Abstract
The severity of the toxic side effects of chemotherapy shows a great deal of interindividual variability, and much of this variation is likely genetically based. Simple DNA tests predictive of toxic side effects could revolutionize the way chemotherapy is carried out. Due to the challenges in identifying polymorphisms that affect toxicity in humans, we use Drosophila fecundity following oral exposure to carboplatin, gemcitabine and mitomycin C as a model system to identify naturally occurring DNA variants predictive of toxicity. We use the Drosophila Synthetic Population Resource (DSPR), a panel of recombinant inbred lines derived from a multiparent advanced intercross, to map quantitative trait loci affecting chemotoxicity. We identify two QTL each for carboplatin and gemcitabine toxicity and none for mitomycin. One QTL is associated with fly orthologs of a priori human carboplatin candidate genes ABCC2 and MSH2, and a second QTL is associated with fly orthologs of human gemcitabine candidate genes RRM2 and RRM2B. The third, a carboplatin QTL, is associated with a posteriori human orthologs from solute carrier family 7A, INPP4A&B, and NALCN. The fourth, a gemcitabine QTL that also affects methotrexate toxicity, is associated with human ortholog GPx4. Mapped QTL each explain a significant fraction of variation in toxicity, yet individual SNPs and transposable elements in the candidate gene regions fail to singly explain QTL peaks. Furthermore, estimates of founder haplotype effects are consistent with genes harboring several segregating functional alleles. We find little evidence for nonsynonymous SNPs explaining mapped QTL; thus it seems likely that standing variation in toxicity is due to regulatory alleles.
Keywords: pharmacogenomics, quantitative trait loci, chemotoxicity, Drosophila Synthetic Population Resource, Multiparent Advanced Generation Inter-Cross (MAGIC), Multiparental populations, MPP, advanced intercross line, complex traits, genetic heterogeneity, DSPR
CHEMOTHERAPEUTIC agents are among the most toxic medications administered to humans (Alley et al. 2002). The toxicity caused by these medications may become severe enough that patients are forced to adjust dosing or switch to a different chemotherapeutic medication, while the disease progresses. Although diet, medical history, age, and other environmental factors of the patient may explain a portion of the toxicity (Gajewski et al. 1989; Meirow and Nugent 2001; Rothenberg et al. 2003; Watters and McLeod 2003; Lee et al. 2005), we have shown that there is a significant genetic component governing the toxic effect of several drugs in Drosophila melanogaster (Kislukhin et al. 2012). Here we use the D. melanogaster model system to dissect the genetic basis of toxicity for three front-line chemotherapeutics: carboplatin, gemcitabine hydrochloride (gemcitabine), and mitomycin C.
Carboplatin is a platinum-containing compound used primarily to treat ovarian cancer. It is also sometimes used to treat lung, breast, bladder, and endometrial cancer; head and neck cancer; cancer of the cervix and testicles; certain types of brain cancer; Wilms’ tumor; neuroblastoma; and retinoblastoma (Wheate et al. 2010). Gemcitabine is an antimetabolite used primarily in combination with other chemotherapy drugs to treat ovarian cancer and breast cancer that has not improved or that has worsened after treatment with other medications. It is also used in combination with other chemotherapy drugs treat non-small-cell lung cancer that has spread to other parts of the body and cannot be treated with surgery or cancer of the pancreas that has spread to other parts of the body and has not improved or worsened after treatment with another medication (Iaffaioli et al. 2000). Mitomycin C is an antitumor antibiotic used primarily to treat adenocarcinoma of the stomach and pancreas. Its other uses include Fanconi anemia (Yao et al. 2013); squamous cell carcinoma of the head and neck, lungs and cervix; adenocarcenoma of the colon and rectum; adenocarcinoma and duct cell carcinoma of the breast; and bladder cancer (Siddique et al. 2010). All three of these drugs have a litany of toxic side effects.
Studying the genetic basis of chemotherapy toxicity in humans is challenging. An ideal experiment would involve carefully observing thousands of patients that undergo treatment from start to finish with only one chemotherapeutic medication without switching the dose, even if side effects become severe. Of course it would be unethical to do so, because the goal of cancer treatment is to eradicate the disease while trying to minimize the harm to the rest of the body. In our previous work we show that D. melanogaster is a powerful model system for studying the heritable toxic side effects of chemotherapy agents (Kislukhin et al. 2012; Kislukhin et al. 2013). In Drosophila we can orally administer a single drug to a group of flies and count the number of offspring that they produce post-treatment; toxicity is then measured as the post-treatment drop in fecundity of female flies. Fecundity is a sensitive measure of the effect of chemotherapy drugs on the physiology of flies, since oogenesis (and spermatogenesis) are the only rapidly dividing cells in adult flies and chemotherapy drugs are designed to stop cell division. We observed that many chemotherapy drugs have measurable effects on fecundity at doses orders of magnitude smaller than those required to cause death (Kislukhin et al. 2012). In addition, D. melanogaster has been proposed as a model system for assessing potential toxic side effects influencing reproduction (Avanesian et al. 2009). Similarities between the reproductive system in D. melanogaster and humans include overall similarity between female reproductive structures, conserved sexual development genes, and the existence of sex hormones (Avanesian et al. 2009). In addition, chemotherapy drugs often target basal cell-level pathways, which often show one-to-one gene conservation between D. melanogaster and humans (Bier 2005), making D. melanogaster a powerful model for identifying candidate genes influencing chemotoxicity in humans.
In this study we employ the Drosophila Synthetic Population Resource (DSPR), created as a community resource for the genetic dissection of complex traits (http://FlyRILs.org; King et al. 2012a,b), to identify the genetic factors underlying chemotherapeutic agent toxicity. This resource consists of >1700 recombinant inbred lines (RILs) of D. melanogaster, generated from two different eight-way, 50 generation synthetic populations. This panel represents a stable resource that allows us to perform exactly the kinds of powerful experiments that are not possible in human studies. Because the RILs are genetically homogeneous, we can measure the effects of multiple drugs on multiple replicates of the same set of genetically identical RILs. Previous studies have also demonstrated the potential for both high statistical power and high mapping resolution in this resource (King et al. 2012b).
Here we identify two QTL each for carboplatin and gemcitabine toxicity. Two of the QTL are associated with a priori human candidate genes ABCC2, MSH2, RRM2, and RRM2B believed to play a role in toxicity of these drugs. ABCC2 is an efflux transporter of carboplatin, and is a major determinant of chemoresistance in tumor cells (Tian et al. 2012). Genetic polymorphisms at MSH2 have been shown to affect treatment response to platinum-based chemotherapy (Cheng et al. 2010). RRM2 and RRM2B encode ribonucleotide reductase proteins. Gemcitabine has been shown to inhibit those enzymes, which are required for DNA synthesis (Li et al. 2012). The two other QTL are associated with novel candidate genes worthy of additional study. One of those QTL colocalize with a QTL for methotrexate (MTX) toxicity, with human ortholog GPx4, identified in previous work (Kislukhin et al. 2013). Exhaustive association scans of all SNPs under the peaks are unable to pinpoint causative sites. The most parsimonious explanation for this result is that mapped QTL are due to several segregating functional alleles at these genes in the DSPR.
Materials and Methods
Mapping population
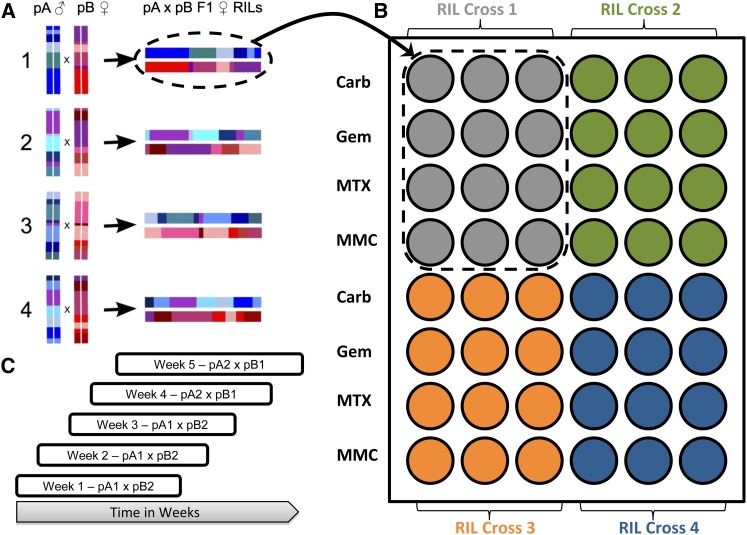
We mapped QTL for all three drug toxicities using RILs from the DSPR (http://FlyRILs.org). The DSPR consists of >1700 D. melanogaster RILs derived from a multiparent, advanced intercross. The RILs are derived from one of two synthetic populations: pA or pB. Each synthetic population was founded by eight inbred lines, seven unique lines (A1–A7 and B1–B7), and one line common to both populations (AB8). Each synthetic population was maintained as two independent replicate subpopulations (pA1 and pA2 or pB1 and pB2), kept at a large population size, and allowed to freely recombine for 50 generations. At generation 50, each subpopulation gave rise to ∼500 RILs via 25 generations of full-sib mating. The genomes of the original 15 inbred founder lines have been completely resequenced and the complete underlying founder haplotype structure of all RILs in the panel has been determined via a hidden Markov model (HMM). Complete details of the development of the DSPR, founder whole genome resequencing, and RIL genotyping are described in King et al. (2012a). The hidden Markov model we used to infer the mosaic structure of the RILs and the power and mapping resolution of the DSPR for QTL mapping are described in King et al. (2012b). All of the raw genomic data and inferred founder genotype data are freely available at http://FlyRILs.org. In addition, the raw sequencing data have been deposited in the NCBI short-read archive (RIL RAD genotyping, SRA051306; founder resequencing, SRA051316) and the inferred founder genotype data have been deposited in the Dryad repository (http://doi.org/10.5061/dryad.r5v40). For this experiment, we generated trans-heterozygote F1 individuals by performing matched crosses between pB1 females and pA2 males or pB2 females and pA1 males (Figure 1A) and phenotyped these individuals to minimize the risk mapping QTL for inbreeding depression (see details below). For a detailed comparision of mapping using a pA–pB cross vs. phenotyping inbred RILs directly, see King et al. (2012b).
Figure 1.
(A) The pA–pB crossing design. For each cross, a pA male was crossed to a pB female producing genetically identical trans-heterozygote F1 individuals. (B) The condo layout. Each cross seeded 12 cells, 3 per drug. There were a total of four crosses per condo. (C) The entire assay was split into five blocks, with start dates on consecutive weeks. Each block took 8 weeks to complete. F1 hybrids from populations A1 × B2 were assayed over the first 3 weeks, and F1 hybrids from populations A2 × B1 were assayed in weeks 4 and 5.
The drug toxicity phenotype
In previous experiments, we established that the toxicity of all three chemotherapeutics that we discuss in this article could be quantified as a decrease in D. melanogaster fecundity (Kislukhin et al. 2012). Furthermore, the decrease in fecundity is proportional to the concentration of the drug orally administered to the flies. Once we established drug concentrations that knock down fecundity by ∼50% relative to a control mock drug treatment, we used fecundity as a measure of toxicity in drug-treated flies. Carboplatin (LKT Laboratories C0171), gemcitabine (LKT Laboratories G1745), and mitomycin C (LKT Laboratories M3377) were orally administered to D. melanogaster at 0.24, 0.76, and 0.043 mM concentrations, respectively. Detailed methods for drug administration and dosing determination are given in Kislukhin et al. (2013).
We performed 396 pA1 male to pB2 female crosses and 302 pA2 male to pB1 females crosses and placed three female and three male F1 hybrids into each fly condo cell. Condos are a matrix of six by eight cells, where each cell is intended to mimic a 1-in.-diameter fly vial. For ease of handling large numbers of flies, we designed the condos to consist of two halves, each half the height of a regular fly vial. This way, food or drug trays are easily replaced by placing the condo meshside down (food tray up) on a CO2 tray, sedating the flies, detaching the old food/drug tray (which is now on top), and attaching a fresh food/drug tray. As a condo has 48 cells, we were able to test three replicate cells of four independent RIL crosses (genotypes) for four drugs (Figure 1B) per condo. In this article we focus on three drugs (carboplatin, gemcitabine, and mitomycin C), having previously reported the results of the fourth drug, MTX (Kislukhin et al. 2013).
The three replicate cells per genotype/drug combination each contained three males and females; thus per-cell fecundity is associated with a pool of three females. We used an already well-tested 3-day exposure method, as described in Kislukhin et al. (2013), where flies were exposed to a drug–food mixture for a 3-day period, then recovered on standard fly food for 24 hr, and laid eggs for a 48-hr period. Subsequently, the original adults were removed from the condos and offspring were allowed to develop into adults for 14 days at 23°. The adult offspring were frozen at −20° and then removed and transferred to a “sandwich” of two GBC SelfSeal repositionable letter size laminating sheets, 3 mil. These fecundity imprints (mirror images of the original condo layout) are stored at −20°. We triple counted each cell and used the mean of the counts as the phenotype measure of the drug’s toxicity.
In addition to the drug-exposure condos, we had control condos. Each control condo contained three cells of three males and three females each for each of 16 RIL crosses. Control flies were obtained from the same cross of pA1 males by pB2 females or pA2 males by pB1 females as the experimental flies. Control condos were handled the same way as exposure condos with two exceptions: (1) the “exposure” tray contained filter paper with only liquid food and DMSO (i.e., a mock drug treatment) and (2) adult flies were removed after a 24-hr layout period, unlike the 24-hr recovery and 48-hr layout period of the exposure condos. After the offspring were frozen, we visually inspected each condo cell for presence of flies. Control condos were used to identify crosses that resulted in poor female fecundity—<50 offspring and those RIL crosses were removed from further analysis. All of the phenotype data described here are available at http://FlyRILs.org and have been deposited in the Dryad Repository: http://dx.doi.org/10.5061/dryad.ct70q.
From cross to count each experiment spanned 6 weeks. We attempted 120 crosses each week; however, not all crosses produced enough offspring to proceed to the next step, leading to ∼100 successful crosses per week. To cover all crosses, we had 8 starting weeks (Figure 1C). In the first 3 weeks we attempted 396 pA1*pB2 crosses and in weeks 4 and 5 we attempted 302 pA2*pB1 crosses. Weeks 6–8 were used to repeat crosses that failed the first time around. There was a highly significant block effect of week on chemotoxicity for all three drugs (see data analysis; carboplatin, χ1 = 317.1 P < 0.0001; gemcitabine, χ1 = 41.5 P < 0.0001; mitomycin C, χ1 = 84.3 P < 0.0001). For all three of the drugs, the last 3 weeks (6–8) all had very low knockdown of offspring, with the number of offspring produced approaching the condo saturation level, that is, close to the number of offspring observed per cell in mock-treated flies, reflecting the upper limit of our assay’s dynamic range (Kislukhin et al. 2013). We observed a similar effect in our previously reported results for MTX (Kislukhin et al. 2013). Given the universal failure to achieve the appropriate knockdown in these 3 weeks, we eliminated them from our analysis. We also eliminated weeks 1, 3, and 4 for carboplatin and week 3 for mitomycin C because these weeks showed very high knockdown with many females producing zero or near zero offspring. Supporting Information, Figure S1 shows the distribution of fecundity by week and drug, showing the weeks we eliminated. In general, we included only weeks that achieved a mean fecundity knockdown near 50% to avoid including data that did not represent our target phenotype. The elimination of these weeks resulted in a reduced number of crosses for each drug (carboplatin, N = 186; gemcitabine, N = 407; mitomycin C, N = 372). In many cases the drug concentration in solution was near saturation level; we currently hypothesize that failed weeks involved drugs coming out of solution or precipitants being administered to flies.
A priori candidate genes
We identified a priori human candidate genes for all three drugs by utilizing what is known about the genes involved in each drug’s cellular pathway. The carboplatin cellular pathway includes genes involved in the drug’s influx into the cell, mismatch repair, excision repair, and efflux (Marsh et al. 2009). The gemcitabine cellular pathway includes genes involved in intracellular transport of the drug, phosphorylation, ribonucleotide reductase, and DNA repair (Whirl-Carrillo et al. 2012). We identified additional nonpathway candidate genes for each drug via PubMed searches with the following words for each drug:
carboplatin: carboplatin, toxicity, and polymorphism;
gemcitabine: gemcitabine, toxicity, and polymorphism; and
mitomycin C: mitomycin C, toxicity, and polymorphism.
These searches resulted in 26, 45, and 11 publications, respectively. Mitomycin C is an alkylating and crosslinking agent, which mean that all genes involved in DNA repair could be considered candidate genes. We thus attempted to identify additional candidate genes that are likely specific to mitomycin C via a PubMed search for “mitomycin C,” “pathway,” and “genes,” resulting in 153 publications. Furthermore, since mitomycin C is often used in treatment of Fanconi anemia, we performed an additional PubMed search using the words “Fanconi,” “mitomycin C,” and “genes,” resulting in 124 publications. Once we had a list of publications for each drug, each publication was manually curated to include only germline (not tumor) gene polymorphisms and studies that showed a significant association between polymorphism and drug toxicity. After identifying the candidate genes, we used http://www.ensembl.org to identify D. melanogaster gene orthologs for each of the genes (Table S1).
Data analysis
All data analysis was performed in R (R Development Core Team 2013). We used a mixed-model ANOVA using the lme function in the nlme package (Pinheiro et al. 2013) to test for an effect of week (block) on the toxicity of the three drugs. Cross ID and week were random effects with cross ID nested within week. All further analyses were performed on the reduced data set after dropping weeks that did not achieve a 50% knockdown (see above).
Heritability and QTL mapping
We estimated the broad sense heritability of drug toxicity by estimating the genetic and phenotypic variance components from a linear mixed model using the lme and VarCorr functions in the nlme package (Pinheiro et al. 2013). Note that because all three replicates were reared in the same condo (although different cells), these heritability estimates include some microenvironmental effects. We estimated the heritability of cross means as the estimated genetic variance component over the total variance of cross means. Heritability estimates were obtained from the censored collection of crosses after dropping weeks that did not achieve 50% knockdown (see above).
The general analytical framework for QTL mapping in the DSPR is described in King et al. (2012a,b) and the specific procedure for mapping QTL for this experiment is described in Kislukhin et al. (2013). Briefly, for each drug, we regressed mean female fecundity for each cross on the 16 additive probabilities corresponding to the probabilities that the paternal RIL was derived from each of the pA founders and the probabilities that the maternal RIL was derived from each of the pB founders and included subpopulation as a covariate. The model is
where µ is the grand mean, S is subpopulation, GA,i are the genotype probabilities for the paternal RIL, GB,i are the genotype probabilities for the maternal RIL, and βs, βA,i, and βB,i are the corresponding effect estimates. Some founder genotypes were poorly represented in the crosses we assayed at some locations in the genome (i.e., fewer than five crosses have that founder genotype at that genomic position). We found that including these terms, with little representation, in the model could lead to inflated P-values. Therefore, at a given position, if the sum of the probabilities across all the crosses assayed for a given founder genotype probability was <4.8, we dropped that term from the model. Summing the probabilities for a given founder genotype is equivalent to counting the number of RILs of that founder genotype. A result of this decision is that the model degrees of freedom vary with genomic position. Including week as a block effect gave very similar results for all drugs (data not shown) and thus, for simplicity, we present the results from the model without the effect of week here. At each position, we calculated the F-statistic for the overall effect of genotype and obtained −log10(P-values). We then used the loess smoothing function in R with a span of 0.005 to smooth the –log10(P-values) across genetic distance to temper any highly localized fluctuations (cf. Paterson et al. 1988).
We estimated the effects of each founder genotype at each QTL separately for each population (pA and pB). To do this, we fit the model , where yr are the residuals after correcting for subpopulation, Gi are the founder genotype probabilities, and βi are the corresponding effect estimates. Once again, only founder genotype probabilities whose sum across all crosses was >4.8 were included.
We performed 1000 permutations of the data separately for each drug to determine genome-wide significance thresholds (Churchill and Doerge 1994) for several false-positive rates. For each permutation, we smoothed the resulting –log10(P-values) as we did for the observed data with the same loess smoothing function. We then used the peak finder function msPeakSimple from the msProcess library (Gong et al. 2012) in R with a span of 50 and a signal-to-noise threshold of 1 to identify distinct peaks across the genome. For a wide range of potential –log10(P-value) thresholds, we quantified the number of distinct peaks per genome scan for each permutation that exceeded that threshold. We could then calculate the number of observed peaks exceeding a given −log10(P-value) threshold per genome scan and determine a range of false-positive rates. For example, the –log10(P-value) threshold that corresponds to 0.05 peaks per genome scan is our threshold corresponding to a 5% genome-wide false-positive rate. We show false-positive rates ranging from 0.05 to 3 expected peaks per genome scan for each drug.
We identified and localized peaks of interest using the smoothed –log10(P-values) as described above. We considered a peak to be a peak of interest if one of the following conditions was met: 1) the peak co-localized with a D. melanogaster ortholog of a known drug toxicity candidate gene identified a priori (see a priori candidate genes section) and the –log10(P-value) exceeded the 3 genome-wide false positive rate threshold, or 2) the –log10(P-value) exceeded our 0.5 (50%) genome-wide false positive rate threshold. To obtain confidence intervals on these peaks, we performed a genome scan without dropping founder genotype terms with poor representation and we converted the resulting F-statistic to a LOD score (Broman and Sen 2009). We did not drop founder genotype terms when determining confidence intervals to keep the degrees of freedom for the model constant as variations in the degrees of freedom alter the relationship between LOD scores and P-values and alters the amount of LOD drop corresponding to a given percent confidence interval (Manichaikul et al. 2006; Broman and Sen 2009; King et al. 2012b). We used these LOD scores to calculate confidence intervals in two ways. First, we used a traditional 2 LOD drop interval. However, while 2 LOD intervals are conservative for two line crosses, the necessary LOD drop increases for larger degrees of freedom in the model (Manichaikul et al. 2006) and we have previously shown they are overly liberal for our 8-way crosses (King et al. 2012b). We also calculated Bayes credible intervals, for which 95% coverage is more consistent for different sample sizes, experimental designs, and effect sizes (Manichaikul et al. 2006; Broman and Sen 2009).
Testing the effects of SNPs candidate gene regions
The four QTL peaks identified (Table 1) were associated with seven candidate genes: CA with CG9413, CG42271, and na; CB with MRP and spel1; GA with RnrS; and GB with PHGPx. For each candidate gene we somewhat arbitrarily defined the candidate gene interval using the coding region of the next gene up- or down-stream of the candidate gene. We simply defined the boundaries of the candidate gene interval as the start of the next gene’s up- or down-stream coding region rounded to the nearest kilobase. These candidate gene regions are given in Table S2. Within each candidate gene region we identified three types of biallelic genetic polymorphisms: non-synonymous SNPs (i.e., SNPs predicted to encode an amino acid variant), other SNPs (including both synonymous SNPs, SNP in UTRs, and SNPs outside the coding regions), and segregating TE insertions. For details on how we called SNPs in the founder lines, see King et al. (2012a, http://FlyRILs.org). To determine the effect of these SNPs, we inferred the probability each RIL harbored the minor allele and assigned a genotype value to each cross by adding the paternal and maternal probabilities. In the case of perfect certainty, genotype values are: 2 = AA, 1 = Aa, and 0 = aa. We then fit the following single marker model for each possible amino acid variant:
where S is subpopulation, M is the cross genotype at the marker, and βs and βm are the corresponding effect estimates. We also performed all the above tests after correcting for kinship among the crosses using the GRAMMAR method (Aulchenko et al. 2007). We observed no substantial difference and report the results without the correction for simplicity. We additionally had access to a set of annotations of transposable element insertions segregating in the founders (Cridland et al. 2013), of which there were six segregating in candidate gene intervals (in CA: X:14485885, X:14167279; in CB: 2L:12725321, 2L:12747238, 2L:12748174, and 2L:14375490). Four of these TEs (in CA: X:14485885; in CB: 2L:12725321, 2L:12748174, and 2L:14375490), were actually segregating at appreciably frequency in the DSPR RILs. This is not surprising, as regional loss of single founder haplotypes is not at all uncommon (King et al. 2012a). In addition, all six TEs were private to a single founder and thus any statistical test is ultimately a test of the effect of an entire founder haplotype. Thus even if a TE is significantly associated with a phenotype it is impossible to say that event is causative vs. in linkage disequilibrium with another SNP private to that same haplotype.
Table 1. Details of identified QTL.
| Name | Peak location (Mb) | −log10(P-value) | 2-LOD CI (Mb)a | 2-LOD CI (cM)a | BCIb (Mb) | BCIb (cM) | H2 (%)c |
|---|---|---|---|---|---|---|---|
| CA | X 14.29 | 3.32 | 14.05–14.55 | 46.37–48.20 | 14.10–14.79 | 46.56 – 49.01 | 23 |
| CB | 2L 12.67 | 3.23 | 12.48–14.64 | 46.63–50.09 | 13.53–14.23 | 48.48 – 49.54 | 17 |
| GA | 2R 8.22 | 2.13 | 6.60–8.79 | 62.72–66.92 | 6.92–9.40 | 63.34 – 68.19 | 9 |
| GB | 3L 3.40 | 4.97 | 3.23–3.62 | 6.24–7.74 | 3.29–3.53 | 6.47 – 7.39 | 18 |
2-LOD CI are 2-LOD support intervals.
Bayesian credible intervals.
Percentage of H2 refers to the percentage of broad sense heritability of cross means.
Results and Discussion
Heritability
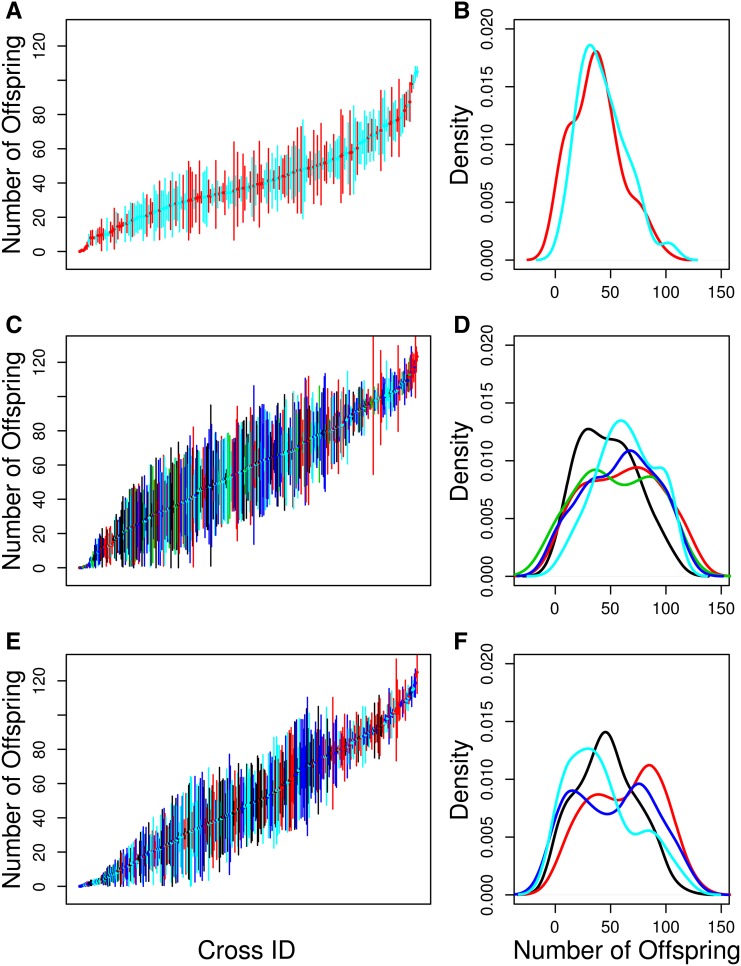
There was substantial variation among genotypes for chemotoxicity as measured by the number of offspring produced following drug exposure (Figure 2). We estimated the broad sense heritability of chemotoxicity for each drug (carboplatin: 42%; gemcitabine: 38%; mitomycin C: 52%). For QTL mapping, we used the mean chemotoxicity (over the three replicate vials per cross) for each drug and thus we also estimated the broad sense heritability of genotype mean chemotoxicity for each drug (carboplatin: 68%; gemcitabine: 63%; mitomycin C: 76%). These estimates are lower than the narrow sense heritabilities we estimated for these same drugs in a previous study using a similar assay (Kislukhin et al. 2012). The heterogeneity in the degree of fecundity knockdown across experimental blocks likely indicates that compared to our previous work (Kislukhin et al. 2012), the dosing of the drugs in this experiment was not as consistent. This dosing inconsistency over time effect required us to discard several experimental blocks in which the flies were obviously being over- or under-dosed. It is very likely the dosing within blocks was also more heterogeneous than in our previous work, and believe this to be the reason for our lower heritability estimates. Nonetheless these are highly heritable traits that are suitable for QTL mapping (cf. King et al. 2012b).
Figure 2.
The distribution of fecundity for carboplatin (A and B), gemcitabine (C and D), and mitomycin C (E and F). (A, C, and E) Means and standard errors of the number of offspring produced by each F1 female from each pA–pB cross. Different colors correspond to different weeks (blocks) of the experiment after removing weeks with poor dosing: black, week 1; red, week 2; green, week 3; blue, week 4; cyan, week 5. (B, D, and F) Density plot of the mean number of offspring produced by the three replicate females from each RIL. Different colors correspond to different weeks (blocks) of the experiment: black, week 1; red, week 2; green, week 3; blue, week 4; cyan, week 5.
Chemotoxicity QTL
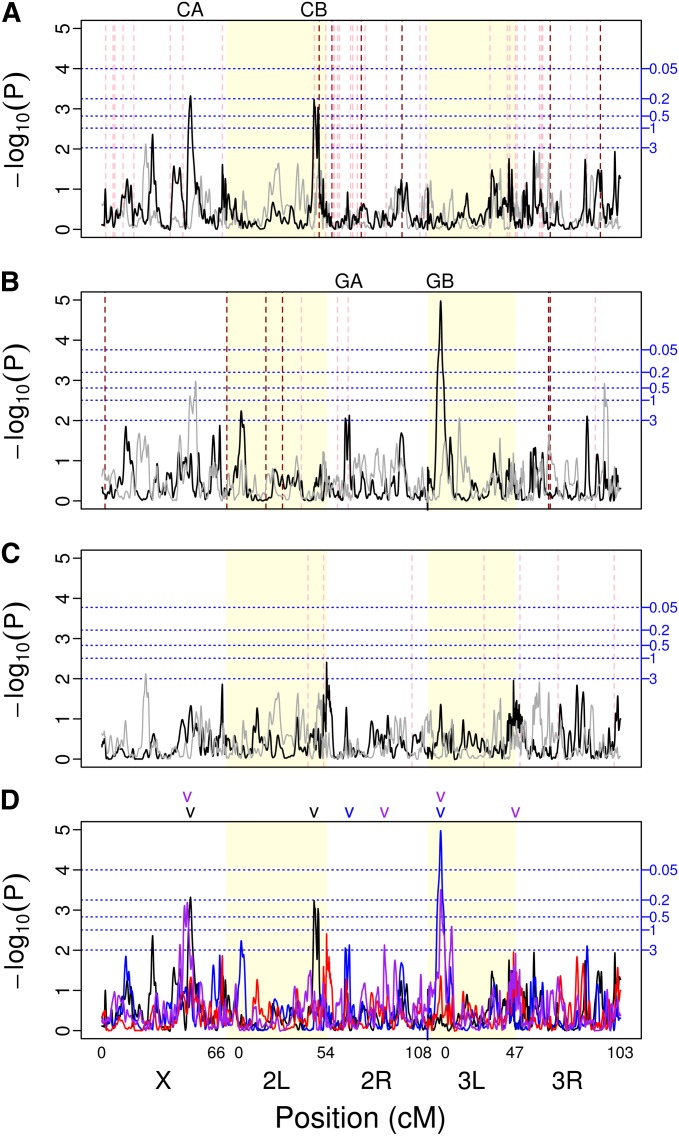
We identified four QTL of interest (Table 1), two for carboplatin toxicity (CA and CB; for carboplatin QTL A and B; Figure 3A; Figure S2, A and B) and two for gemcitabine toxicity (GA and GB; for gemcitabine QTL A and B; Figure 3B; Figure S2, C and D). We did not identify any QTL for mitomycin C toxicity (Figure 3C), despite mitomycin C toxicity having a large heritable component. Three of the mapped QTL (CA, CB, and GB) met our first criteria for identification, exceeding the 0.5 genome-wide false positive rate threshold with QTL peak GB also exceeding the 0.05 genome-wide false positive rate. QTL CB also aligns with an a priori candidate gene, and thus satisfies both our criteria for identification. QTL GA satisfied our second criteria for identification. It aligns with an a priori candidate gene and had a marginal P-value of 0.007.
Figure 3.
Genome scans for (A) carboplatin, (B) gemcitabine, (C) mitomycin C, and (D) all four drugs assayed in this experiment (black, carboplatin; blue, gemcitabine; red, mitomycin C; purple, methotrexate). The major chromosome arms are delineated by different background shading (white/yellow). The black line is the scan of the observed data. To give an example of the results obtainable by chance alone, the gray line is a scan of a single permutation of the observed data. Horizontal blue dotted lines indicate thresholds for various false-positive rates (number of expected peaks per genome scan) obtained via permutations (shown in A–C only). Vertical dashed red lines indicate the location of fly orthologs of previously identified candidate genes for each drug (shown in A–C only). The darker red indicates a gene in the drug pathway while the lighter red indicates all other candidate genes. The locations of mapped QTL are indicated above each plot. In the genome scan with all four drugs (D), different colors indicate QTL mapped for the different drugs (black, carboplatin; blue, gemcitabine; red, mitomycin C; purple, methotrexate).
The CA and CB QTL explain 23 and 17% of the genetic variation for carboplatin toxicity, respectively and together they explain 40% of the total genetic variation (Table 1). The GA and GB QTL explain 9 and 18% of the genetic variation for gemcitabine toxicity, together explaining 27% of the total genetic variation (Table 1). It should be noted, however, that QTL effect estimates have a known upward bias (i.e., the Beavis effect), whose magnitude is proportional to the number of statistical tests carried divided by the number of inbred lines under study (Xu 2003). Therefore, this bias is expected to be more severe for the estimates for carboplatin (number of crosses = 186) than for those of gemcitabine (number of crosses = 407). Regardless, the variance explained for both drugs should be considered upper limits. All QTL were resolved to ∼1–5 cM (∼240–2500 kb; Table 1), with more significant QTL mapped with higher resolution.
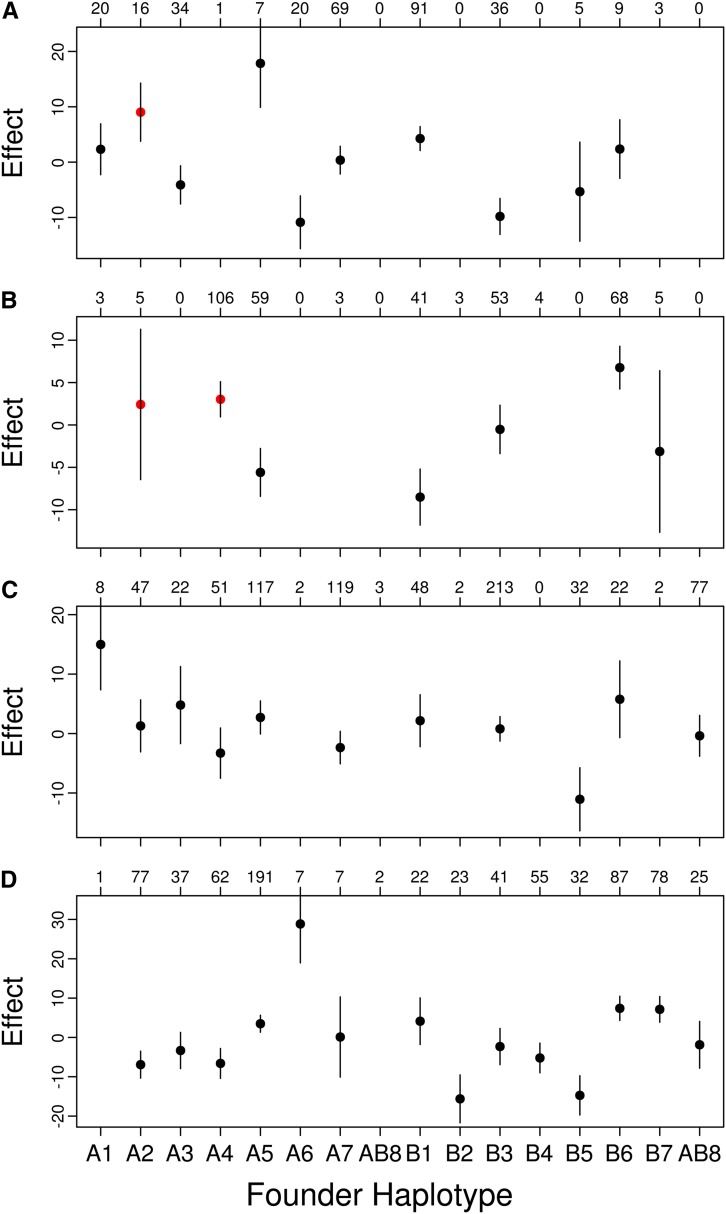
We also estimated the phenotype effects of each founder haplotype on chemotoxicity for all of our identified QTL. The estimated effects of each founder genotype do not show a pattern suggesting simple biallelism for any of the QTL (Figure 4). There are factors such as multiple linked QTL in the region, multiple alleles for a single causative gene, nonadditivity of founder genotypes, or variation in the frequency of founder genotypes in the two populations that could make the effects at our QTL difficult to estimate or interpret. All these factors are collectively examples of heterogeneity, a factor that makes genes difficult to identify using an association study approach (Thornton et al. 2013).
Figure 4.
Standardized founder haplotype means and standard errors at (A) QTL CA, (B) QTL CB, (C) QTL GA, and (D) QTL GB. The number of each confidently assigned (probability >0.95) founder genotype is listed above each plot. Only means with at least five observations are plotted. In A and B, the founder haplotypes harboring a transposable element are red.
QTL CA covers a novel region with no a priori identified candidate genes. The closest a priori candidate gene to this peak is CG1681 (at X:13301019) and it falls well outside the confidence interval for QTL CA. The region spanned by QTL CA includes 60 Drosophila genes, 35 of which have one or more human orthologs. Based on a manual curation effort (see Materials and Methods) for this region we were able to identify three potentially interesting genes that may be involved in carboplatin toxicity, CG9413, CG42271, and na. D. melanogaster gene CG9413 has three human gene orthologs: SLC7A9, SLC7A11, and SLC7A13 (all members of solute carrier family 7A). Human gene SLC7A11 plays a role in maintaining cellular glutathione levels (Dai et al. 2007). Its expression negatively correlates with drug potency across the National Cancer Institute’s 60 cell lines for compounds susceptible to glutathione-mediated chemoresistance (Dai et al. 2007). Although no literature supports that SLC7A11 is directly involved in carboplatin resistance, glutathione detoxification has been broadly implicated in resistance to chemotherapy (Calvert et al. 1989). CG42271 has two human orthologs: INPP4A and INPP4B. Weigman et al. (2012) studied copy number mutations of a region including INPP4B in immortalized human mammary epithelial cell lines and compared their findings to patient survival data. This group found a positive correlation between expression and copy number of INPP4B and showed that INPP4B expression is negatively correlated with relapse-free survival and overall survival using patient data. Therefore, they conclude that loss of the region containing this gene may sensitize tumors to classes of DNA-damaging agents including cisplatin and carboplatin. na in D. melanogaster is a one-to-one ortholog of human gene NALCN. A single nucleotide mutation (SNP) of the NALCN gene at position 100849091 on chromosome 13 has been correlated with decreased patient survival postchemotherapy treatment that included platinum-based treatment. Patients with AA genotype had worse survival rates than GA genotype, and patients with the GG genotype had the best rates of survival in a non-small-cell lung cancer genome-wide association study (Lee et al. 2013). We note that na was not an a priori candidate, since its effect is associated with “platinum-based treatment” as opposed to carboplatin specifically, so it was not picked up using our keyword searches.
QTL CB covers two a priori identified candidate genes: MRP and spellchecker 1 (spel1). This QTL appears as a “double peak,” and given the presence of two candidate genes under this peak possibly represents two closely linked QTL. However, linkage limits our ability to statistically distinguish two QTL in such close proximity and double peaks are sometimes seen with randomized data. MRP is a D. melanogaster one-to-one ortholog of the human gene ABCC2, which is also known as multidrug resistance protein 2 (MRP2) in humans. In the carboplatin cellular pathway, ABCC2 regulates the efflux of platinum products out of the cell. When expressed in the nuclear membrane of an ovarian carcinoma cell culture system this gene has been shown to confer resistance to cisplatin (another platinum-based chemotheraputic agent; Surowiak et al. 2006). Drosophila spel1 is a one-to-one ortholog of the human gene MSH2. MSH2 is involved in mismatch repair; thus it is expected that upregulating MSH2 reduces the cell’s sensitivity to carboplatin-induced point mutations. Fink et al. (1997) showed that an embryonic cell line with a MSH2 knockout is 1.7 times more sensitive to carboplatin than a wild-type cell line.
The gemcitabine toxicity genome scan identified two significant QTL peaks, labeled GA and GB (Figure 3B and Table 1). QTL GA is located on chromosome 2R and includes the a priori identified candidate gene RnrS. This QTL also has a double-peak architecture. However, this is a fairly shallow peak with a wide CI encompassing both portions of the double peak and linkage similarly prevents us from distinguishing multiple QTL within this region. Drosophila gene RnrS is a one:many ortholog of the human genes ribonucleoside–diphosphate reductase subunit M2 (RRM2 and RRM2B). The diphosphate form of gemcitabine is a potent inhibitor of the ribonucleotide reductase activity (needed for DNA synthesis) coded for by these two genes (Whirl-Carrillo et al. 2012). Bhutia et al. (2013) showed that overexpression of RRM2 drives the chemoresistance of pancreatic cancer to gemcitabine. Additionally, it has been shown that prolonged exposure of cancer cells to triapine, an inhibitor of ribonucleotide reductase, enhances gemcitabine activity in vitro (Mortazavi et al. 2013).
QTL GB is located on chromosome 3L (Figure 3B and Table 1). This region includes 40 genes, including fly gene PHGPx, a one-to-one ortholog of human gene GPx4. GPx4 is a gene in the human gene family GPx; however, only GPx4 is considered to be the ortholog of D. melnogaster gene PHGPx. Interestingly, this QTL maps to the same location as a QTL we identified in a previous study dissecting the genes underlying MTX toxicity (Kislukhin et al. 2013; Figure 3D). No studies to date have related GPx4 to gemcitabine toxicity. However, GPx4 encodes a glutathione peroxidase protein shown to have reduced activity in the presence of another cytidine analog, cyterabine. Esfahani et al. (2012) suggest that an increase in the levels of GPx may reduce cytidine efficacy.
Association of chemotoxicity with SNPs and transposable elements
We performed gene-centric association studies for all candidate gene regions under QTL peaks. For all SNPs segregating among the founders we inferred genotypes in the RILs and performed a test for association with chemotoxicity. By focusing on small genomic intervals containing only a handful of SNPs, the statistical threshold for significance is greatly reduced. These gene-centric association scans are depicted in Figure S3 and Figure S4, and the subset of SNPs reaching Bonferroni significance are presented in Table S3 (Bonferroni threshold: P = 3.4 × 10−5). These included one SNP in the CG9413 gene region under QTL CA, explaining as much as 15% of the genetic variation in carboplatin toxicity and nine linked SNPs in the MRP gene region under QTL CB, explaining as much as 13% of the genetic variation in carboplatin toxicity (Figure S3 and Table S3). Only a single SNP associated with gemcitabine toxicity (in gene region PHPGx) reached Bonferroni significance. This SNP may explain as much as 7% of the genetic variance in gemcitabine toxicity (Figure S4 and Table S3). The above being said, it is not uncommon for SNPs under QTL peaks but outside of candidate gene intervals to explain as much of the variation as the SNPs identified above. So these SNPs are only of interest insomuch as the candidate gene is a true positive.
We also identified transposable elements (TE) residing on founder haplotypes in candidate gene regions (Table S2). None of the four TEs segregating at appreciable frequency in the RILs shows a phenotypic effect estimate that is a large outlier, as would be expected if the QTL peak were largely due to the transposable element (Figure 4) and none of the TEs reached Bonferroni significance (Bonferroni threshold, P = 3.4 × 10−5; X:14485885, P = 0.14; 2L:12725321, P = 0.04; 2L:12748174, P = 0.28; 2L:14375490, P = 0.10). In addition, each TE is present only in a single founder haplotype so the effect of the TE cannot be distinguished from the overall effect of the haplotype. Overall, these TEs could be contributors to the signal at QTL CA and CB, but no single TE explains a substantial amount of the variation due to this QTL.
None of the significant SNPs are predicted to encode amino acids, and therefore, our identified QTL cannot be explained primarily by coding SNPs, which have been the focus of most past chemotoxicity studies (cf. Monzó et al. 2008; Schneider et al. 2012). Even in the case where we find significant SNPs under a peak, no single SNP is able to explain the complete linkage signal. This fact, combined with the observation that our estimated haplotype effects do not conform to a simple biallelic pattern (Figure 4), suggests that more than two variants are segregating at each candidate gene. We therefore hypothesize that our QTL are likely due to multiple SNPs, the majority of which may be noncoding.
Pleiotropy for chemotoxicity
Figure 3D depicts a summary of the QTL mapping results for the three drugs of this article as well as our scan for MTX toxicity genes from a previous article (Kislukhin et al. 2013). In most cases, identified peaks are different for the different drugs, consistent with the genetic basis of toxicity being largely independent across drugs. The observation that peaks for one drug do not tend to be associated with subtle peaks for a second drug suggests true independence of the underlying genetics of these characters and not just low power to map QTL. That being said, two QTL for carboplatin and MTX on the X chromosome (QTL CA and MTX QTL A; Kislukhin et al. 2013) are very close to one another. It could be hypothesized that the two peaks represent a single pleiotropic gene. The Bayesian credible intervals for these two peaks do overlap with a common region from X: 13.48 (Mb) to X: 14.10 (Mb). However, we identified a very strong candidate gene for the MTX QTL A, CG32626, an ortholog of the human gene adenosine monophosphate deaminase 1 (AMPD1). The location of this candidate gene (X:13724774) falls outside the confidence interval for carboplatin QTL CA (Table 1). We therefore conclude that these peaks are likely due to independent genes that just happen to be located in close proximity.
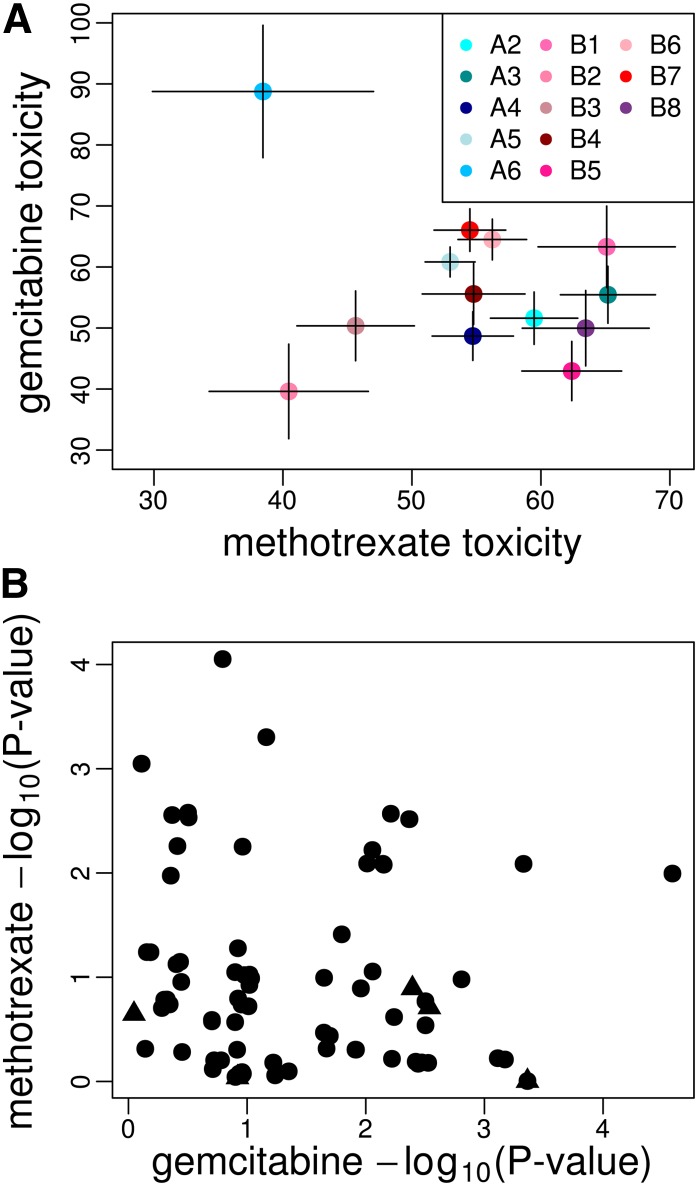
QTL GB and MTX peak C (Kislukhin et al. 2013), located toward the telomeric end of 3L, are likely the same gene. Their Bayesian credible intervals include a large region of overlap [3L: 6.47 (Mb)–3L: 7.39(Mb)]; in fact the entire interval for GB falls within the interval for MTX QTL C. This peak thus seems to represent a fly gene that affects the toxicity of gemcitabine and MTX, but not carboplatin or mitomycin C. Although there are no a priori candidate genes under this peak, the fly gene PHGPx, a one-to-one ortholog of human gene GPx4, is a good a posteriori candidate gene. GPx4 encodes a glutathione peroxidase, a major reactive oxygen species (ROS)-scavenging enzyme. Because one mechanism by which MTX and gemcitabine induce cell damage is via an increase in ROS in treated cells (Koulajian et al. 2013), it is possible that fly genotypes with increased PHGPx expression tolerate these drugs better than low-expression genotypes. The estimated founder means at PHGPx are negatively correlated; however, this correlation is driven entirely by founder A6, which has only N = 6 representative genotypes of which three appear to have very low fecundity knockdown for gemcitabine (Figure 5A, Figure 4D, and Kislukhin et al. 2013, Figure 4C). Excluding founder haplotype A6, the founder means appear to be weakly positively correlated.
Figure 5.
(A) Founder haplotype means and standard errors at QTL GB/MTX QTL D. The estimates are the standardized means after correcting for subpopulation added to the average fecundity over all crosses for each drug. Different colors correspond to different haplotypes. Only means with at least five observations are plotted. (B) The −log10(P-value) for single-marker scans for gemcitabine toxicity vs. methotrexate toxicity. Triangles are nsSNPs.
We also attempted to identify SNPs in or near PHGPx that are strongly associated with toxicity in both MTX and gemcitabine-treated flies. Sixteen SNPs are significant at P < 0.01 for both drugs, of which two are also significant at P < 0.001 for gemcitabine, and none of these SNP are nonsynonymous (Figure 5B and Table S4). This being said, several SNPs are significant for one drug but not the other, consistent with different SNPs affecting the different drugs, or perhaps just limited power to implicate the causative SNPs themselves (King et al. 2012b). Also, as stated above, no single SNP in PHGPx explains the entire mapping signal for either carboplatin or MTX, so by extension it is equally difficult to identify a single site affecting both drugs as causative.
Are candidate genes enriched under QTL peaks?
In our previous study of MTX toxicity we identify three QTL, of which two are directly over 2 of 15 a priori candidate genes (Kislukhin et al. 2013). In this study we identified two QTL for carboplatin (covering 2.66 Mb total) associated with 2 of 32 candidate genes, and two QTL for gemcitabine (covering 2.58 Mb total) associated with 1 of 9 candidate genes (Table S1 and Table S2). Despite QTL peaks being much more narrowly defined in the DSPR than previous QTL mapping experiments in Drosophila (Mackay 2001; King et al. 2012b), cumulatively, they still cover megabase size regions of the genome. Furthermore, despite candidate genes being extremely helpful, chemotherapy drugs have broad enough targets that in many cases a plethora of excellent candidate genes exist. It is therefore important to determine if the association between candidate genes and peaks in our data sets is greater than we expect by chance alone. For any given genome scan the probability of observing the NQTL or greater candidate genes under peaks out of NT total is 1 − ppois(NQTL − 1, NT *LQTL/LT), where ppois is the poisson cumulative distribution, LQTL is the total length of the genome under QTL peaks, and LT is the total length of the fly genome with normal levels of recombination to which we can localize QTL(= 82 Mb; X:2.5–21, 2L:1–17.5, 2R:7–19, 3L:1–19, 3R:7–24). For MTX, carboplatin, and gemcitabine these probabilities are 7.5, 28, and 24%, respectively. Although these probabilities are suggestive only for any individual drug, over the three drugs for which we have mapped QTL the overall probability of observing the correspondence with a priori candidate genes by chance is 0.5%. Although some of the candidate genes associated with peaks are likely false positives, the overall correspondence between Drosophila orthologs of human candidate genes is quite striking.
Conclusion
We have shown that we can use the reduction in female fecundity following oral exposure to chemotherapy drugs as a measure of chemotoxicity. Furthermore, this fecundity knockdown in flies often has a large heritable component (Kislukhin et al. 2012; Kislukhin et al. 2013; this article) and the genetic basis of the knockdown is polygenic. We have used the DSPR as a tool to map genes affecting this fecundity knockdown following oral chemotherapy phenotype and identify a handful of QTL for each drug examined (excluding mitomycin C). We observe that roughly half of all mapped QTL are associated with fly orthologs of an a priori set of human candidate genes believed to be important in chemotoxicity. This suggests that Drosophila can be efficiently used to achieve a greater understanding of toxicity, especially for these genes. In the cases where mapped QTL are not associated with a priori candidate genes they are often localized well enough that we identify new candidate genes. Validating candidate genes that affect fecundity is especially challenging because knockdowns may often phenocopy a decrease in fecundity. Future studies will attempt to validate these novel candidates using tissue-targeted knockdowns with carefully designed controls, but these genes should be examined simultaneously in clinical settings as soon as is reasonably possible. It is of interest that one QTL mapped for both gemcitabine and MTX likely represents a single gene affecting the toxicity of both drugs. We are unaware of any such genes in humans, making PHGPx/GPx4 of particular interest.
An important property of the DSPR is that 15 different wild founder alleles are segregating in the 1700 RILs that are available for QTL mapping. This allows estimates of the effect of each founder chromosome on the toxicity phenotype at the most likely location of mapped QTL. When we estimate these effects for the drugs studied to date, the distribution of founder haplotype effects suggests that the mapped QTL are segregating several, as opposed to two, functional alleles affecting chemotoxicity. This polyallelism leads to a genetic region having a large effect in a mapping context, yet makes it difficult to identify individual causative SNPs in an association study framework. Consistent with the genetic basis of chemotoxicity being due to a few genes of large effect each segregating several smaller-effect functional alleles, association scans under the peaks fail to identify single SNPs or transposable elements that explain the entire linkage signal.
Finally, the association signals with nonsynonymous SNPs are generally weaker than with noncoding SNPs, perhaps suggesting that the overall genetic basis of toxicity as measured by a knockdown in female fecundity following oral exposure to chemotherapy drugs is largely due to changes in gene regulation, as opposed to changes in the protein structure itself. Future experiments could utilize the RNAseq and DNAse I hypersensitivity “seq” data sets associated with the DSPR lines to identify SNPs contributing to variation in gene expression at these candidate genes. Clearly it is of great value to estimate the fraction of chemotoxicity genetic variation associated with regulatory vs. coding changes.
Supplementary Material
Acknowledgments
We acknowledge Alyssa Skala for her contribution to acquiring phenotypic data, Dr. Mahtab Jafari for her contribution to preliminary studies of chemotherapeutic drug delivery to D. melanogaster, and Dr. Stuart J. Macdonald for support.This work was supported by National Institutes of Health grants R01 GM085251 (to A.D.L.), R01 RR024862 (to A.D.L. and Stuart J. Macdonald), and F32 GM099382 (to E.G.K.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.161968/-/DC1
Communicating editor: B. E. Huang
Literature Cited
- Alley E., Green R., Schuchter L., 2002. Cutaneous toxicities of cancer therapy. Curr. Opin. Oncol. 14: 212–216 [DOI] [PubMed] [Google Scholar]
- Aulchenko Y. S., de Koning D.-J., Haley C., 2007. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 177: 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanesian A., Semnani S., Jafari M., 2009. Can Drosophila melanogaster represent a model system for the detection of reproductive adverse drug reactions? Drug Discov. Today 14: 761–766 [DOI] [PubMed] [Google Scholar]
- Bhutia Y. D., Hung S. W., Krentz M., Patel D., Lovin D., et al. , 2013. Differential processing of let-7a precursors influences RRM2 expression and chemosensitivity in pancreatic cancer: role of LIN-28 and SET oncoprotein. PLoS ONE 8: e53436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., 2005. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6: 9–23 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Sen S., 2009. A Guide to QTL Mapping with R/qtl. Springer, New York [Google Scholar]
- Calvert A. H., Newell D. R., Gumbrell L. A., O’Reilly S., Burnell M., et al. , 1989. Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J. Clin. Oncol. 7: 1748–1756 [DOI] [PubMed] [Google Scholar]
- Cheng H., Sun N., Sun X., Chen B., Li F., et al. , 2010. Polymorphisms in hMSH2 and hMLH1 and response to platinum-based chemotherapy in advanced non-small-cell lung cancer patients. Acta Biochim. Biophys. Sin. 42: 311–317 [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., MacDonald S. J., Long A. D., Thornton K. R., 2013. Abundance and distribution of transposable elements in two QTL resources. Mol. Biol. Evol. 30(10): 2311–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Huang Y., Sadee W., Blower P., 2007. Chemoinformatics analysis identifies cytotoxic compounds susceptible to chemoresistance mediated by glutathione and cystine/glutamate transport system xc-. J. Med. Chem. 50: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Esfahani A., Ghoreishi Z., Nikanfar A., Sanaat Z., Ghorbanihaghjo A., 2012. Influence of chemotherapy on the lipid peroxidation and antioxidant status in patients with acute myeloid leukemia. Acta Med. Iran. 50: 454–458 [PubMed] [Google Scholar]
- Fink D., Nebel S., Aebi S., Nehme A., Howell S., 1997. Loss of DNA mismatch repair due to knockout of MSH2 or PMS2 results in resistance to cisplatin and carboplatin. Int. J. Oncol. 11: 539–542 [DOI] [PubMed] [Google Scholar]
- Gajewski J. L., Ho W. G., Nimer S. D., Hirji K. F., Gekelman L., et al. , 1989. Efficacy of intensive chemotherapy for acute myelogenous leukemia associated with a preleukemic syndrome. J. Clin. Oncol. 7: 1637–1645 [DOI] [PubMed] [Google Scholar]
- Gong, L., W. Constantine, and Y. A. Chen, 2012 msProcess: Protein Mass Spectra Processing, R package version 1.0.7. http://CRAN.R-project.org/package=msProcess
- Iaffaioli R. V., Tortoriello A., Santangelo M., Turitto G., Libutti M., et al. , 2000. Phase I dose escalation study of gemcitabine and paclitaxel plus colony-stimulating factors in previously treated patients with advanced breast and ovarian cancer. Clin. Oncol. 12: 251–255 [DOI] [PubMed] [Google Scholar]
- King E. G., MacDonald S. J., Long A. D., 2012a Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics 191: 935–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S., et al. , 2012b Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislukhin G., King E. G., Walter K. N., Macdonald S. J., Long A. D., 2013. The genetic architecture of methotrexate toxicity is similar in Drosophila melanogaster and humans. G3 (Bethesda): 3: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislukhin G., Murphy M. L., Jafari M., Long A. D., 2012. Chemotherapy-induced toxicity is highly heritable in Drosophila melanogaster. Pharmacogenet. Genomics 22: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulajian, K., A. Ivovic, K. Ye, T. Desai, A. Shah et al., 2013 Overexpression of glutathione peroxidase 4 (GPx4) prevents β-cell dysfunction induced by prolonged elevation of lipids in vivo. Am. J. Physiol. Endocrinol. Metab. 305: E254–E262 [DOI] [PubMed] [Google Scholar]
- Lee W., Lockhart A. C., Kim R. B., Rothenberg M. L., 2005. Cancer pharmacogenomics: powerful tools in cancer chemotherapy and drug development. Oncologist 10: 104–111 [DOI] [PubMed] [Google Scholar]
- Lee Y., Yoon K.-A., Joo J., Lee D., Bae K., et al. , 2013. Prognostic implications of genetic variants in advanced non-small cell lung cancer: a genome-wide association study. Carcinogenesis 34: 307–313 [DOI] [PubMed] [Google Scholar]
- Li L., Schaid D. J., Fridley B. L., Kalari K. R., Jenkins G. D., et al. , 2012. Gemcitabine metabolic pathway genetic polymorphisms and response in patients with non-small cell lung cancer. Pharmacogenet. Genomics 22: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339 [DOI] [PubMed] [Google Scholar]
- Manichaikul A., Dupuis J., Sen S., Broman K. W., 2006. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics 174: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S., McLeod H., Dolan E., Shukla S. J., Rabik C. A., et al. , 2009. Platinum pathway. Pharmacogenet. Genomics 19: 563–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirow D., Nugent D., 2001. The effects of radiotherapy and chemotherapy on female reproduction. Hum. Reprod. Update 7: 535–543 [DOI] [PubMed] [Google Scholar]
- Monzó M., Navarro A., Ferrer G., Artells R., 2008. Pharmacogenomics: a tool for improving cancer chemotherapy. Clin. Transl. Oncol. 10: 628–637 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Ling Y., Martin L. K., Wei L., Phelps M. A., et al. , 2013. A phase I study of prolonged infusion of triapine in combination with fixed dose rate gemcitabine in patients with advanced solid tumors. Invest. New Drugs 31: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Lander E. S., Hewitt J. D., Peterson S., Lincoln S. E., et al. , 1988. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335: 721–726 [DOI] [PubMed] [Google Scholar]
- Pinheiro, J. C., D. M. Bates, S. Debroy, D. Sarkar, and R Development Core Team, 2013 nlme: linear and nonlinear mixed effects models, R package version 3.1-109. Available at: http://CRAN.R-project.org/package=nlme
- R Development Core Team, 2013 R: A language and environment for statistical computing R Foundation for Statistical Computing Vienna, Austria. http://www.R-project.org/
- Rothenberg M. L., Oza A. M., Bigelow R. H., Berlin J. D., Marshall J. L., et al. , 2003. Superiority of oxaliplatin and fluorouracil-leucovorin compared with either therapy alone in patients with progressive colorectal cancer after irinotecan and fluorouracil-leucovorin: interim results of a phase III trial. J. Clin. Oncol. 21: 2059–2069 [DOI] [PubMed] [Google Scholar]
- Schneider B. P., Shen F., Miller K. D., 2012. Pharmacogenetic biomarkers for the prediction of response to antiangiogenic treatment. Lancet Oncol. 13: e427–e436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique Y. H., Ara G., Beg T., Gupta J., Afzal M., 2010. Assessment of cell viability, lipid peroxidation and quantification of DNA fragmentation after the treatment of anticancerous drug mitomycin C and curcumin in cultured human blood lymphocytes. Exp. Toxicol. Pathol. 62: 503–508 [DOI] [PubMed] [Google Scholar]
- Steffensen K. D., M. Waldstrøm, and A. Jakobsen, 2009. The relationship of platinum resistance and ERCC1 protein expression in epithelial ovarian cancer. Int. J. Gynecol. Cancer 19: 820–825 [DOI] [PubMed] [Google Scholar]
- Sugiyama E., Lee S.-J., Lee S. S., Kim W.-Y., Kim S.-R., et al. , 2009. Ethnic differences of two non-synonymous single nucleotide polymorphisms in CDA gene. Drug Metab. Pharmacokinet. 24: 553–556 [DOI] [PubMed] [Google Scholar]
- Surowiak P., Materna V., Kaplenko I., Spaczynski M., Dolinska-Krajewska B., et al. , 2006. ABCC2 (MRP2, cMOAT) can be localized in the nuclear membrane of ovarian carcinomas and correlates with resistance to cisplatin and clinical outcome. Clin. Cancer Res. 12: 7149–7158 [DOI] [PubMed] [Google Scholar]
- Thornton K. R., Foran A. J., Long A. D., 2013. Properties and modeling of GWAS when complex disease risk is due to non-complementing, deleterious mutations in genes of large effect. PLoS Genet. 9: e1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Ambrosone C. B., Darcy K. M., Krivak T. C., Armstrong D. K., et al. , 2012. Common variants in ABCB1, ABCC2 and ABCG2 genes and clinical outcomes among women with advanced stage ovarian cancer treated with platinum and taxane-based chemotherapy: a Gynecologic Oncology Group study. Gynecol. Oncol. 124: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters J. W., Mcleod H. L., 2003. Cancer pharmacogenomics: current and future applications. Biochim. Biophys. Acta 1603: 99–111 [DOI] [PubMed] [Google Scholar]
- Weigman V. J., Chao H.-H., Shabalin A. A., He X., Parker J. S., et al. , 2012. Basal-like breast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy, and patient survival. Breast Cancer Res. Treat. 133: 865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheate N. J., Walker S., Craig G. E., Oun R., 2010. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 39: 8113–8127 [DOI] [PubMed] [Google Scholar]
- Whirl-Carrillo M., McDonagh E. M., Hebert J. M., Gong L., Sangkuhl K., et al. , 2012. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92: 414–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., 2003. Theoretical basis of the Beavis effect. Genetics 165: 2259–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C. J., Du W., Zhang Q., Zhang F., Zeng F., et al. , 2013. Fanconi anemia pathway: the way of DNA interstrand cross-link repair. Pharmazie 68: 5–11 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.