Abstract
Inappropriate expression or activation of transcription factors can drive patterns of gene expression leading to the malignant behavior of breast cancer cells. We have found that the transcriptional repressor BCL6 is highly expressed in breast cancer cell lines, and its locus is amplified in about half of primary breast cancers. To understand how BCL6 regulates gene expression in breast cancer cells, we utilized ChIP-seq to identify the BCL6 binding sites on a genomic scale. This revealed that BCL6 regulates a unique cohort of genes in breast cancer cell lines compared to B cell lymphomas. Furthermore, BCL6 expression promotes the survival of breast cancer cells, and targeting BCL6 with a peptidomimetic inhibitor leads to apoptosis of these cells. Finally, combining a BCL6 inhibitor and a STAT3 inhibitor provided enhanced cell killing in triple negative breast cancer cell lines, suggesting that combination therapy may be particularly useful. Thus, targeting BCL6 alone or in conjunction with other signaling pathways may be a useful therapeutic strategy for treating breast cancer.
Keywords: Breast cancer, Transcription modulators, Targeted therapy, Gene expression
Introduction
One hallmark of cancer cells is aberrant gene expression leading to phenotypes promoting malignancy such as proliferation, evasion of apoptosis, self-renewal, and metastasis. Inappropriate expression or activation of transcription factors drives these gene expression patterns, and cancer cells are often dependent on their continued activation. Understanding how these oncogenic transcription factors regulate gene expression and phenotypic characteristics of cancer cells can shed light on tumor pathogenesis, and provide insight into strategies for rationally designed targeted therapies.
Several lines of evidence suggest that BCL6, a transcriptional repressor that is oncogenic in B cell lymphomas, may play a role in the pathogenesis of breast cancer. In addition to maintaining the survival and blocking the terminal differentiation of lymphoma cells, BCL6 has been shown to prevent differentiation of mammary cells [1]. Through its effects on gene regulation, BCL6 controls cellular processes including cell cycle progression, DNA damage sensing, and protein ubiquitination [2, 3]. BCL6 is directly regulated by oncogenic STAT transcription factors, and its expression is enhanced by inappropriately activated STAT3, which occurs in a majority of breast cancers [4, 5]. While BCL6 is rarely detected by immunohistochemistry in normal mammary epithelium, it is commonly expressed in breast cancers, and has been found in 68% of high grade ductal carcinomas [1, 6]. Whether BCL6 regulates gene expression and contributes to the survival and biological properties of breast cancer cells remains unknown.
BCL6 binds to canonical DNA sequences in the regulatory region of target genes. Once bound, it recruits corepressor complexes that introduce repressive chromatin marks, thereby repressing transcription [7]. For example BCL6 can recruit SMRT, NCOR, and BCOR through its BTB domain. These repressor complexes are then able to repress a subset of BCL6 target genes involved in cellular proliferation and survival [8–12]. BCL6 can also interact with MTA3 through its second repression domain (RD2). Through MTA3 BCL6 represses genes involved in terminal differentiation [13, 14]. Finally, BCL6 also recruits CtBP, through which it can repress its own expression [15, 16]. Thus, BCL6 can mediate complex and diverse effects on gene expression.
Given these distinct interactions, the activity of BCL6 can be inhibited in specific ways. A peptidomimetic inhibitor, RI-BPI, can prevent the recruitment of SMRT, NCoR, and BCoR by binding to the BTB domain of BCL6 [9, 17]. Following treatment with RI-BPI, BCL6 is still able to bind to DNA, but it can no longer repress expression of genes normally regulated by those complexes. Importantly, RI-BPI kills diffuse large B cell lymphoma cell lines and patient samples [9, 17], demonstrating that genes derepressed by RI-BPI are important for the survival of DLBCL cells. In addition, RI-BPI treatment is effective in B cell lymphoma models in vivo, while having no apparent toxicity in mice [17]. These findings suggest that BCL6 is an appealing therapeutic target.
Given the potential role of BCL6 in the biology of breast cancer cells, we analyzed the genomic binding patterns of BCL6 in breast cancer cell lines. We also determined the role of BCL6 on gene expression and survival of breast cancer cells, and assessed its potential as a therapeutic target in breast cancer.
Results
BCL6 is amplified in primary breast cancers
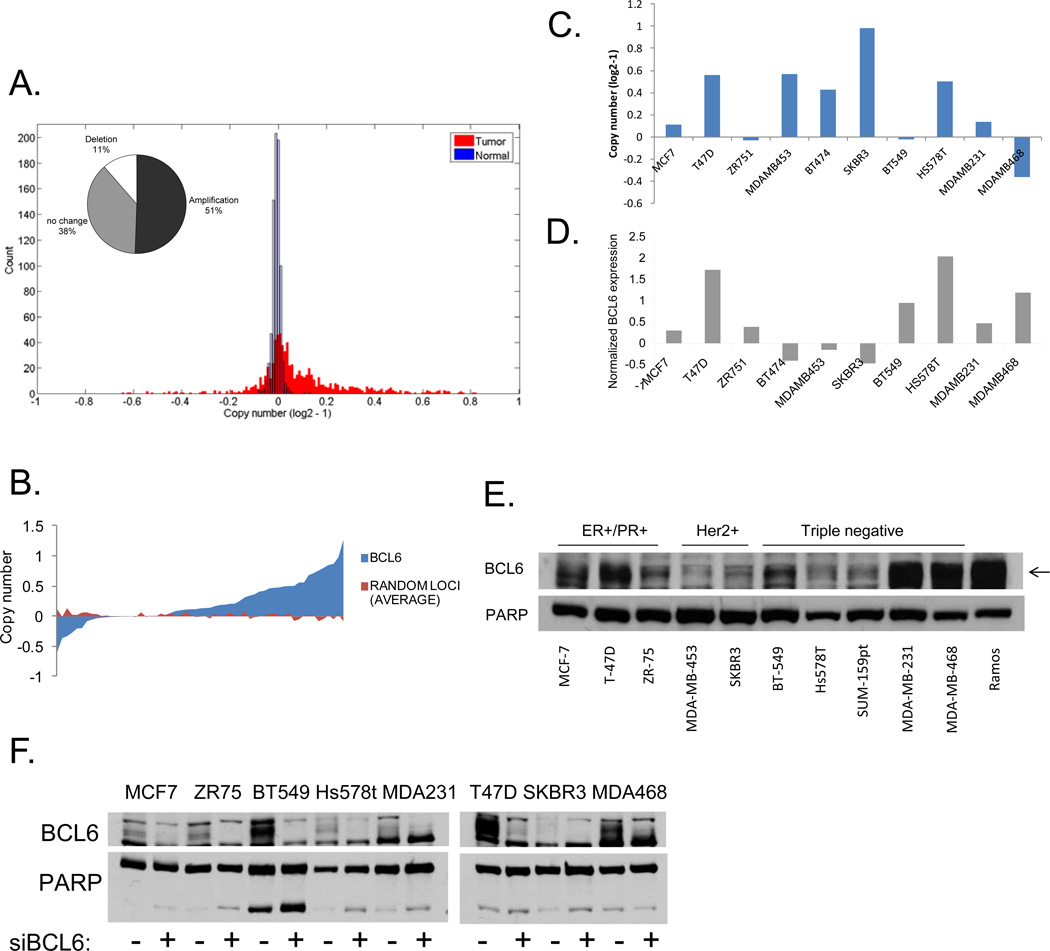
Given the suggestion that BCL6 might play a role in breast cancer pathogenesis, we wanted to determine if the BCL6 locus was amplified in primary human breast tumors. Using data derived from The Cancer Genome Atlas (TCGA), we found that 51% of the 790 breast tumors analyzed showed amplification of the BCL6 locus, while only a small subset (11%) showed a reduction of this locus (Figure 1a). The skewing towards increased copy number showed a nominal p value of .056 compared to 1,000 randomly chosen sites. This finding raised the possibility that overexpression of BCL6 may be important in breast cancer pathogenesis. Next, we used data from the Cancer Cell Line Encyclopedia [18] to analyze BCL6 copy number and mRNA expression in breast cancer cell lines. We found that the majority of breast cancer cell lines also displayed amplification of this locus (p=0.01) (Figure 1b), and this was evident in lines derived from all three subtypes of breast cancer (Supplementary Figure 1a). In addition, greater than 75% of the cell lines showed enhanced expression of BCL6. Of the small number of cell lines showing a loss at this locus, most were from triple negative breast tumors. Interestingly, however, even among this group, the majority of lines showed increased BCL6 mRNA expression, suggesting that there is a strong selective pressure for expression of this gene (Supplementary Figure 1a). To better understand the role BCL6 plays in breast cancer, we chose a panel of breast cancer cells containing both amplifications and deletions of this locus, and which represented the three major classes of breast cancer: hormone receptor positive (ER+/PR+), Her2/ErbB2 amplified (Her2+), and ER−/PR−/Her2− (triple negative) (Figure 1c). We then analyzed the expression of BCL6 mRNA (Figure 1d) and protein (supplementary figure 1b) in these lines, compared to non-tumorigenic mammary epithelial lines. We found that BCL6 was expressed to varying degrees in 10 of the 11 breast cancer cells lines analyzed and there was generally less expression of BCL6 in non-tumorigenic breast lines compared to the breast cancer cell lines (Supplementary Figure 1b). Expression levels did not correlate with a specific breast cancer subtype. We further analyzed levels of nuclear BCL6 expression and found that BCL6 was present to varying degrees in the nucleus of all ten breast cancer cell lines tested (Figure 1e). BCL6 becomes post-translationally modified [19] and runs across a range of molecular weights. To ensure analysis of the correct band from protein extracts, we treated cells with siRNA to BCL6 or control and performed immunoblots to BCL6 on nuclear extracts. This confirmed that BCL6 is expressed in all eight of the breast cancer cell lines analyzed (Figure 1f). Thus, BCL6 is expressed in nearly all breast cancer cell lines tested, and the locus is amplified in approximated half of primary human breast tumors and the majority of breast cancer cell lines. We chose to focus on T-47D, SK-BR-3, and MDA-MB-468 cells for further study as they represent the three different subtypes of breast cancer and also reflect three different levels of BCL6 expression.
Figure 1.
BCL6 is expressed in breast cancer. Copy number for BCL6 was analyzed using data from (A) the Cancer Genome Atlas for breast tumors (inset contains percentages) or (B) from the Cancer Cell Line Encyclopedia for breast cancer cell lines. Copy number (C) and BCL6 gene expression (D) from the Cancer Cell Line Encyclopedia for a subset of breast cancer cell lines. (E) Nuclear lysates were generated from the indicated breast cancer cell lines and analyzed for BCL6 expression. Whole cell extracts of Ramos cells served as a positive control for BCL6 expression. PARP served as a control for nuclear protein. (F) Nuclear lysates were generated from the indicated cell lines after 72 hours of transfection with siRNA to BCL6 (or control). PARP served as a control for nuclear protein.
BCL6 binds to unique genomic sites in breast cancer cells
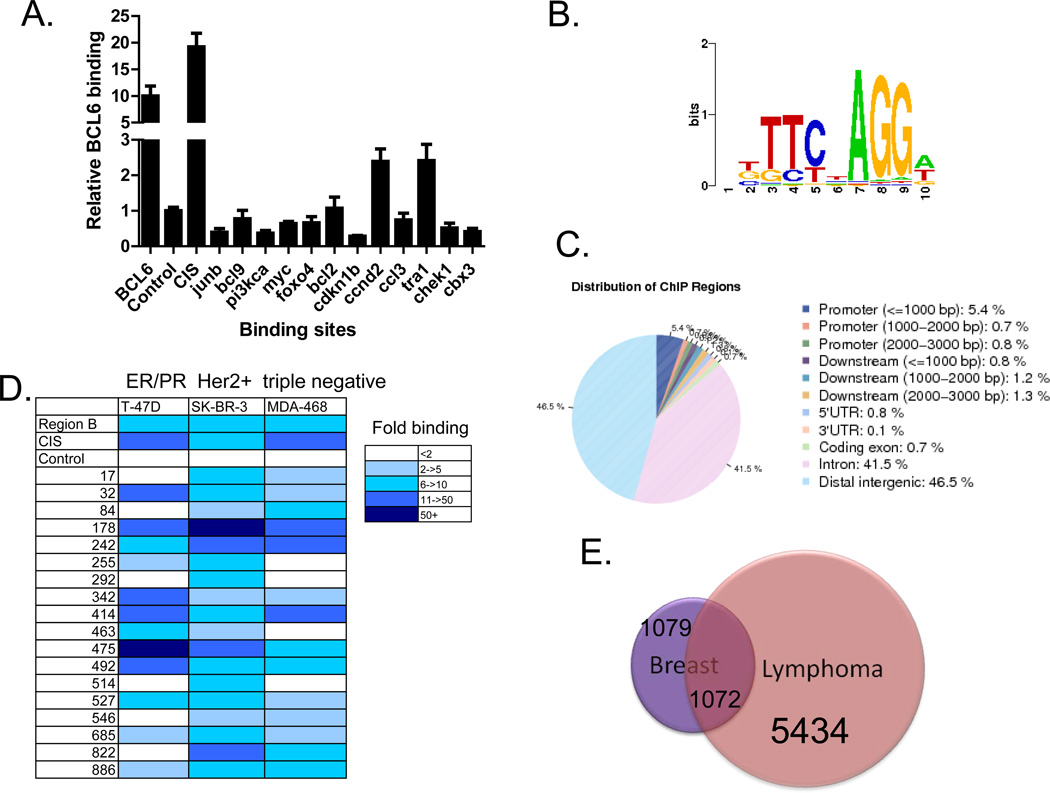
To determine the function of BCL6 in breast cancer cells, we next sought to identify its target genes in this tumor type. Although binding sites for BCL6 have been identified in B cell lymphoma cells [2, 3], it was not known whether BCL6 binding sites would be similar in breast cancer cells. We performed ChIP in SK-BR-3, T-47D, and MDA-MB-468 breast cancer cell lines, and first analyzed previously characterized BCL6 binding sites. We found that BCL6 binds to the well-characterized BCL6 binding site within the BCL6 gene itself, the so-called region B [20] as well as the promoter of the CIS gene (Figure 2a and data not shown). Having confirmed BCL6 localization to these sites, we then analyzed 12 BCL6 binding sites that had been defined in lymphoma cells [2, 3]. We found that only two of these sites were bound by BCL6 in only one of three breast cancer cell lines tested (Figure 2a and data not shown). This suggested that while some of the BCL6 binding sites in breast cancer cells overlap with those identified in lymphoma cells, there was likely to be a unique pattern of binding of BCL6 in breast cancer.
Figure 2.
BCL6 binds to specific genomic sites in breast cancer cells. (A) ChIP was performed in SK-BR-3 cells for BCL6 binding to target sites identified in lymphoma cells. Data are expressed as BCL6 binding relative to background (at a nonbinding region). (B) De novo motif analysis of genomic sequences associated with BCL6 binding sites in breast cancer cells identified the BCL6 binding motif. (C) Genomic distribution of BCL6 binding sites relative to genes in breast cancer cells. (D) BCL6 ChIP was performed in three different breast cancer cell lines, and binding of BCL6 to binding sites was analyzed by qPCR. (E) Genes containing at least one BCL6 binding site were compared from the breast cancer and lymphoma ChIP-seq datasets.
To analyze BCL6 binding in breast cancer on a genomic scale, we performed ChIP followed by deep sequencing (ChIP-seq). Given that we had validated strong binding of BCL6 to four sites in SK-BR-3 cells, we chose to perform ChIP-seq in this cell line. Using a p value of 10−6 as a cutoff, we identified 4118 BCL6 binding sites. Analysis of the sequences within the peaks using a de novo motif search identified the BCL6 motif (Figure 2b). In addition, analysis of known motifs through the TRANSFAC database also identified the BCL6 binding site as one of the top binding sites found within these sequences (p value = 9.33 × 10−18). Together this suggests that this approach identified bona fide BCL6 binding sites. Analysis of BCL6 localization revealed that 42% of the binding sites are within introns of genes and 47% of the sites are located in regions that are greater than 3,000 base pairs from the transcription start site. Only 7% of the sites are within the first 3,000 base pairs 5’ from the start site of genes, the traditional promoter regions (Figure 2c). Similar distribution of binding has been described for other transcriptional modulators [21, 22]. Quantitative ChIP analysis was performed on 20 of the most highly bound sites identified by ChIP-seq (with a false discovery rate of 0). All 20 of these sites (18 new sites and two known sites) were bound by BCL6 in SK-BR-3 cells with at least 2 fold binding over background (i.e., genomic regions known not to be bound by BCL6) (Figure 2d). In addition, comparison of binding of these sites in cell lines representing the three major classes of breast cancer types determined that most, but not all of the sites, were bound by BCL6 in all three cell lines, though to varying levels (Figure 2d). This suggests that some differences in BCL6 function may occur in the different subtypes of breast cancer cells.
To determine whether BCL6 target genes in breast cancer cells were distinct from those in B-cell lymphomas we performed ChIP-seq for BCL6 in the OCI-LY1 diffuse large B-cell lymphoma cell line. We then compared the genes containing at least one peak in each cell line. From this analysis, we determined that one sixth of the genes bound by BCL6 in lymphoma cells also show BCL6 binding in breast cancer cells (Figure 2e). Conversely, about half of the potential BCL6 target genes in breast cancer cells are bound by BCL6 in lymphoma cells, and half are unique to the breast cancer cells. These findings suggest that BCL6 has both similar and distinct functions in breast cancer and lymphoma.
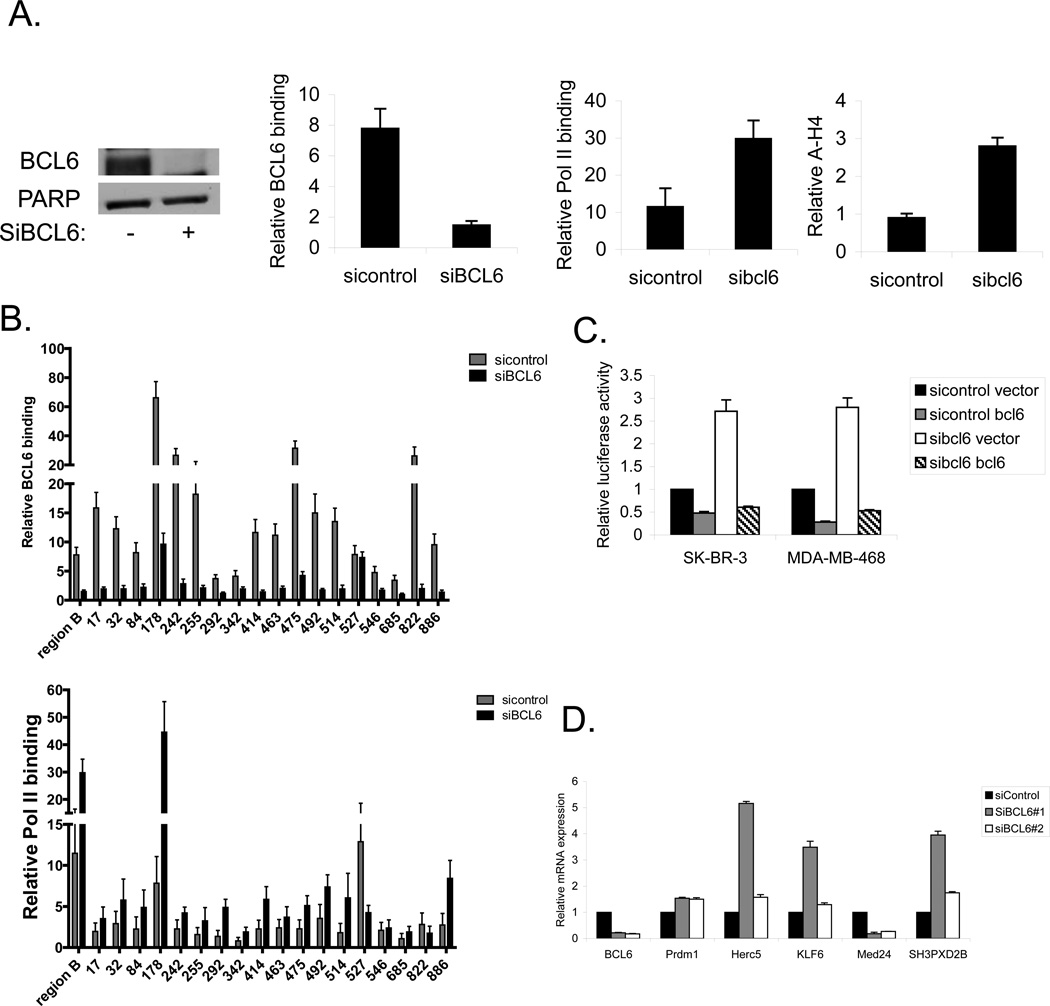
To validate that these sites are bona fide functional BCL6 binding sites important for gene regulation, we knocked down BCL6 with siRNA (Figure 3a). Focusing initially on the BCL6 binding site within the BCL6 locus (region B), we determined that siRNA to BCL6 reduced BCL6 binding to this site by approximately 75% (Figure 3a). In addition, knocking down BCL6 also resulted in an increase of RNA polymerase II binding and an increase in acetylated histone H4, consistent with the loss of a repressive effect of BCL6 on transcription at this site. Similar results were seen in MDA-MB-468 cells, and there was a more modest increase in RNA polymerase II binding in T-47D cells (Supplementary Figure 2a). Analysis of the newly identified BCL6 binding sites demonstrated that binding of BCL6 was reduced at 17 of the 18 sites upon introduction of siRNA to BCL6 (Figure 3b). In addition, RNA polymerase was recruited to most but not all of the sites with depletion of BCL6. These findings provide additional evidence that the sites identified by ChIP-seq in breast cancer cells are likely to be functional BCL6 binding sites.
Figure 3.
BCL6 modulates gene expression in breast cancer cells. (A) SK-BR-3 cells were transfected with siRNA to BCL6 (or control). Extracts were then analyzed for BCL6 expression by immunoblot (left) or by ChIP for binding of BCL6, RNA polymerase II, and acetylated histone H4 to region B of the BCL6 gene (right). (B) SK-BR-3 cells treated with siRNA to BCL6 (or control) were analyzed by ChIP for BCL6 (top) and RNA polymerase II (bottom) binding at the indicated sites. Binding is expressed relative to a negative control region. (C) Breast cancer cells treated with the indicated expression vectors and siRNA constructs were transfected with a BCL6 responsive luciferase plasmid and analyzed for luciferase activity. (D) SK-BR-3 breast cancer cells treated with siRNA to BCL6 or control were analyzed by qRT-PCR for expression of the indicated genes relative to GAPDH.
BCL6 binding is associated with modulation of gene expression
To directly measure the effect of BCL6 binding on gene expression in breast cancer cells, we first transfected a BCL6 responsive luciferase reporter into SK-BR-3, MDA-MB-468, and T-47D breast cancer cells (Figure 3c and supplementary figure 2b). Ectopic expression of BCL6 reduced expression of luciferase, demonstrating that this reporter is repressed by BCL6. Conversely, knocking down BCL6 with siRNA resulted in increased luciferase expression. Finally, forced re-expression of BCL6 in the presence of siRNA to BCL6 again led to repression of luciferase activity. Additionally, the small molecule BCL6 inhibitor 79-6 [23] also resulted in increased luciferase activity in all three cell lines (Supplementary Figure 2c), further demonstrating that BCL6 is active in these cells and that inhibiting BCL6 results in derepression of this reporter construct.
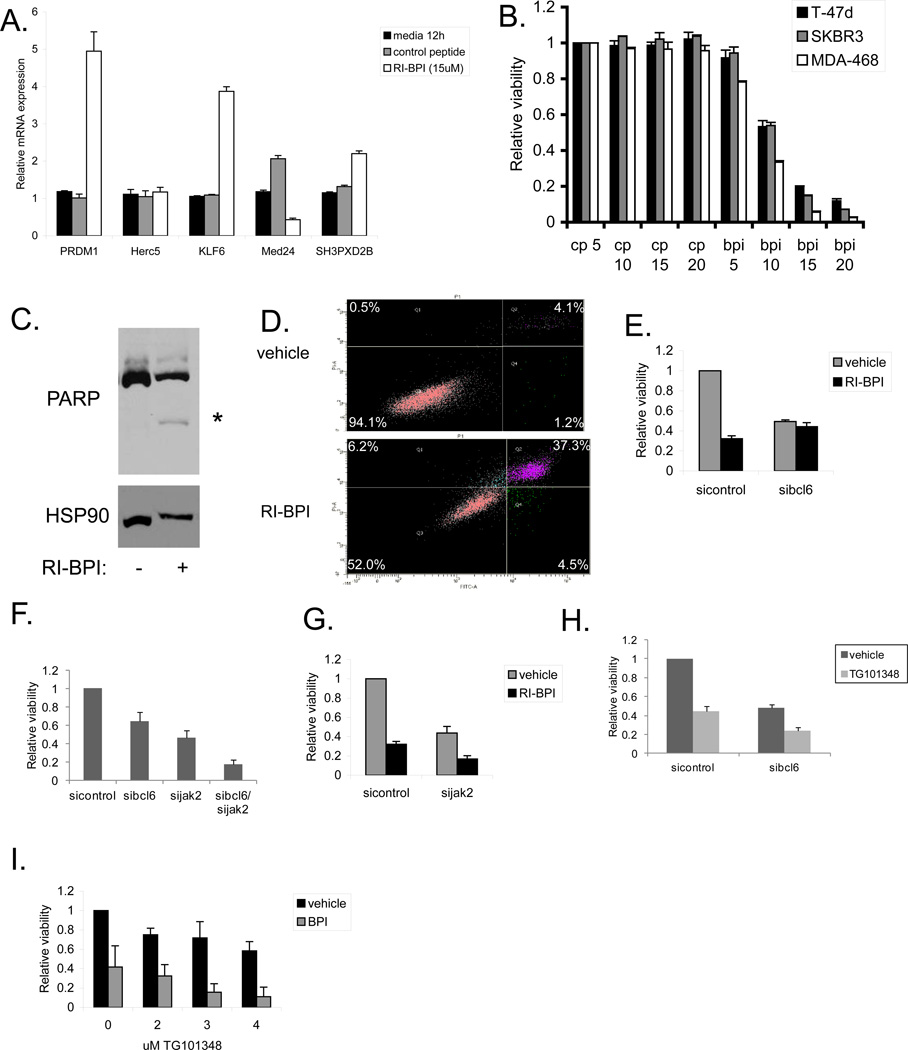
Having determined that BCL6 represses expression of a BCL6 responsive reporter in breast cancer cells, we next determined the effects of BCL6 on endogenous genes. We utilized siRNA to deplete cells of BCL6, then analyzed the expression of target genes identified by ChIP-seq. PRDM1, a well-described BCL6 target gene, served as a control (Figure 3d). Analysis of four of the newly identified BCL6 target genes demonstrated that three of the genes are repressed by BCL6 (HERC5, KLF6, and SH3PXD2B), while the fourth, MED24, is upregulated by BCL6 in SK-BR-3 cells (Figure 3d), confirming these as bona fide BCL6 regulated genes in breast cancer cells. While BCL6 is generally thought to be a transcriptional repressor, BCL6 may also be involved in upregulating a subset of genes such as MED24. In eight additional diverse breast cancer cell lines, depletion of BCL6 led to decreased expression of MED24 in all, and generally increased expression of the other three newly identified target genes (Supplementary Figure 3).
BCL6 is required for survival of breast cancer cells
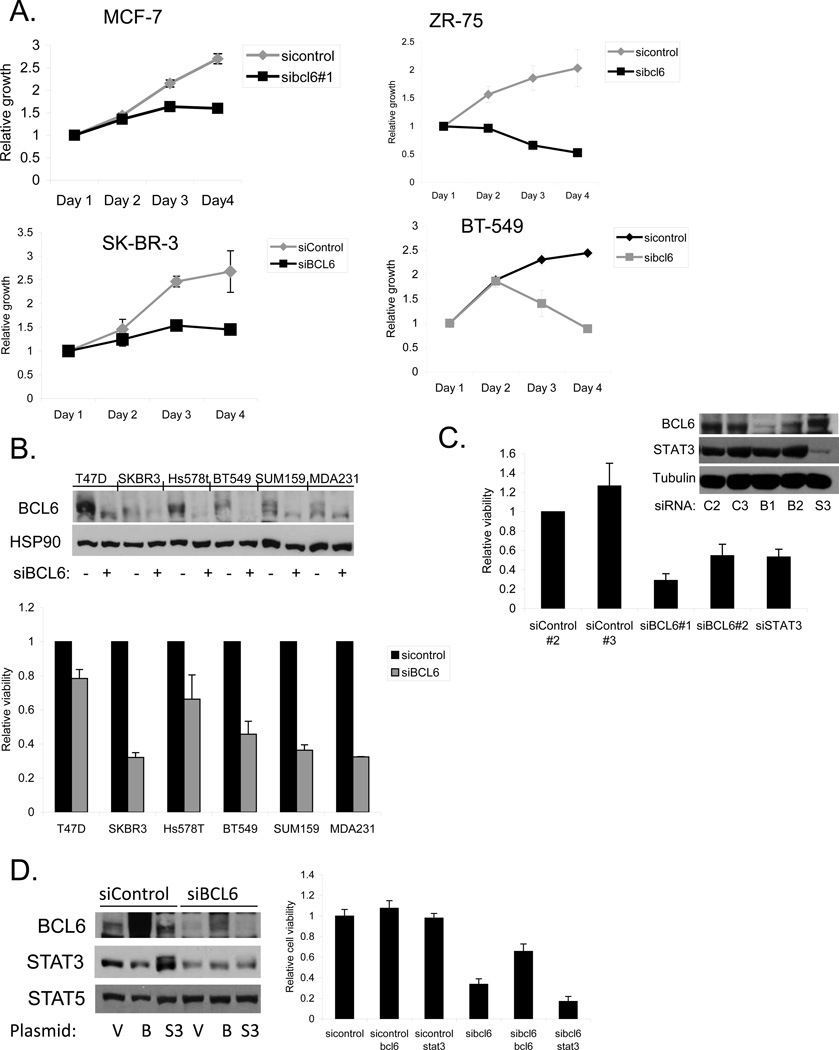
Having found that BCL6 has unique effects on gene expression in breast cancer cells, we wanted to determine the biological effects of BCL6 in these cells. We found that reducing the levels of BCL6 by RNA interference caused a distinct morphologic change in MCF-7 and MDA-MB-468 breast cancer cells, compared to control (Supplementary Figure 4a). The cells appeared to be more vacuolated, and displayed less cell-cell adhesion. We next examined viable cell number over time in culture. Depletion of BCL6 by RNAi led to a lower population doubling rate and a lower plateau density compared to cells treated with control siRNA (Figure 4a). Analysis of a variety of breast cancer cell lines upon BCL6 depletion demonstrated reduced cell number compared to control cells (Figure 4b). To further evaluate the role of BCL6 on breast cancer cell survival and proliferation, we performed replating assays. Cells in which BCL6 had been depleted displayed a significant reduction in plating efficiency (Supplementary Figure 4b), providing further evidence for the important role of this protein.
Figure 4.
BCL6 depletion inhibits breast cancer growth and survival. (A) Breast cancer cells were transfected with siRNA to BCL6 and viable cell number was analyzed by measuring ATP dependent luminescence. (B) The indicated breast cancer cell lines were transfected with siRNA to BCL6 or control. BCL6 knockdown was confirmed by immunoblot (top). Viable cell number was measured at 96 hours after transfection (bottom). (C) MDA-MB-468 cells were transfected with the indicated siRNAs, and BCL6 expression (top) and viable cell number (bottom) were measured 48 hours after transfection. (D) MDA-MB-468 cells treated with control or BCL6-targeting siRNA were transfected with expression vectors for BCL6 or STAT3. Expression of the indicated proteins was measured by immunoblot (left), and viable cell number was measured 48 hours after transfection (right). Although the coding sequence of the BCL6 expression construct was wildtype, high efficiency transfection allowed expression in the presence of the siRNA to BCL6.
BCL6 has a cognate binding site that overlaps with that of the STAT family of transcription factors including STAT3, a transcription factor also known to play an oncogenic role in breast cancer. We have found that STAT3 is activated in the majority of triple negative breast cancer cell lines but not in lines derived from the other subtypes [24]. Therefore, we considered the possibility that BCL6 and STAT3 played complementary roles in triple negative breast cancer cellular function. In MDA-MB-468 breast cancer cells, which contain activated STAT3 (Supplementary Figure 5), we knocked down BCL6 or STAT3 by RNAi and measured cell viability. Knockdown of either BCL6 or STAT3 led to a reduction in relative viability in these cells compared to cells treated with a control siRNA (Figure 4c). Forced over-expression of BCL6 rescued most of the loss of viability caused by knocking down BCL6 (Figure 4d). By contrast, ectopic expression of STAT3 had no effect on the viability of cells in which BCL6 had been knocked down. Taken together, these findings demonstrate that BCL6 plays a unique role in promoting breast cancer cell survival.
Pharmacological targeting of BCL6 decreases viability of breast cancer cells
Since BCL6 promotes breast cancer cell survival, we wanted to determine if targeting BCL6 using the peptidomimetic inhibitor RI-BPI would have therapeutic benefit [17]. To determine if this molecule was exerting a mechanism-specific effect, we first analyzed the expression of BCL6 target genes after RI-BPI treatment. Since RI-BPI disrupts the interaction of corepressors that bind the BTB domain of BCL6, only genes that are repressed through this mechanism should be affected by this compound. Therefore, we treated MDA-MB-468 cells with RI-BPI, and analyzed gene expression. We found that RI-BPI, but not a control peptide, increased expression of PRDM1, KLF6, and SH3PXD2B, (Figure 5a). Furthermore, MED24, which is upregulated by BCL6, showed decreased expression following RI-BPI treatment. Similar results were seen in T-47D and SK-BR-3 cells (Supplementary Figure 6a). These findings demonstrate that, as expected, RI-BPI inhibits some but not all BCL6 transcriptional function in breast cancer cells.
Figure 5.
BCL6 inhibition decreases the viability of breast cancer cells. (A) MDA-MB-468 breast cancer cells were treated with RI-BPI or control peptide for twelve hours and then expression of BCL6 target genes were analyzed by qRT-PCR. (B) Breast cancer cells were treated with the indicated doses of RI-BPI or control peptide (µM) for 48 hours after which viable cell number was determined. (C) MDA-MB-468 breast cancer cells were treated with 15 µM RI-BPI for 48 hours and then initiation of apoptosis was determined by immunoblot for PARP cleavage. HSP90 served as a loading control. (D) MDA-MB-468 cells were treated with vehicle or 15 µM RI-BPI for 48 hours after which apoptosis was determined by Annexin V/PI staining and flow cytometry. (E) MDA-MB-468 cells treated with control or BCL6-targeted siRNA were then treated with vehicle or RI-BPI for 48 hours and viable cell number was measured. (F) MDA-MB-468 cells were transfected with siRNA to BCL6 and/or Jak2. Relative cell viability was measured after 72 hours. (G) MDA-MB-468 cells treated with control or Jak2-targeted siRNA were treated with 10 µM RI-BPI for 48 hours and viable cell number was determined. (H) MDA-MB-468 cells transfected with siRNA to BCL6 were treated with the Jak2 inhibitor TG101348 (3 µM) for 48 hours and cell viability was measured. (I) MDA-MB-468 cells were treated with 10 µM RI-BPI (BPI) and the indicated doses of TG101348 for 48 hours and then cell viability was measured.
We next treated cells derived from each major subtype of breast cancer with RI-BPI and measured cell viability. All of the lines showed a similar dose dependent loss of viability in response to RI-BPI with an IC50 of approximately 10 µM, whereas treatment with a control peptide had no effect (Figure 5b). In addition, treatment of breast cancer cells with RI-BPI led to apoptosis as measured by PARP cleavage (Figure 5c and Supplementary Figure 6b) and annexin V/PI positivity (Figure 5d). Importantly, after knocking down BCL6 by siRNA, RI-BPI caused no additional loss of viability. This supports the hypothesis that the effect of RI-BPI is mediated solely through BCL6 (Figure 5e). Treatment of additional triple negative breast cancer cells demonstrated that they were all sensitive to RI-BPI to varying degrees (Supplementary Figure 6c). The degree of sensitivity to RI-BPI did not always correlate with levels of nuclear BCL6 expression (Figure 1e), and may relate to other cell-specific factors such as such as the binding of BCL6 to specific binding sites, differences in intracellular accumulation of the inhibitors, or disparate distributions of BCL6 among the cells in a given cell line. The small molecule BCL6 inhibitor 79-6 also reduced the cell viability of a variety of breast cancer cell lines, indicating that this is a mechanism-specific effect (Supplementary Figure 7).
About 70% of breast tumors contain activated STAT3, which is known to promote survival, proliferation, and self-renewal of breast cancer cells [4, 24, 25]. Therefore, we determined if combinations of a STAT3 inhibitor and the BCL6 inhibitor RI-BPI showed enhanced effects in killing breast cancer cells. Treatment of MDA-MB-468 breast cancer cells with the STAT3 inhibitor nifuroxazide [26] resulted in a decrease in STAT3 target gene expression (Supplementary Figure 8a) and a decrease in cell viability (Supplementary Figure 8b). Combining nifuroxazide with RI-BPI resulted in an additive loss of cell viability (Supplementary Figure 8b). In addition, reducing the levels of BCL6 with siRNA also sensitized breast cancer cells to nifuroxazide, providing further evidence that inhibiting STAT3 and BCL6 may be an effective combination for breast cancer therapy, particularly in triple negative tumors (Supplementary Figure 8c). Importantly, this effect was seen for the triple negative cell line MDA-MB-468, but there was no effect for the ER/PR positive cell line T-47D and only a slight enhancement for Her2 positive SK-BR-3 cells, which parallels the STAT3 phosphorylation status of these cells [24].
The activation of STAT3 in breast cancer cells is commonly dependent on Jak2, which is also a potential target in breast cancer therapy. Thus, we determined if depleting Jak2 also had an effect on cell viability in combination with BCL6 inhibition. We found that reducing the levels of Jak2 in MDA-MB-468 cells with siRNA inhibited STAT3 target gene expression (Supplementary Figure 8d) and led to an approximately 50% decrease in viable cell number (Figure 5f). Dual knockdown of BCL6 and Jak2 lead to enhanced loss of viability compared to either knockdown alone (Figure 5f). In addition, combining knockdown of Jak2 with the BCL6 inhibitor RI-BPI led to approximately an 80% reduction in viable cell number (Figure 5g). Next, we assessed the effects of the Jak2 inhibitor TG101348 [27, 28] with BCL6 inhibition. As expected, TG101348 inhibited STAT3 target gene expression in MDA-MB-468 cells (Supplementary Figure 9a). Treatment of breast cancer cells transfected with siRNA to BCL6 with TG101348 resulted in enhanced loss of cell viability of MDA-MB-468 cells (Figure 5h) and SK-BR-3 cells (Supplementary Figure 9b). Similarly, combining TG101348 with RI-BPI resulted in significant loss of viability compared to either treatment alone (Figure 5i). As expected, the Jak inhibitor TG101348 had little effect on T-47D cells, which lack constitutive STAT3 phosphorylation (Supplementary Figure 9b). Taken together, these findings demonstrate that BCL6 is important for breast cancer cell survival, and that inhibition of BCL6 with RI-BPI, alone or in combination with other therapies, may be an important therapeutic approach for breast cancer.
Discussion
We have demonstrated that BCL6 is expressed in most breast cancer cells lines and that its genetic locus is amplified in approximately 50% of breast tumor samples and the majority of breast cancer cell lines. In addition, BCL6 is important for breast cancer cell survival. Targeting BCL6 with peptidomimetic or small molecule inhibitors kills breast cancer cells alone and in combination with STAT3 or Jak2 inhibitors.
Performing whole genome ChIP-seq allowed us to identify BCL6 binding sites in breast cancer cells. When the genes associated with these sites were compared to the genes bound by BCL6 in B cell lymphomas, there was only limited overlap between the two groups (Figure 2e). This suggests that BCL6 has both overlapping and distinct roles in these two cell types. Why BCL6 does not bind to the same sites in the two different cell types raises several interesting hypotheses, and clearly indicates that DNA sequence alone is not sufficient to specify transcription factor binding in the context of chromatin. Other sources of tissue specific differences may include alterations in DNA itself, such as methylation, or modifications of histones by methylation or acetylation. Further analysis of these factors may provide important information about the mechanism by which BCL6 regulates gene expression differently in distinct tissues.
The interaction of BCL6 with corepressors may also be distinct in breast cancer compared to other tissue types. The well characterized BCL6 target gene PRDM1 has been shown to be repressed by BCL6 interaction with MTA3 in lymphoma cells [13]. MTA3 is also expressed in breast cancer cells and is important for regulating invasiveness [29]. However, in breast cancer cells, we have found that the BCL6 inhibitor RI-BPI, which prevents the interaction with SMRT, leads to upregulation of PRDM1 (Figure 5a and Supplementary Figure 6a). This suggests that SMRT is involved in regulation of PRDM1 in breast cancer cells. Blimp1, the protein encoded by the PRDM1 gene, has been shown to be involved in breast cancer and represses ERα [30], whereas in B cells Blimp1 promotes plasma cell differentiation [13]. The reason for difference in the regulation of PRDM1 between these two cell types is unclear and warrants further study.
We have found that BCL6 is expressed in 10 of 11 breast cancer cell lines tested (Supplementary Figure 1b). Interestingly, expression of BCL6 was not restricted to any one breast cancer cell type. Therefore, we analyzed BCL6 binding in all three types of breast cancer cells and determined that while many of the sites are shared between the three major classes of breast cancer, there are also distinct differences between the cells. Additionally, the responses to siRNA and drug treatment also had some distinct differences, with the triple negative MDA-MB-468 and BT-549 cells responding more than the other cell types. This raises the possibility that triple negative breast tumors may be more sensitive to targeting BCL6, and therefore, may benefit more strongly from treatment with a BCL6 inhibitor. This may be particularly important as these tumors often become resistant to chemotherapy.
Treating breast cancer cells with RI-BPI leads to loss of cell viability and induction of apoptosis (Figure 5c and d, and Supplementary Figure 6b). This suggests that targeting BCL6 may be an important new strategy for cancer therapy. In addition, the combination of RI-BPI with inhibition of another transcription factor, STAT3, also showed promise (Supplementary Figure 8b). Over 70% of breast tumors have activation of STAT3 [4, 25], and STAT3 activation is highly associated with triple negative breast tumors [24]. Therefore, triple negative breast tumors may particularly benefit from the combination of STAT3 and BCL6 inhibitors. STAT3 can also directly increase expression of BCL6 in breast cancer cells [4]. Thus, this combination may not only show efficacy by inhibiting the function of each transcription factor alone, but inhibiting STAT3 may also reduce the levels of BCL6 expression, providing a further level of inhibition of BCL6 in these triple negative breast cancer cells. Additionally, Jak inhibitors are being tested in clinical trials for triple negative breast cancer. We have found that the combination of the Jak2 inhibitor TG101348 with BCL6 inhibition reduces viability of breast cancer cells more than either approach alone (Figure 5h and i). Therefore, combinations of Jak inhibitors and RI-BPI or other BCL6 inhibitors may also be an effective treatment strategy for breast cancer. Ultimately, it will be important to confirm these findings in in vivo systems.
In conclusion, we have demonstrated that BCL6 is an important transcriptional regulator in breast cancer, and that targeting BCL6 induces apoptosis in these cells. About half of the genes to which BCL6 binds in breast cancer cells are unique when compared to lymphoma cells, indicating that BCL6 likely mediates some unique effects in breast cancer. As BCL6 is expressed in all three major classes of breast cancer, developing targeted therapies to BCL6 may be an important new avenue for breast cancer therapy.
Materials and Methods
Cells and treatments
MDA-MB-468 cells (kindly provided by Myles Brown, DFCI), T-47D cells (ATCC), MCF-7 (kindly provided by Francis Kern, Southern Research Institute, Birmingham, AL), were maintained in DMEM with 10% FBS. SK-BR-3 cells (kindly provided by Lyndsay Harris, Yale) were maintained in RPMI with 10% FBS. These cells were authenticated prior to manuscript submission by STR DNA analyses at Genetica, DNA Laboratories, Inc. IMECs (kindly provided by Myles Brown) were maintained as described [31]. Additional breast cancer cells were obtained and maintained as described [24]. Ramos cells were maintained in RPMI with 10% FBS. Breast cancer cells were treated with control peptide or RI-BPI at the indicated doses [17], or with vehicle or the indicated doses of 79-6 (EMD Millipore, Billerica, MA). Cells were treated with nifuroxazide (Chembridge, San Diego, CA) or TG101348 (VWR, Radnor, PA) at the indicated doses.
Copy number analysis
Level 3 Affymetrix Genome-Wide SNP Array 6.0 data for breast invasive carcinoma was downloaded from The Cancer Genome Atlas (cancergenome.nih.gov). The data spanned 790 tumor samples and 786 matched normal breast samples. Copy number (log2 − 1) at the BCL6 locus was determined for each sample by identifying the value in column “seg.mean” at chromosome 3, position 188,929,714, corresponding to the center of the BCL6 gene. The extent of rightward skewness of the BCL6 copy number distribution in tumor was quantified using Pearson’s median skewness coefficient, defined as
The statistical significance of the skewness coefficient was calculated by determining the skewness at 1000 randomly selected locations in the genome. An empirical p-value was derived by determining the number of locations with skewness equal to or greater than that at the BCL6 locus. Similar analyses were performed using data from the Cancer Cell Line Encyclopedia (CCLE) [18] for breast cancer cell lines. For breast specific subtypes, the cell lines were matched to subtype based on expression of ER, PR, and Her2 expression reported from [32–34]. The statistical significance of the skewness coefficient was calculated as above using 100 randomly selected locations in the genome. Gene expression was also obtained from the CCLE.
RNA interference
Breast cancer cells were reverse transfected with siRNAs using Lipofectamine RNAiMax (Invitrogen) at 10 nM unless otherwise indicated for the indicated times. RNAi included siControl (D-001210-03, Dharmacon), siBCL6#1 (HSS100968, Invitrogen), siBCL6#2 (HSS100966, Invitrogen), siSTAT3 (M-003544-02, Dharmacon), and siJak2 (M-003146-02-0005, Dharmacon). Rescue experiments were performed with the combination transfection of siRNA and expression plasmids for pRcCMV STAT3 or pCDNA3 BCL6.
Chromatin Immunoprecipitation
ChIP was performed essentially as described [35]. Cells were fixed in 1% formaldehyde for 10 minutes, sonicated, and lysates were immunoprecipitated overnight with 2 µg of antibodies to BCL6 (sc-858) and RNA polymerase II (sc-9001) from Santa Cruz Biotechnology or acetyl H4 from Cell Signaling Technology. ChIP product was measured using Quant-iT™ dsDNA HS Assay (Invitrogen), and equal amounts of product and input were analyzed by quantitative PCR. Data are expressed as fold binding over background (using both input and genomic regions known not to be bound by BCL6, including the rhodopsin gene and a previously defined control region found within the BCL6 gene [20]). For siRNA ChIPs, 7.5×106 SK-BR-3, T-47D, or MDA-MB-468 cells were plated and ChIP was performed 72 hours after reverse transfection. ChIP product was analyzed by qPCR using the indicate primers (Supplementary Table 1 or as described [3]). OCI-Ly1 ChIP was performed essentially as above except cells were lysed in RIPA buffer (150 mM NaCl, 1% v/v Nonidet P-40, 0.5% w/v deoxycholate, 0.1% w/v SDS, 50 mM Tris pH 8, and 5 mM EDTA).
ChIP-seq
Libraries for ChIP-seq were created from 10 ng of BCL6 ChIP and 10 ng of corresponding input in SK-BR-3 cells according to the manufacture’s protocol (Illumina), with size selection of 250–400 base pairs. ChIP-seq was performed with a SR36bp run. Sequences were aligned to the human genome (UCSC hg18) using ELAND. Peaks were defined using MACS [36]. Motifs were identified using both the de novo motifs search and the TRANSFAC database using the SeqPos motif tool [37]. Binding site analysis and annotation was performed using CEAS [38]. Analysis tools were from the Galaxy/Cistrome website (http://cistrome.org). ChIP-seq was performed essentially as above for the OCI-LY1 cells. For comparisons between BCL6 binding in breast cancer and lymphoma cells, only sequences mapping uniquely to the genome with not more than two mismatches were used, and peaks were assigned using ChIPseeqer [39] for both datasets. Each dataset was compared to its own input control.
Luciferase reporter analysis
Breast cancer cells were reverse transfected with siRNA. The following day, (BCL6)4-TK-Luc was transfected into breast cancer cells in combination with renilla luciferase using Lipofectamine 2000. These were alone or with vector or pCDNA3-BCL6. Luciferase was analyzed as described [20]. For cells treated with 79-6, breast cancer cells were transfected with BCL6-Luc for 6 hours and then treated with 300 µM 79-6 for 16 hours.
RT-PCR
RT-PCR was performed as described [4]. mRNA primer sequences are as indicated (Supplementary Table 2).
Immunoblots and nuclear extracts
Immunoblotting was performed as described [26] using the following antibodies: PARP (9542, Cell Signaling); tubulin (T-5168; Sigma); BCL6 (sc-858; Santa Cruz Biotechnology), HSP90 (sc-13119, Santa Cruz Biotechnology), P-STAT3 (9131, Cell Signaling Technology), STAT3 (sc- 482, Santa Cruz Biotechnology), and actin (Sigma). Nuclear extracts were generated using the Nuclear Extract Kit from Active Motif (Carlsbad, CA) according to the manufacturer’s protocol.
Viable cell number quantitation and clonogenic assays
Breast cancer cells were plated in 384 or 96 well plates for viability in duplicate or quadruplicate. Cells were treated with drug for the indicated time and viable cell number was measured using an ATP dependent luciferase assay (CellTiterGlo, Promega), and expressed relative to control or vehicle treated cells. For siRNA knockdown, breast cancer cells were reverse transfected and then analyzed on the indicated day. For growth assays, cell number was measured daily by CellTiterGlo and normalized to day one values. For clonogenic assays, cells were reverse transfected with siRNA. The following day, 500 cells were seeded in 6 well dishes. After 10–14 days, colonies were visualized by staining with a 6% glutaraldehyde and 0.5% crystal violet solution.
Apoptosis assay
Breast cancer cells were treated with vehicle or 15 µM BPI for 48 hours and analyzed for annexin/PI staining (BD Biosciences) by flow cytometry.
Supplementary Material
Acknowledgements
Grant Support
This work was supported by a BCRF-AACR grant for Translational Breast Cancer Research, and grants from the National Cancer Institute (R01-CA160979), Susan G. Komen for the Cure, the Brent Leahey Fund, and Friends of the Dana-Farber Cancer Institute.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, Bobrow L, et al. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiaiton. Oncogene. 2003;22:5572–5578. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- 2.Polo JM, Juszczynski P, Monti S, Cerchietti L, Ye K, Greally JM, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proceedings of the National Academy of Sciences of the United States of America. 2007 Feb 27;104(9):3207–3212. doi: 10.1073/pnas.0611399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ci W, Polo JM, Cerchietti L, Shaknovich R, Wang L, Yang SN, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009 May 28;113(22):5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, et al. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009 Jun;7(6):966–976. doi: 10.1158/1541-7786.MCR-08-0238. [DOI] [PubMed] [Google Scholar]
- 5.Walker SR, Nelson EA, Yeh JE, Pinello L, Yuan GC, Frank DA. STAT5 outcompetes STAT3 to regulate the expression of the oncogenic transcriptional modulator BCL6. Molecular and cellular biology. 2013 May 28; doi: 10.1128/MCB.01620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos R, van Diest PJ, van der Groep P, Greijer AE, Hermsen MA, Heijnen I, et al. Protein expression of B-cell lymphoma gene 6 (BCL-6) in invasive breast cancer is associated with cyclin D1 and hypoxia-inducible factor-1alpha (HIF-1alpha) Oncogene. 2003 Dec 4;22(55):8948–8951. doi: 10.1038/sj.onc.1206995. [DOI] [PubMed] [Google Scholar]
- 7.Polo JM, Ci W, Licht JD, Melnick A. Reversible disruption of BCL6 repression complexes by CD40 signaling in normal and malignant B cells. Blood. 2008 Aug 1;112(3):644–651. doi: 10.1182/blood-2008-01-131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Molecular cell. 2003 Dec;12(6):1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 9.Polo JM, Dell'Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nature medicine. 2004 Dec;10(12):1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 10.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes & development. 2000 Jul 15;14(14):1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 11.Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998 Nov 12;17(19):2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- 12.Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Prive GG. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Molecular cell. 2008 Feb 15;29(3):384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parekh S, Polo JM, Shaknovich R, Juszczynski P, Lev P, Ranuncolo SM, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007 Sep 15;110(6):2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004 Oct 1;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Mendez LM, Polo JM, Yu JJ, Krupski M, Ding BB, Melnick A, et al. CtBP is an essential corepressor for BCL6 autoregulation. Molecular and cellular biology. 2008 Apr;28(7):2175–2186. doi: 10.1128/MCB.01400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Li Z, Naganuma A, Ye BH. Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas. Proceedings of the National Academy of Sciences of the United States of America. 2002 Nov 12;99(23):15018–15023. doi: 10.1073/pnas.232581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, et al. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009 Apr 9;113(15):3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012 Mar 29;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nature genetics. 2002;32(4):606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 20.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007 Jan 11;26(2):224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 21.Welboren W-J, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FCGJ, Span PN, et al. ChIP-Seq of ERalpha] and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28(10):1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim H-P, Oh J, Tunyaplin C, et al. Analysis of Interleukin-21-Induced Prdm1 Gene Regulation Reveals Functional Cooperation of STAT3 and IRF4 Transcription Factors. Immunity. 2009;31(6):941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer cell. 2010 Apr 13;17(4):400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. The Journal of clinical investigation. 2011 Jun 1; doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez JV, Febbo PG, Ramaswamy S, Loda M, Richardson A, Frank DA. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer research. 2005 Jun 15;65(12):5054–5062. doi: 10.1158/0008-5472.CAN-04-4281. [DOI] [PubMed] [Google Scholar]
- 26.Nelson EA, Walker SR, Kepich A, Gashin LB, Hideshima T, Ikeda H, et al. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008 Dec 15;112(13):5095–5102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, et al. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer cell. 2008 Apr;13(4):311–320. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Lasho TL, Tefferi A, Hood JD, Verstovsek S, Gilliland DG, Pardanani A. TG101348, a JAK2-selective antagonist, inhibits primary hematopoietic cells derived from myeloproliferative disorder patients with JAK2V617F, MPLW515K or JAK2 exon 12 mutations as well as mutation negative patients. Leukemia. 2008 Sep;22(9):1790–1792. doi: 10.1038/leu.2008.56. [DOI] [PubMed] [Google Scholar]
- 29.Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003 Apr 18;113(2):207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Belguise K, O'Neill CF, Sanchez-Morgan N, Romagnoli M, Eddy SF, et al. RelB NF-kappaB represses estrogen receptor alpha expression via induction of the zinc finger protein Blimp1. Molecular and cellular biology. 2009 Jul;29(14):3832–3844. doi: 10.1128/MCB.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiRenzo J, Signoretti S, Nakamura N, Rivera-Gonzalez R, Sellers W, Loda M, et al. Growth factor requirements and basal phenotype of an immortalized mammary epithelial cell line. Cancer research. 2002 Jan 1;62(1):89–98. [PubMed] [Google Scholar]
- 32.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PloS one. 2009;4(7):e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 2011 Jul;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer cell. 2006 Dec;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. The Journal of biological chemistry. 2004 Dec 24;279(52):54724–54730. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, et al. Nucleosome dynamics define transcriptional enhancers. Nature genetics. 2010;42(4):343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics (Oxford, England) 2009 Oct 1;25(19):2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 39.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.