Abstract
One approach to understanding the genetic basis of speciation is to scan the genomes of recently diverged taxa to identify highly differentiated regions. The house mouse, Mus musculus, provides a useful system for the study of speciation. Three subspecies (M. m. castaneus, M. m. domesticus, and M. m. musculus) diverged ∼350 KYA, are distributed parapatrically, show varying degrees of reproductive isolation in laboratory crosses, and hybridize in nature. We sequenced the testes transcriptomes of multiple wild-derived inbred lines from each subspecies to identify highly differentiated regions of the genome, to identify genes showing high expression divergence, and to compare patterns of differentiation among subspecies that have different demographic histories and exhibit different levels of reproductive isolation. Using a sliding-window approach, we found many genomic regions with high levels of sequence differentiation in each of the pairwise comparisons among subspecies. In all comparisons, the X chromosome was more highly differentiated than the autosomes. Sequence differentiation and expression divergence were greater in the M. m. domesticus–M. m. musculus comparison than in either pairwise comparison with M. m. castaneus, which is consistent with laboratory crosses that show the greatest reproductive isolation between M. m. domesticus and M. m. musculus. Coalescent simulations suggest that differences in estimates of effective population size can account for many of the observed patterns. However, there was an excess of highly differentiated regions relative to simulated distributions under a wide range of demographic scenarios. Overlap of some highly differentiated regions with previous results from QTL mapping and hybrid zone studies points to promising candidate regions for reproductive isolation.
Keywords: Mus musculus, demography, speciation, selection, reproductive isolation, expression
UNDERSTANDING the genetic basis of speciation is a fundamental goal of evolutionary biology. This problem has primarily been approached in two ways: through laboratory studies using crosses and through studies of genetic variation in natural populations. Laboratory studies control for genetic background and environment, and they make it possible to connect genotype and phenotype. These types of studies have produced some spectacular successes including the identification of individual genes underlying postzygotic isolation in Drosophila (e.g., Ting et al. 1998; Presgraves et al. 2003; Brideau et al. 2006; Masly et al. 2006), Arabidopsis (Bomblies et al. 2007; Bikard et al. 2009), Mus (Mihola et al. 2009), and others (reviewed in Presgraves 2010 and Nosil and Schluter 2011).
Studies of natural populations rely on the idea that regions of the genome that are important in reproductive isolation may be more differentiated than other regions of the genome. Therefore, by studying patterns of differentiation, one may gain insight into the genomic regions that underlie isolation. The idea that the genomes of closely related species are mosaics of differentiated and less differentiated regions is not new and first emerged in the literature on hybrid zones (e.g., Key 1968; Harrison 1986; Tucker et al. 1992; Rieseberg et al. 1999; reviewed in Harrison 2012). The advent of genomic methods has fueled a renewed interest in studying patterns of differentiation between closely related species, including work on mosquitoes (Turner et al. 2005; Lawniczak et al. 2010; Neafsey et al. 2010), mice (Harr 2006), Drosophila (Kulathinal et al. 2009), Heliconius butterflies (Nadeau et al. 2012), flycatchers (Ellegren et al. 2012), crickets (Andrés et al. 2013), sunflowers (Renaut et al. 2013), and others.
Despite their appeal, genome scans present a number of challenges. One is correctly identifying genomic regions that show unexpectedly high levels of differentiation. This has typically been done either by specifying an appropriate null demographic model against which an observed distribution can be compared or by simply identifying extreme values as potential candidate regions. Another challenge is interpreting the biological meaning of a genomic region showing a high level of differentiation. Shared polymorphism can result from retained ancestral variation or from gene flow; conversely, differentiation can result from sorted ancestral variation (due to drift or selection) or from absence of gene flow. Charlesworth (1998) pointed out that reduced variation within a population will inflate estimates of differentiation, such as Fst, that are based on both within- and between-population components of variation. As a result, background selection (Charlesworth 1993) and genetic hitchhiking (Maynard Smith and Haigh 1974) may lead to localized high values of Fst even for regions that are not involved in reproductive isolation (Cruickshank and Hahn 2014). Therefore, genomic “islands of differentiation” may reflect (1) stochastic variation in lineage sorting; (2) regions of reduced gene flow; (3) regions in which the effects of selection at linked sites are more pronounced, regardless of involvement with reproductive isolation; or (4) some combination of these processes.
House mice provide a valuable system for the study of speciation. Mus musculus consists of three subspecies that are distributed parapatrically: M. m. domesticus in western Europe, M. m. musculus in eastern Europe and northern Asia, and M. m. castaneus in southeast Asia. These subspecies are believed to have diverged in allopatry at roughly the same time—∼350,000 years ago (Bonhomme et al. 2007; Geraldes et al. 2008, 2011; White et al. 2009)—and come into secondary contact much more recently (e.g., Cucchi et al. 2005; Duvaux et al. 2011). Each subspecies meets and hybridizes with the other two species where their ranges come into contact (e.g., Tucker et al. 1992; Boursot et al. 1993; Duvaux et al. 2011), although only the hybrid zone between M. m. musculus and M. m. domesticus is well-studied. Differences in estimated Ne among the subspecies provide an opportunity to investigate the effects of demography on patterns of differentiation. While estimates of effective population size (Ne) are large for M. m. castaneus (200,000–733,000), estimates are smaller for M. m. domesticus (58,000–200,000) and M. m. musculus (25,000–120,000; Salcedo et al. 2007; Geraldes et al. 2008, 2011; Halligan et al. 2010).
The degree of reproductive isolation differs in pairwise comparisons among house mouse subspecies. While significant reductions of F1 male fertility are seen in crosses between M. m. domesticus and M. m. musculus (e.g., Good et al. 2008; White et al. 2011), significant infertility is not observed until the F2 in crosses between M. m. castaneus and M. m. domesticus, and the degree of infertility is not as severe (White et al. 2012). There are no published studies documenting reduced fertility in lab crosses between M. m. castaneus and M. m. musculus. In fact, a cross between M. m. castaneus and M. m. musculus was used for a recombination mapping study and infertility was not observed (Dumont and Payseur 2011).
In this study, we used short-read sequencing of the testis transcriptomes of wild-derived inbred lines of M. m. castaneus, M. m. domesticus, and M. m. musculus to characterize genome-wide patterns of sequence differentiation in pairwise comparisons between each subspecies. Although this study was primarily designed to investigate sequence differentiation, we also investigated patterns of differential gene expression among the subspecies.
Materials and Methods
Samples
All mice came from wild-derived inbred strains (Supporting Information, Table S1). We sequenced eight lines of M. m. castaneus, seven of M. m. domesticus, and eight of M. m. musculus. Wild-derived laboratory strains of Mus spretus and Mus caroli were included for use as outgroups (She et al. 1990; Suzuki et al. 2004; Tucker et al. 2005). Mice were killed and testes were dissected under RNAse-free conditions. Testes samples were kindly provided by François Bonhomme, Polly Campbell, Courtney Clayton, Matt Dean, and Annie Orth. Testes were placed in RNAlater at 4° overnight and then transferred to −80° for storage. RNA was extracted from frozen tissue using Quiagen’s RNAeasy Plus Mini Kit.
Sequencing
Single-end 76-bp reads were sequenced from the mRNA of each individual on an Illumina GAIIx. For most lines, between 0.80 and 1.68 Gb of sequence was obtained (Table S2). One wild-derived inbred line of M. musculus and two wild-derived lines selected from outgroup taxa were sequenced at higher coverage (1.92–3.64 Gb). Reads containing <20 high-quality bases (phred ≥ 20) were removed prior to mapping. Sequence data can be accessed via National Center for Biotechnology Information BioProject PRJNA252743. TopHatv1.2 (Trapnell et al. 2009) was used with default settings to map reads to the C57BL/6 reference genome, and only reads that mapped uniquely were retained. Finally, sites with a depth <6× of high-quality sequence (phred ≥ 20) were removed from the analysis. These filters left between 12.01 and 22.70 Mb of sequence per line. Genomic sequence data (>20×) were available from the Wellcome Trust Mouse Genomes Project for two of the lines included in our study (SPRET/EiJ and PWK/PhJ; Yalcin et al. 2012), and genomic sequence data were used to augment transcriptome sequencing. We compared genotype calls between our data and the Wellcome Trust data in regions of overlap (Table S3). Although both data sets were obtained via shotgun sequencing, the higher coverage of the Wellcome Trust data allows for an assessment of the possible risks of sequencing and mapping using our lower-coverage approach. Mismatches were rare, occurring at rates ranging from 1 in ∼325,000 to 1 in ∼420,000 coding sites (Table S3). In addition, we included data from two lines (CAST/EiJ and WSB/Eij) sequenced only by the Wellcome Trust Mouse Genomes Project (Yalcin et al. 2012).
Previous analyses have shown that wild-derived inbred lines can contain large introgressed segments from other subspecies (Yang et al. 2011). STRUCTURE analysis (Pritchard et al. 2000) was used to test for admixture in the wild-derived inbred lines sequenced in this study. We found that two lines of M. m. castaneus and one line of M. m. musculus were highly admixed, and we excluded them from the study (Table S1). After excluding these lines, the remaining subspecies formed three distinct groups corresponding to the three subspecies, and each line was assigned with most support to the expected subspecies (Table S4). After removing the admixed lines and including the lines sequenced by the Wellcome Trust, six M. m. castaneus, eight M. m. domesticus, and seven M. m. musculus were used in all analyses.
SAMtools was used with default settings to call bases and all SNPs within and among subspecies were identified using custom PERL scripts (Li et al. 2009; File S1). Inbred lines are expected to be homozygous. Observed heterozygosity may reflect true residual heterozygosity in inbred lines or errors in sequencing; distinguishing between these two possibilities is difficult with low-coverage data. When heterozygosity was inferred using SAMtools, the site was masked and not included in further analyses. In addition, indels and sites with more than two segregating alleles were excluded. After filtering, we identified >32,000 SNPs within and among subspecies of M. m. musculus from 4705 genes with an average of 6.12 SNPs per gene. This represents ∼20% of the genes in the genome.
Measures of sequence differentiation
There are many statistics for measuring differentiation, and these capture different aspects of the data. Here, we calculated Fst (Hudson et al. 1992), Dxy (Nei 1987), and δ, the absolute value of the difference in allele frequencies (see Renaut et al. 2010; Gagnaire et al. 2012; equations given in File S2) for each SNP for each pairwise comparison: M. m. castaneus–M. m. domesticus (hereafter CD), M. m. castaneus–M. m. musculus (hereafter CM), and M. m. domesticus–M. m. musculus (hereafter DM). To account for unequal sample sizes of the subspecies, we subsampled five lines per subspecies 10,000 times at each site with sufficient data, and the average value of a given statistic was used for all subsequent analyses. Measures of differentiation were highly correlated in our data set (Table 1); thus we chose to use δ for subsequent analyses, although similar results were obtained using other measures. We defined SNPs as highly differentiated if average resampled values of δ per site were ≥0.8. In such cases, the two subspecies are one allele or fewer from fixation of alternate nucleotides. We defined highly differentiated as δ ≥ 0.8 rather than using an approach based on the distributions of statistics (e.g., the upper 5% of values) because it allowed us to compare among the three pairwise analyses. We then asked how many sites were fixed in a single subspecies but polymorphic in both of the other two subspecies. Finally, we identified all fixed, derived sites in each subspecies using comparisons to the outgroup taxa, M. spretus and M. caroli.
Table 1. Summary statistics describing patterns of differentiation at all SNPs in pairwise comparisons of the subspecies of M. musculus.
| Subspecies | Subspecies | Chromosome | No. of SNPs | (SD) | (SD) | (SD) | % fixed differences | % private polymorphisms | % shared polymorphisms | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. m. castaneus | M. m. domesticus | Autosomes | 24,136 | 0.22 (0.32) | 0.41 (0.28) | 0.40 (0.29) | 0.99 | 0.98 | 0.98 | 9.14 | 83.66 | 7.20 |
| X | 226 | 0.32 (0.41) | 0.53 (0.34) | 0.53 (0.35) | 0.99 | 0.99 | 1.00 | 23.89 | 73.91 | 2.21 | ||
| M. m. castaneus | M. m. musculus | Autosomes | 23,709 | 0.26 (0.38) | 0.44 (0.30) | 0.43 (0.31) | 0.97 | 0.98 | 0.99 | 12.50 | 81.55 | 5.95 |
| X | 237 | 0.35 (0.39) | 0.54 (0.33) | 0.53 (0.33) | 0.97 | 0.98 | 0.99 | 18.14 | 73.00 | 8.86 | ||
| M. m. domesticus | M. m. musculus | Autosomes | 21,598 | 0.38 (0.41) | 0.53 (0.35) | 0.52 (0.35) | 0.98 | 0.99 | 0.99 | 24.01 | 70.96 | 5.03 |
| X | 246 | 0.46 (0.44) | 0.57 (0.38) | 0.57 (0.38) | 0.99 | 0.99 | 1.00 | 30.08 | 68.29 | 1.63 |
Sliding windows
To identify genomic regions with groups of sites that are highly differentiated, we performed two kinds of sliding-window analyses. First, sliding-window analyses of δ (100-kb windows with a 25-kb step size) were used to identify regions of the genome that were highly differentiated among subspecies. We defined regions as highly differentiated if average values of δ were ≥ 0.8 across all sites in a window. All SNPs were included in these analyses, and windows were evaluated only when there were three or more SNPs in the window.
Private polymorphisms (i.e., those segregating in just one species) can lower the average level of differentiation in a region as measured by Fst, Dxy, and δ. The presence of private polymorphisms does not mean that such regions are not potentially relevant to speciation, only that they are less likely, on average, to have experienced recent coalescent events. Analyses that do not distinguish between shared and private polymorphisms may fail to identify many regions that are fully sorted. To address this problem, we tracked the ratio of fixed differences to shared polymorphisms plus fixed differences using 100-kb windows with a 25-kb step size. This ratio can take on values between zero and one and is defined for all regions that contain an informative site. High values indicate reciprocally monophyletic gene genealogies while low values indicate populations that harbor ancestral polymorphism or are experiencing gene flow (e.g., Carneiro et al. 2013). For this window analysis, we included only windows with at least three topologically informative SNPs.
In both sliding-window approaches, regions were delimited by joining overlapping or adjacent windows with the same classification, and overall levels of diversity and differentiation were estimated by averaging across all SNPs in a delimited region. The average number of SNPs in these regions is given in Table 2. We also adopted a third approach to defining regions of differentiation by delimiting runs of fixed differences uninterrupted by shared polymorphisms. The results of these analyses were very similar to the two sliding-window analyses and are given in File S2, Figure S1, Table S5, and Table S6.
Table 2. Summary statistics describing regions identified as highly differentiated with a sliding-window analysis vs. all other regions in pairwise comparisons of the subspecies of M. musculus.
| Subspecies | Subspecies | Chromosome | Window typea | nb | Average size of region (bp)c (SD) | Average no. of SNPs (SD) | (SD) | (SD) | (SD) | (SD) | (SD) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. m. castaneus | M. m. domesticus | Autosomes | Highly differentiated | 63 | 133,333* (37,567) | 5.13* (2.85) | 0.80* (0.10) | 0.87* (0.06) | 0.87* (0.06) | 0.11* (0.12) | 0.05* (0.07) |
| Autosomes | All others | 1651 | 235,933* (122,354) | 13.74* (13.67) | 0.21* (0.14) | 0.40* (0.12) | 0.39* (0.13) | 0.26* (0.11) | 0.16* (0.09) | ||
| X | Highly differentiated | 5 | 160,000 (33,541) | 4.60 (2.30) | 0.84 (0.15) | 0.90 (0.09) | 0.90 (0.09) | 0.11 (0.15) | 0.03 (0.04) | ||
| X | All others | 27 | 169,444 (37,553) | 5.11 (2.87) | 0.31 (0.23) | 0.45 (0.20) | 0.45 (0.20) | 0.19 (0.17) | 0.13 (0.11) | ||
| All | All | 1746 | 230,985 (121,119) | 13.27 (13.45) | 0.24 (0.18) | 0.42 (0.15) | 0.41 (0.16) | 0.25 (0.12) | 0.15 (0.10) | ||
| M. m. castaneus | M. m. musculus | Autosomes | Highly differentiated | 105 | 129,762* (35,878) | 5.31* (2.84) | 0.80* (0.09) | 0.87* (0.06) | 0.87* (0.06) | 0.11* (0.10) | 0.05* (0.07) |
| Autosomes | All others | 1625 | 229,708* (114,947) | 13.55* (13.46) | 0.24* (0.15) | 0.43* (0.12) | 0.41* (0.13) | 0.27* (0.11) | 0.14* (0.11) | ||
| X | Highly differentiated | 6 | 154,167 (33,229) | 3.67 (1.03) | 0.86 (0.17) | 0.92 (0.09) | 0.92 (0.09) | 0.03 (0.08) | 0.12 (0.14) | ||
| X | All others | 26 | 168,269 (43,335) | 5.38 (3.54) | 0.32 (0.18) | 0.49 (0.14) | 0.48 (0.15) | 0.21 (0.17) | 0.16 (0.12) | ||
| All | All | 1762 | 222,588 (113,626) | 12.90 (13.14) | 0.28 (0.20) | 0.46 (0.16) | 0.45 (0.17) | 0.25 (0.11) | 0.13 (0.11) | ||
| M. m. domesticus | M. m. musculus | Autosomes | Highly differentiated | 287 | 135,279* (38,250) | 5.98* (3.62) | 0.81* (0.09) | 0.87* (0.06) | 0.87* (0.06) | 0.07* (0.07) | 0.07* (0.09) |
| Autosomes | All others | 1561 | 216,944* (111,486) | 11.99* (11.47) | 0.34* (0.17) | 0.49* (0.14) | 0.48* (0.14) | 0.18* (0.10) | 0.15* (0.11) | ||
| X | Highly differentiated | 4 | 162,500 (25,000) | 4.75 (0.50) | 0.90 (0.09) | 0.93 (0.07) | 0.93 (0.07) | 0.04 (0.04) | 0.05 (0.09) | ||
| X | All others | 33 | 170,455 (36,150) | 4.97 (2.58) | 0.38 (0.22) | 0.50 (0.19) | 0.50 (0.19) | 0.11 (0.09) | 0.15 (0.11) | ||
| All | All | 1885 | 203,581 (106,856) | 10.94 (10.79) | 0.41 (0.23) | 0.55 (0.19) | 0.54 (0.19) | 0.16 (0.10) | 0.14 (0.11) |
P < 10−10 in two-sided t-tests comparing highly differentiated autosomal regions and all other autosomal regions in each pairwise comparison.
Highly differentiated regions defined as average δ ≥ 0.8.
Number of delimited regions.
Regions were delimited by joining overlapping or contiguous windows with the same classification. The resolution of individual windows is limited to the sliding-window increment of 25,000.
and refer to nucleotide diversity (Nei and Li 1979) in the first and second subspecies, respectively.
Demographic simulations
We used coalescent simulations (Hudson 2002) to compare observed patterns of differentiation to those expected under different demographic scenarios (Table S7). Parameters in these models included divergence time, current and ancestral Ne, and migration rates in each direction and were based on maximum-likelihood estimates obtained using the program Isolation with Migration (IM) (Nielsen and Wakeley 2001) in a previous study (Geraldes et al. 2011). We assumed no recombination within loci and free recombination among loci. This assumption is reasonable given that linkage disequilibrium decays over distances of 10–50 kb in house mice (Laurie et al. 2007). For each pairwise split, we simulated 100,000 gene genealogies, given five chromosomes from each subspecies, and assumed a scaled θ value of 1.33 based on estimates of the mutation rate (4 × 10−9; Waterston et al. 2002), ancestral population size, and the approximate average number of sites surveyed per locus. Because the program ms (Hudson 2002) simulates individual loci, we then compared the distribution of δ from the simulations to the observed measures across individual genes in our data set. We also explored a wider range of demographic parameters to better match simulated distributions to observed values (see Results).
Recombination and inversions
Regions of low recombination are expected to be more highly differentiated than other regions of the genome (Noor et al. 2001; Rieseberg 2001; Nachman and Payseur 2012). For example, a recent study of sunflowers showed that regions of greater differentiation were strongly associated with reduced recombination (Renaut et al. 2013). We used the revised genetic map to estimate the recombination rate for 5-Mb intervals of the mouse genome (Shifman et al. 2006; Cox et al. 2009). We defined low-recombination regions as intervals with recombination rates falling in the bottom 10% of the genome and high-recombination regions as intervals falling in the top 10% of the genome. We then asked whether levels of differentiation differed between regions of low and high recombination. One limitation of this approach is that the genetic map derives from M. m. domesticus and there is some evidence that recombination rate varies among subspecies (Dumont and Payseur 2011; Dumont et al. 2011). This likely limits the power to detect differences if they exist. To compare these results with those from a previous study (Geraldes et al. 2011), we repeated the analyses estimating recombination rates over 10-Mb intervals. We also repeated the analyses defining high- and low-recombination regions as those falling in the upper or lower 5, 15, and 20% of the distribution of recombination rate.
Inversions may suppress recombination. Inversion data for M. m. castaneus and M. m. musculus relative to the reference mouse genome (C57BL/6) are available from the Wellcome Trust (Yalcin et al. 2012). C57BL/6 is primarily of M. m. domesticus origin but contains small introgressions from other subspecies. We used the Mouse Phylogeny Viewer (Wang et al. 2012) to eliminate regions not of M. m. domesticus origin from the inversion data for M. m. castaneus and M. m. musculus. The location of inversions between M. m. castaneus and M. m. musculus was determined by identifying and removing inversions in both lines that overlap and therefore represent inversions relative to M. m. domesticus. Many inversions were identified between each pair of subspecies, but most were small (CD: , range = 99–19,005, n = 398; CM: , range = 63–19,752, n = 620; DM: , range = 63–23,239, n = 479). Therefore, variant SNPs in inversions were rare in our data set. However, many runs of fixed differences spanned inversions. To investigate the relationship between inversions and differentiation, we asked whether runs of fixed differences were more likely to overlap with inversions than expected by chance. To determine the expected overlap, we randomly generated the same number of regions across the genome sampled with replacement from the same size distribution as the runs data and determined the overlap with inversions. We did this 10,000 times and determined the percentile rank of the observed data.
Gene expression differences
The primary motivation for generating transcriptome data was to provide a set of common loci at which patterns of sequence differentiation could be analyzed. Nonetheless, these data also provide an opportunity to study gene expression differences and to compare expression divergence with sequence differentiation.
All mice were unmated, reproductively mature males, but they differed in age. In addition, M. m. domesticus individuals were reared in one facility while M. m. castaneus and M. m. musculus individuals were reared in another. We used two approaches to assess whether differences in rearing conditions might bias expression analysis. First, we calculated the average count of transcripts mapped for each gene in each subspecies correcting for differences in sequencing effort. Pairwise comparisons between subspecies showed that expression patterns were highly correlated (> 0.99, d.f. = 15,123, P < 10−15; > 0.99, d.f. = 15,123, P < 10−15;> 0.99, d.f. = 15,123, P < 10−15). Second, we compared our results to those of another study on gene expression in M. m. domesticus and M. m. musculus (M. Nachman, unpublished data). In that study, testis transcriptomes were sequenced for three individuals of one inbred line of M. m. domesticus (LEWES) and three individuals of one inbred line of M. m. musculus (PWK). All mice were unmated, reproductively mature males of the same age, and all were housed in the same room of a single animal care facility. Average mean counts of transcripts mapped per gene for each subspecies after normalization were highly correlated in the two data sets ( = 0.98, d.f. = 11,671, P < 10−15; = 0.98, d.f. = 11,671, P < 10−15). In addition, the log2fold change in expression for each gene between the two subspecies was significantly correlated between the two studies (= 0.71, d.f. = 11,671, P < 10−15). Although the power of the two studies differs due to design, the majority of genes (∼80%) identified in the smaller study as having significantly different expression between the two subspecies after correction for multiple testing (α = 0.01) were identified as significantly differently expressed in this study after correction for multiple testing given a less conservative cutoff (α = 0.05). These analyses suggest that expression patterns in this study were not strongly biased by differences in rearing conditions.
We identified genes that were differentially expressed in each pairwise comparison of subspecies. Given results from TopHat (see above), HTseq (Anders et al. 2014) was used to create tables of counts of reads mapped for all represented genes. All genes with an average read count of ≤10 in more than one subspecies were removed from the analysis. The DESeq package in R (Anders and Huber 2010) was used to further filter the data and identify genes with significant differential expression using a binomial test. We first normalized counts to account for differences in sequencing effort among individuals. We then filtered out the bottom 20% of the data based on sums of the counts across all subspecies. This left 12,098 genes with sufficient data for analysis. We estimated dispersions for each subspecies and then used a binomial test to identify differentially expressed genes between each pair of subspecies. P-values were adjusted via a Benjamini–Hochberg correction with a false discovery rate of 0.01 (Benjamini and Hochberg 1995). Sites were filtered in each pairwise test if the average normalized read count was ≤10 across both subspecies.
We estimated the correlation between measures of sequence differentiation on a gene-by-gene basis and the absolute value of the log2fold change in expression for each pair of subspecies. Sequence differentiation and expression differentiation might be correlated, particularly if differences in expression are due to sequence changes at or near the gene itself (i.e., cis-regulatory changes). In another study, patterns of allele-specific expression in the testes of F1 hybrids of M. m. domesticus and M. m. musculus were used to identify genes in which differences in expression between the two subspecies were due to cis-regulatory changes (M. Nachman, unpublished data) as in Wittkopp et al. 2004. We used those data and repeated the correlation analysis in the DM comparison, restricting it to genes identified as having cis-regulatory changes. Similar data were not available for the other two pairwise comparisons.
Testis-specific expression
Genes involved in reproduction are known to evolve quickly (e.g., Begun et al. 2000; Wyckoff et al. 2000; Good and Nachman 2005), and genes that are tissue-specific have higher rates of evolution than others in mammals (Duret and Mouchiroud 2000). We asked whether regions containing testis-specific genes were more highly differentiated than other regions in the window analysis based on all SNPs. Genes with testis-specific expression were identified using data from Su et al. (2004) available via BioGPS (Wu et al. 2009). Expression data were reduced to those tissues with support for independent expression, and expression values were averaged over the available measurements for a given tissue (Winter et al. 2004). Genes were considered testis-specific if the proportion of total expression in the testis compared to overall expression was ≥ 0.1 (Winter et al. 2004; File S3). Measures of differentiation for all regions containing testis-specific genes were then compared to measures for all other regions using one-sided t-tests.
Identifying candidate regions for reproductive isolation
One approach to identifying candidate genes for reproductive incompatibilities is to look for overlap between the results of genomic scans and other methods such as QTL mapping studies and cline analyses in hybrid zones. There are many reasons to expect that the results of such studies will not overlap: QTL analyses focus on specific traits, hybrid zone data may track more recent processes, and genomic scans of differentiation will identify many regions that do not contribute to reproductive isolation. Nevertheless, intervals that are identified consistently across different methods are good candidates for additional study.
There are no published QTL mapping data of traits relevant to reproductive isolation for crosses between M. m. castaneus and M. m. musculus, nor are there any detailed hybrid zone studies between these taxa. However, there are published QTL mapping results for the other two pairwise comparisons (White et al. 2011, 2012) and many studies of the hybrid zone between M. m. domesticus and M. m. musculus (e.g., Vanlerberghe et al. 1986, 1988; Tucker et al. 1992; Prager et al. 1993; Munclinger et al. 2002; Macholan et al. 2007; Teeter et al. 2008, 2010; Janoušek et al. 2012). For QTL, we identified overlap between 1.5-LOD intervals associated with sterility phenotypes and highly differentiated regions in the window analysis based on all SNPs (White et al. 2011, 2012). We combined QTL into a single region for analysis if the QTL 1.5-LOD intervals were overlapping. For comparison with patterns in the musculus–domesticus hybrid zone, we used the study by Janoušek et al. (2012) in which candidate Bateson–Dobzhansky–Muller incompatibility (BDMI) loci were identified from patterns of introgression and epistasis in two different transects. We identified overlap between a 2-MB window centered on the candidate BDMI loci of Janoušek et al. (2012) and highly differentiated regions in the window analysis based on all SNPs. For both types of comparisons, we compared the observed overlap to a distribution created using 10,000 simulated data sets of genomic regions from the same size distribution as those identified as highly differentiated in our study. We repeated all analyses identifying overlap between genes that were differentially expressed in our data and the results of previous studies. For these analyses, we compared the observed overlap to a distribution created using 10,000 simulated data sets. Simulations were conducted by sampling with replacement from among those genes for which there were expression data.
We used the coordinates of all SNPs in the Ensembl mouse genome assembly GRCm38 to identify genes found in overlapping regions, and we used the Mouse Genome Database to identify phenotypes in laboratory lines associated with mutations in these genes (Eppig et al. 2012). When highly differentiated regions were flanked by regions with fewer than three SNPs, we expanded the query regions to the next region with data or 2 MB, whichever was smaller. We did not include regions from the X chromosome as the QTL associated with male infertility in both crosses encompassed most of the chromosome.
Results
Measures of sequence differentiation
Over 20,000 SNPs were segregating in each pairwise comparison, but many were private, segregating at low-to-moderate frequency within a single subspecies in a given comparison (Table 1). Different measures of differentiation (Fst, Dxy, and δ) were highly correlated (Table 1). Average levels of differentiation varied among pairwise comparisons, and all measures of differentiation were consistently higher in DM than in either of the other two pairwise comparisons. In addition, all measures of differentiation were higher on the X chromosome than on the autosomes (Table 1). Nonsynonymous sites showed slightly lower levels of differentiation than synonymous sites in each pairwise comparison (Table S8).
There were 9529 individual SNPs that were highly differentiated (δ ≥ 0.80) in at least one of the three pairwise comparisons (CD: 4223 from 1970 genes; CM: 5338 from 2343 genes; DM: 7423 from 2783 genes). Fewer SNPs were fixed in M. m. castaneus but segregating in both of the other lines (744 from 437 genes) than in either M. m. domesticus (1084 from 651 genes) or M. m. musculus (1396 from 881 genes). In addition, many fewer sites represented derived states relative to the other subspecies and both outgroups in M. m. castaneus (372 from 263 genes) than in either M. m. domesticus (770 from 554 genes) or M. m. musculus (1570 from 1099 genes).
Sliding-window analyses
The first sliding-window approach (based on average values of δ) included ∼385–403 Mb in each pairwise comparison after filtering (∼15% of the genome). We identified many windows with an average value of δ ≥ 0.80 in each pairwise comparison between subspecies (Figure 1 and Table 2). Highly differentiated regions were characterized by higher measures of Fst and Dxy and lower measures of within-subspecies variation than other regions (Table 2). Strikingly, regions of high differentiation represent a much larger part of the total surveyed transcriptome in the DM comparison than in either of the other two comparisons (Table 2). In each pairwise comparison, >65% of genes sampled in highly differentiated regions contained at least one fixed difference. Approximately half of those genes contained at least one nonsynonymous fixed difference (CD: 57.4% of genes; CM: 46.6% of genes; DM: 42.4% of genes). Although this implies that approximately half of these genes have no nonsynonymous fixed differences, it is important to bear in mind that coverage was incomplete for many genes and thus some nonsynonymous changes may have been missed.
Figure 1.
Sliding-window analysis showing average values of δ throughout all chromosomes for each pairwise subspecies comparison (CD, M. m. castaneus vs. M. m. domesticus; CM, M. m. castaneus vs. M. m. musculus; DM, M. m. domesticus vs. M. m. musculus). Each dot marks the start of a delimited region, and red dots represent regions for which the average value of δ is ≥ 0.8.
The second sliding-window approach (based on the ratio of fixed differences to shared polymorphisms plus fixed differences), after filtering, included ∼89–146 Mb in each pairwise comparison. This analysis required at least three topologically informative SNPs per window and thus covered much less of the genome than the first sliding-window approach. We identified many fully sorted windows in each pairwise comparison (Table 3; Figure S2, A–C). Even when including private polymorphisms, autosomal regions that were fully sorted were characterized by higher measures of Fst, Dxy, and δ and by lower measures of within-subspecies variation than other regions (Table 3). Notably, all regions on the X chromosome were fully sorted in all pairwise comparisons. Fully sorted regions represent a much larger part of the total surveyed transcriptome in the DM comparison than in either of the other two comparisons (Table 3). As expected, a much higher proportion of the surveyed genome was identified as fully sorted in this analysis than was identified as highly differentiated in the analysis averaging across all variable sites in a window.
Table 3. Summary statistics describing patterns of differentiation in fully sorted regions vs. all other regions in pairwise comparisons of the subspecies of M. musculus.
| Subspecies | Subspecies | Chromosome type | Window typea | nb | Average size of region (bp) (SD) | Average no. of SNPs (SD) | (SD) | (SD) | (SD) | (SD) | (SD) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. m. castaneus | M. m. domesticus | Autosomes | Fully sorted | 223 | 159,193 (38,840) | 15.05** (9.17) | 0.48*** (0.16) | 0.59*** (0.13) | 0.59*** (0.13) | 0.15*** (0.10) | 0.11*** (0.07) |
| Autosomes | All others | 321 | 157,243 (46,855) | 19.36** (12.56) | 0.20*** (0.13) | 0.43 *** (0.10) | 0.39*** (0.11) | 0.29*** (0.09) | 0.20*** (0.09) | ||
| X | Fully sorted | 5 | 175,000 (0) | 5.20 (1.92) | 0.80 (0.12) | 0.85 (0.09) | 0.85 (0.09) | 0.06 (0.06) | 0.05 (0.04) | ||
| M. m. castaneus | M. m. musculus | Autosomes | Fully sorted | 317 | 172,003** (52,573) | 16.54 (10.86) | 0.47*** (0.17) | 0.60*** (0.13) | 0.60*** (0.13) | 0.20*** (0.10) | 0.07*** (0.06) |
| Autosomes | All others | 287 | 154,355** (52,334) | 18.20 (12.42) | 0.19*** (0.14) | 0.42*** (0.11) | 0.38*** (0.13) | 0.28*** (0.09) | 0.21*** (0.10) | ||
| X | Fully sorted | 3 | 175,000 (0) | 3.67 (1.15) | 1.00 (0.00) | 1.00 (0.00) | 1.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | ||
| M. m. domesticus | M. m. musculus | Autosomes | Fully sorted | 624 | 178,766 *** (55,386) | 13.93 (9.50) | 0.57*** (0.17) | 0.66*** (0.13) | 0.66*** (0.13) | 0.12*** (0.07) | 0.08*** (0.06) |
| Autosomes | All others | 234 | 140,171*** (51,005) | 14.50 (10.45) | 0.27*** (0.16) | 0.48*** (0.12) | 0.44*** (0.14) | 0.22*** (0.09) | 0.23*** (0.10) | ||
| X | Fully sorted | 10 | 16,000 (24,152) | 5.50 (1.27) | 0.70 (0.18) | 0.76 (0.15) | 0.76 (0.15) | 0.07 (0.06) | 0.05 (0.06) |
P < 0.05, ** P < 10−3, *** P < 10−7 in two-sided t-tests comparing highly differentiated autosomal regions and all other autosomal regions in each pairwise comparison.
Fully sorted regions are defined as those in which: (see Materials and Methods).
Number of delimited regions.
and refer to nucleotide diversity (Nei and Li 1979) in the first and second subspecies, respectively.
Demographic simulations
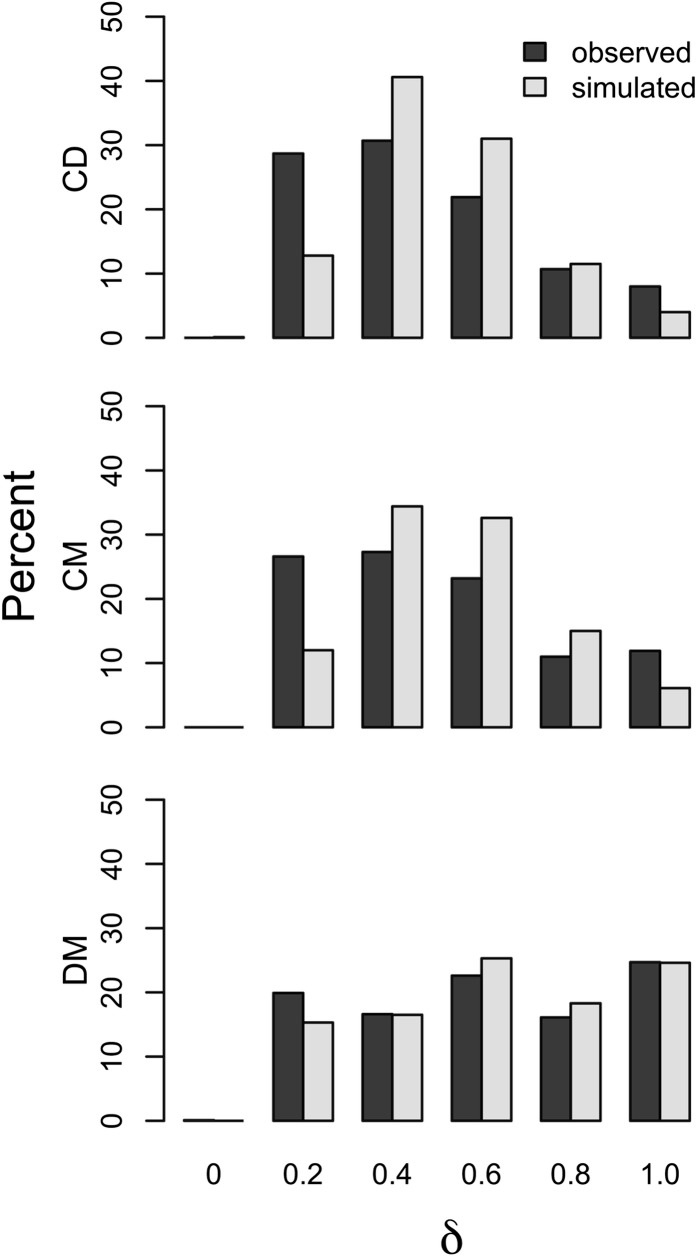
We conducted coalescent simulations based on demographic parameters estimated in a previous study (Geraldes et al. 2011; Table S7). The simulations predicted the greatest overall levels of differentiation in the DM comparison and the lowest overall levels in the CD comparison (Figure 2), a pattern consistent with the observed data. However, for all three pairwise comparisons, the observed distribution of δ was flatter than the simulated distribution, resulting from a larger-than-predicted proportion of genes with extreme values. Differences between the observed and the simulated distributions were significant in all pairwise comparisons (Kolmogorov–Smirnov test; CD: D2-sided = 0.23, P < 1 × 10−10; CM: D2-sided = 0.21, P < 1 × 10−10; DM: D2-sided = 0.19, P < 1 × 10−10). This pattern was most pronounced in the CD and CM comparisons. An excess of genes with low values of differentiation is consistent with higher-than-simulated rates of gene flow, whereas an excess of genes with high levels of differentiation is consistent with longer-than-simulated divergence times, lineage-specific positive selection, and/or barriers to gene flow. However, on average, the simulated loci had more variable sites than were surveyed in the observed data (Table S7). We repeated the simulations choosing values of current and ancestral Ne and divergence time to try to match more closely the number of SNPs in the simulated and observed data (Table S9). Overall patterns were similar under both demographic scenarios (Figure S3 and Figure S4) with more loci falling in the extremes of the distribution than expected based on the simulations.
Figure 2.
The distribution of δ values in the observed data and in the simulated data based on demographic parameters from Geraldes et al. (2011).
We further explored the simulation parameter space to determine if increasing gene flow and divergence times could generate patterns similar to those observed in the data. We started with the original parameter values (Table S7) but with a common divergence time of 325 KYA. We then explored different values of gene flow for each pairwise comparison until the proportion of genes with low values of differentiation (0 < δ ≤ 0.2) in the simulations matched the proportion observed in the data. The levels of gene flow required were high, ranging from 7 to 15 times the values originally simulated (Table S10). In all cases, increasing gene flow to the level required resulted in an even larger excess of highly differentiated genes in the observed data relative to the simulations than under the original demographic scenario (Figure S4). Next, we tested whether increasing divergence times in tandem with gene flow could result in a distribution more similar to the one observed. We increased the divergence time first to 425 KYA and then to 825 KYA. Increasing divergence time had little effect on the proportion of genes falling in the extreme tails of the distribution for both the CD and CM comparison (Figure S5 and Figure S6). Increasing divergence times given such high levels of gene flow tended to increase the proportion of simulations for which the average value of δ was low with little effect on the proportion of highly differentiated loci. In the DM comparison, increasing divergence time did increase the proportion of the distribution that was highly differentiated to levels close to or exceeding those observed (Figure S5 and Figure S6). However, in both simulations with older divergence times, the proportion of simulated loci with low values of differentiation was much smaller than observed. We did not exhaustively explore the effects of uncertainty in estimates of effective population size, recombination rate, or mutation rate, any of which might affect the expected distribution of differentiation. Nonetheless, taken together, the simulations suggest that differences in demography can account for some of the observed patterns, such as increased differentiation in the DM comparison, but also that some regions of high differentiation result from either lineage-specific positive selection or barriers to gene flow.
Recombination and inversions
Levels of differentiation were generally higher in regions of low recombination than in regions of high recombination, but the difference was significant only in the CM comparison. Results were similar among different measures of differentiation; we report results for δ (Table 4). Repeating the analysis with 10-Mb windows or with different cutoff values for high- and low-recombination regions yielded qualitatively similar results (data not shown). We found no evidence of greater-than-expected overlap between inversions and runs of fixed differences in the CM and CD comparisons, but we did find significant overlap in the DM comparison, with the observed overlap falling in the extreme tail of the simulated distribution (P = 0.015; Table S11)
Table 4. Average sequence differentiation in regions of low and high recombination.
| Subspecies | Subspecies | (SD) | (SD) | t | P1-tailed | n |
|---|---|---|---|---|---|---|
| M. m. castaneus | M. m. domesticus | 0.29 (0.12) | 0.28 (0.06) | 0.22 | 0.83 | 88 |
| M. m. castaneus | M. m. musculus | 0.36 (0.10) | 0.31 (0.05) | 2.72 | <0.01 | 88 |
| M. m. domesticus | M. m. musculus | 0.38 (0.13) | 0.35 (0.07) | 1.17 | 0.12 | 88 |
Gene expression differences
We identified many more significantly differentially expressed genes in the DM comparison than in either of the other two comparisons (CD: 594 of 12,078; CM: 685 of 12,078; DM: 1049 of 12,081; File S4). Average δ per gene and log2fold change in expression were significantly positively correlated in all pairwise comparisons of subspecies, although the correlation coefficients were small (CD: = 0.05, d.f. = 2483, P = 0.01; CM: = 0.06, d.f. = 2471 P = 0.001; DM: = 0.05, d.f. = 2356, P = 0.02). Results were very similar for other measures of differentiation (data not shown). Restricting the data to genes identified in another study as being significantly differentially transcribed due to cis-regulatory changes (M. Nachman, unpublished data) strengthened the correlation in the DM comparison although the significance was reduced given less power (= 0.09, d.f. = 439, P = 0.05).
Testis-specific expression
Approximately 30% of regions surveyed in the analyses including all SNPs contained at least one testis-specific gene (Table S12). Overall, testis-specific genes were significantly more common in highly differentiated regions than in other regions ( = 3.74, n = 9822, P1-tailed = 0.03). Regions containing testis-specific genes were more differentiated than other regions in the CM and DM comparisons, but these differences were consistently significant only in the DM comparison (Table S12).
Identifying candidate regions for reproductive isolation between M. m. castaneus and M. m. domesticus
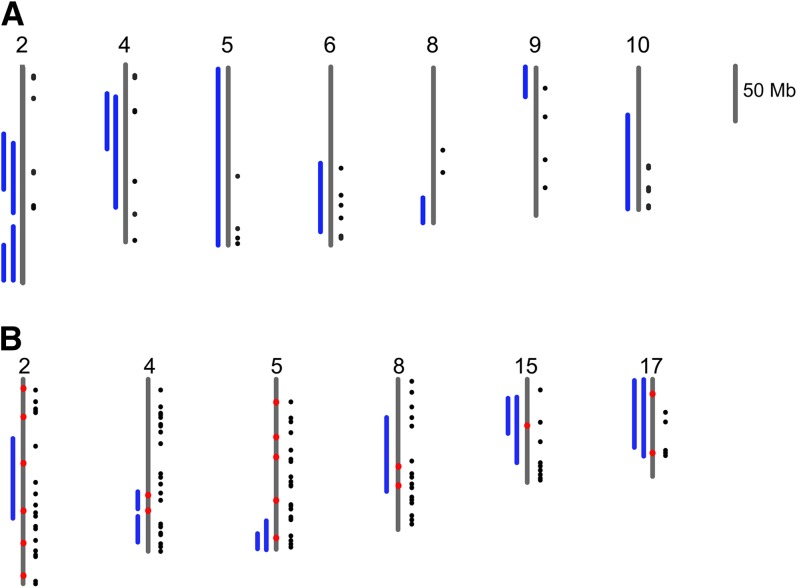
In comparisons between M. m. castaneus and M. m. domesticus, we did not observe significant overlap between differentially expressed genes and QTL associated with hybrid male infertility. However, we did observe significant overlap between highly differentiated regions and QTL (White et al. 2012; Figure 3A). Of the nine QTL intervals, seven contained peaks of high differentiation, and the observed overlap ranked in the 97th percentile of the simulated distribution.
Figure 3.
Candidate regions for reproductive incompatibilities. (A) Overlap between autosomal regions identified as QTL associated with male sterility in a cross between M. m. castaneus and M. m. domesticus (blue bars) (White et al. 2012) and regions identified as highly differentiated in our scan based on all SNPs (black dots). (B) Overlap between QTL associated with male infertility (blue bars) (White et al. 2011), regions identified as contributing to BDMIs between M. m. domesticus and M. m. musculus in two-hybrid zones in central Europe (red dots) (Janoušek et al. 2012) and regions identified as highly differentiated in our scan based all SNPs (black dots).
Regions of overlap on the autosomes contained 221 protein-coding regions (Table S13). Across all 221 autosomal genes in regions of overlap, 20 genes were testis-specific (BC049635, Bps9, Catsper2, Ccdc53, Ccl27a, Ccl27b, Eif3j1, Faf1, Gm13306, Lin7a, Lrrc57, Nup37, Parpbp, Psmc3, Sord, Sycp3, Tex26, Trim69, Tsc22d4, Ttbk2), 11 genes had functional annotations and/or phenotypes associated with male fertility (Arhgap1, Bps9, Cdkn2c, Celf1, Duox2, Ehd4, Igf1, Illra1, Nr1h3, Pmch, and Pparg), and 3 genes were both testis-specific and had mutational variants associated with male infertility (Catsper2, Sord, Sycp3). Nine genes in regions of overlap showed significant expression differences (padj <0.05; 2700089E24Rik, Atg7, B2m, Capn3, Cdkn2c, Igf1, Nup37, Ppip5k1, Shf). Two of those, Cdkn2c and Igf1, are associated with male fertility, and one, Nup37, is testis-specific.
In general QTL are large, while regions of high differentiation are relatively small, potentially helping to narrow QTL intervals. For example, one QTL on chromosome 9 associated with amorphous sperm heads encompasses 26.7 Mb and contains ∼220 protein-coding genes (White et al. 2012). It overlaps with only one highly differentiated region that contains only one protein-coding gene, Bbs9. Bbs9 has gene ontology (GO) terms relating to cilia and is testis-specific. In humans, Bbs9 mutations are associated with Bardet–Biedl syndrome, a disease with multiple phenotypic effects including reduced testis size. Expression levels at Bbs9 were different in M. m. castaneus and M. m. domesticus, but this difference was not significant after correction for multiple testing (P < 0.025, Padj = 0.16). We observed three silent and no replacement fixed differences between M. m. castaneus and M. m. domesticus at Bbs9. Not all sites were surveyed, and thus observed patterns of differentiation may reflect linkage to functionally important unsurveyed sites. It is also important to bear in mind that not all genes in QTL intervals were surveyed.
Identifying candidate regions for reproductive isolation between M. m. domesticus and M. m. musculus
In comparisons between M. m. domesticus and M. m. musculus, the overlap between highly differentiated regions or differentially expressed genes and QTL associated with male infertility (White et al. 2011) was not more than expected by chance. The overlap between differentially expressed genes and candidate BDMIs loci from the hyrbid zone study of Janoušek et al. (2012) was also not more than expected by chance. However, the overlap between highly differentiated regions and the candidate BDMI loci ranks in the 92nd percentile of simulated data (Janoušek et al. 2012). Regions of overlap between all three kinds of studies (QTL, candidate BDMI loci from the hybrid zone, and regions of high differentiation) are particularly promising candidates for reproductive isolation. Importantly, six autosomal regions were identified in which candidate BDMIs and regions of high differentiation overlap precisely or are contiguous and fall within QTL intervals (Figure 3B). These regions collectively contain 242 genes, and the number of genes found in each specific region ranges from 17 to 97 (Table S14).
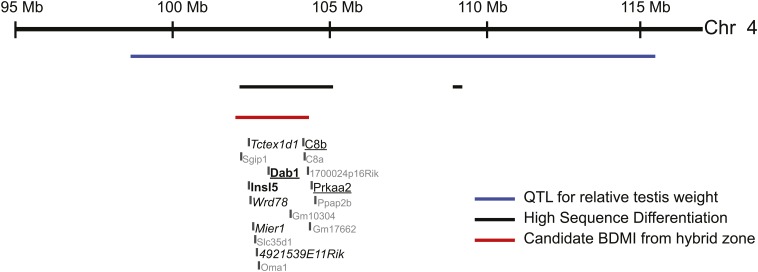
Two regions fall in relatively small QTL intervals. The first is on chromosome 4. This QTL is associated with relative testis weight (White et al. 2011) and contains only 16 genes (Figure 4). Of these 16 genes, 4 are testis-specific (4921539E11Rik, Mier1, Tctex1d1, and Wdr78) and two (Insl5 and Dab1) are associated with male infertility. We found that three genes (C8b, Dab1, and Prkaa2) in this interval are differentially expressed between M. m. domesticus and M. m. musculus after correction for multiple testing (α = 0.05).
Figure 4.
A region on chromosome 4 in which a QTL for relative testis weight (White et al. 2011), a candidate BDMI (Janoušek et al. 2012), and a highly differentiated region identified in this study overlap in the DM comparison. Testis-specific genes are given in italics, differentially expressed genes are underlined, and genes known to be related to male fertility are given in boldface type. All other genes are shown in grey.
The second small QTL interval with precise overlap is found on chromosome 5. This QTL is associated with both total abnormal sperm and distal bent-tail phenotypes. This interval contains 97 genes and is relatively well sampled in our study. There are 14 testis-specific genes in this interval (Fbxo24, Mcm7, Mepce, Muc3, Myl10, Ppp1r35, Rabl5, Srrt, Stag3, Taf6, Tmem184a, Tsc22d4, Znhit1, and Zscan21). Two of these genes (Stag3 and Zscan21) have GO annotations and/or phenotypes relating to male fertility. Myl10 and Rabl5 were differentially expressed between M. m. domesticus and M. m. musculus after correction for multiple testing (Padj < 0.05). There are 10 additional genes in the region with known functional annotations and/or phenotypes relating to male fertility (Ache, Cnpy4, Cux1, Fam20c, Pdgfa, Smok3a, Smok3b, Sun1, Vgf, and Zan).
Discussion
Genome-wide patterns of sequence differentiation
We used a transcriptomic approach to characterize genome-wide patterns of differentiation between the three subspecies of house mice and discovered many highly differentiated regions in all pairwise comparisons. By comparing three subspecies that split from one another at approximately the same time but that have different estimated effective population sizes, we were able to study the influence of demography on the early stages of speciation and divergence. In this case, we found higher levels of sequence differentiation between M. m. domesticus and M. m. musculus than between the other pairs of subspecies. This result is consistent with estimates of the demographic history of these species; both M. m. domesticus and M. m. musculus are believed to have undergone significant bottlenecks resulting in a current Ne of ∼1/10 to 1/2 of the ancestral Ne. M. m. castaneus, on the other hand, is estimated to have a population size very similar to the ancestral population (Geraldes et al. 2011). Lineage-specific changes observed in this study support those expectations. The fewest lineage-specific changes were assigned to M. m. castaneus, the subspecies with the highest Ne, and the most were assigned to M. m. musculus, the subspecies with the smallest Ne. More generally, the coalescent simulations performed here recapitulated the broad patterns of differentiation seen among the three subspecies, suggesting that many of the observed patterns can be explained by differences in Ne and levels of gene flow (Figure 2).
At the same time, greater reproductive isolation is seen in laboratory crosses between M. m. domesticus and M. m. musculus than between M. m. castaneus and M. m. domesticus or M. m. castaneus and M. m. musculus (Dumont and Payseur 2011; White et al. 2011, 2012). This observation, by itself, leads to the prediction of greater differentiation in the DM comparison than in the other two comparisons. Because this pattern is also predicted by demographic differences among the subspecies, it is difficult to disentangle the relative contribution of differences in demography and differences in reproductive isolation to the observed patterns. It is also unclear whether differences in demography are the cause of the differing levels of reproductive isolation. For example, if most BDMI alleles were neutral on their own genetic background, then subspecies with smaller Ne would be expected to accumulate more BDMI differences due to drift and would therefore show greater reproductive isolation. However, most BDMI genes in other systems seem to show evidence of positive selection, suggesting that drift is not the predominant process fixing alleles involved in BDMIs (Coyne and Orr 2004; Presgraves 2010).
Differentiation on the X chromosome
The X chromosome was significantly more differentiated than the autosomes in all pairwise comparisons (Table 1). In principle, this pattern is expected for two reasons. First, the smaller estimated Ne of the X chromosome should result in faster lineage sorting. Second, this pattern is consistent with the large X effect, that is, the disproportionate accumulation of reproductive incompatibilities on the X chromosome (e.g., Coyne and Orr 1989). In this case, the greater level of differentiation appears to be more than can be explained by differences in the X to autosome ratio of Ne. At migration-drift equilibrium, assuming constant bidirectional migration, a sex ratio of 1, and equal migration of males and females, Fst = 1/(4Nm + 1) for the autosomes and Fst = 1/(3Nm + 1) for the X chromosome. If Nm is ∼0.1 (Table S7), then the expected X:autosome ratio of Fst is 1.08 and the observed ratios are 1.45 (CD), 1.35 (CM), and 1.21 (DM) (Table 1). This model is clearly overly simplistic. For example, there is some evidence of male-biased dispersal in this system (Pocock et al. 2005). However, these rough calculations suggest that differences in Ne alone cannot account for the greater differentiation seen on the X chromosome.
On the other hand, our observations are consistent with previous work suggesting a large X effect. For example, hybrid zone studies of M. m. domesticus and M. m. musculus indicate reduced introgression on the X (Tucker et al. 1992; Dod et al. 1993), and IM analysis of a limited number of loci in all three subspecies suggests that gene flow on the X chromosome is lower than that of autosomes (Geraldes et al. 2008, 2011). In laboratory crosses, hybrid male sterility phenotypes map to the X chromosome in crosses between M. m. domesticus and M. m. musculus (Storchová et al. 2004; Good et al. 2008; White et al. 2011) and M. m. castaneus and M. m. domesticus (White et al. 2012). Our findings here demonstrate that elevated differentiation on the X chromosome is a general pattern, is observed in allopatric populations, and extends to all pairs of subspecies.
Recombination
Several recent models suggest that chromosomal rearrangements may lead to reduced gene flow via their effect on suppressing recombination (Noor et al. 2001; Rieseberg 2001; Navarro and Barton 2003). Recombination can also influence differentiation by amplifying the effects of genetic hitchhiking and background selection within lineages (Maynard Smith and Haigh 1974; Charlesworth 1993), reducing variation within lineages and thus leading to increased differentiation between lineages. We found weak support for a negative relationship between differentiation and recombination, consistent with these models. However, the power of this approach may be limited by the absence of data on recombination rate variation across the genome in all three subspecies.
Testis-specific expression
Regions of high differentiation contained a significantly higher proportion of testis-specific genes than other regions. This result is unexpected if highly differentiated regions simply reflect stochastic variation in lineage sorting. In contrast, this result is consistent with (1) reduced gene flow due to BDMIs associated with testis-specific genes, (2) hitchhiking effects associated with positive selection at testis-specific genes, (3) or both. Importantly, this observation suggests that some proportion of highly differentiated regions is associated with functional differences within or between nascent species.
Candidate regions for reproductive incompatibilities
It would be incorrect to claim that all regions of high differentiation contribute to reproductive isolation when many such regions are expected simply as a consequence of drift in small populations. In addition, some regions of high differentiation are likely the result of lineage-specific selection that may not contribute to reproductive isolation. Nonetheless, the observed data differed from demographic simulations in one major way: the distribution of differentiation statistics was flatter, with more values in the extremes of the distribution. This is consistent with more gene flow and more differentiation than expected. Even when exploring a broad range of values for gene flow and divergence time, we were unable to find a demographic scenario that recapitulated observed patterns. Therefore, some highly differentiated regions likely result either from lineage-specific positive selection and/or from barriers to gene flow at loci underlying incompatibilities.
One approach to prioritizing candidate reproductive isolation loci is to identify overlap between the results of genome scans, laboratory crosses, and hybrid zone analyses. We identified several areas of overlap between QTL associated with male sterility in a cross of M. m. castaneus and M. m. domesticus (White et al. 2012) and highly differentiated regions identified in our study. The overlap was more than expected by chance, but in most cases QTL were large, making the overlap difficult to interpret (e.g., chromosome 5, Figure 3A). However, in other cases, the QTL intervals were narrower, there was reasonable coverage in our data set, and relatively few genes were found in the overlap. For example, on chromosome 9, there is just one gene in a region of high differentiation that falls in a moderately sized QTL. Even though >200 genes fall in all of the areas of overlap, this number is considerably smaller than the total number of genes that fall in QTL intervals (White et al. 2012). Moreover, fewer than 3 dozen of those genes have known phenotypes or GO terms that relate to male fertility and/or are testis-specific as might be expected if they affect male sterility phenotypes measured in the QTL analyses. Of course, GO annotation and documentation of phenotypes associated with mutations or knockouts in mice are far from complete, and additional genes in these regions may be related to fertility.
In the DM comparison, overlap between our results and QTL analyses was considerable, but not more than expected by chance. More promisingly, there was meaningful overlap between candidate BMDIs (Janoušek et al. 2012) and our results. In particular, there were six cases in which regions identified as highly differentiated in our study were directly overlapping or contiguous with a candidate BDMI and fell in a QTL interval. In two of those cases, the overlap was relatively precise, and the region of overlap contains a short list of genes that are testis-specific, differentially expressed, and/or related to male infertility. While there is still much work to be done, the intersection of results from multiple studies is encouraging and highlights the promise of this approach for narrowing QTL intervals.
Supplementary Material
Acknowledgments
We thank F. Bonhomme, P. Campbell, C. Clayton, M. Dean, and A. Orth for providing tissue samples; J. MacDonald for providing access to the source code for the program DNA Slider; and members of the Nachman Lab, D. Begun, D. Matute, and one anonymous reviewer for their thoughtful comments on the manuscript. This research was funded by National Institutes of Health grant R01 GM074245 to M.W.N. This work used the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation grant ACI-1053575.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.166827/-/DC1.
Communicating editor: D. Begun
Literature Cited
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., P. T. Pyl, and W. Huber, 2014 HTSeq: a Python framework to work with high-throughput sequencing data. bioRxiv DOI: 10.1101/002824 [DOI] [PMC free article] [PubMed]
- Andrés J. A., Larson E. L., Bogdanowicz S. M., Harrison R. G., 2013. Patterns of transcriptome divergence in the male accessory gland of two closely related species of field crickets. Genetics 193: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Whitley P., Todd B. L., Waldrip-Dail H. M., Clark A. G., 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156: 1879–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57: 289–300 [Google Scholar]
- Bikard D., Patel D., Metté C. L., Giorgi V., Camilleri C., et al. , 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626 [DOI] [PubMed] [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., et al. , 2007. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme F., Rivals E., Orth A., Grant G. R., Jeffreys A. J., et al. , 2007. Species-wide distribution of highly polymorphic minisatellite markers suggests past and present genetic exchanges among house mouse subspecies. Genome Biol. 8: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot P., Auffray J. C., Britton-Davidian J., Bonhomme F., 1993. The evolution of house mice. Annu. Rev. Ecol. Syst. 24: 119–152 [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Carneiro M., Baird S. J. E., Afonso S., Ramirez E., Tarroso P., et al. , 2013. Steep clines within a highly permeable genome across a hybrid zone between two subspecies of the European rabbit. Mol. Ecol. 22: 2511–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1993. The evolution of sex and recombination in a varying environment. J. Hered. 84: 345–350 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15: 538–543 [DOI] [PubMed] [Google Scholar]
- Cox A., Ackert-Bicknell C. L., Dumont B. L., Ding Y., Bell J. T., et al. , 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 1989. Patterns of speciation in Drosophila. Evolution 43: 362–381 [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Cruickshank T. C., Hahn M. W., 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23: 3133–3157. [DOI] [PubMed] [Google Scholar]
- Cucchi T., Vigne J.-D., Auffray J.-C., 2005. First occurrence of the house mouse (Mus musculus domesticus SCHWARTZ & SCHWARTZ, 1943) in Western Mediterranean: a revision of sub-fossil house mice occurrences using a zooarchaeological critical grid. Biol. J. Linn. Soc. Lond. 84: 429–445 [Google Scholar]
- Dod B., Jermiin L. S., Boursot P., Chapman V. H., Nielsen J. T., et al. , 1993. Counterselection on sex chromosomes in the Mus musculus European hybrid zone. J. Evol. Biol. 6: 529–546 [Google Scholar]
- Dumont B. L., Payseur B. A., 2011. Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet. 7: e1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome Res. 21: 114–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17: 68–070 [DOI] [PubMed] [Google Scholar]
- Duvaux L., Belkhir K., Boulesteix M., Boursot P., 2011. Isolation and gene flow: inferring the speciation history of European house mice. Mol. Ecol. 20: 5248–5264 [DOI] [PubMed] [Google Scholar]
- Ellegren H., Smeds L., Burri R., Olason P. I., Backström N., et al. , 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491: 756–760 [DOI] [PubMed] [Google Scholar]
- Eppig J. T., Blake J. A., Bult C. J., Kadin J. A., Richardson J. E., 2012. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 40: D881–D886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire P.-A., Normandeau E., Bernatchez L., 2012. Comparative genomics reveals adaptive protein evolution and a possible cytonuclear incompatibility between European and American eels. Mol. Biol. Evol. 29: 2909–2919 [DOI] [PubMed] [Google Scholar]
- Geraldes A., Basset P., Gibson B., Smith K. L., Harr B., et al. , 2008. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol. Ecol. 17: 5349–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A., Basset P., Smith K. L., Nachman M. W., 2011. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol. Ecol. 20: 4722–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Nachman M. W., 2005. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol. Biol. Evol. 22: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Good J. M., Dean M. D., Nachman M. W., 2008. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179: 2213–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D. L., Oliver F., Eyre-Walker A., Harr B., Keightley P. D., 2010. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 6: e1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr B., 2006. Genomic islands of differentiation between house mouse subspecies. Genome Res. 16: 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. G., 1986. Pattern and process in a narrow hybrid zone. Heredity 56: 347–359 [Google Scholar]
- Harrison R. G., 2012. The language of speciation. Evolution 66: 3643–3657 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M., Maddison W. P., 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoušek V., Wang L., Luzynski K., Dufková P., Vyskočilová M. M., et al. , 2012. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol. Ecol. 21: 3032–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key K. H. L., 1968. The concept of stasipatric speciation. Syst. Zool. 17: 14–22 [Google Scholar]
- Kulathinal R. J., Stevison L. S., Noor M. A. F., 2009. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie C. C., Nickerson D. A., Anderson A. D., Weir B. S., Livingston R. J., et al. , 2007. Linkage disequilibrium in wild mice. PLoS Genet. 3: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak M. K. N., Emrich S. J., Holloway A. K., Regier A. P., Olson M., et al. , 2010. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330: 512–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. ; 1000 Genome Project Data Processing Subgroup, 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholán M., Munclinger P., Sugerková M., Dufková P., Bímová B., et al. , 2007. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61: 746–771 [DOI] [PubMed] [Google Scholar]
- Masly J. P., Jones C. D., Noor M. A. F., Locke J., Orr H. A., 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313: 1448–1450 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C., Forejt J., 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375 [DOI] [PubMed] [Google Scholar]
- Munclinger P., Boziková E., Sugerková M., Piálek J., Macholán M., 2002. Genetic variation in house mice (Mus, Muridae, Rodentia) from the Czech and Slovak Republics. Folia Zool. (Brno) 51: 81–92 [Google Scholar]
- Nachman M. W., Payseur B. A., 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N. J., Whibley A., Jones R. T., Davey J. W., Dasmahapatra K. K., et al. , 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Barton N. H., 2003. Chromosomal speciation and molecular divergence: accelerated evolution in rearranged chromosomes. Science 300: 321–324 [DOI] [PubMed] [Google Scholar]
- Neafsey D. E., Lawniczak M. K. N., Park D. J., Redmond S. N., Coulibaly M. B., et al. , 2010. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330: 514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., 1987 Molecular Evolutionary Genetics Columbia University Press, New York. [Google Scholar]
- Nei M., Li W. H., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Wakeley J., 2001. Distinguishing migration from isolation: a Markov Chain Monte Carlo approach. Genetics 158: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. A., Grams K. L., Bertucci L. A., Reiland J., 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P., Schluter D., 2011. The genes underlying the process of speciation. Trends Ecol. Evol. 26: 160–167 [DOI] [PubMed] [Google Scholar]
- Pocock M. J. O., Hauffe H. C., Searle J. B., 2005. Dispersal in house mice. Biol. J. Linn. Soc. Lond. 84(3): 565–583 [Google Scholar]
- Prager E. M., Sage R. D., Gyllensten U., Thomas W. K., Hübner R., et al. , 1993. Mitochondrial DNA sequence diversity and the colonization of Scandinavia by house mice from East Holstein. Biol. J. Linn. Soc. Lond. 50: 85–122 [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., Balagopalan L., Abmayr S. M., Orr H. A., 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut S., Nolte A. W., Bernatchez L., 2010. Mining transcriptome sequences towards identifying adaptive single nucleotide polymorphisms in lake whitefish species pairs (Coregonus spp. Salmonidae). Mol. Ecol. 19: 115–131 [DOI] [PubMed] [Google Scholar]
- Renaut S., Grassa C. J., Yeaman S., Moyers B. T., Lai Z., et al. , 2013. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat. Commun. 4: 1827. [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358 [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Whitton J., Gardner K., 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo T., Geraldes A., Nachman M. W., 2007. Nucleotide variation in wild and inbred mice. Genetics 177: 2277–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J. X., Bonhomme F., Boursot P., Thaler L., Catzeflis F., 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linn. Soc. 41: 83–103 [Google Scholar]
- Shifman S., Bell J. T., Copley R. R., Taylor M. S., Williams R. W., et al. , 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 4: e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová R., Gregorová S., Buckiová D., Kyselová V., Divina P., et al. , 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524 [DOI] [PubMed] [Google Scholar]
- Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., et al. , 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Shimada T., Terashima M., Tsuchiya K., Aplin K., 2004. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 33: 626–646 [DOI] [PubMed] [Google Scholar]
- Teeter K. C., Payseur B. A., Harris L. W., Bakewell M. A., Thibodeau L. M., et al. , 2008. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 18: 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter K. C., Thibodeau L. M., Gompert Z., Buerkle C. A., Nachman M. W., et al. , 2010. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution 64: 472–485 [DOI] [PubMed] [Google Scholar]
- Ting C.-T., Tsaur S.-C., Wu M.-L., Wu C.-I., 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P. K., Sage R. D., Warner J., Wilson A. C., Eicher E. M., 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146–1163 [DOI] [PubMed] [Google Scholar]
- Tucker P. K., Sandstedt S. A., Lundrigan B. L., 2005. Phylogenetic relationships in the subgenus Mus (genus Mus, family Muridae, subfamily Murinae): examining gene trees and species trees. Biol. J. Linn. Soc. Lond. 84: 653–662 [Google Scholar]
- Turner T. L., Hahn M. W., Nuzhdin S. V., 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3: e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe F., Dod B., Boursot P., Bellis M., Bonhomme F., 1986. Absence of Y-chromosome introgression across the hybrid zone between Mus musculus domesticus and Mus musculus musculus. Genet. Res. 48: 191–197 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe F., Boursot P., Nielsen J. T., Bonhomme F., 1988. A steep cline for mitochondrial DNA in Danish mice. Genet. Res. 52: 185–193 [DOI] [PubMed] [Google Scholar]
- Wang J. R., de Villena F. P.-M., Lawson H. A., Cheverud J. M., Churchill G. A., et al. , 2012. Imputation of single-nucleotide polymorphisms in inbred mice using local phylogeny. Genetics 190: 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- White M. A., Ané C., Dewey C. N., Larget B. R., Payseur B. A., 2009. Fine-scale phylogenetic discordance across the house mouse genome. PLoS Genet. 5: e1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Genetic dissection of a key reproductive barrier between nascent subspecies of house mice, Mus musculus domesticus and Mus musculus musculus. Genetics 169: 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Stubbings M., Dumont B. L., Payseur B. A., 2012. Genetics and evolution of hybrid male sterility in house mice. Genetics 191: 917–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E. E., Goodstadt L., Ponting C. P., 2004. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 14: 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wu C., Orozco C., Boyer J., Leglise M., Goodale J., et al. , 2009. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff G. J., Wang W., Wu C. I., 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403: 304–309 [DOI] [PubMed] [Google Scholar]
- Yalcin B., Adams D. J., Flint J., Keane T. M., 2012. Next-generation sequencing of experimental mouse strains. Mamm. Genome 23: 490–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.