Abstract
By means of a circadian clock system, all the living organisms on earth including human beings can anticipate the environmental rhythmic changes such as light/dark and warm/cold periods in a daily as well as in a yearly manner. Anticipating such environmental changes provide organisms with survival benefits via manifesting behavior and physiology at an advantageous time of the day and year. Cell-autonomous circadian oscillators, governed by transcriptional feedback loop composed of positive and negative elements, are organized into a hierarchical system throughout the organisms and generate an oscillatory expression of a clock gene by itself as well as clock controlled genes (ccgs) with a 24 hr periodicity. In the feedback loop, hetero-dimeric transcription factor complex induces the expression of negative regulatory proteins, which in turn represses the activity of transcription factors to inhibit their own transcription. Thus, for robust oscillatory rhythms of the expression of clock genes as well as ccgs, the precise control of subcellular localization and/or timely translocation of core clock protein are crucial. Here, we discuss how sub-cellular localization and nuclear translocation are controlled in a time-specific manner focusing on the negative regulatory clock proteins.
Keywords: circadian rhythms, nuclear translocation, phosphorylation, posttranslational modification, O-GlcNAcylation
INTRODUCTION
The molecular clock present in nearly every cell is composed of transcriptional/translational feedback loop, namely TTFL [1]. Although specific components of TTFL are different, the governing rules of TTFL are well conserved from fungi to vertebrate, including humans [2]. Current understanding of the underlying biochemical mechanisms in animal circadian clockworks is largely based on earlier studies using the Drosophila. as a model system [3, 4]. In 1971, the pioneering behavioral geneticists Seymour Benzer and Ron Konopka searched for mutant flies having defects in daily rhythmic eclosion, a process of flies coming out of the pupae that happens mostly early in the morning [5]. During this screening, they identified 3 lines of mutant flies with affected eclosion rhythm in the population. One mutant was arrhythmic; another had a short (~19 hr) period; the third had a long period (~28 hr). These mutants were named per0, perS, and perL, respectively. A decade later, all three mutations turned out to be present on a single gene. This gene was named period after the mutants and became the first "clock gene." Many more clock genes were identified through genetic analysis in the following years in the Drosophila. Also, homology search revealed the mammalian clock genes, except the Clock. Clock (Circadian Locomotor Output Cycles Kaput) was identified in 1994 by Takahashi and his colleagues through forward ENU mutagenesis screen and is homologous to the Drosophila Clk [6, 7].
In the Drosophila, two basic-helix-loop helix and PAS containing transcription factors, dCLOCK (dCLK) and CYCLE (CYC) dimerize and induce the transcription of period (per) and timeless (tim) by binding to the E box (CACGAG) sequence element of genome at midday [8]. The rise in the newly made per and tim mRNA leads to the accumulation of PER and TIM proteins in the cytoplasm as the herterodimer form during the early evening. After a ~4 hr delay in the cytoplasm, PER and TIM translocate to the nucleus, presumably in a separate manner, to repress the transcriptional activity of dCLK/CYC resulting in a down-regulation of their own mRNA levels constituting namely the "core-loop." In the so-called "stabilizing-loop," the expression of dClk is controlled and interlocks with the "core-loop." dCLK-CYC stimulates the expression of two bZip containing transcription factors, vrille (vri) and PAR domain protein 1 ε (PDP1ε). While VRI represses the expression of dClk at early night, PDP1ε mediated stimulation of dClk is followed 3~4 hrs later generating a daily rhythmic oscillation of dClk mRNA levels. Due to this relationship among the proteins in the feedback loops, dClk mRNA levels cycle in an anti-phasic fashion to mRNA levels of per, tim, vri, and pdp1ε in a day. On the other hand, overall daily levels of cyc mRNA manifests no daily oscillation. Another dCLK/CYC downstream clock gene, bHLH orange domain putative transcription factor clock work orange (cwo) works as an oscillator amplifier by repressing and/or activating dCLK/CYC mediated transcription [9, 10, 11, 12].
In the mammalian system, a similar circuitry operates. CLK and CYC homolog BMAL1, posit in the center of interlocked TTFL produce Drosophila per gene homolog mPer1, mPer2, and mPer3. CLK/BMAL1 dimer also turns on the expression of mCryptochrome1 (mCry1) and mCryptochrome2 (mCry2) which dimerize with mPER proteins to inhibit the transcriptional activity of CLK/BMAL [13]. One twist of the mammalian TTFL is that while CRY proteins play a role in transmitting light signals to the molecular clock in Drosophila, the mammalian CRY proteins work as a repressor for mCLK/BMAL1. Although the mammalian TIM protein is also produced, its role in the clock system is not yet evident. In the stabilizing loop, retinoid-related orphan receptors (RORa, b, c) [14] activate and REV-ERBα inhibits the transcription of Bmal1 rather than Clk as in the Drosophila. Clock controlled rhythms in many physiological processes and behavior are generated by a cyclical gene expression governed by this interlocked feedback loop either directly or indirectly [15, 16] (For review, see [17]).
At a systemic level, cell-autonomous oscillator is orchestrated in a hierarchical network of master and peripheral oscillators. In the Drosophila, about 150 neurons in the brain work as a master clock driving its circadian behavior [18]. In mammals, suprachiasmatic nucleus (SCN) is the master clock synchronizing all other peripheral oscillators in various tissues (e.g. liver, heart, lung, etc.) [19, 20]. Although peripheral oscillators generate self-sustained rhythms, without the SCN, as in SCN-lesioned animals, rhythms of peripheral tissues become out of phase to an external LD cycle and to oscillations in other tissues [21]. Thus, the main role of a master clock is considered to be synchronizing peripheral oscillators by sending time cues via hormonal and neural pathway.
Although transcriptional control via interlocked feedback loop posits as a framework for the molecular clock, diverse regulations employed after the clock genes are transcribed also play crucial roles to finely adjust the clock speed to a 24 hr period. Post-translational modification, most notably phosphorylation of the clock proteins, has been extensively studied so far [22, 23, 24, 25, 26]. The first example was the Drosophila PER, which manifested timely progressive phosphorylation and hyper-phosphorylated isoforms degraded through a ubiquitin-proteasome system at the early day leading to the de-repression of dCLK/CYC transactivation [27, 28, 29, 30]. In turn, another round of the cycle could start the next day. Casein kinase 1ε homolog DOUBLETIME (DBT), Glycogen synthase kinase 3β (GSK 3β), casein kinase 2 (CK2), and NEMO (NMO) are identified as kinases for PER to regulate its levels, activity as a repressor, and subcellular localization [31, 32, 33, 34, 35, 36, 37, 38, 39]. TIM is also phosphorylated by Glycogen synthase kinase 3β and CK2 regulating its levels and nuclear entry time [40, 41]. More recently, numerous studies revealed the diverse regulation of molecular clock at the post-transcriptional level. Please refer to the excellent recent review for more information [42].
One important issue in circadian rhythm is to generate oscillation in such a long 24 hr period. Based on a simple oscillator model [43], synthetic feedback loop only generates rhythmic oscillation with a 2 hr period; thus, imposing a time delay between transcriptional activation and repression is inevitable to generate such a long rhythm period [44, 45, 46, 47, 48, 49, 50, 51]. The observation that nuclear accumulation of PER is lagged in both Drosophila and mammals by approximately 4~6 hours with respect to the peak mRNA levels support this notion [47, 52]. There could be various means to impose a time delay between the activation of circadian transcription factors and repression by circadian repressor proteins. Delaying the nuclear entry time of circadian repressor proteins could be employed as a time delay in the clock system. This review will focus on how clock speed is regulated by controlling negative circadian regulator's nuclear entry time.
SUBCELLULAR LOCALIZATION AND NUCLEAR ENTRY REGULATION BY SIGNAL SEQUENCE MOTIF
Traffic between the nucleus and the cytoplasm is carried out through specialized apertures, nuclear pore complexes (NPCs) [53, 54]. Various carrier proteins are involved in the translocation of cargo proteins through NPCs. Cargo proteins are targeted for nuclear import by a short nuclear localization signal (NLS) sequence motif. A well-known NLS is composed of one (monopartite) or two (bipartite) basic amino acid clusters. The classic nuclear import pathway uses importin β1 (Impβ1) as a carrier, which recognizes NLS as cargo and binds through the adaptor molecule importin α (Impα). It is the trimeric cargo containing protein complex Impα/Impβ1/NLS that can enter the nucleus [55, 56, 57].
The Drosophila PER protein has a functional bipartite NLS sequence at the C-terminus. Albeit with a functional NLS, the full-length PER protein expressed in the Drosophila S2 cell resides in the cytoplasm, most likely due to the cytoplasmic localization domain (CLD) at the C-terminal end of its PAS domain [58, 59]. When TIM protein was co-expressed, PER/TIM proteins were both detected in the nucleus indicating that the heterodimer formation is crucial for nuclear entry [59]. PER and TIM consistently accumulated in the cytoplasm in tim0 and per0 mutant flies, respectively [60, 61, 62]. Nonetheless, real-time imaging analysis revealed that PER/TIM complex formed in the cytoplasm of S2 cells dissociates before nuclear translocation and that dPER was detected in the nucleus before TIM in flies' pacemaker lateral neurons, suggesting that presumably PER and TIM independently moved to the nucleus in a short period of time [46, 63, 64]. Subcellular localization of TIM might be regulated in a slightly different manner as to the case of PER [65]. TIM could shuttle independently between the nucleus and cytoplasm both in flies and S2 cells. The role of PER was suggested to be necessary for nuclear retention of TIM in this case. Export from the nucleus to the cytoplasm is mediated through the recognition of nuclear export signal (NES) sequence. The typical NES is characterized as a leucine-rich sequence that is recognized by CRM1/exportin1, which belongs to Impβ family [57, 66]. Through a sequence scan, several putative NES of TIM were provided, although which one might be functional in vivo is not yet proven [65].
Similar to the Drosophila, the mammalian negative regulator's subcellular localization is affected by the interaction with its partner proteins. Exogenously expressed mPER1 or 2 in COS7 and NIH3T3 cells can accumulate in the nucleus in the presence of co-expressed mCRY proteins [67] or mPER3 [68]. Although the mCRY protein is retained in the nucleus when expressed in tissue cultures, the observation that the co-expression of mPER2 lacking NLS motif induced the retention of CRY in the cytoplasm supports the idea that mPER plays an important role in the nuclear localization of CRY as well. Consistently, putative NLS sequence motifs were identified from mPER1, 2, 3 and CRY1, 2 proteins [68, 69, 70, 71, 72, 73]. Nonetheless, the predominant nuclear localization of mPER2 in the liver of mCry1/mCry2 deficient mouse might suggest that the role of CRY is not necessarily for the nuclear entry per se, but rather for stabilizing PER in the nucleus [73]. Taken together, the nuclear accumulation of circadian repressor proteins such as dPER and TIM in Drosophila and mPERs and mCRYs in mammals are inter-dependent on partner proteins. How can interaction between binding partners affect nuclear localization of clock repressor proteins? The interaction between the partner proteins (e.g. PER/TIM, mPER/CRY, mPER1/mPER3, mPER2/mPER3) might adopt the conformation that allow their NLS unmasked, leading to the recognition by their carrier proteins.
TIMELY CONTROL OF NUCLEAR ENTRY BY PHOSPHORYLATION OF CLOCK PROTEINS
The Drosophila PER protein has 250 putative phosphorylation sites which suggest that critical functions of dPER might be controlled by the dynamic regulation of phosphorylation. Indeed, progressive phosphorylation of dPER occurs in a timely manner, and the phosphorylation status of dPER is different depending on the time of day [27]. Numerous reports indicated that phosphorylation of dPER is involved in a tight regulation of nuclear entry time. Glycogen synthase kinase 3β homolog, shaggy (sgg), promotes nuclear entry of dPER via direct phosphorylation of dPER [32] and/or indirect effects by phosphorylating TIM [41]. CK2 also promotes the nuclear entry of dPER in the Drosophilia pacemaker neurons [34, 36, 74]. CK2 is a tetrameric holo-enzyme composed of a catalytic subunit α2 and a regulatory subunit β2. Both α2 subunit mutant Timekeeper (Tik) and β subunit mutant andante flies manifest delayed nuclear entry time of dPER. When GSK 3β or CK2 activity was down-regulated either by mutation or decreased protein expression, circadian periods lengthened with concomitant delayed nuclear entry of dPER in pacemaker cells [34, 36, 41, 74]. On the other hand, DBT hindered the nuclear accumulation of dPER in the pacemaker neurons of Drosophilia. This notion is supported by the observation that in dbtP and dbtAR mutant flies, of which kinase activity is severely compromised, dPER is predominantly present in the nucleus even without TIM [33, 75]. However, dbtS flies which have reduced DBT kinase activity, exhibited delayed nuclear entry of dPER [76] which complicate the role of DBT on dPER nuclear entry. Given that another DBT allele which supposedly has decreased kinase activity-dbtL-manifested lengthened circadian periods [77], we prefer the idea that dbtS and dbtL flies might exert the effects in a more qualitative and not in a quantitative manner; suggesting DBTS and DBTL mutant proteins induced the alteration of phosphorylation sites on dPER, ultimately leading to the different outcomes in the length of the circadian periods. Taken together, while de-novo synthesized, hypo-phosphorylated dPER by DBT at specific sites, is retained in the cytoplasm, and the interaction with TIM would somehow relieve the cytoplasmic retention via antagonizing DBT mediated phosphorylation on dPER in Drosophila [75, 78]. This antagonizing effect could be aforementioned GSK3, CK2 mediated phosphorylation. Nonetheless, the duration of cytoplasmic retention of dPER might also be controlled by other posttranslational modifications (see below).
In mammals, the regulation of nuclear entry via phosphorylation of mPER is inconsistent depending on types of cells and kinds of proteins studied. Casein kinase 1 delta (CKIδ) and CKIε are two paralogs of mammalian CKI, both target mPERs as substrates regulating their stability and subcellular localization [79, 80]. Although some degree of functional redundancy of CKIε and CKIδ was observed, depending on the tissues, one might act more dominant than the other, as shown in the study where the depletion of CKIδ resulted in a long circadian period in the absence of behavioral effect with the depletion of CKIε [81]. In HEK293T cells, ectopically expressed mPER1 enters the nucleus while mPER2 resides in the cytoplasm [72]. This observation provided the idea that there must be a mechanism to prevent the premature nuclear entry of mPER1. This turned out to be a CKIε mediated phosphorylation of mPER1 via masking of NLS motif on mPER1 [72]. Interestingly, the co-expression of mCRY1 in the presence of CKIε and mPER1 brings all three components in the vicinity, and this trimeric complex can enter the nucleus [82]. Thus, mCRY1 is able to negate the CKIε mediated cytoplasmic retention of mPER1. This phenomenon is very closely related to the situation where DBT dependent phosphorylation retards the nuclear entry of dPER in Drosophila lateral neurons. In contrast, in other cell types, e.g. COS7 cells, CKIε mediated phosphorylation of Ser-661 and Ser-663 is a prerequisite for the nuclear entry of mPER1 [83]. Similarly CKIε and CKIδ induced phosphorylation of mPER3 accelerated nuclear translocation in COS7 cells while mPER1 and mPER2 were not affected by the co-expression of these kinases [79]. The inconsistent results regarding the effects of CKI mediated phosphorylation of mPERs in nuclear translocation might be attributed to the in vitro cell culture system of studies.
O-GLCNACYLATION MODULATES CLOCK PROTEIN LOCALIZATION
Recent findings have indicated that the extent of O-GlcNAc modification on dPER was correlated with nuclear translocation of dPER in Drosophila [84]. Aside from phosphorylation, the hydroxyl groups of Ser/Thr residues on proteins can also be modified with O-GlcNAc (O-GlcNAcylation) [85, 86]. Two enzymes mediate reversible addition of the β-N-acetylglucosamine moiety to the hydroxyl side chains of Ser/Thr residues of protein substrates; namely, glycosyltransferase (O-GlcNAc transferase; OGT) and β-N-acetylglucosaminidase (O-GlcNAcase; OGA) [87, 88]. Numerous findings reveal a complex interplay between phosphorylation and O-GlcNAcylation [89, 90]. In the case of MYC, threonine 58 in the transactivation domain (TAD) is either O-GlcNAcylated or phosphorylated by GSK3 in a competitive manner regulating transactivation potential of MYC [91, 92]. Stability of p53 is regulated by other modes of interplay between O-GlcNAcylation and phosphorylation; competitive occupancy at different sites. Treatment of MCF-7 cells with streptozotocin-OGA inhibitor-increased O-GlcNAcylation at serine 149 on p53. This O-GlcNAcylation represses the phosphorylation on threonine 155 leading to the inhibition of the interaction with the UPS system, ultimately resulting in the accumulation of p53 proteins in the cells [93]. O-GlcNAcylation of proteins might lead to an increase in phosphorylation at other sites as seen in the example of IRS-1 [94]. In the mouse liver, O-GlcNAcylation of IRS-1 directly correlates with an increase in serine 307, 632/635 phosphorylation, which are sites known to attenuate insulin signaling [94].
O-GlcNAc modification of dPER was evident in Drosophila S2 cells and in flies. More interestingly, O-GlcNAcylation of the dPER protein is temporally regulated. In flies, peak level in O-GlcNAcylation of dPER was observed during the first half of the night before a massive phosphorylation of dPER occured [84]. Remarkably, genetic manipulation of O-GlcNAc levels by either down- or up-regulating OGT in clock cells speeds up or slows down the pace of circadian behavioral rhythms, respectively. In the key pacemaker neurons in flies, the timing of dPER nuclear translocation is advanced in ogt knockdown flies and delayed in ogt overexpressing flies. Because O-GlcNAcylation of dPER mainly occured when it was retained in the cytoplasm, authors suggested the compelling hypothesis that O-GlcNAcylation gates the timing of when dPER translocates from the cytoplasm to the nucleus possibly via the interplay between phosphorylation and O-GlcNAcylation.
mPER2 is also modified with O-GlcNAc. Consistent with the observation in flies, conditional knockout of OGT shortened the circadian locomotor period [95]. Interestingly, O-GlcNAcylation can occur on S662 always together with O-GlcNAcylation on S671 [95]. O-GlcNAcylation on S662 decreased the phosphorylation on S662 suggesting the possible antagonism between O-GlcNAc and phospho-modification. It has been well known that phosphorylation on S662 is tightly linked to the familial advanced sleep phase syndrome (FASPS) in humans. People having FASPS disorders have a phenotype of early morning awakening and early sleep times together with a shortened circadian rhythm [96]. Two genes have been identified to be related with the FASPS disorder-S662G mutation on dPER or hypomorphic T44A mutation on CKIδ [96, 97]. A later study further revealed that phosphorylation of S662 was necessary for serial phosphorylation of S662-S674 cluster, which somehow leads to the increase in mPer2 transcript levels [95]. Another study suggests different roles for S662 phosphorylation, which is stabilizing mPER2 from degradation by blocking nuclear export [98]. Although the role of S662 phosphorylation on the metabolism of mPER is still controversial, the mPER2 nuclear localization might also be regulated by controlling O-GlcNAc modification of mPER2, as similar to that of the Drosophilia.
CONCLUSION
To be able to sustain a 24 hr rhythm period, timely control of nuclear translocation is crucial in both Drosophila and mammals [99]. Strong nuclear accumulation of major circadian repressor protein-PER- is lagged several hours to the times of peak transcript levels. This delayed nuclear entry was controlled via post-translational modification of PER, namely phosphorylation that may be affected by the interaction with other partner proteins (TIM in Drosophila and mPERs or CRYs in mammals) and/or dynamic interplay with other post-translational modifications (e.g. O-GlcNAcylation) (Fig. 1). One important feature of the circadian clock system is that it may be able to re-synchronize to changes in time-cues when travelling through different time-zones. To be able to re-synchronize, cellular oscillators may be able to easily adapt to the extracellular time-cues. Recently, studies about PTM crosstalk in regulating the function of a protein have been accumulating (for review, [100]). Currently, more than 450 PTMs are listed in the protein data base [101]. As shown in the control of nuclear entry of PER proteins by possible crosstalks between O-GlcNAcylation and phosphorylation, other PTM crosstalks might play crucial roles in controlling nuclear entry of clock proteins and other functions as well. Future studies in this direction will shed light on understanding the detailed biochemical underpinnings of nuclear entry regulation of clock proteins.
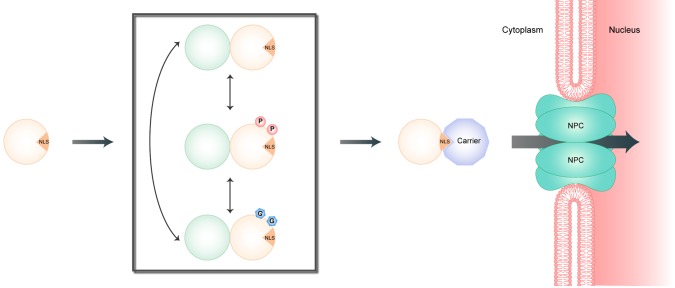
Fig. 1.
A model for the timely nuclear entry of circadian repressor proteins in Drosophila and mammals. Newly synthesized circadian repressor protein (orange circle) is modified in the cytoplasm (gray box) over the course of time resulting in conformational changes. Conformational changes might unmask the NLS, which is recognized by carrier proteins mediating the transport through the nuclear pore complex (NPC). Mainly, phosphorylation (P) of repressor protein seems to control the timely nuclear translocation of repressor protein. Nonetheless, interaction with the other repressor protein (green circle) and crosstalk with other posttranslational modifications (e.g. O-GlcNAcylation, G) might regulate timely phosphorylation of repressor protein (orange circle).
ACKNOWLEDGEMENTS
E.Y.K. was supported by the National Research Foundation of Korea (NRF) grant (NRF-2012R1A2A2A02014188).
References
- 1.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 3.Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- 4.Edery I. Circadian rhythms in a nutshell. Physiol Genomics. 2000;3:59–74. doi: 10.1152/physiolgenomics.2000.3.2.59. [DOI] [PubMed] [Google Scholar]
- 5.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King DP, Vitaterna MH, Chang AM, Dove WF, Pinto LH, Turek FW, Takahashi JS. The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit. Genetics. 1997;146:1049–1060. doi: 10.1093/genetics/146.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, Hardin PE, Tanimura T, Ueda HR. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richier B, Michard-Vanhée C, Lamouroux A, Papin C, Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J Biol Rhythms. 2008;23:103–116. doi: 10.1177/0748730407313817. [DOI] [PubMed] [Google Scholar]
- 13.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 14.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 17.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annu Rev Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 20.Roenneberg T, Merrow M. Circadian clocks - the fall and rise of physiology. Nat Rev Mol Cell Biol. 2005;6:965–971. doi: 10.1038/nrm1766. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi JS, Menaker M. Role of the suprachiasmatic nuclei in the circadian system of the house sparrow, Passer domesticus. J Neurosci. 1982;2:815–828. doi: 10.1523/JNEUROSCI.02-06-00815.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae K, Edery I. Regulating a circadian clock's period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem. 2006;140:609–617. doi: 10.1093/jb/mvj198. [DOI] [PubMed] [Google Scholar]
- 23.Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- 24.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 25.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 26.Diernfellner AC, Schafmeier T. Phosphorylations: making the Neurosporacrassa circadian clock tick. FEBS Lett. 2011;585:1461–1466. doi: 10.1016/j.febslet.2011.03.049. [DOI] [PubMed] [Google Scholar]
- 27.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grima B, Lamouroux A, Chélot E, Papin C, Limbourg-Bouchon B, Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- 30.Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 31.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 32.Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3beta/SGG in circadian clocks. J Neurosci. 2010;30:12664–12675. doi: 10.1523/JNEUROSCI.1586-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 34.Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- 35.Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145:357–370. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 37.Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawathean P, Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- 39.Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meissner RA, Kilman VL, Lin JM, Allada R. TIMELESS is an important mediator of CK2 effects on circadian clock function in vivo. J Neurosci. 2008;28:9732–9740. doi: 10.1523/JNEUROSCI.0840-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 42.Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci. 2013;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 43.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 44.Dunlap JC. Physiology. Running a clock requires quality time together. Science. 2006;311:184–186. doi: 10.1126/science.1122839. [DOI] [PubMed] [Google Scholar]
- 45.Leloup JC, Goldbeter A. A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J Biol Rhythms. 1998;13:70–87. doi: 10.1177/074873098128999934. [DOI] [PubMed] [Google Scholar]
- 46.Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- 47.Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marrus SB, Zeng H, Rosbash M. Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15:6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 49.Scheper TO, Klinkenberg D, van Pelt J, Pennartz C. A model of molecular circadian clocks: multiple mechanisms for phase shifting and a requirement for strong nonlinear interactions. J Biol Rhythms. 1999;14:213–220. doi: 10.1177/074873099129000623. [DOI] [PubMed] [Google Scholar]
- 50.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edery I. Role of posttranscriptional regulation in circadian clocks: lessons from Drosophila. Chronobiol Int. 1999;16:377–414. doi: 10.3109/07420529908998716. [DOI] [PubMed] [Google Scholar]
- 52.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cyert MS. Regulation of nuclear localization during signaling. J Biol Chem. 2001;276:20805–20808. doi: 10.1074/jbc.R100012200. [DOI] [PubMed] [Google Scholar]
- 54.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 55.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 56.Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 57.Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Chang DC, Reppert SM. A novel C-terminal domain of drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol. 2003;13:758–762. doi: 10.1016/s0960-9822(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 59.Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 60.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 61.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 62.Vosshall LB, Price JL, Sehgal A, Saez L, Young MW. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 63.Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shafer OT, Levine JD, Truman JW, Hall JC. Flies by night: effects of changing day length on Drosophila's circadian clock. Curr Biol. 2004;14:424–432. doi: 10.1016/j.cub.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 65.Ashmore LJ, Sathyanarayanan S, Silvestre DW, Emerson MM, Schotland P, Sehgal A. Novel insights into the regulation of the timeless protein. J Neurosci. 2003;23:7810–7819. doi: 10.1523/JNEUROSCI.23-21-07810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kutay U, Güttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 67.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 68.Yagita K, Yamaguchi S, Tamanini F, van Der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 69.Hirayama J, Fukuda I, Ishikawa T, Kobayashi Y, Todo T. New role of zCRY and zPER2 as regulators of sub-cellular distributions of zCLOCK and zBMAL proteins. Nucleic Acids Res. 2003;31:935–943. doi: 10.1093/nar/gkg174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyazaki K, Mesaki M, Ishida N. Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein. Mol Cell Biol. 2001;21:6651–6659. doi: 10.1128/MCB.21.19.6651-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakakida Y, Miyamoto Y, Nagoshi E, Akashi M, Nakamura TJ, Mamine T, Kasahara M, Minami Y, Yoneda Y, Takumi T. Importin alpha/beta mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J Biol Chem. 2005;280:13272–13278. doi: 10.1074/jbc.M413236200. [DOI] [PubMed] [Google Scholar]
- 72.Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin JM, Schroeder A, Allada R. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J Neurosci. 2005;25:11175–11183. doi: 10.1523/JNEUROSCI.2159-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bao S, Rihel J, Bjes E, Fan JY, Price JL. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci. 2001;21:7117–7126. doi: 10.1523/JNEUROSCI.21-18-07117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Preuss F, Fan JY, Kalive M, Bao S, Schuenemann E, Bjes ES, Price JL. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hara T, Koh K, Combs DJ, Sehgal A. Post-translational regulation and nuclear entry of TIMELESS and PERIOD are affected in new timeless mutant. J Neurosci. 2011;31:9982–9990. doi: 10.1523/JNEUROSCI.0993-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002;22:1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:21359–21364. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, Weaver DR. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem. 2002;277:17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takano A, Isojima Y, Nagai K. Identification of mPer1 phosphorylation sites responsible for the nuclear entry. J Biol Chem. 2004;279:32578–32585. doi: 10.1074/jbc.M403433200. [DOI] [PubMed] [Google Scholar]
- 84.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 86.Holt GD, Hart GW. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem. 1986;261:8049–8057. [PubMed] [Google Scholar]
- 87.Dong DL, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 88.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 89.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J Cell Sci. 2010;123:13–22. doi: 10.1242/jcs.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chou TY, Dang CV, Hart GW. Glycosylation of the c-Myc transactivation domain. Proc Natl Acad Sci U S A. 1995;92:4417–4421. doi: 10.1073/pnas.92.10.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 93.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 94.Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 95.Kaasik K, Kivimäe S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptáček LJ, Fu YH. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17:291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptácek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 97.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptácek LJ, Fu YH. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 98.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Curtin KD, Huang ZJ, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 100.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 2014;14:513–524. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 101.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]