Abstract
Meiotic recombination is initiated by the programmed induction of double-strand DNA breaks (DSBs), lesions that pose a potential threat to the genome. A subset of the DSBs induced during meiotic prophase become designated to be repaired by a pathway that specifically yields interhomolog crossovers (COs), which mature into chiasmata that temporarily connect the homologs to ensure their proper segregation at meiosis I. The remaining DSBs must be repaired by other mechanisms to restore genomic integrity prior to the meiotic divisions. Here we show that HIM-6, the Caenorhabditis elegans ortholog of the RecQ family DNA helicase BLM, functions in both of these processes. We show that him-6 mutants are competent to load the MutSγ complex at multiple potential CO sites, to generate intermediates that fulfill the requirements of monitoring mechanisms that enable meiotic progression, and to accomplish and robustly regulate CO designation. However, recombination events at a subset of CO-designated sites fail to mature into COs and chiasmata, indicating a pro-CO role for HIM-6/BLM that manifests itself late in the CO pathway. Moreover, we find that in addition to promoting COs, HIM-6 plays a role in eliminating and/or preventing the formation of persistent MutSγ-independent associations between homologous chromosomes. We propose that HIM-6/BLM enforces biased outcomes of recombination events to ensure that both (a) CO-designated recombination intermediates are reliably resolved as COs and (b) other recombination intermediates reliably mature into noncrossovers in a timely manner.
Keywords: meiosis, recombination, BLM helicase, MutSγ, Caenorhabditis elegans
IN most eukaryotes, accurate segregation of homologous chromosomes during meiosis depends on crossover (CO) recombination events, as COs form the basis of connections known as chiasmata that help homologs orient toward opposite spindle poles at the meiosis I division (Page and Hawley 2003). Multiple mechanisms collaborate to guarantee that COs will form between every homolog pair. One level of regulation governs the initiation of recombination through the programmed formation of double-strand DNA breaks (DSBs), which form in substantial excess of eventual COs. Recent evidence suggests that checkpoint-like feedback mechanisms operate to ensure both that DSB formation continues until each homolog pair has at least one CO-eligible recombination intermediate and that DSB formation will shut down once this condition is met (Rosu et al. 2013; Stamper et al. 2013). Following DSB formation, a subset of the initial recombination intermediates is selected to become COs, recruiting a cohort of CO- promoting (pro-CO) proteins that function to stabilize and protect these CO-designated intermediates (Baudat and De Massy 2007; Kohl and Sekelsky 2013; Lynn et al. 2007). A widely conserved solution for protecting potential CO intermediates involves the MutSγ complex, comprising MSH4 and MSH5, meiosis-specific members of the MutS protein family that can form a sliding clamp on DNA in response to recognition of branched DNA structures (Baudat and De Massy 2007; Lynn et al. 2007; Snowden et al. 2004). In many organisms, MutSγ is initially recruited to multiple sites in excess of eventual COs, but it becomes stabilized at only a subset of these sites through recruitment of other pro-CO factors (Kneitz et al. 2000; Yokoo et al. 2012; Reynolds et al. 2013; Holloway et al. 2014; Qiao et al. 2014). The CO designation process is tightly regulated, yielding a highly nonrandom distribution in which a relatively small number of CO-based connections are formed between homologs, yet chromosome pairs lacking such connections are extremely rare. CO designation per se is not sufficient to ensure CO formation for every chromosome pair, however. Since the number of CO-designated sites in many organisms is on the order of one per chromosome (or one per chromosome arm), eventual resolution of the CO-designated intermediates must occur in a highly biased fashion, such that maturation of each CO-designated intermediate reliably yields a CO. Finally, excess recombination intermediates not destined for the CO fate must be faithfully repaired in a timely fashion to restore integrity of chromosomes prior to the meiotic divisions.
In the current work, we investigate the roles of HIM-6, the Caenorhabditis elegans ortholog of the BLM DNA helicase (Wicky et al. 2004), in promoting successful meiosis. BLM is best known for its “anti-CO” activities and roles in antagonizing recombination in mitotically dividing cells. Mutations in the human Blm gene cause a familial cancer predisposition syndrome known as Bloom syndrome, and a diagnostic feature of Blm mutant patient cells is a highly elevated frequency of COs between sister chromatids (Chaganti et al. 1974; Ellis et al. 1995). Supporting the view of BLM as an anti-CO agent, BLM was identified as part of a protein complex that has an in vitro “dissolution” activity that can dismantle model recombination substrates containing double Holliday junctions in a manner that exclusively yields noncrossover products (Wu and Hickson 2003). Further, anti-CO roles during meiotic recombination have been demonstrated or proposed for BLM orthologs or its protein complex partners in a variety of species, including Saccaromyces cerevisiae, Arabidopsis, mice, and Drosophila (e.g., Rockmill et al. 2003; Jessop et al. 2006; Oh et al. 2007; Chelysheva et al. 2008; Holloway et al. 2010; Kohl et al. 2012). However, this reputation of BLM as an antagonist of crossing over was not readily reconciled with the finding that loss of function of him-6 results in a reduction of COs and chiasmata, implying a pro-CO rather than anti-CO role for BLM in C. elegans meiosis (Zetka and Rose 1995; Wicky et al. 2004).
In the interim since HIM-6 was first identified as C. elegans BLM, substantial progress has been made in the C. elegans system both in identifying meiotic recombination machinery components acting at early and late steps and in development of in situ markers for visualizing ongoing recombination events and other features of meiotic prophase progression. Here, we exploit these advances to revisit the roles of HIM-6/BLM in meiotic recombination. We show that HIM-6/BLM has a role in promoting the formation of MutSγ-dependent COs between homologous chromosomes that manifest itself late in meiotic prophase, and we show that HIM-6/BLM functions to ensure that CO-designated recombination intermediates reliably mature into interhomolog COs. Moreover, we show that in addition to its role in promoting meiotic CO formation, HIM-6/BLM plays a role in eliminating and/or preventing the formation of persistent MutSγ-independent recombination-based interactions between the homologs. Our data suggest that HIM-6/BLM may function in multiple distinct contexts during meiosis, contributing both to ensuring the formation of COs and to restoring integrity of the chromosomes to enable their faithful segregation. This work complements and extends the findings of recent parallel studies investigating the requirements for resolution of meiotic CO intermediates in C. elegans (Agostinho et al. 2013; O’Neil et al. 2013; Saito et al. 2013) and contributes to a growing recognition that BLM can be deployed in a variety of different contexts to affect the structure of multiple classes of recombination intermediates and/or the timing and outcome of their resolution. The prominence of the pro-CO role of BLM during C. elegans meiosis suggests that the C. elegans system may be especially well suited for future studies addressing how the highly CO-biased outcome of resolution at CO-designated sites is accomplished.
Materials and Methods
C. elegans strains
Strains were maintained at 20° under standard conditions. Experiments were performed at 20° unless otherwise noted. Strains used in this study are:
N2
VC193 him-6(ok412) IV (Wicky et al. 2004)
AV630 meIs8[unc-119(+) pie-1promoter::gfp::cosa-1] II (Yokoo et al. 2012)
AV639 him-6(ok412) IV; meIs8[unc-119(+) pie-1promoter::gfp::cosa-1] II; unc-119(ed3) III
AV713 meIs8 II; him-18(tm2181) / qC1dpy-19(e1259) glp-1(q339) nIs189 III
PD4251 ccIs4251[myo-3 promoter::gfp::lacZ-NLS, myo-3 promoter::mito-gfp, dpy-20(+)] I; dpy-20(e1282) IV (Fire et al. 1998)
AV596 cosa-1(tm3298)/ qC1dpy-19(e1259) glp-1(q339) qIs26 III(Yokoo et al. 2012)
AV780 cosa-1(tm3298)/ qC1dpy-19(e1259) glp-1(q339) qIs26 III; him-6(ok412) IV
KK0313 him-14(it23) unc-4(e120)/ mnC1 II(Kemphues et al. 1988)
TY5434 syIs44 [ dpy-20(+), hsp-16 promoter::lacI::gfp, lacO(256)] V [(syIs44 is an array containing multiple copies of the indicated DNAs, integrated into chromosome V (Gonzalez-Serricchio and Sternberg 2006)]
AV806 cosa-1(tm3298)/ qC1dpy-19(e1259) glp-1(q339) qIs26 III; him-6(ok412) IV; syIs44 V
Cytological analyses
Immunofluorescence:
For images in Figure 1, Figure 2, and Figure 4, immunofluorescence analyses and imaging using the DeltaVision deconvolution microscopy system (Applied Precision) were conducted as in Martinez-Perez and Villeneuve (2005), with minor modifications as described in Libuda et al. (2013) (Figure 1, Figure 2C, and Supporting Information, Figure S1), Yokoo et al. (2012) (Figures 2A and 4B), or Martinez-Perez et al. (2008) (Figure 4A). For these analyses, gonads were dissected and fixed 20–24 hr post-L4 stage. For images in Figure 3, immunofluorescence and imaging were conducted as in Nabeshima et al. (2004), with minor modifications as described in Rosu et al. (2013), using worms dissected and fixed at 48 hr post-L4. Extent of the SUN-1 S8Pi-positive zone was quantified as in Mlynarczyk-Evans et al. (2013).
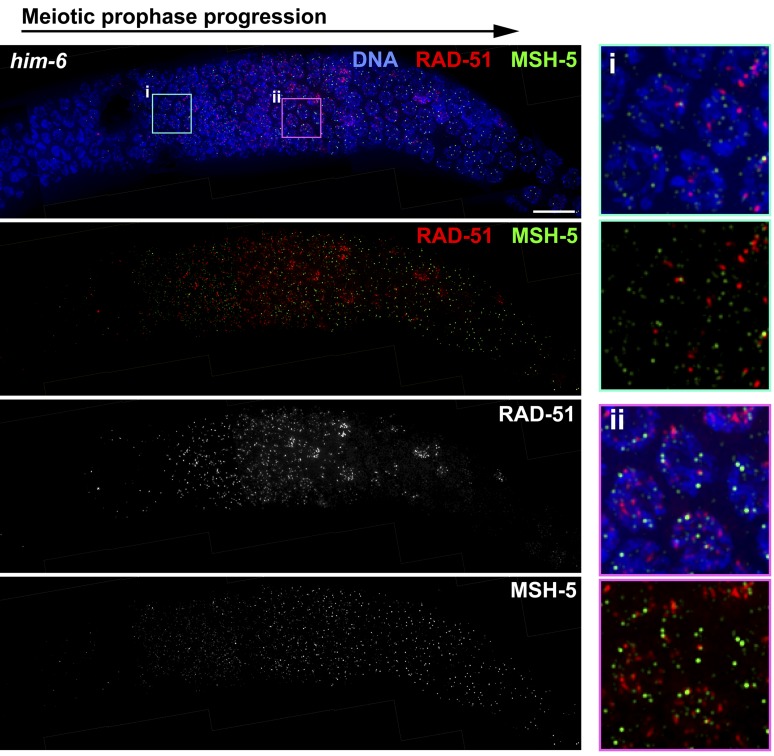
Figure 1.
The him-6 mutant is proficient for timely loading of RAD-51 and MSH-5 at nascent recombination sites. Immunofluorescence images showing RAD-51 and MSH-5 foci in a him-6 mutant gonad, revealing dynamics of RAD-51 and MSH-5 foci that are similar to wild type (shown in Figure S1). Images show a region of the gonad extending from meiotic prophase entry (left) until the end of the pachytene stage (right). RAD-51 foci are abundant during early pachytene, decline during mid-pachytene, and are nearly absent by late pachytene; faint MSH-5 foci appear during early pachytene, become brighter and more abundant during mid-pachytene, and then reduce in number at late pachytene, where they persist at crossover-designated sites. At right, insets of the indicated fields show that although both RAD-51 and MSH-5 foci are present in the same nuclei, they rarely overlap. Scale bar, 10 μm.
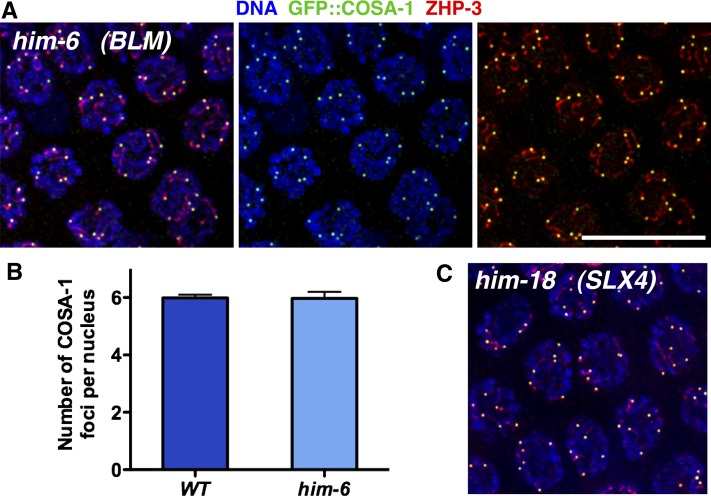
Figure 2.
The him-6 mutant is proficient for crossover designation and crossover regulation. (A) Immunofluorescence images of late pachytene nuclei from a him-6 mutant gonad, showing that each nucleus has six GFP::COSA-1 foci, each associated with a comet-like ZHP-3 signal; the him-6 mutant is cytologically indistinguishable from wild type, where GFP::COSA-1 foci reflect designation of a single cytologically differentiated CO site on each homolog pair (Yokoo et al. 2012). (B) Graph showing quantitation of GFP::COSA-1 foci in late pachytene nuclei from wild-type (n = 81) and him-6 (n = 94) worms; error bars indicate standard deviation. (C) GFP::COSA-1 and ZHP-3 immunofluorescence in late pachytene nuclei from a him-18 mutant, which is defective at a late step in crossover formation. Scale bar, 10 μm.
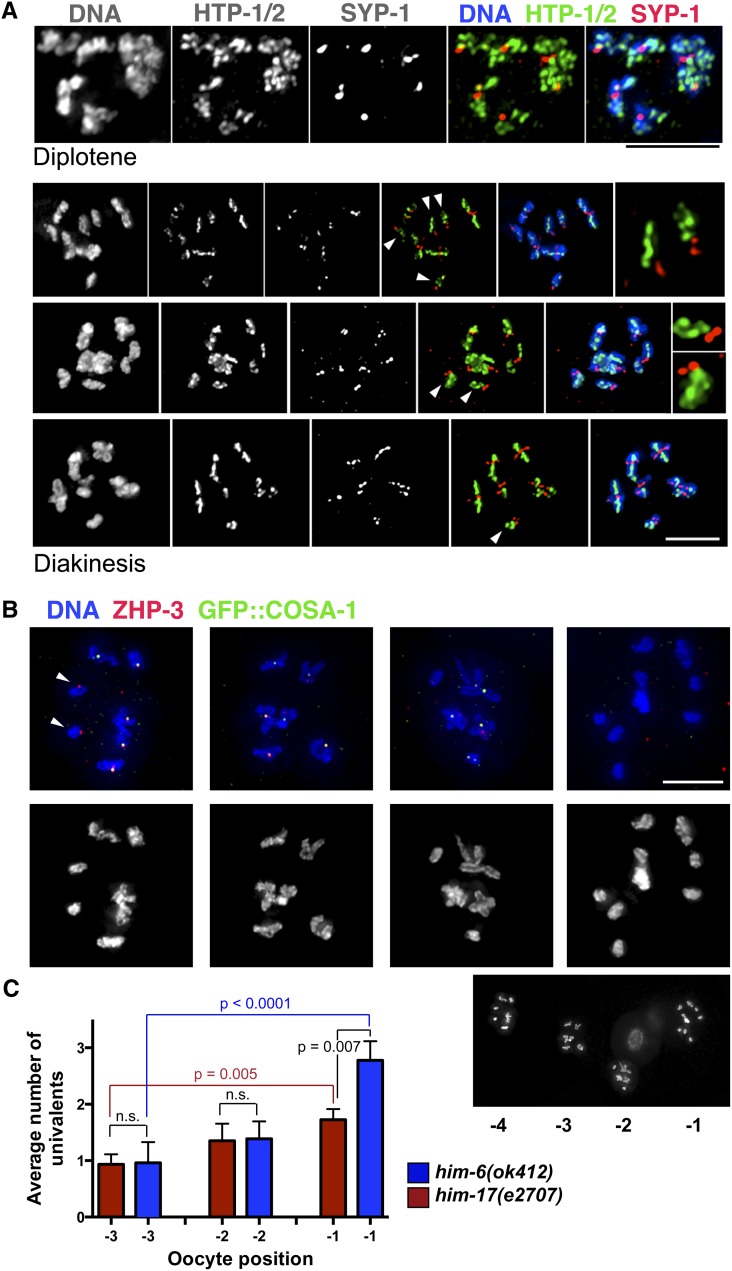
Figure 4.
Dissociation of some chromosome pairs into univalents by the end of diakinesis in him-6 mutants. (A) Diplotene and diakinesis-stage oocytes from the him-6 mutant, stained with antibodies against chromosome axis proteins HTP-1/2 and SC central region protein SYP-1. The chromosomes appear indistinguishable from wild type at the diplotene stage (Martinez-Perez et al. 2008), with the SYP-1 and HTP-1/2 proteins localizing to reciprocal domains as the chromosomes desynapse. By diakinesis, a mixture of bivalents and univalents (arrowheads) are detected; moreover, the univalents exhibit a reciprocal localization pattern for HTP-1/2 and SYP-1 that is normally associated with a crossover/chiasma. Right: For the top two diakinesis nuclei, the insets show selected univalents. (B) Full chromosome complements of individual him-6 diakinesis oocytes (from a single germ line, shown below). Oocyte nuclei from left to right were in the −4, −3, −2, and −1 positions relative to the spermatheca, with the −1 oocyte being the most mature. Arrows indicate univalents that have ZHP-3 foci, which normally mark crossover sites. Scale bars, 5μm. (C) Graph showing that the incidence of univalents in the him-6 mutant increases as oocytes progress through the diakinesis stage. The him-17(e2707) mutant, in which a decrease in DSB formation is responsible for the reduction in crossovers/chiasmata, is used as a control; in the him-17(e2707) control, there is a modest increase in the number of univalents scored during progression from the −3 to −1 position, reflecting improved ability to detect univalents as chromosome compaction increases during oocyte maturation. In contrast, there is a larger and more significant increase in univalents observed between the −3 and −1 oocytes in the him-6 mutant, suggesting the presence of more persistent connections that eventually dissociate (only a subset of P-values is depicted; see text). Error bars indicate SEM. Numbers of nuclei scored: him-17, n = 95; him-6, n = 83.
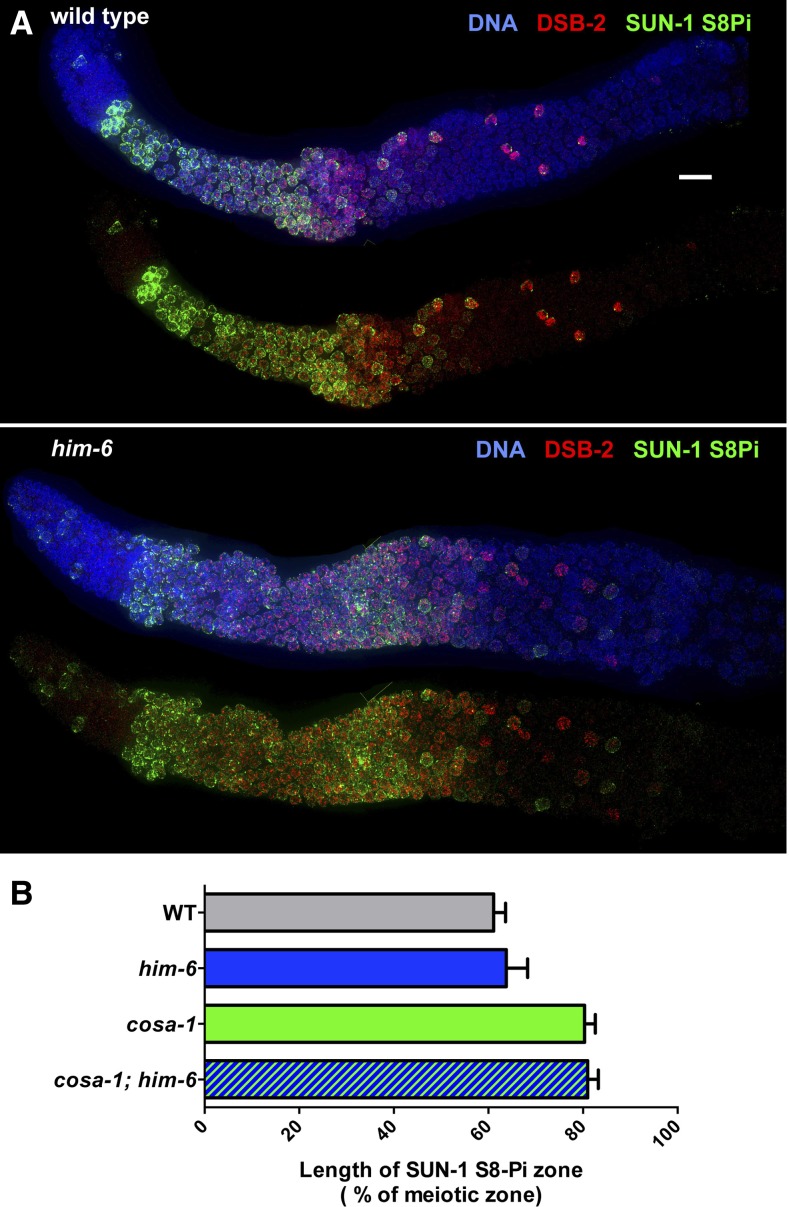
Figure 3.
Timely transition in pachytene progression in the him-6 mutant. (A) Immunoflouorescence images of whole-mount gonads, extending from the distal premeiotic tip to the end of the pachytene region. In both the wild-type and him-6 mutant, phosphorylation of nuclear envelope protein SUN-1 (SUN-1 S8Pi) and association of the DSB-promoting protein DSB-2 with chromatin show similar dynamics: They are detected in germ-cell nuclei at the onset of meiotic prophase and then decline and disappear from most nuclei during mid-pachytene. As SUN-1 S8Pi and DSB-2 persist until late pachytene in multiple mutants that fail to make crossover-eligible recombination intermediates (Rosu et al. 2013; Woglar et al. 2013) (see B), this finding is consistent with the conclusion that him-6 mutants are proficient for generating crossover intermediates. Scale bar, 10 μm. (B) Bar graph showing quantitation of the percentage of the meiotic zone occupied by SUN-1 S8-Pi-positive nuclei in germ lines of indicated genotypes. The presence/absence of SUN-1 S8-Pi signals was assessed in the portion of the germ line extending from the onset of meiotic prophase to the end of the pachytene region. The extent of the SUN-1 S8-Pi-positive zone was defined as the number of contiguous rows of nuclei in which all rows contained two or more nuclei with SUN-1 S8-Pi staining/total rows of nuclei in the scored region. Data are represented as mean ±SD. Whereas the SUN-1 S8-Pi-positive zones were significantly extended in the cosa-1 and cosa-1; him-6 mutants relative to wild type and the him-6 single mutant (P < 0.0001, two-tailed Mann–Whitney tests), no significant difference was observed between wild type and the him-6 mutant (P = 0.10). Numbers of germ lines scored: wild type, 18; him-6, 21; cosa-1, 20; cosa-1; him-6, 17.
For images in Figure 6B and Figure 7A, immunofluorescence and image processing were conducted as in Martinez-Perez and Villeneuve (2005), with the following modifications. For Figure 7, the integrated lacO array on chromosome V was detected in situ with affinity purified, bacterially expressed LacI–His6–GFP fusion protein (Darby and Hine 2005). LacI–His6–GFP, diluted 1:250 in PBST, was added to slides prior to the antibody incubation steps; slides were incubated for 1.5 hr at room temperature, followed by 3× 10-min washes in PBST. Incubation times for subsequent primary and secondary antibody steps were reduced to 1 hr each to minimize background. Images were acquired at 512 × 512 pixel dimensions, as Z-stacks at 0.2-μm intervals on an Applied Precision OMX imaging system in wide-field mode using a 100× objective. Subsequent deconvolution, alignment, and projection steps were carried out using Applied Precision softWoRx software. Images were viewed and converted to TIFF format using ImageJ.
Figure 6.
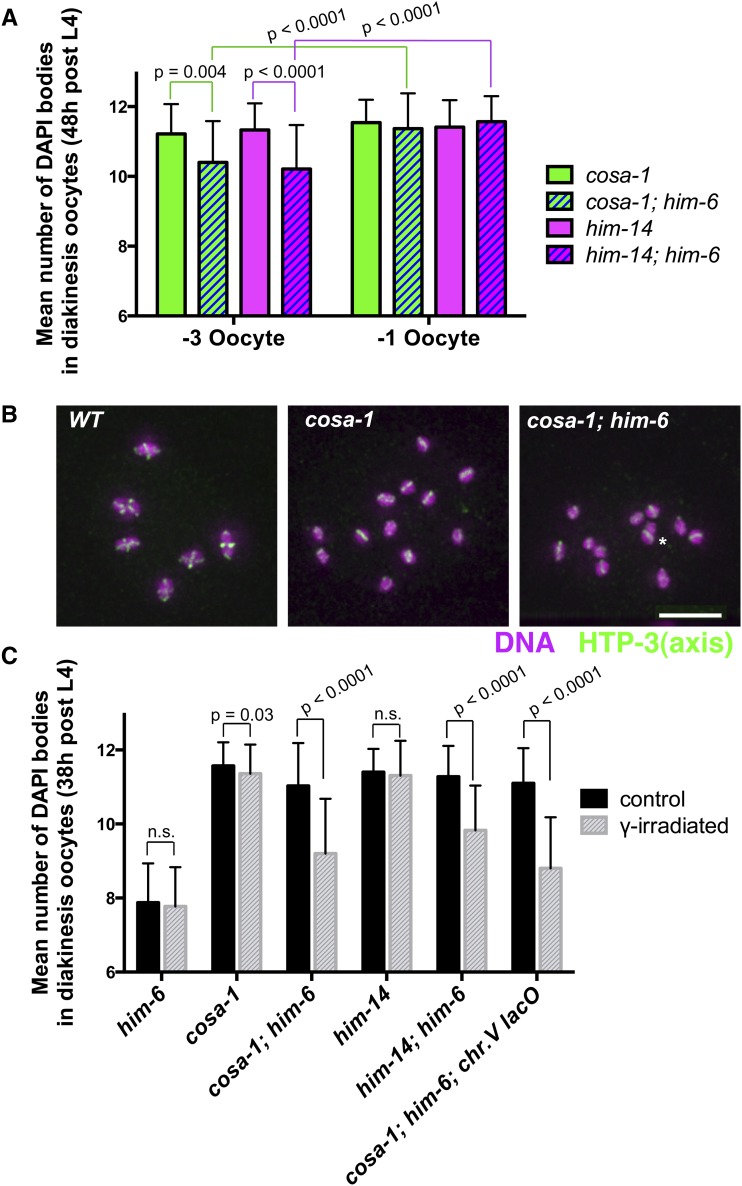
Quantitation of late-prophase interchromosomal associations in mutants lacking activity of conserved meiotic crossover factors COSA-1 and MutSγ. (A and C) Graphs showing quantitation of DAPI bodies in diakinesis-stage occytes in worms of the indicated genotypes; error bars indicate standard deviation. In this assay, wild-type oocytes have 6 DAPI bodies (reflecting chiasmata connecting all 6 chromosome pairs), and 12 DAPI bodies indicates a complete lack of chiasmata; the assay tends to overestimate the incidence of connections between chromosomes, as some univalents lie too close together to be resolved unambiguously. (A) Worms were fixed for DAPI staining at 48 hr post-L4 stage, and the data for oocytes in the −3 (less mature) and −1 (more mature) positions were graphed separately. For oocytes in the −1 positions, no significant differences were observed between the cosa-1 single mutant and cosa-1; him-6 double mutant or between the him-14 single mutant and him-14; him-6 double mutant, consistent with all six homolog pairs lacking chiasmata and CO at the end of prophase in the double mutants. However, significant differences between the numbers of DAPI bodies detected in the −3 vs. −1 oocytes were observed for the cosa-1; him-6 and him-14; him-6 double mutants (P < 0.0001), but not for the cosa-1 and him-14 single mutants (P = 0.19, P = 0.52). In combination, these data suggest the presence of persistent associations between homologs in the cosa-1; him-6 and him-14; him-6 double mutants that were ultimately resolved prior to ovulation. Numbers of oocyte nuclei scored: him-6, 60; cosa-1; him-6, 193; cosa-1, 71; him-14, 137; him-14; him-6, 129. (B) Immunofluorescence images depicting the full chromosome complements of individual −2 diakinesis oocytes of the indicated genotypes, stained for DNA (purple), and chromosome axis protein HTP-3 (green) following fixation at 38 hr post-L4. HTP-3 cruciform structures (reflecting the presence of chiasmata) are readily detected on bivalents in wild-type oocytes, but infrequent in cosa-1 and cosa-1; him-6 oocytes (none in the images shown). Images are projections of 3-D data stacks encompassing whole nuclei; asterisk indicates univalents that appear to overlap in the projection but are actually from different focal planes. Scale bar, 5 μm. (C) Graph depicting the effect of exposure to 5 krad γ-irradiation on the mean number of DAPI bodies in diakinesis oocytes (fixed 18 hr after IR treatment at 20 hr post-L4). Data for oocytes in the −3, −2, and −1 position were combined. Whereas IR treatment had no detectable impact on the him-6 single mutant (P = 0.41) or the him-14 single mutant (P = 0.91) and only a marginal effect on the cosa-1 single mutant (P = 0.03), IR treatment resulted in highly significant reductions (P < 0.0001) in the number of DAPI bodies detected in cosa-1; him-6 oocytes, him-14; him-6 oocytes, and cosa-1; him-6; syIs44 oocytes (which carry a high-copy transgene array containing multiple copies of the lacO sequence integrated into chromosome V), indicating an increase in the incidence of connections between chromosomes. Numbers of oocyte nuclei scored: him-6 control, 122; him-6 irradiated,140; cosa-1 control, 126; cosa-1 irradiated, 132; cosa-1; him-6 control, 137; cosa-1; him-6 irradiated, 115; him-14 control, 126; him-14 irradiated, 137; him-14; him-6 control, 117; him-14; him-6 irradiated, 139; cosa-1; him-6; syIs44 control, 90; cosa-1; him-6; syIs44 irradiated, 81.
Figure 7.
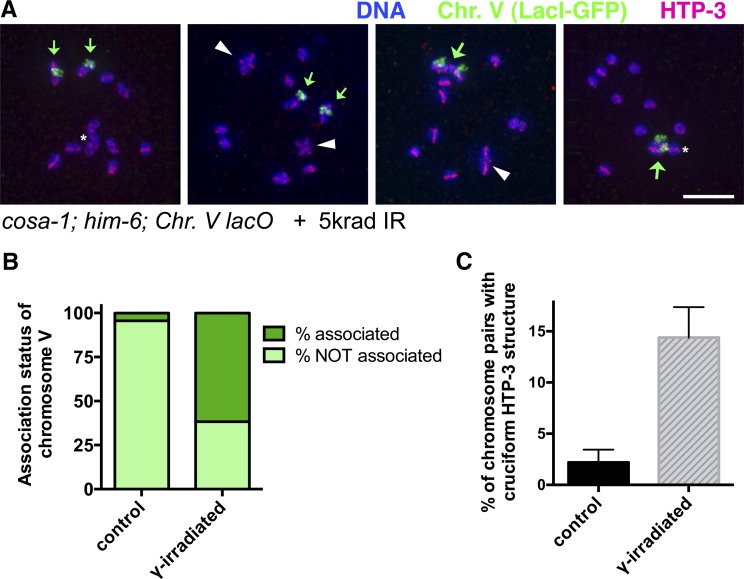
IR induces persistent interhomolog associations in cosa-1; him-6 mutant oocytes. (A) Immunofluorescence images depicting the full chromosome complements of individual diakinesis oocytes from cosa-1; him-6; syIs44 V worms (in which chromosome V is tagged by an integrated array of lacO repeats, indicated as Chr. V lacO), following exposure to 5 krad of γ-irradiation at 20 hr post-L4 and dissection and fixation at 38 hr post-L4. Small green arrows indicate unassociated chromosome V univalents, large green arrows indicate associated chromosome V homolog pairs, and white arrowheads indicate HTP-3 cruciform structures. Images are projections of 3-D data stacks encompassing whole nuclei; asterisk indicates univalents that appear to overlap other chromosomes in the projection but are actually in different focal planes. Oocyte nuclei (l to r) were in the −2, −2, −2, and −1 positions. Scale bar, 5 μm. (B) Graph depicting quantitation of association status of chromosome V in oocytes in the −3, −2, and −1 positions. Numbers of nuclei scored: control, 90; irradiated, 81. (C) Quantitation of the percentage of chromosome pairs (of six total per nucleus) for which a cruciform HTP-3 structure was detected in diakinesis nuclei (−3, −2, and −1 positions) from control and γ-irradiated cosa-1; him-6; syIs44 worms. n = 540 chromosome pairs for both conditions; error bars indicate 95% confidence interval.
For quantitation of GFP::COSA-1 foci in late pachytene nuclei, late L4 hermaphrodites were picked and shifted to 25° for 24 hr prior to dissection and fixation for immunofluorescence; foci were quantified as in Yokoo et al. (2012).
The following primary antibodies were used at the indicated dilutions in PBST with 0.5% BSA: rat anti-RAD-51 (1:250) (Rosu et al. 2013), rabbit anti-MSH-5 (1: 10,000) (SDIX), guinea pig anti-ZHP-3 (1:500) (Bhalla et al. 2008), rabbit anti-GFP (1:1000) (Yokoo et al. 2012), guinea pig anti-SUN-1 S8Pi (1:1000) (Penkner et al. 2009), rabbit anti-DSB-2 (1:5000)(Rosu et al. 2013), rabbit anti-HTP-1/2 (1:200)(Martinez-Perez et al. 2008), guinea pig anti-SYP-1 (1:50) (MacQueen et al. 2002), and chicken anti-HTP-3 (1:250) (MacQueen et al. 2005).
Chromosome painting:
Chromosome painting was conducted as in Nabeshima et al. (2011), using the two-color X chromosome probe.
Quantitative analyses of DAPI bodies in diakinesis oocytes:
For Figure 4C and Figure 6, A and C, numbers of DNA bodies present in diakinesis oocytes (in the −3, −2, and −1 positions relative to the spermatheca) were assessed in intact adult hermaphrodites fixed in ethanol and stained with 4′,6-diamidino-2-phenylindole (DAPI) as in Bessler et al. (2007). Note that this analysis tends to overestimate the incidence of interhomolog connections, as sometimes univalents lie too close to each other to be resolved unambiguously. For Figure 4C and Figure 6A, worms were fixed at 48 hr post-L4. For Figure 6C, worms were exposed to 5 krad γ-irradiation from a 137Cs source at 20 hr post-L4, and irradiated worms and age-matched controls were fixed 18 hr later (38 hr post-L4). For Figure 4C, data were transformed to “average number of univalents” to more clearly illustrate the increase in univalents detected in him-6 mutants as apparent bivalents dissociate as oocytes progress through diakinesis (e.g., 6 DAPI bodies = 0 univalents, 7 DAPI bodies = 2 univalents, 8 DAPI bodies = 4 univalents). For experiments in Figure 6, where mutants lacking activity of canonical meiotic pro-CO factors (COSA-1 or MutSγ) were analyzed, data are presented as “average number of DAPI bodies.” Mann–Whitney tests were used for statistical analyses of diakinesis data; two-sided P-values were calculated using GraphPad Prism software.
Use of a chromosomally integrated lacO array to assess IR-induced interhomolog associations:
Gonads of cosa-1(tm3298); him-6(ok412); syIs44[lacO array] V hermaphrodites that had been exposed to 5 krad γ-irradiation at 20 hr post-L4 (and age-matched controls) were dissected and fixed at 38 hr post-L4. For quantitating numbers of DAPI-stained bodies, frequencies of association between chromosome V homologs, and incidence of cruciform HTP-3 structures, scoring was performed using a Zeiss Axioimager microscope; oocytes in the −1, −2 and −3 positions were scored. For images in Figure 7A and for more detailed evaluation of chromosome organization, image stacks acquired on the OMX imaging system were used.
Measurement of genetic map distance in oocyte meiosis and detection of exceptional gamete types:
To measure recombination frequencies in the dpy-3unc-3 interval specifically during oocyte meiosis, control dpy-3unc-3/++ hermaphrodites and him-6; dpy-3unc-3/++ hermaphrodites were crossed with ccIs4251/+ males, which carry a chromosomally integrated transgene insertion expressing GFP under control of the myo-3 promoter (Fire et al. 1998). Hermaphrodites were transferred daily to fresh plates, and cross progeny were identified on the basis of expression of GFP in body wall muscle; only male cross progeny, which have a single X chromosome, were used to assess recombination frequencies. Numbers of recombinant (Dpy non-Unc and Unc non-Dpy) and parental Dpy Unc male progeny types were scored, and recombination frequencies were calculated as: (recombinants)/(recombinants + 2 × Dpy Uncs). Since him-6 oocytes produce significant numbers of nullo-X ova (reflecting loss or nondisjunction of X chromosomes during chromosome segregation; see below), non-Dpy non-Unc male cross progeny (which could have inherited their X chromosome from either their mother or their father) were excluded from this analysis. For the control, 221 recombinant and 233 Dpy Unc male progeny were scored; for the him-6 mutants, 94 recombinant and 185 Dpy Unc male progeny were scored.
The frequency of nullo-X ova produced by him-6 mutant hermaphrodites in these crosses was calculated as “number of male progeny with a paternally derived X”/“total number of progeny with a paternally-derived X,” where the numerator N is estimated as the excess number of non-Dpy non-Unc males (relative to Dpy Unc males), and the denominator D = N + the total number of non-Dpy non-Unc hermaphrodites. Our calculated frequency of nullo-X ova was 6.2% for him-6(ok412) (D = 607), which closely matches the value of 6.4% previously reported for him-6(e1423) (Hodgkin et al. 1979).
These crosses also allowed us to detect exceptional XX GFP+ cross progeny (produced by him-6 mutant mothers) that lacked a paternally derived X chromosome but had inherited two maternally derived X chromosomes bearing recessive markers. We detected 5 Dpy Unc hermaphrodites (dpy-3unc-3/ dpy-3unc-3), 1 Unc non-Dpy hermaphrodite (+ unc-3/ dpy-3unc-3), and 2 Dpy non-Unc hermaphrodites (dpy-3 +/dpy-3unc-3). The types of exceptional XX ova giving rise to these progeny classes represented 1.6% of the ova produced by him-6 mutant hermaphrodites. Their frequency was calculated as “number of exceptional XX cross progeny showing recessive phenotypes”/“total number of cross progeny derived from nullo-X sperm (corrected for inviability of nullo-X zygotes resulting from fertilization of nullo-X ova),” where the denominator = (2× Dpy Unc males + recombinant males + 2× exceptional XX progeny)/0.938. The incidence of these exceptional XX progeny types among directly scorable progeny arising from nullo-X sperm differed significantly (P = 0.007; Fisher’s exact test) between wild type (0/687) and the him-6 mutant (8/472). We note that the classes of exceptional XX ova detected by this assay are not consistent with expectations for a simple meiosis I nondisjunction of nonrecombinant X chromosomes followed by normal equational segregation of sister chromatids at meiosis II; this type of segregation pattern would yield diplo-X gametes heterozygous for all markers, which would not be detected by the assay. However, meiosis I nondisjunction of nonrecombinant chromosomes likely does also occur in him-6 mutants, as products consistent with meiosis I nondisjunction in the him-6(e1423) mutant were previously detected by a different assay (Hodgkin et al. 1979).
Results
To elucidate the nature of the defects responsible for the achiasmate chromosomes observed at the end of meiotic prophase in him-6 mutants, we evaluated multiple markers of ongoing recombination events and other aspects of prophase progression.
him-6 mutants are proficient for formation of early recombination intermediates and loading of MutSγ
Figure 1 shows simultaneous immunostaining for DNA strand-exchange protein RAD-51 (which loads onto DNA at resected DSBs) and conserved CO-promoting factor MSH-5 (a component of the MutSγ heterodimer) in a him-6 mutant germ line. In wild-type germ lines, RAD-51 foci are abundant in early–mid-pachytene nuclei and then decline and disappear from most nuclei by late pachytene (Alpi et al. 2003; Colaiacovo et al. 2003; Rosu et al. 2013; and Figure S1). MSH-5 is first detected as faint foci in early pachytene nuclei; MSH-5 foci become brighter and more abundant during mid-pachytene and then decline in number upon transition to late pachytene, where their localization becomes restricted to designated CO sites (Yokoo et al. 2012 and Figure S1). Our images showed an overall temporal/spatial pattern of appearance and disappearance of RAD-51 foci during pachytene progression in the him-6 mutant that was roughly similar to the wild type, with foci abundant in early and mid-pachytene, but absent from most late-pachytene nuclei (Figure 1); this pattern is consistent with previous reports (Saito et al. 2009; O’Neil et al. 2013), albeit quantitation in those studies revealed a modest increase in numbers of foci in him-6 mutants. Likewise, the temporal/spatial pattern of appearance of MSH-5 foci and the late pachytene decline in their numbers in him-6 mutant germ lines were also comparable to the wild type (Figure 1). Together, these data support the conclusion that him-6 mutants are substantially proficient for formation and resection of DSBs and for licensing of potential CO intermediates through loading of MutSγ.
Interestingly, although the MutSγ complex is presumed to function at recombination intermediates after resection of DSBs, we did not observe a class of “early” nuclei that contained only RAD-51 foci but no MSH-5 foci. In both wild-type and him-6 mutant germ lines (Figure S1 and Figure 1), essentially all nuclei that have RAD-51 foci also have MSH-5 foci. Moreover, when both RAD-51 foci and MSH-5 foci are present in the same nucleus, they rarely colocalize (see Figure S1 legend). This finding indicates either that MSH-5 does not load until after the majority of RAD-51 has been removed at a nascent recombination event or that MSH-5 foci form predominantly at sites that did not acquire high levels of RAD-51 in the first place. Importantly, these colocalization experiments suggest that previous analyses of RAD-51 foci alone are likely to underestimate the number of ongoing recombination events.
him-6 mutants are proficient for crossover designation and crossover regulation:
Immunofluorescence analyses indicated that CO designation and CO regulation are apparently normal in him-6 mutants. We assessed CO designation and regulation by immunolocalization of GFP::COSA-1 (a cyclin-like protein that is conserved specifically in metazoa and is required for CO formation) and ZHP-3, another conserved CO-promoting protein (Jantsch et al. 2004; Bhalla et al. 2008; Yokoo et al. 2012). During wild-type meiosis, GFP::COSA-1 foci mark CO-designated recombination sites beginning in the late pachytene stage, and ZHP-3 initially localizes along the lengths of the synaptonemal complexes (SCs) earlier in the pachytene stage and then gradually reduces its chromosomal localization during the late pachytene stage until it colocalizes with COSA-1 at CO sites (Jantsch et al. 2004; Bhalla et al. 2008; Yokoo et al. 2012). Further, as C. elegans exhibits very strong CO interference (Hillers and Villeneuve 2003), each of the six chromosome pairs normally receives only a single GFP:COSA-1 focus (Yokoo et al. 2012). As in wild-type meiosis, we detected precisely six COSA-1 foci per nucleus during the late-pachytene stage in him-6 mutant germ lines (Figure 2, A and B). Our quantitation of COSA-1 foci dovetails with independent quantitation of ZHP-3 foci in the him-6 mutant (Agostinho et al. 2013) and indicates that him-6 mutants are proficient for concentrating CO-promoting factors at a single site per chromosome pair, implying that both CO designation and CO interference mechanisms are operational. We similarly detected six COSA-1 foci in the him-18 mutant (Figure 2C), which is defective for the C. elegans ortholog of SLX4, which has been implicated in the resolution of CO-designated intermediates at the end of the recombination process to yield mature CO products (Saito et al. 2009). These data are consistent with HIM-6/BLM being required to accomplish a late step in the process of crossover formation.
Markers of meiotic progression appear normal in him-6 mutants:
Several recent studies have provided evidence for a checkpoint-like negative feedback network that operates during C. elegans meiosis to couple multiple aspects of meiotic prophase progression to the formation of crossover-eligible recombination intermediates (Rosu et al. 2011; Rosu et al. 2013; Stamper et al. 2013; Woglar et al. 2013). These studies suggested that germ cells have a capacity to sense whether CO-eligible recombination intermediates have formed on all six chromosome pairs; fulfillment of this requirement enables nuclei to shut down early processes and progress in a timely manner to the late-pachytene stage, when pro-CO factors become concentrated at a single CO site per homolog pair. Conversely, lack of CO-eligible intermediates on one or more homolog pairs delays multiple aspects of prophase progression, which can be detected cytologically as prolonged persistence of several early prophase markers, including association of meiotic DSB-promoting protein DSB-2 on chromatin and phosphorylation of nuclear envelope protein SUN-1 (SUN-1 S8Pi) (Rosu et al. 2013; Woglar et al. 2013).
Immunostaining for DSB-2 and SUN-1 S8Pi showed that this progression delay is not elicited in him-6 mutant germ lines (Figure 3, A and B). In contrast to the prolonged persistence of DSB-2 and SUN-1 S8Pi observed in mutants defective for DSB formation, DSB processing, strand exchange, or formation of CO-specific intermediates (Rosu et al. 2013; Woglar et al. 2013), immunofluorescence analysis using these markers indicates that the timing of meiotic prophase progression in him-6 mutant germ lines is similar to that in wild-type controls. This finding further supports our inference that him-6 mutants are proficient for multiple steps in recombination leading up to and including formation of CO-specific recombination intermediates, and thus the HIM-6/BLM protein is dispensable for these steps.
Recombination events at a subset of CO-designated sites fail to mature into chiasmata in a him-6 mutant
Despite the presence of one COSA-1-marked CO-designated site on each homolog pair, suggesting designation of the necessary number of COs to allow accurate chromosome segregation, some achiasmate chromosome pairs (univalents) are detected in him-6 mutant oocytes at diakinesis, the last stage of meiotic prophase (Wicky et al. 2004; Agostinho et al. 2013). We investigated how these achiasmate chromosomes arise by analyzing chromosome organization following exit from the pachytene stage. We found that at the diplotene stage, when homologous chromosomes desynapse, chromosome organization in the him-6 mutant still appeared comparable to wild type (Figure 4A and Figure S2). Specifically, asymmetric disassembly of the SC and relocalization of chromosome axis proteins HTP-1/2 and SC central region protein SYP-1 to reciprocal chromosomal domains, processes that are normally coupled to and dependent on (nascent) CO events (Nabeshima et al. 2005; Martinez-Perez et al. 2008), were also observed for all six chromosome pairs in him-6 mutant diplotene oocytes, as previously reported (Agostinho et al. 2013).
By the diakinesis stage, however, abnormalities were readily apparent in the him-6 mutant. Specifically, we observed univalents that exhibited clear reciprocal localization of HTP-1/2 and SYP-1 into two distinct domains or that bore markers of CO-designated sites despite lack of any evident connections to their homologs (Figure 4, A and B). Further, even in some cases where the chromosomes did appear connected, the configurations of the apparent bivalents were atypical, suggesting that they may have been in the process of dissociating.
To test the hypothesis that chromosomes that appear to be connected in early diakinesis eventually dissociate into univalents in him-6 mutants, we quantitated the numbers of univalents detected in him-6 mutant oocytes in the −3, −2, and −1 positions in the gonad, where −1 is the most mature oocyte, adjacent to the spermatheca (the sperm storage compartment through which oocytes are ovulated) (Figure 4C). The him-17(e2707) mutant, which has a reduction in crossovers/chiasmata that is caused by a decrease in DSB formation (Reddy and Villeneuve 2004), served as a control for this analysis. In the him-17(e2707) control, there was a modest increase in the number of univalents detected during progression from the −3 to −1 position (P = 0.049 for −2 vs. −1 oocyte; P = 0.0046 for −3 vs. −1 oocyte); this increase in the him-17(e2707) mutant reflects the fact that as chromosomes compact during oocyte maturation, the ability to resolve/detect univalents improves. In contrast, in the him-6 mutant, we observed a larger and more significant increase in the number of univalents detected during progression from the −3 to −1 position (P = 0.0046 for −2 vs. −1 oocytes; P < 0.0001 for −3 vs. −1 oocytes). Further, whereas the numbers of univalents detected in the −3 and −2 positions did not differ significantly between him-17(e2707) and the him-6 mutant, the number of univalents detected in the −1 oocytes was significantly higher in the him-6 mutant (P = 0.0068). These data are consistent with the presence of more persistent connections in the him-6 mutant that eventually dissociate.
Correspondence between presence of achiasmate X chromosomes and reduced frequency of X chromosome crossovers in the him-6 mutant
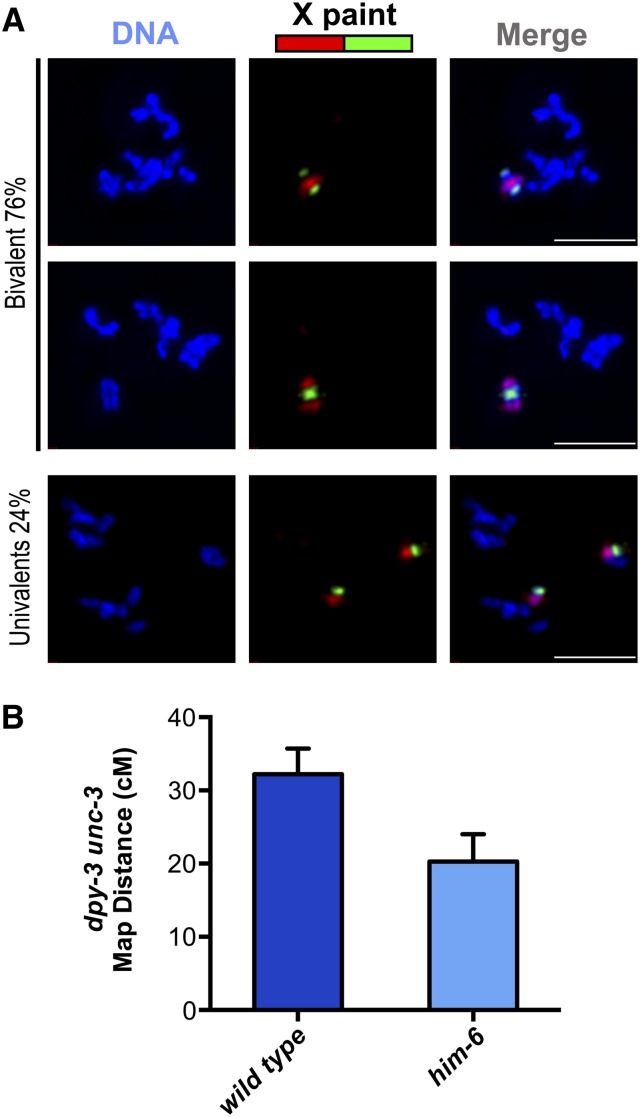
The presence of cytological markers of COs on achiasmate chromosomes in diakinesis-stage oocytes could potentially reflect either of two possibilities: (1) that COs had formed, but did not result in chiasmata, or (2) that CO-designated intermediates had formed, but these intermediates did not mature into COs. We infer that lack of COs is the most parsimonious explanation for the majority of achiasmate chromosomes observed in him-6 mutant oocytes, based on a combination cytological and genetic analyses. Specifically, we evaluated both the incidence of connections between homologs in diakinesis oocytes and the frequency of crossing over during oocyte meiosis for the same chromosome pair (Figure 5). Using chromosome paints to identify the X chromosomes (Figure 5A), we found that the X chromosomes were present as univalents in 24% of him-6 mutant diakinesis oocytes examined (n = 107). Further, by assessing CO frequencies specifically during oocyte meiosis for the dpy-3–unc-3 genetic interval spanning the majority of the length of the X chromosome, we detected a 37% reduction in the frequency of crossing over in the him-6 mutant (Figure 5B and Materials and Methods). These data are consistent with previous studies assessing CO frequencies in self-fertilizing hermaphrodites, which likewise reported significant reductions in recombination frequencies in him-6 mutants (Zetka and Rose 1995; Wicky et al. 2004; Agostinho et al. 2013). Thus, we conclude that the presence of chromosome pairs lacking chiasmata in him-6 mutant oocytes reflects a deficit of CO recombination events between the homologs and that HIM-6/BLM is required to ensure the CO outcome of repair at CO-designated recombination sites.
Figure 5.
Occurrence of achiasmate X chromosomes in him-6 mutant oocytes correlates with reduced crossover frequency during oocyte meiosis. (A) Each column shows the full karyotype of a single diakinesis oocyte from a him-6 mutant worm, with the X chromosomes visualized by chromosome paint. In the top two oocytes, the X chromosome pairs comprise a bivalent, whereas in the bottom oocyte, the X chromosomes clearly lack a chiasma and are present as univalents. (The different configurations of the red and green paint signals on the X chromosome bivalents in the top two images likely reflect different CO positions.) Scale bars, 5 μm. (B) Graph showing the genetic map distances measured for oocyte meiosis for the dpy-3 unc-3 interval on the X chromosome; error bars indicate 95% confidence intervals. Although the reduction in CO frequency in the tested interval in the him-6 mutant appears nominally higher than the incidence of achiasmate X chromosomes observed using the paint assay, based on the experimental error associated with both types of measurements, there is insufficient statistical power to conclude that this difference is significant. Thus, these data are not considered as part of the evidence for persistent connections between noncrossover chromosomes.
Meiotic prophase defects in him-6 mutants impair chromosome segregation, resulting predominantly in chromosome loss but also in nondisjunction events that yield gametes with two copies of a given chromosome (Hodgkin et al. 1979; Haack and Hodgkin 1991). The same crosses used to measure CO frequency in the dpy-3–unc-3 interval during him-6 oocyte meiosis also enabled us to identify several classes of exceptional XX hermaphrodite cross progeny resulting from fertilization of diplo-X ova (with two maternally derived X chromosomes) by nullo-X male sperm. These were identified on the basis of phenotypes indicating homozygosity for recessive markers and included worms both with and without a CO in the dpy-3–unc-3 interval (see Materials and Methods). The types of exceptional XX ova giving rise to these progeny classes represented 1.6% of the ova produced by him-6 mutant hermaphrodites. Importantly, the composition of these exceptional XX ova is inconsistent with them having arisen by meiosis I nondisjunction of nonrecombinant homologs, which would be expected to cause heterozygosity for all loci. Thus, we conclude that HIM-6/BLM has additional roles that contribute to proper chromosome segregation during meiosis beyond its role in promoting interhomolog COs.
HIM-6 antagonizes COSA-1/MutSγ-independent interhomolog connections
In several organisms where an anti-crossover role has been demonstrated or proposed for HIM-6/BLM orthologs, loss of BLM function can suppress the deficit of COs associated with loss of meiotic pro-crossover factors (Jessop et al. 2006; Oh et al. 2007; Kohl et al. 2012). Although most of the data presented thus far indicate a pro-CO role for HIM-6/BLM in C. elegans meiosis, these prior findings in other species nevertheless prompted us to test whether loss of him-6 function might suppress the lack of chiasmata observed in mutants defective for either cosa-1 or him-14/msh-4 (which encodes the MSH-4 component of MutSγ) (Zalevsky et al. 1999; Yokoo et al. 2012). Specifically, we scored the numbers of DAPI-stained bodies in diakinesis oocytes, where 6 DAPI bodies (as observed in wild-type oocytes) indicates that all chromosome pairs are connected and 12 DAPI bodies indicates that all chromosomes are achiasmate univalents.
Analysis of DAPI-stained bodies at 48 hr post L4 (Figure 6A) demonstrated that chiasmata were not substantially restored in him-14; him-6 or cosa-1; him-6 double mutants; however, it also suggested a role for HIM-6 in antagonizing persistent COSA-1/MutSγ-independent interhomolog connections. Specifically, when we considered oocytes in the −1 position (i.e., the most mature oocytes), we observed no significant differences between the numbers of DAPI bodies detected in cosa-1; him-6 and him-14; him-6 double mutants and in the corresponding cosa-1 and him-14 single mutants, indicating that loss of HIM-6/BLM did not bypass the requirements for COSA-1 and MutSγ in chiasma formation. However, we found that in the cosa-1; him-6 and him-14; him-6 double mutants (but not in the cosa-1 and him-14 single mutants), the average numbers of DAPI bodies scored in −3 (i.e., less mature) oocytes were significantly lower than those in −1 oocytes (P < 0.0001). Likewise, when only the −3 oocytes were considered, the numbers of DAPI bodies detected in the cosa-1; him-6 and him-14; him-6 double mutants were significantly lower than those in the single mutant controls. Together these analyses indicate the presence of a low level of persistent COSA-1/MutSγ-independent connections between homologs in the −3 oocytes; however, these connections appear to be distinct from normal chiasmata in that they are usually eliminated by the end of prophase (in a HIM-6-independent manner).
Immunostaining of chromosome axis component HTP-3 in diakinesis oocytes at 38 hr post-L4 provided additional evidence that loss of him-6 function does not substantially suppress the chiasma deficit caused by loss of cosa-1 function (Figure 6B). When all six chromosome pairs are connected by chiasmata (i.e., during wild-type meiosis) and the full complement of chromosomes in a diakinesis nucleus is viewed from a given perspective, approximately half of the bivalents are oriented in a way that allows HTP-3 staining to be visible in a clear cruciform configuration. Whereas cruciform HTP-3 structures were detected on 43.7% of bivalents in wild-type oocytes in the −3 to −1 positions (n = 558 homolog pairs), cruciform HTP-3 structures were very infrequent in both the cosa-1 single mutant (0.4%, n = 540) and cosa-1; him-6 double mutant oocytes (1.8%, n = 330), consistent with a severe defect in chiasma formation in both cases.
Our observation of temporary COSA-1/MutSγ-independent connections in the cosa-1; him-6 and him-14; him-6 double mutants (Figure 6A) suggested a possible role for HIM-6 either in (a) promoting the timely removal of COSA-1/MutSγ-independent interhomolog connections that should have been eliminated earlier in prophase or in (b) preventing the formation of inappropriate interchromosomal connections. We hypothesized that the observed COSA-1/MutSγ-independent connections reflect the presence of recombination intermediates at sites that would normally be destined for repair via a noncrossover pathway. To test this hypothesis, we assessed numbers of DAPI bodies in cosa-1; him-6 and him-14; him-6 diakinesis oocytes following exposure to a 5-krad dose of ionizing radiation (IR) (Figure 6C), reasoning that increasing the number of DSBs should increase the frequency of such connections if they were recombination based. We found that IR treatment had no detectable impact on the number of DAPI bodies in the him-6 single mutant (presumably because the majority of chromosome pairs are already connected as bivalents even in the absence of IR; P = 0.41) or the him-14 single mutant (P = 0.91) and only a marginal effect on the cosa-1 single mutant (P = 0.03). However, IR treatment resulted in a highly significant reduction in the number of DAPI bodies detected in cosa-1; him-6 oocytes (P < 0.0001) and him-14; him-6 oocytes (P < 0.0001), indicating a substantial increase in the incidence of connections between chromosomes. Further, the numbers of DAPI bodies detected following IR treatment of cosa-1; him-6 and him-14; him-6 double mutants were likewise much lower than in the IR-treated cosa-1 and him-14 single mutants (P < 0.0001 for both comparisons), indicating that loss of him-6 function is responsible for these IR-induced interchromosomal connections.
To further investigate the nature of the IR-induced interchromosomal connections, we used cosa-1; him-6; syIs44 V worms, in which chromosome V is tagged by a large integrated transgene array containing thousands of copies of the lacO repeat, to simultaneously assess both the incidence of association between homologs and the frequency of HTP-3 cruciforms detected following IR treatment (Figure 7). This analysis demonstrated that the interchromosomal connections induced by IR occur predominantly between homologs (Figure 7B). Specifically, whereas apparent associations between the chromosome V homologs were infrequent in diakinesis oocytes of untreated controls (4%, n = 90), the chromosome V homologs were associated in 62% (n = 81) of diakinesis oocytes of irradiated cosa-1; him-6; syIs44 worms. Further, when the chromosome V’s were not associated with each other, they were usually discernable as clearly resolved univalents (Figure 7A) (albeit occasional associations among nonhomologous chromosomes could not be excluded). It is likely that comparable high frequencies of interhomolog associations were also induced for the other homolog pairs, as the average of 8.8 DAPI bodies in diakinesis oocytes observed following IR treatment of this strain (Figure 6C) is consistent with induced associations occurring between 53% of chromosome pairs; this “expected” frequency of homolog association does not differ significantly from that measured for chromosome V.
Interestingly, in addition to inducing persistent associations between homologous chromosomes, IR treatment of cosa-1; him-6; syIs44 worms also resulted in a substantial increase in the frequency of cruciform HTP-3 structures detected (P < 0.0001, Fisher exact test; Figure 7, A and C). However, it was not possible to discern whether these cruciforms reflected the presence of bona fide chiasmata (i.e., containing mature crossover products) or “pseudo-chiasmata” in which chromosome axes had become reorganized into a cross-shaped structure around the site of unresolved recombination intermediates. Further, the frequency of induced cruciforms observed was significantly lower than would be expected if chiasmata/pseudochiasmata were present at all IR-induced associations (P < 0.0001, χ2 test), indicating that they occur at only a subset of the induced interhomolog associations.
Taken together, these analyses indicate that in addition to its role in promoting the formation of meiotic COs via the COSA-1/MutSγ-dependent pathway, HIM-6 functions to prevent and/or eliminate other recombination-based connections between homologs.
Discussion
HIM-6/BLM function in promoting meiotic COs
To ensure the formation of COs in meiosis, two conditions must be met: (1) It is necessary to license potential CO sites, to designate a subset of sites for maturation as COs and to protect recombination intermediates at designated CO sites from proteins that might dismantle them prematurely/inappropriately, and (2) resolution must occur in a biased fashion to guarantee the CO outcome.
C. elegans him-6 mutants are clearly competent to meet most of the former condition. First, they form and resect DSBs and load MutSγ in a timely fashion. Second, they are competent to form later recombination intermediates that can be recognized as fulfilling the requirements of a “crossover assurance” checkpoint that couples meiotic prophase progression to formation of CO-eligible recombination intermediates (Rosu et al. 2013; Stamper et al. 2013; Woglar et al. 2013). Third, him-6 mutants are proficient to recruit COSA-1 to a subset of potential CO sites, to concentrate other pro-CO proteins at those sites, and to reorganize chromosome structural proteins in response (Agostinho et al. 2013). Moreover, they are proficient for the robust regulation of CO designation, reliably generating precisely one COSA-1 marked site per chromosome pair.
Despite successfully concentrating pro-CO factors at one CO-designated site per chromosome pair, however, him-6 mutants fail to convert the intermediates at a subset of these into actual CO products and chiasmata. This late-manifesting CO defect suggests that HIM-6/BLM may function as a pro-CO factor predominantly in the “end game” of CO formation during C. elegans meiosis, primarily at the level of biasing the outcome of resolution at CO-designated sites to favor/guarantee the CO outcome. This could be strictly a late role, with HIM-6/BLM operating directly at CO sites after the CO designation step. Alternatively, loss of CO bias in a him-6 mutant might be a downstream consequence of an earlier defect leading to accumulation of recombination intermediates with aberrant structures. We note that under this latter scenario, however, the hypothesized aberrant intermediates would have to be close enough to normal to trick the CO assurance checkpoint and thereby enable meiotic prophase progression.
Our idea that the main role of C. elegans HIM-6/BLM in promoting meiotic COs is to confer/enforce bias to the resolution process is compatible with the findings of a recent study investigating the potential roles of several different enzymes in meiotic CO resolution in C. elegans (Agostinho et al. 2013). Based on their analysis of a large collection of double and single mutants affecting various structure-specific endonucleases and/or him-6, Agostinho et al. (2013) proposed that two major activities resolve meiotic CO intermediates, one provided by XPF-1 and HIM-6 and another provided by MUS-81 and SLX-1. Based on our own data and a reevaluation of the data from Agostinho et al. (2013), we propose a revised model in which HIM-6 is required both (1) to promote efficient resolution by the XPF-1-mediated pathway and (2) to confer a CO-biased outcome to resolution events mediated by either the XPF-1 or MUS-81 pathway. Our reasoning is as follows: Nuclease single-mutant phenotypes suggest that either pathway is capable of mediating resolution of recombination intermediates to yield COs and chiasmata in a manner that usually yields a CO/chiasma for every homolog pair. In contrast, him-6 single mutants are apparently competent to resolve CO-designated intermediates by the end of prophase, but produce a mixture of bivalents and univalents and a reduced incidence of COs, indicating loss of the resolution bias that ensures the CO outcome. Further, whereas most chromosome pairs in mus-81; xpf-1 oocytes showed evidence of unresolved recombination intermediates (reflecting loss of both the XPF-1 and MUS-81 resolvase activities), chromosomes with unresolved intermediates were a minority class in mus-81; him-6 oocytes and were not detected in xpf-1; him-6 oocytes. Together, the data suggest that at least in the context of meiotic crossing over, HIM-6 is not strictly required for either resolvase activity, although it does appear to enhance the activity of XPF-1. Moreover, they further imply that both the XPF-1- and MUS-81-dependent resolvase pathways exhibit a strong bias toward the CO outcome and that HIM-6 is responsible for conferring this bias. As it has been suggested that CO bias may be an inherent feature of the structure of the CO intermediates upon which resolvases act, we hypothesize that HIM-6 may confer CO bias by affecting the structure of the intermediates present at CO-designated sites.
The idea that a BLM helicase might be responsible for conferring CO resolution bias during meiosis represents an important conceptual shift in thinking about how crossover assurance might be achieved. Whether BLM orthologs also contribute to biasing the resolution outcome at CO-designated sites in other organisms warrants further investigation. Differences among organisms regarding the relative balance between pro-CO and anti-CO roles for BLM may tend to obscure a possible role in CO bias. Several features of the C. elegans system, including the ability to quantify CO-designated sites in late pachynema, chiasmata at diakinesis, and CO frequencies in progeny enabled us to discover this role in CO bias, as we could see that normal numbers of CO sites had been designated, yet not all designated sites had yielded COs.
Two recent articles have proposed pro-CO roles for Sgs1, the S. cerevisiae ortholog of BLM, but it remains to be seen whether the pro-CO roles revealed for the BLM orthologs worms and yeast are analogous or distinct. De Muyt et al. (2012) proposed an early pro-CO role for Sgs1, in channeling recombination intermediates into the MutSγ-dependent CO pathway; a directly analogous role seems unlikely for C. elegans HIM-6/BLM since the him-6 mutant appears highly proficient at populating the MutSγ-dependent CO pathway by multiple different criteria. However, we acknowledge that the structure of the intermediates formed in the him-6 mutant might be abnormal despite their ability to recruit canonical pro-CO factors and to respond to CO regulation. Zakharyevich et al.(2012) proposed a late pro-CO role for Sgs1 during budding yeast meiosis, in promoting resolution of recombination intermediates by the MutLγ (Mlh1–Mlh3) complex. Whereas MutLγ is inferred to be the predominant resolvase pathway yielding meiotic COs in budding yeast, C. elegans lacks MutLγ and therefore must generate COs using distinct resolvases (Saito et al. 2009, 2013; Agostinho et al. 2013; O’Neil et al. 2013).
HIM-6/BLM function in antagonizing COSA-1/MutSγ-independent interhomolog associations
The BLM helicase is best known for its roles in antagonizing recombination, originally based on its role in inhibiting sister chromatid exchange and interhomolog COs in mitotically dividing cells. More recently, anti-CO roles during meiosis have been demonstrated or inferred for BLM orthologs in a variety of species. For example, in S. cerevisiae and Drosophila, loss of BLM orthologs during meiosis suppressed the CO deficits in mutants lacking “anti-anti-CO” factors, i.e., MutSγ in budding yeast and the mei-MCM complex in flies (Jessop et al. 2006; Oh et al. 2007; Kohl et al. 2012). Further, loss of BLM function in mouse germ cells was associated with elevated numbers of chiasmata (Holloway et al. 2010).
The prominent role of C. elegans HIM-6/BLM in promoting COs and chiasmata has tended to obscure the extent to which it might also contribute to repair of meiotic DSBs not destined to become COs. By eliminating the “complication” of CO-specific recombination intermediates protected by COSA-1 and MutSγ, we uncovered a separate role for HIM-6/BLM during C. elegans meiosis, in antagonizing COSA-1/MutSγ-independent associations between homologs. We found that interhomolog connections persisted later in diakinesis in the cosa-1 and him-14 mutant backgrounds when HIM-6 was eliminated, indicating a role for HIM-6 in removing such connections and/or in preventing their formation. Further, our demonstration that we could increase such connections by increasing the number of DSBs by γ-irradiation implies that these connections reflect DSB-dependent recombinational interactions.
Together our data indicate that in addition to promoting COs, HIM-6 activity can contribute to the timely repair of recombination intermediates at sites not designated as interhomolog meiotic CO. However, the extent to which HIM-6 is required to do so during wild-type meiosis and the nature of the intermediates on which it may operate remain unclear. The interhomolog associations observed in the cosa-1; him-6 and him-14; him-6 double mutants imply that HIM-6 can operate on repair intermediates that form between homologs, and we suggest that some of these associations may reflect the presence of aberrant DNA joint molecules similar to those observed in S. cerevisiae sgs1 mutants (Oh et al. 2007). Additionally, the classes of exceptional XX ova that we observed in the him-6 mutant are most consistent with meiosis II nondisjunction, suggesting that HIM-6 may also contribute to resolving and/or preventing the formation of aberrant recombination intermediates between sister chromatids that might impede their segregation at meiosis II.
Dependence of COs/chiasmata on MutSγ and COSA-1
Our work also has implications regarding the role of MutSγ and its partner proteins in promoting meiotic COs. While it has been suggested that MutSγ promotes COs primarily by antagonizing the functions of helicases that would dismantle CO intermediates (Kohl and Sekelsky 2013), we have found that in the context of C. elegans meiosis, the requirements for MutSγ and its partner COSA-1 in chiasma formation are not bypassed by eliminating either HIM-6/BLM (this work) or RTEL-1 (Yokoo et al. 2012). We suggest that instead of (or in addition to) antagonizing anti-CO helicases, MutSγ and other pro-CO factors may be needed to antagonize inappropriate resolvase activities, to recruit appropriate resolvases, to recruit HIM-6/BLM, and/or to preserve the structure of the intermediates in a form that confers biased resolution.
Although loss of him-6 function alone was not sufficient to bypass the requirement for MutSγ and COSA-1 in chiasma formation, we found that a fivefold increase in DSB levels induced by γ-irradiation in a cosa-1; him-6 mutant did in fact result in significant levels of chiasma-like structures. Thus, it may be possible to generate substantial levels of MutSγ-independent COs/chiasmata even in C. elegans, given high enough levels of DSBs and interhomolog joint molecules. Perhaps a major role of MutSγ and cofactors is to minimize the threat to genomic integrity imposed by meiosis by enabling reliable CO/chiasma formation with lower levels of potentially dangerous intermediates.
Supplementary Material
Acknowledgments
We thank M. Colaiacovo, the Caenorhabditis Genetics Center (funded by National Institutes of Health, NIH, Office of Research Infrastructure Programs P40 OD010440), and the Mitani lab/C. elegans National BioResource Project (Ministry of Education, Culture, Sports, Science and Technology in Japan) for strains. We thank N. Bhalla, A. Dernburg, and V. Jantsch for antibodies. This work was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship to M.S., Katharine D. McCormick Advanced Postdoctoral Fellowship and NIH K99 award HD076165 to D.E.L., Leukemia and Lymphoma Society Postdoctoral Fellowship 5381-12 to K.A.Z., Basil O’Connor Starter Scholar Research Award Grant 5-FY07-666 from the March of Dimes Foundation to K.N., and NIH grants R01GM67268 and R01GM53804 to A.M.V. B.J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.161513/-/DC1.
Communicating editor: J. Sekelsky
Literature Cited
- Agostinho A., Meier B., Sonneville R., Jagut M., Woglar A., et al. , 2013. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi A., Pasierbek P., Gartner A., Loidl J., 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16 [DOI] [PubMed] [Google Scholar]
- Baudat F., De Massy B., 2007. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 15: 565–577 [DOI] [PubMed] [Google Scholar]
- Bessler J. B., Reddy K. C., Hayashi M., Hodgkin J., Villeneuve A. M., 2007. A role for Caenorhabditis elegans chromatin-associated protein HIM-17 in the proliferation vs. meiotic entry decision. Genetics 175: 2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Wynne D. J., Jantsch V., Dernburg A. F., 2008. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4: e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R. S., Schonberg S., German J., 1974. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71: 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Vezon D., Belcram K., Gendrot G., Grelon M., 2008. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet. 4: e1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo M. P., Macqueen A. J., Martinez-Perez E., McDonald K., Adamo A., et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474 [DOI] [PubMed] [Google Scholar]
- Darby R. A., Hine A. V., 2005. LacI-mediated sequence-specific affinity purification of plasmid DNA for therapeutic applications. FASEB J. 19: 801–803. [DOI] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., et al. , 1995. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 83: 655–666 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Serricchio A. S., Sternberg P. W., 2006. Visualization of C. elegans transgenic arrays by GFP. BMC Genet. 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack H., Hodgkin J., 1991. Tests for parental imprinting in the nematode Caenorhabditis elegans. Mol. Gen. Genet. 228: 482–485 [DOI] [PubMed] [Google Scholar]
- Hillers K. J., Villeneuve A. M., 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13: 1641–1647 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J. K., Morelli M. A., Borst P. L., Cohen P. E., 2010. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 188: 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J. K., Sun X., Yokoo R., Villeneuve A. M., Cohen P. E., 2014. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J. Cell Biol. 205: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V., Pasierbek P., Mueller M. M., Schweizer D., Jantsch M., et al. , 2004. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell. Biol. 24: 7998–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Rockmill B., Roeder G. S., Lichten M., 2006. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Kusch M., Wolf N., 1988. Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics 120: 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B., Cohen P. E., Avdievich E., Zhu L., Kane M. F., et al. , 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14: 1085–1097 [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Sekelsky J., 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda D. E., Uzawa S., Meyer B. J., Villeneuve A. M., 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A., Soucek R., Borner G. V., 2007. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15: 591–605 [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., Colaiacovo M. P., Mcdonald K., Villeneuve A. M., 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla N., Weiser P., Villeneuve A. M., et al. , 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Villeneuve A. M., 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19: 2727–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Schvarzstein M., Barroso C., Lightfoot J., Dernburg A. F., et al. , 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22: 2886–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S., Roelens B., Villeneuve A. M., 2013. Evidence that masking of synapsis imperfections counterbalances quality control to promote efficient meiosis. PLoS Genet. 9: e1003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., Hillers K. J., 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168: 1275–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., Colaiacovo M. P., 2005. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 168: 683–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Mlynarczyk-Evans S., Villeneuve A. M., 2011. Chromosome painting reveals asynaptic full alignment of homologs and HIM-8-dependent remodeling of X chromosome territories during Caenorhabditis elegans meiosis. PLoS Genet. 7: e1002231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Lao J. P., Hwang P. Y., Taylor A. F., Smith G. R., et al. , 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil N. J., Martin J. S., Youds J. L., Ward J. D., Petalcorin M. I., et al. , 2013. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789 [DOI] [PubMed] [Google Scholar]
- Penkner A. M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., et al. , 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139: 920–933 [DOI] [PubMed] [Google Scholar]
- Qiao H., Prasada Rao H. B., Yang Y., Fong J. H., Cloutier J. M., et al. , 2014. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46: 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K. C., Villeneuve A. M., 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452 [DOI] [PubMed] [Google Scholar]
- Reynolds A., Qiao H., Yang Y., Chen J. K., Jackson N., et al. , 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Fung J. C., Branda S. S., Roeder G. S., 2003. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13: 1954–1962 [DOI] [PubMed] [Google Scholar]
- Rosu S., Libuda D. E., Villeneuve A. M., 2011. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334: 1286–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S., Zawadzki K. A., Stamper E. L., Libuda D. E., Reese A. L., et al. , 2013. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 9: e1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Youds J. L., Boulton S. J., Colaiacovo M. P., 2009. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 5: e1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Lui D. Y., Kim H. M., Meyer K., Colaiacovo M. P., 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R., 2004. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Stamper E. L., Rodenbusch S. E., Rosu S., Ahringer J., Villeneuve A. M., et al. , 2013. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9: e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky C., Alpi A., Passannante M., Rose A., Gartner A., et al. , 2004. Multiple genetic pathways involving the Caenorhabditis elegans Bloom’s syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Mol. Cell. Biol. 24: 5016–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., Daryabeigi A., Adamo A., Habacher C., Machacek T., et al. , 2013. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet. 9: e1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I. D., 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K. A., Nabeshima K., Drake M., Arur S., et al. , 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen A. J., Duffy J. B., Kemphues K. J., Villeneuve A. M., 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M. C., Rose A. M., 1995. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics 141: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.