Abstract
Chromosomal rearrangements and fusion genes play important roles in tumor development and progression. Four high-frequency prostate cancer-specific fusion genes were recently reported in Chinese cases. We attempted to confirm one of the fusion genes, USP9Y-TTTY15, by reverse transcription PCR, but detected the presence of the USP9Y-TTTY15 fusion transcript in cancer samples, nonmalignant prostate tissues, and normal tissues from other organs, demonstrating that it is a transcription-induced chimeric RNA, which is commonly produced in normal tissues. In 105 prostate cancer samples and case-matched adjacent nonmalignant tissues, we determined the expression level of USP9Y-TTTY15 and a previously reported transcription-induced chimeric RNA, SLC45A3-ELK4. The expression levels of both chimeric RNAs vary greatly in cancer and normal cells. USP9Y-TTTY15 expression is neither higher in cancer than adjacent normal tissues, nor correlated with features of advanced prostate cancer. Although the expression level of SLC45A3-ELK4 is higher in cancer than normal cells, and a dramatic increase in its expression from normal to cancer cells is correlated with advanced disease, its expression level in cancer samples alone is not correlated with any clinical parameters. These data show that both chimeric RNAs contribute less to prostate carcinogenesis than previously reported.
Introduction
Gene fusions induced by chromosomal rearrangement, initially identified in hematological malignancies and soft tissue sarcomas, for example the BCR-ABL fusion in chronic myeloid leukemia (Mitelman et al., 2007), have been used for tumor diagnosis, subclassification, prognosis, recurrence monitoring, and therapeutic targets. In addition to the generation of novel fusion proteins, this genomic fusion can lead to the overexpression of oncogenes under the control of the constitutive, or tissue type-specific high activity promoters that they join (Mitelman et al., 2007). Recently, with the development and application of modern genetic technologies, gene fusions have also been frequently identified in carcinoma, the most common type of human malignancy, including prostate (Tomlins et al., 2005), lung (Kohno et al., 2012; Soda et al., 2007), and other cancers (Edwards, 2010; Seshagiri et al., 2012), which have deepened our understanding of the importance of fusion genes in human malignant diseases. The EML4-ALK fusion found in lung cancer (Soda et al., 2007) has also been used as a target for molecular therapy (Pikor et al., 2013).
Prostate cancer is the most common cancer and the second leading cause of cancer-related death in Western men (Jemal et al., 2011). Numerous studies have been carried out to explore the genetic alterations underlying prostate cancer development and progression. The prostate cancer-specific TMPRSS2-ERG fusion, which has been found in approximately 50% of prostate cancer cases, is the most commonly detected fusion gene in human malignancies (Mitelman et al., 2007). In addition to TMPRSS2-ERG fusion, many other fusion genes have also been identified in prostate cancer, although at much lower frequencies (Boyd et al., 2012). Those fusion partners predominantly involve the ETS family genes and androgen-regulated transcripts (Boyd et al., 2012), although there are cases where both partner genes do not belong to these two categories, for example, the ESRP1-RAF1 fusion gene (Palanisamy et al., 2010). The identification of the high frequency TMPRSS2-ERG fusion also promoted research into fusion genes in other human solid tumors.
There is a clear difference in the incidence and mortality of prostate cancer between different populations (Jemal et al., 2011). Many factors, including living and working environment, diet, race, age (>65 y), sexual history, and genetic factors, have been proposed to contribute to this population difference (Gronberg, 2003; McCracken et al., 2007; Shook et al., 2007). We and other researchers have found that the frequency of certain somatic genomic changes in prostate cancers is drastically different between cases from the Western world and East Asian populations (Boyd et al., 2012; Lee et al., 2010; Magi-Galluzzi et al., 2011; Mao et al., 2010; Miyagi et al., 2010; Ren et al., 2012). This includes the TMPRSS2-ERG fusion, which is commonly found in Western prostate cancer but at a much lower frequency in Chinese and other East Asian populations (Lee et al., 2010; Magi-Galluzzi et al., 2011; Mao et al., 2010; Miyagi et al., 2010). Using next generation sequencing of the transcriptome, four new high-frequency fusion genes, USP9Y-TTTY15(19/54), RAD50-PDLIM4(15/54), CTAGE5-KHDRBS3(20/54) and SDK-AMACR(13/54) were identified in prostate cancer cases from China (Ren et al., 2012). Both USP9Y and TTTY15 are located on the Y chromosome (Yq11) within a short genomic distance. The USP9Y-TTTY15 transcript is not predicted to code a functional protein. If translated, the product would be a 96AA (10.56kDa) protein, containing the first 32AA from USP9Y, a region with no known functional domain, and potentially 64AA of TTTY15, also lacking a predicted functional domain. In this study we attempted to confirm the USP9Y-TTTY15 fusion as a Chinese prostate cancer-specific genetic alteration in a separate cohort of Chinese prostate cancer samples. Surprisingly, we detected USP9Y-TTTY15 fusion transcripts not only in prostate cancer samples but also in nonmalignant prostate tissues, as well as nonmalignant tissue from other organs.
Therefore, USP9Y-TTTY15 is a transcription-induced chimeric RNA (an RNA created in normal cells by combining exons from two or more distinctly known parental genes) (Akiva, 2005; Fang et al., 2012; Gingeras, 2009; Kannan et al., 2011; Kim DS et al., 2007, 2012; Kim P et al., 2010; Kim RN et al., 2012; Li et al., 2008; Parra, 2005; Prakash et al., 2010; Thomson et al., 2000; Zhang et al., 2012). Linked to this finding, we also investigated the expression of another transcription-induced chimeric RNA previously studied in prostate cancer from Western countries, SLC45A3-ELK4, of which both partner genes lie on the same chromosome within a short genomic distance (1q32) (Maher et al., 2009; Rickman et al., 2009; Zhang et al., 2012). We found that the roles they play in prostate cancer may not be as important as previously thought.
Materials and Methods
Samples
105 pairs of fresh frozen prostate cancer and their case-matched normal prostate tissue samples from prostate cancer patients who underwent radical prostatectomy, and one kidney, one lung, and one gall bladder tissue sample removed during routine surgery in noncancer patients at the First Affiliated Hospital, Zhejiang University Medical College, Hangzhou, China were collected for this study. In addition, two prostate benign hyperplasia (BPH), four cancer, two high-grade prostatic intraepithelial neoplasia, and seven nonmalignant prostate tissues adjacent to malignant lesions from patients who underwent urological surgery at Barts Health NHS Hospital, London, UK, were obtained. Samples were snap frozen and preserved at −80°C or in liquid nitrogen. Detailed clinicopathological information for all the Chinese cases are available, except diagnostic prostate specific antigen (PSA) levels missing in two patients (Supplementary Table S1).
The tissue morphology and Gleason grade of cancer lesions were confirmed by the pathologist authors. Cancer and gland-rich areas of adjacent normal tissues were macrodissected for RNA extraction. Those surgically removed tissues, remaining after pathological diagnosis, were used for this study with written informed consent from patients. This study, including the content procedures, was approved by the ethical committee of First Affiliated Hospital, Zhejiang University Medical College, China (No: 2012-42) and East London and The City REC Alpha, UK (No. 09/H0704/4). Five prostate cancer cell lines, VCaP, DU145, LNCaP, PC3, and 22RV1, and two immortalized prostate epithelial cell cultures, PNT1a and PNT2-C2, confirmed by STR genotype analysis were also used in this study.
Reverse transcription polymerase chain reaction (RT-PCR) and real time quantitative RT-PCR using SYBR Green technology
Total RNA was extracted from the above samples using Trizol reagent (Invitrogen) and cDNA synthesis was conducted in a 25 μL reaction mixture using Reverse Transcriptase M-MLV (RNase H¯) and random primer as previously described (Noel et al., 2010). As the previously reported primer pairs (USP9Y Forward: CTGTGTCAAGTATGACAGCCATC and TTTY15 Reverse: CCAGTTTTTCCAAGGGCTTT) (Ren et al., 2012) always generate a nonspecific PCR product 9 bp shorter than the fusion product at a range of annealing temperatures we tested, we designed a pair of primers (USP9Y Forward: GCAAAGATCTGTGCTGTGTCAAGTA and TTTY15 Reverse: GCTTAGTTTTCAGTGACTCACAGGT) to detect the reported USP9Y-TTTY15 fusion transcripts, with an expected product of 232 bp using an annealing temperature of 57.6°C. Each RT-PCR product band was gel-excised (E.Z.N.A. Gel Extraction Kit, Omega Bio-tek, Inc. Norcross, GA) for sequencing analysis.
100 ng cDNA was analyzed by quantitative RT-PCR using the SYBR Premix Ex Taq (Perfect Real Time, Takara, Dalian, China) following the manufacturer's instructions. The amplification program consisted of an initial denaturation step at 95°C for 30 sec, followed by 50 cycles of denaturation at 95°C for 5 sec, annealing at 60°C for 30 sec, and extension at 72°C for 15 sec. The total quantity of fluorescent product was measured at the end of every cycle with a single acquisition. Prior to the detection of gene expression, a standard curve for each primer pair was created by performing quantitative RT-PCR on a series of 10-fold dilutions of cDNA templates. Ct values between 8 and 35 were plotted against log10 of the dilution factor and the application efficiency was assessed using the slope of the regression line in the standard curve. The specificity of the primers was determined through a dissociation peak and further confirmed on a 1.5% agarose gel. Each sample was performed in triplicate, and a nontemplate control was included to detect possible contamination. For each triplicate, a mean Ct value was calculated and samples with aberrant values within the triplicate were repeated. Gene expression levels were normalized to the housekeeping gene GAPDH in the same sample by subtracting the Ct value of GAPDH from the Ct value of the target probes (ΔCt). ΔΔCt was calculated as the ΔCt of the target sample subtracted by the ΔCt of the control sample. Fold change compared with the control was calculated by 2-ΔΔCt. The transcript-specific primers used for quantitative RT-PCR are listed in Supplementary Table S2.
Sequencing analysis
PCR products (the corresponding fusion product bands) were cloned into pMD 18-T vector (Takara, Dalian, China) with a TA cloning technique according to the manufacturer's instructions. The molar ratio of vector DNA to insert DNA used for cloning ranged from 1:2–10, and the ligation reaction was performed at 16°C for 50 min. The plasmid was then transformed into competent E.coli and subsequently cultured in LB growth medium containing ampicillin. Monoclones were picked for sequencing analysis using M13+ primers and the ABI3730XL sequencing machine.
Detection of chimeric transcripts in prostate cancer transcriptome sequencing data
Raw next generation sequencing data was curated from three datasets, E-MTAB-567 Chinese dataset (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-567), EGAD00001000305 (https://www.ebi.ac.uk/ega/datasets/EGAD00001000305), and GSE22260 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22260), representing 40 primary prostate cancer and 25 nontumor prostate samples in total. Read sequences were mapped to the human reference genome (hg19) and Ensembl genes using the Bowtie algorithm (Langmead, 2010). Sequence alignments were subsequently processed to nominate chimeric RNAs using ChimeraScan (Iyer et al., 2011) with default parameters. Gene fusion nominations were required to have at least two independent fragments supporting individual chimeras.
Statistical analysis
A cut off value (fold change of these chimeric transcript expression level) was used to categorize the cases into groups and a Chi-squared test was performed. When the chimeric transcript expression level was left ungrouped, paired and unpaired t tests were applied. All statistical analysis was performed using SPSS 16.0 for Windows (SPSS, Chicago, IL, USA) with two tailed tests. A p value of less than 0.05 was considered statistically significant.
Results
The USP9Y-TTTY15 fusion transcript is expressed in all samples including different types of normal tissues
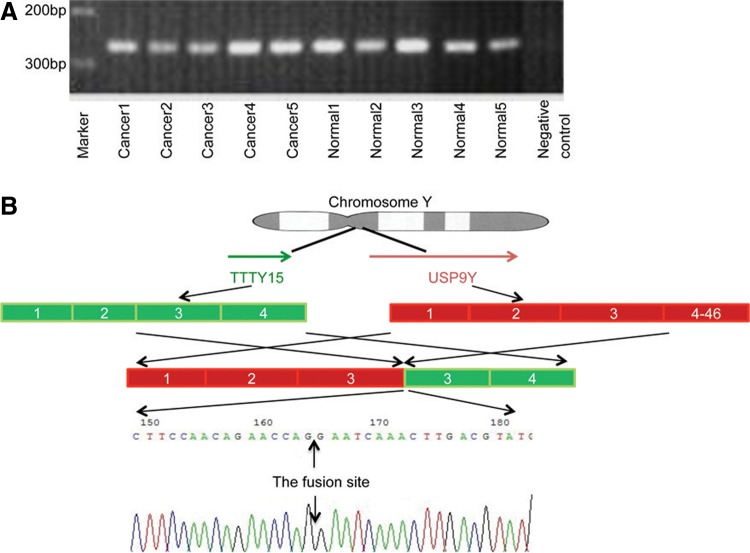
Using the RT-PCR primer pair we designed, we detected the expected 232 bp PCR product, which correlates to the previously reported form of the USP9Y-TTTY15 fusion. However, this fusion sequence was found in all prostate cancer (n=32) and adjacent normal tissue (n=15) samples analyzed (Fig. 1A). The fusion product was confirmed by sequencing analysis in all 21 randomly selected prostate cancer samples and nine adjacent nonmalignant tissues (Fig. 1B). Only one form of the USP9Y-TTTY15 fusion transcript was found using these primers.
FIG. 1.
Detection of the USP9Y-TTTY15 fusion transcript in prostate cancer and adjacent nonmalignant prostate tissue. (A) Examples of the RT-PCR products of the USP9Y-TTTY15 fusion transcript at 232 bp in prostate cancer (lanes 1–5) and case-matched adjacent normal tissue (lanes 6–10) samples along with a negative control without input cDNA template (lane 11). Marker: 1 Kb plus (Life Technology) DNA size marker. (B) Schematic presentation of the alignment of TTTY15 and USP9Y on the Y chromosome and the fusion transcripts based on the sequencing data of the fusion at the end of USP9Y exon 3 and the beginning of TTTY15 exon 3. The bottom panel is a representative image of the fusion sequence from a clinical sample. The position of the nucleotide fusion site is indicated by the black arrows.
We further examined the expression of the USP9Y-TTTY15 chimeric transcript in one BPH, one normal kidney, one lung, and one gall bladder sample by RT-PCR, with subsequent confirmation by sequencing. We found that the fusion transcript existed in all normal human tissues tested (Fig. 2). Therefore, USP9Y-TTTY15 could either be a transcription-mediated chimeric RNA produced in normal human cells, or an artifact generated from template switching during reverse transcription rather than a prostate cancer specific fusion gene (Cocquet et al., 2006).
FIG. 2.
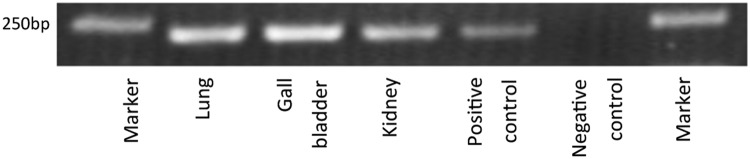
Detection of the USP9Y-TTTY15 fusion transcript by RT-PCR in normal lung, gall bladder, and kidney tissues. Gel image showing the USP9Y-TTTY15 fusion transcript detected at the expected size of 232 bp. A positive control using a prostate cancer cDNA sample and a negative control without cDNA template were also included. Marker: 1 Kb plus (Life Technology) DNA size marker.
In order to rule out the possibility of template switching-induced artifact fusion, we checked whether there are short duplicated sequences at the exon–intron and intron–exon boundaries, which has been proposed as the mechanism for template switching during reverse transcription (Cocquet et al., 2006). We did not find any short duplicated sequences near the junctions. In addition, the junction is canonical (GT-AG), further supporting that this chimeric sequence is a transcription-mediated chimeric RNA generated by trans or cis splicing. We also analyzed two BPH and a number of prostate cancer and adjacent nonmalignant samples from UK patients, as well as five prostate cancer cell lines (VCaP, DU145, LNCaP, PC3, and 22RV1) and two immortalized prostate epithelial cell cultures (PNT1a and PNT2-C2). We identified the fusion transcript in all of them, although at different expression levels (Fig. 3), indicating that expression of this chimeric transcript is not specific to the Chinese population either.
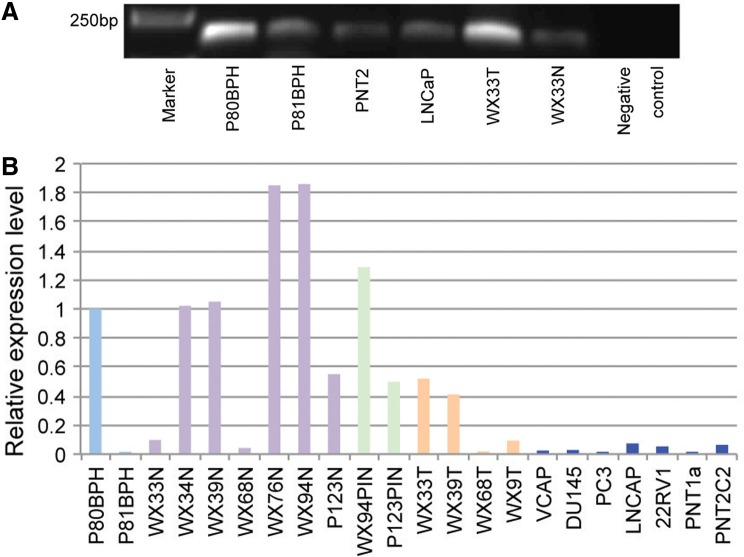
FIG. 3.
USP9Y-TTTY15 expression in UK prostate samples. (A) Examples of the RT-PCR products of the USP9Y-TTTY15 fusion transcript at 232 bp in two BPHs, one prostate cancer sample (WX33T), together with its adjacent normal tissue (WX33N) from an UK patient and two prostate cell lines (PNT2 and LNCaP). (B) USP9Y-TTTY15 expression level measured by qRT-PCR in UK samples was presented in N fold change relative to sample P80BPH (setting the expression level at 1). Light blue, BPH samples; purple, normal prostate tissue; green, PIN samples; brown, cancer samples; dark blue, prostate cell lines. BPH, benign prostatic hyperplasia; N, normal; PIN, prostate intraepithelial neoplasia; T, tumor.
Correlation of USP9Y-TTTY15 and SLC45A3-ELK4 expression levels with clinicopathological parameters in Chinese prostate cancer cases
We further analyzed the expression level of the chimeric transcripts USP9Y-TTTY15 and SLC45A3-ELK4 by quantitative RT-PCR in both the cancer and case-matched adjacent nonmalignant prostate tissue samples from 105 Chinese prostate cancer cases, where clinical data were available. The expression levels of USP9Y-TTTY15 in both cancer tissues and adjacent normal tissues vary dramatically, with higher expression in cancer cells than their case-matched normal tissues in 59 cases and lower in 45 cases. There was no statistically significant difference between the overall expression in cancer and adjacent nonmalignant tissues (p>0.05 for both paired and unpaired t test, Fig. 4A). The ratio of USP9Y-TTTY15 expression between cancer and adjacent normal tissues was significantly associated with a lower baseline level of PSA in normal tissues (using PSA cut off at 20 ng/mL, p=0.004), as well as older age at diagnosis (using 70y as cut off, p=0.025). However, USP9Y-TTTY15 expression level changes from normal to cancer samples was not correlated with Gleason score (p=0.781), clinical (TNM) stage (p=0.856), or lymph node metastasis (p=0.799). The correlation between prostate cancer clinicopathological parameters and the changes in USP9Y-TTTY15 expression in cancer samples compared to case-matched normal tissues are shown in Table 1. When the expression levels in cancer samples alone (not considering the expression in adjacent normal tissue) were compared between cases with different clinical parameters, the expression level of USP9Y-TTTY15 was not correlated with any parameters (Table 2). However, a higher expression level in normal prostate tissue was significantly associated with a higher baseline PSA (using 20 ng/ml as cut off, p=0.025) and diagnosis of prostate cancer at a younger age (using 70 years for grouping, p=0.043). The remaining correlations were not statistically significant (Table 2, Fig. 4A).
FIG. 4.
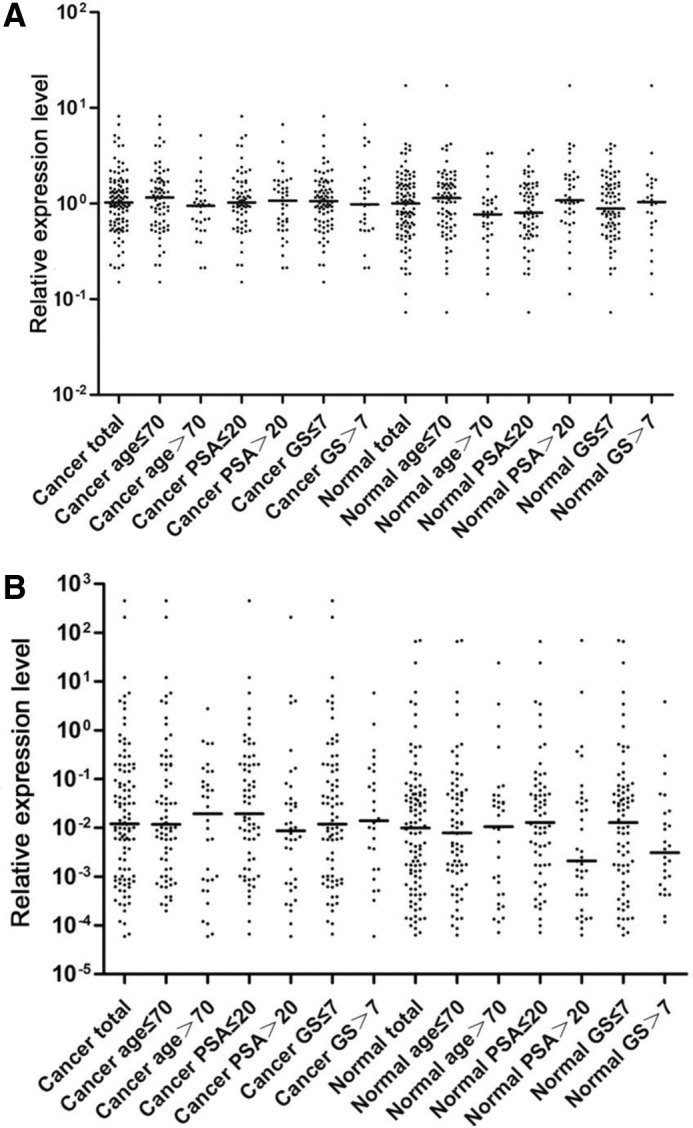
Distribution of the transcription-mediated chimera expression. (A) The distribution of USP9Y-TTTY15 expression level in the Chinese prostate cancer and adjacent nonmalignant tissues overall and in different clinical subgroups. (B) The distribution of SLC45A3-ELK4 expression level in the Chinese prostate cancer and adjacent nonmalignant tissues, split into different groups based on clinical parameters. The expression level of USP9Y-TTTY15 and SLC45A3-ELK4 (y axis) was presented in n fold change relative to sample 31C (setting the expression level at 1). Age, age of the patient at diagnosis; GS, Gleason score; PSA, prostate specific antigen.
Table 1.
Correlation of Clinicopathological Parameters and Expression Changes of USP9Y-TTTY15 in Chinese Prostate Cancer Samples Related to Case-Matched Normal Tissues
| Variable | Folds ≤1 | Folds >1 | p value |
|---|---|---|---|
| Age ≤70 | 37 | 35 | 0.021 |
| Age >70 | 9 | 24 | |
| PSA ≤20 | 22 | 43 | 0.004 |
| PSA >20 | 24 | 14 | |
| GS ≤7 | 34 | 45 | 0.781 |
| GS >7 | 12 | 14 | |
| Clinical stage ≤3 | 40 | 52 | 0.856 |
| Clinical stage >3 | 6 | 7 | |
| Meta negative | 42 | 53 | 0.799 |
| Meta positive | 4 | 6 |
Chi-squared test was performed. Folds ≤1 shows the number of cases where cancer samples express lower or equal levels of USP9Y-TTTY15 than the normal tissues, while Folds >1 shows the number of cases where cancer samples express higher levels of USP9Y-TTTY15 than the normal tissues. GS, Gleason score; meta, metastasis; PSA, prostate specific antigen.
Table 2.
Correlation of Clinicopathological Parameters and Expression Level of USP9Y-TTTY15 and SLC45A3-ELK4 in Prostate Cancer and Normal Tissues
| p values for USP9Y-TTTY15 | p values for SLC45A3-ELK4 | ||||
|---|---|---|---|---|---|
| Clinicopathological parameters | Number of cases | Cancer | Normal | Cancer | Normal |
| Age ≤70 | 72 | 0.216 | 0.043 | 0.249 | 0.706 |
| Age >70 | 33 | ||||
| PSA ≤20 | 65 | 0.607 | 0.025 | 0.183 | 0.058 |
| PSA >20 | 38 | ||||
| GS ≤7 | 79 | 0.904 | 0.818 | 0.815 | 0.150 |
| GS >7 | 26 | ||||
| Clinical stage ≤3 | 92 | 0.990 | 0.501 | 0.535 | 0.908 |
| Clinical stage >3 | 13 | ||||
| Meta negative | 95 | 0.821 | 0.604 | 0.683 | 0.940 |
| Meta positive | 10 | ||||
T test was performed. GS, Gleason score; meta, metastasis; PSA, prostate specific antigen.
We investigated whether stimulation by androgens in the androgen-sensitive prostate cancer cell line LNCaP would increase expression of USP9Y-TTTY15. LNCaP cells grown in androgen free medium (charcoal stripped serum) were treated with the synthetic androgen Mibolerone at 0.1, 1, and 10 nM, together with a 0.01% ethanol-treated control. Mibolerone treatment decreased the USP9Y-TTTY15 expression level in a dose dependent manor (Supplementary Fig. S1).
For the previously well-characterized SLC45A3-ELK4 chimeric transcript, although there are several forms of the fusion, we mainly detected the SLC45A3 exon 1 to ELK4 exon 2 fusion, which is consistent with the previous report (Rickman et al., 2009). This form of the fusion has been previously confirmed experimentally as a cis-splicing mediated chimeric transcript. We also found that the junction is canonical (GT-AG) without short duplicated sequences near the junctions. The expression levels of SLC45A3-ELK4 also vary dramatically in both cancer and adjacent normal tissues. In a pairwise comparison, while in 63 cases the expression in cancer cells was higher than case-matched normal tissues, in many cases (n=42), the expression in cancer cells was lower than their case-matched normal tissue. Overall, there is significantly higher expression of SLC45A3-ELK4 in prostate cancer samples than adjacent normal tissue if a paired t test is used for analysis (p=0.003). However, this significance disappeared (p=0.136) if an unpaired t test was applied due to the great variation in each group and overlapping of SLC45A3-ELK4 expression levels in these two sample groups (Fig. 4B). The expression of SLC45A3-ELK4 in cancer samples or normal prostate tissues alone did not correlate with any clinical parameters.
There was a trend towards higher SLC45A3-ELK4 expression in normal tissues in cases with lower baseline PSA and lower Gleason score, but none of the comparisons were statistically significant (p=0.058 and 0.15, respectively) (Table 2, Fig. 4B). Furthermore, we frequently detected ten times higher SLC45A-ELK4 expression in prostate cancer than in normal samples in cases with higher baseline PSA (≥20 ng/ml; p=0.031), Gleason score (p=0.032), TNM stage (p=0.012), and with lymph node metastasis (p=0.013), indicating that a dramatic increase of SLC45A3-ELK4 expression is associated with the progression of prostate cancer to an advanced disease status (Table 3).
Table 3.
Correlation of Clinicopathological Parameters and Expression Changes of SLC45A3-ELK4 in Chinese Prostate Cancer Samples Related to Case-Matched Normal Tissues
| Variable | Folds ≤10 | Folds >10 | p value |
|---|---|---|---|
| Age ≤70 | 56 | 16 | 0.400 |
| Age >70 | 28 | 5 | |
| PSA ≤20 | 67 | 11 | 0.031 |
| PSA >20 | 15 | 10 | |
| GS ≤7 | 67 | 12 | 0.032 |
| GS >7 | 17 | 9 | |
| Clinical stage ≤3 | 77 | 15 | 0.012 |
| Clinical stage >3 | 7 | 6 | |
| Meta negative | 79 | 16 | 0.013 |
| Meta positive | 5 | 5 |
Chi-squared test was performed. GS, Gleason score; meta, metastasis; PSA, prostate specific antigen.
We analyzed three next generation sequencing datasets of the prostate cancer transcriptome to detect these two chimeric transcripts. We found USP9Y-TTTY15 chimeric transcripts in six of 14 Chinese prostate cancer samples, with reads in individual samples ranging from 2 to 7. We also found USP9Y-TTTY15 chimeric transcripts in two of 14 normal tissues in this Chinese dataset, each sample had three reads. We did not detect this chimeric transcript in any of the samples in the GSE22260 dataset, where the sequencing depth is lower than the other two datasets. In the EGAD00001000305 dataset, where the sequencing depth is several times higher than the other two data sets, the USP9Y-TTTY15 chimeric transcript was detected in six out of eight cancer samples, each with only two or three reads. However, we did not find any SLC45A3-ELK4 positive cases from these three datasets. We also searched for TMPRSS2-ERG fusion transcripts in these three datasets and found TMPRSS2-ERG in four of 14 Chinese prostate cancer samples, 13 of 18 cancer samples in the GSE22260 dataset, and five of eight cancer samples from the EGAD00001000305 dataset, in more than half of the cases, there were more than 50 TMPRSS2-ERG reads supporting this fusion. Adjusted by the gene size, the number of reads for TMPRSS2 and SLC45A3 is 50 times more than that for USP9Y.
Discussion
With recent interest into the study of gene fusions in nonhematological malignancies, fusion genes have now been recognized as a common mechanism in the development and/or progression of many human solid tumors (Edwards, 2010; Mitelman et al., 2007). The four high frequency fusions reported to be specific to Chinese prostate cancer (Ren et al., 2012), once confirmed by independent studies, will change the understanding of fusion genes in prostate cancer. However, using a considerably large number of clinical samples, we found that one of the four fusion genes reported in that study, USP9Y-TTTY15, is not specific to Chinese prostate cancer. It has been detected in different types of human normal tissues and was not confined to the Chinese population either. This highlights the importance of careful examination of the adjacent and other normal tissues for a fusion transcript initially identified in cancer cells.
In fact, transcription-induced chimeric RNAs, generated in normal healthy cells, without corresponding genomic rearrangements have been well recognized in the field of genetic research (Akiva, 2005; Fang et al., 2012; Gingeras, 2009; Kim DS, et al., 2007; Kim DS, et al., 2012; Kim P, et al., 2010; Kim RN, et al., 2012; Li et al., 2008; Parra, 2005; Prakash et al., 2010; Thomson et al., 2000; Zhang et al., 2012). However, the existence of transcription-induced chimeric RNAs has not been well appreciated in the cancer research field, where fusion genes play an important role in disease development, progression, and treatment (Edwards, 2010; Mitelman et al., 2007). Transcription-induced chimeric RNAs are generally expressed at low levels (Prakash et al., 2010).
As the current approach in cancer genetics is mainly focused on fusion gene discovery in cancer samples, some transcription-induced chimeric RNAs can easily be reported as cancer-specific fusions, without thorough gene expression analysis of the normal tissues and genomic analysis to identify the corresponding rearrangements in tumor cells. Even in cases where fusion transcripts without corresponding genomic rearrangements have been identified (Guerra et al., 2008; Li et al., 2008; Maher et al., 2009; Rickman et al., 2009; Zhang et al., 2012), their contributions to tumorigenesis may be overestimated due to the focus on their roles in malignant disease in those studies. Therefore, it is essential to highlight the existence of transcription-induced chimeric RNAs in normal cells in the cancer research field, to prevent future reports of such chimeric transcripts as recurrent cancer specific fusion genes. Here we not only identified USP9Y-TTTY15 as a transcription-induced chimeric RNA with little role in prostate carcinogenesis, but also showed that SLC45A3-ELK4 may not be associated with prostate cancer development and progression as greatly as previously reported.
When we explored the correlation of the chimeric transcript USP9Y-TTTY15 with the clinicopathological data in this cohort of Chinese prostate cancer samples, we found that USP9Y-TTTY15 transcript expression is correlated with PSA levels in the normal tissues and inversely correlated with age at diagnosis, suggesting that USP9Y-TTTY15 expression may be influenced by androgens, that are also associated with PSA level and are at higher levels in younger men in comparison to older men. However, USP9Y-TTTY15 expression was not stimulated by androgen. The slightly reduced expression of USP9Y-TTTY15 may result from the increased expression of androgen controlled genes and relative ratio reduction of non-androgen controlled genes. The lower number of USP9Y reads, compared to that of androgen regulated genes TMPRSS2 and SLC45A3 from the transcriptome sequencing data, also support that USP9Y-TTTY15 is not prostate specific, or under the control of androgens. The above findings, together with the lack of an association between transcript expression levels and any other prostate cancer development or progression features, indicates that USP9Y-TTTY15 expression levels may play little or no role in prostate carcinogenesis.
Following TMPRSS2, SLC45A3 is the second most common 5′ fusion partner reported in prostate cancer in Western countries (Boyd et al., 2012). However, the SLC45A3-ELK4 fusion product is a transcription-induced chimeric RNA, which is generated by transcriptional run over of adjacent genes and cis-splicing in the absence of chromosomal rearrangement (Maher et al., 2009; Rickman et al., 2009; Zhang et al., 2012). It has been found in both prostate cancer and normal tissues (Rickman et al., 2009; Zhang et al., 2012). While transcription-induced chimeric RNAs exist in normal cells, changes in the expression levels of certain chimeric RNAs have the potential to be associated with cancer development and/or progression. The expression of many transcription-induced chimeric RNAs has been detected at a much higher level in prostate cancer samples than non-malignant prostate tissues (Kannan et al., 2011). Therefore, identifying the correlation between the expression level of SLC45A3-ELK4 fusion transcripts and prostate cancer development and progression (Maher et al., 2009; Rickman et al., 2009; Zhang et al., 2012) might still provide a potential diagnostic or prognostic biomarker for prostate cancer.
However, in our study of a large number of case-matched cancer and adjacent normal prostate tissues, SLC45A3-ELK4 expression varied greatly, both in cancer and normal samples. In many cases (42/105), SLC45A3-ELK4 was expressed at a higher level in the case-matched adjacent tissues than in cancer samples. The expression of SLC45A3-ELK4 was significantly higher in prostate cancer compared with adjacent normal tissue, only when a paired t-test was applied. Furthermore, contrary to a previous report (Rickman et al., 2009), SLC45A3-ELK4 expression levels in cancer cells alone were not associated with any features of advanced prostate cancer. While there may be population differences between the association of SLC45A3-ELK4 expression level and prostate cancer progression, careful examination of the data from the previous report showed that the association of SLC45A3-ELK4 expression with high Gleason score, advanced stage, and cancer metastasis were based on the comparison of the expression in advanced disease with nonmalignant tissues, instead of the less advanced cancer cases (Rickman et al., 2009).
SLC45A3 is a prostate-specific androgen responsive gene, which is highly expressed in prostate cancer. Its expression is increased by stimulation with the synthetic androgen R1881 (Tomlins et al., 2007). Treatment with R1881 also increased SLC45A3-ELK4 expression in LNCaP cells by androgen receptor dependent activation (Rickman et al., 2009; Zhang et al., 2012). While this upregulation of SLC45A3-ELK4 increases cell proliferation in cancer cells, it may also play a similar role in normal prostate cells. In our study, we found that the change in SLC45A3-ELK4 expression level was associated with prostate cancer progression, only when its expression was dramatically (ten-fold) increased in cancer cells, compared to case-matched normal tissues. This association included high PSA levels, Gleason score, clinical stage, and lymph node metastasis. Therefore, it appears that the baseline expression level of SLC45A3-ELK4 is not important for prostate carcinogenesis, and only in certain cases where the expression of SLC45A3-ELK4 is dramatically elevated by an unknown mechanism, is it associated with prostate cancer development and progression. These findings indicate that, while further investigations into the contribution of transcription-induced chimeric RNAs in prostate cancer development and progression are required, careful analysis of normal tissues within this process is necessary.
Although the mechanism of formation and the functional roles of chimeric transcripts remain elusive (Kannan et al., 2011; Kim RN, et al., 2012; Parra, 2005), two main mechanisms involved in the generation of transcription-induced chimeric transcripts have been established: 1) Through conjoined genes (two or more closely located genes on the same strand of a chromosome (Kannan et al., 2011; Kim DS, et al., 2012; Kim RN, et al., 2012; Parra, 2005; Prakash et al., 2010), where chimeric transcripts can be generated either by run-off transcription of the upstream gene followed by cis-splicing (Kannan et al., 2011; Prakash et al., 2010; Zhang et al., 2012) or skipping/truncation of the final exon in the upstream gene (Kim RN, et al., 2012); 2) Trans-splicing of RNAs, which generates inter- and intra-chromosomal chimeric transcripts (Fang et al., 2012; Gingeras, 2009; Kannan et al., 2011; Li et al., 2008). The USP9Y and TTTY15 genes are located next to each other in the same direction, meeting the criteria for a conjoined gene. However, the 5′ end of the downstream gene is placed in front of the 3′ end of the upstream gene. Therefore, a trans-splicing or novel mechanism may be used to generate the chimeric transcript in this case. The unique mechanisms used in the formation of transcription-induced chimeric RNAs may play a role in protein evolution or gene regulation (Akiva, 2005; Kannan et al., 2011; Prakash et al., 2010), but the role of those chimeric RNAs in carcinogenesis has to be carefully evaluated.
The genomic changes in many cancers have been shown to differ between populations (Boyd et al., 2012). These differences are also expected for fusion/chimeric transcripts, and different frequencies of the TMPRRS2-ERG fusion gene in prostate cancer have been demonstrated between populations (Boyd et al., 2012; Mao et al., 2010; Ren et al., 2012). The USP9Y-TTTY15 chimeric RNA was initially detected as Chinese prostate cancer specific. Although we have demonstrated in this study that it is expressed in samples from both Chinese and Western populations, there are more reads in the Chinese transcriptome sequencing database than from Western men. Our transcriptome sequencing data analysis reveals that both USP9Y-TTTY15 and SLC45A3-ELK4 are present at a very low abundance, and we failed to identify SLC45A3-ELK4 positive cases from the three transcriptome sequencing datasets. We have detected the previously reported USP9Y-TTTY15 and TMPRSS2-ERG positive cases from the same datasets. Althogh SLC45A3-ELK4 has been identified from the dataset, in each positive sample, only one copy has been detected, which was not considered by our analysis with a minimum criteria of two reads to define a positive case.
Nevertheless, many relatively rare fusion/chimeric transcripts have been identified through whole genome and transcriptome sequencing (Kannan et al., 2011; Maher et al., 2009; Ren et al., 2012). With the increased efficiency and reduced cost of next generation sequencing, more fusion/chimeric transcripts will be identified and population sequencing for different populations will help to understand those fusion events and their potential roles in cancer development and expression. As many of those fusion/chimeric sequences are presented in low abundance, their expression level differences between cancer and normal samples and among populations should be assessed by targetted extensively deep sequencing or quantitative PCR method.
In summary, we found that the USP9Y-TTTY15 chimeric RNA, previously described as a prostate-specific fusion gene in the Chinese population, is in fact a transcription-induced chimeric RNA, expressed in different types of normal human tissues. Using a large cohort of prostate cancer samples, with case-matched adjacent nonmalignant tissues, we investigated the role of two transcription-induced chimeric RNAs. We found that USP9Y-TTTY15 plays a limited role in prostate cancer carcinogenesis. Although SLC45A3-ELK4 expression is slightly higher in cancer than in normal prostate tissues, only a dramatic increase of its expression in cancer cells compared to case-matched normal tissues is associated with advanced disease. Conversely, expression levels of SLC45A3-ELK4 in cancer cells alone did not significantly correlate with any clinical parameters. The existence of transcription-induced chimeric RNAs are largely ignored in the field of cancer research, where people are focused on identifying new fusion genes, as they are always assumed critical to malignant disease development and/or progression. This study suggests that it is essential to carefully examine a new fusion transcript in both cancer and normal tissues when it is identified, and bias towards a positive association between fusion transcripts and carcinogenesis should be avoided.
Conclusion
Using both malignant and nonmalignant samples, we show that USP9Y-TTTY15 is a transcription-induced chimeric RNA, which is expressed in different types of normal human tissues, and both USP9Y-TTTY15 and SLC45A3-ELK4 chimeric RNAs contribute less to prostate carcinogenesis than previously reported. Therefore, although exploring the role of chimeric RNAs in prostate cancer may have the potential to bring new findings to help our understanding and improve clinical management of this disease, it is essential to keep an open mind in this area of research before making the assumption that the presence of chimeric RNAs contribute to carcinogenesis.
Supplementary Material
Acknowledgments
This research has been financed by Orchid (http://www.orchid-cancer.org.uk), Chinese National Key Funding 863 (sq2011SF11B03602), and Zhejiang University Education fund (www.zju.edu.cn). The funding sources had no role in the study design; the collection, analysis or interpretation of the data; the preparation of the manuscript; or the decision to submit the manuscript for publication.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Akiva P. (2005). Transcription-mediated gene fusion in the human genome. Genome Res 16, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd LK, Mao X, and Lu YJ. (2012). The complexity of prostate cancer: Genomic alterations and heterogeneity. Nat Rev Urol 9, 652–664 [DOI] [PubMed] [Google Scholar]
- Cocquet J, Chong A, Zhang G, and Veitia RA. (2006). Reverse transcriptase template switching and false alternative transcripts. Genomics 88, 127–131 [DOI] [PubMed] [Google Scholar]
- Edwards PA. (2010). Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol 220, 244–254 [DOI] [PubMed] [Google Scholar]
- Fang W, Wei Y, Kang Y, and Landweber LF. (2012). Detection of a common chimeric transcript between human chromosomes 7 and 16. Biol Direct 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR. (2009). Implications of chimaeric non-co-linear transcripts. Nature 461, 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronberg H. (2003). Prostate cancer epidemiology. Lancet 361, 859–864 [DOI] [PubMed] [Google Scholar]
- Guerra E, Trerotola M, Dell' Arciprete R, et al. (2008). A bicistronic CYCLIN D1-TROP2 mRNA chimera demonstrates a novel oncogenic mechanism in human cancer. Cancer Res 68, 8113–8121 [DOI] [PubMed] [Google Scholar]
- Iyer MK, Chinnaiyan AM, and Maher CA. (2011). ChimeraScan: A tool for identifying chimeric transcription in sequencing data. Bioinformatics 27, 2903–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, et al. (2011). Global cancer statistics. CA: A Cancer Journal for Clinicians 61, 69–90 [DOI] [PubMed] [Google Scholar]
- Kannan K, Wang L, Wang J, et al. (2011). Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci USA 108, 9172–9177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Huh JW, and Kim HS. (2007). HYBRIDdb: A database of hybrid genes in the human genome. BMC Genomics 8, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Kim DW, Kim MY, et al. (2012). CACG: A database for comparative analysis of conjoined genes. Genomics 100, 14–17 [DOI] [PubMed] [Google Scholar]
- Kim P, Yoon S, Kim N, et al. (2010). ChimerDB 2.0—A knowledge base for fusion genes updated. Nucleic Acids Res 38, D81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RN, Kim A, Choi SH, et al. (2012). Novel mechanism of conjoined gene formation in the human genome. Funct Integr Genomics 12, 45–61 [DOI] [PubMed] [Google Scholar]
- Kohno T, Ichikawa H, Totoki Y, et al. (2012). KIF5B-RET fusions in lung adenocarcinoma. Nat Med 18, 375–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. (2010). Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11, Unit 1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Chae JY, Kwak C, Ku JH, and Moon KC. (2010). TMPRSS2-ERG gene fusion and clinicopathologic characteristics of Korean prostate cancer patients. Urology 76, 1268e1267–1213 [DOI] [PubMed] [Google Scholar]
- Li H, Wang J, Mor G, and Sklar J. (2008). A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 321, 1357–1361 [DOI] [PubMed] [Google Scholar]
- Magi-Galluzzi C, Tsusuki T, Elson P, et al. (2011). TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate 71, 489–497 [DOI] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, et al. (2009). Transcriptome sequencing to detect gene fusions in cancer. Nature 458, 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Yu Y, Boyd LK, et al. (2010). Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res 70, 5207–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken M, Olsen M, Chen MS Jr., et al. (2007). Cancer incidence, mortality, and associated risk factors among Asian Americans of Chinese, Filipino, Vietnamese, Korean, and Japanese ethnicities. CA Cancer J Clin 57, 190–205 [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, and Mertens F. (2007). The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7, 233–245 [DOI] [PubMed] [Google Scholar]
- Miyagi Y, Sasaki T, Fujinami K, et al. (2010). ETS family-associated gene fusions in Japanese prostate cancer: Analysis of 194 radical prostatectomy samples. Modern Pathol 23, 1492–1498 [DOI] [PubMed] [Google Scholar]
- Noel EE, Yeste-Velasco M, Mao X, et al. (2010). The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am J Pathol 176, 2607–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. (2010). Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 16, 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G. (2005). Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res 16, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikor LA, Ramnarine VR, Lam S, and Lam WL. (2013). Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer 82, 179–189 [DOI] [PubMed] [Google Scholar]
- Prakash T, Sharma VK, Adati N, et al. (2010). Expression of conjoined genes: Another mechanism for gene regulation in eukaryotes. PLoS One 5, e13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Liu X, Mao X, et al. (2012). Identification of frequent BRAF copy number gain and alterations of RAF genes in chinese prostate cancer. Genes Chromosomes Cancer 51, 1014–1023 [DOI] [PubMed] [Google Scholar]
- Ren S, Peng Z, Mao J-H, et al. (2012). RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res 22, 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman DS, Pflueger D, Moss B, et al. (2009). SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res 69, 2734–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, et al. (2012). Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook SJ, Beuten J, Torkko KC, et al. (2007). Association of RNASEL variants with prostate cancer risk in Hispanic Caucasians and African Americans. Clin Cancer Res 13, 5959–5964 [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, et al. (2007). Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 [DOI] [PubMed] [Google Scholar]
- Thomson TM, Lozano JJ, Loukili N, et al. (2000). Fusion of the human gene for the polyubiquitination coeffector UEV1 with Kua, a newly identified gene. Genome Res 10, 1743–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, et al. (2007). Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448, 595–599 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, et al. (2005). Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gong M, Yuan H, et al. (2012). Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Disc 2, 598–607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.