Abstract
We aimed to identify peripheral blood mononuclear cell (PBMC) gene expression profiles predictive of poor outcomes in idiopathic pulmonary fibrosis (IPF) by performing microarray experiments of PBMCs in discovery and replication cohorts of IPF patients. Microarray analyses identified 52 genes associated with transplant-free survival (TFS) in the discovery cohort. Clustering the microarray samples of the replication cohort using the 52-gene outcome-predictive signature distinguished two patient groups with significant differences in TFS. We studied the pathways associated with TFS in each independent microarray cohort and identified decreased expression of “The costimulatory signal during T cell activation” Biocarta pathway and, in particular, the genes CD28, ICOS, LCK, and ITK, results confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). A proportional hazards model, including the qRT-PCR expression of CD28, ICOS, LCK, and ITK along with patient’s age, gender, and percent predicted forced vital capacity (FVC%), demonstrated an area under the receiver operating characteristic curve of 78.5% at 2.4 months for death and lung transplant prediction in the replication cohort. To evaluate the potential cellular source of CD28, ICOS, LCK, and ITK expression, we analyzed and found significant correlation of these genes with the PBMC percentage of CD4+CD28+ T cells in the replication cohort. Our results suggest that CD28, ICOS, LCK, and ITK are potential outcome biomarkers in IPF and should be further evaluated for patient prioritization for lung transplantation and stratification in drug studies.
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic and progressive fibrosing interstitial lung disease with an unknown etiology. Diagnosis of IPF is based on clinical and radiological features and, when available, findings of usual interstitial pneumonia on lung biopsy. IPF patients have an overall median survival of 3 to 3.5 years (1). The disease is more prevalent and probably more lethal among males (2, 3). With the exception of lung transplantation, no therapy has been proven beneficial for IPF. The course of IPF is highly variable and largely unpredictable among individual patients. Disease progression in current clinical practice is monitored by pulmonary function tests, including forced vital capacity (FVC) and diffusion capacity for carbon monoxide (DLCO), high-resolution computed tomography scans, and measures of oxygenation. Previous studies have demonstrated that changes in dyspnea score, total lung capacity, and FVC over 12 months or scores calculated on the basis of age, gender, FVC, and DLCO at presentation seem to correlate with disease severity or outcome in IPF (2–6). Although these advances allow for staging of patients with IPF, they do not address the difficulty of predicting outcomes for patients with very similar clinical presentation or provide insight into molecular mechanisms of disease.
Current evidence suggests that plasma protein concentrations or changes in blood cells may be informative of disease presence, severity, and prognosis in IPF patients (7–12). Recently, a difference in the peripheral blood transcriptome was shown between IPF patients and healthy controls (13, 14); however, the ability of the transcriptome to predict outcome was not assessed. Given the evidence that peripheral blood mononuclear cell (PBMC) gene expression is informative of disease presence and outcomes in other clinical entities, such as multiple sclerosis (15, 16), heart transplant rejection (17), pulmonary hypertension associated with scleroderma (18), and lung cancer (19), among others, we hypothesized that PBMC gene expression patterns may be predictive of poor outcomes in IPF patients. For this purpose, we examined PBMC gene expression in two independent cohorts and identified a signature of 52 genes significantly associated with transplant-free survival (TFS) in both cohorts. Decreased expression of genes belonging to “The costimulatory signal during T cell activation” Biocarta pathway, in particular, CD28, ICOS, LCK, and ITK, was associated with shorter TFS, findings confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The addition of these genes to an outcome prediction model improved its performance compared to a model that only included clinical parameters. Our findings suggest that PBMC gene expression may improve outcome prediction in IPF.
RESULTS
Patient population and clinicopathology
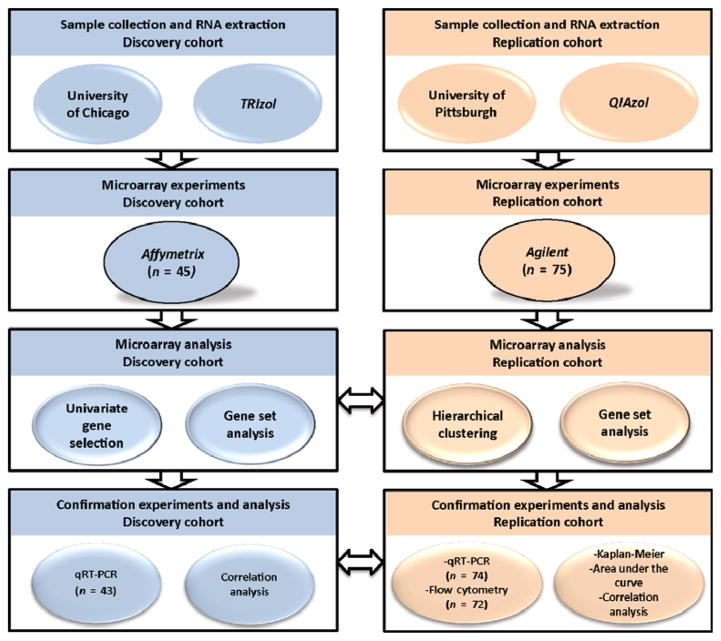
IPF patients included in this prospective cohort study were followed in clinics (at 3- to 4-month intervals) from blood draw until death or completion of the study. The time-to-event outcome analyzed was TFS; in this analysis, transplants and deaths were both counted as events. Figure 1 provides information regarding the cohorts and study design. The discovery (n = 45) and replication (n = 75) cohorts were similar with respect to age, smoking status, pulmonary function tests, diagnostic strategy, and use of immunosuppression with the exception of gender, race, and lung transplants (table S1). Although 75.5 and 68% of subjects in the discovery and replication cohort were Caucasian males, respectively, the discovery cohort patients had a more diverse ethnic background. Females were more represented in the replication than in the discovery cohort (30.7 versus 11.1%, respectively). The rate of lung transplants was higher in the replication cohort (20%) compared to the discovery cohort (4%).
Fig. 1. Study design and cohorts.
The outline summarizes the studied cohorts, the experiments performed in each cohort, and the statistical analyses used. The horizontal arrows represent the confirmation of microarray and qRT-PCR experiments in both cohorts.
Microarray analysis of the discovery cohort
RNA was isolated from the PBMCs of patients (n = 45), labeled, and hybridized to GeneChip Human 1.0 exon ST arrays at the University of Chicago. Using significance analysis of microarrays (SAM), we identified 52 genes that were significantly [false discovery rate (FDR) <5%, Cox score ≥2.5 and ≤−2.5] associated with TFS in this cohort. Increased expression of 7 genes (genes with a Cox score ≥2.5) and decreased expression of 45 genes (genes with a Cox score ≤−2.5) were correlated with shorter TFS times (Table 1).
Table 1. A 52-gene signature associated with TFS in the discovery cohort.
Expression data were collected for genes with a Cox score ≥2.5 and ≥−2.5 (FDR <5%). A positive Cox score indicates that higher expression correlates with shorter TFS, whereas lower expression indicates longer TFS time. A negative score indicates that higher expression correlates with longer TFS time, whereas lower expression correlates with shorter TFS time.
| Gene | Gene symbol | Cox score |
|---|---|---|
| Phospholipase B domain containing 1 | PLBD1 | 3.32 |
| Tyrosylprotein sulfotransferase 1 | TPST1 | 3.25 |
| Chromosome 19 open reading frame 59 (mast cell–expressed membrane protein 1) | C19orf59 (MCEMP1) | 3.13 |
| Interleukin-1 receptor, type II | IL1R2 | 3.05 |
| Haptoglobin | HP | 2.91 |
| FMS-related tyrosine kinase 3 | FLT3 | 2.90 |
| S100 calcium-binding protein A12 | S100A12 | 2.89 |
| Lymphocyte-specific protein tyrosine kinase | LCK | −2.50 |
| Calcium/calmodulin-dependent protein kinase IIδ | CAMK2D | −2.50 |
| Nucleoporin 43 kD | NUP43 | −2.51 |
| SLAM family member 7 | SLAMF7 | −2.52 |
| Leucine-rich repeat containing 39 | LRRC39 | −2.52 |
| Inducible T cell costimulator | ICOS | −2.53 |
| CD47 molecule | CD47 | −2.54 |
| Limb bud and heart development | LBH | −2.55 |
| SH2 domain containing 1A | SH2D1A | −2.55 |
| CCR4-NOT transcription complex, subunit 6–like | CNOT6L | −2.56 |
| Methyltransferase-like 8 | METTL8 | −2.56 |
| V-ets erythroblastosis virus E26 oncogene homolog 1 | ETS1 | −2.58 |
| Chromosome 2 open reading frame 27A | C2orf27A | −2.60 |
| Purinergic receptor P2Y, G protein–coupled, 10 | P2RY10 | −2.60 |
| T cell receptor–associated transmembrane adaptor 1 | TRAT1 | −2.61 |
| Butyrophilin, subfamily 3, member A1 | BTN3A1 | −2.62 |
| La ribonucleoprotein domain family, member 4 | LARP4 | −2.63 |
| Tandem C2 domains, nuclear | TC2N | −2.63 |
| G protein–coupled receptor 183 | GPR183 | −2.65 |
| MORC family CW-type zinc finger 4 | MORC4 | −2.67 |
| Signal transducer and activator of transcription 4 | STAT4 | −2.67 |
| Lysophosphatidic acid receptor 6 | LPAR6 | −2.67 |
| Chromosome 7 open reading frame 58 (cadherin- like and PC-esterase domain containing 1) | C7orf58 (CPED1) | −2.68 |
| Dedicator of cytokinesis 10 | DOCK10 | −2.69 |
| Rho GTPase-activating protein 5 | ARHGAP5 | −2.71 |
| Major histocompatibility complex, class II, DPα1 | HLA-DPA1 | −2.72 |
| Baculoviral IAP repeat containing 3 | BIRC3 | −2.73 |
| G protein–coupled receptor 174 | GPR174 | −2.73 |
| CD28 molecule | CD28 | −2.73 |
| Utrophin | UTRN | −2.76 |
| CD2 molecule | CD2 | −2.76 |
| Major histocompatibility complex, class II, DPβ1 | HLA-DPB1 | −2.77 |
| ADP-ribosylation factor–like 4C | ARL4C | −2.78 |
| Butyrophilin, subfamily 3, member A3 | BTN3A3 | −2.79 |
| Chemokine (C-X-C motif) receptor 6 | CXCR6 | −2.81 |
| Dynein cytoplasmic 2 light intermediate chain 1 | DYNC2LI1 | −2.84 |
| Butyrophilin, subfamily 3, member A2 | BTN3A2 | −2.84 |
| IL-2–inducible T cell kinase | ITK | −2.85 |
| Small nucleolar RNA host gene 1 | SNHG1 | −2.94 |
| CD96 molecule | CD96 | −3.03 |
| Guanylate binding protein 4 | GBP4 | −3.03 |
| Sphingosine-1-phosphate receptor 1 | S1PR1 | −3.06 |
| Nucleosome assembly protein 1–like 2 | NAP1L2 | −3.10 |
| Kruppel-like factor 12 | KLF12 | −3.15 |
| Interleukin-7 receptor | IL7R | −3.48 |
To determine the pathways associated with TFS, we performed a survival gene set analysis (GSA) in the discovery cohort. GSA identified 18 pathways (FDR <5%) associated with TFS (table S2). Among them, “The costimulatory signal during T cell activation” Biocarta pathway (Table 2 and table S2) was the top-ranked pathway with a maxmean score of −1.91, indicating that lower expression of most genes in this pathway was correlated with shorter TFS. CD28, ICOS, LCK, and ITK were the genes of this pathway with the strongest association with TFS when underexpressed (Cox scores = −3.12, −3.01, −2.77, and −3.2, respectively) (Table 2), and they were also part of the 52-gene outcome-associated signature. Because GSA calculates a partial likelihood Cox score statistic for each gene after fitting a full multivariate model, whereas SAM uses one gene at a time to estimate Cox scores based on univariate models, we observed slight differences between the Cox scores of CD28, ICOS, LCK, and ITK calculated by SAM from those calculated by GSA (Tables 1 and 2) in the discovery cohort; however, they fully agree in direction and magnitude.
Table 2. Genes in “The costimulatory signal during T cell activation” Biocarta pathway associated with TFS in the discovery cohort.
A positive Cox score indicates that higher expression correlates with shorter TFS time, whereas lower expression indicates shorter TFS time. A negative Cox score indicates that higher expression correlates with longer TFS time, whereas lower expression correlates with shorter TFS time.
| Gene | Gene symbol | Cox score |
|---|---|---|
| Growth factor receptor–bound protein 2 | GRB2 | 1.08 |
| CD80 molecule | CD80 | 0.10 |
| Interleukin-2 | IL2 | −0.81 |
| Protein tyrosine phosphatase, nonreceptor type 11 | PTPN11 | −0.92 |
| Cytotoxic T lymphocyte–associated protein 4 | CTLA4 | −1.85 |
| CD3 molecule, ε (CD3-TCR complex) | CD3E | −1.90 |
| CD3 molecule, δ (CD3-TCR complex) | CD3D | −1.99 |
| Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α | PIK3CA | −2.06 |
| Major histocompatibility complex, class II, DRα | HLA-DRA | −2.22 |
| T cell receptor α locus | TRA@ | −2.35 |
| Major histocompatibility complex, class II, DRβ1 | HLA-DRB1 | −2.37 |
| Phosphoinositide-3-kinase, regulatory subunit 1 (α) | PIK3R1 | −2.37 |
| CD247 molecule | CD247 | −2.45 |
| CD3 molecule, γ (CD3-TCR complex) | CD3G | −2.59 |
| CD86 molecule | CD86 | −2.68 |
| Lymphocyte-specific protein tyrosine kinase | LCK | −2.77 |
| Inducible T cell costimulator | ICOS | −3.01 |
| CD28 molecule | CD28 | −3.12 |
| IL-2–inducible T cell kinase | ITK | −3.20 |
Microarray analysis of the replication cohort
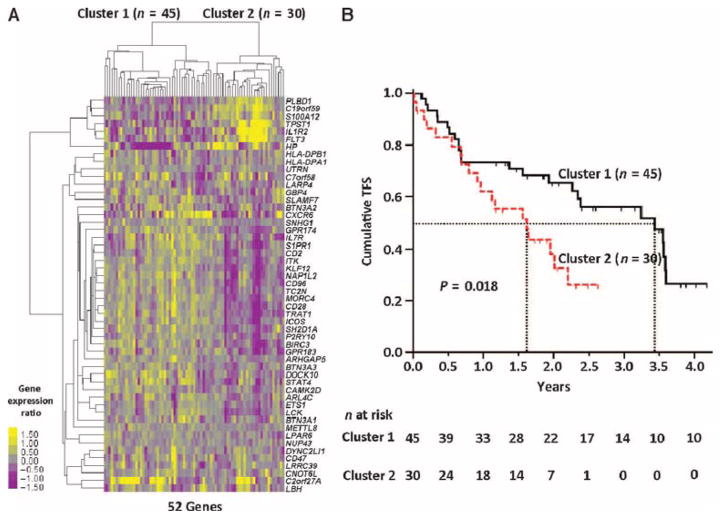
RNA isolated from PBMCs obtained from IPF patients at the University of Pittsburgh was labeled and hybridized to Agilent whole human genome microarrays. To determine whether the 52-gene TFS predictive signature identified in the discovery cohort predicted outcome in the replication cohort, we used hierarchical clustering. Briefly, gene expression values in the replication cohort, of the 52 genes derived from the discovery cohort, were used in a hierarchical clustering algorithm that uses expression values to cluster samples. This clustering algorithm identified two major patient clusters in the replication cohort (Fig. 2A). The patients in the two clusters differed significantly with respect to TFS [hazard ratio, 1.96; 95% confidence interval (CI), 1.01 to 3.8] (Fig. 2B) but did not differ significantly with respect to clinical variables (table S3). The median TFS at the conclusion of the observation period for replication cohort patients in cluster 1 was 3.44 years compared to 1.62 years for patients in cluster 2 (Fig. 1B and table S4).
Fig. 2. Hierarchical clustering discriminates subgroups with outcome differences in the replication cohort.
(A) Hierarchical clustering of IPF patients from the replication cohort (n = 75) based on the 52-gene signature found in the discovery cohort to be associated with TFS (FDR <5%, Cox score ≥2.5 and ≤−2.5). Two major clusters of IPF patients were identified. Every row represents a gene, and every column, a patient. Color scale is shown adjacent to heat map in log2 scale; generally, yellow denotes increase over the geometric mean of samples, and purple, decrease. (B) TFS differs between clusters in the replication cohort; the median survival of each group is depicted in dotted vertical lines; n at risk is the number of IPF patients at risk of death or lung transplant at the beginning of each time point. P value was determined by the log-rank test.
TFS GSA was performed on the Agilent microarray gene expression data, independently obtained from the replication cohort. This analysis yielded “The costimulatory signal during T cell activation” Biocarta pathway as the top-ranked pathway that correlated with TFS with a maxmean score of −1.24 (table S5). Similar to our previous observation in the discovery cohort, CD28, ICOS, LCK, and ITK were also the genes with the lowest Cox score within this pathway in the replication cohort (Table 3).
Table 3. Genes in “The costimulatory signal during T cell activation” Biocarta pathway associated with TFS in the replication cohort.
A positive Cox score indicates that higher expression correlates with shorter TFS time, whereas lower expression indicates shorter TFS time. A negative score indicates that higher expression correlates with longer TFS time, whereas lower expression correlates with shorter TFS time.
| Gene | Gene symbol | Cox score |
|---|---|---|
| Growth factor receptor–bound protein 2 | GRB2 | 1.76 |
| Protein tyrosine phosphatase, nonreceptor type 11 | PTPN11 | 0.71 |
| Major histocompatibility complex, class II, DRα | HLA-DRA | 0.13 |
| CD86 molecule | CD86 | 0.11 |
| Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α | PIK3CA | −0.38 |
| Phosphoinositide-3-kinase, regulatory subunit 1 (α) | PIK3R1 | −0.47 |
| Major histocompatibility complex, class II, DRβ1 | HLA-DRB1 | −0.52 |
| CD247 molecule | CD247 | −0.71 |
| Interleukin-2 | IL2 | −0.71 |
| CD3 molecule, γ (CD3-TCR complex) | CD3G | −0.81 |
| CD80 molecule | CD80 | −0.91 |
| T cell receptor α locus | TRA@ | −1.39 |
| CD3 molecule, δ (CD3-TCR complex) | CD3D | −1.64 |
| CD3 molecule, ε (CD3-TCR complex) | CD3E | −1.67 |
| Cytotoxic T lymphocyte–associated protein 4 | CTLA4 | −2.09 |
| IL-2–inducible T cell kinase | ITK | −2.38 |
| Lymphocyte-specific protein tyrosine kinase | LCK | −2.66 |
| Inducible T cell costimulator | ICOS | −2.77 |
| CD28 molecule | CD28 | −2.99 |
Association of CD28, ICOS, LCK, and ITK with poor IPF outcomes
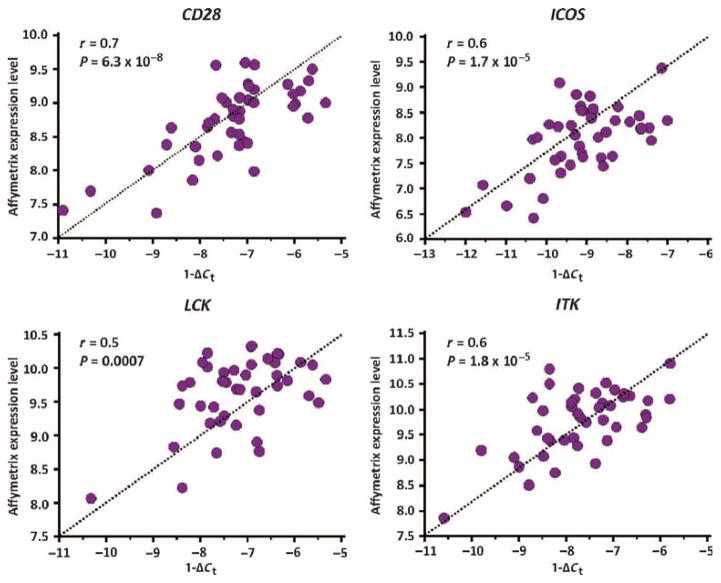
To confirm the microarray findings in the discovery cohort, we designed a custom SmartChip qRT-PCR assay that allowed us to simultaneously measure the expression of CD28, ICOS, LCK, and ITK as well as housekeeping genes in multiple samples. SmartChip expression values (reflected by 1 − ΔCt) from the discovery cohort (n = 43) were significantly correlated with the Affymetrix microarray gene expression values for CD28 (r = 0.71; 95% CI, 0.53 to 0.83), ICOS (r = 0.6; 95% CI, 0.38 to 0.77), LCK (r = 0.5; 95% CI, 0.23 to 0.70), and ITK (r = 0.6; 95% CI, 0.37 to 0.77) (Fig. 3).
Fig. 3. qRT-PCR confirms microarray findings in the discovery cohort.
Correlation between log2-transformed microarray gene expression values and corresponding SmartChip qRT-PCR expression levels for CD28, ICOS, LCK, and ITK in patients (n = 43) from the discovery cohort. P values were determined by Student’s t distribution for Pearson correlation.
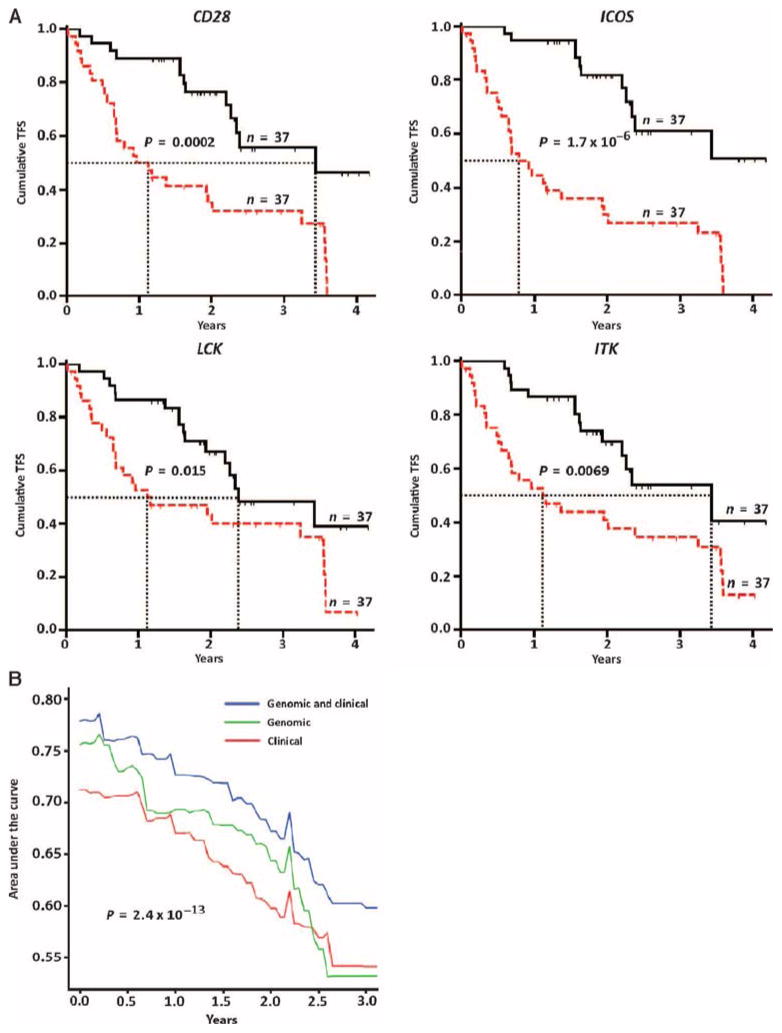
In the replication cohort (n = 74), decreased expression of CD28, ICOS, LCK, and ITK (split at 4.858, 6.303, 4.333, and 5.069 cycles, respectively) was significantly associated with decreased TFS (Fig. 4A). At the end of the observation period, TFS of patients with low CD28 expression was 22% compared with 65% among patients with high CD28 expression. In patients with low ICOS expression, TFS was 16% compared to 70% among patients with high ICOS expression. In patients with low ITK expression, TFS was 24% compared to 62% among patients with high ITK expression. In patients with low LCK expression, TFS was 30% compared to 57% among patients with high LCK expression. A decrease in CD28, ICOS, LCK, or ITK expression was individually associated with median TFS that ranged from 0.92 to 1.17 years, and increased expression was associated with longer median TFS, ranging from 2.39 to 3.44 years (table S4). The unadjusted hazard ratios for CD28 (3.2; 95% CI, 1.73 to 5.92), ICOS (4.52; 95% CI, 2.42 to 8.42), LCK (2.1; 95% CI, 1.14 to 3.86), and ITK (2.3; 95% CI, 1.25 to 4.23) were between 2.1 and 4.5, indicating that low levels of expression of these genes (when split by their median value) at evaluation were associated with a two- to fourfold higher risk of dying or having a lung transplant. TFS prediction was also significant after adjusting continuous ΔCt values of each individual gene to age, gender, and percent predicted FVC (FVC%) (tables S6 to S9).
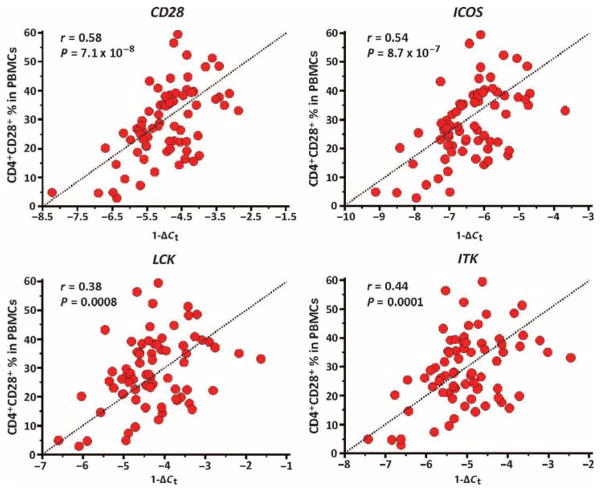
Fig. 4. CD28, ICOS, LCK, and ITK are potential IPF outcome biomarkers.
(A) TFS analysis in the replication cohort (n = 74) with available qRT-PCR data for CD28, ICOS, LCK, and ITK. In the Kaplan-Meier plots for each gene, the red lines are patients with expression levels above the ΔCt median value (representing a decrease in gene expression); the black lines are patients with expression levels below the ΔCt threshold (representing an increase in gene expression); the median survival of each group is depicted in dotted vertical lines. P values were determined by the log-rank test. (B) AUC of time-dependent ROC analysis for TFS based on clinical and/or genomic models in replication cohort subjects with all available variables (n = 72). Genomic model included continuous ΔCt values of CD28, ICOS, LCK, and ITK. Clinical model included age, gender, and FVC%. P values were determined by the Wilcoxon signed rank test.
We compared the area under the receiver operating characteristic (ROC) curve (AUC) of a genomic model (qRT-PCR expression of CD28, ICOS, LCK, and ITK), a clinical model (age, gender, and FVC%), and a combined genomic and clinical model (qRT-PCR expression of CD28, ICOS, LCK, and ITK along with age, gender, and FVC%). The highest AUC of all tested Cox proportional hazard models was observed at 2.4 months (0.2 years). The AUC for the combined genomic and clinical model at this time point was higher (78.5%) than that for the genomic model alone (76.6%) or the clinical model alone (70.9%) (Fig. 4B and table S10). The AUC differences between these models were statistically significant.
Changes in CD4+CD28+ T cells and gene expression findings
To evaluate the potential cellular source of the PBMC gene expression changes, we correlated the qRT-PCR expression level (reflected by 1 −ΔCt) of CD28, ICOS, LCK, and ITK with the percentage of CD4+CD28+ T cells in PBMCs, in replication cohort patients with simultaneous assays (n = 72). CD28 (r = 0.58; 95% CI, 0.41 to 0.72), ICOS (r = 0.54; 95% CI, 0.35 to 0.69), LCK (r = 0.39; 95% CI, 0.17 to 0.57), and ITK (r = 0.44; 95% CI, 0.23 to 0.61) were significantly correlated with the percentage of CD4+CD28+ T cells in PBMCs (Fig. 5), suggesting that a decreased number of these cells may explain, at least in part, the decreased expression of these genes. Along these lines, decreased percentage of CD4+CD28+ T cells in PBMCs (split at the median percentage or 27.8%) was associated with decreased TFS in the replication cohort (fig. S1A). TFS prediction was significant after adjusting the CD4+CD28+ T cell percentages to age, gender, and FVC% (table S10). Predictive models for death or lung transplant, including the percentage of CD4+CD28+ T cells in PBMCs, demonstrated an outcome prediction that was lower than predictive models, using qRT-PCR expression of CD28, ICOS, LCK, and ITK (fig. S1B and table S11).
Fig. 5. Expression levels of CD28, ICOS, LCK, and ITK correlate with the number of circulating CD4+CD28+ T cells.
Correlation between the percentage of CD4+CD28+ T cells in PBMCs and their corresponding 1 − ΔCt SmartChip qRT-PCR expression levels of CD28, ICOS, LCK, and ITK in (n = 72) patients from the replication cohort. P values were determined by Student’s t distribution for Pearson correlation.
There were no statistically significant differences (P = 0.52, Fisher’s exact test) in the use of immunosuppression between the patients with high versus low percentage of CD4+CD28+ T cells in PBMCs (split by the median) in the replication cohort; we also did not find immunosuppression use as an independent predictor of TFS in the discovery and replication cohorts (P = 0.59 and 0.23, respectively, Cox proportional hazard model). CD28, ICOS, LCK, or ITK expression levels did not correlate (P > 0.05 for each gene, Student’s t distribution for Pearson correlation) with the absolute number of peripheral blood lymphocytes in IPF patients (n = 35) from the discovery cohort that had this measure at the time of PBMC extraction.
Given the reported associations of increased number of CD4+CD28null T cells in IPF patients with poor outcomes (11), we measured the protein expression by flow cytometry of the T cell costimulatory protein ICOS, the T cell receptor complex protein CD3ε, and the tyrosine kinases LCK and ITK among paired autologous CD4+CD28+ and CD4+CD28null T cells in patients with IPF from the replication cohort. Although these proteins were significantly decreased in CD4+CD28null T cells (figs. S2 and S3), the percentage of CD4+CD28null T cells in PBMCs was not significantly correlated with the expression of CD28, ICOS, LCK, and ITK genes in IPF patients from the discovery cohort (n = 72) with simultaneous assays (P > 0.05 for each gene, Student’s t distribution for Pearson correlation).
DISCUSSION
Here, we identified changes in the expression of genes and pathways in PBMCs that correlated with poor IPF outcomes in two independent cohorts from different academic institutions, using different microarray platforms. We initially identified a signature of 52 genes as significantly associated with shorter TFS in the discovery cohort. Using this signature, we clustered the patients in the replication cohort to look for TFS differences between the patients in the major clusters and identified two clusters of patients with significant differences in TFS. Analysis of gene sets associated with shorter TFS showed decreased expression of most of the genes of “The costimulatory signal during T cell activation” Biocarta pathway in both cohorts. The genes CD28, ICOS, LCK, and ITK were members of the 52-gene signature and had the lowest Cox score when performing GSA, thus having the highest association with shorter TFS in this pathway when underexpressed. qRT-PCR confirmed that IPF patients with decreased expression of CD28, ICOS, LCK, and ITK had shorter TFS. A combined genomic and clinical prediction model including ΔCt expression of CD28, ICOS, LCK, and ITK along with age, gender, and FVC% provided better outcome prediction than using the clinical predictors alone.
Recognition that the course of IPF is variable and unpredictable has generated substantial interest in molecular biomarkers. Increases in the concentrations of peripheral blood proteins such as Krebs von den Lungen-6 (KL-6), surfactant protein A (SP-A), chemokine ligand 18 (CCL18), matrix metalloproteinase 7 (MMP7), intercellular adhesion molecule 1 (ICAM), and interleukin-8 (IL-8) (7, 10, 12, 20) have all been associated with decreased survival in IPF patients. However, these studies rarely contained a replication cohort and were limited in their discovery potential because they only tested a small, predefined set of markers. Recently, a study reporting a comparison of whole-blood transcriptomes of patients with IPF to healthy controls demonstrated the potential wealth of information available in the peripheral blood of patients with IPF (13); however, this study did not contain any information about outcome-associated genes or the potential cellular source of the gene expression changes. Thus, the attributes that distinguish our study from previous work are the focus on an unbiased genome-scale screening for predictors of outcomes, the use of a discovery and a replication cohort, and the attempt to outline the cellular source of the signature. Our unbiased screen led us to discover that decreases in molecules and pathways rarely studied in IPF, such as the T cell costimulatory proteins CD28 and ICOS, the tyrosine kinases LCK and ITK, as well as other members of the Biocarta pathway “The costimulatory signal during T cell activation,” are indicative of more severe outcomes in IPF. Decreases in gene expression of CD28, ICOS, LCK, and ITK may be related to a decrease in the number of CD4+CD28+ T cells in the peripheral blood—a finding that has not been previously reported in IPF and warrants detailed mechanistic follow-up.
The clinical implications of predicting outcome in IPF are substantial. The only effective therapy currently available for IPF patients is lung transplantation. The timing of transplantation is determined by the clinical evaluation, combined with the lung allocation score (21). Pretrans-plant evaluations are cost-intensive and not accurate enough to establish optimal timing (22). Shortage of organs is also a limitation. Hence, adding information about the expression of CD28, ICOS, LCK, and ITK to clinical parameters could be useful in determining who should be referred for pretransplantation assessments and specifically, given the ability of the model to predict early outcomes, to prioritize organ allocations to those who have been evaluated. The ability to predict TFS is also important for drug studies in IPF. In a relatively uncommon disease, to show an effect of a drug on mortality, investigators need to recruit patients who are likely to progress during the course of the study. It is possible that patients from a certain risk strata end up randomly and disproportionately assigned to one of the experimental groups, leading to spurious results. The significantly increased AUC of the combined genomic and clinical model in comparison to the clinical model alone may suggest that adding CD28, ICOS, LCK, and ITK expression levels to clinical parameters may help recruiting patients who are likely to progress.
It is important to consider several limitations of our study. First, despite the inclusion of two independent cohorts, the size and diversity of our cohorts are limited. Larger studies on more ethnically and clinically diverse populations will be required to determine the applicability of our markers to the general IPF population. Second, our study was designed to capture only mortality or transplant as outcomes. It would be beneficial to include in the model other IPF outcomes, such as acute exacerbations and disease progression, as reflected by declines in pulmonary functions. In this context, assessing gene expression changes during disease progression would be a highly useful tool to evaluate shifts in their patient risk profiles. Finally, although our study supports the emerging notion that proteins, gene transcripts, and cells in the blood are informative with regard to pathogenesis and outcomes in IPF—a disease previously considered to be limited to the lung—it does not provide information whether changes in peripheral blood gene expression have an added or different utility than bloodstream proteins. Future work should assess all likely markers in parallel and determine their relative value as biomarkers.
In summary, in our study, a microarray-derived 52-gene expression profile or qRT-PCR of CD28, ICOS, LCK, and ITK, members of this signature, was sufficient to identify IPF patients destined for poor outcomes. Combining gene expression data with clinical parameters enhanced outcome prediction; thus, our results could have considerable value in clinical evaluations and management of patients with this devastating lung disease. Naturally, despite the reproducibility of our findings across two cohorts, additional and larger studies focused on validating our results will be required before PBMC gene expression can be used clinically for prognosis in IPF.
MATERIALS AND METHODS
Study design: Patients and cohorts
Patients were recruited from the University of Chicago (discovery cohort; n = 45) and the University of Pittsburgh (replication cohort; n = 75). IPF diagnosis was established by a multidisciplinary group at each institution with the American Thoracic Society/European Respiratory Society criteria (23) and was consistent with recent guidelines (24). Patients were excluded in the study if they had evidence of autoimmune syndromes, malignancies, infections, drugs, or occupational exposures known to cause lung fibrosis. The studies were approved by the institutional review boards at the two institutions, and informed consent was obtained from all patients. Demographic and clinical information was collected in all patients at the time of blood draw. Spirometric data and diffusion capacity of the lung for carbon monoxide (DLCO) obtained within 3 months of blood draw were available, with the exception of four IPF patients of the replication cohort who did not have DLCO values available within this time range.
The time-to-event outcome analyzed was TFS. Patients were followed in clinics (at 3- to 4-month intervals) from blood draw until death or completion of the study on 5 February 2011. In this analysis, transplants and deaths were both counted as events. Transplant and vital status could not be confirmed in three patients evaluated at the University of Pittsburgh who were lost to follow-up; these patients were censored at their last visit day.
Microarray experiments and data preprocessing
Microarray expression was determined in two cohorts: a discovery cohort of IPF patients evaluated at the University of Chicago (n = 45) and a replication cohort of IPF patients evaluated at the University of Pittsburgh (n = 75). Microarray experiments were compliant with MIAME (Minimum Information About a Microarray Experiment) guidelines. The complete data sets are available in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE28221. For the discovery cohort, PBMC samples were obtained by density centrifugation. RNA was extracted with TRIzol (Invitrogen), and labeling reactions were performed with GeneChip WT cDNA Synthesis and Amplification Kit, followed by hybridization with GeneChip Human 1.0 exon ST arrays (Affymetrix) following the manufacturer’s protocol. A microarray experiment was run for every subject’s sample in the discovery cohort, and these experiments were performed at the University of Chicago. Data were processed and normalized with dChip software (25). For the replication cohort, PBMC samples were obtained by density centrifugation. Total RNA was extracted with QIAzol (Qiagen), and labeling reactions were performed with Agilent Quick Amp labeling kit, one-color, followed by hybridization with Whole Human Genome Oligo Microarray, 4 × 44K (G4112F, Agilent Technologies) following the manufacturer’s protocol. A microarray experiment was run for every subject’s sample in the replication cohort, and these experiments were performed at the University of Pittsburgh. To normalize the gProcessedSignal, we performed cyclic loess as previously described (26). Please see Supplementary Methods for more information regarding sample collection, RNA extraction, and microarray experiments.
Given the differences in microarray technologies between the studied cohorts (discovery cohort used Affymetrix, replication cohort used Agilent), we matched the gene probes across platforms in each microarray expression data set. In brief, after each microarray platform normalization, we matched the Affymetrix gene probes (n = 44,280 probes) with the Agilent gene probes (n = 29,807 probes) by their corresponding gene IDs (http://www.ncbi.nlm.nih.gov/gene). Because there are multiple replicated probes for the same gene in each platform studied, we selected only the unique probes with the highest interquartile range variation across the arrays and generated two independent data sets (Affymetrix and Agilent), each with n = 17,417 unique gene symbols. Last, for univariate gene selection and GSA in the discovery cohort, we applied a minimum fold change filter to the previously matched Affymetrix data set (n = 17,417) to exclude noninformative gene probes; for this step, we selected only the Affymetrix gene probes where 10% of the expression values of a given gene probe had at least a fold change of 1.25 from the median expression value of that probe, resulting in a set of n = 11,991 unique gene probes. The corresponding n = 11,991 unique gene probes in the Agilent data set were used for analyses in the replication cohort. SAM was used to test the association between PBMC microarray gene expression and TFS in IPF patients from the discovery cohort, as described in Supplementary Methods.
Hierarchical clustering of samples by TFS-associated genes identified in the discovery cohort microarrays was performed in the replication cohorts’ microarrays with Cluster 3.0 software (27). The samples were hierarchically clustered with median normalization of the genes and centroid linkage, and the similarity metric used was Pearson correlation.
Statistical analyses
Differences between IPF patients
Differences in age and pulmonary function tests between IPF patients were evaluated with an unpaired, two-tailed t test. Differences in gender, smoking status, diagnostic strategy, and use of immunosuppressive therapy were evaluated with Fisher’s exact test. Significance was defined as P < 0.05.
qRT-PCR—Discovery cohort
Affymetrix log2-transformed microarray gene expression values were correlated with their corresponding SmartChip qRT-PCR expression levels (1 − ΔCt) with Pearson correlation. P values were derived with Student’s t distribution. Significance was defined as P < 0.05.
TFS analyses—Replication cohort
For the qRT-PCR outcome cohort analyses, we used the survival (28) and risksetROC (29) packages of the R environment (30). When performing Cox proportional hazard models, we applied the stepAIC (31) approach for variable selection and included all variables as continuous covariates with the exception of gender. In brief, qRT-PCR ΔCt values of CD28, ICOS, LCK, and ITK as well as the percentage of CD4+CD28+ T cells in PBMCs were split by their median value into high- and low-risk ranges, and TFS differences were calculated with Kaplan-Meier curves and the log-rank test. The predictive significance of each gene as well as the percentage of CD4+CD28+ T cell for TFS was evaluated with Cox proportional hazard models after adjusting for clinical covariates known to be associated with poor IPF outcomes (age, gender, and FVC%). Finally, to evaluate which Cox proportional hazard model resulted in higher outcome prediction, we fit five different Cox proportional hazard models in subjects with all available variables (n = 72), as follows: genomic and clinical (ΔCt of CD28, ICOS, LCK, ITK, age, gender, and FVC%), genomic (ΔCt of CD28, ICOS, LCK, and ITK), clinical (age, gender, and FVC%), CD4+CD28+ % (percentage of CD4+CD28+ T cells), and CD4+CD28+ % and clinical (percentage of CD4+CD28+ T cells, age, gender, and FVC%). To plot the differences between the analyzed Cox proportional hazard models, we used time-dependent ROC for censored data (32) and AUC. When deriving the AUC estimates, we performed a 10-fold cross-validation procedure to handle any potential bias. In addition, we compared prediction accuracies (bias-controlled AUCs) of any two Cox regression models with a Wilcoxon signed rank test. Significance was defined as P < 0.05.
Flow cytometry analyses—Replication cohort
The correlation between 1 −ΔCt expression of CD28, ICOS, LCK, and ITK with the percentage of CD4+CD28+ T cells in PBMCs in the replication cohort was performed with Pearson correlation. P values were derived with Student’s t distribution. The comparison of the T cell costimulatory protein ICOS, the T cell receptor complex protein CD3ε, and the tyrosine kinases LCK and ITK between CD4+CD28+ and CD4+CD28null cells was performed with the Wilcoxon test for paired samples. Significance was defined as P < 0.05.
Cross-validation for AUC—Replication cohort
For the 10-fold cross-validation, the whole data set was randomly divided in data sets (folds) of similar size. A test set was randomly selected among one of the 10 folds, and the remaining nine sets were used to train the validation model. Subsequent iterations of training and validation were performed, and within each iteration, a different fold of the data was held out for validation, whereas the remaining folds were used for learning, a procedure that was repeated for a total of 10 times, thus estimating 10 AUCs from each test data set. The final AUC value was estimated from the average of the 10 resulting AUCs at each specific time point. The SE was calculated from the variation of the 10 resulting AUCs at each time point.
Supplementary Material
Acknowledgments
We thank L. Chensny, M. Klesen, and T. Black for their help in patient recruitment, sample collection and preparation, and database management, and A. Sperling for invaluable scientific input, criticism, and advice.
Funding: The Dorothy P. and Richard P. Simmons Endowed Chair for Pulmonary Research, HL0894932, HL108642, and HL095397 (N.K.); HL073241 and HL107172 (S.R.D.); HL101740, HL080513, Pulmonary Fibrosis Foundation, and Coalition for Pulmonary Fibrosis (I.N.); and HL98050, HL101740, and HL105371 (J.G.N.G.).
Footnotes
Author contributions: Conception and design: J.D.H.-M., I.N., K.F.G., J.G.N.G., S.D.S., and N.K. Patient recruitment, diagnosis ascertainment, and quality control: I.N., K.F.G., K.O.L., R.V., B.M.J.-G., J.D.H.-M., and N.K. RNA extraction, labeling, and microarray hybridization: B.M.J.-G., S.-F.M., and J.D.H.-M. Analysis of microarray data and intellectual contribution: E.F., S.K., J.D.H.-M., B.M.J.-G., G.C.T., S.-F.M., Y.L., Y.H., J.G.N.G., and N.K. qRT-PCR analysis, statistical modeling, and analysis: E.F., S.K., T.J.R., S.D.S., J.D.H.-M., and N.K. Flow cytometry experiments and analyses: J.X., S.R.D., and J.D.H.-M. Manuscript preparation: J.D.H.-M., E.F., S.R.D., I.N., B.M.J.-G., S.R.D., G.C.T., K.F.G., J.G.N.G., and N.K.
Competing interests: J.D.H.-M., I.N., T.J.R., and N.K. have a patent application, in conjunction with the University of Pittsburgh, titled “Marker panels for idiopathic pulmonary fibrosis diagnosis and evaluation.” J.D.H.-M., I.N., Y.H., J.G.N.G., and N.K. have a patent application, in conjunction with the University of Chicago. S.R.D. has a patent application, in conjunction with the University of Pittsburgh, for use of T cell characteristics as biomarkers in IPF and other chronic lung diseases. N.K. was a consultant to Sanofi-Aventis and Stromedix, and currently consults for InterMune, Vertex, Promedior, Takeda, and Actelion. N.K. is a recipient of research grants from Centocor in the past and presently Gilead and Celgene.
Data and materials availability: The complete data sets are available in the Gene Expression Omnibus database (accession number GSE28221). All materials were generated in the laboratories of N.K. and I.N., and they are available upon request in accordance to institutional regulations and policies.
www.sciencetranslationalmedicine.org/cgi/content/full/5/205/205ra136/DC1
Methods
Fig. S1. CD4+CD28+ T cells predict TFS.
Fig. S2. CD4+CD28null T cells have decreased protein expression of T cell markers.
Fig. S3. CD4+CD28null cells have decreased protein expression of selected T cell markers.
Table S1. Clinicopathological characteristics of the IPF patients in the two cohorts.
Table S2. Significant gene sets associated with TFS in the discovery cohort.
Table S3. Clinicopathological characteristics of the IPF patients in the two major clusters of the replication cohort.
Table S4. Median TFS times and CIs.
Table S5. Significant gene sets associated with TFS in the replication cohort.
Table S6. Multivariate Cox proportional hazard model including CD28 and clinical variables.
Table S7. Multivariate Cox proportional hazard model including ICOS and clinical variables.
Table S8. Multivariate Cox proportional hazard model including LCK and clinical variables.
Table S9. Multivariate Cox proportional hazard model including ITK and clinical variables.
Table S10. AUCs and SEs for TFS.
Table S11. Multivariate Cox proportional hazard model including the percentage of CD4+CD28+ T cells and clinical variables.
REFERENCES AND NOTES
- 1.Fernández Pérez ER, Daniels CE, Schroeder DR, St Sauver J, Hartman TE, Bartholmai BJ, Yi ES, Ryu JH. Incidence, prevalence, and clinical course of idiopathic pulmonary fibrosis: A population-based study. Chest. 2010;137:129–137. doi: 10.1378/chest.09-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz DA, Helmers RA, Galvin JR, Van Fossen DS, Frees KL, Dayton CS, Burmeister LF, Hunninghake GW. Determinants of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 1994;149:450–454. doi: 10.1164/ajrccm.149.2.8306044. [DOI] [PubMed] [Google Scholar]
- 3.King TE, Jr, Tooze JA, Schwarz MI, Brown KR, Cherniack RM. Predicting survival in idiopathic pulmonary fibrosis: Scoring system and survival model. Am J Respir Crit Care Med. 2001;164:1171–1181. doi: 10.1164/ajrccm.164.7.2003140. [DOI] [PubMed] [Google Scholar]
- 4.Zappala CJ, Latsi PI, Nicholson AG, Colby TV, Cramer D, Renzoni EA, Hansell DM, du Bois RM, Wells AU. Marginal decline in forced vital capacity is associated with a poor outcome in idiopathic pulmonary fibrosis. Eur Respir J. 2010;35:830–836. doi: 10.1183/09031936.00155108. [DOI] [PubMed] [Google Scholar]
- 5.Ley B, Ryerson CJ, Vittinghoff E, Ryu JH, Tomassetti S, Lee JS, Poletti V, Buccioli M, Elicker BM, Jones KD, King TE, Jr, Collard HR. A multidimensional index and staging system for idiopathic pulmonary fibrosis. Ann Intern Med. 2012;156:684–691. doi: 10.7326/0003-4819-156-10-201205150-00004. [DOI] [PubMed] [Google Scholar]
- 6.Collard HR, King TE, Jr, Bartelson BB, Vourlekis JS, Schwarz MI, Brown KK. Changes in clinical and physiologic variables predict survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2003;168:538–542. doi: 10.1164/rccm.200211-1311OC. [DOI] [PubMed] [Google Scholar]
- 7.Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, Lokshin AE, Lindell KO, Cisneros J, Macdonald SD, Pardo A, Sciurba F, Dauber J, Selman M, Gochuico BR, Kaminski N. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med. 2008;5:e93. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moeller A, Gilpin SE, Ask K, Cox G, Cook D, Gauldie J, Margetts PJ, Farkas L, Dobranowski J, Boylan C, O’Byrne PM, Strieter RM, Kolb M. Circulating fibrocytes are an indicator of poor prognosis in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;179:588–594. doi: 10.1164/rccm.200810-1534OC. [DOI] [PubMed] [Google Scholar]
- 9.Prasse A, Probst C, Bargagli E, Zissel G, Toews GB, Flaherty KR, Olschewski M, Rottoli P, Müller-Quernheim J. Serum CC-chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;179:717–723. doi: 10.1164/rccm.200808-1201OC. [DOI] [PubMed] [Google Scholar]
- 10.Kinder BW, Brown KK, McCormack FX, Ix JH, Kervitsky A, Schwarz MI, King TE., Jr Serum surfactant protein-A is a strong predictor of early mortality in idiopathic pulmonary fibrosis. Chest. 2009;135:1557–1563. doi: 10.1378/chest.08-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilani SR, Vuga LJ, Lindell KO, Gibson KF, Xue J, Kaminski N, Valentine VG, Lindsay EK, George MP, Steele C, Duncan SR. CD28 down-regulation on circulating CD4 T-cells is associated with poor prognoses of patients with idiopathic pulmonary fibrosis. PLoS One. 2010;5:e8959. doi: 10.1371/journal.pone.0008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards TJ, Kaminski N, Baribaud F, Flavin S, Brodmerkel C, Horowitz D, Li K, Choi J, Vuga LJ, Lindell KO, Klesen M, Zhang Y, Gibson KF. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;185:67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang IV, Luna LG, Cotter J, Talbert J, Leach SM, Kidd R, Turner J, Kummer N, Kervitsky D, Brown KK, Boon K, Schwarz MI, Schwartz DA, Steele MP. The peripheral blood transcriptome identifies the presence and extent of disease in idiopathic pulmonary fibrosis. PLoS One. 2012;7:e37708. doi: 10.1371/journal.pone.0037708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herazo-Maya JD, Kaminski N. Personalized medicine: Applying ‘omics’ to lung fibrosis. Biomark Med. 2012;6:529–540. doi: 10.2217/bmm.12.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ottoboni L, Keenan BT, Tamayo P, Kuchroo M, Mesirov JP, Buckle GJ, Khoury SJ, Hafler DA, Weiner HL, De Jager PL. An RNA profile identifies two subsets of multiple sclerosis patients differing in disease activity. Sci Transl Med. 2012;4:153ra131. doi: 10.1126/scitranslmed.3004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Achiron A, Gurevich M, Friedman N, Kaminski N, Mandel M. Blood transcriptional signatures of multiple sclerosis: Unique gene expression of disease activity. Ann Neurol. 2004;55:410–417. doi: 10.1002/ana.20008. [DOI] [PubMed] [Google Scholar]
- 17.Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA, Bogaev RC, Elashoff B, Baron H, Yee J, Valantine HA. IMAGE Study Group, Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–1900. doi: 10.1056/NEJMoa0912965. [DOI] [PubMed] [Google Scholar]
- 18.Risbano MG, Meadows CA, Coldren CD, Jenkins TJ, Edwards MG, Collier D, Huber W, Mack DG, Fontenot AP, Geraci MW, Bull TM. Altered immune phenotype in peripheral blood cells of patients with scleroderma-associated pulmonary hypertension. Clin Transl Sci. 2010;3:210–218. doi: 10.1111/j.1752-8062.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Showe MK, Vachani A, Kossenkov AV, Yousef M, Nichols C, Nikonova EV, Chang C, Kucharczuk J, Tran B, Wakeam E, Yie TA, Speicher D, Rom WN, Albelda S, Showe LC. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res. 2009;69:9202–9210. doi: 10.1158/0008-5472.CAN-09-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama A, Kondo K, Nakajima M, Matsushima T, Takahashi T, Nishimura M, Bando M, Sugiyama Y, Totani Y, Ishizaki T, Ichiyasu H, Suga M, Hamada H, Kohno N. Prognostic value of circulating KL-6 in idiopathic pulmonary fibrosis. Respirology. 2006;11:164–168. doi: 10.1111/j.1440-1843.2006.00834.x. [DOI] [PubMed] [Google Scholar]
- 21.Egan TM, Murray S, Bustami RT, Shearon TH, McCullough KP, Edwards LB, Coke MA, Garrity ER, Sweet SC, Heiney DA, Grover FL. Development of the new lung allocation system in the United States. Am J Transplant. 2006;6:1212–1227. doi: 10.1111/j.1600-6143.2006.01276.x. [DOI] [PubMed] [Google Scholar]
- 22.Trulock EP, Edwards LB, Taylor DO, Boucek MM, Keck BM, Hertz MI. Registry of the International Society for Heart and Lung Transplantation: Twenty-second official adult lung and heart-lung transplant report—2005. J Heart Lung Transplant. 2005;24:956–967. doi: 10.1016/j.healun.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 23.American Thoracic Society; European Respiratory Society, American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med. 2002;165:277–304. doi: 10.1164/ajrccm.165.2.ats01. [DOI] [PubMed] [Google Scholar]
- 24.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, Lynch DA, Ryu JH, Swigris JJ, Wells AU, Ancochea J, Bouros D, Carvalho C, Costabel U, Ebina M, Hansell DM, Johkoh T, Kim DS, King TE, Jr, Kondoh Y, Myers J, Müller NL, Nicholson AG, Richeldi L, Selman M, Dudden RF, Griss BS, Protzko SL, Schünemann HJ. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis, An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.http://www.bioinformatics.org/dchip
- 26.Wu W, Dave N, Tseng GC, Richards T, Xing EP, Kaminski N. Comparison of normalization methods for CodeLink Bioarray data. BMC Bioinformatics. 2005;6:309. doi: 10.1186/1471-2105-6-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 28.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer; New York: 2000. [Google Scholar]
- 29.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 30.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Statist. 1996;5:299–314. [Google Scholar]
- 31.Venables WN, Ripley BD. Modern Applied Statistics with S. 4. Springer; New York: 2002. [Google Scholar]
- 32.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 33.Feghali-Bostwick CA, Tsai CG, Valentine VG, Kantrow S, Stoner MW, Pilewski JM, Gadgil A, George MP, Gibson KF, Choi AM, Kaminski N, Zhang Y, Duncan SR. Cellular and humoral autoreactivity in idiopathic pulmonary fibrosis. J Immunol. 2007;179:2592–2599. doi: 10.4049/jimmunol.179.4.2592. [DOI] [PubMed] [Google Scholar]
- 34.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.http://www.biocarta.com/
- 38.Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stümpflen V, Mewes HW. CORUM: The comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L. Reactome: A knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.http://www.sigmaaldrich.com/
- 43.Saunders B, Lyon S, Day M, Riley B, Chenette E, Subramaniam S, Vadivelu I. The Molecule Pages database. Nucleic Acids Res. 2008;36:D700–D706. doi: 10.1093/nar/gkm907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.http://stke.sciencemag.org/
- 45.http://www.sabiosciences.com/
- 46.Efron B, Tibshirani R. On testing the significance of set of genes. Ann Appl Stat. 2007;1:107–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.