Abstract
Comprehensive application of solution NMR spectroscopy to studies of macromolecules remains fundamentally limited by the molecular rotational correlation time. For proteins, molecules larger than 30 kDa require complex experimental methods, such as TROSY in conjunction with isotopic labeling schemes that are often expensive and generally reduce the potential information available. We have developed the reverse micelle encapsulation strategy as an alternative approach. Encapsulation of proteins within the protective nano-scale water pool of a reverse micelle dissolved in ultra-low viscosity nonpolar solvents overcomes the slow tumbling problem presented by large proteins. Here, we characterize the contributions from the various components of the protein-containing reverse micelle system to the rotational correlation time of the encapsulated protein. Importantly, we demonstrate that the protein encapsulated in the reverse micelle maintains a hydration shell comparable in size to that seen in bulk solution. Using moderate pressures, encapsulation in ultra-low viscosity propane or ethane can be used to magnify this advantage. We show that encapsulation in liquid ethane can be used to reduce the tumbling time of the 43 kDa maltose binding protein from ~23 ns to ~10 ns. These conditions enable, for example, acquisition of TOCSY-type data resolved on the adjacent amide NH for the 42 kDa encapsulated maltose binding protein dissolved in liquid ethane, which is typically impossible for proteins of such size without use of extensive deuteration or the TROSY effect.
Keywords: encapsulated proteins, reverse miclles, low viscosity fluids, triple resonance NMR, resonance assignment, structure determination
INTRODUCTION
Though solution NMR has become a powerful tool for structural characterization of biological acromolecules, particularly proteins, it is fundamentally limited by the so-called ‘tumbling problem’ (Cavanagh 2007). Large proteins tumble relatively slowly in solution producing greatly broadened spectral lines as a result of fast transverse relaxation. Increased transverse relaxation not only broadens peaks but also decreases magnetization transfer efficiency, which drastically impedes collection of multidimensional data that is often needed for efficient analysis. Several strategies have been developed to mitigate these problems including the use of isotopic labeling schemes to minimize cross-relaxation and simplify spectra (Gardner et al. 1998), the TROSY effect to reduce the detrimental effects of averaged transverse relaxation (Pervushin et al. 1997) and the CRINEPT strategy for more optimal polarization transfer (Riek et al. 2000). Unfortunately, limitations inherent to these approaches have restricted the precision of structural models for proteins of significant size.
The Stokes-Einstein relation often satisfactorily describes molecular tumbling in solution and links a spherical particle’s rotational correlation time (τm) to its volume and the solvent viscosity (η). In this context, we consider the hydrated radius (rH) of a protein such that:
| (1) |
where T and kB have their usual meaning. For many solution NMR studies, reduced tumbling times are achieved by increasing the temperature but this is an obviously limited approach. The reverse micelle encapsulation strategy attempts to take advantage of the contribution of viscosity to the rate of molecular tumbling.
Reverse micelles are inverted micellar complexes which spontaneously form from the mixture of appropriate surfactants, bulk nonpolar solvents such as the alkanes, and small amounts of aqueous solution (De and Maitra 1995). Historically, reverse micelle encapsulation of proteins has been studied using viscous alkane solvents and at micromolar concentrations of encapsulated protein. We have adopted this approach for studies of encapsulated proteins in low viscosity solvents and at near millimolar concentrations {Wand, 1998 #138}. This has required the development of a library of cationic, anionic and nonionic surfactants that allow encapsulation of a wide range of proteins at concentrations and in solvents suitable for multidimensional high resolution NMR spectroscopy (Lefebvre et al. 2005; Peterson et al. 2005; Shi et al. 2005). The basic capabilities of encapsulation of proteins with high structural fidelity and of controlling transverse relaxation within an encapsulated protein by simply changing the bulk solvent was demonstrated early on (Babu et al. 2001; Wand et al. 1998). The inability to generally prepare solutions of encapsulated proteins beyond 0.25 mM is somewhat mitigated by the fact that such solutions are non-lossy and do not degrade the performance of cryogenically cooled probes (Flynn et al. 2000)
In recent years, reverse micelle encapsulation has been employed as a multidimensional NMR-based tool to characterize integral membrane proteins (Kielec et al. 2009; Van Horn et al. 2008), peripheral anchored membrane proteins (Valentine et al. 2010) and proteins of marginal stability (Peterson et al. 2004). It has also found application in protein and nucleic acid biophysics in a variety of contexts (Babu et al. 2004; Nucci et al. 2011; Pometun et al. 2006; Simorellis and Flynn 2006; Van Horn et al. 2005; Workman and Flynn 2009). The key parameter in most of these studies is the effectiveness of macromolecular tumbling. Here we characterize the components of the reverse micelle system with respect to the tumbling behavior of the protein. Such a detailed analysis has not been previously reported. Importantly, we demonstrate that under the reverse micelle encapsulation conditions employed the protein maintains a hydration shell comparable that seen in aqueous solution. This has important ramifications for the distribution of excess water between protein-containing and “empty” reverse micelles and the corresponding influence upon the tumbling of protein-containing reverse micelle particles.
MATERIALS AND METHODS
Protein Purification
Six proteins were used in this study: human ubiquitin (8.5 kDa), flavodoxin from Cyanobacterium anabaena (19 kDa, bound to flavin mononucleotide), human dihydrofolate reductase (DHFR, 23.5 kDa, in complex with folic acid and NADP+), maltose-binding protein from Escherichia coli (MBP, 43 kDa, bound to β-cyclodextrin) and malate synthase G from Escherichia coli, strain K12 (MSG, 81 kDa). Ubiquitin (Wand et al. 1996), flavodoxin (Liu et al. 2001), MBP (Gardner et al. 1998), and MSG (Howard et al. 2000) were expressed and purified via previously described methods. The gene for human dihydrofolate reductase (DHFR) (American Type Culture Collection #10324984, Manassas, VA) was cloned into the pET15b vector (Novagen) to generate an N-terminal histidine tag, similarly to previous methods (Tai et al. 2002). The protein was expressed in BL21 (DE3) RIL E. coli cells grown in M9 minimal media containing 15NH4Cl as the sole nitrogen source. The cells were lysed by sonication and the protein was purified on a Ni-NTA Superflow column (Qiagen) using standard methods. Resuspension of the cells, sonication, and column chromatography were performed in the presence of 7 mM β-mercaptoethanol. Folic acid was included in excess (5 mM) in lysis and elution buffers. After elution from the column, the protein was exchanged to 20 mM Tris, 100 mM NaCl, 10 mM Folic acid, pH 8.5, with 10 mM DTT and 5 mM NADP+ and concentrated as required.
Reverse Micelle Encapsulation
All isotopically-labeled solvents and surfactants were obtained from Cambridge Isotopes Laboratories (Andover, MA). Unlabeled chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Unless otherwise noted, all reverse micelle samples were prepared in 99% deuterated pentane (d-12). Ubiquitin was used to examine the effects of varied water content or surfactant composition on molecular tumbling of reverse micelle-encapsulated proteins. The typical encapsulation conditions for ubiquitin (Babu et al. 2001) are composed of 75 mM bis(2-ethylhexyl)-sulfosuccinate (AOT) with a molar ratio of water to AOT surfactant (“water loading” or W0) of 10. An appropriate volume of aqueous protein solution (~25 mM) to achieve a W0 of 10 was injected into a solution of 75 mM AOT in d-pentane. Higher water loadings were achieved by the initial preparation at a W0 of 8 and subsequent addition of the volume of buffer needed to increase the W0 to a desired value. At W0 greater than 22, a separate aqueous phase appears. After each addition, the sample was mixed by gentle inversion and allowed to equilibrate for 5 minutes before measurement. Ubiquitin was also encapsulated in 150 mM AOT, 150 mM CTAB with 800 mM hexanol, or a mixture of 105 mM dodecyl tetraethylene glycol (C12E4) and 45 mM AOT using the same injection procedure. For the AOT and AOT/C12E4 reverse micelles the buffer was 50 mM sodium acetate, pH 5, with 50 mM NaCl while for the CTAB/hexanol reverse micelles the buffer was 50 mM Tris at pH 8.3. Unless otherwise stated, a W0 of 10 was used for all three conditions.
In order to examine the protein size and molecular tumbling under reverse micelle-encapsulation, five proteins were encapsulated under identical surfactant conditions: 75 mM CTAB with 400 mM hexanol, at the W0 of optimum sample stability (target W0: ubiquitin = 10, flavodoxin = 10, DHFR = 17.5, MBP = 15, MSG = 22). Optimum stability is defined as that condition under which the protein remains encapsulated and properly folded, as determined by 15N-HSQC spectra, for weeks or longer. To make the reverse micelle samples, an appropriate volume of protein solution in optimized buffer was injected into a mixture of surfactants in d-pentane (ubiquitin – as above; flavodoxin – 5 mM in 50 mM sodium phosphate, pH 5.8 (Lefebvre et al. 2005); DHFR – DHFR - 5 mM in 10 mM Tris, 50 mM NaCl, 5 mM Folic acid, pH 8.5, 5 mM DTT, 5 mM NADP+; MBP – 5 mM in 20 mM sodium phosphate, pH 7.5, with 5 mM EDTA and 7 mM β-cyclodextrin; MSG – 4 mM in 20 mM tris, pH 8.0, with 20 mM MgCl2 and 5 mM DTT, yielding final protein concentrations of 337 μM, 68.2 μM, 119 μM, 102 μM, and 120 μM, respectively). Each sample was mixed by inversion and insoluble material permitted to settle fully before measurement. Actual W0 values were directly measured by integration of the 1H signals of the water and surfactant and were determined to be: ubiquitin = 10, flavodoxin = 12, DHFR = 15, MBP = 15, MSG = 18.5. The tumbling behavior of each of these proteins was also measured in aqueous solution at ~ 1 mM in the same buffer as that used for encapsulation.
To examine the effect of the viscosity of the non-polar solvent, MBP was encapsulated in d - pentane, d-propane (d8 - 98%), or d-ethane (d6 - 98%). Samples in propane or ethane included 5% pentane by volume to improve encapsulation stability and required pressurization to liquefy the main nonpolar solvent. These samples were prepared and measured using apparatus from Daedalus Innovations, LLC (Philadelphia, PA) according to previous methods (Ehrhardt et al. 1999; Peterson et al. 2005; Peterson and Wand 2005). Encapsulation conditions used for propane and ethane samples were identical to those used in pentane. Encapsulation pressures are as noted below.
To demonstrate the advantages of carrying out multidimensional triple resonance NMR spectroscopy on encapsulated proteins, maltose binding protein in buffer as described above was prepared at 0.2 mM in ethane with 75 mM CTAB, 450 mM d-hexanol at 4,500 p.s.i.
NMR Spectroscopy and characterization of macromolecular reorientation
Unless otherwise stated, all samples were uniformly 15N-labeled, and data was collected at 25°C and 500 MHz (1H) on a Bruker AVANCE III spectrometer equipped with a TXI cryoprobe. The TRACT method (Lee et al. 2006) was used to rapidly estimate the effective molecular reorientation correlation time of encapsulated proteins. A gradient-selected version of the TRACT experiment (gs-TRACT) was used to suppress artifacts resulting from aliphatic hydrogens of the surfactant and nonpolar solvent. The sensitivity-enhanced, gradient-selected, 15N-resolved TROSY-HSQC (Zhu et al. 1999) was modified by replacing the 15N chemical shift evolution period, including the low-power gradients, with a variable relaxation delay that included a centered 15N 180x pulse to refocus 15N chemical shift in the pulse sequence for collection of the 15Nα transverse relaxation. For 15Nβ transverse relaxation, the phase of the first 15N 90 pulse was phase shifted by 180°.
Each TRACT measurement incorporated at least 20 relaxation delay times for both the 15NαHα and 15NβHα states. Spectra were processed and the downfield portion (~8 – 10 p.p.m.) of the 15N-filtered one-dimensional amide hydrogen envelope was integrated in a manner designed to avoid contributions from highly dynamic side chain signals. The decay of this integral was fitted to a three-parameter, single exponential function. The rates determined from the decay fits were then used to find the rotational correlation time as described (Lee et al. 2006), which assumes an isotropic tumbling model. The effective 15N–1H internuclear distance and the axially symmetric 15N chemical shift tensor used were 1.04 Å and 170 ppm, respectively. The errors in tumbling times as calculated from the fitting error were generally less than 1%. The systematic 2–5 ns offset arising from the rigid body approximation is not reflected in this error estimate and is assumed to be similar for encapsulated proteins. This assumption is based on a previous measurement of backbone dynamics of ubiquitin in reverse micelles, which showed minimal difference from bulk solution (Simorellis and Flynn 2006).
Modeling the Reverse Micelle-Protein Particle
The size of the reverse micelle encapsulated water pool has been shown to be directly dependent on the W0 of the reverse micelle mixture (De and Maitra 1995). In order to predict the W0-dependence of ubiquitin tumbling in AOT reverse micelles, first the water pool radius was calculated using the previously determined dependence on W0 (Kinugasa et al. 2002). In order to account for the effect of the protein on the reverse micelle size, the water pool radius was increased by the volume percentage of the protein in the aqueous portion of the mixture. Under the present conditions, approximately 30% of reverse micelles in the mixture contained an ubiquitin molecule at W0 of 10, thus ubiquitin was calculated to contribute an additional twelve percent to the volume of the aqueous phase of the mixture. This volume contribution was calculated by considering ubiquitin to be a 8.5 kDa sphere of uniform density (1.4 g/cm3). The thickness of the surfactant layer, 10.5 Å (Vandijk et al. 1989), was added to the scaled water pool radii for each W0 tested, yielding a predicted average radius of each reverse micelle in the mixture. This radius was used to calculate the tumbling of the particle with the Stokes-Einstein equation (Eq. 1). This method of calculating reverse micelle tumbling is referred to as the “W0-dependent model.”
The radius of the reverse micelle particle was also calculated by using the radius of hydrated protein derived directly from the measured rotational correlation time in aqueous solution, essentially solving Eq. 1 for the radius of the sphere given the rotational correlation time of each protein. To this radius, the contour length of the surfactant layer was added. For AOT, 10.5 Å was used (Vandijk et al. 1989). The contour length for CTAB has been empirically estimated to be 15.4 Å (Palazzo et al. 2003). No published value could be found for C12E4, so the values for AOT and CTAB were compared to the length of the fully extended surfactant molecule and the ratio of these distances was used to estimate the thickness of the C12E4/AOT layer to be 18.1 Å. Using these radii for the various surfactant conditions and the hydrated radii of the proteins as calculated from the aqueous tumbling times, the experimentally measured tumbling behavior of encapsulated proteins was well reproduced as a function of both protein size and solvent viscosity. This approximation of the reverse micelle complex is referred to below as the “hydrated radius model.”
RESULTS
In an effort to understand the hydrodynamic performance of reverse micelles containing protein molecules we have employed the TRACT method, which measures the relaxation rate of the α and β states of amide 15N to estimate the protein’s effective rotational correlation time (Lee et al. 2006). This requires approximation of the protein as a rigid-body and generally results in an underestimate of the rotational correlation time. The rotational correlation time of each of the five model proteins was measured in aqueous solution using TRACT and compared to published rotational correlation times determined using a comprehensive suite of backbone relaxation measurements (Lee and Wand 1999; Mauldin et al. 2009; Tugarinov and Kay 2003; Yang and Kay 1999; Zhang et al. 1997). This comparison showed that TRACT-estimated tumbling times were 2–5 ns shorter than those obtained using the more rigorous method.
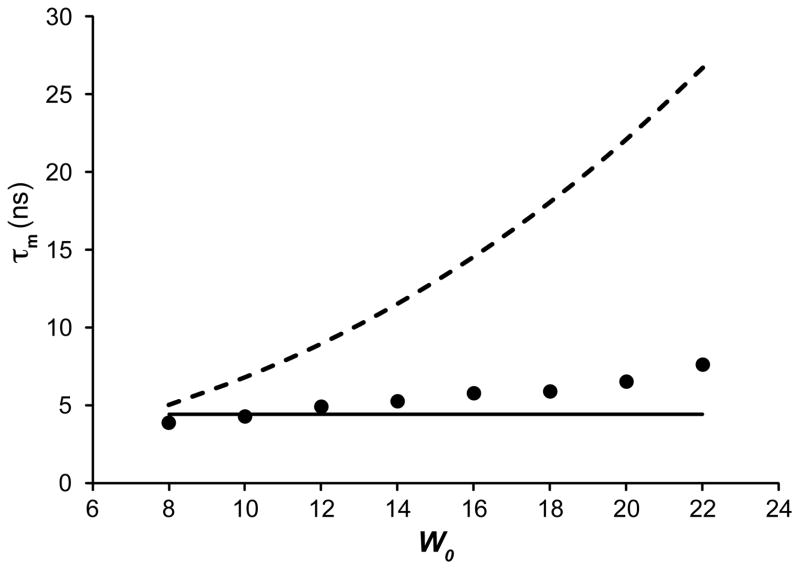
Protein-containing reverse micelle mixtures are composed of four main components: protein, a small amount of aqueous solvent, surfactant, and nonpolar bulk solvent. Ubiquitin was used to test the effects of the water loading (W0) and surfactant composition on the rotational correlation time of the protein. Ubiquitin was encapsulated in AOT in pentane at a low water loading (W0 = 8) and the rotational correlation time was measured using the TRACT method. The W0 was then increased by direct addition of additional water and a TRACT estimate of the protein’s rotational correlation time made. The obtained estimates of rotational correlation times clearly indicate that the effective tumbling of the protein-containing reverse micelles shows minimal dependence on the total W0 of the reverse micelle mixture (Figure 1). This finding suggested that the W0 does not dominate the tumbling behavior of the encapsulated protein, thus an alternative model was developed. The hydrated radius model predicts the protein tumbling based on a summation of the aqueous hydrated radius for the protein and the previously determined thickness of the hydrated surfactant layer. This model was used to predict tumbling for all other variables tested.
Figure 1.
The dependence upon water loading of the effective isotropic molecular reorientation correlation time of encapsulated ubiquitin dissolved in pentane. Solid symbols correspond to correlation times estimated using TRACT analysis of 15N-labeled ubquitin encapsulated in AOT reverse micelles at the indicated water loading (W0). The dashed line was calculated using the W0-dependent model (see Methods). The solid line was calculated using the hydrated radius model (see Methods) and shows better agreement suggesting that the water incorporated into protein containing reverse micelles is dictated by the hydration layer of the protein and is not simply defined by an equal distribution of water between protein containing and “empty” reverse micelles.
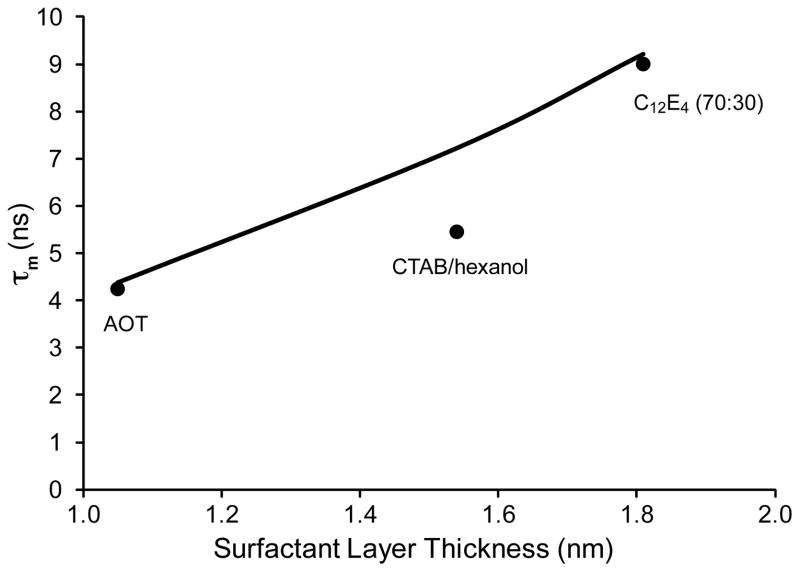
It has been previously shown that ubiquitin can also be encapsulated in a mixture of AOT and the non-ionic surfactant C12E4 (Peterson et al. 2005). We have determined conditions under which ubiquitin can be encapsulated in CTAB/hexanol reverse micelles. This permits the direct comparison of the hydrodynamic performance of different surfactant compositions using the same protein and W0. TRACT estimates of rotational correlation times for ubiquitin in these reverse micelle surfactant systems in pentane at water loading of 10 are well represented by the hydrated radius model and its prediction of the dependence of hydrodynamic performance on surfactant contour length (Figure 2). The close agreement of the hydrated radius model with these data indicates that it may offer a more accurate representation of the protein-containing reverse micelles than the W0-dependent model provides (Figure 2).
Figure 2.
The dependence upon surfactant composition of the effective isotropic molecular reorientation correlation time of encapsulated proteins dissolved in pentane. Symbols correspond to correlation times estimated using TRACT analysis of 15N-labeled ubquitin encapsulated in reverse micelles of the indicated surfactant composition at a water loading of 10. Ubiquitin was encapsulated in 150 mM AOT, 150 mM CTAB with 800 mM hexanol, or a mixture of 105 mM dodecyl tetraethylene glycol (C12E4) and 45 mM AOT using the injection procedure. The solid line was calculated using the hydrated radius model (see Methods).
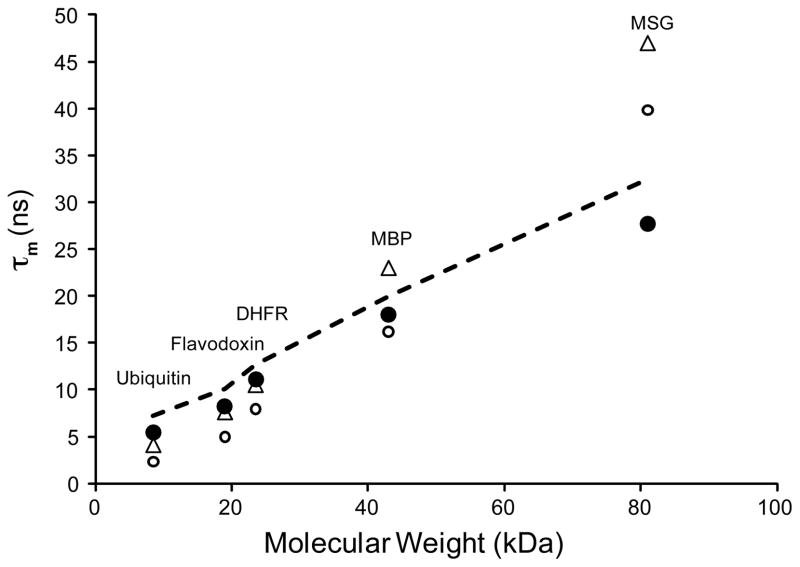
The slight dependence of tumbling on W0 and moderate dependence on the nature of the surfactant layer suggest that molecular tumbling of proteins in reverse micelles may be largely determined by the two remaining components of the system: the protein and the nonpolar solvent viscosity. To determine the dependence on the size of the encapsulated protein, five proteins of various molecular weights were encapsulated under identical surfactant conditions in liquid pentane and their rotational correlation times measured and compared to those in aqueous solution (Figure 3). The hydrated radius model reflects the molecular weight dependence of the measured tumbling values very well including the expected 2–5 ns offset that largely results from the rigid body approximation used to interpret the TRACT data. Importantly, these data also show that a tumbling advantage for reverse micelles dissolved in pentane over bulk aqueous solution is obtained for proteins larger than ~ 40 kDa. For MSG, the largest protein tested here, a tumbling advantage of greater than 30% is achieved in pentane.
Figure 3.
Effective isotropic molecular reorientation correlation times of proteins in free aqueous solution (○, △) and encapsulated in reverse micelles dissolved in liquid pentane (●) at 25 °C. Estimates of τm for the proteins in solution derived from TRACT (○) and from previously published 15N backbone relaxation measurements (Lee and Wand 1999; Mauldin et al. 2009; Tugarinov and Kay 2003; Yang and Kay 1999; Zhang et al. 1997) (△) are shown. All of the backbone relaxation values were collected at the same temperature as the TRACT measurements performed here (25 °C) except that for MSG, which was extrapolated from measurements at 37 °C using the Stokes-Einstein equation and the known temperature dependence of the bulk solvent viscosity. For the reverse micelle samples, each protein was prepared at its empirically determined optimal water loading (ubquitin, W0 = 10; flavodoxin, W0 = 12; Dihydrofolate reductase ternary complex, W0 = 15; maltose binding protein binary complex with β-cyclodextrin, W0 = 15; malate synthase G, W0 = 18.5). The estimated tumbling times for these samples are shown from the TRACT measurement (●) and from the hydrated radius model (dashed line). Though this model includes no explicit consideration of the W0, it accurately predicts the tumbling behavior for all five proteins despite the varied W0 between the samples. This result strongly indicates that the reverse micelles that contain protein incorporate only the appropriate amount of water to maintain the aqueous solution hydration shell of the proteins.
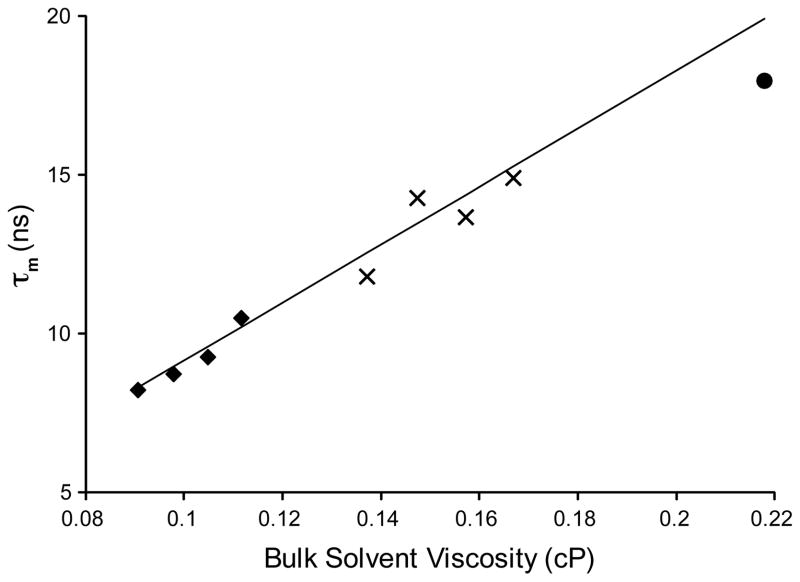
We have shown previously that variation in the bulk solvent from iso-octane through to liquid propane results in a roughly linear increase in transverse 15N relaxation times for encapsulated ubiquitin (Wand et al. 1998). Here we extend this to liquid ethane and use the significantly larger maltose binding protein (MBP). MBP was encapsulated in identical CTAB/hexanol mixtures in liquid pentane, propane and ethane. In both propane and ethane, the viscosity is pressure dependent, so simply varying the pressure of the sample can be used to change the bulk viscosity. Molecular reorientation time of encapsulated MBP in each of these samples was measured using TRACT from low viscosity to high (Figure 4). The hydrated radius model for reverse micelle hydrodynamics predicts the observed molecular reorientation times rather well, indicating that the model applies in all three short chain alkane solvents used. Under ideal encapsulation conditions at the lowest pressure in liquid ethane a molecular reorientation time of 8 ns was measured for MBP. Even if the TRACT measurement is an underestimate of the actual rotational correlation time by 2–5 ns, as illustrated above, an improvement in the tumbling time of more than 2-fold is achieved.
Figure 4.
Effective isotropic molecular reorientation correlation times of encapsulated MBP as a function of bulk solvent viscosity. The TRACT estimates for tumbling of encapsulated MBP dissolved in liquid ethane (◆), propane (⊥) and pentane (●) are shown. The viscosity of the ethane and propane was adjusted by varying the pressure of the sample (ethane: 4500, 5500, 6500, 7500 psi; propane: 3000, 4000, 5000, 6000 psi). The pressure dependence of the alkane solvent viscosity was taken from (Younglove and Ely 1987). The solid line is the viscosity dependence predicted by the hydrated radius model and does not include contribution from the 400 mM hexanol in these samples. Hexanol has previously been shown to raise the bulk viscosity of CTAB reverse micelle mixtures only slightly (Palazzo et al. 2003). Under optimal conditions in liquid ethane, the 43 kDa MBP exhibits a tumbling time in the range of ~ 8–10 ns, a vast improvement over bulk solution value of 23 ns (Yang and Kay 1999).
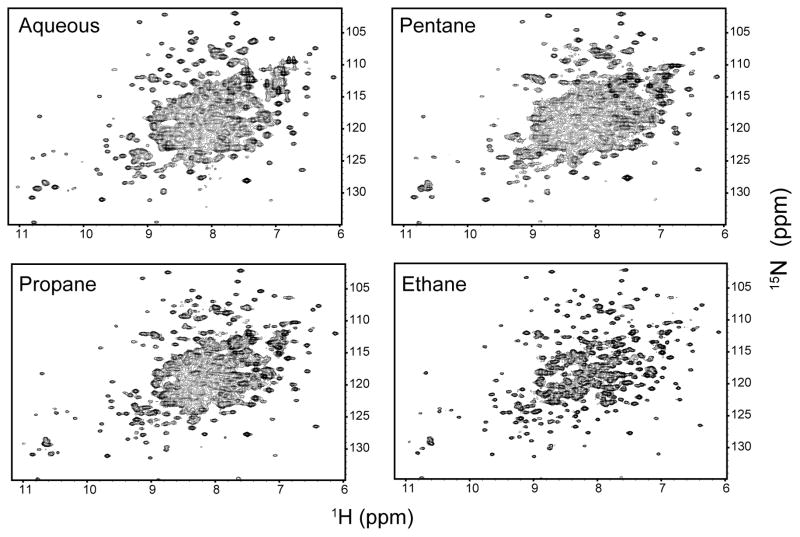
Solution NMR of large proteins in aqueous solution is problematic because slow tumbling leads both to broadened spectral lines, hindering resolution, and to poor transfer efficiency, all of which compound to result in poor sensitivity in multidimensional experiments. A main purpose of the reverse micelle encapsulation approach is to permit the acquisition of high quality long-distance correlations in large proteins, thereby making them more accessible to the full range of experiments available for smaller proteins. In Figure 5 shows standard (i.e. non-TROSY) 15N-HSQC spectra of MBP in aqueous solution and in reverse micelles in the three different short chain alkane solvents. The improvement in tumbling is clearly manifested as drastic improvements in the linewidth, resolution and signal-to-noise in the HSQC spectra (Figure 5).
Figure 5.
15N-HSQCs of uniformly 15N-labeled MBP in aqueous solution and in CTAB/hexanol reverse micelles in various bulk alkane solvents. No deuteration or TROSY effect was employed. Spectra were collected with identical parameters (128 complex 15N increments) and were processed identically except that the aqueous spectrum was collected with 64 scans while all others were collected with 16 scans. All spectra were recorded at 500 MHz (1H) and 25 °C. The propane and ethane samples were pressurized to 3000 and 4500 psi, respectively. The improved tumbling behavior shown in Figure 4 with reduced viscosity is manifested here as improved resolution and signal-to-noise.
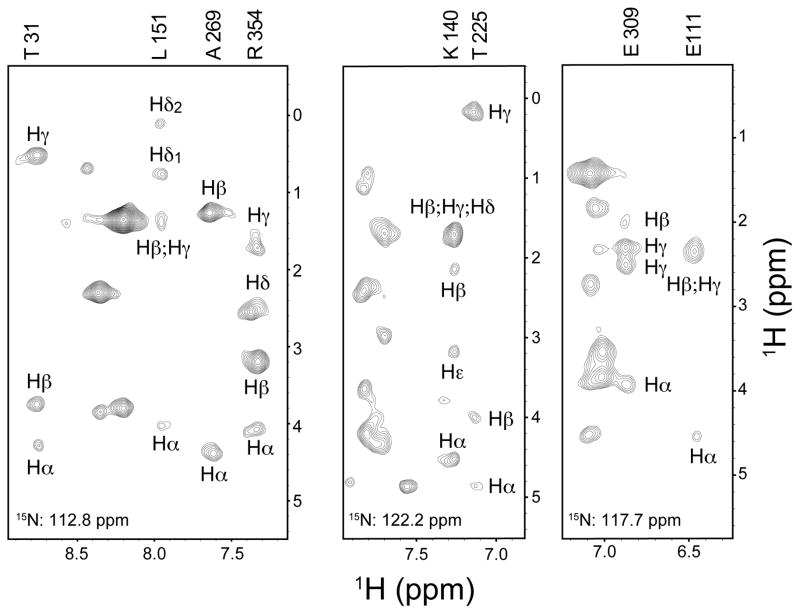
To more fully illustrate the advantages offered by carrying out multidimensional NMR of encapsulated proteins in liquid ethane, we prepared a uniformly 15N,13C-labeled sample of the β-cyclodextrin complexed maltose binding protein, which is a monomer of 42 kDa. The encapsulation pressure was 4,500 p.s.i. A 1H-evolved, 15N-resolved HCC(CO)NH-TOCSY (Lyons and Montelione 1993; Montelione et al. 1992) spectrum employing a 16 ms isotropic mixing time was obtained at 600 MHz (1H). This type of experiment perhaps represents the most efficient means for comprehensive side chain assignments. However, as noted when introduced, they are particularly sensitive to transverse relaxation and are generally only used for very small protein systems because of this. Encapsulation of the 42 kDa within reverse micelles dissolved in liquid ethane clearly relieves this restriction (Figure 6).
Figure 6.
15N planes of a H(CC)(CO)NH TOCSY spectrum of uniformly 15N,13C-labeled MBP in CTAB/hexanol reverse micelles dissolved in liquid ethane at 4,500 psi. The 42 kDa monomeric protein was 0.2 mM in 75 mM CTAB and 450 mM d-hexanol reverse micelles with a water loading of 15. The sample was prepared from a stock solution of 5 mM MBP in 20 mM sodium phosphate, pH 7.5, with 5 mM EDTA and 7 mM β-cyclodextrin. The spectrum was acquired at 600 MHz (1H) at 20 °C using a cryoprobe. The data set was obtained using 128 scans per FID and included 24 complex increments in the nitrogen dimension, 42 complex increments in indirect hydrogen dimension. The DIPSI carbon TOCSY mixing period was 16 ms. Various 15N planes are shown to illustrate the richness of the remote correlations obtained. Indicated assignments were mapped from those obtained using a triple resonance assisted main chain directed assignment strategy (Xu et al. 2006).
DISCUSSION
The physical behavior of reverse micelle systems not containing macromolecules has been extensively studied, particularly for AOT and CTAB surfactant mixtures. The size of such “empty” reverse micelles is directly controlled by the W0 of the mixture (De and Maitra 1995; Kinugasa et al. 2002; Palazzo et al. 2003). The studies presented here suggest a quite different scenario for reverse micelles containing a protein molecule. Reverse micelles containing ubiquitin exhibit only a slight increase in size with increasing W0 of the mixture. Using accepted values for the AOT aggregation number at the surfactant concentration and W0 used here, we estimate that approximately 30% of the reverse micelles in the system contain an ubiquitin molecule. The observed W0-dependence on molecular reorientation of encapsulated ubiquitin then suggest that protein-containing reverse micelles resist uptake of water beyond that required for normal protein hydration and that protein-free reverse micelles accommodate excess water. It is interesting to note that at W0 greater than 22, a separate aqueous phase appears. In the absence of protein, the water maximum solubilization capacity of AOT reverse micelles in pentane is not reached until W0 values greater than ~40 (Nucci and Vanderkooi 2005; Tingey et al. 1990). The lower water solubilization capacity of protein-containing reverse micelle solutions is consistent with the hydrated radius model which predicts that the lowest free energy state of the protein-containing reverse micelles will take up only as much water as necessary to hydrate both the surfactant layer and the encapsulated protein. Under these conditions, only non-protein-containing reverse micelles will take up excess aqueous solution added to the system. As there will be fewer of these ‘empty’ reverse micelles in the presence of protein, the total aqueous solution volume that may be solubilized by the reverse micelle ensemble will be dramatically reduced. It should be noted that although this model predicts that the reverse micelle mixture at very high W0 will be bidisperse, the 1H water NMR signal remains a single peak and shows minimal W0-depenence in linewidth (data not shown). However, the exchange of aqueous contents between reverse micelles has been placed in the 103 to 106 s−1 regime (Verbeeck and Deschryver 1987) and thus it is unsurprising that a lack of monodispersity is not revealed.
The accuracy of the hydrated radius model for predicting encapsulated protein tumbling was most extensively tested as a function of protein size (Figure 3). Each of the five proteins examined in CTAB/hexanol reverse micelles was prepared at the W0 of optimal stability, thus the W0 values varied from protein to protein. Despite the fact that the hydrated radius model includes no explicit consideration of water content, the trend observed in the measured data is well reproduced. The range of stable W0 values is quite small for several of these proteins, especially the largest ones tested. MBP and MSG are stable only at W0 values within ~±1.5 of those used here. Higher or lower water content than the optimal value either prevents encapsulation, reduces the lifetime of the sample or results in encapsulation of unfolded protein (data not shown). This indicates that the optimal encapsulation W0 value corresponds to that water content that yields a homogeneous population of protein-containing reverse micelles with a water pool that approximately matches the hydrated radius of the encapsulated protein in aqueous solution.
The finding that the reverse micelles that contain a protein molecule tune their water content to match the hydrated radius of the protein has several important implications for the NMR spectroscopy of encapsulated proteins. This property will tend to automatically balance appropriate hydration of the protein, maintaining structural and functional fidelity, with minimal size of the protein-containing reverse micelle particle, providing optimal tumbling behavior. In addition to this serendipitous advantage, the maintenance of the hydration shell seen in bulk aqueous solution provides strong evidence that the solvation environment of the protein in the reverse micelle is comparable to that in bulk solution. We have recently shown that reverse micelle encapsulation permits site-resolved, time-resolved measurement of hydration dynamics through measurement of protein-water dipolar magnetization exchange via the nuclear Overhauser effect (Nucci et al. 2011). Maintenance of the bulk solution hydration shell in the reverse micelle lends added credence to such measurements. Finally, the reverse micelle has been extensively used in recent years as a model for the nanoscale confines of the cell (Fayer and Levinger 2010; Moilanen et al. 2007; Thompson and Gierasch 1984; Valdez et al. 2001). It is now well known that the intracellular milieu is packed with macromolecular surfaces and the majority of the water inside the cell is confined in nanoscale spaces between these surfaces (Bagchi 2005; Ball 2008; Medalia et al. 2002). The tendency of the reverse micelle-encapsulated proteins to maintain their optimal hydration shell suggests an important energetic relevance for the size of the hydration shell that may prove to have biological implications, particularly if future investigations show that this shell is also maintained in vivo.
Though the reverse micelle methodology provides a useful means for studying the effects of nanoscale confinement, the primary goal of developing this technology for solution NMR of proteins has been to make typically challenging proteins, particularly large ones, more accessible to traditional NMR experiments. Though the tumbling advantage over aqueous solution seen for MSG in reverse micelles dissolved in pentane is considerable, it is insufficient to permit collection of typical high-resolution, high-sensitivity, multi-dimensional solution NMR data. Using previously developed technology and apparatus we encapsulated the 42 kDa MBP protein in the ultra-low viscosity liquids propane and ethane. Use of this solvent requires pressurization of the sample both for initial mixing and for measurement, though the pressures used are generally well below those required to produce destabilization of protein structure (Akasaka 2006; Royer 2002). The viscosities of these solvents are themselves pressure-dependent, providing a convenient means to examine the viscosity dependence of the tumbling of the encapsulated protein.
Encapsulation of MBP in CTAB reverse micelles dissolved in liquid ethane produces an experimental condition in which long-range TOCSY-type magnetization transfer becomes possible. This demonstrates the utility of reverse micelle encapsulation in ultra-low viscosity fluids for the structural characterization of large proteins without use of the TROSY effect and without employing perdeuteration or complex labeling schemes.
Acknowledgments
Supported by NSF grants MCB-0842814 and DMR05-200020 and NIH grant GM 085120 and NIH postdoctoral fellowship GM087099 to N.V.N. J.D. acknowledges financial support from the Wenner-Gren Foundations.
Footnotes
A.J.W. and R.W.P. declare a financial conflict of interest as Members of Daeadalus Innovations, LLC, a manufacturer of high-pressure and reverse micelle NMR apparatus.
References
- Akasaka K. Probing conformational fluctuation of proteins by pressure perturbation. Chem Rev. 2006;106:1814–1835. doi: 10.1021/cr040440z. [DOI] [PubMed] [Google Scholar]
- Babu CR, Flynn PF, Wand AJ. Validation of protein structure from preparations of encapsulated proteins dissolved in low viscosity fluids. J Am Chem Soc. 2001;123:2691–2692. doi: 10.1021/ja005766d. [DOI] [PubMed] [Google Scholar]
- Babu CR, Hilser VJ, Wand AJ. Direct access to the cooperative substructure of protein and the protein ensemble via cold denaturation. Nature Struct Mol Biol. 2004;11:352–357. doi: 10.1038/nsmb739. [DOI] [PubMed] [Google Scholar]
- Bagchi B. Water dynamics in the hydration layer around proteins and micelles. Chem Rev. 2005;105:3197–3219. doi: 10.1021/cr020661+. [DOI] [PubMed] [Google Scholar]
- Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- De TK, Maitra A. Solution behavior of aerosol Ot in nonpolar-solvents. Adv Colloid Interface Sci. 1995;59:95–193. [Google Scholar]
- Ehrhardt MR, Flynn PF, Wand AJ. Preparation of encapsulated proteins dissolved in low viscosity fluids. J Biomol NMR. 1999;14:75–78. doi: 10.1023/a:1008354507250. [DOI] [PubMed] [Google Scholar]
- Fayer MD, Levinger NE. Analysis of water in confined geometries and at interfaces. Ann Rev Anal Chem. 2010;3:89–107. doi: 10.1146/annurev-anchem-070109-103410. [DOI] [PubMed] [Google Scholar]
- Flynn PF, Mattiello DL, Hill HDW, Wand AJ. Optimal use of cryogenic probe technology in NMR studies of proteins. J Am Chem Soc. 2000;122:4823–4824. [Google Scholar]
- Gardner KH, Zhang XC, Gehring K, Kay LE. Solution NMR studies of a 42 KDa Escherichia coli maltose binding protein beta-cyclodextrin complex: Chemical shift assignments and analysis. J Am Chem Soc. 1998;120:11738–11748. [Google Scholar]
- Howard BR, Endrizzi JA, Remington SJ. Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 angstrom resolution: Mechanistic implications. Biochemistry. 2000;39:3156–3168. doi: 10.1021/bi992519h. [DOI] [PubMed] [Google Scholar]
- Kielec JM, Valentine KG, Babu CR, Wand AJ. Reverse micelles in integral membrane protein structural biology by solution NMR spectroscopy. Structure. 2009;17:345–351. doi: 10.1016/j.str.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa T, Kondo A, Nishimura S, Miyauchi Y, Nishii Y, Watanabe K, Takeuchi H. Estimation for size of reverse micelles formed by AOT and SDEHP based on viscosity measurement. Colloids and Surfaces. 2002;204:193–199. [Google Scholar]
- Lee AL, Wand AJ. Assessing potential bias in the determination of rotational correlation times of proteins by NMR relaxation. J Biomol NMR. 1999;13:101–112. doi: 10.1023/a:1008304220445. [DOI] [PubMed] [Google Scholar]
- Lee D, Hilty C, Wider G, Wuthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Lefebvre BG, Liu W, Peterson RW, Valentine KG, Wand AJ. NMR spectroscopy of proteins encapsulated in a positively charged surfactant. J Magn Reson. 2005;175:158–162. doi: 10.1016/j.jmr.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Liu WX, Flynn PF, Fuentes EJ, Kranz JK, McCormick M, Wand AJ. Main chain and side chain dynamics of oxidized flavodoxin from Cyanobacterium anabaena. Biochemistry. 2001;40:14744–14753. doi: 10.1021/bi011073d. [DOI] [PubMed] [Google Scholar]
- Lyons BA, Montelione GT. An HCCNH triple-resonance experiment using C-13 isotropic mixing for correlating Backbone amide and side-chain aliphatic resonances in isotopically enriched proteins. J Magn Reson Ser B. 1993;101:206–209. [Google Scholar]
- Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: modulation of dynamics within a structural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Moilanen DE, Levinger NE, Spry DB, Fayer MD. Confinement or the nature of the interface? Dynamics of nanoscopic water. J Am Chem Soc. 2007;129:14311–14318. doi: 10.1021/ja073977d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione GT, Lyons BA, Emerson SD, Tashiro M. An efficient triple resonance experiment using C-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J Am Chem Soc. 1992;114:10974–10975. [Google Scholar]
- Nucci NV, Vanderkooi JM. Temperature dependence of hydrogen bonding and freezing behavior of water in reverse micelles. J Phys Chem B. 2005;109:18301–18309. doi: 10.1021/jp051068+. [DOI] [PubMed] [Google Scholar]
- Nucci NV, Pometun MS, Wand AJ. Site-resolved measurement of water-protein interactions by solution NMR. Nature Struct Mol Biol. 2011;18:245–U315. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo G, Lopez F, Giustini M, Colafemmina G, Ceglie A. Role of the cosurfactant in the CTAB/water/n-pentanol/n-hexane water-in-oil microemulsion. 1. Pentanol effect on the microstructure. J Phys Chem B. 2003;107:1924–1931. [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Nat Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RW, Anbalagan K, Tommos C, Wand AJ. Forced folding and structural analysis of metastable proteins. J Am Chem Soc. 2004;126:9498–9499. doi: 10.1021/ja047900q. [DOI] [PubMed] [Google Scholar]
- Peterson RW, Lefebvre BG, Wand AJ. High-resolution NMR studies of encapsulated proteins in liquid ethane. J Am Chem Soc. 2005;127:10176–10177. doi: 10.1021/ja0526517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RW, Pometun MS, Shi Z, Wand AJ. Novel surfactant mixtures for NMR spectroscopy of encapsulated proteins dissolved in low-viscosity fluids. Protein Sci. 2005;14:2919–2921. doi: 10.1110/ps.051535405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RW, Wand AJ. Self-contained high-pressure cell, apparatus, and procedure for the preparation of encapsulated proteins dissolved in low viscosity fluids for nuclear magnetic resonance spectroscopy. Rev Sci Inst. 2005;76 doi: 10.1063/1.2038087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pometun MS, Peterson RW, Babu CR, Wand AJ. Cold denaturation of encapsulated ubiquitin. J Am Chem Soc. 2006;128:10652–10653. doi: 10.1021/ja0628654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R, Pervushin K, Wuthrich K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem Sci. 2000;25:462–468. doi: 10.1016/s0968-0004(00)01665-0. [DOI] [PubMed] [Google Scholar]
- Royer CA. Revisiting volume changes in pressure-induced protein unfolding. Biochim Biophys Acta-Protein Struct Mol Enzym. 2002;1595:201–209. doi: 10.1016/s0167-4838(01)00344-2. [DOI] [PubMed] [Google Scholar]
- Shi Z, Peterson RW, Wand AJ. New reverse micelle surfactant systems optimized for high-resolution NMR spectroscopy of encapsulated proteins. Langmuir. 2005;21:10632–10637. doi: 10.1021/la051409a. [DOI] [PubMed] [Google Scholar]
- Simorellis AK, Flynn PF. Fast local backbone dynamics of encapsulated ubiquitin. J Am Chem Soc. 2006;128:9580–9581. doi: 10.1021/ja061705p. [DOI] [PubMed] [Google Scholar]
- Tai NW, Ding YY, Schmitz JC, Chu E. Identification of critical amino acid residues on human dihydrofolate reductase protein that mediate RNA recognition. Nucleic Acids Res. 2002;30:4481–4488. doi: 10.1093/nar/gkf562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KF, Gierasch LM. Conformation of a peptide solubilizate in a reversed micelle water pool. J Am Chem Soc. 1984;106:3648–3652. [Google Scholar]
- Tingey JM, Fulton JL, Smith RD. Interdroplet attractive forces in AOT water-in-oil microemulsions formed in subcritical and supercritical solvents. J Phys Chem. 1990;94:1997–2004. [Google Scholar]
- Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- Valdez D, Le Huerou JY, Gindre M, Urbach W, Waks M. Hydration and protein folding in water and in reverse micelles: Compressibility and volume changes. Biophys J. 2001;80:2751–2760. doi: 10.1016/S0006-3495(01)76243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine KG, Peterson RW, Saad JS, Summers MF, Xu X, Ames JB, Wand AJ. Reverse micelle encapsulation of membrane-anchored proteins for solution NMR studies. Structure. 2010;18:9–16. doi: 10.1016/j.str.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn WD, Simorellis AK, Flynn PF. Low-temperature studies of encapsulated proteins. J Am Chem Soc. 2005;127:13553–13560. doi: 10.1021/ja052805i. [DOI] [PubMed] [Google Scholar]
- Van Horn WD, Ogilvie ME, Flynn PF. Use of reverse micelles in membrane protein structural biology. J Biomol NMR. 2008;40:203–211. doi: 10.1007/s10858-008-9227-5. [DOI] [PubMed] [Google Scholar]
- Vandijk MA, Joosten JGH, Levine YK, Bedeaux D. Dielectric Study of Temperature-Dependent Aerosol Ot/Water Isooctane Microemulsion Structure. J Phys Chem. 1989;93:2506–2512. [Google Scholar]
- Verbeeck A, Deschryver FC. Fluorescence quenching in inverse micellar systems - possibilities and limitations. Langmuir. 1987;3:494–500. [Google Scholar]
- Wand AJ, Urbauer JL, McEvoy RP, Bieber RJ. Internal dynamics of human ubiquitin revealed by 13C-relaxation studies of randomly fractionally labeled protein. Biochemistry. 1996;35:6116–6125. doi: 10.1021/bi9530144. [DOI] [PubMed] [Google Scholar]
- Wand AJ, Ehrhardt MR, Flynn PF. High-resolution NMR of encapsulated proteins dissolved in low-viscosity fluids. Proc Natl Acad Sci USA. 1998;95:15299–15302. doi: 10.1073/pnas.95.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman H, Flynn PF. Stabilization of RNA oligomers through reverse micelle encapsulation. J Am Chem Soc. 2009;131:3806–3807. doi: 10.1021/ja8084753. [DOI] [PubMed] [Google Scholar]
- Xu YQ, Zheng Y, Fan JS, Yang DW. A new strategy for structure determination of large proteins in solution without deuteration. Nature Methods. 2006;3:931–937. doi: 10.1038/nmeth938. [DOI] [PubMed] [Google Scholar]
- Yang D, Kay LE. Improved 1HN-detected triple resonance TROSY-based experiments. J Biomol NMR. 1999;13:3–10. doi: 10.1023/A:1008329230975. [DOI] [PubMed] [Google Scholar]
- Younglove BA, Ely JF. Thermophysical properties of fluids. II. Methane, ethane, propane, isobutane, and normal butane. J Phys Chem Ref Data. 1987;16:577–798. [Google Scholar]
- Zhang P, Dayie KT, Wagner G. Unusual lack of internal mobility and fast overall tumbling in oxidized flavodoxin from Anacystis nidulans. J Mol Biol. 1997;272:443–455. doi: 10.1006/jmbi.1997.1266. [DOI] [PubMed] [Google Scholar]
- Zhu G, Kong XM, Sze KH. Gradient and sensitivity enhancement of 2D TROSY with water flip-back, 3D NOESY-TROSY and TOCSY-TROSY experiments. Journal of Biomolecular NMR. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]