Abstract
Gene conversions and crossovers are related products of the repair of double-stranded DNA breaks by homologous recombination. Most previous studies of mitotic gene conversion events have been restricted to measuring conversion tracts that are <5 kb. Using a genetic assay in which the lengths of very long gene conversion tracts can be measured, we detected two types of conversions: those with a median size of ∼6 kb and those with a median size of >50 kb. The unusually long tracts are initiated at a naturally occurring recombination hotspot formed by two inverted Ty elements. We suggest that these long gene conversion events may be generated by a mechanism (break-induced replication or repair of a double-stranded DNA gap) different from the short conversion tracts that likely reflect heteroduplex formation followed by DNA mismatch repair. Both the short and long mitotic conversion tracts are considerably longer than those observed in meiosis. Since mitotic crossovers in a diploid can result in a heterozygous recessive deleterious mutation becoming homozygous, it has been suggested that the repair of DNA breaks by mitotic recombination involves gene conversion events that are unassociated with crossing over. In contrast to this prediction, we found that ∼40% of the conversion tracts are associated with crossovers. Spontaneous mitotic crossover events in yeast are frequent enough to be an important factor in genome evolution.
Keywords: yeast, mitotic recombination, gene conversion, DNA damage repair, loss of heterozygosity
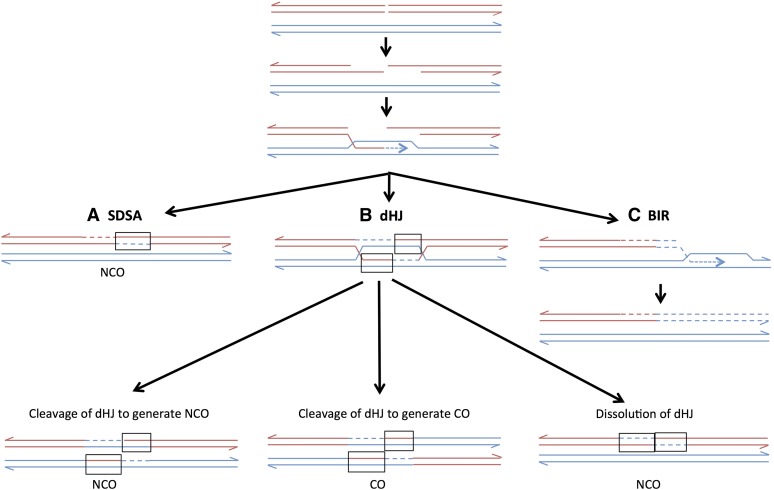
IN the yeast Saccharomyces cerevisiae, double-stranded DNA breaks (DSBs) in mitotically dividing cells are usually repaired by homologous recombination using an unbroken DNA molecule as a template (Symington 2002). In G2 diploid cells, sister chromatids, rather than the homolog, are the favored substrate for repair (Kadyk and Hartwell 1992). Associated with the repair of most DSBs, there is a region of DNA transferred nonreciprocally from the unbroken chromosome to the broken chromosome; when this process alters the sequence of the recipient chromosome, it is called “gene conversion” (Petes et al. 1991; Paques and Haber 1999). In the current models of recombination, conversion events are produced through several different pathways (Figure 1). In one pathway (synthesis-dependent strand annealing, or SDSA), one broken end invades the homologous template and primes DNA synthesis. The invading end is then ejected from the template and reanneals to the other broken end (Figure 1A). Mismatches within the resulting heteroduplex (boxed regions in Figure 1) can be repaired to generate the conversion event. This pathway produces conversions unassociated with crossovers [noncrossover (NCO) conversions]. An NCO conversion can also be generated by dissolution of a double Holliday junction (dHJ) intermediate. Cleavage of the dHJ can produce either a conversion event associated with a crossover (CO) conversion or with an NCO conversion (Figure 1B). The resolution of the dHJ is biased toward the crossover, rather than toward the noncrossover pathway, in mitosis (Mitchel et al. 2010). Finally, a DSB can be repaired by a nonreciprocal process in which one broken end invades and copies the homolog to the end of the chromosome, and the other chromosome end is lost. This pathway is break-induced replication (BIR, Figure 1C) (Llorente et al. 2008). In S. cerevisiae, in both meiosis (Allers and Lichten 2001) and mitosis (Mitchel et al. 2010), most NCO gene conversion events reflect the SDSA pathway rather than processing of a dHJ.
Figure 1.
Pathways of repair of DNA breaks by homologous recombination. Two recombining double-stranded DNA molecules are shown as paired red and blue lines. Dashed lines indicate DNA synthesis. Recombination events are initiated by a DSB, followed by 5′ to 3′ processing of the broken ends. All pathways are initiated by invasion of one end of the broken chromosome into the unbroken chromosome, followed by formation of a D loop caused by DNA synthesis from the invading 3′ strand. (A) Synthesis-dependent strand annealing (SDSA). Following DNA synthesis, the invading strand disassociates from the template and reassociates with the other broken end. The net result is a heteroduplex on one side of the position of the original DSB with flanking markers in the original configuration (NCO). The region of the heteroduplex is boxed. Repair of mismatches within the heteroduplex can result in a gene conversion (two blue strands) or a restoration event (two red strands). (B) Double Holliday junction (dHJ). The non-invading broken end anneals with the D loop forming two junctions. Depending on the mode of cleavage of the junctions, this structure can be resolved as a noncrossover (left) or a crossover (center). Alternatively, the structure can be dissolved without junction cleavage (right). (C) Break-induced replication (BIR). The end from the left portion of the broken chromosome sets up a moving D loop that replicates the intact chromosome by conservative replication. The right portion of the broken chromosome is lost.
Spontaneous mitotic recombination events are ∼104-fold less frequent than meiotic events (Esposito and Wagstaff 1981). Consequently, analysis of spontaneous mitotic gene conversion events requires a selective system or induction of events by DNA damage or site-specific meganucleases such as HO or I-SceI (Paques and Haber 1999). The most common method of detecting spontaneous conversions is to use auxotrophic heteroalleles. For example, in one study in which one homolog had a leu2-K allele and the other a leu2-R allele, Leu+ conversion events were detected at a rate of ∼1 × 10−7/division (Lichten and Haber 1989). The rates of conversion in most heteroallelic studies on homologs vary between 10−7 and 10−6/division (Jinks-Robertson and Petes 1986; Steele et al. 1991; Nickoloff et al. 1999; Lettier et al. 2006). In one study, conversion rates involving heteroallelic genes located close together were higher: 4 × 10−6/division (Aguilera and Klein 1989).
Previous studies of the lengths of mitotic gene conversion tracts have yielded disparate results. In a study of mitotic gene conversion between homologs, Judd and Petes (1986) found that ∼20% of conversion tracts were <2 kb, but at least 40% were >4 kb. In this study, conversion tracts >4 kb could not be measured because of the lack of flanking markers. Similarly, in a study of HO-induced recombination, Nickoloff et al. (1999) found that ∼40% of the conversion tracts were ≤2 kb. The other conversion tracts were at least 3.3 kb in length, but the lack of flanking markers prevented an accurate determination of tract length. In a plasmid-chromosome recombination assay (Mitchel et al. 2010), NCO conversions were usually <400 bp, and CO conversions often extended to the limits of the homology, a distance of >500 bp from the initiating DNA lesion. In two other studies in which conversion events were classified as either short (<1 kb) or long (>1 kb), long tracts were more frequently associated with crossovers (Aguilera and Klein 1989; Ho et al. 2010).
The use of heteroalleles to monitor the frequency of conversion and the length of conversion tracts has a substantial limitation. If conversion is a consequence of heteroduplex formation followed by mismatch repair and if mismatches in a heteroduplex are usually repaired using the same template strand, a conversion event that includes both mismatches will not generate a wild-type allele. Thus, estimates of the frequency of conversion based on heteroalleles will usually underestimate the frequency of heteroduplex formation and will underestimate tract size. We have recently used microarrays or related methods to measure the conversion tract lengths associated with recombination between homologs using a system that does not involve heteroalleles (Lee et al. 2009; St. Charles et al. 2012; St. Charles and Petes 2013; Yin and Petes 2013). Most of these studies examined CO conversion tracts. The median size of spontaneous CO conversion tracts was ∼10 kb; NCO conversion tracts were not examined in these studies. In a study of ultraviolet radiation (UV)-induced recombination events, NCO tracts were significantly shorter than CO tracts with median sizes of 4.9 and 7.6 kb, respectively (Yin and Petes 2013).
One controversial issue concerning mitotic gene conversion in yeast is the relative frequencies of CO and NCO conversion events. Mitotic crossing over has the negative effects of allowing harmful recessive mutations to become homozygous, and ectopic crossovers cause changes in chromosome structure. It has been suggested, therefore, that most mitotic repair events should be associated with NCO conversions rather than CO conversions. In yeast, however, the percentage of CO conversion events varies greatly in different studies: 10% (Jinks-Robertson and Petes 1986), 12% (Inbar and Kupiec 1999), 17% (Chua and Jinks-Robertson 1991), 20% (Nickoloff et al. 1999), 25% (Malkova et al. 1996), 25% (Haber and Hearn 1985), 33% (Yin and Petes 2013), 50% (Aguilera and Klein 1989), and 50% (Welz-Voegele and Jinks-Robertson 2008). Most of these studies involved heteroallelic recombination, DNA damage-induced events, and/or ectopic recombination.
Below, we describe experiments to measure the frequency of spontaneous gene conversion between homologous chromosomes, the lengths of gene conversion tracts, and the fraction of CO and NCO conversions. We utilize a system that allows detection of very long (>100 kb) conversion tracts and that does not have the biases associated with heteroallelic conversion.
Materials and Methods
Strain construction and genetic methods
Experiments were conducted using the diploid strain EY7. The construction of EY7 is described in Supporting Information, Table S1, and primers used for strain construction and sequence analysis are listed in Table S2. EY7 was generated by mating two haploids isogenic with W303-1A and YJM789. The resulting diploid is heterozygous for ∼55,000 single nucleotide polymorphisms (SNPs) (Lee et al. 2009). These polymorphisms allow the mapping of gene conversion events. Aside from these markers, the relevant genotype of EY7 is MATa/MATα::natMX4 leu2-3,112/LEU2his3-11,15/HIS3ade2-1/ade2-1 ura3-1/ura3-p trp1-1/TRP1can1-100/CAN1GAL2/gal2ho/ho::hisG IV957578::hphMX4/IV957578 IV1013217::URA3/IV1013217::ura3-e IV1510386::SUP4-o/IV1510386. The mutations in ura3-1, ura3-p, and ura3-e are the following: G to A at position 701 of URA3, G-to-A change at position 608, and T-to-A alteration at position 170, respectively.
Transformation, mating, media preparation, and tetrad dissection were performed according to standard methods (Guthrie and Fink 1991). Cells were grown at 30°. As explained in the Results, a complete analysis of the conversion events in EY7 required meiotic analysis. Although EY7 has a disruption of the MATα locus, which prevents sporulation under normal conditions, sporulation can be induced in sporulation medium with 5 mM nicotinamide (St. Charles and Petes 2013). The sporulation medium also contained 25 μg/ml uracil.
UV was used in one experiment (described elsewhere) to determine coupling relationships of markers on the two homologs. For these experiments, cells from overnight cultures were placed on solid medium (modified SD-Arg, Barbera and Petes 2006) at a density of ∼1000 cells/plate. The cells were then treated with UV (15 J/m2 using a TL-2000 Translinker) and allowed to form colonies. About 20 red/white sectored colonies were obtained per plate. The analysis of these colonies is described in the Results.
Identification of strains with gene conversion events at the URA3 gene on chromosome IV
A gene conversion event in which information is transferred between ura3-e and the wild-type URA3 gene on chromosome IV in EY7 results in derivatives that are resistant to 5-fluoroorotic acid (5-FOAR), hygromycin-resistant (HygR), and tryptophan prototrophs (Trp+) and that form pink colonies. To select 5-FOAR derivatives of EY7, we made patches of individual colonies of EY7 grown on rich medium (YPD) on solid medium containing 5-FOA (Boeke et al. 1987). We purified the resulting 5-FOAR colonies nonselectively and determined their phenotypes by replica-plating to media lacking uracil or tryptophan or containing hygromycin. Only one 5-FOAR colony per patch was examined to ensure that the observed events were independent. The color of the colony was determined by allowing the colonies to grow for 3 days at 30°, followed by a 1-day incubation at 4°. We also measured the rate of 5-FOAR derivatives by measuring the frequency of 5-FOAR in 39 independent cultures and then calculating the rate by using the method of the median (Lea and Coulson 1949).
Use of microarrays to measure conversion tract length
Mitotic gene conversion events in EY7 result in an interstitial region of loss of heterozygosity (LOH) flanked by heterozygous markers. To determine the extent of LOH, we used DNA microarrays capable of distinguishing whether the strain was homozygous for a SNP specific for the W303-1A-derived homolog, homozygous for a SNP specific for the YJM789-derived homolog, or heterozygous. The design of a microarray to detect LOH for SNPs between W303-1A and YJM789 was described in detail by St. Charles et al. (2012). In brief, four 25-base oligonucleotides centered on each SNP of interest were designed, two representing Watson and Crick sequences for one homolog and two representing Watson and Crick sequences for the other. Under the appropriate hybridization conditions, the detected hybridization of the genomic DNA to the perfectly matched oligonucleotides is stronger than the detected hybridization to oligonucleotides that have a mismatch, allowing detection of patterns of LOH. Additional details about the microarray analysis are provided in, File S1.
Of the 59 samples isolated from 5-FOAR HygR Trp+ pink colonies, most had LOH events involving at least five adjacent SNPs. For those samples with no LOH event detectable by microarrays, we sequenced the ura3 gene on the W303-1A-derived chromosome IV. For this analysis, we sporulated the diploid and isolated DNA from spore cultures. We identified which spores contained the W303-1A-derived homolog (as described in the Results). We amplified the ura3 gene on chromosome IV using the primer pairs URA3-EY F and URA3-EY R (Table S2) and sequenced the resulting product.
The microarrays used for mapping do not contain all of the SNPs that distinguish the two homologs. For conversion events that had a breakpoint in a region sparsely represented by oligonucleotides on the microarray, we refined the mapping using a different method (Lee et al. 2009). This method is described in File S1 and Table S3.
Results
The purposes of these experiments are the following: (1) to measure the frequency of mitotic gene conversion in a system that is not limited by the length of sequence homology; (2) to determine what fraction of mitotic gene conversion events are associated with crossovers; and (3) to analyze the length distribution of mitotic gene conversion tracts. The gene conversion events in the current study fall into two categories: those with a median length of ∼6 kb and those with a median length of >50 kb. Even the short class of conversions is longer than most of the tracts described by previous studies. As discussed in the Introduction, the short tracts observed in previous studies likely reflect the use of heteroalleles or systems involving ectopic recombination between repeats with limited homology.
Experimental rationale
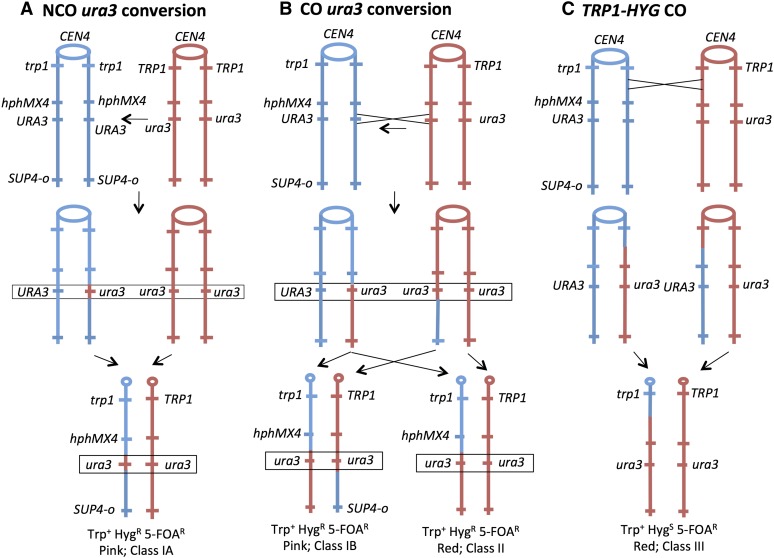
The strain used for this analysis is shown in Figure 2. The diploid EY7 was constructed by crossing two sequenced-diverged haploids (W303-1A and YJM789) resulting in a diploid that is heterozygous for ∼55,000 SNPs. Similar strains have been used previously to map spontaneous and UV-induced mitotic crossovers (Lee et al. 2009; St. Charles et al. 2012; St. Charles and Petes 2013; Yin and Petes 2013). In our experiments, we monitor conversion and crossover events on chromosome IV. In our description of the approximate location of diagnostic markers or the boundaries of recombination breakpoints, we use Saccharomyces Genome Database (SGD) coordinates. The first base on the left end of chromosome IV is assigned the coordinate 1 bp, and the last base on the right end is coordinate 1531933 bp. The diploid is heterozygous or hemizygous for the following markers (SGD coordinate of the midpoint of the gene or position of insertion rounded to a kilobase value): TRP1 (462 kb), hphMX4, which encodes an antibiotic that confers resistance to hygromycin (958 kb); URA3 (1013 kb); and SUP4-o (1510 kb); the centromere is located at SGD coordinate 450 kb. The diploid is also homozygous for the ade2-1 ochre mutation. Diploids with the ade2-1 mutation form white, pink, and red colonies in the presence of two, one, or no copies of the SUP4-o ochre suppressor gene, respectively (Barbera and Petes 2006).
Figure 2.
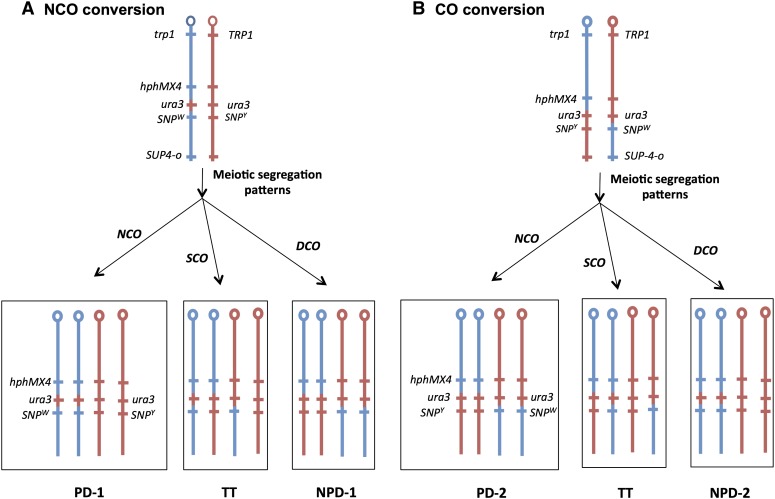
Genetic system for the detection of gene conversion and crossover events on chromosome IV. Conversion and crossover events are shown as occurring between replicated chromatids. Centromeres are depicted as ovals, and red and blue lines indicate the YJM789- and the W303-1A-derived homologs, respectively. The hphMX4 and the SUP4-o genes are located on only one of the two homologs. Recombination events are selected on plates containing 5-FOA that selects for loss of the wild-type URA3 gene. Diploid cells with zero, one, and two copies of SUP4-o form red, pink, and white colonies, respectively. (A) Gene conversion without an associated crossover (NCO, class IA). In this event, wild-type URA3 sequences are replaced by mutant sequences as indicated by the horizontal arrow. As shown by the pair of arrows, one type of segregation will result in a Trp+ HygR 5-FOAR pink colony with the flanking markers in the original coupling arrangement. (B) Conversion with an associated crossover (CO, class IB). Following the crossover, if the recombinant chromatids cosegregate (left), a Trp+ HygR 5-FOAR pink colony will be observed as in A. If one recombinant and one nonrecombinant chromatid cosegregate (right), a Trp+ HygR 5-FOAR red colony will be formed. (C) Crossover centromere-proximal to the hphMX4 marker. In this event, the 5-FOAR derivative is generated without a conversion. Following the crossover, if one of the recombinant chromosomes cosegregates with the nonrecombinant YJM789-derived homolog, a Trp+ HygS 5-FOAR red colony will be generated.
We selected for conversion events in which information is transferred from the mutant ura3-e allele to the wild-type allele using solid medium containing 5-FOA that selects against cells with wild-type URA3 activity (Boeke et al. 1987). To distinguish conversion events from other events that would result in a 5-FOAR derivative, we also examined phenotypes of the other markers on chromosome IV. As shown in Figure 2A, a gene conversion event unassociated with a crossover (NCO conversion) would be expected to result in a 5-FOAR colony that is Trp+ HygR and pink (class IA). A conversion associated with a crossover (CO conversion) could produce colonies of two different phenotypes (Figure 2B). If the recombinant products cosegregated (Figure 2B, left, class IB), then the 5-FOAR colony would have the same phenotype as the conversion unassociated with the crossover (Trp+ HygR pink). If the recombinant products do not cosegregate (Figure 2B, right, class II), then the 5-FOAR product would be Trp+ HygR and red. A 5-FOAR derivative could be generated by a crossover between the URA3 genes and centromere without a conversion involving URA3 (Figure 2C). For example, a crossover between the TRP1 marker (closely linked to the centromere) and the heterozygous hphMX4 insertion would generate a 5-FOAS Trp+ red colony (Figure 2C, class III). Since the length of a mitotic gene conversion tract is expected to be small relative to the distance between the TRP1 marker and URA3 marker (∼500 kb), we expect the event depicted in Figure 2C to be much more common than the events shown in Figure 2, A and B. In addition, a class III strain could result from a BIR event in which a break occurring between trp1 and hphMX4 on the blue chromosome is repaired using the red chromosome as a template. Crossovers and BIR events are indistinguishable without recovery of both daughter cells derived from the cell in which the recombination event occurred (Barbera and Petes 2006).
Other than the mitotic recombination events shown in Figure 2, several other genetic changes could result in a 5-FOAR strain. Chromosome loss would generate a 5-FOAR Trp+ HygS red colony. A point mutation or deletion of the wild-type URA3 gene would result in a 5-FOAR Trp+ HygR pink colony.
In summary, we expect three common phenotypic classes of 5-FOAR derivatives of EY7: class I (5-FOAR Trp+ HygR pink), class II (5-FOAR Trp+ HygR red), and class III (5-FOAR Trp+ HygS red). Of a total of 625 independent 5-FOAR derivatives, the numbers of derivatives observed for each class were the following: 59 in class I, 46 in class II, and 520 in class III. Our subsequent analysis concerns only class I strains. There are four expected subclasses of class I: conversion in which information is transferred from ura3-e to URA3 unassociated with a crossover (class IA); conversion between ura3-e and URA3 associated with a crossover (class IB); a point mutation inactivating the wild-type URA3 allele (class IC); and deletion of the wild-type URA3 allele (class ID).
Measurements of gene conversion tract lengths
For both class IA and class IB events, the minimum conversion tract length would be 1 base. We showed previously, however, that spontaneous and damage-induced mitotic gene conversion events have a median tract length of 5–10 kb (St. Charles and Petes 2013; Yin and Petes 2013). Thus, we expect that most of the conversion events detected as class I events will result in LOH for multiple SNPs located near the mutant ura3-e allele. In our previous studies, we used oligonucleotide-containing microarrays to identify regions of LOH (St. Charles et al. 2012; St. Charles and Petes 2013; Yin and Petes 2013). The details of this method are described in Materials and Methods and in File S1.
Not all SNPs on chromosome IV near the URA3 insertion are represented on the microarray. To refine the mapping of the breakpoints, for some events, we used a different method of determining LOH termed “SPA” (single-nucleotide-polymorphism PCR analysis) (St. Charles et al. 2010). In brief (details in File S1 and Table S3), we identified polymorphisms in the genomic region of interest in which one homolog had a restriction site that the other lacked. By PCR, we generated a small (<700 bp) fragment from genomic DNA of the strain of interest and treated the fragment with the diagnostic restriction enzyme. By examining the resulting products on an agarose gel, we could distinguish which allele was present. Gene conversion tracts were primarily examined by microarrays, but a few were also examined by SPA.
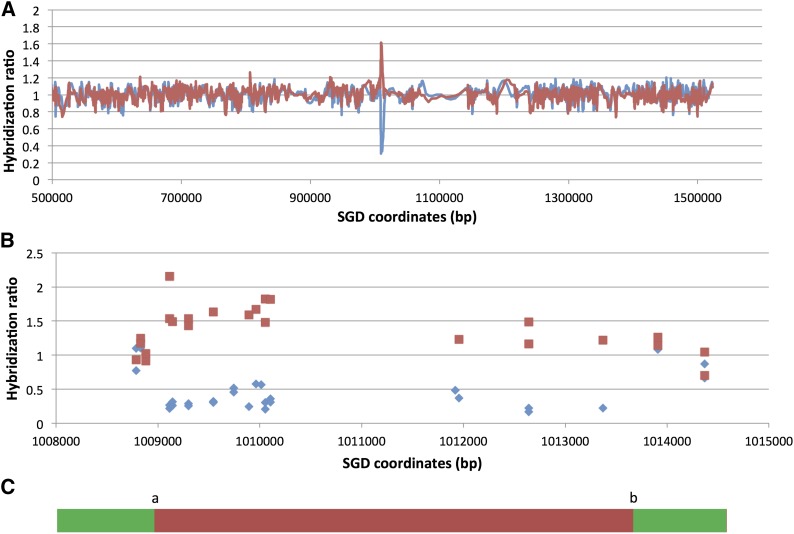
Figure 3 shows a microarray analysis of one class I strain (EY7-40). In this strain, the conversion event is flanked by heterozygous sites at SGD coordinates 1008881 and 1013909, and the homozygous sites closest to the transitions are located at SGD coordinates 1009116 and 1013370. The lengths of conversion tracts in such strains were estimated by averaging the distance between the heterozygous sites and the homozygous sites and adding the size of the inserted URA3 gene (∼1.1 kb). The tract length of the event shown in Figure 3 is 5.8 kb.
Figure 3.
Microarray analysis of the extent of a gene conversion event in EY7-40. The strain EY7-40 has the phenotype indicative of a gene conversion event (class I, 5-FOAR Trp+ HygR pink). DNA was isolated and hybridized to a SNP-specific microarray, and the ratio of hybridization (EY7-40 vs. DNA from a control heterozygous strain) to W303-1A-specific and YJM789-specific SNPs was measured. The red lines or boxes show hybridization to YJM789-specific SNPs, and the blue lines or diamonds indicate hybridization to W303-1A-specific SNPs. (A) Low-resolution analysis. The values on the x-axis are SGD coordinates in base pairs. The URA3 insertion is located between bases 1013217 and 1013218. The ratio was calculated in a moving window of 9 SNPs. (B) High-resolution analysis. In this depiction, each square and diamond shows the hybridization signal to a specific oligonucleotide on the microarray. (C) Schematic depiction of the conversion event showing the transitions between heterozygous and homozygous SNPs (green indicating heterozygous SNPs and red showing the region homozygous for the YJM789-derived SNPs). The “a” transition is located between coordinates 1008881 and 1009116, and the “b” transition is between 1013370 and 1013909 (Table S4).
Of the class I strains examined by microarray, most had undergone LOH for one or more SNPs flanking the URA3 gene on chromosome IV. The coordinates for the transitions between heterozygous and homozygous SNPs for all conversion events are in Table S4. Most of the conversion events had only two transitions between heterozygous and homozygous regions. In Table S4, the centromere-proximal and centromere-distal transitions are called “a” and “b,” respectively. There were two conversion events with additional transitions (EY7-63 and EY7-69). These events are described in File S1.
In five of the class I strains, no flanking SNPs had undergone LOH. There are three mechanisms that could produce such strains: (1) short allelic conversion events that failed to include flanking markers, (2) new mutations within the URA3 gene on the W303-1A-derived homolog, and (3) ectopic gene conversion between the URA3 gene on chromosome IV and one of the mutant ura3 genes (ura3-1 or ura3-p) located on chromosome V. To distinguish among these possibilities, we sporulated these five diploid strains and identified spores that had the mutant ura3 gene on the W303-1A-derived homolog. We PCR-amplified the ura3 gene on chromosome IV from these haploid strains and sequenced the resulting product. In one of these strains (EY7-8), the mutant allele generated by conversion had the same ura3-e allele that was originally heterozygous in EY7; this strain, therefore, had a short gene conversion tract (<1 kb). The other four strains (EY7-19, -24, -32, -68) that did not have coconversion of flanking markers had one or more new mutations in URA3. The mutational positions in URA3 (1 being the first base of the initiating ATG) and types of alterations in these strains were the following: 175, A to G (EY7-19); 62, T to C (EY7-24); 345, G to A (EY7-32); 84, G to A; 90, A to T; 91, A to T (EY7-68).
Finally, the one strain (EY7-3) had a deletion of the wild-type URA3 allele on the W303-1A-derived homolog, resulting in hemizygosity for the ura3-e allele. The heterozygous/homozygous transitions for the deletion were at SGD coordinates 980838 and 993113 (centromere-proximal) and at coordinates 1079615 and 1089446 (centromere-distal); the resulting deletion is ∼100 kb.
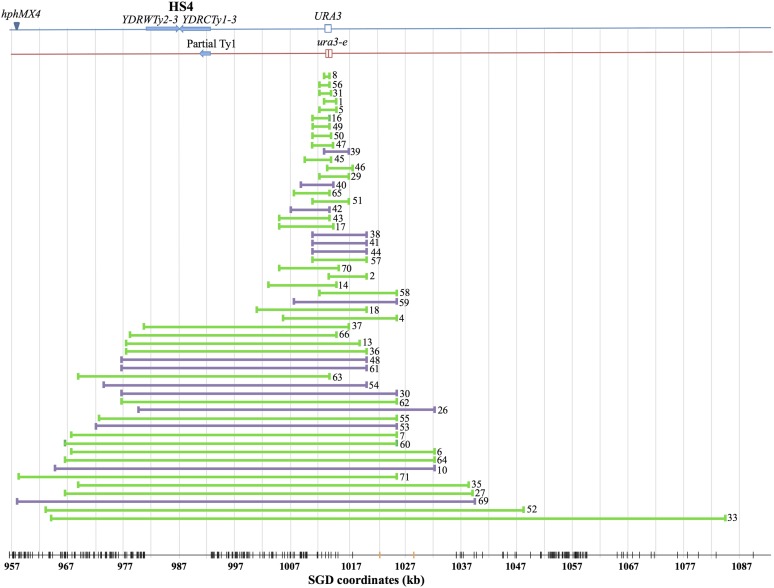
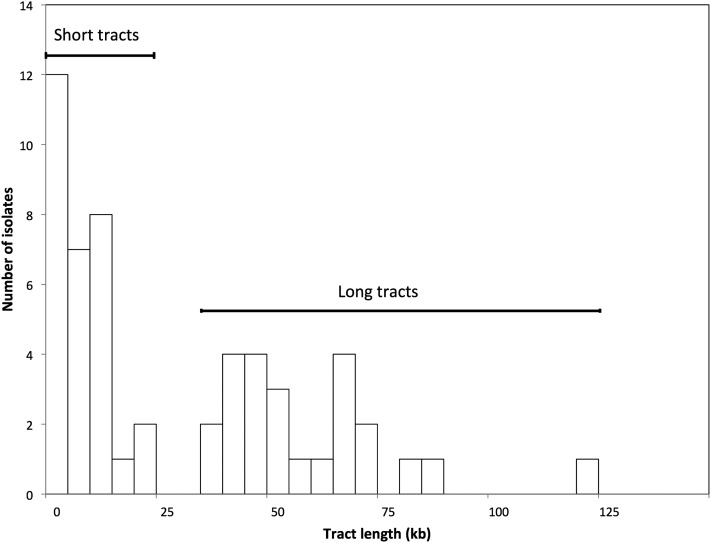
In summary, of the 59 class I strains, 54 were gene conversions (classes IA and IB), 4 were new ura3 point mutations (class IC), and 1 was a large deletion (class ID). The locations of the 54 conversion tracts are in Figure 4, and the tract sizes are shown in a histogram in Figure 5. It is apparent that about half of the tracts were very long (>25 kb) and the remainder had a median length of <10 kb. Considering all of the data, the median tract length was 16.6 kb. The tracts shorter or longer than 25 kb had median lengths of ∼6.4 kb (4.4–10 kb, 95% confidence limits) and 54 kb (45–69 kb), respectively. Table S4 lists all conversion tract lengths.
Figure 4.
Map locations of gene conversion events. Our mapping of 54 conversion events that include the ura3-e mutation is summarized. The blue and red lines indicate the homologs derived from W303-1A and YJM789, respectively. The lengths of independent conversion events are shown by horizontal lines labeled with the number of the EY7 isolate. Green and purple lines indicate NCO conversions and CO conversions, respectively. The vertical black lines at the bottom of the figure show the distribution of SNPs on the microarray, and the vertical orange lines show the location of SNPs examined by SPA.
Figure 5.
Histogram of gene conversion tract lengths. Based on the analysis of Table S4, the 54 conversion events appear to have two distinct size distributions. All of the events with tracts lengths >25 kb include HS4.
Rate of gene conversion
By measuring the frequency of 5-FOAR derivatives in multiple independent cultures of EY7 and applying the method of the median (Lea and Coulson 1949), we calculated a rate of 5-FOAR of 1.5 × 10−5/division (95% confidence limits of 0.9–2.0 × 10−5). From the analysis described above, of 625 independent 5-FOAR derivatives of EY7, 54 resulted from gene conversion. The rate of allelic conversion in which information is transferred from the ura3-e allele to the wild-type URA3 allele on the other homolog is ∼1.3 × 10−6/division [54/625 × (1.5 × 10−5)].
Association of crossovers with conversions
As shown in Figure 2, A and B, conversion events at the URA3 locus could be unassociated with crossovers (NCO) or crossover-associated (CO). By microarray analysis, NCO and CO conversions are indistinguishable. The distinction can be made by examining the coupling of heterozygous markers flanking the conversion tract. If the conversion event is NCO, then heterozygous markers flanking the tract have the same coupling arrangement as in the original EY7. In contrast, if the conversion event is associated with a crossover, the coupling of markers flanking the tract is reversed. We determined the coupling arrangements by meiotic analysis of all strains with a conversion event (Figure 6). For this analysis, we used the hphMX4 insertion as the heterozygous marker centromere-proximal to the tract. The centromere-distal markers were SNPs that were close to the conversion tract and could be diagnosed by SPA analysis as described above. The SNP that was used for the meiotic analysis for each conversion strain is given in Table S5. In Figure 6, we show the heterozygous SNP used in the analysis as SNPW for the W303-1A-derived SNP and SNPY for the allelic YJM789-derived SNP.
Figure 6.
Meiotic analysis of the coupling of markers flanking a mitotic gene conversion event. To distinguish whether the conversion was unassociated (NCO) or associated (CO) with a crossover, we examined class I strains by tetrad analysis. We analyzed the patterns of segregation for the centromere-proximal hphMX4 marker and a SNP located centromere-distal to the conversion tract. The red and blue lines signify chromosome regions derived from the YJM789-derived and W303-1A-derived homologs, respectively. (A) Meiotic segregation patterns expected for class I strains with an NCO conversion. If there is no meiotic crossover (NCO), we expect a PD tetrad: two HygR SNPW spores and two HygS SNPY spores. A single crossover (SCO) between the hphMX4 marker and the diagnostic SNP would produce a tetratype tetrad (TT): one HygR SNPW spore, one HygR SNPY spore, one HygS SNPW spore, and one HygS SNPY spore. Finally, a four-stranded double-crossover (DCO) between the hphMX4 marker and the diagnostic SNP would produce an NPD: two HygR SNPY spores and two HygS SNPW spores. (B) Meiotic segregation patterns expected for class I strains with a CO conversion. Similar segregation patterns to those observed in A are expected, but the coupling of the markers in the PD and NPD tetrads would be reversed from those observed in A.
If two markers in yeast are closely linked, most of the tetrads will be of two classes: parental ditype (PD; all four spores with markers in the original parental coupling relationship,) or tetratype (TT; two spores with markers in the parental configuration and two markers with the recombinant arrangement). Nonparental ditype tetrads (NPD; all four spores with markers in recombinant arrangements) for linked markers are rare since such tetrads require four-strand double crossovers (Petes et al. 1991). For most of the conversion events, the distance between the hphMX4 marker and the centromere-distal end of the conversion tract is ≤80 kb. On chromosome IV, the average association between the genetic and the physical distance is ∼0.3 cM/kb (Saccharomyces Genome Database). Therefore, the genetic distance between the hphMX4 marker and the centromere-distal heterozygous SNP is <25 cM. For a 25-cM distance, PD and TT tetrads are expected to be about equally frequent and there would be very few NPD tetrads. As shown in Figure 6, the genotypes of spores that constitute PD and NPD segregation patterns are reversed for NCO and CO conversion events. We will refer to the PD and NPD patterns of the strains with an NCO conversion event as PD-1 and NPD-1, and the patterns of the strains with CO conversions as PD-2 and NPD-2.
For each strain with a conversion event, we analyzed all spores derived from two to eight tetrads with four viable spores for the segregation of the hphMX4 marker and the centromere-distal SNP. If at least two of the tetrads had a PD-1 segregation pattern and no tetrads had the PD-2 segregation pattern, we concluded that the conversion was an NCO conversion. Similarly, if at least two of the tetrads had a PD-2 segregation pattern and no tetrads had the PD-1 segregation pattern, we concluded that the event was a CO conversion. By this criterion, we showed that 39 of 54 conversion events were NCO, and 15 were CO (summarized in Table S4 and Table S5).
In strains with very long conversion tracts, the procedure for determining the coupling of markers by tetrad dissection was tedious since most of the tetrads had tetratype segregation. We developed a second method of examining coupling relationships by mitotic recombination using the 5-FOAR derivative EY7-33 that had a very long (121 kb) conversion tract. Both class IA (NCO) and class IB (CO) strains are heterozygous for the hphMX4 and SUP4-o markers (Figure S1). We treated EY7-33 with ultraviolet light (details in Materials and Methods) to stimulate an additional mitotic crossover event on chromosome IV. As shown in Figure S1A, a crossover between the hphMX4 marker and CEN4 in strains with the coupling relationship shown in class IA strains could generate a red/white sectored colony in which the cells in the white sector would be HygR and the cells in the red sector would be HygS. In contrast, if the strain has the markers in the reverse coupling relationship (Figure S1B), the cells in the white and red sectors would be HygS and HygR, respectively. Of 76 red/white sectored colonies examined, 29 showed cosectoring of the hygromycin-resistance phenotype. In all 29 of these colonies, the white sector was HygR and the red sector was HygS, as expected if the EY7-33 conversion event was unassociated with a crossover.
Discussion
In this study, we examined spontaneous mitotic gene conversion in a diploid strain in which conversion events could be measured without selecting against long conversion tracts. Our analysis showed two distinct size classes of events likely generated by two different mechanisms. About 40% of the conversions were associated with crossovers, demonstrating that there is not a strong bias against the generation of crossovers in mitosis.
Frequency of gene conversion
The rate of allelic gene conversions in this study was ∼1.3 × 10−6/division, similar to rates of allelic conversions in other studies that vary between 10−7 and 10−6 (Petes et al. 1991; Paques and Haber 1999). Although the rate estimates obtained in heteroallelic experiments are usually lower than observed in our study (presumably because of the more restrictive system for detecting events), rates of conversion as high or higher than ours were observed in some heteroallelic studies (Golin and Esposito 1984). These high rates may be the result of a marker being located near a mitotic recombination hotspot. It should also be pointed out that, although the rate of mitotic recombination events is much less than the rate of meiotic recombination events, these events are still likely to be important in generating novel combinations of alleles. It has been estimated that there are ∼30,000 clonal (mitotic) generations to one sexual (meiotic) cycle for yeast (Magwene et al. 2011).
Lengths of gene conversion tracts
In Figure 5, there appear to be two discrete size classes of conversion tracts: those with a median size of ∼6.4 kb (range 1–21 kb) and those with a median size of 54 kb (range 36–121 kb). The smaller size class is similar in length to the conversion events that we have examined in other studies (Lee et al. 2009; St. Charles et al. 2012; St. Charles and Petes 2013; Yin and Petes 2013). Our classification of the conversion tracts into two classes is based on three considerations: (1) using R software and the Kolmogorov–Smirnov test, the distribution of tract lengths is significantly different from a uniform distribution (P < 0.001) and a normal distribution (P = 0.01); (2) there are no tracts between 21 and 36 kb in length, although there are 30 tracts <21 kb and 24 tracts >36 kb; and (3) all of the tracts >36 kb overlap the inverted Ty elements located centromere-proximal to ura3 (Figure 4). The asymmetric distribution of the long conversion tracts relative to the ura3 gene argues strongly that these long conversion tracts are inherently different from the short conversion tracts.
In our previous mapping of unselected crossovers on the right arm of chromosome IV (St. Charles and Petes 2013), the region containing the inverted Ty elements was associated with a high frequency of crossovers and was termed HS4 (HotSpot4). We previously noted that the crossover-associated conversion events involving HS4 had long tracts with a median size of 48 kb. Our current analysis confirms that this estimate that was based on a smaller number of events and, in addition, demonstrates that NCO conversions involving HS4 are also very long (median length of 59 kb). In our previous study, we showed that deletion of one of the Ty elements composing HS4 or expanding the distance between the two Ty elements resulted in loss of hotspot activity (St. Charles and Petes 2013). We argued, therefore, that the hotspot was a consequence of cleavage of a hairpin structure formed when a single-stranded gap was generated near the center of HS4. Because both the CO and NCO long conversion tracts in the current study overlap with HS4, we assume that they are also initiated as a consequence of processing the hairpin structure associated with the inverted Ty elements.
Several other points are relevant concerning the HS4-associated gene conversion events. First, we previously showed that CO conversions involving HS4 were a consequence of the repair of two sister chromatids broken at approximately the same position, likely reflecting a DSB formed in G1 of the cell cycle (St. Charles and Petes 2013). In the current study, in which we recover only one of the two daughter cells with recombinant products, we cannot determine whether the HS4-associated events are associated with a G1- or G2-initiated DNA lesion. Second, HS4 is homolog-specific (St. Charles and Petes 2013). The W303-1A-derived chromosome has the inverted pair of Ty elements, whereas the YJM789-derived chromosome has only a portion of one Ty element (Figure 4). Thus, for the broken ends of the W303-1A homolog to invade the YJM789 homolog, the ends would have to be resected by ∼10 kb to expose flanking homology. This extent of resection is much less than the median length of the long conversion tracts (54 kb). Although in the current study any conversion tracts initiated at HS4 must extend ∼28 kb to include the URA3 marker, most of the tracts are considerably >28 kb. Third, the very long tracts associated with HS4 are not typical, but are also not unique. Long conversion events are also associated with a second pair of inverted Ty elements on chromosome IV (HS3, St. Charles and Petes 2013), and a mitotic recombination hotspot associated with the trinucleotide repeats (GAA) results in conversion tracts with a median length of 20 kb (Tang et al. 2011). In addition, conversion tracts >30 kb have been observed in multiple other studies (Lee et al. 2009; St. Charles and Petes 2013), although these events are usually <10% of the conversion events. One interpretation of the data is that most mitotic recombination events are initiated by random DNA lesions that are processed to produce conversion tracts with a median length of ∼6 kb. The other class of recombination events is initiated at sequences capable of forming secondary DNA structures and is processed to produce very long conversion tracts.
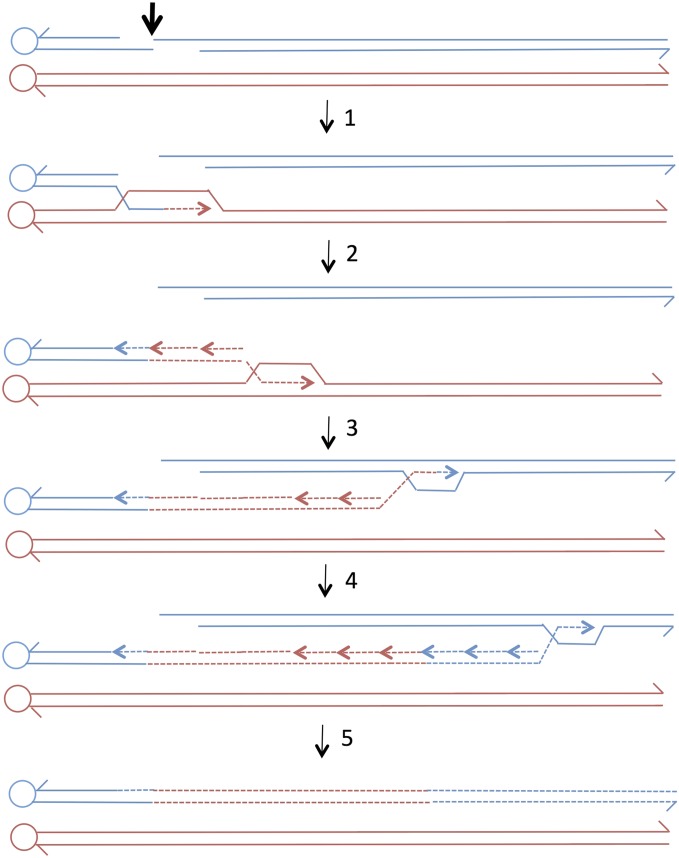
An important question is whether the mechanism of generating the short tracts is the same as that which generates the long tracts. Previous studies have demonstrated that small mitotic tracts (≤1 kb) are a consequence of heteroduplex formation, followed by mismatch repair as predicted by the models in Figure 1 (Nickoloff et al. 1999; Mitchel et al. 2010). Although it is possible that the very long conversion tracts reflect the formation of very long heteroduplexes and concerted repair of the resulting mismatches, there are two other plausible alternatives. First, these tracts could be a consequence of the repair of a very long double-stranded DNA gap (Lee et al. 2009). Second, these events could represent a BIR event that copies a region of one homolog before switching to the acentric chromosome fragment (Figure 7, NCO conversion). A long CO conversion could be generated by processing of the BIR structure (shown as the intermediate between steps 1 and 2 in Figure 7) to yield a recombinant chromosome and a broken centromere-associated red chromosome. Invasion of this broken red chromosome into an unbroken blue chromosome, followed by BIR, would produce a long gene conversion tract associated with a crossover. Production of a crossover by two BIR events has been proposed previously (Figure S4 in Lee et al. 2009). Template switching during BIR is common (Smith et al. 2007), and the generation of long conversion tracts by BIR has been suggested previously (Voelkel-Meiman and Roeder 1990; Llorente et al. 2008; Lee et al. 2009; Chandramouly et al. 2013; Yin and Petes 2013). It is important to distinguish BIR events that proceed to the end of the chromosome (Voelkel-Meiman and Roeder 1990) from those in which a long interstitial conversion event is generated; the latter class requires a template switch.
Figure 7.
Generation of a long gene conversion tract by a double BIR event. We show two double-stranded recombining DNA molecules with the centromere shown as a circle. Dotted lines show DNA synthesis. The conversion tract is formed by two BIR events in the following steps: step 1—a DSB occurs on the blue chromosome (thick arrow) with 5′ to 3′ resection of the broken ends; step 2—the left end invades the red chromosome and initiates DNA synthesis (dotted red line); step 3—synthesis continues in a moving D-loop mode with second strand synthesis occurring on the displaced strand; step 4—the chromosome involved in the BIR event disengages from the red chromosome and invades the centromere-distal fragment of the blue chromosome to begin a second BIR event; and step 5—the BIR event continues to the end of the chromosome, and the acentric blue chromosome fragment is lost.
Analysis of the genetic requirements for the HS4-associated gene conversion tracts could help distinguish among the mechanisms discussed above. Mutations in POL32 and PIF1 reduce the frequency of BIR (Llorente et al. 2008; Chung et al. 2010) and would be expected to reduce the frequency of long conversion tracts if BIR is involved. However, since Pol32p is a general processivity factor for DNA polymerase and since gene conversion by the SDSA pathway requires extensive DNA synthesis primed from the invading chromosome end, it is not clear that the pol32 mutation would affect BIR exclusively. Ho et al. (2010) showed that pol32 strains had shorter conversion tracts in a standard mitotic recombination assay.
Relationship between conversion events and crossovers
In the past, it was assumed that CO and NCO conversions reflect alternative patterns of cleavage of the double Holliday junction (Szostak et al. 1983). It is now clear that most NCO conversions in meiosis (Allers and Lichten 2001) and mitosis (Mitchel et al. 2010) are a consequence of SDSA and/or dHJ dissolution pathways (Figure 1) rather than cleavage of a dHJ. In our study, 15 of 54 conversions (28%) were crossover-associated and 39 of 54 (72%) were NCO. In our system, we screen for those conversion events in which the recombinant chromosomes cosegregate. Since events in which one recombinant and one nonrecombinant chromosome cosegregate are equally frequent (Chua and Jinks-Robertson 1991), we conclude that 43% of the conversions were CO [(15 × 2)/(54 + 15)] and 57% were NCO. In earlier studies (summarized in the Introduction), NCO conversions often substantially exceeded CO conversions. These results may have been influenced by the heteroallelic systems used to select the conversion or by limited amounts of homology available for ectopic recombination. Our findings argue strongly that the mitotic repair of recombinogenic lesions in yeast is often associated with crossovers. In other genetic systems such as Drosophila, however, NCO conversions are much more frequent than CO conversions (Andersen and Sekelsky 2010).
In our previous analysis of UV-induced conversion events, we showed that CO tracts were significantly longer than NCO tracts (Yin and Petes 2013). In our current study, however, the numbers of CO and NCO events for the short tracts (7 and 23, respectively) and long conversions (8 and 16, respectively) were not significantly different (Fisher exact test, P = 0.54). Within the short-tract category, the nine shortest tracts (those <4 kb) were all NCO events. The comparison with the short tracts >4 kb (4–22 kb), however, was not statistically significant (P = 0.07). The difference between conversion tracts generated by UV and our current results may reflect the difference in the recombinogenic lesions. Alternatively, the ratio of NCO to CO events could be affected by chromosome context. Mancera et al. (2008) reported such differences for meiotic events in yeast.
In conclusion, our analysis of spontaneous mitotic gene conversion events demonstrates the existence of two types of conversions distinguishable by their size. For both types of conversions, CO and NCO events are recovered with approximately equal frequencies.
Supplementary Material
Acknowledgments
We thank members of the Petes and Jinks-Robertson labs for useful discussions; Y. Yin, S. Jinks-Robertson, A. Guo, and E. Hum for comments on the manuscript; and Y. Yin for help with the statistical analysis. The research was supported by National Institutes of Health grants GM24110 and GM52319 to T.D.P., and K.E.O. was supported by a Graduate Research Fellowship Program fellowship from the National Science Foundation (1106401).
Footnotes
Available freely online through the author-supported open access option.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167395/-/DC1.
Communicating editor: N. M. Hollingsworth
Literature Cited
- Aguilera A., Klein H. L., 1989. Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics 123: 683–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 225–231 [DOI] [PubMed] [Google Scholar]
- Andersen S. L., Sekelsky J., 2010. Meiotic vs. mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic vs. mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays 32: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera M. A., Petes T. D., 2006. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 12819–12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R., 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Chandramouly G., Kwok A., Huang B., Willis N. A., Xie A., et al. , 2013. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat. Commun. 4: 2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P., Jinks-Robertson S., 1991. Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics 129: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. H., Zhu Z., Papusha A., Malkova A., Ira G., 2010. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 6: e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M. S., Wagstaff J. E., 1981. Mechanisms of mitotic recombination, pp. 341–370 in The Molecular Biology of Yeast Saccharomyces: Life Cycle and Inheritance, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Golin J. E., Esposito M. S., 1984. Coincident gene conversion during mitosis in Saccharomyces. Genetics 107: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego [Google Scholar]
- Haber J. E., Hearn M., 1985. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics 111: 7–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K., Mazon G., Lam A. F., Symington L. S., 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell 40: 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar O., Kupiec M., 1999. Homology search and choice of homologous partner during mitotic recombination. Mol. Cell. Biol. 19: 4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Petes T. D., 1986. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114: 731–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd S. R., Petes T. D., 1986. Physical lengths of meiotic and mitotic gene conversion tracts in Saccharomyces cerevisiae. Genetics 118: 401–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H., 1992. Sister chromatids are preferred over homologs as substrates for recombination repair in Saccharomyces cerevisiae. Genetics 132: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1949. The distribution of number of mutants in a bacterial population. J. Genet. 49: 264–285 [DOI] [PubMed] [Google Scholar]
- Lee P. S., Greenwell P. W., Dominska M., Gawel M., Hamilton M., et al. , 2009. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5: e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettier G., Feng Q., de Mayolo A. A., Erdeniz N., Reid R. J., et al. , 2006. The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet. 2: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Haber J. E., 1989. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123: 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B., Smith C. E., Symington L. S., 2008. Break-induced replication: What is it and what is it for? Cell Cycle 7: 859–864 [DOI] [PubMed] [Google Scholar]
- Magwene P. M., Kayikci O., Granek J. A., Reininga J. M., Scholl Z., et al. , 2011. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 1987–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Ivanov E. L., Haber J. E., 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93: 7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K., Zhang H., Welz-Voegele C., Jinks-Robertson S., 2010. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol. Cell 38: 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Sweetser D. B., Clikeman J. A., Khalsa G. J., Wheeler S. L., 1999. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics 153: 665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Malone R. E., Symington L. S., 1991. Recombination in yeast, pp. 407–521 in The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by Broach J. R., Pringle J. R., Jones E. W. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Smith C. E., Llorente B., Symington L. S., 2007. Template switching during break-induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- St. Charles J., Petes T. D., 2013. High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 mb arm of yeast chromosome IV. PLoS Genet. 9: e1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Charles J., Hazkani-Covo E., Yin Y., Andersen S. L., Dietrich F. S., et al. , 2012. High-resolution genome-wide analysis of irradiated (UV and gamma-rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 190: 1267–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele D. F., Morris M. E., Jinks-Robertson S., 1991. Allelic and ectopic interactions in recombination-defective yeast strains. Genetics 127: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W., 1983. The double-strand-break repair model for recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- Tang W., Dominska M., Greenwell P. W., Harvanek Z., Lobachev K. S., et al. , 2011. Friedreich’s ataxia (GAA)n•(TTC)n repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 7: e1001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Roeder G. S., 1990. Gene conversion tracts stimulated by HOT1-promoted transcription are long and continuous. Genetics 126: 851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz-Voegele C., Jinks-Robertson S., 2008. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics 179: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Petes T. D., 2013. Genome-wide high-resolution mapping of UV-induced mitotic recombination events in Saccharomyces cerevisiae. PLoS Genet. 9: e1003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.