Abstract
Maize is an especially well-suited species for studying the effects of aneuploidy on plant development. We used B-A translocations and testers that were crossed seven times into inbred W22 to generate a dosage series for 14 chromosome arms. This is the first report of dosage effects on maize morphogenesis using inbred B-A stocks and inbred tester stocks. We compared plants containing one dose or three doses of each of the 14 chromosome arms with plants containing two doses for seven measured traits. These were leaf width, leaf length, plant height, ear height, internode length, ear node circumference, and tassel branch number. We observed the typical maize aneuploid syndrome wherein one dose was more widespread and more severe in its effects than three doses. All but two of the one-dose effects were negative, and all of the three-dose effects were negative. The occurrence of positive responses by hyperploid plants in our earlier B-A-A study and the absence of any positive responses among the hyperploids reported for the 14 simple B-A translocations tested for dosage effects in the present study and previously may reflect gene dosage interaction between the two chromosome arm segments present in the B-A-A translocations. The overall congruence of our results with those of previous studies suggests that the traits measured are quantitative traits controlled by multiple genes whose activities provide a balanced regulation that transcends individual inbred lines or diverse genetic backgrounds and that such genes may be especially abundant in chromosome arm 1L.
Keywords: maize, B-A chromosomes, translocations, plant morphogenesis, dosage analysis
HERE we report the results of our studies on the effects of varying the chromosome arm dosage on maize plant morphogenesis. These studies have been facilitated by the discovery of nondisjunction of the maize B chromosome during pollen development by Roman (1947). The isolation of B-A and A-B chromosomes resulting from the reciprocal exchange of chromosome arm segments between a B chromosome arm and an arm of a standard A chromosome (Roman 1947) enabled the assembly of a collection of B-A chromosome stocks (Beckett 1991, 1993). This collection contains reciprocal exchanges for all 20 of the maize chromosome arms except for chromosome arms 2S, 2L, and 8S (Sheridan and Auger 2008; James Birchler, personal communication). The maize B-A chromosomes have been used to study the dosage effects of individual genes as well as to determine the chromosome arm location of many recessive mutations, particularly kernel mutations (Neuffer and Sheridan 1980; Beckett 1983; Scanlon et al. 1994). In addition, B-A translocations have been used to study the effects of hypoploidy of individual chromosome arms on endosperm development and kernel size (Lin 1982; Beckett 1983; Birchler and Hart 1987; Birchler 1993).
Two studies that have used B-A translocations to examine the effects of chromosome arm dosage on maize development are of particular interest. Lee et al. (1996) reported the effects on 12 plant traits for 14 maize chromosome arms. These investigators measured morphological traits in F1 hybrid plants produced by crossing B-A translocations in a B73 inbred background onto Mo17 inbred ears. The resulting hybrid progeny were identified as hyperploid, euploid, or hypoploid for the chromosome arm of interest by assessing pollen sterility and restriction length polymorphisms.
The second study of dosage effects on plant morphology was performed using compound B-A translocations, also known as B-A-A translocations (Sheridan and Auger 2008), utilizing a large collection of newly constructed B-A-A translocations (Sheridan and Auger 2006). The former study reported on the effects that the two A segments of the B-A-A translocations had on seven plant morphological traits when hyperploid plants (carrying three copies of the A chromosome segments of interest) were compared with nonhyperploid plants. The study with the B-A-A translocations was performed with genetic stocks that did not involve inbred lines so that the simple B-A translocation stocks and the A-A translocation stocks that they were crossed onto to obtain the rare B-A-A recombinant stocks were not genetically homogeneous (Sheridan and Auger 2006). This was also the case for the tester stocks used as females for crosses in producing the ploidy series of plants for trait measurements. Because of this lack of uniformity in genetic background of the B-A-A translocation stocks and the tester stocks, the interpretation of the observed effects on plant traits was more difficult than if all of the stocks had been in an inbred line background.
Here we report the results of our investigation of the effects on seven morphological traits (Table 1) of chromosome arm dosage variation where both the simple B-A translocations and the tester stocks had been converted to a W22 inbred background. In both the studies of Lee et al. (1996) and of Sheridan and Auger (2008), plants hypoploid for chromosome arm segments generally exhibited a reduction in the morphological trait examined. In both earlier studies there were numerous cases where plants hyperploid for chromosome arm segments exhibited a reduction in the morphological trait examined. However, Sheridan and Auger (2008) reported several cases where hyperploidy resulted in an increase in morphological traits. Here we report that, as in the two earlier studies, hypoploidy for chromosome arms is generally associated with a reduction in morphological traits. However, in contrast to the earlier studies, we have found that hyperploidy for chromosome arm segments resulted in many fewer cases where hyperploidy exhibited a reduction in a morphological trait and in only a single instance where it resulted in an increase. We consider the implications of these data obtained with inbred materials regarding the previous results obtained with hybrids (Lee et al. 1996) or compound B-A-A translocations and testers in diverse backgrounds (Sheridan and Auger 2008).
Table 1. Phenotypes examined and the description of where on the plant the measurements were obtained.
| Phenotype | Description |
|---|---|
| Leaf width | Width of primary ear leaf |
| Leaf length | Length of primary ear leaf |
| Plant height | Distance from soil to base of flag leaf |
| Ear height | Distance from soil to primary ear node |
| Internode length | Length of the internode below primary ear |
| Primary ear node circumference | Circumference of node below primary ear |
| Tassel-branch number | No. of lateral branches present in tassel |
Materials and Methods
The 14 simple B-A translocation stocks used in this study are listed in Table 2. These are the same B-A translocations as those used by Lee et al. (1996) and Sheridan and Auger (2008) except that we did not use TB-2Sa, TB6-Sa, TB-7Sc, or TB 10Sc because these were not available in the W22 inbred background. Furthermore, whereas Lee et al. (1996) used TB-4Lc and TB-5La and Sheridan and Auger (2008) used TB-4Lf and TB-5La, we used TB-4Lb and TB-5Lb, but these 4L and 5L B-A translocations have equivalent breakpoints (Beckett 1991, 1993). The breakpoints in the A chromosome involved in the interchange with the B chromosome are described as the proportional distance from the centromere to the end of the normal chromosome arm where the exchange is located. For example, TB-1Sb has a breakpoint at 1S.05, which means that the chromosome 1 centromere retains only 5% of the original short arm, the remainder being replaced by a portion of the long arm of the B chromosome. The remaining 95% of 1S is attached to the long arm of the B chromosome.
Table 2. Chromosome breakpoints, dominant markers, and recessive testers for each of the B-A translocations.
| B-A stock | Breakpoint in A chromosome | Dominant marker located on chromosome arm | Tester for propagating B-A |
|---|---|---|---|
| TB-1Sb | 1S.05 | P-vv | r:m3 |
| TB-1La | 1L.20 | Bz2 | bz2 |
| TB-3Sb | 3S.50 | Ac | r:m3 |
| TB-3La | 3L.10 | A1 | a1 |
| TB-4Sa | 4S.25 | Dt6 | a1-m1-rtd |
| TB-4Lb | 4L.15 | C2 | c2 |
| TB-5Sc | 5S.30 | A2 | a2 |
| TB-5Lb | 5L.10 | Pr | pr |
| TB-6Lc | 6L.11 | Dt 2 | a1-m1-rtd |
| TB-7Lb | 7L.30 | Dt 3 | a1-m1-rtd |
| TB-8Lc | 8L.24 | Ac2 | bz2-m |
| TB-9Sd | 9S.08 | C1 | c1 |
| TB-9Lc | 9L.10 | Ac | r:m3 |
| TB-10L19 | 10L.cent | Rscm2 | r:m3 |
The set of 14 B-A translocation stocks were created by James Birchler and kindly provided by him after they and their tester stocks had been crossed seven times with the inbred W22. The use of these B-A translocations depends on their being marked with autonomous transposable elements (Ac or Dt) that act as dominant markers or colored aleurone dominant alleles (Bz2, A1, C2, A2, C1, R1-scm2; Birchler and Alfenito 1993). These B-A stocks are propagated by crossing them as the pollen parent onto the tester stocks carrying all of the appropriate dominant alleles for aleurone and embryo color as well as the reporter allele for the dominant marker borne on the A chromosome segment of the B-A translocation. For example, TB-1Sb is marked by the presence of the P-vv allele containing the autonomous Ac transposable element on its A segment, and its tester carries r1:mutable3, which responds to the presence of Ac in the same cell by transposition of a nonautonomous Ds element at the r1 locus with the accompanying reversion of the allele to the dominant R1 condition and the presence of purple spots in the aleurone and/or the embryo. Similarly, TB-1La is marked by the presence of the dominant Bz2 allele on its A segment, and its tester carries bz2, which responds to the presence of the Bz2 allele by synthesis of anthocyanin in the aleurone and/or embryo.
Selection of kernels to obtain four types of plants
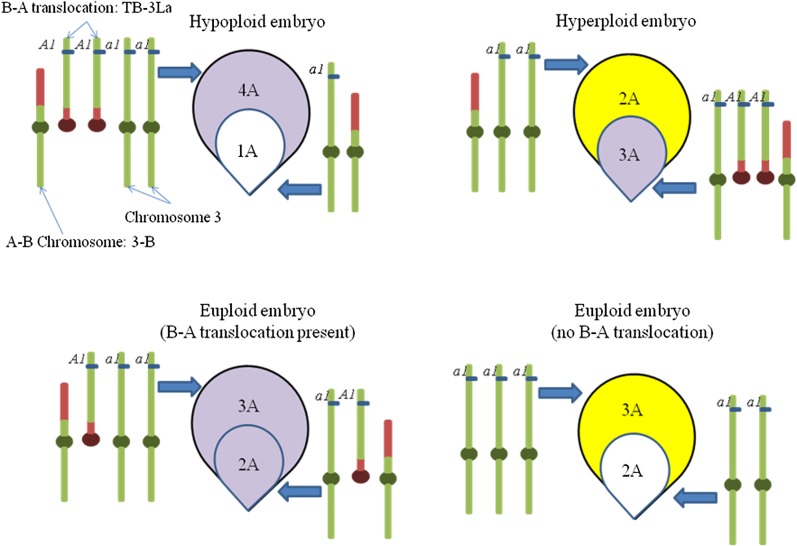
The use of the B-A translocation stocks depends on being able to distinguish kernels of four different genetic constitutions with regard to the dosage of the A chromosome arm segment borne on the B-A chromosome. The four types can be distinguished by the presence or absence of anthocyanin (purple) coloration of their aleurone and embryo tissues: (type 1) kernels with a colored or spotted (hyperploid) aleurone, containing four doses of the A chromosome and a colorless (hypoploid) embryo containing one dose of the A chromosome segment; (type 2) kernels with a colorless (hypoploid) aleurone containing two doses of the A chromosome segment and a colored or spotted (hyperploid) embryo containing three doses of the A chromosome segment; (type 3) kernels with a colored or spotted (euploid) endosperm containing three doses of the A chromosome segment and a colored or spotted (euploid) embryo containing two doses of the A chromosome segment; and (type 4) kernels with a colorless (euploid) aleurone containing three doses of the A chromosome segment and a colorless (euploid) embryo containing two doses of the A chromosome segment. These four kernel types are illustrated in Figure 1. Because the inbred, W22, is Y1/Y1, the endosperm is yellow except for pigmentation that may be present in the aleurone layer of the endosperm.
Figure 1.
Identification of the four kernel types. These kernel types were identified and used to plant the four different dosage types of plants. The B-A chromosome TB-3La and chromosome 3 are illustrated. All other chromosomes are not shown, but still are present in their euploid amounts. TB-3La has a dominant marker, A1, to produce purple, and the complementary homologous segment on chromosome 3 has a recessive marker, a1, that does not produce purple.
The B-A stocks are usually maintained as heterozygous stocks that contain two copies of the B-A and one copy of the A-B chromosomes and one normal A chromosome. They are propagated by planting kernels with hyperploid embryos, which are colored or spotted as shown for kernel type 2, and crossed onto colorless tester stocks that are of the same genetic and chromosomal constitution as the colorless aleurone and the colorless embryo kernel type 4 in Figure 1. Because all four of the kernel types can be readily distinguished, they can be used for planting to produce plants that are hypoploid for the A chromosome segment of interest (type 1 kernels); plants that are hyperploid for the A chromosome segment of interest (type 2 kernels); plants that are euploid but contain one B-A chromosome and one A-B chromosome and therefore carry one dose of the A segment of interest on a B-A chromosome and a second dose on the corresponding normal A chromosome (type 3 kernels); and plants that are euploid and contain two doses of the A chromosome segment of interest, both of which are on normal A chromosomes (type 4 kernels). Two additional systems of plant verification of chromosomal constitution were pollen examination and examination of the color pattern of the kernels on pollinated ears of the measured plants. Hypoploid plants can be assayed by examining freshly shed pollen; 50% pollen abortion confirms hypoploidy.
Planting plan
For each of the 14 B-A stocks, the four types of kernels were planted in families of 16 kernels in the summer of 2010 at a nursery in Grand Forks, North Dakota. Two replicates were planted: the first on May 22 and the second on May 29.The kernels were planted 9 in. apart in 60-ft long rows that were spaced 50 in. apart. The replicates consisted of 14 rows in which each row was dedicated to a particular B-A translocation. Each row had four 16-kernel families planted with regard to embryo genotype and two 8-kernel families of the appropriate tester in the pattern: tester family, hyperploid family, euploid (no B-A chromosome present) family, hypoploid family, euploid (B-A chromosome present) family and tester family. The replicates were enclosed by border rows.
Selection and measuring of phenotypic traits
The tester traits that were measured include the seven morphological traits evaluated by Lee et al. (1996) and Sheridan and Auger (2008). These were (1) leaf width, (2) leaf length, (3) plant height, (4) ear height, (5) internode length, (6) stalk circumference, and (7) tassel branch number (Table 1). The morphological trait measurements were performed after the plants had completed flowering. A 2-m-long measuring stick and a tape measure calibrated to millimeters were used to obtain the measurements. The measurements were recorded to the nearest millimeter and transferred to an Excel spreadsheet. Calculations were performed to determine the total, the mean value, the standard deviation, and the standard error of the mean.
Data analysis
The analysis of the data included three methods: principle component analysis, multivariate analysis of variance (MANOVA), and an analysis of variance (ANOVA). The MANOVA indicated that the seven morphological phenotypes showed significant difference in relation to replicate, chromosome arm, and dosage, including all combinations between these three factors. Therefore, the two replicates did not show consistent differences between the chromosome arms and dosages and the two replicates could not be combined for analysis as was originally planned. In reference to chromosome arm and dosage, these analyses indicate that each chromosome arm and dosage combination showed different effects on the seven phenotypes, as was expected.
Results
An ANOVA was run for each of the seven characteristics separately and separately for each replicate to determine the differences between chromosome arms and dosages. The chromosome arms and dosages showed significant differences between their means. This result allowed for a post hoc Tukey’s comparison test to determine which hypoploid plants, hyperploid plants, euploid plants with no B-A chromosome present, and euploid plants with B-A present because of failure of nondisjunction means were significantly different from each other for each of the chromosome arms. The Tukey’s comparison test indicated that euploids with no B-A present and euploids with B-A present because of failure of nondisjunction, with the exception of chromosome arm 5S in the first replicate, showed no significant differences between them. This result indicates that the presence of a B chromosome had no detectable effect on the traits that were measured. Therefore, the euploid plants were used as the basis of comparison for the hypoploid plants and hyperploid plants; however, these two euploid groups are kept separate in the presentation of data. To distinguish them, the euploid plants no B-A present are designated “E euploid plants” (grown from type 4 kernels) and the euploid plants with B-A present because of failure of nondisjunction are designated “F euploid plants” (grown from type 3 kernels).
Comparison of dosage effects between hypoploids and hyperploids
Leaf width was significantly reduced when 10 of the 14 chromosome arms were hypoploid plants: 1S, 1L, 3L, 4L, 5S, 5L, 6L, 7L, 8L, and 9L (see Supporting Information, Table S1 and Table S2). The only increase in measurement of morphological traits in hypoploid plants was an increase in leaf width in replicate 2 for plants hypoploid for chromosome arm 3S as compared with the F euploid plants. The leaf width of plants was significantly reduced when 4 of the 14 chromosome arms were hyperploid: 1L, 3L, 8L, and 10L. Hypoploidy for all of these 4 arms except 10L resulted in significant reduction in leaf width.
Leaf length was shorter when 9 of 14 chromosome arms were hypoploid: 1S, 1L, 3L, 4L, 6L, 7L, 8L, 9S, and 9L (Table S3 and Table S4). The leaf length of hyperploid plants was significantly reduced for only chromosome arm 6L.
Plant height was significantly reduced when 12 of the 14 chromosome arms were hypoploid: 1S, 1L, 3L, 4L, 5S, 5L, 6L, 7L, 8L, 9S, 9L, and 10 L (Table S3 and Table S4). The plant height was significantly reduced when 5 of the 14 chromosome arms were hyperploid: 1S, 1L, 5S, 8L, and 9L (Table S5 and Table S6). All 5 of these arms also resulted in plant height reduction when they were hypoploid.
Ear height was significantly reduced when 11 of the 14 chromosome arms were hypoploid: 1S, 1L, 3L, 4L, 5S, 5L, 6L, 7L, 8L, 9L, and 10L (Table S7 and Table S8). This is a nearly complete parallel pattern to the set of 12 arms (all except 9S) that reduced plant height when the arms were hypoploid. The ear height was significantly reduced when 3 of the chromosome arms were hyperploid: 1S, 1L, and 8L (Table S7 and Table S8). These 3 chromosome arms also reduced ear height when they were hypoploid. The only instance when hyperploidy resulted in an increase in a plant morphological trait was in the case of plants hyperploid for chromosome arm 9S, but only in reference to the E euploid set in replicate 2.
Internode length was significantly reduced when 8 of the 14 chromosome arms were hypoploid: 1S, 3L, 4L, 5S, 7L, 8L, 9S, and 10L (Table S9 and Table S10). The internode length was significantly reduced when 4 of the 14 chromosome arms were hyperploid: 1S, 1L, 3L, and 5S. These same 4 chromosome arms also reduced internode length when they were hypoploid.
Primary ear node circumference was significantly reduced when 9 of the 14 chromosome arms were hyperploid: 1S, 1L, 3L, 4L, 5S, 5L, 7L, 8L, and 9L (Table S11 and Table S12). The primary ear node circumference was significantly reduced when 2 of the 14 chromosome arms were hyperploid: 1L and 8L. Both of these arms also reduced primary ear node circumference when they were hypoploid.
Tassel branch number was significantly reduced when 6 of the 14 chromosome arms were hypoploid: 1L, 3L, 4L, 5S, 7L, and 9S (Table S13 and Table S14). The tassel branch number was significantly reduced when 1 of the 14 chromosomes arms was hyperploid: 8L. It did not reduce tassel branch number when it was hypoploid.
Dosage sensitivity
The dosage-sensitive responses of the seven morphological traits are shown in Table 3 and Table 4. Table 3 summarizes the significant differences between hypoploid plant data sets for the 14 tested chromosome arms when they were compared to both sets of euploid data. If either euploid set in a replicate indicates a difference in one of the phenotypes, then a “–” or a “+” mark indicates that the hypoploid plant data sets were significantly lower or higher in the measurement for that trait. Table 4 summarizes the significant differences between the hyperploid plant data sets when they were compared to both euploid data sets. (See Table S1, Table S2, Table S3, Table S4, Table S5, Table S6, Table S7, Table S8, Table S9, Table S10, Table S11, Table S12, Table S13, and Table S14 for the detailed data on which Table 3 and Table 4 are based.) The most obvious effect of varying the chromosome arm dosage was a significant decrease in the trait when measured in the hypoploid or the hyperploid plants as compared with the euploid plants. It is also apparent that the negative effects of hypoploidy are generally more widespread among the chromosome arms and more severe than those of hyperploidy.
Table 3. Summary of significant differences between hypoploid and euploid plants.
| Phenotype: | Leaf width |
Leaf length |
Plant height |
Primary ear height |
Internode length |
Primary ear node circumference |
Tassel branch number |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Replicate: | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd |
| 1S | − | − | − | − | − | − | − | − | − | − | − | |||
| 1L | − | − | − | − | − | − | − | − | − | − | − | |||
| 3S | + | |||||||||||||
| 3L | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 4S | ||||||||||||||
| 4L | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 5S | − | − | − | − | − | − | − | − | − | − | − | |||
| 5L | − | − | − | − | − | − | ||||||||
| 6L | − | − | − | − | − | − | − | |||||||
| 7L | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 8L | − | − | − | − | − | − | − | − | − | |||||
| 9S | − | − | − | − | − | − | − | − | ||||||
| 9L | − | − | − | − | − | − | − | |||||||
| 10L | − | − | − | − | − | − | ||||||||
A “−” indicates that the hypoploid plants were significantly lower in a measurement by P < 0.05. A “+” indicates that the hypoploid plants where significantly higher in a measurement by P < 0.05. And a blank box indicates “no significant difference between the hypoploid plant data and either set of euploid plants.”
Table 4. Summary of significant differences between hyperploid and euploid plants.
| Phenotype: | Leaf width |
Leaf length |
Plant height |
Primary ear height |
Internode length |
Stalk circumference |
Tassel branch number |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Replicate: | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd | 1st | 2nd |
| 1S | − | − | − | − | ||||||||||
| 1L | − | − | − | − | − | − | − | |||||||
| 3S | ||||||||||||||
| 3L | − | − | − | |||||||||||
| 4S | ||||||||||||||
| 4L | ||||||||||||||
| 5S | − | − | − | |||||||||||
| 5L | ||||||||||||||
| 6L | − | |||||||||||||
| 7L | ||||||||||||||
| 8L | − | − | − | − | − | |||||||||
| 9S | + | |||||||||||||
| 9L | − | − | ||||||||||||
| 10L | − | |||||||||||||
A “−” indicates that the hyperploid plants were significantly lower in a measurement by P < 0.05. A “+” indicates that the hyperploid plants where significantly higher in a measurement by P < 0.05. And a blank cell indicates “no significant difference between the hyperploid plant data and either set of euploid plants.”
Some chromosome arms affect more traits than other chromosome arms
In Table 5 we present a side-by-side comparison of the effects of hypoploidy (one dose) and hyperploidy (three doses) of each of the 14 chromosome arms on the morphological traits. The table was constructed by indicating a significant effect if a significant effect was obtained for either the first or the second replicate of data. The arrangement of the data as shown in Table 5 readily reveals that there is a wide range of frequency of effects on the seven plant morphological traits among the 14 chromosome arms tested. The three chromosome arms 3L, 4L, and 7L affected all seven of the plant traits when hypoploid, but when hyperploid affected only two, none, and none of these seven traits, respectively. Both chromosome arms 1L and 5S affected six of the seven plant traits when hypoploid and when hyperploid affected five and two of the plant traits, respectively. All three chromosome arms 1S, 8L, and 9L affected five plant traits when hypoploid and when hyperploid affected three, three, and one of the plant traits, respectively. Chromosome arms 5L, 6L and 9S when hypoploid affected four plant traits and when hyperploid affected zero, one, and one respectively. The positive hyperploid effect of chromosome arm 9S was one of only two positive effects obtained in this study. The other positive effect occurred in plants hypoploid for chromosome arm 3S. Chromosome arm 3S showed no effect as a hyperploid. Chromosome arm 10L when hypoploid affected three plant traits and when hyperploid affected one plant trait. Only one chromosome arm, 4S, had no effect on plant traits when hypoploid or hyperploid.
Table 5. Comparison of the effects of one dose and three doses of each of the chromosome arm on the morphological traits.
| Chr arm | Leaf-width dosage |
Leaf-length dosage |
Plant-height dosage |
Ear-height dosage |
Internode length dosage |
Ear-node-circumference dosage |
Tassel-branch-number dosage |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | |
| 1S | ** | ** | ** | ** | ** | ** | ** | * | ** | ** | ||||
| 1L | ** | * | ** | ** | ** | ** | ** | ** | * | ** | ** | |||
| 3S | (*) | |||||||||||||
| 3L | ** | ** | ** | ** | ** | ** | * | ** | ** | |||||
| 4S | ||||||||||||||
| 4L | ** | ** | ** | ** | ** | ** | ** | |||||||
| 5S | ** | ** | ** | ** | * | ** | ** | ** | ||||||
| 5L | ** | ** | ** | ** | ||||||||||
| 6L | ** | ** | ** | ** | ** | |||||||||
| 7L | ** | ** | ** | ** | * | ** | * | |||||||
| 8L | * | * | ** | ** | ** | ** | ** | * | ** | ** | ||||
| 9S | * | ** | * | (*) | ** | * | ||||||||
| 9L | ** | ** | ** | * | ** | ** | ||||||||
| 10L | * | ** | ** | ** | ||||||||||
An asterisk indicates that the mean value of the measured trait of the plants with one dose or three doses of the chromosome arm was significantly lower than the value of plants with two doses. *P-value = 0.05–0.01; **P-value ≤ 0.01. “(*)” indicates that the measured trait was significantly higher than the plants with two doses at a P-value from 0.05 to 0.01.
Chromosome arms differed in their hypoploid vs. hyperploid effects
Some of the chromosome arms affected plant morphological traits more frequently both under hypoploid conditions and under hyperploid conditions. Most noteworthy is chromosome arm 1L, which in a single dosage affected all seven traits except tassel branch number and in a triple dosage affected all seven traits except leaf length and tassel branch number (Table 5). Similarly, chromosome arm 1S affected six traits in a single dosage and three traits in a triple dosage; chromosome arm 3L affected seven traits in a single dosage and two traits in a triple dosage; chromosome arm 5S affected six traits in a single dosage and two traits in a triple dosage; and chromosome arm 8L affected five traits in a single dosage and three traits in a triple dosage. Among the remaining chromosome arms, three arms (6L, 9S, and 9L) affected only a single trait as hyperploids and three arms (4L, 5L, and 7L) affected four to seven traits as hyperploids but none of the morphological traits as hyperploids. Chromosome arm 3S affected only one trait as a single dose and affected none of the traits as a hyperploid. Chromosome arm 4S affected none of the traits in either a single or a triple dosage.
The data in Table 5 show that there were 67 instances where a single dose of a chromosome arm resulted in a significant effect on a morphological trait, only one of which (3S affecting leaf width) had a positive effect while there were 21 instances where three doses of a chromosome arm resulted in a significant effect on a morphological trait, only one of which (9S affecting primary ear height) had a positive effect.
Not only were the effects of hypoploidy more widespread among the chromosome arms and among the morphological traits affected, but hypoploidy usually had a more severe negative effect on the traits. For example, among the 12 chromosome arms for which hypoploidy resulted in a reduction in plant height, in 5 of these chromosome arms hyperploidy resulted in a reduction in plant height; in 3 of these 5 arms, the reduction resulting from hypoploidy was significantly greater than that resulting from hyperploidy. This same difference in severity of effect is evident for the relative effects of the 11 chromosome arms for which hypoploidy resulted in a reduction in ear height. Three of these 11 chromosome arms resulted in a reduction in ear height when hyperploid, and, for 2 of these 3 arms, the reduction resulting from hypoploidy was significantly greater than that resulting from hyperploidy (see Table S3 and Table S4 for the detailed data regarding these comparisons).
Discussion
B-A translocations in an inbred background are well suited to measure dosage effects
The B-A translocations and tester stocks used in this study were in a W22 inbred background. Previous investigators have used B-A translocations to evaluate the effects of aneuploidy for individual chromosome arms on maize plant morphological traits. Their results have been similar to what we have found in the present study. When the dose of chromosome arms was individually increased, the traits in the resulting hyperploid plants were modestly affected (Chang 1984; Lee et al. 1996; Neuffer et al. 1997); but when the dose of chromosome arms was individually decreased, the traits in the resulting hypoploid plants were more severely altered, as was their relative vigor (Chang et al. 1987; Beckett 1991; Lee et al. 1996; Neuffer et al. 1997). These results were obtained when using B-A translocation stocks in diverse genetic backgrounds (not in an inbred background) or, in the case of Lee et al. (1996), the B-A translocations had been introgressed into the B73 Ht inbred and crossed onto the Mo17 Ht inbred to produce F1 hybrids.
As far as we are aware, the results presented here on the effects of aneuploidy for individual chromosome arms on maize plant morphological traits are the first reported where the B-A translocation stocks and testers as well as the progeny evaluated were all in a uniform inbred background. Because, except for Lee et al. (1996), the previous studies used B-A stocks and testers such that the parents and progeny varied in their genetic constitutions, we believe that the effects of aneuploidy described here are more accurate reflections of the actual effects of dosage differences. We believe that this is the case because the possible variations in the traits evaluated resulting from allelic heterozygosity and the segregation of traits among the progeny should be minimized when all of the genetic stocks employed in the study have been introgressed into a common inbred line.
Results of previous dosage analysis of simple B-A translocations
In the study of Chang et al. (1987), 15 B-A translocation stocks were crossed four times with A619 and then crossed onto genetic testers for identification of hypoploids. They measured plant height and leaf width and noted that “the plant height of hypoploids is consistently smaller than those of normal sibs,” but TB-1Sb, TB-1La, and TB-9Lc had greater effects than others on plant height and leaf width. Chang et al. (1987) did not identify the genotype of the testers onto which they crossed their B-A stocks, but they refer to them as genetic testers and were probably of diverse, noninbred backgrounds. We found that a single dose of chromosome arm 1S, 1L, 3L, 7L, and 10L had the greatest reduction in plant height while a single dose of chromosome arm 1S, 1L, 3L, 5S, and 9L had the greatest reduction in leaf width.
A comparison of the effects of hyperploidy among 17 of the maize chromosome arms was performed by Chang (1984). The smallest plant height was observed in plants hyperploid of TB-1Sb, TB-5Sc, and TB-9Lc. The genetic background of the B-A translocation and the tester stocks was not identified but were, presumably, genetic stocks of noninbred backgrounds.
We report here a total of 67 cases where chromosome hypoploidy and 21 cases where hyperploidy resulted in a significant effect on a morphological trait. These represent fewer dosage effects than those reported by Lee et al. (1996) who found 88 cases of hypoploidy and 63 cases of hyperploidy resulting in significant effects on maize morphological traits [see Table S15 for a comparison of our Table 5 with table 5 in Lee et al. (1996)]. When we examine the effects of varying the dosage of the individual chromosome arms in the two data sets, we observe that, in general, the results are congruent. For example, among those seven chromosome arms that we found to affect the most traits as hypoploids, five were included among the eight arms that Lee et al. (1996) observed to affect the most traits as hypoploids. Furthermore, 58 of the 67 cases of a hypoploid effect observed by us were also reported by Lee et al. (1996). When the hyperploid data were compared, we found 21 cases where the hyperploid condition resulted in a significant effect on plant morphological traits. Among the 20 of these 21 cases where Lee et al. also obtained data, 19 also had a significant effect on morphological traits in their report (the exception was internode length on chromosome arm 3L). Overall, our results in most cases are congruent with those of Lee et al. (1996). To the extent that the analyses of Chang (1984), Chang et al. (1987), Lee et al. (1996), and our results reported here are comparable, the congruence of our results suggest that the traits are quantitative traits controlled by multiple genes whose activities provide a balanced regulation that transcends individual inbred lines or diverse genetic backgrounds.
We previously reported on chromosome segmental dosage analysis of maize morphogenesis using B-A-A translocations (Sheridan and Auger 2008). These compound B-A translocations enabled us to simultaneously assess the possible dosage effects of two nonhomologous chromosome segments on plant development. We compared hyperploid plants with nonhyperploid plants for the effects of hyperploidy on the same seven traits examined in the present study. We noted that the effects of hyperploidy on plant phenotypes obtained with numerous B-A-A translocations are more severe than those obtained with simple B-A translocations in noninbred stocks. We suggested that the more severe effects on plant morphological traits obtained when two chromosome segments are simultaneously increased in dosage either are an additive effect that is the sum of the hyperploid effects observed with hyperploidy of the simple B-A translocations involving the individual chromosome arm segments or are the result of the interactions of genetic information borne on those two segments.
It is noteworthy that among the 57 dosage-sensitive responses reported in the B-A-A translocation study, 8 of these were positive results involving leaf width (four cases), leaf length (one case), stalk circumference (two cases), and tassel branch number (one case). These increases in the measured trait values were associated with hyperploidy for five different B-A-A translocations; all five of these translocations included a segment of chromosome arm 1L. As noted above, we found in the present study that changes in chromosome arm 1L dosage affected a larger number of traits in both a single dose and a triple dose than in any of the other arms tested. We suggest that the occurrence of the positive responses in the B-A-A study of hyperploid plants may reflect gene dosage interaction between the two arm segments present in the B-A-A translocations and that chromosome arm 1L (the genetically longest chromosome arm) may be especially abundant in genes that participate in dosage interactions.
Aneuploidy and trans-acting dosage effects on gene expression in maize
Since the reports were made by Birchler (1979) and Birchler and Newton (1981) it has been evident that the levels of expression of specific proteins in both the maize embryo and endosperm can remain constant or can increase or decrease when the chromosome arm bearing the gene coding for the protein is varied in dosage. The same was shown to be true for proteins coded for by genes borne on chromosome arms other than the arm being varied.
In their seminal article on trans-acting dosage effects on the transcript levels of nuclear-encoded genes in maize aneuploids, Guo and Birchler (1994) noted that genes whose dosage was not altered (genes located on chromosome arms other than the hypoploid or hyperploid arm of the B-A chromosome) were affected by aneuploidy of unlinked segments (located on the A segment of the B-A chromosome). These authors suggested that the reduction in vigor in monosomics and trisomics results from the limiting effects of many gene products not only encoded on the respective chromosome being varied in dosage, but throughout the genome. Because Guo and Birchler (1994, p. 2001) observed that the varied chromosomal regions tend to exhibit multiple trans effects enhancing or inhibiting the expression of genes elsewhere, they hypothesized that “this spectrum of effects results from altering the dosage of the individual components of the gene regulatory system that affect each structural gene.”
The same set of B-A translocations used in the present study were used by Auger et al. (2001) to examine their effects on the transcript levels of six mitochondrial protein genes. The messenger RNA levels of the six genes of both the embryo and the endosperm were measured. Whereas Guo and Birchler (1994) found that dosage effects on the embryo were predominantly negative and the effects on the endosperm were predominantly positive, Auger et al. (2001) found in their study of dosage effects on mitochondrial gene expression that negative effects predominate in both the embryo and the endosperm. In both the Guo and Birchler (1994) and Auger et al. (2001) articles, each gene was affected by the dosage series of several different chromosome arms. Auger et al. (2001) suggest that the large number of dosage effects most likely reflects a hierarchy of dosage-dependent processes. They also suggest that genes at the top of the hierarchy are able to affect the phenotype because they act through a series of dosage-dependent steps to the ultimate monitored phenotype. They note that “this concept unifies the polygenic nature of additive quantitative traits and the extensive effects of aneuploidy on any one character” (Auger et al. 2001, p. 1719).
In their recent review, Birchler and Veitia (2012) hypothesized that gene dosage balance—the presence of the relative dosage of the euploid chromosome arm segments throughout the genome—is critical for normal development and phenotypic characteristics. They suggest that the changes observed when gene balance is altered “result from stoichiometric differences among members of macromolecular complexes, the interactome, and signaling pathways” (Birchler and Veitia 2012, p. 14746). The general observation that hypoploid maize plants exhibit more severe reduction in morphological traits is likely a manifestation of haploinsufficiency. This is known to be the case for many morphogenetic regulators in mice (Boell et al. 2013), and this suggests that negative effects of hypoploidy in the aneuploidy syndromes are the cumulative result of such haploinsufficiency.
In a more recent report from the same laboratory, Yao et al. (2013) observed that heterosis is subject to dosage effects and that this observation suggests that molecular models of heterosis should consider the diversity of alleles and how they interact across the genome. The studies cited above suggest the presence of a genome-wide dosage-sensitive gene regulatory system. In maize, such a system may consist of many genes on each chromosome arm whose transcription products regulate gene expression on other chromosome arms and thereby affect quantitative traits, mostly in a negative direction (see Guo and Birchler 1994).
A recent report (Letourneau et al. 2014) on genome-wide gene expression dysregulation in human cells trisomic for chromosome 21 may be useful for modeling a genome-wide dosage-sensitive regulatory system function in maize. The results of a genome-wide transcriptome analysis of trisomic 21 cells differed from normal diploid cells inasmuch as the trisomy condition appears to have a leveling effect such that chromosomal domains normally exhibiting a high transcript level are decreased and those domains normally exhibiting a low level are increased in their transcript level (Letourneau et al. 2014). These researchers also found that the chromosomal domains exhibiting dysregulation of transcription are conserved in the Ts65Dn mouse model of Down syndrome.
The research on aneuploidy effects on maize discussed above provides some constraints on a feasible model of a maize genome-wide dosage-sensitive gene regulatory system. Any such model must accommodate the genome-wide trans-acting dosage effects resulting from an increase or a decrease in the copy number of one (B-A) or two (B-A-A) chromosome segments; it must explain how dosage-sensitive regulatory effects are tissue-specific; it must explain how different phenotypic effects occur depending on which chromosome segment is varied; and it should provide insight into the different phenotypic effects observed with different B-A-A translocations and with those B-A-A translocations where trait measurements were increased. The genome-wide gene expression regulatory system described by Letourneau et al. (2014) provides a model that may be suitable in a modified form to help explain the maize aneuploidy results. However, the modified model would need to allow for the selective modulation of transcription rates, in particular chromosome transcription domains, so as to accommodate tissue specificity as well as the different phenotypic effects resulting from increased and decreased dosage of different chromosome segments. Similarly, our results and discussion may be useful to other researchers investigating the effects of aneuploidy in a wide range of organisms.
Supplementary Material
Acknowledgments
We thank J. B. Beckett, M. T. Chang, and E. H. Coe for permission to cite their Maize Newsletter articles; James Birchler for providing the B-A translocation stocks and tester stocks; Don Auger, James Birchler, Jan Clark, and two anonymous reviewers for helpful comments on the manuscript; and Robert Newman for assistance with the statistical analyses.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.166330/-/DC1.
Communicating editor: A. Houben
Literature Cited
- Auger D. L., Newton K. J., Birchler J. A., 2001. Nuclear gene dosage effects upon the expression of maize mitochondrial genes. Genetics 157: 1711–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett J. B., 1983. Kernel weight effects and transmission of a partial trisome involving the long arm of chromosome 5 in maize. Can. J. Genet. Cytol. 25: 346–353 [Google Scholar]
- Beckett J. B., 1991. Cytogenetic, genetic and breeding applications of B-A translocations in maize, pp. 491–527 in Chromosome Engineering in Plants, edited by Tsuchia T., Gupta P. K. Elsevier, Amsterdam [Google Scholar]
- Beckett J. B., 1993. Locating recessive genes to chromosome arm with B-A translocations, pp. 315–327 in The Maize Handbook, edited by Freeling M., Walbot V. Springer-Verlag, New York [Google Scholar]
- Birchler J. A., 1979. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics 92: 1211–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., 1993. Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27: 181–204 [DOI] [PubMed] [Google Scholar]
- Birchler J. A., Alfenito M. G., 1993. Marker systems for B-A translocations in maize. J. Hered. 84: 135–138 [Google Scholar]
- Birchler J. A., Hart J. A., 1987. Interaction of endosperm size factors in maize. Genetics 117: 309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Newton K. J., 1981. Modulation of protein levels in chromosomal dosage series of maize: the biochemical basis of aneuploidy syndromes. Genetics 99: 247–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Veitia R. A., 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 109: 14746–14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boell L. L., Pallares F., Brodski C., Chen Y., Christian J., et al. , 2013. Exploring the effects of gene dosage on mandible shape in mice as a model for studying the genetic basis of matural variation. Dev. Genes Evol. 223: 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.-T., 1984. Characterization of hyperploid B-A translocations. Maize Genet. Coop Newsl. 58: 63–64 [Google Scholar]
- Chang M.-T., Coe E. H., Beckett J. B., 1987. Genetic effects of hypoploidy on kernel weight, plant height and leaf width. Maize Genet. Coop. Newsl. 61: 91–92 [Google Scholar]
- Guo M., Birchler J. A., 1994. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266: 1999–2002 [DOI] [PubMed] [Google Scholar]
- Lee E. A., Darrah L., Coe E., 1996. Dosage effects on morphological and quantitative traits in maize aneuploids. Genome 39: 898–908 [DOI] [PubMed] [Google Scholar]
- Letourneau A., Santoni F. A., Bonilla X., Sallani M. R., Gonzalex D., et al. , 2014. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508: 345–350 [DOI] [PubMed] [Google Scholar]
- Lin B.-Y., 1982. Association of endosperm reduction with parental imprinting in maize. Genetics 100: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer M. G., Sheridan W. F., 1980. Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95: 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer M. G., Coe E. H., Wessler S. R., 1997. Mutants of Maize. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Roman H., 1947. Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32: 391–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon M. J., Stinard P. S., James M. G., Myers A. M., Robertson D. S., 1994. Genetic analysis of 63 mutations affecting maize kernel development isolated from Mutator stocks. Genetics 136: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan W. F., Auger D. L., 2006. Construction and uses of new compound B-A-A maize chromosome translocations. Genetics 174: 1755–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan W. F., Auger D. L., 2008. Chromosome segmental dosage analysis of maize morphogenesis using B-A-A translocations. Genetics 180: 755–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Gray A. D., Auger D. L., Birchler J. A., 2013. Genomic dosage effects on heterosis in triploid maize. Proc. Natl. Acad. Sci. USA 110: 2665–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.