Abstract
Little is known about the functional domain architecture of long RNA molecules, mainly because of a relative paucity of suitable methods to analyze RNA function at a domain level. Here we describe domain-specific chromatin isolation by RNA purification (dChIRP), a scalable technique to dissect pairwise RNA-RNA, RNA-protein, and RNA-chromatin interactions in living cells. dChIRP of roX1, a lncRNA essential for Drosophila X-chromosome dosage compensation, reveals a “three-fingered hand” ribonucleoprotein topology. Each RNA finger binds chromatin and the Male-Specific Lethal (MSL) protein complex, and can individually rescue male lethality in roX-null flies, thus defining a minimal RNA domain for chromosome-wide dosage compensation. dChIRP improves RNA genomic localization signal by >20-fold relative to previous techniques, and these binding sites are correlated with chromosome conformation data, indicating that most roX-bound loci cluster in a nuclear territory. These results suggest dChIRP can reveal lncRNA architecture and function with new precision and sensitivity.
Long noncoding RNAs (lncRNAs) are a recently recognized class of molecules that participate in diverse biological processes. Many lncRNAs act at the interface of chromatin-modifying machinery and the genome, and regulate homeotic gene expression, epigenetic imprinting and dosage compensation of entire chromosomes1,2. Although thousands of lncRNAs have been discovered with tissue- and disease-specific expression, the biological functions of the vast majority remain unknown or have not been mechanistically characterized3,4. One prevailing theory states that the functional diversity of lncRNAs is achieved through modularity of specific RNA domains that coordinate combinatorial RNA-RNA, RNA-DNA, and RNA-protein interactions5.
Many existing protein-centric technologies can detect the interaction of RNAs with other biomolecules. Cross-linking immunoprecipitation (CLIP) and related methods identify RNAs bound to specific RNA binding proteins (RBP)6,7. Multiple CLIP experiments are required to reveal multivalent interactions of one RNA with multiple RBP. Separate experiments are required to map RNA-RNA interactions via proximity ligation8. By contrast, chromatin isolation by RNA purification (ChIRP) and related strategies are RNA-centric techniques for exploring chromatin-associated lncRNA function9,10,11. Specifically, ChIRP enables the genome-wide identification of RNA-chromatin binding sites and has been used to provide insights into the mechanisms of dosage compensation, cancer progression, viral pathogenesis, and FMR1 gene silencing9,11,12,13,14. But how do lncRNAs interact with chromatin-modifying complexes? Do these chromatin-associated lncRNAs have modular domains – much like their protein counterparts, transcription factors – that are responsible for their varied functions?
Here we describe domain ChIRP (dChIRP), a technique that dissects lncRNAs domain-by-domain to discover functional elements. We demonstrate the utility of the method by identifying functional domains in the roX1 lncRNA. The roX RNAs are essential for dosage compensation in male flies, wherein gene expression from the single male X chromosome is doubled to match that of females’ two15. This X up-regulation is directed by the male-specific lethal (MSL) ribonucleoprotein (RNP) complex, composed of roX1 and roX2 lncRNAs and five MSL proteins (MSL1-3, MLE, and MOF), which spread in cis along the X chromosome and deposit activating histone marks at defined loci16,17,18,19. Both RNAs are known to interact specifically with the X chromosome with roX2 and MSL3 co-occupying the same sites, called chromatin entry sites (CES)9,17. CLAMP, a zinc finger protein, directly binds to the MSL recognition element (MRE) within CES and somehow links the roX-MSL complex to DNA20. The two roX RNAs are functionally redundant and individually dispensable, despite sharing limited sequence homology, differing in size by an order of magnitude (roX1 ~3.8kb, roX2 ~600bp), and having different developmental expression patterns21. Although these RNAs have been studied through phylogenetic studies, genetic screens and genomic assays, their biochemical role in dosage compensation remains a poorly defined9,10,22,23,24. Here, application of dChIRP uncovers several features of roX RNA’s architecture and function.
RESULTS
Concept of dChIRP
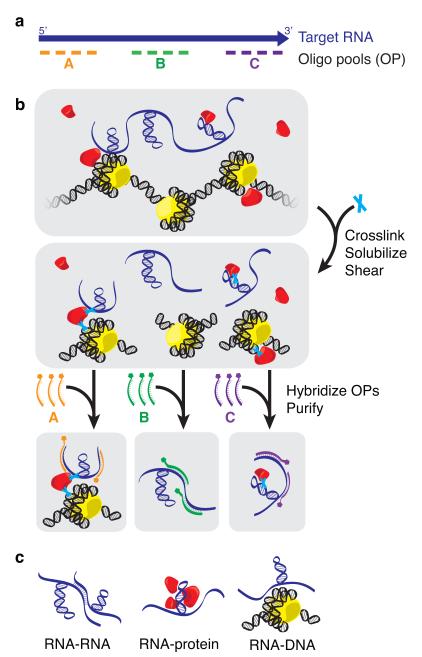
The goal of a dChIRP experiment is to dissect the functional domains of an RNA of interest within its native cellular context. For a target RNA, dChIRP can simultaneously map domain-level RNA-RNA, RNA-protein, and RNA-chromatin interactions, as well as identify genomic binding sites with increased sensitivity. First, biotinylated antisense 20-mer oligos are designed with non-overlapping and non-redundant sequences, avoiding regions of low complexity or high occurrence in the reference genome (Fig. 1a). Instead of dividing the oligos into two equal groups (“even” and “odd” pools) that tile the whole RNA as with traditional ChIRP experiments, in dChIRP the oligos are divided into domain-specific oligo pools (OPs), such that each OP targets a distinct RNA domain. The targeted RNA regions may be devised arbitrarily (such as even subdivision of the RNA length) or defined by biochemical, genetic, or conservation-based functional evidence.
Figure 1.
dChIRP uses antisense oligos to purify specific RNA domains and associated RNAs, proteins, and chromatin. (a) dChIRP oligo design strategy. Biotinylated antisense oligos are designed to tile specific regions of the target RNA. (b) dChIRP workflow. To prepare chromatin, whole cells are cross-linked to preserve protein-nucleic acid interactions. Sonication is used to solubilize the nuclear fraction and shear nucleic acids. Next, the chromatin is subdivided into equal samples. OPs are added to each sample, which hybridize to the targeted RNA fragments. The biotinylated oligos, RNA targets, and cross-linked biomolecules are then purified on magnetic streptavidin beads, and unbound material is washed away. (c) RNA-, protein-, and DNA-sensitive modalities of dChIRP. RNA, protein, and DNA fractions are extracted from each dChIRP sample. Intra- or inter- molecular RNA-RNA, RNA-protein, and RNA-DNA interactions may be measured by RT-qPCR, immunoblotting, and qPCR or sequencing, respectively.
Next, whole cells are cross-linked to preserve protein-nucleic acid interactions (Fig. 1b). We have found that fixation with 1% glutaraldehyde or 1% formaldehyde followed by 3% formaldehyde cross-linking, as used in Capture Hybridization Analysis of RNA Targets (CHART)9,10 gave the best results. The nuclei from fixed cells are then extracted and lysed. Sonication is used to solubilize the chromatin fraction and shear nucleic acids. It is important to fragment DNA to ~500bp for sequencing, and RNAs should be sheared to roughly the size of the target RNA regions (200-500nt) such that domain-specific interactions can be independently purified. The sheared chromatin is then divided into equal samples. OPs are added to each sample and allowed to hybridize under stringent conditions. After hybridization, the biotinylated oligos, hybridized RNA and associated biomolecules are purified on magnetic streptavidin beads and washed thoroughly to remove nonspecific interactions.
The recovered material from each dChIRP sample is further divided for RNA, DNA and protein extraction and analyzed. The RNA fraction can be analyzed by RT- (reverse transcription) qPCR (quantitative polymerase chain reaction) with primers designed to amplify the targeted RNA regions or other RNAs species. This analysis is used to confirm efficient, domain-specific RNA recovery, and identify potential intramolecular or intermolecular RNARNA interactions. The protein fraction may also be analyzed by immunoblotting against suspected RNA-associated proteins, thus identifying relevant protein-binding RNA domains. In this way, dChIRP is the reciprocal of CLIP6,7. Lastly, analyzing the DNA by qPCR reveals domain-level RNA-DNA or RNA-chromatin interactions. Recovered DNA may also be sequenced to identify RNA-occupied sites genome-wide. Thus, in one in situ experiment, dChIRP can map RNA-, DNA-, and protein-interacting domains of an RNA simultaneously (Fig. 1c).
The roX1 D domains form topological “fingers”
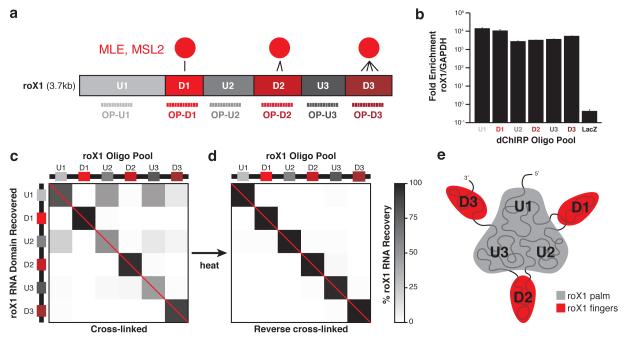
We tested and validated the dChIRP method using roX1 lncRNA. We have previously reported by CLIP that MLE and MSL2 directly contact roX1 RNA at three distinct domains (denoted D1, D2 and D3) whereas the intervening domains (U1, U2 and U3) exhibit very limited binding (Fig. 2a)18. Using these empirically-determined domains as a guide, we designed six dChIRP OPs, each comprised of twelve distinct biotinylated oligos that tile roughly equal lengths of roX1 (OP-U1 to OP-D3). We performed dChIRP in chromatin prepared from 1%+3% formaldehyde cross-linked Clone 8 cells (a male D. melanogaster line) using the six roX1 OPs and a negative control OP against the absent LacZ mRNA.
Figure 2.
dChIRP RNA co-recovery reveals roX1’s topological architecture. (a) Schematic representation of known roX1 domain interactions with MLE protein and dChIRP OP design strategy. MLE directly contacts the three D domains (D1, D2, and D3). The three intervening U domains (U1, U2, and U3) exhibit minimal binding. Six OPs were designed to target and recover each domain. (b) roX1 dChIRP specifically enriches for roX1 RNA. roX1 RNA is >1000-fold enriched over the abundant GAPDH mRNA in roX1 dChIRP samples. LacZ ChIRP does not enrich for roX1 over GAPDH. Average of technical triplicates +s.d. shown. (c, d) roX1 RNA recovery by dChIRP. To confirm that roX1 dChIRP successfully recovers the targeted RNA domain, the RNA fraction of each dChIRP sample was analyzed by RT-qPCR, using primers within each domain of roX1. Within each sample, roX1 domain recovery was quantified against input and normalized to total roX1 RNA recovery (% roX1 RNA recovery). As expected, each OP best enriches for the target roX1 domain (b, red diagonal). Whereas domains D1, D2, and D3 were recovered independently, domains U1, U2, and U3 were co-recovered. To demonstrate that the co-recovery of domains U1, U2, and U3 is cross-linking-dependent, fixed chromatin was thermally de-cross-linked and dChIRP was performed (c). Each of the domains of roX1 is independently recovered. (e) Schematic representation of roX1 intramolecular topology. Domains U1, U2, and U3 are topologically proximal to one another, forming the core “palm” of roX1. Domains D1, D2, and D3 extend as “fingers” and are distant from one another and the intervening U domains.
To confirm that dChIRP could recover the intended fragments of roX1 RNA, we purified the RNA fraction from the dChIRP samples and analyzed RNA recovery by RT-qPCR, using primers for each of the six roX1 domains and GAPDH, a control mRNA that should not be enriched by roX1 dChIRP. RNA recovery of each domain was quantitated against input RNA. We confirmed that roX1 dChIRP specifically retrieved roX1 RNA (>1000-fold enriched over GAPDH mRNA), whereas LacZ ChIRP did not enrich for roX1 RNA (Fig. 2b). For each dChIRP sample, we normalized roX1 RNA fragment recovery to total roX1 RNA recovery (% roX1 RNA recovery), and found that each dChIRP OP best enriched for its targeted RNA fragment (Fig. 2c, along the diagonal). D1, D2 and D3 dChIRP recovered their target fragment nearly exclusively and independently, whereas U1, U2 and U3 dChIRP predominantly co-recovered the U domains. This is unexpected because the U domains are discontiguous and distant in one-dimensional space. For example, U3 dChIRP efficiently retrieves the U3 fragment without the neighboring D2 and D3 fragments; yet, the more distant U1 and U2 domains are retrieved. The co-recovery suggests that the U3 domain is associated with both U1 and U2, whereas the neighboring D2 and D3 domains are sheared off during chromatin preparation.
To determine if the co-recovery of U domains is cross-linking-dependent, we performed roX1 dChIRP in thermally reverse cross-linked chromatin (Fig. 2d). Here, each roX1 RNA fragment was uniquely recovered; co-recovery of the U domains disappears. Furthermore, to confirm that the observed co-recovery was not caused by cross-hybridization of oligos within each OP, we subdivided the six OPs into twelve non-overlapping even-odd paired OPs (Supplementary Fig. 1a). In these subdivided OPs, the pattern of roX1 U domain co-recovery is reproduced in the U domain even-odd pairs, demonstrating that co-recovery is not an artifact of oligos from one OP miss-hybridizing to other roX1 RNA fragments (Supplementary Fig. 1b, c). dChIRP of human HOTAIR lncRNA from MDA-MB-231 breast cancer cells retrieved domain-specific RNA regions (Supplementary Fig. 2), highlighting the generality of the dChIRP approach and the unique architecture of roX1.
The cross-linking-dependent co-recovery of roX1’s U domains indicates that these three domains are topologically associated in three-dimensional space, cross-linked together possibly through accessory RNA-binding proteins, RNA-RNA interactions or both. Conversely, the unique recovery of domains D1, D2 and D3 suggests that these domains are physically distant from all other domains. One topological model consistent with this pattern is a “three-fingered hand” architecture, such that the U domains form a “palm” from which the D domains individually extend like “fingers” (Fig. 2e).
We wanted to know if these U domain interactions were mediated by base-pairing and used Mfold to produce in silico models of secondary structures of roX125. The structure modeling did not predict substantial secondary structures or complementary sequences between or within the U domains. This suggests that the U1-U2-U3 interaction is not likely to be caused by Watson-Crick base-pairing, but rather by tertiary RNA structures or interacting proteins. Mfold did, however, predict that the three D domains adopt long, linear stem-loops (Supplemental Fig. 3), not unlike those observed in roX218,26. These putative structures are coincident with MLE-bound residues and roX-boxes, a repeated 8-nucleotide motif in roX1 and roX2. The D3 structures were previously validated by biochemical structure mapping18.
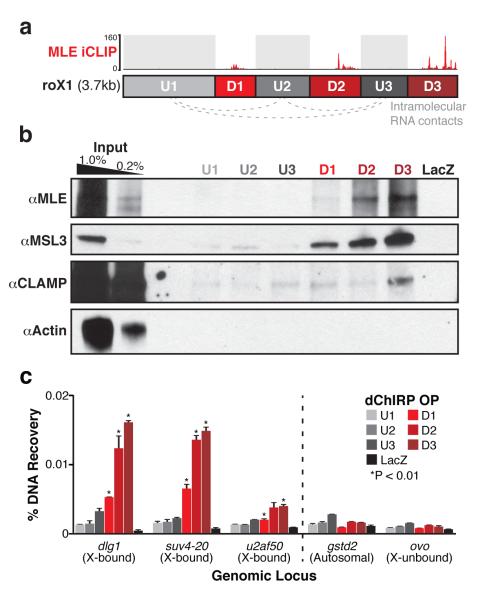
roX1 fingers bind the MSL complex and chromatin
Having mapped intramolecular interactions within roX1, we next used dChIRP to verify the previous iCLIP finding that MLE protein directly contacts roX1’s D domains (Fig. 3a)18. From each dChIRP sample, we extracted the protein fraction and performed Western blotting with MLE, MSL3, CLAMP and Actin antibodies (Fig. 3b). We found that U1, U2 and U3 recovered little or no detectable MLE or MSL3, whereas the D1, D2 and D3 domains recovered substantially more with distinct efficiencies (D3>D2>D1). This hierarchy is consistent with the iCLIP binding pattern18 and demonstrates that dChIRP is sensitive to such differences in affinity. Furthermore, the roX1 D domain interactions with MSL3 suggest that the entire core MSL complex interacts with the roX RNAs en masse, although by dChIRP alone we cannot establish whether roX1 contacts MSL3 directly or indirectly via MSL2. By contrast, only D3 could recover CLAMP, albeit substantially less than MLE or MSL3. This weak recovery suggests that the interaction between roX1 and CLAMP may be less direct or of lower affinity than the roXMSL interaction. As negative controls, the LacZ control OP recovered no proteins and Actin was not detected in any sample. roX1 dChIRP from reverse cross-linked chromatin recovered substantially less protein, indicating that protein recovery is cross-linking-dependent (Supplementary Fig. 4a). To further investigate CLAMP’s association with the MSL complex, we performed immunoprecipitation (IP) of CLAMP in untreated, DNase- or RNase-treated chromatin. MLE was co-precipitated regardless of nuclease treatment, suggesting that protein-mediated interaction with MLE may link CLAMP to the core MSL complex (Supplementary Fig. 4b).
Figure 3.
roX1 D domains interact with the MSL complex and chromatin on the X. (a) Schematic representation of roX1 interactions. The three D domains (D1, D2, and D3) directly contact MLE by iCLIP18. The three intervening U domains do not contact MLE, but are topologically associated (gray dotted lines). (b) dChIRP-Western blot confirms known MLE-bound domains of roX1. The protein fraction from each roX1 dChIRP sample was analyzed by immunoblotting against MLE, MSL3, CLAMP, and Actin. roX1 domains D1, D2, and D3 efficiently recovered MLE and MSL3 proteins. D3 recovered more protein than D2, and D2 recovered more than D1. Domains U1, U2, and U3 recovered minimal or undetectable MLE and MSL3. Only D3 recovered CLAMP appreciably, albeit very weakly. LacZ ChIRP recovered no detectable protein. Actin was not detected in any sample. (c) The three D domains of roX1 are associated with chromatin at dosage compensated loci on the X chromosome. DNA fractions from each roX1 dChIRP sample were analyzed by qPCR and normalized to input. Five genomic loci were investigated: three MSL-bound X-linked loci (dlg1, suv4-20, u2af50), one locus from an autosome (gstd2), and an unbound X-linked locus (ovo). dChIRP of domains D1, D2, and D3 significantly enrich for X-bound loci relative to control loci (*P-value < 0.01, t-test). Domains D2 and D3 recover significantly more X-bound DNA than D1 or the three U domains. LacZ ChIRP fails to recover substantial DNA from any locus. Average of technical triplicates +s.d. shown.
Next, to discover which domains of roX1 most closely contact chromatin, we analyzed the DNA fraction from each dChIRP sample by qPCR. We used primers against three known MSL- and roX2-bound loci on the X chromosome (dlg1, suv4-20, u2af50), as well as against two negative control loci, one autosomal (gstd2) and one on the X chromosome (ovo). As expected, roX1 dChIRP significantly enriches for X-bound loci relative to the control loci (Fig. 3c). The D1, D2 and D3 domains recover more X-bound DNA than the U1, U2 and U3 domains. This suggests that the D domains of roX1, which exclusively interact with MSL proteins, are more closely associated with chromatin than the U domains. D2 and D3 also significantly enrich for each X-bound locus relative to D1, recapitulating the protein-binding affinity hierarchy. The negative control LacZ OP does not enrich for X-bound loci.
Incorporating these results with the roX1 RNA architecture, we conclude that each of the roX1 D domain “fingers” can independently bind to the MSL proteins to form a RNP complex, which together grasp chromatin at hyper-expressed loci on the X chromosome.
dChIRP maps the genome-wide binding sites of roX1
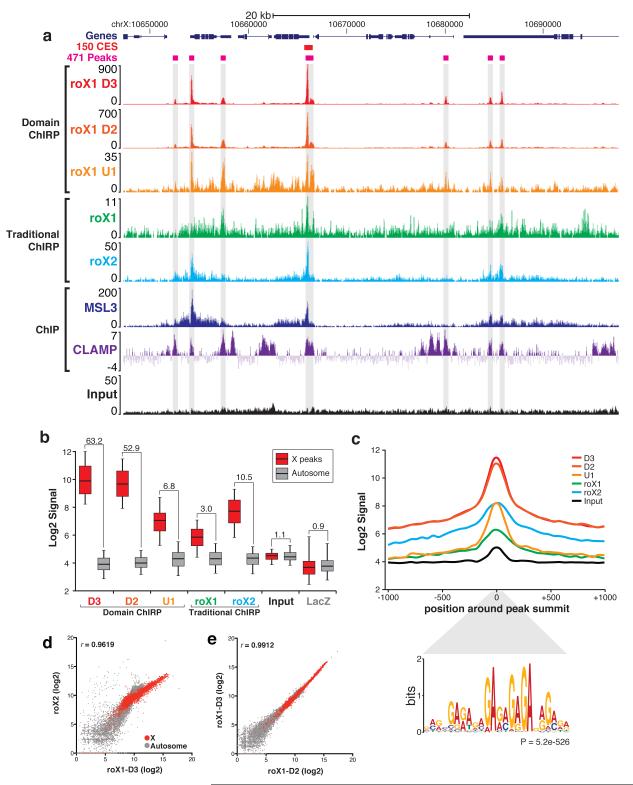
As dChIRP of roX1 D domains recovered comparatively high amounts of DNA (Fig. 3c), we suspected that this domain-specific strategy could improve the signal from ChIRP-sequencing (-seq) experiments and thus facilitate better identification of genome-wide lncRNA-associated loci. To directly compare the two techniques, we performed both traditional ChIRP and dChIRP in Clone 8 cells. We used 11 different OPs: even and odd OPs for full-length roX2 (12 oligos each), full-length roX1 (75 each), roX1 domain U1 (9 each), roX1 domain D2 (9 each) and roX1 D3 (9 each), as well as a single OP for LacZ (12 oligos) as a control (Supplementary Fig. 1a, asterisks). We prepared sequencing libraries from the DNA fractions and sequenced each. Even and odd lanes were aligned separately and normalized to mappable reads. The even-odd pairs were then merged and plotted, as previously described9.
We observed that the roX1 dChIRP and ChIRP tracks showed clear peaks that aligned precisely with known roX2-, MSL3- and CLAMP-binding sites9,17,20 (Fig. 4a). Most prominently, the peaks from roX1 D3 and D2 are much higher in magnitude and focally tighter than those from roX1 U1 dChIRP or traditional roX1 ChIRP (the y-axes, or ChIRP signal, are not equally scaled).
Figure 4.
dChIRP boosts signal-to-noise ratio relative to traditional ChIRP-sequencing. (a) Genomic tracks of dChIRP-seq results at representative X-linked locus. Sequencing tracks from roX1 dChIRP (U1, D2, D3) and traditional ChIRP (roX1, roX2). roX1 peaks (gray highlight) align with peaks from roX2, MSL3 ChIP, and CLAMP ChIP17,20. (b) Comparison of signal from the X and noise on autosomes. Signal was calculated at 457 peaks on the X and 457 random loci on autosomes. Box-plot represents 1st, 2nd, and 3rd quartiles; whiskers denote 5th and 95th percentiles. Signal-to-noise ratio (X peak mean to autosome mean) is indicated above each sample. (c) Average peak diagram of 457 peaks on X. roX1 dChIRP produces higher signal and more focal peaks than traditional ChIRP. The MRE GA-repeat motif is significantly enriched within peaks (P = 5.2e-526, MEME36) and is located specifically at peak summits (P = 5.3e-182, CentriMo36). (d, e) Correlation between ChIRP-sequencing experiments. (d) roX1-D3 dChIRP and roX2 ChIRP signal are highly correlated (r=0.9619), especially on the X chromosome (red). (e) roX1-D2 and -D3 dChIRP are very highly correlated (r=0.9912). roX1 D2, D3, and roX2 co-occupy the same loci on the X.
We next sought to quantify dChIRP’s improvement in signal-to-noise ratio (SNR) in the sequencing data relative to ChIRP. First, we used the MACS and ZINBA software suites to identify peaks and locate summits from the sequencing data. We then filtered the peaks based on signal magnitude, even-odd pair correlation and enrichment score, yielding 471 peaks, 457 of which were on the X (>97%). By contrast, MSL3 ChIP and roX2 ChIRP identified 150 and 308 CES, respectively. To represent background (noise), we randomly selected an equal number of autosomal sites with non-zero signal. We then calculated the average signal over each site from the peaks on the X chromosome and the autosomes (Fig. 4b). The SNR was calculated by dividing the X peak signal by autosomal background. Traditional ChIRP of roX1 produced especially noisy results (SNR = 3.0), whereas dChIRP of roX1 domain U1 – the most weakly chromatin-associated roX1 domain – increased SNR over traditional ChIRP (SNR = 6.8), more than two-fold. dChIRP of D2 and D3 further increased the SNR relative to traditional ChIRP (52.9, and 63.2, respectively).
We believe that this improvement in SNR is the result of two factors. First, using fewer oligos per OP (e.g. 9 for dChIRP, versus 75 for traditional ChIRP) decreases the likelihood of recovering DNA in an RNA-independent manner through direct oligo-DNA hybridization. Additionally, using fewer oligos decreases the risk of having two oligos in opposite pools with homologous sequences, which may produce false-positive peaks. Second, traditional roX1 ChIRP further dilutes signal by targeting domains of the RNA that are not involved in chromatin interaction, such as the U domains; this is observed in the SNR boost between U1 and D3 dChIRP (from 6.8 to 63.2). By minimizing OPs and targeting strongly chromatin-associated domains, dChIRP can improve SNR >20-fold over traditional ChIRP.
To further demonstrate the increase in SNR achievable by dChIRP, we plotted the average signal around X peaks in 50bp bins (Fig. 4c). Notably, roX1 dChIRP peaks have higher amplitude and are more focal than peaks from traditional roX1 or roX2 ChIRP. All 457 roX1 peaks on the X contain the MRE motif, which is significantly enriched directly at the peak summit. This motif is virtually indistinguishable from the motifs identified by roX2 ChIRP, MSL3 ChIP and CLAMP ChIP9,17,20.
dChIRP of roX1 also reveals that roX1 occupies both its own genomic locus as well as the roX2 locus with equal signal intensity; similarly, roX2 occupies the roX1 locus (Supplementary Fig. 5). roX1 dChIRP also identified 11 autosomal sites that are weakly occupied by roX1, predominantly at the transcriptional start site of genes (Supplementary Fig. 6). The bound sites contain the MRE motif and are co-occupied by CLAMP, though not MSL3. These sites may not be related to canonical dosage compensation and possibly represent misguided roX RNAs. roX1 dChIRP-seq also allowed us to resolve similarities in chromatin occupancy between roX1 and roX2. Signal from roX1 D3 dChIRP and roX2 ChIRP are strongly correlated, especially on the X chromosome (Fig. 4d), indicating that these two RNAs bind the same loci with equivalent relative affinities. roX1 D2 and D3 dChIRP are also highly correlated and therefore bind the same loci (Fig. 4e). These findings support the observation that roX1 and roX2 are genetically redundant, and that roX1 exhibits internal redundancy.
CES cluster in a dosage compensation territory
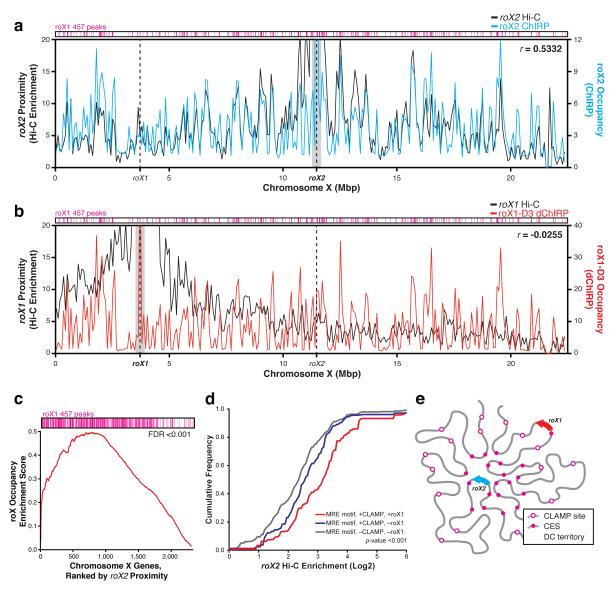
We next wanted to determine whether roX RNA occupancy is related to three-dimensional chromosome conformation to better understand how the roX RNAs spread along the X chromosome. Overlaying Hi-C enrichment data of roX1 and roX2 gene loci27 with the roX ChIRP data revealed two notable patterns (Fig. 5a, b). Regions proximal to the roX2 locus and roX2 RNA occupancy are correlated (R=0.53), indicating that the roX2 gene locus and CES of dosage compensated genes are spatially proximal and reside within the same chromosome territory (Fig. 5a). This is consistent with previous DNA FISH experiments that show three CES cluster in an MSL2-occupied nuclear territory in a male-specific, MSL-dependent manner28. By contrast, the roX1 locus makes few long-range contacts with distant chromosomal regions and the correlation between roX1 RNA occupancy and roX1 proximity is poor (R=-0.03, Fig. 5b). Gene Set Enrichment Analysis (GSEA) showed that authentic CES sites are significantly enriched for spatial proximity to roX2 locus (Fig. 5c, FDR<0.001). Furthermore, CLAMP-bound sites that are proximal to the roX2 locus are significantly more likely to be co-occupied by roX2 RNA and MSL3 than other CLAMP-bound sites not in roX2 proximity (Fig. 5d, p<0.001). Thus, the roX2 locus and CES (but not the roX1 locus) cluster into a “dosage compensation territory” formed by large-scale chromosomal conformation (Fig. 5e).
Figure 5.
CES cluster together in a dosage compensation territory of the nucleus. (a, b) Correlation between (a) roX2 occupancy by ChIRP and roX2 proximity by Hi-C, and (b) roX1-D3 occupancy by ChIRP and roX1 proximity by Hi-C27. roX2 RNA occupancy is correlated with roX2 proximity (r=0.5332); roX1 RNA occupancy is not correlated with roX1 proximity (r=-0.0255). roX1 457 peaks (magenta) are clustered at sites of high roX RNA occupancy. 400kb around the roX gene loci were excluded (gray mask) for correlation calculation (Pearson’s r), so as to exclude signal from direct ChIRP oligo-DNA recovery and one-dimensionally proximal chromosome sites. (c) Gene set enrichment analysis (GSEA) of roX occupied genes. Genes that are occupied by roX RNAs are significantly more likely to be proximal to the roX2 locus (FDR<0.001). (d) Instances of the MRE motif that are more proximal to the roX2 locus are significantly more likely to be bound by CLAMP and co-occupied by roX RNAs. P-value<0.001 by Kolmogorov-Smirnov test. (e) Model of X chromosome conformation. The roX2 locus and CES are clustered in a dosage compensation (DC) territory. The roX1 locus lies outside of the DC territory.
roX1’s D domains are independent RNA modules
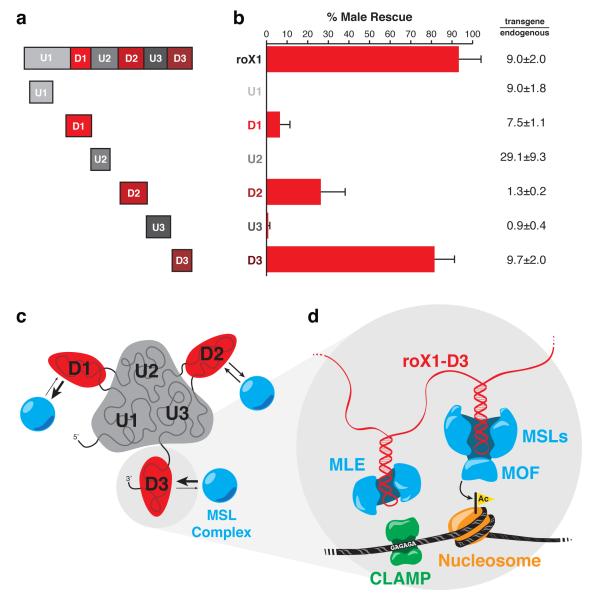
The three D domains of roX1 are topologically independent and interact with MSL proteins and chromatin with distinct affinities (D3>D2>D1). These findings suggest that the D domains are independently functional RNA modules, and that each may suffice for dosage compensation. To test this hypothesis, we over-expressed single U or D domains of roX1 as tubulin-GAL4-driven transgenes inserted in position 65B2 of chr3L in roX-null flies and tested their ability to rescue male lethality (Fig. 6a). None of the U domain constructs appreciably rescued males, but all three of the D domain constructs rescued roX deficiency, albeit with different efficiencies (Fig. 6b). The D domain constructs’ rescue efficiency echoes the previously observed affinity hierarchy (D3>D2>D1, Fig. 3). The D3 construct alone was able to rescue male lethality as efficiently as full-length roX1. We do not observe a direct correlation between rescue and transgene expression relative to endogenous roX1 in wild-type males.
Figure 6.
roX1’s D domains are independent, functional RNA subunits. (a) Transgene designs. Transgenic constructs of full-length roX1 and the six individual domains were cloned, chromosomally integrated, and expressed under the tubulin-GAL4 promoter in roX-null flies. (b) Rescue of male lethality by roX1 transgenes. Transgenic males surviving to adulthood were counted and normalized to females. Only the D domains rescued males appreciably. Rescue by D3 is not significantly different from that of full-length roX1 (t-test, P-value=0.20). Average of three separate crosses +s.d. shown (on average, n=800). roX transgene expression was quantified and normalized to endogenous roX1 expression in wild-type males, represented as relative fold (transgene/endogenous) ±s.d. (c, d) Integrated interaction map of the dosage compensation complex with chromatin. (c) roX1 RNA is topologically organized such that the three U domains form a core palm and each of the D domains extends independently as a finger. Each D domain finger directly binds to proteins of the MSL complex, with domain D3 having the highest affinity and D1 the weakest. (d) CLAMP binds the GAGA motif at X-linked CES, and associates with MLE. MLE binds to stem-loops on roX1, which tethers MLE to the core MSL complex. MOF of the MSL complex recognizes and acetylates H4K16 of adjacent nucleosomes.
To test whether multiple D domains can enhance rescue male lethality, we expressed a D1-D3 fusion, but we did not observe a significant change in rescue efficiency over D3 alone (Supplementary Figure 7a). Only when the transgenes are driven by a weaker promoter (daughterless-GAL4) at near-endogenous levels does the two-domain fusion increase male rescue (Supplementary Figure 7b). This suggests that the multiple D domains act cooperatively, increasing the RNA’s functional output at lower concentrations. When one of the putative stem-loops in D2 is disrupted by truncation (D2ΔSL and D2ΔSL-D3), male rescue is greatly diminished relative to D2, indicating that this sequence is essential to transgene function (Supplementary Figure 7).
DISCUSSION
dChIRP is an RNA-centric technology for dissecting RNA functional domains involved in RNA-RNA, RNA-protein, and RNA-chromatin interaction. dChIRP is a generally applicable method for dissecting lncRNAs of sufficient length. As traditional ChIRP has been successfully applied to diverse RNAs with a wide range of abundances, the improved sensitivity and information content of dChIRP should expand the use of this technology9,12,13,14. dChIRP traps endogenous RNA-chromatin interactions in living cells, then breaks the RNA apart to decipher which part of the RNA is doing what job. dChIRP interrogates domain-level interactions with protein, RNA and chromatin. The identified domains are then appropriate subjects for further dissection by additional methods, such as secondary structure probing by SHAPE. The lower limit of dChIRP resolution is ~200-500bp as determined by RNA shearing, and different regions may be targeted and iteratively refined. Targeted regions may be determined arbitrarily or based on existing knowledge of the RNA’s biology.
Currently the standard approach to dissect RNA function involves generation of many deletion mutants; each mutant is individually tested for physical interaction or function. This is both laborious and suffers from many potential caveats involving unintended changes in expression, folding, stability, or cytotopic localization. By contrast, dChIRP stabilizes the endogenous interactions by cross-linking, then dissects the RNA domains involved in situ. No mutant constructs are initially required, the number of configurations tested is readily scaled to the number of oligonucleotide pools desired and multiple types of RNA-mediated interactions can be tested simultaneously.
We applied dChIRP to roX1 RNA to dissect the nature of the interactions between roX1, MSL proteins, chromatin and CLAMP, elucidating an integrated interaction model. First, roX1 is topologically organized such that the three U domains form a core, or “palm,” from which each of the D domains extends independently as “fingers” (Fig. 6c). The U domains exhibit neither chromatin nor MSL binding and are genetically dispensable, implying that these domains and their association are not essential to dosage compensation. The D domains contain double stem-loops and the roXbox motif that we previously found to be the target of MLE and MSL2 interaction17. We found that each D domain finger independently binds the MSL proteins and chromatin, for which D1 has the weakest affinity, and D3 has the strongest. MLE and the core MSL complex bind to double-stranded regions within roX1’s D domains at or near roX-boxes (Fig. 6d). CLAMP binds to the MRE motif (GA-repeat) at X-linked CES and is associated with MLE. MLE is tethered to the core MSL complex via roX1 RNA binding. The MSL proteins bind chromatin via MOF, which acetylates H4K16 of adjacent nucleosomes19. Finally, these findings allowed us to design hypothesis-driven genetic mutants that proved the D domains are each minimally sufficient for dosage compensation. Despite being approximately one-tenth the size of wild-type roX1, D3 can rescue roX-null male flies as efficiently as the wild-type gene, defining to our knowledge the smallest RNA unit sufficient for chromosome-wide dosage compensation. Additional D domains may enhance D3 function, as suggested by prior genetic studies23 and supports the idea that roX1 contains multiple D domains that act cooperatively and are functionally equivalent.
In addition to studying individual RNA domains, dChIRP improves the signal-to-noise ratio of sequencing experiments by more than an order of magnitude, enabling unbiased genome-wide mapping of RNA occupancy with greater precision and confidence. We used dChIRP-seq to map the genomic binding sites of roX1, which are nearly identical to roX2 and MSL3 binding sites, providing molecular evidence of redundant function between roX1 and roX2. The signal improvement is most relevant for longer RNAs, where the use many oligos to tile the target RNA increases false-positives and background, while sacrificing true signal by unproductively targeting nonfunctional RNA regions. This strategy is an example of RNA partitioning, wherein “functional” interactions are partitioned from the nonfunctional, therefore sequencing depth needs not be wasted on nonfunctional elements. Just as genome partitioning technologies like exome sequencing have revolutionized human genetics, this RNA partitioning technology may catalyze advances in RNA genetics and genomics.
We found that many roX1 and roX2 target sites (including the roX2 locus) cluster in a “dosage compensation territory,” extending an idea suggested by previous DNA FISH experiments28. Because autosomally-integrated roX transgenes can still target the X and rescue male lethality18, fly dosage compensation represents a strikingly different strategy of sex chromosome targeting as compared to that of mammals. Recent studies in mammal suggest that XIST targeting depends on the chromosomal location of the XIST gene locus11,29. One important consequence of CES clustering is an increase in the local density of target sites, which may increase the avidity of the dosage compensation complex for CES and distinguish the X chromosome from autosomes. Our findings with the roX RNAs are reminiscent of mammalian lncRNAs such as HOTTIP and some enhancer-like RNAs that connect chromosome conformation to three-dimensionally proximal gene activation12,30,31. Nonetheless, the existing Hi-C data is from mixed-sex embryos, and reflects a gender-averaged map of the X. The relationship between chromosome conformation, the dynamics of roX1 vs. roX2 spreading, and dosage compensation merits further investigation in the future.
MATERIALS AND METHODS
dChIRP oligo design
Biotinylated 20-mer antisense oligos were designed according to ChIRP using Stellaris single-molecule FISH probe designer (biosearch.com) following Chu et al., 2012 (Ref. 32). ChIRP oligos are listed in Supplementary Resources.
Tissue culture
Clone 8 cells (Drosophila Genomics Resource Center, dgrc.cgb.indiana.edu) were grown in M3 insect medium (Sigma) supplemented with 5μg/mL human insulin (Sigma), 1X Penicillin-Streptomycin (Gibco), 2% heat-inactivated fetal bovine serum (HyClone) and 2.5% fly extract and maintained at 27°C. Cultures were split every 5 days to a concentration of 5×106 cells/mL. MDA-MB-231 breast cancer cell line overexpressing HOTAIR were cultured according to Gupta et al., 2010 (Ref. 33).
Chromatin preparation
Two-step 1%-3% formaldehyde cross-linking was performed as previously described10. Chromatin was prepared and sonicated according to ChIRP32 with the exception that chromatin was sheared by sonication until the bulk of nucleic acids was between 200-400bp. MDA-MB-231 HOTAIR cells were cross-linked in 1% glutaraldehyde according to Chu et al., 2012 (Ref.32). For thermally reverse cross-linked samples, chromatin was heated at 65°C for 4hrs.
ChIRP
ChIRP was performed as previously described32 using OPs listed in Supplementary Resources. RNA and DNA were extracted and quantified as described, using RT-qPCR and qPCR primers listed in Supplementary Resources. Western blots were performed following Ilik et al., 2013 (Ref. 18) using MLE and MSL3, CLAMP (1:1000; courtesy of Erica Larschan20) and Actin (ab1801, Abcam) primary antibodies.
Sequencing and data analysis
High-throughput sequencing libraries were constructed as previously described9 and sequenced on Genome Analyzer IIx or HiSeq 2000 (Illumina) with read lengths of 36 or 50, respectively.
dChIRP-seq bioinformatics analysis was performed as previously described9. dChIRP peaks were defined by MACS34, peak summits were identified by ZINBA35. We filtered the resulting raw peaks by maximum signal (>10,000), median peak signal (>0) and P-value (>130), yielding 471 total peaks. For the correlation analysis, the D. melanogaster genome (dm3 assembly) was divided into 33739 5kb windows. For each sample, the normalized number of reads that fall into each window was calculated, log2-transformed and plotted pairwise as scatter plots (roX1 D2 vs. roX1 D3 dChIRPs; roX1 D3 dChIRP vs. roX2 ChIRP). Motif analysis of the peaks was performed using MEME36. Hi-C data showing chromosome confirmation in mixed male/female embryos was obtained from Sexton et al., 2012 (Ref. 27, GSM849422) at a resolution of 80kb. The proximities between all 80kb chromosome bins on X chromosome with roX1 and roX2 loci were defined as the observed counts divided by the expected counts in Hi-C experiment. roX2 ChIRP and roX1 dChIRP occupancy (log2-transformed number of reads within each 80kb window) versus roX2 and roX1 Hi-C proximities, respectively were shown and Pearson correlations between each pair were calculated. Signals within a 400kb window around the roX1 and roX2 loci were masked before calculating the Pearson correlation to avoid bias due to strong correlations of ChIRP/dChIRP signals with Hi-C signals at these loci. The dosage compensated gene set was defined by X-linked genes containing a roX1-D3 dChIRP peak and enrichment of this gene set of roX2 chromosomal proximity was estimated by GSEA analysis37, with FDR<0.001. 15051 loci with GAGA motif were obtained by searching for perfect match of GAGAGA sequences on X chromosome. These loci were then ranked by their occupancy of CLAMP signals. The top 2000 loci were defined as GAGA motif with CLAMP signal and the bottom 1000 loci were defined as GAGA motif without CLAMP signal. Loci with CLAMP signal were further segregated into two groups, those with or without roX1-D3 dChIRP peaks. Cumulative frequencies of roX2 proximity of these three subgroups (GAGA motif without CLAMP signal, GAGA motif with CLAMP signal and without roX1 peak and GAGA motif with CLAMP signal and roX1 peak) were obtained and significance of the difference was estimated using Kolmogorov-Smirnov test.
Genetic mutants
Fly work has been done essentially as described in Ilik et al., 2013 (Ref. 18). Briefly, all roX1 constructs were cloned into pUASattB vector and transgenic flies were created using phiC31 integrase-mediated germ-line transformation as previously described38. To score male viability, roX1SMC17A, roX2Δ; tubGal4/TM6Tb or daGAL4 virgin females were crossed to UAS-roX1*U1, D1, U2, D2, U3, D3, D1-D3 or D2-D3 males. Male and female adult flies from at least three independent crosses were counted daily for a period of 10 days from the start of eclosion, without blinding. The total number of non-Tb males was divided by the total number of non-Tb females that eclosed during the 10-day period, which was used as an internal control for 100% viability.
For gene expression analysis, wandering 3rd instar larvae of the correct genotype were homogenized in TRIzol (Qiagen) and total RNA was extracted from these lysates using the Direct-zol kit (Zymo) as per manufacturer’s instructions. Total RNA was then reverse-transcribed using Superscript III (Life Tech.) and random hexamers, after which transcript abundances were calculated using qPCR and the 2−ΔΔCt method.
Supplementary Material
Acknowledgments
We thank members of the Chang and Akhtar labs, P. Sharp and X. Wu for meaningful discussion and E. Larschan for CLAMP antibody. Supported by National Institutes of Health R01-CA118750 and R01-ES023168 (H.Y.C.), Max Planck Society (A.A.) and Bio-X Fellowship (J.J.Q.). This work was supported by DFG funded SFB992, SFB746 and EU funded EpiGeneSys awarded to A.A. H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute; A.A. is part of the BIOSS excellence initiative.
Footnotes
Author Contributions J.J.Q., I.A.I., P.G., C.C., H.Y.C. and A.A. designed the research; J.J.Q., I.A.I. and P.G. performed the research; K.Q. and J.J.Q. performed bioinformatics analyses; J.J.Q. and H.Y.C. wrote the manuscript; all authors discussed the results and reviewed the manuscript.
Accession Raw sequencing reads, merged ChIRP lanes and called peaks can be accessed at GEO (www.ncbi.nlm.nih.gov/geo) using accession number GSE53020.
REFERENCES
- 1.Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JT, Bartolomei MS. X-Inactivation, Imprinting, and Long Noncoding RNAs in Health and Disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Hafner M, et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent non-canonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveals Principles of RNA-Chromatin Interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engreitz JM, et al. The Xist lncRNA Exploits Three-Dimensional Genome Architecture to Spread Across the X Chromosome. Science. 2013;341:6147. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P, Pari GS. Regulation of Viral and Cellular Gene Expression by Kaposi’s Sarcoma-Associated Herpesvirus Polyadenylated Nuclear RNA. J. Virol. 2013;87:5540–5553. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colak D, et al. Promoter-Bound Trinucleotide Repeat mRNA Drives Epigenetic Silencing in Fragile X Syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad T, Akhtar A. Dosage Compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat. Rev. Genet. 2012;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 16.Straub T, Gilfillan G, Maier VK, Becker PB. The Drosophila MSL complex activates the transcription of target genes. Genes and Dev. 2005;19:2284–2288. doi: 10.1101/gad.1343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alekseyenko AA, et al. A Sequence Motif within Chromatin Entry Sites Directs MSL Establishment on the Drosophila X Chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilik IA, et al. Tandem Stem-loops in roX RNAs Act Together to Mediate X Chromosome Dosage Compensation in Drosophila. Mol. Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kind J, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 20.Soruco MML, et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes and Dev. 2013;27:1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meller VH, Rattner BP. The roX genes encode redundant Male-Specific-Lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SW, et al. An evolutionarily conserved domain of roX2 RNA is sufficient for induction of H4-Lys19 Acetylation on the Drosophila X chromosome. Genetics. 2007;177:1429–1437. doi: 10.1534/genetics.107.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuckenholz C, Meller VH, Kuroda MI. Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics. 2003;164:1003–1014. doi: 10.1093/genetics/164.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley RL, Lee OK, Shim YK. Transcription rate of noncoding roX1 RNA controls local spreading of the Drosophila MSL chromatin remodeling complex. Mech. Dev. 2008;125:1009–1019. doi: 10.1016/j.mod.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maenner S, Müller M, Fröhlich J, Langer D, Becker PB. ATP-Dependent roX RNA Remodeling by the Helicase maleless Enables Specific Association of MSL Proteins. Mol. Cell. 2013;51:174–184. doi: 10.1016/j.molcel.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Grimaud C, Becker PB. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes & Devel. 2009;23:2490–2495. doi: 10.1101/gad.539509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon MD, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013 doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu C, Quinn JJ, Chang HY. Chromatin isolation by RNA purification (ChIRP) J. Vis. Exp. 2012;61:e3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Intl. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groth AC, Fish M, Nusse R, Calos MP. Construction of Transgenic Drosophila by Using the Site-Specific Integrase from Phage PhiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.