ADP-glucose pyrophosphorylase) and accumulate less than 1% of the starch compared with the wild-type under nitrogen-depleted conditions [5]. This deficiency impacts carbon metabolism and flow which may induce accumulation of noticeable levels of TAG in sta6-1 mutants [6]. It has been proven that adaptive mutation plays an important role in the evolution of microorganisms [7]. Adaptive mutations have been reported in some microbes, such as bacteria and yeast, but not well-known for microalgae [8]. It has been reported that the evolution of the microalgae Dictyosphaerium chlorelloides has caused it to adapt to an environment containing the highly toxic material 2,4,6,-trinitrotoluene (TNT) [7]. We therefore studied the mechanism of adaptation by C. reinhardtii wild-type and sta6-1 mutant strains under nitrogen-depleted and -replete conditions using flow cytometry. A study by Ramanan et al. [9] proposed a separate chloroplast pathway for TAG synthesis in starchless mutants of C. reinhardtii strains. We performed adaptive evolution of wild-type CC124 and sta6-1 mutants from a TAG accumulation perspective which included nitrogen starvation, time course, cell density-dependent adaptive evolution, and the overall yields of lipids, elemental composition, and proteomic analyses.
Flow cytometry enables the analysis of the different features or physiological states of microalgae at the single-cell level. Cells with a specific characteristic can be separated from the heterologous population for growth or analysis using fluorescence-activated cell sorting (FACS) [10]. Enhanced production of lipid bodies can be achieved by optimization of the production processes of microalgae, or the selection of strains with improved features or overproducers [11]. The unique ability of microalgae to adapt their metabolism to various culture conditions provides opportunities to modify and maximize the lipid production [11]. For example adaptive responses, which help microalgae to survive under environmental stresses, can cause the algae to maximize the lipid content including polyunsaturated fatty acids [12]. Therefore, gaining more detailed information on the underlying regulatory mechanisms is necessary to devise and implement such production strategies [13]. More recently we have examined and compared intracellular lipid body accumulation in C. reinhardtii wild-type and mutant sta6-1 strains by flow cytometry [14]. Using different fluorescent dyes in combination with FACS, we can isolate microalgal strains that produce high levels of lipid in certain stress conditions or after treating them with different mutagens. Doan and Obbard [15], and Montero et al. [16] isolated lipid-overproducing Nannochloropsis and T. suecica strains, respectively, using lipophilic fluorescent dye Nile Red in combination with FACS. Recently, we have demonstrated that the fluorescent dye BODIPY 505/515, in combination with FACS, allows single-cell level isolation and regeneration of algal strains that possess a high lipid content [14].
The recent genome sequencing of C. reinhardtii helps us to understand the molecular mechanisms of lipid body formation, TAG synthesis, and accumulation in microalgae [2,6]. Comprehensive proteomics analysis provides clear insight into the mechanisms of microalgal lipid biosynthesis. Proteomics analyses have been reported for chloroplast, mitochondria, and lipid bodies [17-20]. However, proteomics analysis of the total proteome of C. reinhardtii has so far been lacking. In this study, our primary focus is developing a flow cytometry-based strategy to address adaptive evolution and strains development with respect to high lipid accumulation. To this effect, we established a unique flow cytometry-based adaptive evolution procedure for C. reinhardtii wild-type and mutant sta6-1 strains that have been subjected to nitrogen limitation conditions for over 50 days. Specifically, we analyzed the total proteomics of C. reinhardtii strains to determine their metabolic response to the adaptive evolution.
Results and discussion
Physiological behavior of wild-type and mutant sta6-1 during adaptive evolution under nitrogen-depleted and -replete conditions
The primary objective of this study was to specifically select and regenerate the high-lipid content cells of C. reinhardtii strains from a heterogeneous population of cells that contain a wide range of lipid contents (Figure 1). Microalgal cellular growth and metabolic pathways respond in a highly dynamic way to environmental conditions, especially nutrient availability [21]. To obtain a healthy population (red-color region in Figure 1), the seed cultures of wild-type CC124 and mutant sta6-1 were obtained from the late-log phase in nitrogen-supplemented Tris Acetate Phosphate liquid Medium (TAP) medium. The initial seed cultures for inoculum were maintained around approximately 2.0 × 106 cells ml1 in TAP medium. The lipophilic fluorescent dye BODIPY 505/515 was used to stain the wild-type CC124 and mutant sta6-1 for flow cytometry screening and cell sorting as described previously [14]. Prior to sorting (0 hours), samples were taken immediately after re-suspension in TAP medium. Since our aim is to isolate lipid overproducing strains, 25,000 cells from the high-lipid content population of BODIPY 505/515-stained seed cells of CC124 and sta6-1 were sorted directly into TAP medium and were re-suspended in nitrogen-depleted or -replete TAP medium separately (in triplicate).
Figure 1.
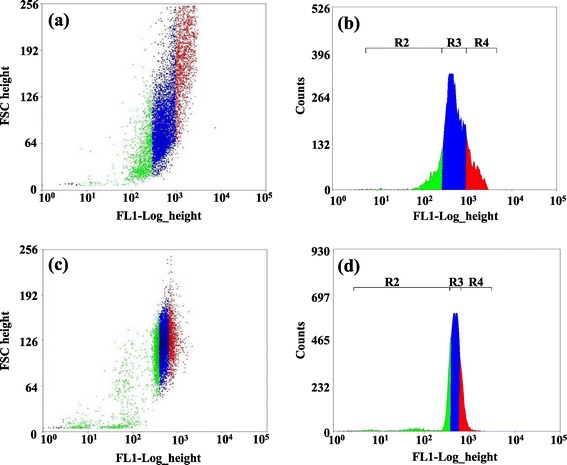
Two dimensional dot plots (a, c) and flow cytograms (b, d) of BODIPY 505/515-stained 0 hour seed cells of CC124 (a, b) and sta6-1 (c, d). Region R2 (green), R3 (blue), and R4 (red) in flow cytograms represent low, medium, and high lipid content cells, respectively. A total of 25,000 cells from the top 2 to 3% of R4 region were sorted and regenerated in nitrogen depletion and repletion TAP medium, separately. The mean values of the histograms of the seed cells of CC124 and sta6-1 were 662.11 and 494.35, respectively. Abbreviations: BODIPY, 4,4-difluoro-1,3,5,7-tetramethyl-r-bora-3a,41-diaza-s-indacene.
At the beginning of evolution period, the wild-type CC124 was found to reach high cell density (or late-stationary phase) by day 19 (Figure 2). The mutant sta6-1 showed high cell density (or late-stationary phase) by day 14 (Figure 2). Cells from day 19 and day 14 cultures of strains CC124 and sta6-1 respectively were considered as first generation cultures and prepared for next consecutive rounds of cell sorting. A total of 25,000 cells from the top 2% were sorted using flow cytometry. Sorted cells from each round (at indicated intervals) were used for inoculation of a new culture and the remaining portions of the cells were used for the measurement of total lipid content, microscopic analysis, elemental analysis, and proteomics analysis. After three rounds of sorting, the wild-type CC124 was shown to have relatively slow growth compared with earlier sorting periods, and finally reached high cell density by day 62. A possible reason for the slow growth at this time point (from day 32 to day 62) is that during the sorting, cells may partially affect by the fluid acceleration, electrical or mechanical shock, and optical stress and that caused the slower growth of sorted cells at the initial stage. However, the cells showed healthy growth after the initial stage and reached high cell density by day 62. In our adaptive evolution experiments, we kept the 0 hour seed and first generation cultures as positive controls. The lipid accumulation and microalgal growth performance of further generations were compared with 0 hour (seed cultures) and first generation positive controls. The cell concentrations of CC124 and sta6-1 in the presence and absence of nitrogen were measured and shown in Figure 2. The starchless strain sta6-1 had attained the highest cell concentrations in nitrogen-depleted condition rather than nitrogen-replete conditions during the adaptive evolution period. This indicates that the rate of arrest of cell division in nitrogen-depleted conditions was relatively lower than nitrogen-replete conditions during the adaptive evolution period. The average highest cell concentration for both strains were found to be approximately 25 × 106 cells ml−1 in nitrogen-depleted condition during the adaptive evolution period (Figure 2). The wild-type CC124 showed overall similar cell concentrations in nitrogen-depleted and -replete conditions at all indicated times (Figure 2a). This indicates that wild-type CC124 cell divisions and regenerations are similar in both media regardless of whether nitrogen is present or not. However, the starchless mutant sta6-1 showed a different growth pattern, as relatively fewer cell concentrations were observed in nitrogen-replete conditions at all indicated times (Figure 2b). It is interesting to observe that the mutant strains have superior growth in nitrogen-depleted conditions than nitrogen-replete conditions.
Figure 2.
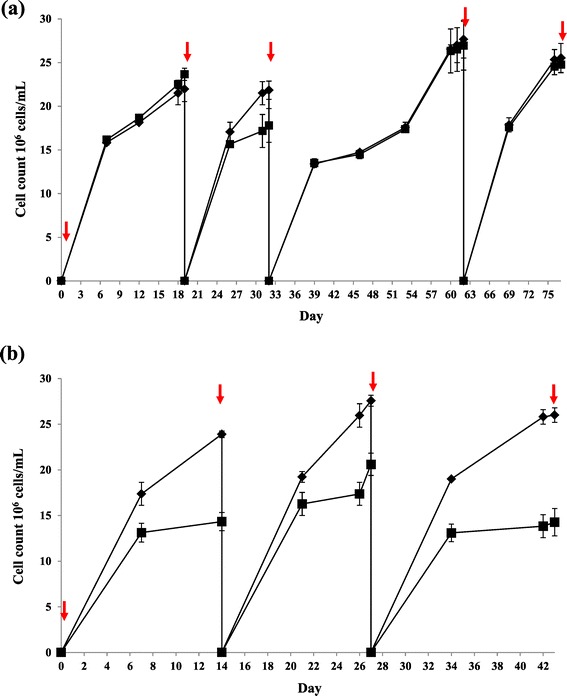
Growth curve (cell count) for CC124 (a) and the sta6-1 (b) strains, in nitrogen-depletion (diamond) and -repletion (square) TAP medium. Each data point represents three replicates. Arrows indicate the inoculation points of the 25,000 cells from the top 2% of cells sorted using flow cytometry (Day 0 indicates inoculation of 25,000 cells from the top 2% of 0 hour seed culture). Abbreviations: TAP, Tris Acetate Phosphate liquid Medium.
Lipid accumulation in C. reinhardtii strains during adaptive evolution
C. reinhardtii CC124 and mutant sta6-1 were periodically collected during the course of adaptive evolution at indicated times and were stained with BODIPY 505/515 for FACS analysis of intracellular lipid bodies. The lipophilic dye BODIPY 505/515 can be used as a vital stain for the detection of neutral lipid bodies in the C. reinhardtii strains. Recently, BODIPY-flow cytometry-based protocol has been used to evaluate the lipid accumulation performance between wild-type and transgenic mutants of the diatom Thalassiosira pseudonana [22]. Figure 3 shows the autofluorescence intensities for unstained cells, along with the FL1 signals of BODIPY 505/515-stained CC124 and sta6-1. Flow cytograms from each sorting round for CC124 and sta6-1 are representative of the green fluorescence (FL1) from the first to last rounds of sorting for CC124 and sta6-1 during the adaptive evolution period in nitrogen-depleted conditions. We found that wild-type CC124 required a longer time to attain highest lipid accumulation than the starchless mutant sta6-1. The mean fluorescence intensity (FL1) from BODIPY staining of CC124 peaked on day 77 and similar autofluorescence intensity results were observed in nitrogen-depleted conditions (Figure 3a). The mutant sta6-1 attained the highest lipid content more rapidly compared to CC124, as it only took 43 days to reach peak autofluorescence intensity, which was observed throughout nitrogen-depleted conditions (Figure 3b). However, after further sorting and regeneration, CC124 and sta6-1 strains were found to not be accumulating more lipids (data not shown). This could be due to the fact that the maximum level of lipid accumulation had been achieved at or near those days during adaptive evolution, and spontaneous mutations might have caused cells to return to their original phenotype under some selective conditions [8]. Figure 3 presents a sequence of autofluorescence and fluorescence histograms related to the increased lipid accumulation in CC124 and sta6-1 during adaptive evolution. It is evident that there was a well-defined shift of autofluorescence as well as BODIPY 505/515-stained cells from the initial day to the final day of adaptation. This indicates the progressive movement of selected microalgal populations towards a significantly higher level of lipid accumulation in nitrogen starvation conditions during the adaptive evolution period. However, wild-type CC124 and mutant sta6-1 were unable to achieve progressive lipid accumulation in nitrogen-replete conditions (FACS data not shown, please refer to the lipid content measurement data in nitrogen-replete conditions in Table 1).
Figure 3.
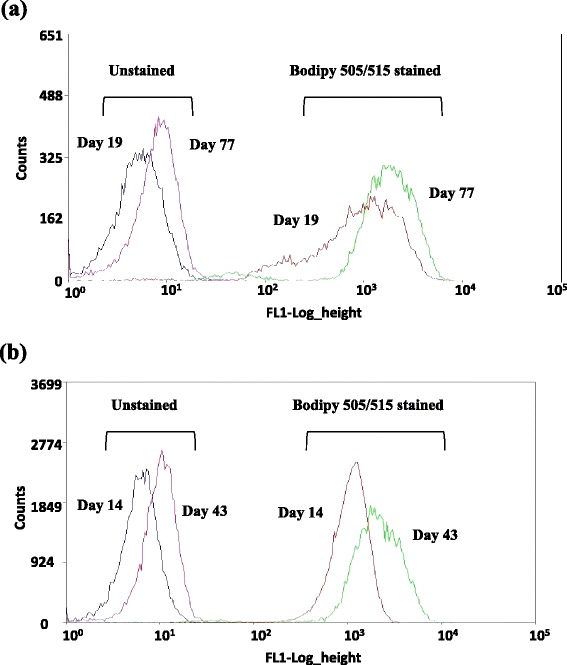
Flow cytograms of adaptive evolved strains of CC124 (a) and sta6-1 (b) in nitrogen-depletion TAP medium. (a) CC124 - Blue and purple representing unstained cells of day 19 and 77, respectively. Red and green representing BODIPY 505/515-stained cells of day 19 and 77, respectively. (b) sta6-1 - Blue and purple representing unstained cells of day 14 and 43, respectively. Red and green representing BODIPY 505/515-stained cells of day 14 and 43, respectively. The mean values of histograms of unstained and BODIPY 505/515-stained cells of CC124 of day 19 and 77 were 6.7, 10.98, 1724.45, and 2206.01, respectively. The mean values of histograms of unstained and BODIPY 505/515-stained cells of sta6-1 of day 14 and 43 were 5.2, 9.54, 1138.34, and 2427.78, respectively. Abbreviations: TAP, Tris Acetate Phosphate liquid Medium; BODIPY, 4,4-difluoro-1,3,5,7-tetramethyl-r-bora-3a,41-diaza-s-indacene.
Table 1.
GC quantification of FAME content of adaptive evolved strains of CC124 and sta6-1 at different stages
| Strain | Different stages (Day) | Nitrogen-repletion conditions | Nitrogen-depletion conditions | Fatty acid (% of total fatty acids in nitrogen-depletion conditions) a | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total lipid (%) | Total lipid (%) | C14:0 | C14:1 | C15:0 | C15:1 | C16:0 | C16:1 | C17:1 | C18:0 | C18:1-3 (n) b | ||
| CC124 | 19 | 6.6 | 10.5 | 0.3 | 0.2 | 0.2 | 0.3 | 39.3 | 2.7 | 2.7 | 5.4 | 45.3 |
| 32 | 5.9 | 13.5 | 0.3 | 0.0 | 0.2 | 0.2 | 40.1 | 4.5 | 2.8 | 3.4 | 45.3 | |
| 62 | 5.3 | 11.4 | 0.2 | 0.0 | 0.2 | 0.4 | 51.7 | 0.7 | 2.0 | 8.5 | 34.8 | |
| 77 | 7.0 | 15.9 | 0.2 | 0.0 | 0.2 | 0.4 | 40.7 | 0.5 | 1.2 | 8.9 | 46.4 | |
| sta6-1 | 14 | 5.8 | 5.9 | 0.4 | 2.0 | 0.6 | 0.8 | 85.3 | 2.7 | 0.7 | 5.2 | 45.0 |
| 27 | 6.7 | 8.1 | 1.1 | 0.0 | 0.5 | 0.7 | 40.6 | 1.2 | 1.8 | 5.0 | 47.0 | |
| 43 | 7.4 | 14.0 | 0.2 | 0.0 | 0.1 | 0.3 | 48.9 | 0.6 | 2.4 | 11.3 | 35.8 | |
aAll data are expressed as mean quantity of triplicate experiments.
bC18:1n9c, C18:2n6t, C18:3n6c. Abbreviations: GC, Gas chromatography; FAME, Fatty acid methyl esters.
To confirm the conclusions drawn from the FACS analysis, the fatty acid methyl esters (FAME) contents of adaptive evolved CC124 and sta6-1 strains were quantified at all indicated time points by gas chromatography. Table 1 shows the separation by GC of FAME, prepared from total lipids of C. reinhardtii strains, comparison of the fatty acid composition of the TAG fraction, and total lipids. During the adaptive evolution period, the major fatty acid components observed in CC124 and sta6-1 cells were C16:0, C16:1, C18:0, and C18:1-3 (Table 1). Sequential increase of lipid content in the adaptive evolution period indicates the intergenerational stability of lipid levels of sorted CC124 and sta6-1 strains. The presence of palmitic, palmitoleic, stearic, and linoleic acids indicated a stable fatty acid composition (Table 1). C18:0 and C18:1-3 profiles of adaptive evolution strains of CC124 and sta6-1 showed a gradual increase throughout the adaptation. After adaptive evolution, CC124 and sta6-1 showed a 50% and 175% increase (10.52 to 15.9% and 5.94 to 14.04%) of lipids, respectively. As shown in FACS analysis (Figure 3), the mutant sta6-1 achieved the highest lipid (15.9%) content within 43 days. On the other hand, under nitrogen-replete conditions, the mutant sta6-1 showed relatively little increase in lipid content (5.84 to 7.42%), and wild-type CC124 showed no increase in lipid content.
The lipid accumulation in the cells that went through adaptive evolution was analyzed with confocal microscope. Figure 4 shows confocal images of BODIPY 505/515-stained cells of CC124 and sta6-1 after adaptive evolution. The lipophilic property of the dye facilitates the BODIPY 505/515 accumulation in intracellular lipid compartments of the cell [23]. The strong red autofluorescence allowed the chlorophyll content to be easily distinguished from the lipid content (Figure 4). The confocal microscopy showed that the numbers of intracellular lipid bodies massively increased at the end of adaptive evolution (more than 22 intracellular lipid bodies were found per cell in adapted CC124 and sta6-1) in nitrogen-depleted condition (Figure 4a and c). Recently, Goodson et al. [1] reported that sta6 mutants had a range of 6 to 25 lipid bodies per cell in nitrogen starvation conditions, with an acetate boost. The large increase in numbers of intracellular lipid bodies in the adaptively evolved CC124 and sta6-1 strains under nitrogen-depleted conditions is a new observation. Meanwhile, the numbers of intracellular lipid bodies did not increase in CC124 and sta6-1 under nitrogen-replete condition (Figure 4b and d).
Figure 4.
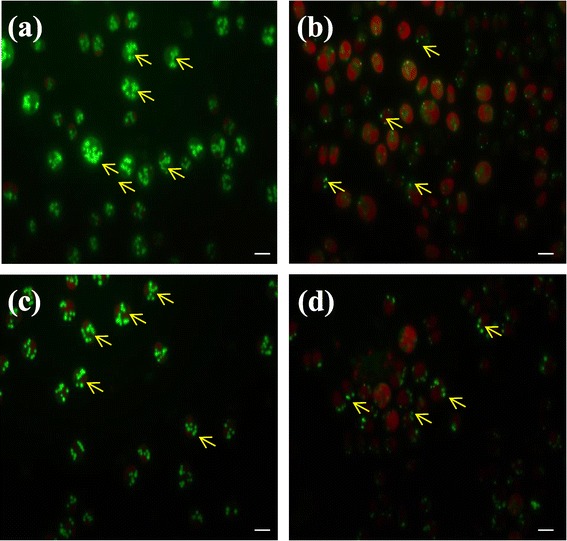
Confocal images of BODIPY 505/515-stained adaptive evolved Chlamydomonas reinhardtii strains. (a) CC124 in nitrogen-depleted conditions at day 77; (b) CC124 in nitrogen-replete conditions at day 77; (c) sta6-1 in nitrogen-depleted conditions at day 43; (d) sta6-1 in nitrogen-replete conditions at day 43. Arrows indicate lipid droplets. Each scale bar indicates 5 μm.
As shown in Figure 4b and 4d, the chlorophyll content in the adapted cells of CC124 and sta6-1 was found to be at a steady level during cultivation under nitrogen-replete conditions. These observations are in agreement with recent findings regarding the chlorophyll content in CC124 and sta6 in a nitrogen-replete medium [5]. Meanwhile, the chlorophyll content was significantly reduced in cells of CC124 and sta6-1 in nitrogen-depleted medium at the end of the adaptive evolution period (Table 2). Several researchers have also reported that the chlorophyll content of C. reinhardtii decreased under nitrogen limitation conditions [5,21,24,25]. It is probable that the accelerated loss of chlorophyll in the starchless mutants of C. reinhardtii is associated with the more severely attenuated O2 evolution activities observed in these strains [5]. Due to reduced levels of chlorophyll, less light energy is absorbed to be used for carbon fixation and cell division [21].
Table 2.
Autofluorescence of chloroplast of adaptive evolved strains CC124 and sta6-1 at different stages
| Strain | Different stages (Day) | Mean fluorescence intensity (FL5 channel) (arbitrayunits) a | |
|---|---|---|---|
| Unstained | BODIPY 505/515 stained | ||
| CC124 | 19 | 24.8 | 22.2 |
| 77 | 17.0 | 16.4 | |
| sta6-1 | 14 | 15.9 | 16.3 |
| 43 | 11.7 | 11.4 | |
aAll data are expressed as mean quantity of triplicate experiments. Abbreviations: BODIPY, 4,4-difluoro-1,3,5,7-tetramethyl-r-bora-3a,41-diaza-s-indacene.
Elemental analysis revealed that the cellular carbon to nitrogen ratio (C:N) increased at the end of adaptive evolution period (from day 19 to day 77 and day 14 to day 43, in CC124 and sta6-1, respectively) (Table 3). At day 19 and day 77 for CC124 strain, the C:N ratio was 9.5:1 and 11.1:1, respectively. At day 14 and day 43 for sta6-1 strain, the C:N ratio was 10.3:1 and 11.8:1, respectively (Table 3). This increase in C:N could also be explained in the context of an increase in lipid accumulation [26]. A unicellular marine diatom Phaeodactylum tricornutum reported an increase in C:N ratio as the intracellular lipid accumulation increased [26]. However this C:N ratio did not influence specific intracellular cellular carbohydrates [26].
Table 3.
Elemental analysis of adaptive evolved strains CC124 and sta6-1 at different stages
| Strain | Different stages (Day) | Biochemical elements a | C:N ratio | |||
|---|---|---|---|---|---|---|
| C | N | H | S | |||
| CC124 | 19 | 47.8 ± 0.14 | 5.0 ± 0.03 | 7.1 ± 0.1 | 0.3 ± 0.01 | 9.5:1 |
| 32 | 49.2 ± 0.1 | 5.3 ± 0.01 | 7.2 ± 0.004 | 0.4 ± 0.001 | 9.2:1 | |
| 62 | 50.3 ± 0.13 | 4.6 ± 0.02 | 7.5 ± 0.02 | 0.3 ± 0.002 | 10.9:1 | |
| 77 | 51.0 ± 0.13 | 4.6 ± 0.04 | 7.5 ± 0.02 | 0.3 ± 0.003 | 11.1:1 | |
| sta6-1 | 14 | 49.2 ± 0.04 | 4.8 ± 0.01 | 7.2 ± 0.03 | 0.3 ± 0.01 | 10.2:1 |
| 27 | 47.2 ± 0.1 | 5.9 ± 0.0004 | 6.9 ± 0.002 | 0.3 ± 0.01 | 8.0:1 | |
| 43 | 52.5 ± 0.2 | 4.5 ± 0.02 | 7.8 ± 0.03 | 0.3 ± 0.01 | 11.8:1 | |
aAll data are expressed as mean quantity of triplicate experiments. Abbreviations: C:N, Carbon:Nitrogen ratio; C, Carbon; N, Nitrogen, H, Hydrogen; S, Sulfur.
Molecular response of C. reinhardtii for adaptive evolution
To understand the molecular mechanisms of adaptive evolution under nitrogen starvation, we performed comprehensive proteomics analyses of the adaptively evolved cells of CC124 and sta6-1. At all indicated time points, lyophilized cells (nitrogen-depleted conditions) of adaptive evolved CC124 and sta6-1 were used for total protein extraction. The extracted protein materials were separated by two-dimensional gel electrophoresis using two different pH ranges (pH 3 to 10 and 4 to 7), separately. All scanned two-dimensional gel images of proteomes are provided in supplementary information (Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, and Additional file 4: Figure S4). Interestingly, we observed that the numbers of protein spots expressed between pH range 4 to 7 gradually increased in line with the adaptive evolution period for CC124 and sta6-1 (Table 4, Additional file 3: Figure S3, and Additional file 4: Figure S4). A recent report by Choi et al. [27] studied the comparative proteomic response of lipid metabolism in C. reinhardtii between pH range 4 to 10 using lipid over- and underproducing strains. In addition, the majority of earlier reports have been heavily centered at various specific targets (proteomic characterization of mitochondria [17], chloroplast [18], and lipid bodies [19]). While preparing this paper, Wase et al. [28] reported the protein profiling of nitrogen stress response in the C. reinhardtii strain using iTRAQ (isobaric Tags for Relative and Absolute Quantitation) methodology. However, the protein profiling of C. reinhardtii strains had not been characterized between different pH ranges. Thus, the observation of high amounts of accumulation of protein spots between acidic to neutral ranges (pH 4 to 7) is a new observation for C. reinhardtii CC124 and mutant sta6-1.
Table 4.
List of numbers of protein spots identified between pH range 4 to 7 for adaptive evolved cells of CC124 and sta6-1
| Strain | Different stages (Day) | Number of protein spots |
|---|---|---|
| CC124 | 19 | 578 |
| 32 | 785 | |
| 62 | 805 | |
| 77 | 876 | |
| sta6-1 | 14 | 598 |
| 27 | 759 | |
| 43 | 969 |
An extensive two-dimensional gel image analysis was carried out using PDQuest (Version 7.0, Bio-Rad, Hercules, CA, USA) software and differentially expressed protein profiles were identified at different time points during the adaptive evolution. After an extensive examination of image analysis results, proteins which exhibited a fold change difference of greater or less than 3-fold, at different time points of adaptive evolution, were considered to be significantly up- or down-regulated. We selected 60 significantly up- or down-regulated spots (common spots for both wild-type CC124 and mutant sta6-1) for protein identification using MALDI-TOF (Matrix-Assisted Laser Desorption-Ionization Time-of-Flight), and out of these we were able to identify 44 protein spots (Figures 5 and 6). The spots were selected based on their gradual differential expression patterns and specific differences in response to the adaptive evolution period; therefore it is conceivable that the differential expression was due to adaptive evolution. Among 44 spots, glutathione S-transferase (GST), Rubisco activase (RuBA), isocitrate lyase (ICL), mitochondrial translation factor Tu (mtTF-Tu), periplasmic L-amino acid oxidase catalytic subunit (LAO1), and mitochondrial carbonic anhydrase β-type (mtCA) were found twice. Among 44 spots, four proteins including protein phosphates (PPI), Ran-like small GTPase (GTPase), mitochondrial translation factor (mtTF-Tu), and mitochondrial carbonic anhydrase (mtCA) were found to be down-regulated, but all the other proteins including esterase (EST), pyruvate-formate lyase (PFL), and Rubisco exhibited up-regulation during adaptation (Figures 5 and 6). All spot images of the identified proteins during the adaptations are provided in Additional file 5: Figure S5 and Additional file 6: Figure S6.
Figure 5.
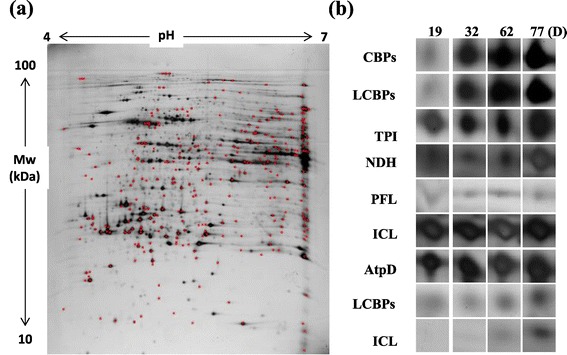
Proteome profiles of CC124 during adaptive evolution period. (a) The proteins showing differentially expressed levels during the adaptive evolution period are indicated as red points. (b) Of these, nine zoomed in spot areas highlighted from the day 19 profile gel are compared to corresponding protein spots of the day 32, 62, and 77 profile gels (comparison of all 20 proteins spots is provided in Additional file 5: Figure S5). The abbreviations for the enzymes included are as follows: CBP, chlorophyll-ab-binding proteins; LCBP, light harvesting proteins; TPI, triose phosphate isomerase; NDH, NADP depended-malate dehydrogenase; PFL, pyruvate-formate lyase; ICL, isocitrate lyase; AtpD, ATP synthase CF1 beta subunit.
Figure 6.
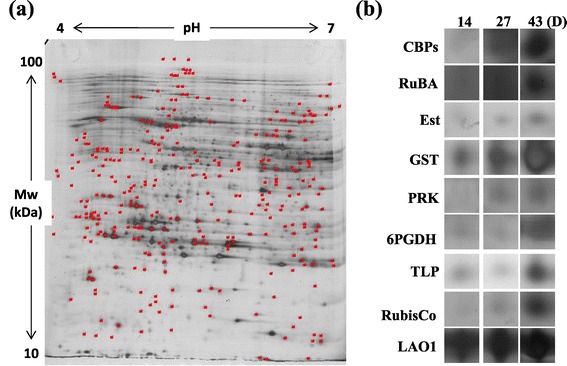
Proteomic profiles of sta6-1 during adaptive evolution period. (a) The proteins showing differentially expressed levels during the adaptive evolution period are indicated as red points. (b) Of these, nine zoomed in spot areas highlighted from the day 14 profile gel are compared to corresponding protein spots of the day 27 and 43 profile gels (comparison of all 24 proteins spots are provided in Additional file 6: Figure S6). The abbreviations for the enzymes included are as follows: CBP, chlorophyll-ab-binding proteins; RuBA, rubisco activase; Est, esterase; GST, glutathione-S-transferases; PRK, Phosphoribulokinase Rubisco activase; 6PGDH, 6-phosphogluconate dehydrogenase; TLP, thylakoid lumen protein; RubisCo, Ribulose-1,5-biphosphate carboxylase/oxygenase large subunit; LAO1, periplasmic L-amino acid oxidase catalytic subunit.
Functional classification showed that the differentially expressed proteins were involved in photosynthesis, mitochondrial metabolism, lipid metabolism, vesicle trafficking, and other functions (Additional file 7: Table S1). Most of the identified proteins were chloroplast, mitochondrial, and lipid-related proteins [19,20]. Observations of an increased number of differentially expressed chloroplast proteins (especially higher abundance and up-regulation of Calvin cycle enzymes) revealed the potential role of carbon uptake and fixation pathways during lipid accumulation in C. reinhardtii strains (Figure 7). It is clear that stress conditions significantly modulate photosynthesis and anabolic processes in the C. reinhardtii strains [5]. Under some circumstances, the intracellular mechanisms are activated to sense total nitrogen levels in the medium in order to balance the use of nitrogen for protein or nucleotide biosynthesis [21]. In contrary to the reduction in chlorophyll content that was observed (Table 2), heightened regulation of the major chlorophyll enzymes and the enzymes involved in carbon fixation and carbon uptake were found, especially chlorophyll-ab-binding proteins (CBP), light harvesting proteins (LCBP), dehydroascorbate reductase (DHAR), Rubisco, and Rubisco activase (RuBA) (Additional file 5: Figure S5, Additional file 6: Figure S6, and Additional file 7: Table S1). A recent report by Valenzuela et al. [26] revealed that multiple carbon fixation pathways were activated during lipid accumulation in the marine diatom P. tricornutum. Valenzuela et al. [26] reported that when cell growth slowed and dissolved inorganic carbon (DIC) became more available during the nutrient-depleted conditions, carbon concentrating mechanism (CCM) may not be needed, but biophysical CCM could still increase CO2 flux to Rubisco by facilitating bicarbonate transport into chloroplast. This mechanism would potentially concentrate CO2 in the lumen surrounding the plastid for increased delivery to Rubisco. Some molecular components associated with the CCM have been identified [29,30], however, the association between the Rubisco function and CCM activity in Chlamydomonas had not previously been proven. Recently, Meyer et al. [31] reported that α-helices of Rubisco small subunit control pyrenoid formation in Chlamydomonas and the line between the pyrenoid and a CCM improves the operating efficiency of carbon assimilation and overcomes diffusive limitations in aquatic photosynthesis. In evolutionary terms, the activity of a CCM in Chlamydomonas involves intermediate specificity of Rubisco [31]. It has been suggested that the operational efficiency of CCM in Chlamydomonas can be determined or correlated with the evolution of Rubisco [31]. In addition, 6-phosphogluconate dehydrogenase (6PGDH), involved in the oxidative pentose-phosphate pathway (OPPP), was found to be overexpressed (Additional file 7: Table S1). 6PGDH reportedly oxidizes the glucose-6-phosphate into 6-phosphoglucolactone and further yields 6-phosphogluconate followed by Calvin cycle protein ribulose-5-phosphate (Ru5P) [4]. Interestingly, triose phosphate isomerase (TPI) was up-regulated, which is key enzyme that converts the triose phosphate (G3P) of Calvin cycle into dihydroxyacetone phosphate (DHAP). This reversible reaction (conversion of G3P into DHAP) is a key step for glycolysis and gluconeogenesis in chloroplasts [4]. These observations strongly support the hypothesis that detailed knowledge on carbon partitioning as nutrient conditions change is important to predict carbon flow for lipid synthesis [26]. When microalgal strains were allowed for adaptation under nutrient starvation conditions, multiple changes occurred in proteins involved in carbon, nitrogen, and energy metabolism. These proteins were involved in both cellular development and lipid accumulation metabolisms. For example, under continuous nitrogen starvation conditions, carbon metabolism in C. reinhardtii shifted from glucose synthesis to utilization and storage as starch [28]. It has been reported that four central enzymes were involved in microalgal carbon fixation, highly regulated under continuous light stress conditions [32]. These included phosphoribulokinase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, and triose phosphate isomerase [32]. It is notable that triose phosphate isomerase (TPI) was also significantly overexpressed in our study. The abundance of carbon fixation proteins during constant stress conditions increased lipid synthesis and storage. Furthermore, this lipid accumulation caused slower growth and reduction in photosynthetic pigments (note the reduction in chloroplast fluorescence under nitrogen starvation). However, the details of this process or linkage are still unknown and it has recently been suggested that a deeper understanding of carbon flux throughout the full range of cellular processes is necessary to optimize both growth and lipid accumulation in microalgal cultures [22]. In our study, the enzymes involved in nitrogen/carbon balance metabolism are up-regulated during adaptive evolution. The key control protein (periplasmic L-amino acid oxidase (LAO1)) of carbon-nitrogen integration was specifically overexpressed (Figure 6). This enzyme reportedly releases ammonium in addition to providing a supply of carbon backbones that feed back into the TCA cycle and acetyl-CoA [21]. A very recent study by Wase et al. [28] also found that the abundance of LAO1 highly increased in C. reinhardtii during nitrogen starvation conditions. It is suggested that the ammonia released by LAO1 could be transported into the plastid and assimilated to produce glutamate. Glutamate is one of the key molecules that connect the amino acid metabolic pathways to the carbohydrate and lipid biosynthetic pathways [28]. The majority of our proteomics data is in line with a recent iTRAQ based proteomics study of C. reinhardtii under nitrogen starvation conditions [28], which also finds a higher regulation of enzymes involved in carbon and nitrogen cycles, especially 6PGDH, TPI, and ALD. If we combine these results, it is conceivable that predicting the exact role of elements involved in an algal CO2-concentrating mechanism (CCM) can be used to enhance lipid productivity in algae. In addition, the major enzyme involved in glyoxylate shunt was also found to be overexpressed; the overexpression of glyoxylate shunt enzyme such as isocitrate lyase (ICL), and the overexpression of NADP dependent-malate dehydrogenase (NDH) enzyme which plays crucial role in chloroplast C4 cycle, supports the increase of the capacity to produce carbon backbone substrates [21]. Glutathione-S-transferases (GST) and esterase (EST), which are enzymes involved in lipid metabolism, were overexpressed during adaptive evolution. Most of the other identified proteins from our study were identical with those proteins identified from lipid-body proteomics [19,20]. These include Ran-like small GTPase (GTPase), ribosomal protein Sa (RPSA), peptidyl-prolyl cis-trans isomerase (PPlase), Rab GDP dissociation inhibitor protein (Rab GDP), aldehyde dehydrogenase (ALD), gamma-hydroxybutyrate dehydrogenase (GHB), and α and β-tubulin proteins (α-tub/β-tub; Additional file 7: Table S1). We are currently studying the changes and correlations between genetic (inheritable) alterations and the physiological acclimation of microalgal strains during adaptive evolution period. Overall, these molecular results from our adaptive evolution study demonstrate the potential role of photosynthesis performance in line with carbon partitioning, flux, fixation, and carbon/nitrogen metabolism during lipid accumulation in microalgae under nitrogen starvation.
Figure 7.
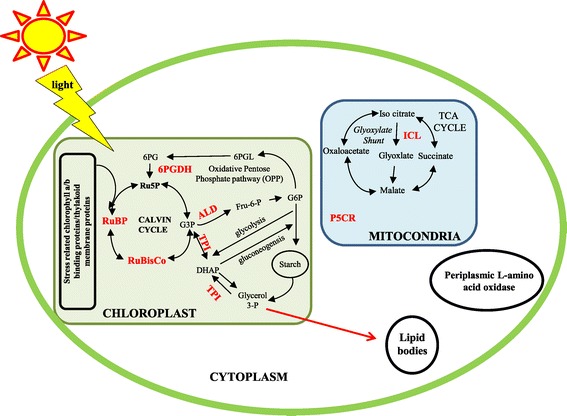
Schematic diagram of up-regulated genes in their respective pathways in Chlamydomonas reinhardtii strains during adaptive evolution. The up-regulated proteins in the proteomic study are indicated in red and bold.
Conclusions
C. reinhardtii strains CC124 and sta6-1 have been subjected to adaptive evolution to understand the molecular mechanisms of lipid accumulation under nitrogen-depleted conditions. The algal population that exhibited an enhanced accumulation of lipids were specifically selected and enriched through several cycles of adaptive evolution using FACS. The adaptive evolution under nitrogen starvation through sorting and regeneration of high-lipid content cells led to the generation of lipid overproducing populations of C. reinhardtii CC124 and sta6-1. The results on lipid composition, elemental analysis, confocal microscopic analysis, and chlorophyll analysis reported here highlight the characterization of lipid accumulation with carbon/nitrogen metabolic pathways. Studies on proteomics further support the phenotypic characterization and reveal the potential role of carbon/nitrogen metabolic pathways in line with lipid accumulation. These results demonstrate that the adaptive evolution of microalgae can be a new tool that could be utilized to develop C. reinhardtii strains, or other microalgal strains with desired phenotypes, such as high lipid accumulation.
Materials and methods
Strains and culturing conditions
C. reinhardtii (wild-type CC124) was kindly provided by the Korea Research Institute of Bioscience and Biotechnology and C. reinhardtii mutant (cc-4348 sta6-1) was purchased from Chlamydomonas Resource Center, University of Minnesota, St. Paul, MN, USA. Both strains were grown to late-log phase in nitrogen-replete tris acetate phosphate liquid medium (TAP) [33] (approximately 2.0 × 106 cells ml−1) and considered as 0 hour seeds. Zero hour seeds were re-suspended in a fresh TAP medium and taken immediately after re-suspension for FACS sorting (details available in flow cytometry and cell sorting section). Cultures were maintained at 27°C under a light intensity of 125 μmol/m2s with an agitation speed of 120 rpm. Samples for analysis were taken at the indicated times. Optical density was measured using a UV spectrophotometer (Beckman-Coulter DU 730 Life Science, California, United States) and cell counts were measured by hemocytometer (Neubauer-improved bright line, Marienfeld, Lauda-Konigshofen, Germany).
BODIPY staining
BODIPY 505/515 (4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacen), was purchased from Sigma Aldrich (St. Louis, Missouri, USA). BODIPY 505/515 staining was performed as reported previously [13]. A 5 mM BODIPY 505/515 stock was prepared by dissolving in dimethyl sulfoxide (Sigma Aldrich, St. Louis, Missouri, United States) and stored in the dark. For efficient staining of the intracellular lipid bodies of C. reinhardtii strains, an aliquot of 0.2% DMSO (vol/vol) was added to the microalgal suspension. A total of 2 μL of BODIPY 505/515 stock solution was added into the l ml of algal-DMSO suspension and agitated for 1 minute on a vortex mixer (Scientific Industries Inc., Bohemia, New York, USA). Samples were then incubated in darkness for 5 minutes at room temperature. After the incubation period, the samples were directly used for FACS and microscopic studies.
Flow cytometric analysis and cell sorting
A high speed flow cytometer, MoFlo XDP (Beckman Coulter, Fullerton, California, United States) was used for the analysis of cell staining and cell sorting. The fluorescence reading was obtained using an excitation of 488 nm with an argon laser (Beckman Coulter, Fullerton, California, United Sates). The emission signal was measured in channels upon excitation (FL1 channel centered at 530/40 nm and FL5 channel centered at 740 LP). The samples mean fluorescence intensity values and images were analyzed using SUMMIT Software Version 5.2 (Beckman Coulter, Fullerton, California, United Sates). The FACS settings of all channels were the same for all sorting procedures. Coulter Isoton II Diluent fluid (Beckman Coulter, Brea, California, United States) was used in all experiments as the flow cytometry sheath fluid. Cell sorting was carried out using cell sort precision mode, with a 70 μm nozzle. The 0 hour seed cell population was divided into three groups (in Figure 1: green, blue, and red color indicating low, average, and high lipid content cells, respectively) in the plot obtained based on two-dimensional dot plot (FSC, represented cell size) versus green fluorescence (FL1 represented neutral lipids; Figure 1). A total of 25,000 cells of the top 2% of the total population (from red group R4 regions) were sorted directly into the sterilized tubes containing 1 ml of nitrogen-supplemented TAP broth and re-suspended in nitrogen-replete or nitrogen-depleted TAP medium, separately. Samples were taken for cell count analysis at different intervals and cells were maintained until they reached high cell density (or late-stationary phase). A portion of high cell density (or late stationary phase) cells were screened using flow cytometry and 25,000 cells from the top 2% cells were sorted into the sterilized tubes containing 1 ml of TAP broth. Cultures were maintained at 27°C under a light intensity of 125 μmol/m2s with an agitation speed of 120 rpm. Triplicates were maintained for both wild-type CC124 and mutant sta6-1 strains, separately. The same protocol was followed for all indicated times.
Total lipid and fatty acid analysis
The total lipids were extracted from the 10 mg of lyophilized biomass with a chloroform-methanol (2:1 v/v) solvent mixture (Merck, Darmstadt, Germany) using a procedure similar to the Folch’s method [34]. Fatty acid methyl esters (FAMEs) were produced from the extracted lipid by a transesterification reaction. Briefly, methanol was added to the extracted lipid with sulfuric acid as a catalyst and a transesterification reaction was allowed to occur at 100°C for 10 minutes. After the reaction, 1 ml of deionized water was added and the organic phase was separated from water phase by centrifugation at 4000 rpm for 10 minutes. A total of 1 ml of chloroform containing 0.5 mg of heptadecanoic acid (C17:0; Sigma Aldrich, St. Louis, MO, USA) was added to each tube as an internal standard. The FAMEs in organic phase were analyzed by gas chromatography (HP5890, Agilent, Santa Clara, CA, USA) with a flame ionized detector (FID) and INNOWAX capillary column (Agilent, Santa Clara, United States, 30 m × 0.32 mm × 0.5 μm).
Microscopic determination of lipids in microalgal cells using BODIPY 505/515
A laser scanning confocal microscope (Eclipse C1si, Nikon, Kanagawa, Japan) with the excitation set at 488 nm and the emission set at 570 to 590 nm was used for selectively detecting microalgal lipid bodies stained with BODIPY 505/515 fluorescent dye. In order to calculate the number of lipid bodies in the cells, a liquid portion of cultures were stained by BODIPY 505/515 stain, fixed, and examined under confocal microscopy. Calculations of numbers of lipid bodies were made using micrographs of the intact cells and lipid bodies were scored.
Elemental analysis
The lyophilized microalgal biomasses were used for elemental analysis. Carbon (C), hydrogen (H), nitrogen (N), and sulfur (S) contents were analyzed using FLASH 2000 series (Thermo Scientific, Waltham, Massachusetts, United States).
Protein extraction and two-dimensional gel electrophoresis
C. reinhardtii CC124 and sta6-1 cells collected at indicated intervals were lyophilized and used for total protein preparation. Briefly, cells were harvested by centrifugation at 1130 g for 5 minutes at 4°C in pre-weighed tubes. The cell pellets were collected and washed twice with sterilized distilled water and frozen at −80°C, followed by vacuum freeze drying for 3 days. Lyophilized (150 mg of CC124 and 100 mg of sta6-1) samples were suspended in Hepes-KOH buffer (25 mM, pH 7.5) containing 1 mM PMSF (PhenylMethylSulfonyl Fluoride) and protease cocktail. The suspension was finely homogenized with a sterilized motor-pistol and centrifuged (13000 g for 30 minutes at 4°C). The supernatant was carefully collected and mixed with 100% acetone (Junsei Chemical Co. Ltd., Tokyo, Japan) (1:3 ratio) for protein precipitation. The mixture was incubated at −24°C for 12 hours. After incubation, the mixture was centrifuged (13000 g for 40 minutes at 4°C), and the protein pellet was collected and air dried until complete evaporation of acetone. The pellet was re-suspended in a minimum amount of rehydration buffer (8 M urea, 2 M thiourea, 4% CHAPS, 5 mM magnesium acetate, DTT and protease inhibitor cocktail). The re-suspended material was centrifuged at 13000 g for 15 minutes at 18°C and the supernatant was carefully collected, protein concentration was determined, and samples were stored at −80°C. The amount of protein was determined with a Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, California, USA) using bovine serum albumin (BSA) (Thermo Scientific, Rockford, IL, USA) as a standard. For two-dimensional gel electrophoresis, 75 μg of protein samples were used. Isoelectric focusing (IEF) was performed using Ettan IPGphorII (Amersham Biosciences, San Francisco, California, United States). Equilibrated strips were inserted on SDS-PAGE for protein separation and silver staining was carried out.
Image analysis
Quantitative analysis of digitized images was carried out using the PDQuest (Version 7.0, BioRad) software according to the protocols provided by the manufacturer. The quantity of each spot was normalized by total valid spot intensity. Protein spots were selected for the significant expression variation deviated over two fold in its expression level compared with control or normal sample.
MALDI-TOF and protein identifications
All chemicals used in the protein identification study were of analytical grade (sodium bicarbonate, 4-Sulfophenyl isothiocyanate, α-cyano-4-hydroxycinnamic acid (CHCA), ammonium bicarbonate (Sigma Aldrich, St. Louis, Missouri, United States)). For protein identification by peptide mass fingerprinting (PMF), protein spots were excised, digested with trypsin (Promega, Madison, Wisconsin, United States), mixed with α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% TFA, and subjected to MALDI-TOF analysis (Microflex LRF 20, Bruker Daltonics, Billerica, USA) as described by Fernandez et al. [35]. Spectra were collected from 300 shots per spectrum over m/z range 600 to 3000 and calibrated by two point internal calibration using Trypsin auto-digestion peaks (m/z 842.5099, 2211.1046). Peak list was generated using Flex Analysis 3.0 (Bruker Daltonics, Billerica, USA). The threshold used for peak-picking was as follows: 500 for minimum resolution of monoisotopic mass, 5 for S/N. The search program MASCOT, developed by The Matrixscience (Matrix Science Ltd, London, UK) was used for protein identification by peptide mass fingerprinting. The following parameters were used for the database search: trypsin as the cleaving enzyme, a maximum of one missed cleavage, iodoacetamide (Cys) as a complete modification, oxidation (Met) as a partial modification, monoisotopic masses, and a mass tolerance of ± 0.1 Da. PMF acceptance criteria is probability scoring.
Statistical analysis
Triplicates of samples were analyzed throughout the experiments. Statistical analyses were performed using SigmaPlot 10.0 software (Systat Software Inc., Chicago, IL, USA). The results were expressed as means of triplicate experiments. The results were expressed as means ± SD (standard deviation). Differences were considered significant at P <0.05.
Acknowledgements
This work was supported by the Advanced Biomass R&D Center (ABC) of Korea Grant funded by the Ministry of Science, ICT and Future Planning (MSIP, grant number: ABC-2013-057282). NV was supported by the BK21 Post-Doctoral Research Fund and MSP was partially supported by the Brain Pool Program of Korea.
Abbreviations
- 6PGDH
6-phosphogluconate dehydrogenase
- ALDH
Aldehyde dehydrogenase
- AtpD
ATP synthase
- CBP
Chlorophyll-ab-binding proteins
- CF1
beta subunit
- DHAR
Dehydroascorbate reductase
- Est
Esterase
- GHB
Gamma-hydroxybutyrate dehydrogenase
- GST
Glutathione-S-transferases
- GTPase
Ran-like small GTPase
- HP
Hypothetical protein
- HSP70C
Heat shock protein 70C
- ICL
Isocitrate lyase
- LAO1
Periplasmic L-amino acid oxidase catalytic subunit
- LCBP
Light harvesting proteins
- mtCA
Mitochondrial carbonic anhydrase β type
- mtTF-Tu
Mitochondrial translation factor Tu
- NDH
NADP dependent-malate dehydrogenase
- P5CR
Pyrroline-5-carboxylate reductase
- PFL
Pyruvate-formate lyase
- PP1
Protein phosphatase 1
- PPlase
Peptidyl-prolyl cis-trans isomerase, FKBP-type
- Pre Prt
Predicted protein
- PRK
Phosphoribulokinase Rubisco activase
- RabGDP
Rab GDP dissociation inhibitor protein
- RPLP0
Acidic ribosomal protein P0
- RPSA
Ribosomal protein Sa, component of cytosolic 80S ribosome and 40S small subunit
- RuBA
Rubisco activase
- RubisCo
Ribulose-1,5-biphosphate carboxylase/oxygenase large subunit
- TLP
Thylakoid lumen protein
- TPI
Triose phosphate isomerase
- α-tub
α -tubulin 1
- β-tub
β-tubulin
Additional files
Two-dimensional gels stained with silver staining (pH 3 to 10) for CC124.
Two-dimensional gels stained with silver staining (pH 3 to 10) for sta6-1.
Two-dimensional gels stained with silver staining (pH 4 to 7) for CC124.
Two-dimensional gels stained with silver staining (pH 4 to 7) for sta6-1.
Proteome profiles of CC124 during adaptive evolution period.
Proteome profiles of sta6-1 during adaptive evolution period.
Complete list of differentially expressed proteins of adaptive evolved Chlamydomonas reinhardtii cells.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NV, MSP, and KJJ designed and conceived the study and drafted the manuscript. NV and MS performed experiments and analyzed data. SSY participated in flow cytometry studies. KJJ, MSP, and JWY contributed to the experimental design, data interpretation, and reviewing the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Natarajan Velmurugan, Email: velmmk@kaist.ac.kr.
Minji Sung, Email: yainoma@kaist.ac.kr.
Sung Sun Yim, Email: vidalsungsun@kaist.ac.kr.
Min S Park, Email: minsungpark0@kaist.ac.kr.
Ji Won Yang, Email: jwyang@kaist.ac.kr.
Ki Jun Jeong, Email: kjjeong@kaist.ac.kr.
References
- 1.Goodson C, Roth R, Wang ZT, Goodenough U. Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot Cell. 2011;10:1592–1606. doi: 10.1128/EC.05242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siaut M, Cuine S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylides C, Li-Beisson Y, Peltier G. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011;11:7–22. doi: 10.1186/1472-6750-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starch-less Chlamydomonas reinhardtii. Eukaryot Cell. 2009;8:1856–1868. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson X, Alric J. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell. 2013;12:776–793. doi: 10.1128/EC.00318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Work VH, Radkovits R, Jnikerson RI, Meuser JE, Elliott LG, Vinyard DJ, Laurens LML, Dismukes GC, Posewitz MC. Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell. 2010;9:1251–1261. doi: 10.1128/EC.00075-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindguist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Villada L, Lopez-Rodas V, Banares-Espana E, Flores-Moya A, Agrelo M, Martin-Otero L, Costas E. Evolution of microalge in highly stressing environments: an experimental model analyzing the rapid adaptation of Dictyosphaerium chlorelloides (Chlorophyceae) from sensitivity to resistance agains 2,4,6-trinitrotoluene by rare preselective mutations. J Phycol. 2002;138:1074–1081. doi: 10.1046/j.1529-8817.2002.01128.x. [DOI] [Google Scholar]
- 8.Foster PL. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu Rev Genet. 1999;33:57–88. doi: 10.1146/annurev.genet.33.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramanan R, Kim BH, Cho DH, Ko SR, Oh HM, Kim HS. Lipid droplet synthesis is limited by acetate availability in starchless mutant of Chlamydomonas reinhardtii. FEBS Lett. 2013;587:370–377. doi: 10.1016/j.febslet.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 10.Mutanda T, Ramesh D, Karthikeyan S, Kumari S, Anandraj A, Bux F. Bioprospecting for hyper-lipid producing microalgal strains for sustainable biofuel production. Bioresour Technol. 2011;102:57–70. doi: 10.1016/j.biortech.2010.06.077. [DOI] [PubMed] [Google Scholar]
- 11.Hyke P, Lickova S, Pribyl P, Melzoch K, Kover K. Flow cytometry for the development of biotechnological processes with microalgae. Biotechnol Adv. 2013;31:2–16. doi: 10.1016/j.biotechadv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Chen F. Effects of salinity on cell growth and docosahexaenoic acid content of the heterotrophic marine microalga Crypthecodinium cohnii. J Ind Microbiol Biotechnol. 1999;23:508–513. doi: 10.1038/sj.jim.2900759. [DOI] [Google Scholar]
- 13.Rosenberg JN, Oyler GA, Wilkinson L, Betenbaugh MJ. A green light forengineered algae: redirecting metabolism to fuel a biotechnology revolution. Curr Opin Biotechnol. 2008;19:430–436. doi: 10.1016/j.copbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Velmurugan N, Sung M, Yim SS, Park MS, Yang JW, Jeong KJ. Evaluation of intracellular lipid bodies in Chlamydomonas reinhardtii strains by flow cytometry. Bioresour Technol. 2013;138:30–37. doi: 10.1016/j.biortech.2013.03.078. [DOI] [PubMed] [Google Scholar]
- 15.Doan TTY, Obbard JP. Improved Nile Red staining of Nannochloropsis sp. J Appl Phycol. 2011;23:895–901. doi: 10.1007/s10811-010-9608-5. [DOI] [Google Scholar]
- 16.Montero MF, Aristizabel M, Reina GG. Isolation of high-lipid content strains of the marine microalga Tetraselmis suecica for biodiesel production by flow cytometry and single-cell sorting. J Appl Phycol. 2011;23:1053–1057. doi: 10.1007/s10811-010-9623-6. [DOI] [Google Scholar]
- 17.Atteia A, Adrait A, Brugiere S, Tardif M, van Lis R, Deusch O, Dagan T, Kuhn L, Gontero B, Martin W, Garin J, Joyard J, Rolland N. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α–proteobacterial mitochondrial ancestor. Mol Biol Evol. 2009;26:1533–1548. doi: 10.1093/molbev/msp068. [DOI] [PubMed] [Google Scholar]
- 18.Joyard J, Myrian F, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N. Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog Lipid Res. 2010;49:128–158. doi: 10.1016/j.plipres.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell. 2010;9:97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen HM, Baudet M, Cuine S, Adriano J–M, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, Li-Beisson Y. Proteomic profiling of oil bodies isolated from the unicellular green microalgae Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics. 2011;11:4266–4273. doi: 10.1002/pmic.201100114. [DOI] [PubMed] [Google Scholar]
- 21.Lee DY, Park JJ, Barupal DK, Fiehn O. System response of metabolic networks in Chlamydomonas reinhardtii to total available ammonium. Mol Cell Proteom. 2012;11:973–988. doi: 10.1074/mcp.M111.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trentacoste EM, Shrestha RP, Smith SR, Gle C, Hartmann AC, Hildebrand M, Gerwick WH. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc Natl Acad Sci U S A. 2014;110:19748–19753. doi: 10.1073/pnas.1309299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper MS, Hardin WR, Petersen TW, Cattolico RA. Visualizing “green oil” in live algal cells. J Biosci Bioeng. 2010;109:198–201. doi: 10.1016/j.jbiosc.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Cakmak T, Angun P, Demiray YE, Ozkan AD, Elibol Z, Tekinay T. Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feed stock production of Chlamydomonas reinhardtii. Biotechnol Bioeng. 2012;109:1947–1957. doi: 10.1002/bit.24474. [DOI] [PubMed] [Google Scholar]
- 25.Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, Gallaher SD, Johnson S, Benning C, Pellegrini M, Grossman A, Merchant SS. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen-starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem. 2012;287:15811–15825. doi: 10.1074/jbc.M111.334052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenzuela J, Mazurie A, Carlson RP, Gerlach R, Cooksey KE, Peyton BM, Fields MW. Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol Biofuels. 2012;5:40–57. doi: 10.1186/1754-6834-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi YE, Hwang H, Kim HS, Ahn JW, Jeong WJ, Yang JW. Comparative proteomics using lipid over-producing or less-producing mutants unravels lipid metabolisms in Chlamydomonas reinhardtii. Bioresour Technol. 2013;145:108–115. doi: 10.1016/j.biortech.2013.03.142. [DOI] [PubMed] [Google Scholar]
- 28.Wase J, Black PN, Stanley BA, DiRusso CC. Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J Proteome Res. 2014;13:1373–1396. doi: 10.1021/pr400952z. [DOI] [PubMed] [Google Scholar]
- 29.Moroney JV, Ma Y, Frey WD, Fusilier KA, Pham TT, Simms TA, DiMario RJ, Yang J, Mukherjee B. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: Intracellular location, expression, and physiological roles. Photosynth Res. 2011;109:133–149. doi: 10.1007/s11120-011-9635-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Duanmu D, Spalding MH. Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynth Res. 2011;109:115–122. doi: 10.1007/s11120-011-9643-3. [DOI] [PubMed] [Google Scholar]
- 31.Meyer MT, Genkov T, Skepper JN, Jouhet J, Mitchell MC, Spreitzer RJ, Griffiths H. Rubisco small-subunit α-helices control pyrenoid formation in Chlamydomonas. Proc Nat Acad Sci U S A. 2012;109:19474–19479. doi: 10.1073/pnas.1210993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM. Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 2012;161:1034–1048. doi: 10.1104/pp.112.206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1966;54:669–1675. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 35.Fernandez J, Gharahdaghi F, Mische SM. Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels or polyvinyl difluoride membranes using matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) Electrophoresis. 1998;19:1036–1045. doi: 10.1002/elps.1150190619. [DOI] [PubMed] [Google Scholar]


